Introduction
The current deployment of the fifth generation of wireless communication technology (5G) has reignited the long-standing debate around the possibility of health effects from the radiation emitted by the existing wireless communication devices and networks and the new ones introduced by the 5G. The opposition of the part of society toward wireless communication technologies is caused by the uncertainty of whether this radiation affects humans as well as fauna and flora.
Some of the population considers themselves sensitive to wireless radiation, the so-called electromagnetic hypersensitivity (EHS). Currently, the existence of EHS has not yet been proven scientifically. However, according to the definition of health of the World Health Organization (1) where “health is a state of complete physical, mental, and social wellbeing and not merely the absence of disease and infirmity”, any person believing his/her health is affected or sensitive to wireless radiation experiences a health effect caused by the wireless radiation. According to the WHO definition of health, just a belief in having EHS and experiencing non-specific symptoms, physiological and/or psychological, is experiencing the health effects of wireless technology. Hence, it is correct to claim that wireless radiation causes health effects.
The existence of individual sensitivity to wireless radiation has not been proven yet, or disproven because to date performed research is inadequate. Of course, it is challenging to prove things with science as an infinite number of potential confounders and covariates need to be considered. However, science should be used to evaluate the balance of probability whether a hypothesis is plausible. Hence, providing an absolute proof might be elusive. Most of the research on EHS was conducted using psychology methods, asking a person, who is concerned that wireless exposure might affect health of the wireless radiation exposure, how the person feels during the real or the sham exposure. Puzzlingly, the frequent observation that the self-declared EHS person can't feel the wireless radiation and can't recognize when the wireless transmitter emits radiation and when it is not transmitting, is considered ultimate proof that the form of individual sensitivity to wireless radiation called EHS is not caused by wireless radiation exposures. This is questionable as no person, sensitive or not, could feel the ionizing radiation or other non-ionizing radiation like ultraviolet in their environment. Thus, research on EHS and individual sensitivity to wireless radiation, in general, has generated scientifically subjective data, unreliable for public health recommendations or radiation safety limits. On the contrary, logically and per analogiam with other environmental factors, individual sensitivity to wireless radiation, which includes EHS, exists as indicated below, and should be studied using biochemical methods.
Environmental sensitivity
It is well established that different individuals can be differently affected by the same agent of chemical or physical nature. This phenomenon of individual sensitivity can vary greatly among different people due to genetic, physiological, and psychological differences. Individual sensitivity is a well-documented phenomenon in the fields of environmental health, toxicology, and epidemiology.
The broad theory of environmental sensitivity (2) proposes that although all people are sensitive to their environment, some individuals tend to be more sensitive than others. The theory of environmental sensitivity suggests that people vary in their sensitivity to the environment due to differences in their ability to perceive and process information coming from the environment. The more sensitive individuals have heightened perception and deeper processing of external and internal environment stimuli due to genetically influenced physiological and neurobiological differences in the functioning of the internal organs and the central nervous system. One also needs to consider factors such as certain drugs, vaccines, prior exposures to radiation, pesticides, mold, viruses, bacteria and chemicals can sensitize people (3).
Understanding the phenomenon of individual sensitivity to environmental factors, chemicals, and radiation requires a multi-faceted approach that encompasses genetic, epigenetic, immunological, and developmental perspectives. Each individual may respond differently based on these factors, leading to varying health outcomes or lack of these. Individual sensitivity to environmental factors is regulated by:
• Variability in genetic makeup where genetic polymorphisms can affect how individuals metabolize and respond to chemicals and environmental factors. For example, variations in genes responsible for detoxification, such as glutathione S-transferase (GST) genes, can influence susceptibility to cancer from environmental toxins (4).
• Epigenetic factors where epigenetic modifications, which can be influenced by environmental factors, may cause individuals to react differently to exposures. For instance, prenatal exposure to certain environmental toxins has been linked to changes in DNA methylation patterns that impact health outcomes (5).
• Age and developmental factors where younger populations, particularly infants and children, often show higher sensitivity to environmental toxins. Developmental stages can influence how the body processes and responds to these chemicals (6).
Here are a few examples of health effects regulated by the variations in individual sensitivity to the environment:
• Immune system variability where individual differences in immune system function can lead to varying responses to environmental chemicals and radiation. For example, some individuals may exhibit allergic reactions or more severe effects from pollutants, which can be due to genetic predisposition or previous exposure history (7).
• Allergies and asthma where some individuals are highly sensitive to environmental allergens such as pollen, dust mites, mold, and pet dander, leading to allergic reactions or asthma. Genetic predisposition plays a significant role in this sensitivity (8).
• Chemical sensitivity, where some individuals experience multiple chemical sensitivity (MCS), where exposure to low levels of various chemicals in the environment leads to symptoms affecting multiple organ systems. This sensitivity can vary widely among individuals (9).
• Environmental stressors and mental health, where factors such as noise pollution, urban overcrowding, and exposure to crime can significantly affect individuals differently, particularly in terms of stress and anxiety levels. Vulnerable populations may be more sensitive to these environmental stressors (10).
• Sunlight and skin sensitivity, where individuals with certain skin types or genetic conditions (such as xeroderma pigmentosum) are particularly sensitive to ultraviolet (UV) radiation, increasing their risk of skin damage and cancers (11).
• Temperature sensitivity, where some individuals have a heightened sensitivity to temperature changes, can be particularly notable in conditions like fibromyalgia, where environmental temperature can exacerbate symptoms (12).
One of the known individual sensitivities affecting human health is radiation sensitivity:
• Where certain individuals, due to genetic factors or pre-existing health conditions, may have a higher risk of developing radiation-induced health issues, such as cancer (13).
It is known that even a slightly elevated level of environmental sensitivity may lead to worse ratings of the environment with no clear relation to the real environment (14). Consequently, environmental sensitivity should be considered as a confounding factor in environmental exposure studies. The independence from real exposure levels is in line with the results from studies showing that the differences in environmental ratings are also driven by psychological factors (14). Hence, both psychology and physiology research methods need to be used when studying individual sensitivities.
Radiation sensitivity
Individual sensitivity to radiation is a well-known and scientifically established phenomenon. Because of the genetic and epigenetic differences between people, different persons may respond physiologically in different ways to exposure to the same radiation exposure. The phenomenon of individual sensitivity to radiation has been described for ionizing radiation (15–17) and for non-ionizing radiation, e.g., ultraviolet radiation (18, 19) or ultrasound (20).
The recently published opinion/review (21) has concluded that while it is well known that different persons can react differently to the same ionizing radiation exposure, the validated biomarkers of either tissue or stochastic effects have not been identified to date.
Search for biomarkers of ionizing radiation (hyper)-sensitivity is ongoing because about 5–10% of radiotherapy patients show severe damage sensitivity in healthy tissue irradiated collaterally during irradiation of pathological tissue (22). The nature and prediction of ionizing radiation hypersensitivity is not possible due to a lack of research that would identify specific biomarkers. The pre-therapeutic identification of radiosensitive patients would allow improvement of individual patients' treatment because hypersensitive patients could be excluded from high-dose radiation treatments and different therapies could be proposed whereas non-sensitive patients could profit even more from enhanced radiation therapy (23–25). Several studies were performed to find biomarkers of ionizing radiation hypersensitivity. However, current studies are difficult to compare with each other due to a lack of standardization of methods and high inter-laboratory variations. Despite these technical problems, the results of these studies support the notion that it will be possible to find the biomarkers and improve patient therapy, and the search for biomarkers of hypersensitivity to ionizing radiation should proceed using high-throughput screening techniques (23).
Individual sensitivity to wireless radiation
One of the recent additions to the list of environmental pollutants is electromagnetic radiation emitted by wireless communication devices and networks (wireless radiation). Wireless radiation can induce various biological effects in cells grown in vitro, in experimental animals, and in humans. The reasons why the epidemic of such health problems like e.g., brain cancer did not materialize as predicted by some scientists might be that wireless-radiation-induced biological effects might affect health only in some particularly sensitive individuals, a minority of the total population. Also, changes in technology, such like the relocation of the mobile phone antenna from the top to the bottom of the phone set, changed how the brain tissue is irradiated.
Research into individual sensitivity to wireless radiation has garnered significant attention as the prevalence of mobile devices and wireless technology increases. Individuals report experiencing various symptoms they attribute to wireless radiation exposure, a phenomenon often referred to as electromagnetic hypersensitivity (EHS). Electromagnetic hypersensitivity is characterized by a collection of symptoms that individuals attribute to exposure to wireless radiation, including headaches, fatigue, stress, sleep disturbances, and skin symptoms. However, a clear scientific consensus on the existence and mechanisms of EHS remains elusive. Studies indicate variability in prevalence rates of self-reported EHS ranging from 1 to 10%. This diversity can partly be attributed to differing levels of public awareness and reporting.
Scientific research on EHS consists of three types of studies: (i) survey studies, (ii) provocation studies, and (iii) biochemical and physiological studies. The three types of studies have major overarching drawbacks (26). Firstly, researchers do not know whether the self-declared EHS persons volunteering in research projects have EHS because there are no diagnostic criteria for determining it. The group of self-declared EHS persons participating in the research study might be contaminated by the misdiagnosed EHS persons. In extreme situations, none of the self-declared EHS volunteers suffers from EHS. Secondly, scientists analyze solely the effects of exposures to wireless radiation and do not address, simultaneously occurring in real life, co-exposures to other environmental pollutants, like chemicals, particulate matter, or radiations other than wireless, which might have synergistic effects. Thirdly, all the experimental data obtained with the help of self-declared EHS volunteers is subjective and prone to bias caused by the beliefs and opinions of the volunteers (26). Conclusions of the provocation studies performed using psychology methods might be affected or even invalidated because of the existence of the placebo and nocebo phenomena. Placebo and nocebo indicate the ability of the human mind to affect the physiology of the human body. There is a well-known phenomenon among medical students of the “medical students' disease”. It is a condition frequently reported in medical students, who perceive themselves to experience symptoms of a disease they are studying. This condition is associated with the fear of contracting the disease in question. The same is likely happening when researchers show the study subjects' films presenting the dangers of wireless radiation exposure. It is obvious and expected that some persons will afterward “experience” some of the symptoms presented in the film. Furthermore, all volunteers have preconceived opinions on EMF and health. Thus, claims that news media reports and dissemination of information about precautionary measures to avoid exposure to wireless radiation cause a rise in the occurrence of EHS is incorrect and was shown to be so (27). The responses of the self-diagnosed EHS persons given during the provocation experiments are influenced by their pre-existing opinions about EHS. The data collected in the psychological provocation studies is not only subjective but is affected to an unknown degree by pre-existing opinions.
Search for sensitive individuals, most commonly using provocation studies where experimentally controlled exposures are followed by inquiries about acutely occurring symptoms and feelings, has failed to detect any sensitivity to wireless radiation. The reason might be that provocation exposures combined with psychological inquiries might be not enough sensitive to detect individual sensitivity to a single agent present in a mix of other environmental agents (26).
Some of the problems with provocation studies are:
• Capturing general sensitivities but not testing for them in the crossover study,
• Use of a generalized test approach that expects all EHS to respond in the same way,
• Testing EHS people with frequencies they report they are not sensitive to,
• Not always including objective biological tests,
• Insufficient recovery time between exposures,
• No accounting for latent effects (EHS people are not like light switches),
• Not tracking symptoms progression or remission over time,
• Pooling results what may hide sensitive individuals in group of non-sensitives—what calls for more personalized tests examining each person individually.
The practical problems of research on individual sensitivity and biomarkers of response to ionizing radiation (21) very closely resemble the problems of the research on non-ionizing radiation emitted by wireless communication devices (26, 28).
The scientific evidence surrounding individual sensitivity to EMF is complex and often inconclusive. While some studies suggest that reported symptoms in EHS individuals may not correlate with objective measures of wireless radiation exposure, the psychological and psychosomatic factors may play significant roles. Continued research is essential to deepen our understanding of EHS, its symptoms, and the underlying mechanisms at play (26).
Due to inter-individual biochemical differences, physiological and biochemical experiments on human volunteers will likely be necessary to determine whether some individuals react differently to wireless exposures.
Biochemical individuality
Another scientific concept that supports the notion that individual sensitivity to wireless radiation needs to be studied using biochemical methods is the biochemical individuality concept. Biochemical individuality is the concept that each person has a unique biochemistry that influences how they respond to nutrients, drugs, and other factors. It suggests that individuals may have differing nutritional requirements, metabolic processes, differing responses to medications, and environmental factors based on their genetic makeup and environmental influences. This idea highlights the importance of personalized approaches to health and wellness, taking into account the individual differences in biochemistry among people. The concept of biochemical individuality was established by Roger J. Williams and it states that there is no such thing as an average person (29). We are all genetically and biologically unique. When sperm fertilizes the egg, our characteristics are not locked in stone. The concept of biochemical individuality argues that genes do not necessarily cause disease by themselves, but that nutrition and environmental factors can alter the outcome. The concept of biochemical individuality explains why some of us are better at detoxifying drugs and chemicals, why cancer genes respond in different ways to diet and environment, why some people are alcoholics or diabetics, why low-fat diets cause some people to gain weight, or why one person needs higher levels of a nutrient than another to stay healthy.
A new research approach is needed to study individual sensitivity to wireless radiation
The to-date proposed biomarkers of EHS are not known/proven to be affected by RF-EMF exposures. Therefore, use of these “biomarkers” identifies persons with some health problem but it is not known what causes it. Attempts of EHS diagnosis using current physiological and biochemical tests are inadequate. In some research studies (30) biochemical tests analyzed biological endpoints thought to correlate with the symptoms experienced by self-diagnosed EHS persons: high-sensitivity C-reactive protein (hs-CRP), vitamin D2-D3, histamine, IgE, protein S100B, nitrotyrosine (NTT), heat shock protein 70 (HSP70), heat shock protein 27 (HSP27), anti-O-myelin autoantibodies, hydroxy-melatonin sulfate, 6-Ω-creatinine. In addition to the biochemical tests, the blood flow in the temporal lobes of the brain was examined with a non-invasive method of ultrasonic tomosphygmography.
The observed changes in expression of the examined biochemical endpoints did occur only in the minority of the self-declared EHS persons what is not surprising in a multifactorial ailment.
Only 40% of self-declared EHS persons had an increase in histamine level. None of the proposed biomarkers was prevalent in EHS persons: hs-CRP increased in 15% of EHS, vitamin D2–D3 declined in 23.2% of EHS, histamine increased in 40% of EHS, IgE increased in 22% of EHS, protein S100B increased in 15.5% of EHS, nitro-tyrosine (NTT) increased in 29% of EHS, Hsp27, Hsp70 detected in 7–19% of EHS, antibody to O-myelin detected in 17–29% of EHS, melatonin to creatinine ratio declined in EHS but the variation was too large to provide a specific number for the ratio.
Brain blood flow, examined with ultrasonic tomosphygmography, was claimed to decline in 50.5% of self-declared EHS persons, but the actual results of the tests were never shown.
Most importantly, there is no evidence that any of the proposed biomarkers was affected by wireless radiation exposure in vitro or in humans. Only two of the examined and proposed biomarkers, Hsp70 and Hsp27, are known to be affected by wireless exposures in cells grown in the laboratory. However, it is not known if the same occurs in living humans. Hence, the diagnostic value of Hsp70 and Hsp27 remains unknown.
Lastly, there are no blinded studies examining a group of volunteers, consisting of sensitive and non-sensitive persons, where the scientists would pinpoint sensitive persons after examining the proposed biomarkers.
The same problem is with the numerous biomarkers and diagnostic criteria for EHS proposed by the European Academy for Environmental Medicine (EUROPAEM). The proposed biomarkers are based on the symptoms claimed by the self-declared EHS persons but they lack proof that the symptoms were caused by wireless radiation exposures and that the wireless radiation exposures can cause changes in the expression of the proposed biomarkers (31).
There is a need for human volunteer studies where the already proposed, and other potentially useful biomarkers, would be examined in groups of sensitive and non-sensitive persons, ethically exposed to wireless radiation.
This newly designed research should be a combination of human provocation studies and examination of molecular-level responses in wireless-radiation-exposed people. Because it is not possible to reliably identify sensitive persons, the group of volunteers should consist of self-declared sensitive and self-declared non-sensitive persons. The self-declarations of volunteers in the group should be blinded from the researchers until the full data is analyzed. Such research should look for corroborating evidence that individual sensitivity to wireless radiation exists and look for the molecular targets of exposures that are proven to be affected by wireless radiation exposures. Regrettably, the to-date performed biomarker studies relied solely on anecdotal evidence from self-diagnosed EHS persons and did not determine whether the biomarkers are indeed in any way affected by the RF-EMF exposures (30).
The way forward in EHS research is to discover biomarkers of EHS, molecules that are affected by wireless radiation exposure, by research using high-throughput screening techniques of proteomics, transcriptomics, and metabolomics (32, 33). For the start, proteomics might be the most promising of these methods.
Proteomics
Proteomics investigates the interactions, function, composition, and structures of proteins and their cellular activities. Proteomics provides a better understanding of the structure, function, and physiology of the organism than genomics or transcriptomics. Changes in gene expression do not affect the physiology of the organism for as long as the gene expression changes are not translated into changes in the expression and activity of proteins. The level of transcription of a gene gives only a rough estimate of its level of expression into a protein because of the physiological regulation of mRNA production, translation, and degradation. Therefore, proteomics provides a much more robust and representative picture of the functioning cell than other forms of large-scale biology, such as the sequencing of genomes or the global analysis of gene expression. At present, strategies for proteomics research can be divided into discovery proteomics and targeted proteomics. Discovery proteomics is more concerned with protein screening and dynamics, while targeted proteomics focuses more on detecting target proteins/peptides to achieve absolute quantification. In the biomedical field, proteomics finds widespread application in cancer research and diagnosis, stem cell studies, and the diagnosis and research of infectious and non-infectious diseases. In addition, it plays a pivotal role in drug discovery and the emerging frontier of personalized medicine. The limitations of proteomics include complexity in analysis, lack of standardization in sample processing, risk of high false positivity, and dynamic range of sample limits the estimation of low abundance of proteins, or failure in the validation of biomarkers in a larger number of patients due to lack of antibodies.
Despite the mentioned limitations, proteomics is being increasingly used for identifying and validating biomarkers for various diseases, like e.g., cancer, cardiovascular diseases, or neurodegenerative disorders. Modern proteomics, and other high-throughput screening techniques, allow global evaluation of the changes in the cellular proteome, metabolome, transcriptome, and genome that will reveal biological effects of wireless radiation exposures that are impossible to predict based on the currently available knowledge. High-throughput screening techniques are already widely used in clinical research in the search for biomarkers of diseases (34–39) and environmental toxicology (40–43).
The search and identification of biomarkers play an essential role in disease diagnosis, prognosis, and the development of personalized medicine. Biomarkers in proteomics refer to measurable substances, typically proteins, which indicate a biological state or condition of an organism/organ and can be used for disease diagnosis, prognosis, or therapeutic monitoring. The high-throughput scale of proteomics allows researchers to rapidly identify and quantify a large variety of potential protein biomarkers, present in various biological samples. Hence, proteomics has become a vital tool in the search for biomarkers—measurable indicators of specific biological states or conditions.
Applications of proteomics in biomarker discovery research:
• Understanding of the mechanisms of diseases: proteomics allows for the identification of protein alterations that occur in diseases. These alterations can serve as potential biomarkers for early diagnosis and understanding of disease progression (44–48).
• Identification of diagnostic biomarkers: comparing the proteomes of healthy individuals with those of patients, permits identification of specific proteins that are differentially expressed (49–59).
• Biomarkers for therapeutic applications: proteomics can identify biomarkers that predict response to therapies, particularly in cancer treatment, enabling personalized medicine (60, 61).
• Biomarkers for prognostic applications: proteomic profiles can serve as prognostic markers to inform of the future potential disease outcomes and patient survival (62).
• Integration of protein biomarker discovery with genomics and metabolomics approaches: combining proteomics with genomics and metabolomics can enhance biomarker discovery and provide a more comprehensive understanding of disease (63).
In summary, proteomics is paving the way for the identification and validation of biomarkers across a spectrum of diseases. Proteomics is invaluable in identifying and validating biomarkers, contributing to advancements in diagnosis, prognosis, and treatment personalization. It is astounding that the research on biological and health effects of the exposures to man-made wireless radiation does not use more efficiently proteomics, and other high-throughput omics technologies, to discover the potential biological and health effects. As technology evolves, the integration of proteomic data with other omics will further enhance the biomarker discovery process, potentially leading to better patient outcomes.
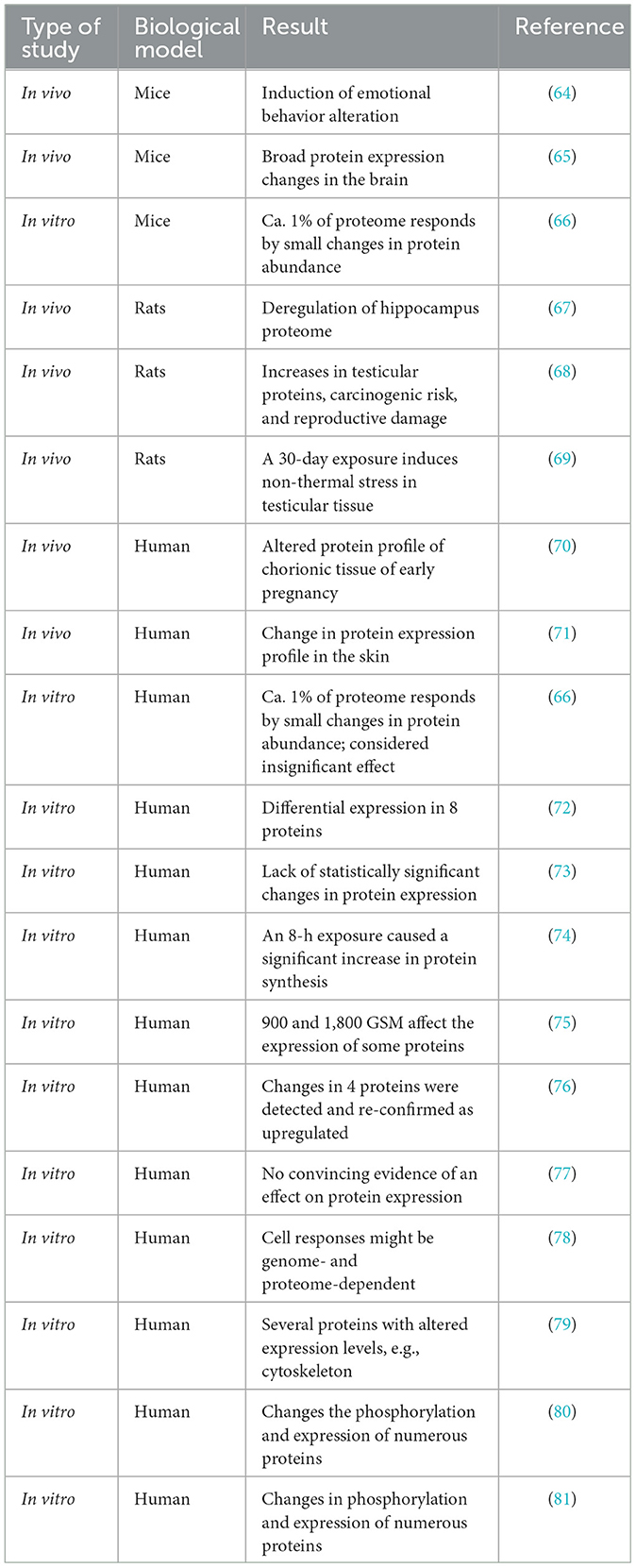
The reasons why proteomics is not used to study the physiological effects of wireless radiation exposures in humans are difficult to understand and comprehend. Despite the advantages of research using proteomics methodology, over the last 20 years, only a few proteomics studies have examined proteome changes in response to wireless radiation exposures. Of these studies, only two were performed on human volunteers (Table 1). Other proteomics studies were performed using animal models and cell lines grown in the laboratory. A review of these studies shows that the knowledge about the physiological impact of long and short-term exposures to wireless radiation is very scarce. The knowledge about the effects of wireless radiation exposures on the physiological processes within the human body is close to non-existent. Proteomics data suggests possible effects but the evidence is too limited for any practical use. This lack of knowledge about the physiological impact of wireless radiation exposure on the human body, at a time when the vast majority of people use mobile phones and are exposed to wireless radiation, is astonishing.
There are numerous advantages of using proteomics in the search for biomarkers of individual sensitivity to wireless radiation (82–84):
• Comprehensiveness of analysis—proteomics allows for the simultaneous analysis of thousands of proteins in a sample. This comprehensive approach can help identify specific proteins involved in the cellular response to radiation, which might not be captured by other techniques like genomics or transcriptomics.
• Providing functional insights—proteins are the functional molecules in the cell, and their levels and modifications can provide direct information about biological processes, pathways, and mechanisms. This functional insight is critical for understanding how individuals might respond differently to radiation exposure.
• Analysis of the dynamic range of response—the proteome can change rapidly in response to stimuli, such as radiation exposure. Proteomics can capture these dynamic changes in protein expression and post-translational modifications more effectively than methods that assess static genetic information.
• Integration of proteomics with other omics techniques, like genomics and metabolomics, will give a more holistic view of the biological response to radiation. This multi-omics approach can elucidate complex interactions and pathways relevant to individual sensitivity.
• Identifying biomarkers in wireless radiation exposed persons—through proteomic studies, researchers can identify potential biomarkers that correlate with sensitivity to radiation. This can facilitate the identification of sensitive individuals, and personalized medicine approaches, and help in predicting treatment responses.
Summary conclusion
In conclusion, it is logical to conclude that the individual sensitivity to wireless radiation emitted by wireless communication devices and networks exists and impacts the health of sensitive persons. Clearly, the to-date unsuccessfully used methods of provocation studies were either too crude or too much affected by the perceptions and preexisting opinions of study volunteers. A combination of psychological inquiry and physiological/biochemical examination of the responses to wireless radiation exposures are necessary to detect and define diagnostic criteria of individual sensitivity to wireless radiation. In physiological/biochemical testing, while no single method can fully capture the complexities of biological responses to radiation, proteomics offers unique insights that are crucial for understanding individual sensitivity. It should be viewed as an integral part of a broader set of tools used to study radiation effects. Proteomics and other high-throughput “omics” screening techniques should and must be broadly introduced to wireless radiation bio-effects and bio-markers research.
Author contributions
DL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author was supported by a personal scientific research grant from The Finnish Electrosensitivity Foundation, Helsinki, Finland.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Available at: https://www.who.int/about/governance/constitution (accessed January 3, 2025).
2. Pluess M. Individual differences in environmental sensitivity. Child Dev Perspect. (2015) 9:138–43. doi: 10.1111/cdep.12120
3. Genuis SJ, Lipp CT. Electromagnetic hypersensitivity: fact or fiction? Sci Total Environ. (2012) 414:103–12. doi: 10.1016/j.scitotenv.2011.11.008
4. Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. (1998) 154:47–85. doi: 10.1007/978-3-642-46870-4_4
5. Bollati V, Baccarelli A. Environmental epigenetics. Heredity. (2010) 105:105–12. doi: 10.1038/hdy.2010.2
6. Landrigan PJ, Miodovnik A. Children's health and the environment: an overview. Mt Sinai J Med. (2011) 78:1–10. doi: 10.1002/msj.20236
7. Korrea S. Role of genetic susceptibility in environmental exposure induced diseases. In:Mothersill C, Mosse I, Seymour C, , editors. Multiple Stressors: A Challenge for the Future (2007). p. 103-123. doi: 10.1007/978-1-4020-6335-0_8
8. Haahtela T, Valovirta E, Saarinen K, Jantunen J, Lindström I, Kauppi P, et al. The Finnish Allergy Program 2008-2018: Society-wide proactive program for change of management to mitigate allergy burden. J Allergy Clin Immunol. (2021) 148:319–26.e4. doi: 10.1016/j.jaci.2021.03.037
9. Cullen MR, Redlich CA. Significance of individual sensitivity to chemicals: elucidation of host susceptibility by use of biomarkers in environmental health research. Clin Chem. (1995) 41:1809–13. doi: 10.1093/clinchem/41.12.1809
10. Cohen S, Hamrick N. Stable individual differences in physiological response to stressors: implications for stress-elicited changes in immune related health. Brain Behav Immun. (2003) 17:407–14. doi: 10.1016/S0889-1591(03)00110-7
11. Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Arch Dermatol. (1988) 124:869–71. doi: 10.1001/archderm.124.6.869
12. Yunus MB. Central sensitivity syndromes: a new paradigm and group nosology for fibromyalgia and overlapping conditions, and the related issue of disease versus illness. Semin Arthritis Rheum. (2008) 37:339–52. doi: 10.1016/j.semarthrit.2007.09.003
13. Kerns SL, Ostrer H, Rosenstein BS. Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer Discov. (2014) 4:155–65. doi: 10.1158/2159-8290.CD-13-0197
14. Reichherzer A, Wargocki P, Mayer F, Norrefeldt V, Herbig B. Increased self-reported sensitivity to environmental stimuli and its effects on perception of air quality and well-being. Int J Hygiene Environ Health. (2022) 246:114045. doi: 10.1016/j.ijheh.2022.114045
15. Foray N, Colin C, Bourguignon M. 100 years of individual radiosensitivity: how we have forgotten the evidence. Radiology. (2012) 264:627–31. doi: 10.1148/radiol.12112560
16. Bourguignon MH, Gisone PA, Perez MR, Michelin S, Dubner D, Di Giorgio M, et al. Genetic and epigenetic features in radiation sensitivity. Part I: cell signalling in radiation response. Eur J Nucl Med Mol Imaging. (2005) 32:229–46. doi: 10.1007/s00259-004-1730-7
17. Bourguignon MH, Gisone PA, Perez MR, Michelin S, Dubner D, Di Giorgio M, et al. Genetic and epigenetic features in radiation sensitivity. Part II: implications for clinical practice and radiation protection. Eur J Nucl Med Mol Imaging. (2005) 32:351–68. doi: 10.1007/s00259-004-1731-6
18. Rees JL. The genetics of sun sensitivity in humans. Am J Hum Genet. (2004) 75:739–51. doi: 10.1086/425285
19. Kelly DA, Young AR, McGregor JM, Seed PT, Potten CS, Walker SL. Sensitivity to sunburn is associated with susceptibility to ultraviolet radiation–induced suppression of cutaneous cell–mediated immunity. J Exp Med. (2000) 191:561–6. doi: 10.1084/jem.191.3.561
20. Barnett SB, Rott HD, ter Haar GR, Ziskin MC, Maeda K. The sensitivity of biological tissue to ultrasound. Ultrasound Med Biol. (1997) 23:805–12. doi: 10.1016/S0301-5629(97)00027-6
21. Rajaraman P, Hauptmann M, Bouffler S, Wojcik A. Human individual radiation sensitivity and prospects for prediction. Ann ICRP. (2018) 47:126–41. doi: 10.1177/0146645318764091
22. Baumann M. Impact of endogenous and exogenous factors on radiation sequelae. In:Dunst J, Suaer R, , editors. Late Sequelae in Oncology Medical Radiology, Diagnostic Imaging and Radiation Oncology. Heidelberg: Springer Verlag. (1995). doi: 10.1007/978-3-642-46794-3_1
23. Greve B, Bölling T, Amler S, Rössler U, Gomolka M, Mayer C, et al. Evaluation of different biomarkers to predict individual radiosensitivity in an inter-laboratory comparison - lessons for future studies. PLoS ONE. (2012) 7:e47185. doi: 10.1371/journal.pone.0047185
24. Bese NS, Hendry J, Jeremic B. Effects of prolongation of overall treatment time due to unplanned interruptions during radiotherapy of different tumor sites and practical methods for compensation. Int J Radiat Oncol Biol Phys. (2007) 68:654- 661 doi: 10.1016/j.ijrobp.2007.03.010
25. Tucker SL, Geara FB, Peters LJ, Brock WA. How much could the radiotherapy dose be altered for individual patients based on a predictive assay of normal-tissue radiosensitivity? Radiother Oncol. (1996) 38:103–13 doi: 10.1016/0167-8140(95)01669-4
26. Leszczynski D. The lack of international and national health policies on electromagnetic hypersensitivity (EHS). Rev Environ Health. (2022) 39:163–89. doi: 10.1515/reveh-2022-0108
27. Boehmert C, Verrender A, Pauli M, Wiedemann P. Does precautionary information about electromagnetic fields trigger nocebo responses? An experimental risk communication study. Environ Health. (2018) 17:36. doi: 10.1186/s12940-018-0377-y
28. Leszczynski D. The grand challenge: use of a new approach in developing policies in the area of radiation and health. Front Public Health. (2014) 2:50. doi: 10.3389/fpubh.2014.00050
29. Williams RJ. Biochemical Individuality: The Key to Understanding What Shapes your Health. New Canaan, CT: Keats Publishing (1998).
30. Belpomme D, Carlo GL, Irigaray P, Carpenter DO, Hardell L, Kundi M, et al. The critical importance of molecular biomarkers and imaging in the study of electrohypersensitivity. A scientific consensus international report. Int J Mol Sci. (2021) 22:7321. doi: 10.3390/ijms22147321
31. Belyaev I, Dean A, Eger H, Hubmann G, Jandrisovits R, Kern M, et al. EUROPAEM EMF Guideline 2016 for the prevention, diagnosis and treatment of EMF-related health problems and illnesses. Rev Environ Health. (2016) 31:363–97. doi: 10.1515/reveh-2016-0011
32. Leszczynski D. Review of the scientific evidence on the individual sensitivity to electromagnetic fields (EHS). Rev Environ Health. (2021) 37:423–50. doi: 10.1515/reveh-2021-0038
33. Leszczynski D. Radiation proteomics: a brief overview. Proteomics. (2014) 14:481–8. doi: 10.1002/pmic.201300390
34. Rohlff C. Proteomics in molecular medicine: applications in central nervous systems disorders. Electrophoresis. (2000) 21:1227–34. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1227::AID-ELPS1227>3.0.CO;2-L
35. Toda M, Ono SJ. Genomics and proteomics of allergic disease. Immunology. (2022) 106:1–10. doi: 10.1046/j.1365-2567.2002.01407.x
36. Wilson KE, Ryan MM, Prime JE, Pashby DP, Orange PR, O'Beirne G, et al. Functional genomics and proteomics: application in neurosciences. J Neurol Neurosurg Psychiatry. (2004) 75:529–38. doi: 10.1136/jnnp.2003.026260
37. Alvarez-Chaver P, Otero-Estévez O, de la Cadena MP, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol. (2014) 20:3804–24. doi: 10.3748/wjg.v20.i14.3804
38. Hanash S, Schliekelman M. Proteomic profiling of the tumor microenvironment: recent insights and the search for biomarkers. Genome Med. (2014) 6:12. doi: 10.1186/gm529
39. Russell C, Rahman A, Mohammed AR. Application of genomics, proteomics and metabolomics in drug discovery, development and clinic. Ther Deliv. (2013) 4:395–413. doi: 10.4155/tde.13.4
40. Albertini RJ. Developing sustainable studies on environmental health. Mutat Res. (2001) 48:317–31. doi: 10.1016/S0027-5107(01)00191-9
41. Hodgson E. The future of human health risk assessment of environmental chemicals. Prog Mol Biol Transl Sci. (2012) 112:307–22. doi: 10.1016/B978-0-12-415813-9.00011-8
42. Sun B, He QY. Chemical proteomics to identify molecular targets of small compounds. Curr Mol Med. (2013) 13:1175–91. doi: 10.2174/1566524011313070010
43. Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. (2013) 121:756–65. doi: 10.1289/ehp.1205784
44. Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. (2016) 537:347–55. doi: 10.1038/nature19949
45. Hanash S, Taguchi A. The grand challenge to decipher the cancer proteome. Nat Rev Cancer. (2010) 10:652–60. doi: 10.1038/nrc2918
46. Stopfer LE, Flower CT, Gajadhar AS, Patel B, Gallien S, Lopez-Ferrer D, et al. High-density, targeted monitoring of tyrosine phosphorylation reveals activated signaling networks in human tumors. Cancer Res. (2021) 81:2495–509. doi: 10.1158/0008-5472.CAN-20-3804
47. Vogel C, Marcotte EM. Insight into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. (2012) 13:227–32. doi: 10.1038/nrg3185
48. Yeat NC, Lin C, Sager M, Lin J. Cancer proteomics: developments in technology, clinical use and commercialization. Expert Rev Proteomics. (2015) 12:391–405. doi: 10.1586/14789450.2015.1051969
49. Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers - blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. (2011) 8:142–50. doi: 10.1038/nrclinonc.2010.220
50. Fernando RC, de Carvalho F, Leme AFP, Colleoni GWB. Tumor microenvironment proteomics: Lessons from multiple myeloma. Front Oncol. (2021) 11:563384. doi: 10.3389/fonc.2021.563384
51. Cho WCS. Proteomics in translational cancer research: biomarker discovery for clinical applications. Expert Rev Proteomics. (2014) 11:131–3. doi: 10.1586/14789450.2014.899908
52. Marto JP, Carvalho AS, Mollet I, Mendonça M, Salavisa M, Meira B, et al. Proteomics to identify new blood biomarkers for diagnosing patients with acute stroke. J Am Heart Assoc. (2023) 12:e030021. doi: 10.1161/JAHA.123.030021
53. Bai B, Vanderwall D, Li Y, Wang X, Poudel S, Wang H, et al. Proteomic landscape of Alzheimer's disease: novel insights into pathogenesis and biomarker discovery. Mol Neurodegener. (2021) 16:55. doi: 10.1186/s13024-021-00474-z
54. Wallis RS, Maeurer M, Mwaba P, Chakaya J, Rustomjee R, Migliori GB, et al. Tuberculosis - advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect Dis. (2016) 16:e34–46. doi: 10.1016/S1473-3099(16)00070-0
55. Hristova VA, Chan DW. Cancer biomarker discovery and translation: proteomics and beyond. Expert Rev Proteomics. (2019) 16:93–103. doi: 10.1080/14789450.2019.1559062
56. Israr MZ, Heaney LM, Suzuki T. Proteomic Biomarkers of Heart Failure. Heart Fail Clin. (2018) 14:93–107. doi: 10.1016/j.hfc.2017.08.010
57. Wang H, Dey KK, Chen PC, Li Y, Niu M, Cho JH, et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer's disease. Mol Neurodegener. (2020) 15:43. doi: 10.1186/s13024-020-00384-6
58. Zhang X, Liu F, Li Q, Jia H, Pan L, Xing A, et al. proteomics approach to the identification of plasma biomarkers for latent tuberculosis infection. Diagn Microbiol Infect Dis. (2014) 79:432–7. doi: 10.1016/j.diagmicrobio.2014.04.005
59. Ngo D, Benson MD, Long JZ, Chen ZZ, Wang R, Nath AK, et al. Proteomic profiling reveals biomarkers and pathways in type 2 diabetes risk. JCI Insight. (2021) 6:e144392. doi: 10.1172/jci.insight.144392
60. Doll S, Gnad F, Mann M. The Case for proteomics and phospho-proteomics in personalized cancer medicine. Proteomics Clin Appl. (2019) 13:e1800113. doi: 10.1002/prca.201800113
61. Goetz LH, Schork NJ. Personalized medicine: motivation, challenges, and progress. Fertil Steril. (2018) 109:952–63. doi: 10.1016/j.fertnstert.2018.05.006
62. Raza A, Bourouba M, Dermime S. Editorial: Genomics, proteomics and immunological signatures as diagnostic, predictive, and prognostic biomarkers in head and neck cancers. Front Immunol. (2023) 14:1122736. doi: 10.3389/fimmu.2023.1122736
63. MacMullan MA, Dunn ZS, Graham N, Yang L, Wang P. Quantitative proteomics and metabolomics reveal biomarkers of disease as potential immunotherapy targets and indicators of therapeutic efficacy. Theranostics. (2019) 9:7872–88. doi: 10.7150/thno.37373
64. Qin T, Liu L, Wang X, Guo L, Lin J, Du J, et al. Ding G. Combined effects of EMP and RF field on emotional behavior in mice. Front Public Health. (2023) 11:1087161. doi: 10.3389/fpubh.2023.1087161
65. Fragopoulou AF, Samara A, Antonelou MH, Xanthopoulou A, Papadopoulou A, Vougas K, et al. Brain proteome response following whole body exposure of mice to mobile phone or wireless DECT base radiation. Electromagn Biol Med. (2012) 31:250–74. doi: 10.3109/15368378.2011.631068
66. Kuzniar A, Laffeber C, Eppink B, Bezstarosti K, Dekkers D, Woelders H, et al. Semi-quantitative proteomics of mammalian cells upon short-term exposure to non-ionizing electromagnetic fields. PLoS ONE. (2017) 12:e0170762. doi: 10.1371/journal.pone.0170762
67. Singh KV, Arya R, Nirala P, Sahu D, Nanda RK, Rajamani P. Effects of mobile phone electromagnetic radiation on rat hippocampus proteome. Environ Toxicol. (2022) 37:836–47. doi: 10.1002/tox.23447
68. Sepehrimanesh M, Kazemipour N, Saeb M, Nazifi S, Davis DL. Proteomic analysis of continuous 900-MHz radiofrequency electromagnetic field exposure in testicular tissue: a rat model of human cell phone exposure. Environ Sci Pollut Res. (2017) 24:13666–73. doi: 10.1007/s11356-017-8882-z
69. Sepehrimanesh M, Kazemipour N, Saeb M, Nazifi S. Analysis of rat testicular proteome following 30-day exposure to 900 MHz electromagnetic field radiation. Electrophoresis. (2014) 35:3331–8. doi: 10.1002/elps.201400273
70. Luo Q, Jiang Y, Jin M, Xu J, Huang HF. Proteomic analysis on the alteration of protein expression in the early-stage placental villous tissue of electromagnetic fields associated with cell phone exposure. Reprod Sci. (2013) 20:1055–61. doi: 10.1177/1933719112473660
71. Karinen A, Heinävaara S, Nylund R, Leszczynski D. Mobile phone radiation might alter protein expression in human skin. BMC Genomics. (2008) 9:77. doi: 10.1186/1471-2164-9-77
72. Zhang Y, Yao K, Yu Y, Ni S, Zhang L, Wang W, et al. Effects of 18 GHz radiofrequency radiation on protein expression in human lens epithelial cells. Hum Exp Toxicol. (2013) 32:797–806. doi: 10.1177/0960327112472353
73. Nylund R, Kuster N, Leszczynski D. Analysis of proteome response to the mobile phone radiation in two types of human primary endothelial cells. Proteome Sci. (2010) 8:52. doi: 10.1186/1477-5956-8-52
74. Gerner C, Haudek V, Schandl U, Bayer E, Gundacker N, Hutter HP, et al. Increased protein synthesis by cells exposed to a 1,800-MHz radio-frequency mobile phone electromagnetic field, detected by proteome profiling. Int Arch Occup Environ Health. (2010) 83:691–702. doi: 10.1007/s00420-010-0513-7
75. Nylund R, Kuster N, Leszczynski D. Proteomic Analysis of the Response of Human Endothelial Cell Line EA.hy926 to 1800 GSM Mobile Phone Radiation. J Proteomics Bioinform. (2009) 2:455–62. doi: 10.4172/jpb.1000105
76. Li HW, Yao K, Jin HY, Sun LX, Lu DQ Yu YB. Proteomic analysis of human lens epithelial cells exposed to microwaves. Jpn J Ophthalmol. (2007) 51:412–6. doi: 10.1007/s10384-007-0483-9
77. Zeng Q, Chen G, Weng Y, Wang L, Chiang H, Lu D, et al. Effects of global system for mobile communications 1800 MHz radiofrequency electromagnetic fields on gene and protein expression in MCF-7 cells. Proteomics. (2006) 6:4732–8. doi: 10.1002/pmic.200600234
78. Remondini D, Nylund R, Reivinen J, Poulletier de Gannes F, Veyret B, Lagroye I, et al. Gene expression changes in human cells after exposure to mobile phone microwaves. Proteomics. (2006) 6:4745–54. doi: 10.1002/pmic.200500896
79. Nylund R, Leszczynski D. Mobile phone radiation causes changes in gene and protein expression in human endothelial cell lines and the response seems to be genome- and proteome-dependent. Proteomics. (2006) 6:4769–80. doi: 10.1002/pmic.200600076
80. Nylund R, Leszczynski D. Proteomics analysis of human endothelial cell line EAhy926 after exposure to GSM 900 radiation. Proteomics. (2004) 4:1359–65. doi: 10.1002/pmic.200300773
81. Leszczynski D, Joenväärä S, Reivinen J, Kuokka R. Non-thermal activation of the hsp27/p38MAPK stress pathway by mobile phone radiation in human endothelial cells: Molecular mechanism for cancer and blood-brain barrier-related effects. Differentiation. (2002) 70:120–9. doi: 10.1046/j.1432-0436.2002.700207.x
82. Unger K. Integrative radiation systems biology. Radiat Oncol. (2014) 9:21. doi: 10.1186/1748-717X-9-21
83. Azimzadeh O, Atkinson MJ, Tapio S. Proteomics in radiation research: present status and future perspectives. Radiat Environ Biophys. (2014) 53:31–8. doi: 10.1007/s00411-013-0495-4
Keywords: electromagnetic hypersensitivity, biomarkers, environmental sensitivity, radiation sensitivity, individual sensitivity, provocation studies, proteomics, omics technologies
Citation: Leszczynski D (2025) Wireless radiation and health: making the case for proteomics research of individual sensitivity. Front. Public Health 12:1543818. doi: 10.3389/fpubh.2024.1543818
Received: 11 December 2024; Accepted: 30 December 2024;
Published: 10 January 2025.
Edited by:
George Louis Carlo, Longwood University, United StatesReviewed by:
Victor Alan Leach, Oceania Radiofrequency Scientific Advisory Association, AustraliaCopyright © 2025 Leszczynski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dariusz Leszczynski, YmxvZ2JyaHBAZ21haWwuY29t
 Dariusz Leszczynski
Dariusz Leszczynski