- 1Clinical Medicine (Surgery), Emergency Medicine and Social Medicine, Private Practice, Düsseldorf, Germany
- 2Department of Evolutionary Genetics and Biosystematics, Faculty of Biology, University of Gdansk, Gdansk, Poland
- 3Norwich Medical School, University of East Anglia, Norwich, United Kingdom
Introduction: Facemasks were widely mandated during the recent SARS-CoV-2 pandemic. Especially the use by the general population is associated with a higher risk of improper handling of the mask and contamination and potential adverse microbiological consequences.
Methods: We investigated and quantified bacterial accumulation in facemasks used by the general population, using 16S rRNA (Sanger Sequencing), culture and biochemical analysis along with Rose Bengal staining. Additionally, a systematic overview of the literature on face mask contamination was undertaken.
Results: We found an average bacterial load of 4.24 × 104 CFU recovered/mask, with a maximum load of 2.85 × 105 CFU. This maximum is 310 times higher than the limit value for contamination of ventilation system outlet surfaces specified by the German standard VDI 6022. Biochemical and molecular identification predominantly found Staphylococcus species (80%), including Staphylococcus aureus, along with endospore-forming Bacillus spp. Literature reports also indicate contamination of masks by bacterial and fungal opportunists of the genera Acinetobacter, Aspergillus, Alternaria, Bacillus, Cadosporium, Candida, Escherichia, Enterobacter, Enterococcus, Klebsiella (including K. pneumoniae), Micrococcus, Microsporum, Mucor, Pseudomonas, Staphylococcus and Streptococcus. Bacterial counts increase linearly with wearing duration.
Discussion: Prolonged use may affect the skin and respiratory microbiomes, promoting consequential eye, skin, oral and airway conditions. These aspects underscore the urgent need for further research and a risk-benefit analysis in respect of mask use, particularly given their unproven efficacy in disrupting the transmission of respiratory viruses and their adverse social consequences.
Introduction
Facemasks covering the entrances to the airways were widely mandated during the recent SARS-CoV-2 pandemic, not only for healthcare workers but also for the general population (1). Professions with frequent human contact were obligated to wear them for long periods as were schoolchildren (1–6).
This raises reasonable concerns: first, because use by the general population is associated with a higher risk of improper handling of the mask (7–11); secondly because their efficacy against respiratory viral infections is unproven by high quality trials, which indicate little or no effect (12, 13) and thirdly, because masks are assumed only to have positive effects (14–16). In reality there is strong evidence that masks pose various risks, especially for pregnant women, children and adolescents, as well as older adults and the unwell (14, 16–19). They have several demonstrably adverse effects, affecting physiology (14, 16, 19–23), psychology (16, 24) and, most obviously, social interactions (25–35). Effects on childhood development are a particular concern. These adverse effects have been recently summarised as the so-called mask-induced exhaustion syndrome MIES (14, 16, 19). Interestingly, Spira (36) and Fögen (37) found significantly higher SARS-CoV-2 infection and mortality rates in the mask-wearing cohorts: explanations are uncertain, but viral trapping and recycling are plausible.
A further concern, encompassed within MIES, relates to the potential adverse microbiological consequences of wearing face masks. Owing to the creation of a warm, moist micro-environment (38–41), bacteria, fungi and even viruses may accumulate on both sides of the worn masks (42–46). So far, these aspects have not been evaluated in depth. The aim of our pilot study was to assess, visualise and categorise the general ability of masks to accumulate bacteria when used by the general population. This also with regard to a risk assessment, using worst-case consideration which is necessary in such a protective approach (47). Accordingly, we undertook a microbiological exploration with random samples of face masks as used by members of the general population, together with a systematic rapid literature review. This combined holistic approach with 16S rRNA (Sanger sequencing), culture and biochemical analysis along with Rose Bengal staining plus systematic literature analysis has not been performed before and is the first of its kind.
Materials and methods
Rose Bengal staining and visualisation of contamination
Staining with Rose Bengal sodium salt was used to detect contamination of masks, as described previously (45). Figure 1 illustrates the area of the mask analysed.

Figure 1. Rose Bengal staining of worn face masks. The area analysed is marked by the red frame. The mask dimensions indicated by the manufactures (175 × 95 mm) exclude folds, which enlarge the surface area.
Microbiological mask study design
In this pilot sample study surgical face masks were collected in March 2022 (during the pandemic obligation) from 15 random willing volunteers (employees of the Gdansk University Department aged 19–65 years), who had worn them for periods from 15 min to 12 h. Wearer details were not further recorded as this did not appear to be crucial for our pilot study, which was intended to show the possible contamination of masks used by the working general population. However, with our random sample, we have captured a realistic usage profile with typical temporal fluctuations due to different users from the general population. Each mask was stored in a separate plastic bag until examination. The masks, excluding the ear loops, were then aseptically cut in several pieces using sterile scissors in a laminar flow cabinet. These pieces were transferred to tubes containing 15 mL of sterile phosphate-buffered saline (PBS), equilibrated for 1 min at room temperature, then vortexed for 30 s. Three unused, clean, surgical masks (Shandong KaiBo Medicinal Packaging Co., Ltd., China) were processed identically as negative controls.
To determine bacterial counts, the suspensions were diluted 10- and 100-fold, then 100-μl volumes were spread on Columbia Agar containing 5% sheep blood (Graso Biotech, Owidz, Poland). Plates were incubated aerobically overnight at 37°C, then colonies were counted. The bacterial load was determined as colony forming units per ml (CFU/mL) of suspension, then rebased as CFU/mask (38). Ten colonies per worn mask were re-plated, grown on Tryptic Soy Broth (Graso Biotech, Owidz, Poland), then stored in 15% glycerol stock solutions (v/v) at −70°C pending molecular identification.
Identification of isolates by sanger sequencing of the 16S rRNA gene
Forty isolates were identified by PCR and Sanger sequencing of the 16S rRNA gene. Briefly, bacterial colonies were suspended in 30 μL of sterile water and lysed in 95°C, followed by centrifugation at 13,000×g for 2 min. The supernates were used for PCR. Primers were: forward F27 5′-AGAGTTTGATCMTGGCTCAG-3′ and reverse R1492 5′-CTACGGYTACCTTGTTACGACTT-3′ (48, 49). The reaction mixture (25 μL) contained: 0.1 μM of each primer, 1 μL of bacterial supernatant, 0.6 U of Taq polymerase (EURx, Gdansk, Poland), 0.2 mM dNTPs and Taq Polymerase buffer (EURx), containing 15 mM MgCl2. Cycling conditions involved 94°C for 5 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 1.5 min and a final step at 72°C for 5 min. Sanger sequencing was performed at Macrogen Europe (Amsterdam, The Netherlands) on a 3730xl DNA Analyzer (Thermofisher Scientific, Waltham, MA, USA). PCR amplification was as described by Monciardini et al. (50). The sequencing data were analysed by FinchTV 1.4 (Geospiza, Inc.; Seattle, WA, USA),1 the ends of sequenced reads were trimmed, and the resulting assemblies were blasted in the NCBI database. Sequencing data are available in Figshare at https://doi.org/10.6084/m9.figshare.23614797 (accessed on 2 July 2023).
Biochemical characterisation of isolates
All sequenced isolates were re-plated on Columbia Blood Agar with 5% sheep blood for evaluation of haemolysis, and on Mannitol Salt Agar (Graso Biotech, Owidz, Poland) for the preliminary identification of Staphylococcus spp. Staphylococci were further tested using the STAPH LATEX KIT (Prolex™, Pro-Lab Diagnostics, Bromborough, UK) to distinguish S. aureus from other species.
Systematic literature search
We systematically searched for peer-reviewed, scientific studies, up until June 2023, that quantitatively analysed colonisation or contamination of cloth, surgical, N95 and similar masks by bacteria and fungi. The search was performed using PubMed and MEDLINE and included both qualitative and quantitative evaluations. Search terms were created according to the criteria defined in the PICO scheme (51). The non-specific search term “mask” was omitted, as it also includes respirators and anaesthesiologic ventilation masks. Instead, specific terms were chosen: “((face mask) OR (facemask) OR (surgical mask) OR (FFP1) OR (FFP2) OR (FFP3) OR (N95) OR (KF94) OR (KN95)) AND ((microbial contamination) OR (bacteria) OR (fungi)).” Two independent researchers identified and screened eligible studies. Qualitative inclusion criteria were: valid reproducible presentation of the microbial contamination, comprehensible collection of evaluated masks, credibility of the results and clear focus. Quantitative inclusion criteria were: appropriate and precise methods, valid measurement of outcomes, representative selection of evaluated masks and reproducible detection/analytical methods. Selected papers were checked by at least three of the present authors for potential eligibility. Study design, methodology, analytical and experimental methods and outcomes were evaluated. Exclusions and reasons were documented. For included studies, the following data were extracted into tables: author and year, method and type of study, sample size and mask type(s), mask wearing duration(s), outcomes/examined microorganisms, content and main species. Simple mathematical calculations and graphics were performed with Libre-Office Calc, a free and open-source office package from The Document Foundation (52).
Results
Abundance and types of bacteria on worn masks
Contamination of worn masks was visible, macroscopically, after staining with Rose Bengal (Figure 2). This dye binds to bacteria, fungi and tissue cells along with debris with the colour intensity suggested to reflect the degree of contamination (53–57).
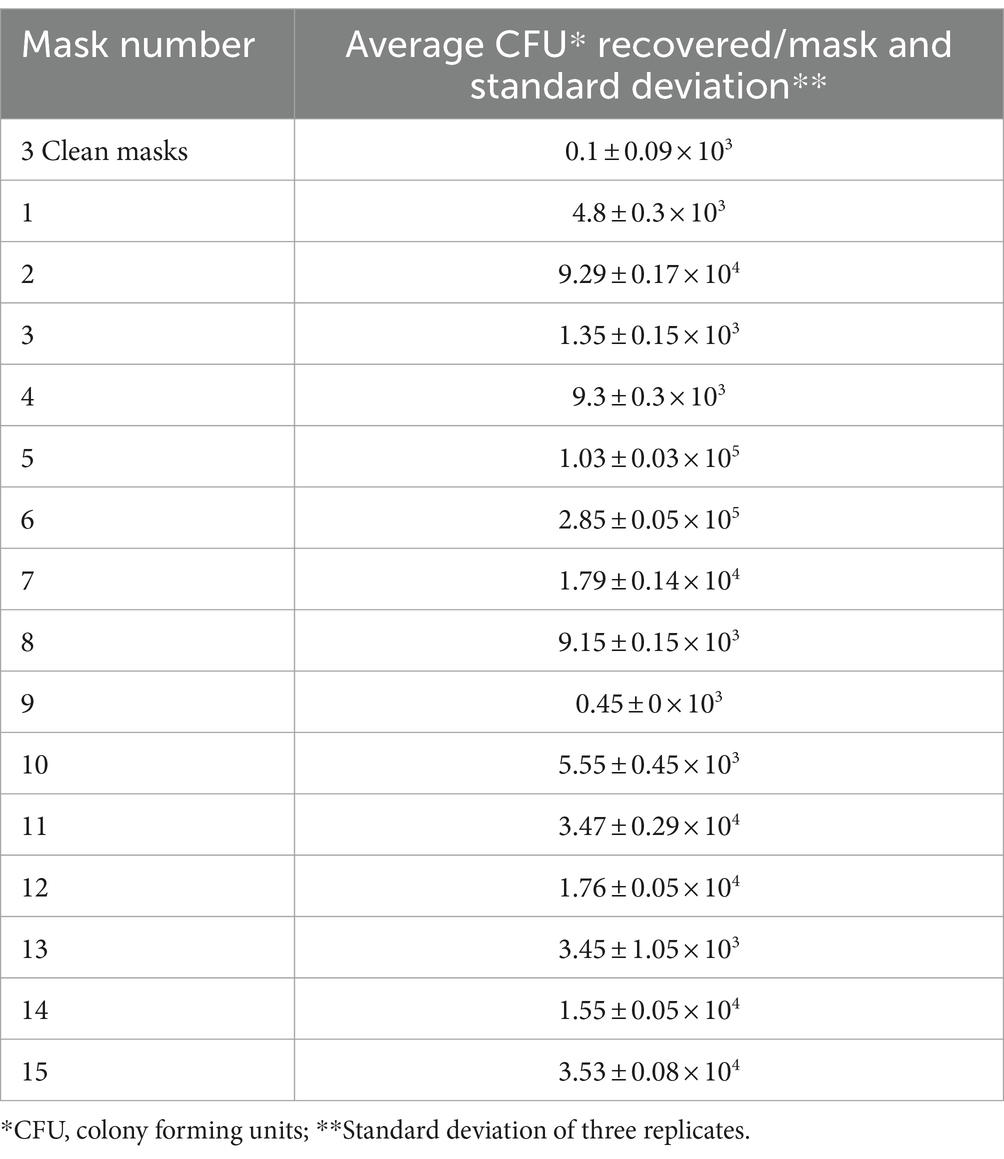
Based upon culture, the average bacterial load of clean, never-used surgical face mask was 0.1 × 103 CFU recovered/mask whereas the arithmetic mean load on used masks was 4.24 × 104 CFU recovered/mask (geometric mean 1.3 × 104). Bacteria were most abundant on worn masks 5 and 6, with 1.03 × 105 and 2.85 × 105 CFU recovered/mask, respectively (Table 1). Biochemical and molecular identification revealed staphylococcal species on both these latter masks, including S. aureus, S. warneri and S. epidermidis (Supplementary Table 2). Although colony morphology differed between masks, the dominant phenotypes, in almost all cases including the unused masks, were the small white colonies typical of S. epidermidis and other coagulase-negative staphylococci (Supplementary Figure 1).
Identification of isolates by sanger sequencing of 16S rRNA gene
Out of 52 colonies subjected to PCR we chose the 40 with the most efficient product amplification for sequencing. Detailed BLAST results are presented in Supplementary Table 1.
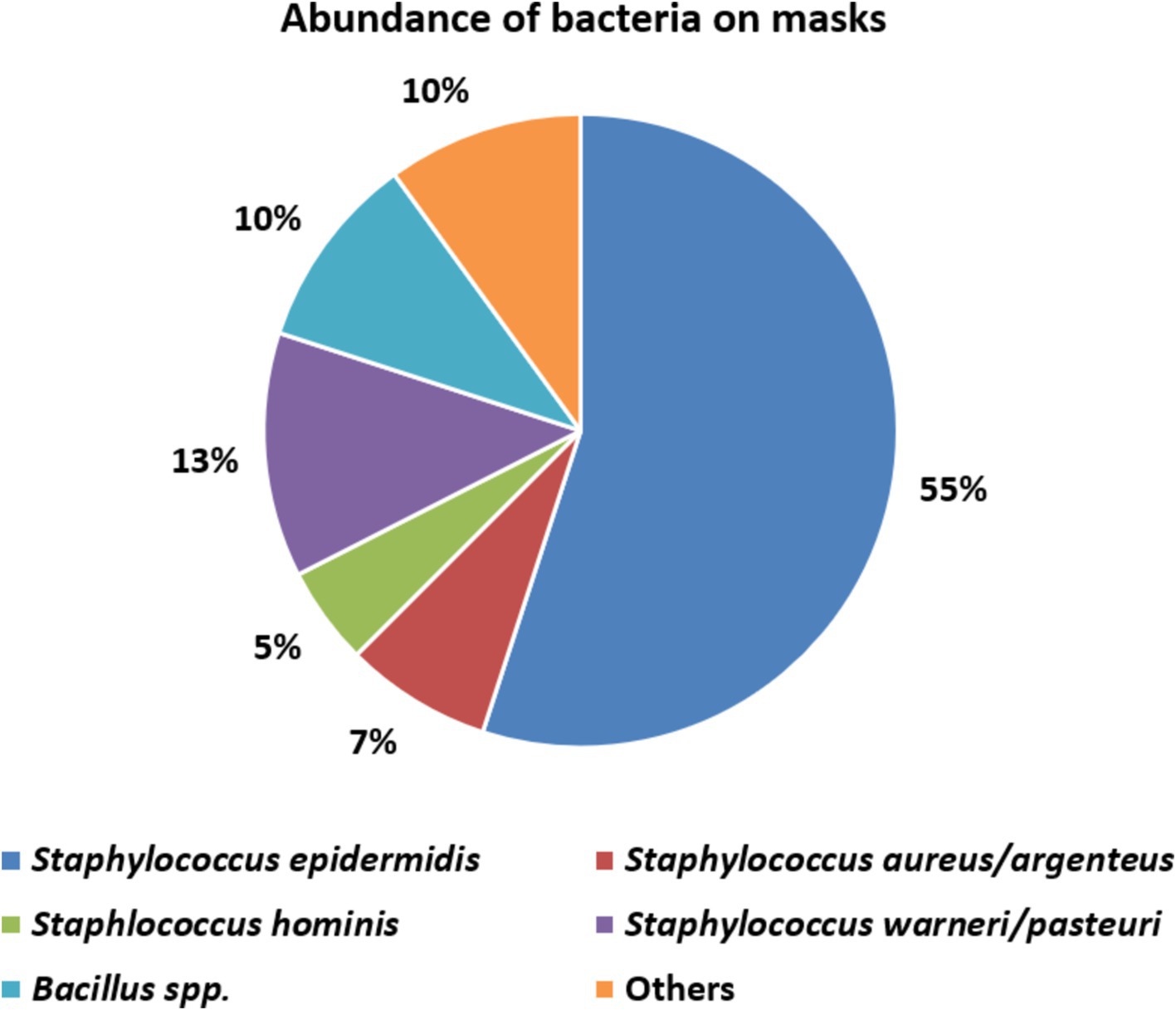
The great majority (32, 80%) of these 40 belonged to the genus Staphylococcus confirming phenotypic identifications. We identified four coagulase-negative species: S. epidermidis (the most abundant), S. warneri, S. pasteuri and S. hominis, all of which belong to the normal human skin and nasal microbiota (Supplementary Table 2) (58). On mask 5 we confirmed coagulase-positive Staphylococcus (Supplementary Table 2) along with S. aureus and S. argenteus.
A further four sequenced colonies comprised endospore-forming Bacillus species, namely B. cereus, B. thuringiensis, B. altitudinis, B. megaterium and others (Supplementary Tables 1, 2), which are soil bacteria (59). Among the four remaining identified colonies (‘Others’ in Figure 3) we found Sporosarcina newyorkensis, another endospore-forming Gram-positive rod, occasionally recovered from human bacteraemias and cow’s milk (60). The sole Gram-negative species found was the pseudomonad Psychrobacter faecalis (Supplementary Table 2), a psychrophilic species associated with pigeon faeces (61) and reported also from human samples (62). We did not isolate streptococci, although these are a major component of the human oral microbiota. Perhaps, their survival rates on the masks is low, or their recovery requires CO2-enriched incubation, not air incubation as used here.
Biochemical identification of isolates
The same 40 colonies were subjected to biochemical identification, yielding results consistent with the sequencing. Haemolysis was detected for almost all these bacteria (Supplementary Table 2) though its intensity was very variable (Supplementary Table 2; Supplementary Figure 1). Most of the bacteria showed halotolerance but only five fermented mannitol: these latter were tested for coagulase and protein A and three, all from mask 5, proved positive for both characters, confirming identification as S. aureus (Supplementary Table 2); all had morphology typical of the species (Supplementary Figure 1, Mask 5).
Systematic literature search
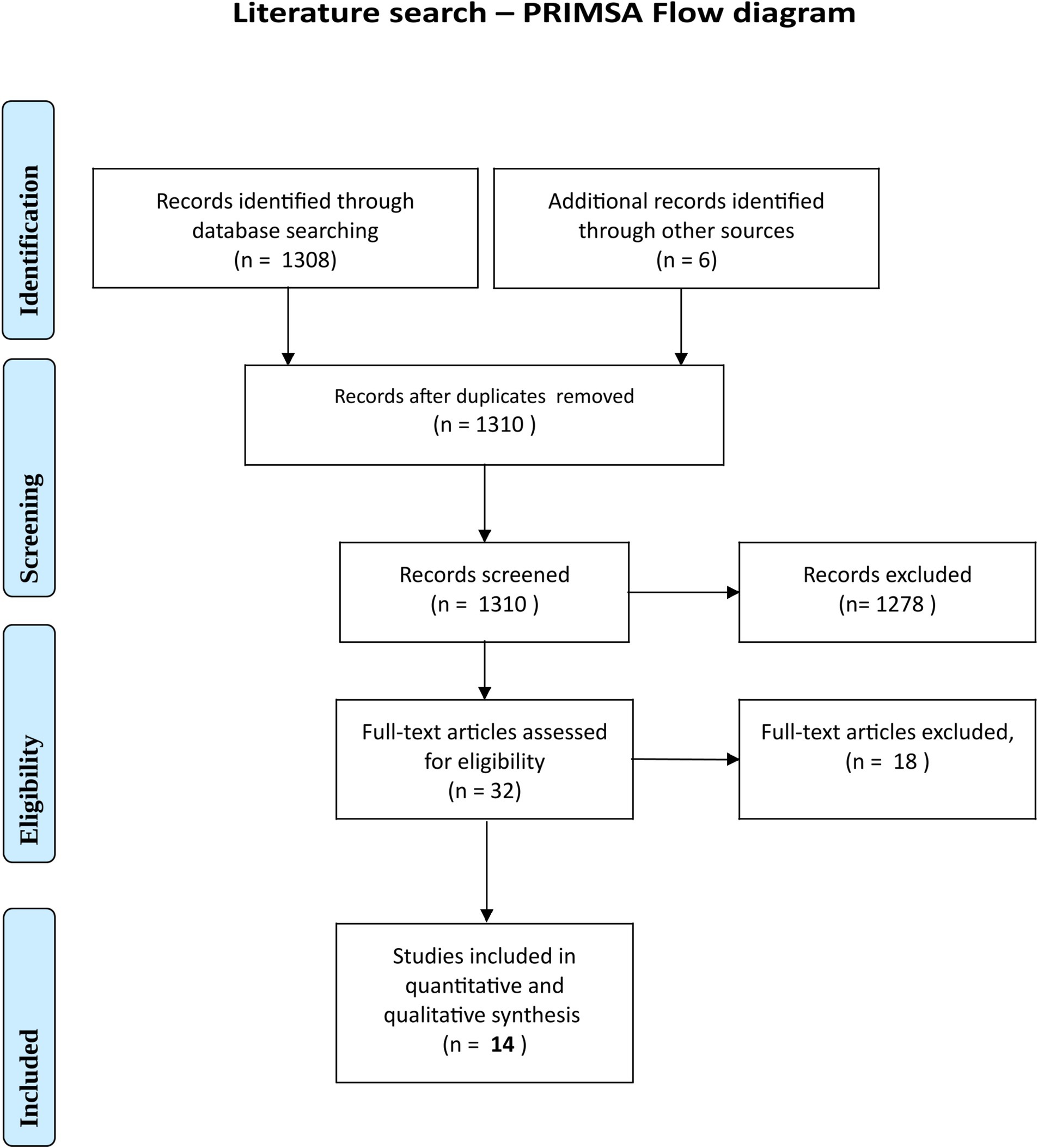
The literature search initially yielded 1,310 results. This was narrowed (see PRISMA diagram, Figure 4) to 14 studies evaluating bacterial and fungal contamination of cloth, surgical and N95 masks, worn for periods ranging from 5 min to 3 days. Eleven studies considered bacteria, five fungi, and three both (Table 2). Four studies were for the general population whereas 10 were for healthcare workers (HCWs) (38, 41, 42, 44, 46, 63–71). Six were for surgical units (one specifically performing orthopaedic surgery) and five for dental practices (44, 64–67). Only two provided exact quantification and bacterial identification by 16S rRNA; these both investigated the general population (38, 63). Results of the literature search are summarised in the extraction (Table 2).

Table 2. Microbiological findings of the literature search (mask contamination with bacteria and fungi).
Discussion
We found heavy bacterial contamination of surgical masks worn by the general population, with up to 2.85 × 105 CFU/mask (average 4.24 × 104).
Unfortunately, there are no microbiological standards for worn masks against which to review these findings; in the EU the only relevant bioburden requirement is EN 14683 for new masks, requiring ≤ 30 CFU/g. Nevertheless, since masks amount to a filtering system upstream of the respiratory tract, the limit values for ventilation systems are pertinent, notably the German standard for surfaces of ventilation and air-conditioning, VDI 6022, part 4 (72). This specifies counts of 25 to 100 CFU/25 cm2 as ‘borderline’, whilst surfaces with counts > 100 CFU/25 cm2 require immediate action or replacement.
A disposable surgical mask has a one-side surface area of ca. 230 cm2 (73), meaning that in our worst case (2.85 × 105 CFU/mask = 3.09 × 104 CFU/25 cm2), the upper limit of VDI 6022 was exceeded by ca. 310-fold (average 46-fold) (Table 1). Values from a comparable study show 166-fold exceedance with cotton masks (38); another study, for healthcare workers with surgical masks worn for an unspecified period, indicated > 2,000-fold exceedance (Table 2) (44). It should be added that the bacterial burden of a mask lies directly in front of the respiratory tract whereas the vent of an air-conditioning system typically lies several metres away.
The EN 14683 requirements for new masks also were widely exceeded for worn items (Table 2), based upon weights of ca. 3 g for a surgical mask and 4 g for N95/FFP2 masks (74); exceedance of this requirement was evident even for the unworn masks (Table 1).
The heavy general contamination of worn masks was further demonstrable by Rose Bengal staining (Figure 2).
Bacteria detected: potential clinical implications
Microbiological investigation of used mask predominantly revealed coagulase-negative skin staphylococci and endospore-forming soil bacteria (Bacillus spp.) on used (Figure 3). This predominance of staphylococci is in line with other studies on contaminated face masks in the general populace and healthcare workers (42, 44, 64–66, 68). One mask (no. 5) was contaminated with S. aureus, a well-known and versatile pathogen (Figure 3; Table 1) (75–78). Up to 30% of the population carry nasal S. aureus without symptoms (79) though with an increased risk for autoinfection (75). Contingent contamination of masks may facilitate dissemination of S. aureus and, plausibly skin infection (75). An association between nasal carriage and surgical as well as KN95 mask contamination was shown previously for S. aureus and even for the non-carriers, the organism was frequently detected on KN95 masks (p = 0.04, Fisher’s exact test) implying exogenous sources of contamination (hands, environment and external droplet containing air streams etc.) (75). In support of this, some authors note S. aureus contaminates on the external as well as internal surfaces of masks (75).
Several authors have associated the use of face masks skin eruptions, some involving S. aureus (80) including new occurrence or exacerbation of acne, rosacea, and seborrhoeic dermatitis (81). Other authors note enrichment of the normal eye microbiota with S. aureus from exhaled breath and droplets while wearing a mask contributing to the development of eyelid inflammation (chalazion) (82, 83) and infections of the cornea (84), also deeper eye infections in the context of treatments (endophthalmitis following vitrectomy) (85). There is also some evidence that S. aureus can increase replication of the SARS-CoV-2 virus by 10- to 15-fold (86), though this seems more pertinent in the upper nose than on a mask, where the virus is unlikely to be replicating.
Among other staphylococci, we predominantly found S. epidermidis (Figure 3). On one hand this is a normal and harmless component of the skin microbiota; on the other, it may be a hazard for vulnerable immunosuppressed individuals (87–89). Even in healthy individuals, coagulase-negative staphylococci, at high abundance, may contribute to inflammatory skin conditions such as atopic dermatitis and acne vulgaris (58, 90–92) with evidence that wearing a mask significantly increased the incidence of acne in particular (93–101).
We also found Bacillus spp. in the masks, including species that produce enterotoxins (59). Although bacterial growth in masks may be possible (see below) we saw no evidence that growth attained the levels—typically >106/g—associated with toxins in food (102). Moreover, wearers (except maybe children) are unlikely to chew on their masks, meaning that these organisms can be dismissed as a risk.
Literature review on mask contamination
Our literature review showed that all relevant mask types (surgical, N95, cloth) become increasingly contaminated with microorganisms during wear (Table 2; Figure 5) (38, 40, 41, 46, 65, 67, 71).
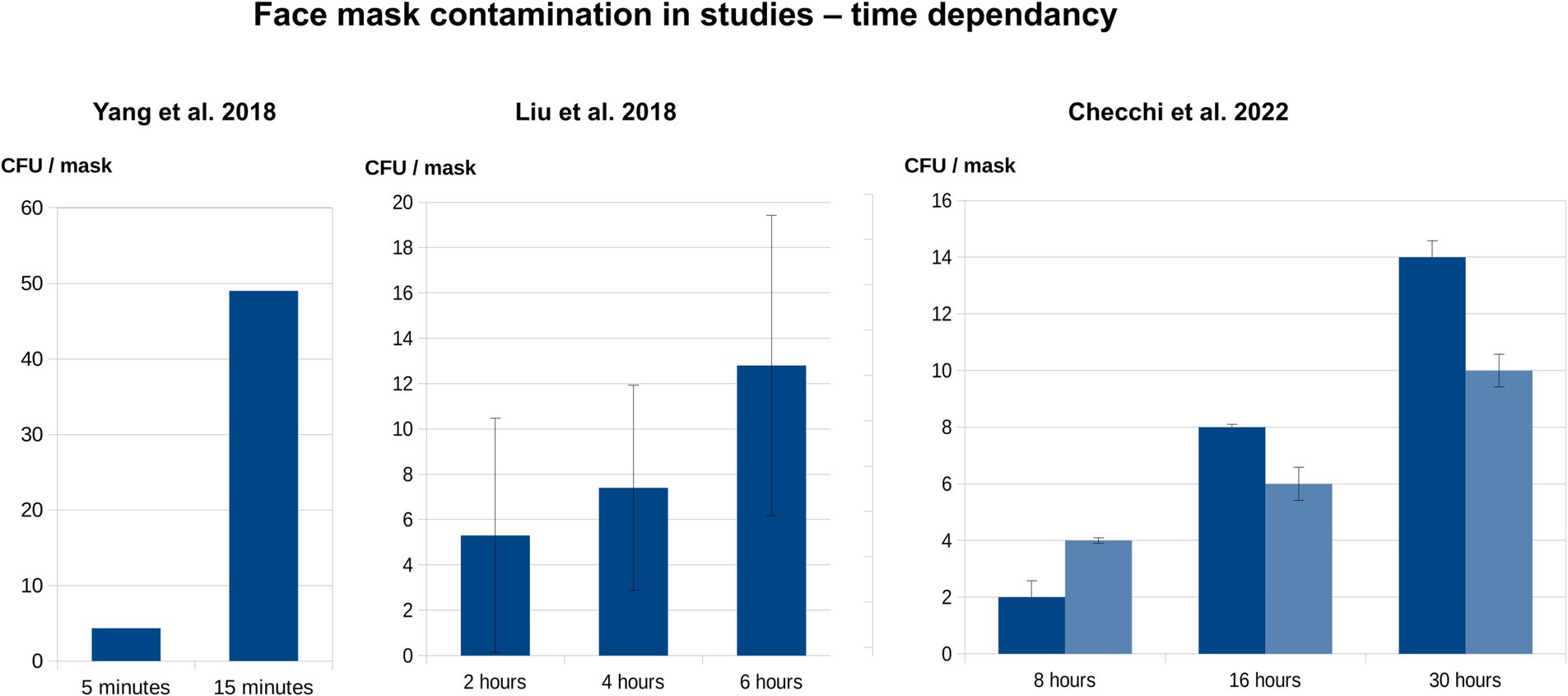
Figure 5. Time dependency of face mask contamination during wear, based upon literature data (Table 2). The diagrams indicate the association between CFU/mask and wearing duration, based on mean values from three publications (41, 46, 67). If included in the primary studies, the standard deviations are also shown. Yang et al. investigated the inner surfaces of masks worn by the general population, whereas both Liu et al. and Checchi et al. examined the outer layers of masks worn by HCW.
The literature reports contamination by bacteria of the genera Acinetobacter, Bacillus, Escherichia (specifically, E. coli, a faecal organism), Enterobacter, Enterococcus (another faecal organism), Klebsiella (including K. pneumoniae), Micrococcus, Pseudomonas, Staphylococcus (including S. aureus) and Streptococcus and by fungi of the genera Aspergillus, Alternaria, Candida, Cadosporium, Microsporum and Mucor (Table 2). These organisms are nourished by human saliva, nebulised oral biofilm and exhaled breath condensate, creating an underestimated biosafety concern.
In the general population, internal mask contamination typically exceeds external for bacteria—and perhaps, although this varies with the study—also for fungi (Table 2) (63, 70). For the healthcare workers using surgical masks, on the other hand, external contamination exceeds internal contamination both for bacteria and fungi (p < 0.001) (42, 44, 64) and correlates with microbiological air quality in the areas where these staff were working (42). For N95 masks, however, internal bacterial contamination appears higher than external even in healthcare settings (68). Moreover, the total bacterial contamination of worn N95 masks exceeded that of similarly worn surgical masks (68).
Fungal contamination is seen up to 70–88% of used masks (70, 71), and can be also higher inside than outside the mask (70). This is surprising, given that fungi must come from outside the mask (63).
A comparison of maximal bacterial face mask counts for healthcare workers and the general population, based on Table 2 data and wearing/using times between 5 min and 3 days, showed a high variance in data due to the variance in wearing times and users and environmental factors. There is a tendency for higher bacterial loads in the general population (Table 2). These findings may reflect wider inappropriate and extended usage in the general population (7, 8). Due to the small number of similar studies, a meta-analytical statistical evaluation was not carried out.
Face mask contamination—contributing factors
Masks are a good matrix for microbial accumulation and potentially, growth, retaining an above-ambient temperature (103–107), moisture, and nutrient-rich debris (38–41, 45, 108). Besides substances sucked in from the outside, nutrients comprise exhaled proteins and other debris, exfoliated and dead epithelial cells. Condensing droplets in the exhaled breath contain non-volatile metabolites, salts, lipids and proteins along with intact and degraded bacteria and viruses (109). This organic richness was visualised in our Bengal Rose staining. Growth, rather than mere survival (38, 39, 41, 45, 108, 110) of bacterial and fungal colonies is revealed by scanning electron microscopy of face masks (FFP2) worn for several hours (40).
The dead-space of rigid N95 masks provide a particularly warm, wet environment (103) with a relative humidity 1.5–2.6 times higher than externally (41) rising to 100% after 60 min of use (40). This may create a particularly attractive breeding ground for bacteria (41) explaining the findings (above) that the N95 masks become more heavily contaminated than surgical masks and that, in healthcare settings, internal contamination exceeded external, reversing the pattern seen for surgical masks (68).
Microorganisms trapped and incubated in the mask may be distributed to the wearer, the environment and to others (16, 111–113). If leakage, owing to defect or poor fit, affects 1% of the mask area, the filtration efficiency is reduced by 50%; if the gap is 2% of the mask area, efficiency is reduced by 75% (114). Moreover, the exhalation filtration efficiency is significantly lower than the theoretical filtration efficiency—being 12.4 and 46.3% for surgical and N95 masks, respectively (115). In operating theatres, the recommended wearing duration is limited to few hours (116) as surgical masks lose effectiveness over time (117). Whereas a fresh mask almost completely prevented bacterial contamination of an agar plate held 10–12 cm from the mouth, this effectiveness was measurably reduced within 30 min and negligible after 2 h (118). This brief period of filtration efficiency was further reduced if the mask was poorly fitted (114, 119) or wetted (119).
Penetration of microorganisms between mask layers is possible, through capillary action depending on humidity and the specific organisms among other factors (120). This in turn, may facilitate the formation of tiny organism-laden droplets. These then may be projected or inhaled with every breath (16, 111, 114, 115, 121–123). In this context, we underscore the predominantly oral breathing while wearing a mask (16, 124), in contrast to normal unimpeded breathing, which is largely via the nose, with greater filtration. Oral breathing increases the hazard of directly inhaling microorganisms from the mask into the deeper airways (125). In a human study with a radiolabelled aerosol and average particle diameters of 4.4 μm (range 3.8–5.1 μm) scientists found a large increase in deposition in the lungs (+37%) when breathing orally compared with via the nose (75% vs. 38%) (126). Additionally, masks—and especially the N95 type—impair natural mucociliary clearance of the upper airways, further enhancing inhalation and distribution of bacteria (127).
Finally, in context, face masks contain plastics, to which microorganisms can adsorb (40, 128). Consequently, as well as aerosols, plastic micro-particles may also be released by masks (129–133), acting as carriers for the distribution of pathogenic bacteria and fungi (134). Interestingly, there is hardly any surface or material, not even the bare skin, that ensures such survival and long-term preservation of infectivity for the viruses as the plastic-polypropylene network of the masks, in which SARS-CoV-2 viruses are stored and remain infectious for up to 2 weeks—even when dried (135).
Face mask contamination—potential clinical implications
In a pre-COVID cross-sectional study on 710 individuals, the wearing (for religious reasons) of cloth facial coverings by Saudi women, drawn from the general population, was associated with statistically increased incidences of ‘common cold’ and asthma (17). Elsewhere, pathophysiological skin changes (136) were associated with mask wearing in the general population and healthcare workers (137, 138). Several authors found changes in skin metabolomics, with an increased risk of barrier disruption and inflammation, putatively owing to dysbioses of the skin microbiome (136, 139, 140) leading to—or promoting development of—atopic dermatitis and acne vulgaris (139). In context N95 respirators caused a more significant disorder than surgical masks (139).
Eye conditions also have been associated with mask use (82–85, 121, 141–145), whilst Islam et al. found indirect evidence of changes in the oral microbiome (146). Sukul et al. changes in the gut microbiome (metabolic alterations) (19) whilst Xiang et al. found change of the nasal microbial communities after prolonged mask wearing (110). Lastly, face masks are mentioned as possible factors behind an increase in mucormycosis cases during the COVID-19 pandemic particularly in immunocompromised or otherwise vulnerable individuals (70, 71, 147).
Practices for minimising microbial contamination
There are general considerations for the use of face masks in any situation, along with official advice on their proper use (16, 129, 148). Minimising microbial contamination is critical to ensure their safe use, especially in healthcare. The WHO recommends to avoid touching the mask surface, also that masks should be stored in a clean, dry place away from potential contaminants (6). Disposable masks should be removed after each use and not reused. Training should be provided on how to put on and take off masks so as to prevent microbial spread and self-infection. The WHO further recommends cleaning hands before touching a mask (both before and after removing it). When the mask is removed, it should be stored in a clean plastic bag or disposed of in a waste garbage can (6).
In some situations, a face shield can be used in conjunction with masks to provide an additional barrier against contamination. Lastly, the mask should be worn for as short a time as possible, not only for microbiological reasons (time-dependent contamination of the face mask during wearing), but also for toxicological and physio-metabolic reasons (14, 129).
It is self-evident that large sections of the population, including children, are unable to follow these complex instructions adequately and consistently (148). Alternatives to masks should be researched and prioritised (e.g., ventilation systems, hygiene measures and other).
Findings in context
Long before the pandemic, face masks became widely used in medicine (notably surgery) healthcare and some manufacturing industries (16, 149–151), aiming to prevent or minimise infection or contamination (8, 14, 73, 151–159). Nevertheless, their effectiveness in healthcare settings was debatable long before 2020 (160) and their role in the operating theatre remains controversial (161). Given this history, there has been surprisingly little research on the effects of long-term usage by professional groups. Although masks filter larger debris and aerosol droplets from the air, they carry the microbiological risks outlined here along with toxicological, physiological, psychological and sociological harms (14, 16, 18–35, 129, 162).
The risks and benefits of requiring mask use by populations must be weighed from ethical and medical standpoints (13, 14, 16, 163, 164). For masks to be demanded, the side effects and risks must be lower than the risk of not wearing a mask. A gold-standard Cochrane evaluation, based on clinical trials (12) found no substantive evidence of efficacy in preventing viral respiratory infections and one recent study, albeit with several possible confounders, even found mask-wearing to be associated with an increased risk of COVID-19 infection (165). On the other hand, the potential harms are numerous (2, 3, 5, 14–16, 19–23, 36, 37, 166–172). They include MIES (16), harmful blood-gas alterations (14, 19) and the potential microbiological hazards outlined here. Masks should not be mandated for the general population given this balance of evidence against their use. These points have been raised by many scientists (14, 16, 17, 36, 37, 129, 166, 173–175) including leading breathing experts (176).
Limitations and strengths
The strengths of our paper are the use a precise method—16S rRNA sequencing—to identify the bacteria found. In addition, we undertook a systematic literature overview and discuss the results from holistic microbiological and clinical perspectives. The masks collected in our study were provided by random individuals during daily life, representing a realistic general population sample. Rose Bengal staining strikingly visualised extensive contamination. Both, our limited sample size and rapid literature review should be seen only as a pilot assessment, with further analysis needed. Due to the small numbers of studies of same design, a meta-analysis was not carried out. Rather the strength of this review is qualitative, cataloguing the extensive scientific literature published by many scientists worldwide over several decades, demonstrating experimental evidence of face mask contamination and its risks.
Conclusion
Both our experimental study and the published literature show that face masks accumulate microorganisms, including pathobionts (Tables 1, 2) (38, 41, 42, 44, 46, 63–71, 177), with a microbial load up to several hundred times higher than the German standard VDI 6022 limit for ventilation systems surfaces (72) and the EN 14683 requirements for unused masks. Contamination increases with extended wearing time (Figure 5) (38, 41, 46, 65–67, 70, 71) and is greater for N95 than surgical masks (68). Most contamination was with staphylococci, occasionally including the pathogen S. aureus.
Put simply: (i) the mask act as a filter trap with bacteria accumulating on its external and internal surfaces; (ii) the mask then acts as a “microbiological incubator” at the entrance of the airways; (iii) microorganisms may grow within the mask, nourished by skin debris, mucus and “exhaled breath condensate” (16, 38, 39, 41, 45, 108–110). These trapped organisms/pathogens then may be inhaled, promoting infection of the respiratory tract (17, 37) or, when distributed via air streams (111, 114, 115, 122, 142, 143, 178, 179) the eye (82–85, 121, 142). In addition, the skin microbiome is disrupted, potentially leading to or promoting other infections and allergic conditions (38, 77, 110, 140, 180).
Lastly, accumulated microorganisms may be distributed via leakage (111, 114, 115), amplified by the atomiser effect of the mask (14, 16, 122, 181, 182).
A Cochrane analysis, based solely on the highest grade of evidence, found no evidence that masks reduced the spread of respiratory viral infections in the general population (12). On the other hand, their detriments, over and above those investigated here, are clear. They impede communication (32–34, 94, 183–188). They impede learning, especially for children (2, 3, 5, 14, 26, 35, 148, 162, 171, 174, 177, 189). They are associated with transient hypoxaemia (decreased blood O2), transient hypercarbia (increased blood CO2) (14, 16, 19, 21–23, 171, 172). They deny the wearer of the most basic individuality—of showing their face (26, 27, 30–34, 162, 189). Their long-term imposition is especially harmful for vulnerable members of the population (14, 16, 19). Recent scientific papers indicate toxicological issues via inhalation of plastic particles and cancerogenic organic compounds originating from the mask material (14, 18, 129, 133).
In short, the adverse effects of masks are clear (2, 3, 5, 16, 18, 19, 23, 36, 129, 166–172, 190), whereas the protective antiviral effect in real life scenarios remains doubtful (12–15, 165, 175, 191–209). Given this, together with the microbiological contamination issues highlighted, masking laws and requirements do not meet the basic medical ethic of ‘Do no harm’. Laws and mandates requiring mask use accordingly have no valid place in respiratory pandemic management.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
KK: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. BW: Conceptualization, Data curation, Formal analysis, Investigation, Funding acquisition, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. AZ: Data curation, Investigation, Visualization, Writing – original draft, Writing – review & editing. DL: Formal analysis, Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. AJ-K: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The publication of this article was partially financed by the University of Gdansk.
Acknowledgments
We wish to thank Dr. Jadwiga Gronczewska from the Department of Evolutionary Genetics and Biosystematics, University of Gdańsk, Poland, for her valuable technical support on this project. We also thank Dr. Bermpohl, hygienist and microbiologist, and the ophthalmologist and physicist MPhys Dr. Mengedoht (both Gütersloh, Germany), who have inspired parts of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1460981/full#supplementary-material
Footnotes
References
1. Face covering policies during the COVID-19 pandemic, our world in data. (2023). Available at: https://ourworldindata.org/grapher/face-covering-policies-covid (Accessed December 29, 2022).
2. Ladhani, SN. Face masking for children - time to reconsider. J Infect. (2022) 85:623–4. doi: 10.1016/j.jinf.2022.09.020
3. Thomson, S. Mask mandates for children during the COVID-19 pandemic: an international human rights perspective. Scand J Public Health. (2022) 50:683–5. doi: 10.1177/14034948221081087
4. World Health Organization, U.N.C. Fund (UNICEF). WHO – Advice on the use of masks for children in the community in the context of COVID-19: annex to the advice on the use of masks in the context of COVID-19, 21 August 2020. (2020). Available at: https://apps.who.int/iris/handle/10665/333919 (Accessed November 7, 2020).
5. Schwarz, S, Jenetzky, E, Krafft, H, Maurer, T, and Martin, D. Corona child studies “co-Ki”: first results of a Germany-wide register on mouth and nose covering (mask) in children. Monatsschr Kinderheilkd. (2021) 169:353–65. doi: 10.1007/s00112-021-01133-9
6. World Health Organization. WHO – Advice on the use of masks in the context of COVID-19: interim guidance, 5 June 2020. (2020). Available at: https://apps.who.int/iris/handle/10665/332293 (Accessed November 7, 2020).
7. Cummings, KJ, Cox-Ganser, J, Riggs, MA, Edwards, N, and Kreiss, K. Respirator donning in post-hurricane New Orleans. Emerg Infect Dis J CDC. (2007) 13, 13:700–7. doi: 10.3201/eid1305.061490
8. Gralton, J, and McLaws, M-L. Protecting healthcare workers from pandemic influenza: N95 or surgical masks? Crit Care Med. (2010) 38:657–67. doi: 10.1097/ccm.0b013e3181b9e8b3
9. Kappstein, I. Mund-Nasen-Schutz in der Öffentlichkeit: Keine Hinweise für eine Wirksamkeit. Krankenhaushygiene. (2020) 15:279–95. doi: 10.1055/a-1174-6591
10. Roberge, R. Facemask use by children during infectious disease outbreaks. Biosecur Bioterror. (2011) 9:225–31. doi: 10.1089/bsp.2011.0009
11. Munro, APS, and Hughes, RC. Face coverings have little utility for young school-aged children. Arch Dis Child. (2023) 108:77–8. doi: 10.1136/archdischild-2022-324809
12. Jefferson, T, Dooley, L, Ferroni, E, Al-Ansary, LA, van Driel, ML, Bawazeer, GA, et al. Physical interventions to interrupt or reduce the spread of respiratory viruses. Cochrane Database Syst Rev. (2023) 2023:CD006207. doi: 10.1002/14651858.CD006207.pub6
13. Sandlund, J, Duriseti, R, Ladhani, SN, Stuart, K, Noble, J, and Høeg, TB. Child mask mandates for COVID-19: a systematic review. Arch Dis Child. (2023) 109:e1–7. doi: 10.1136/archdischild-2023-326215
14. Kisielinski, K, Wagner, S, Hirsch, O, Klosterhalfen, B, and Prescher, A. Possible toxicity of chronic carbon dioxide exposure associated with face mask use, particularly in pregnant women, children and adolescents – a scoping review. Heliyon. (2023) 9:e14117. doi: 10.1016/j.heliyon.2023.e14117
15. Coma, E, Català, M, Méndez-Boo, L, Alonso, S, Hermosilla, E, Alvarez-Lacalle, E, et al. Unravelling the role of the mandatory use of face covering masks for the control of SARS-CoV-2 in schools: a quasi-experimental study nested in a population-based cohort in Catalonia (Spain). Arch Dis Child. (2022) 108:131–6. doi: 10.1136/archdischild-2022-324172
16. Kisielinski, K, Giboni, P, Prescher, A, Klosterhalfen, B, Graessel, D, Funken, S, et al. Is a mask that covers the mouth and nose free from undesirable side effects in everyday use and free of potential hazards? Int J Environ Res Public Health. (2021) 18:4344. doi: 10.3390/ijerph18084344
17. Ahmad, EFEM, Mohammed, M, Al Rayes, AA, Al Qahtani, A, Elzubier, AG, and Suliman, FAE. The effect of wearing the veil by Saudi ladies on the occurrence of respiratory diseases. J Asthma. (2001) 38:423–6. doi: 10.1081/JAS-100001497
18. Ryu, H, and Kim, Y-H. Measuring the quantity of harmful volatile organic compounds inhaled through masks. Ecotoxicol Environ Saf. (2023) 256:114915. doi: 10.1016/j.ecoenv.2023.114915
19. Sukul, P, Bartels, J, Fuchs, P, Trefz, P, Remy, R, Rührmund, L, et al. Effects of COVID-19 protective face masks and wearing durations on respiratory haemodynamic physiology and exhaled breath constituents. Eur Respir J. (2022) 60:2200009. doi: 10.1183/13993003.00009-2022
20. Al-Allaff, RGM, Al-Taee, SMY, and Baker, STD. Some immunological impacts of face mask usage during the COVID-19 pandemic. Pak J Biol Sci. (2021) 24:920–7. doi: 10.3923/pjbs.2021.920.927
21. Vakharia, RJ, Jani, I, Yadav, S, and Kurian, T. To study acute changes in brain oxygenation on MRI in healthcare workers using N95 mask and PPE kits for six hours a day. Indian J Radiol Imaging. (2021) 31:893–900. doi: 10.1055/s-0041-1741086
22. Law, CSW, Lan, PS, and Glover, GH. Effect of wearing a face mask on fMRI BOLD contrast. NeuroImage. (2021) 229:117752. doi: 10.1016/j.neuroimage.2021.117752
23. Patel, S, Mohapatra, E, Suganthy, AK, Shah, S, Abraham, J, Nanda, R, et al. A pilot study to evaluate the changes in venous blood gas parameters and hypoxia biomarkers in health care workers using different kinds of masks. Lung India. (2023) 40:134–42. doi: 10.4103/lungindia.lungindia_343_22
24. Prousa, D. Studie zu psychischen und psychovegetativen Beschwerden mit den aktuellen Mund-Nasenschutz-Verordnungen. PsychArchives. (2020) 1–128. doi: 10.23668/psycharchives.3135
25. Pavlova, MA, Carbon, C-C, Coello, Y, Sokolov, AA, and Proverbio, AM. Editorial: impact of face covering on social cognition and interaction. Front Neurosci. (2023) 17:1150604. doi: 10.3389/fnins.2023.1150604
26. Carbon, C-C, Held, MJ, and Schütz, A. Reading emotions in faces with and without masks is relatively independent of extended exposure and individual difference variables. Front Psychol. (2022) 13:856971. doi: 10.3389/fpsyg.2022.856971
27. Schönweitz, F, Eichinger, J, Kuiper, J, Ongolly, F, Spahl, W, Prainsack, B, et al. The social meanings of artefacts: face masks in the COVID-19 pandemic. Front Public Health. (2022) 10:829904. doi: 10.3389/fpubh.2022.829904
28. Villani, C, D’Ascenzo, S, Scerrati, E, Ricciardelli, P, Nicoletti, R, and Lugli, L. Wearing the face mask affects our social attention over space. Front Psychol. (2022) 13:923558. doi: 10.3389/fpsyg.2022.923558
29. Proverbio, AM, and Cerri, A. The recognition of facial expressions under surgical masks: the primacy of anger. Front Neurosci. (2022) 16:864490. doi: 10.3389/fnins.2022.864490
30. Grundmann, F, Epstude, K, and Scheibe, S. Face masks reduce emotion-recognition accuracy and perceived closeness. PLoS One. (2021) 16:e0249792. doi: 10.1371/journal.pone.0249792
31. Mathis, L. The effects of face masks on emotion interpretation in socially anxious individuals. Grad Stud J Psychol. (2023) 20:88–98. doi: 10.52214/gsjp.v20i1.10167
32. Truong, TL, Beck, SD, and Weber, A. The impact of face masks on the recall of spoken sentences. J Acoust Soc Am. (2021) 149:142–4. doi: 10.1121/10.0002951
33. Sönnichsen, R, Llorach Tó, G, Hochmuth, S, Hohmann, V, and Radeloff, A. How face masks interfere with speech understanding of normal-hearing individuals: vision makes the difference. Otol Neurotol. (2022) 43:282–8. doi: 10.1097/MAO.0000000000003458
34. McKenna, VS, Kendall, CL, Patel, TH, Howell, RJ, and Gustin, RL. Impact of face masks on speech acoustics and vocal effort in healthcare professionals. Laryngoscope. (2022) 132:391–7. doi: 10.1002/lary.29763
35. Education recovery in early years providers: spring 2022, GOV.UK. (2022). Available at: https://www.gov.uk/government/publications/education-recovery-in-early-years-providers-spring-2022/education-recovery-in-early-years-providers-spring-2022 (Accessed February 1, 2023).
36. Spira, B. Correlation between mask compliance and COVID-19 outcomes in Europe. Cureus. (2022) 14:e24268. doi: 10.7759/cureus.24268
37. Fögen, Z. The Foegen effect: a mechanism by which facemasks contribute to the COVID-19 case fatality rate. Medicine (Baltimore). (2022) 101:e28924. doi: 10.1097/MD.0000000000028924
38. Delanghe, L, Cauwenberghs, E, Spacova, I, De Boeck, I, Van Beeck, W, Pepermans, K, et al. Cotton and surgical face masks in community settings: bacterial contamination and face mask hygiene. Front Med (Lausanne). (2021) 8:732047. doi: 10.3389/fmed.2021.732047
39. Szostak-Kotowa, J. Biodeterioration of textiles. Int Biodeterior Biodegradation. (2004) 53:165–70. doi: 10.1016/S0964-8305(03)00090-8
40. Buzzin, A, Domènech-Gil, G, Fraschetti, E, Giovine, E, Puglisi, D, and Caputo, D. Assessing the consequences of prolonged usage of disposable face masks. Sci Rep. (2022) 12:16796. doi: 10.1038/s41598-022-20692-9
41. Yang, Q, Li, H, Shen, S, Zhang, G, Huang, R, Feng, Y, et al. Study of the micro-climate and bacterial distribution in the deadspace of N95 filtering face respirators. Sci Rep. (2018) 8:17382. doi: 10.1038/s41598-018-35693-w
42. Luksamijarulkul, P, Aiempradit, N, and Vatanasomboon, P. Microbial contamination on used surgical masks among hospital personnel and microbial air quality in their working wards: a hospital in Bangkok. Oman Med J. (2014) 29:346–50. doi: 10.5001/omj.2014.92
43. Chughtai, AA, Stelzer-Braid, S, Rawlinson, W, Pontivivo, G, Wang, Q, Pan, Y, et al. Contamination by respiratory viruses on outer surface of medical masks used by hospital healthcare workers. BMC Infect Dis. (2019) 19:491. doi: 10.1186/s12879-019-4109-x
44. Monalisa, D, Aruna, CN, Padma, KB, Manjunath, K, Hemavathy, E, and Varsha, D. Microbial contamination of the mouth masks used by post- graduate students in a private dental institution: an in-vitro study. J Dent Med Sci. (2017) 16:61–7. doi: 10.9790/0853-1605046167
45. Kisielinski, K, and Wojtasik, B. Suitability of rose Bengal sodium salt staining for visualisation of face mask contamination by living organisms. AIMSES. (2022) 9:218–31. doi: 10.3934/environsci.2022015
46. Liu, Z, Chang, Y, Chu, W, Yan, M, Mao, Y, Zhu, Z, et al. Surgical masks as source of bacterial contamination during operative procedures. J Orthopaedic Transl. (2018) 14:57–62. doi: 10.1016/j.jot.2018.06.002
47. Directorate-General for Health and Consumers (European Commission) now known as, making risk assessment more relevant for risk management, Publications Office of the European Union, LU. (2013). Available at: https://data.europa.eu/doi/10.2772/34776 (Accessed April 30, 2023).
48. Lane, DJ. 16S/23S rRNA sequencing In: E Stackenbrandt and M Goodfellow, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons (1991). 115–76.
49. Turner, S, Pryer, KM, Miao, VP, and Palmer, JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. (1999) 46:327–38. doi: 10.1111/j.1550-7408.1999.tb04612.x
50. Monciardini, P, Sosio, M, Cavaletti, L, Chiocchini, C, and Donadio, S. New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes1. FEMS Microbiol Ecol. (2002) 42:419–29. doi: 10.1111/j.1574-6941.2002.tb01031.x
51. Huang, X, Lin, J, and Demner-Fushman, D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annu Symp Proc. (2006) 2006:359–63.
52. Calc | LibreOffice – Free Office Suite – Based on OpenOffice – Compatible with Microsoft. (2023). Available at: https://www.libreoffice.org/discover/calc (Accessed March 17, 2023).
53. Feenstra, RPG, and Tseng, SCG. What is actually stained by rose Bengal? Arch Ophthalmol. (1992) 110:984–93. doi: 10.1001/archopht.1992.01080190090035
54. Conn, HJ. Rose Bengal as a general bacterial stain. J Bacteriol. (1921) 6:253–4. doi: 10.1128/jb.6.2.253-254.1921
55. Maneval, WE. Staining bacteria and yeasts with acid dyes. Stain Technol. (1941) 16:13–9. doi: 10.3109/10520294109106189
56. Saha, DC. A rapid staining method for detection of endophytic fungi in turf and forage grasses. Phytopathology. (1988) 78:237. doi: 10.1094/Phyto-78-237
57. Wojtasik, B, Zbawicka, M, Grabarczyk, L, and Juzwa, W. Flow cytometric approach to evaluate the impact of hydro-technical concrete compounds’ release to the freshwater microbiome. Environ Monit Assess. (2021) 193:698. doi: 10.1007/s10661-021-09481-5
58. Byrd, AL, Belkaid, Y, and Segre, JA. The human skin microbiome. Nat Rev Microbiol. (2018) 16:143–55. doi: 10.1038/nrmicro.2017.157
59. Logan, NA. Bacillus and relatives in foodborne illness. J Appl Microbiol. (2012) 112:417–29. doi: 10.1111/j.1365-2672.2011.05204.x
60. Wolfgang, WJ, Coorevits, A, Cole, JA, De Vos, P, Dickinson, MC, Hannett, GE, et al. Sporosarcina newyorkensis sp. nov. from clinical specimens and raw cow’s milk. Int J Syst Evol Microbiol. (2012) 62:322–9. doi: 10.1099/ijs.0.030080-0
61. Kämpfer, P, Albrecht, A, Buczolits, S, and Busse, H-J. Psychrobacter faecalis sp. nov., a new species from a bioaerosol originating from pigeon faeces. Syst Appl Microbiol. (2002) 25:31–6. doi: 10.1078/0723-2020-00099
62. Deschaght, P, Janssens, M, Vaneechoutte, M, and Wauters, G. Psychrobacter isolates of human origin, other than Psychrobacter phenylpyruvicus, are predominantly Psychrobacter faecalis and Psychrobacter pulmonis, with emended description of P. faecalis. Int J Syst Evol Microbiol. (2012) 62:671–4. doi: 10.1099/ijs.0.032631-0
63. Park, A-M, Khadka, S, Sato, F, Omura, S, Fujita, M, Hashiwaki, K, et al. Bacterial and fungal isolation from face masks under the COVID-19 pandemic. Sci Rep. (2022) 12:11361. doi: 10.1038/s41598-022-15409-x
64. Sachdev, R, Garg, K, Singh, G, and Mehrotra, V. Is safeguard compromised? Surgical mouth mask harboring hazardous microorganisms in dental practice. J Family Med Prim Care. (2020) 9:759–63. doi: 10.4103/jfmpc.jfmpc_1039_19
65. Gund, MP, Boros, G, Hannig, M, Thieme-Ruffing, S, Gärtner, B, Rohrer, TR, et al. Bacterial contamination of forehead skin and surgical mask in aerosol-producing dental treatment. J Oral Microbiol. (2021) 13:1978731. doi: 10.1080/20002297.2021.1978731
66. Gund, MP, Naim, J, Hannig, M, Halfmann, A, Gärtner, B, Boros, G, et al. CHX and a face shield cannot prevent contamination of surgical masks. Front Med (Lausanne). (2022) 9:896308. doi: 10.3389/fmed.2022.896308
67. Checchi, V, Montevecchi, M, Valeriani, L, and Checchi, L. Bioburden variation of filtering face piece respirators over time: a preliminary study. Materials. (2022) 15:8790. doi: 10.3390/ma15248790
68. Yousefimashouf, M, Yousefimashouf, R, Alikhani, MS, Hashemi, H, Karami, P, Rahimi, Z, et al. Evaluation of the bacterial contamination of face masks worn by personnel in a center of COVID 19 hospitalized patients: a cross-sectional study. New Microbes New Infect. (2023) 52:101090. doi: 10.1016/j.nmni.2023.101090
69. Nightingale, M, Mody, M, Rickard, AH, and Cassone, M. Bacterial contamination on used face masks among nursing home healthcare personnel. Antimicrob Steward Healthc Epidemiol. (2023) 3:e54. doi: 10.1017/ash.2023.130
70. Keri, VC, Kumar, A, Singh, G, Mandal, A, Ali, H, Ranjan, P, et al. Pilot study on burden of fungal contamination in face masks: need for better mask hygiene in the COVID-19 era. Infez Med. (2021) 29:557–61. doi: 10.53854/liim-2904-8
71. Merad, Y, Belmokhtar, Z, Hadjazi, O, Belkacemi, M, Matmour, D, Merad, Z, et al. Fungal contamination of medical masks among forensic healthcare workers in the COVID19 era. New Microbes New Infect. (2023) 53:101134. doi: 10.1016/j.nmni.2023.101134
72. VDI 6022, VDI. (2023). Available at: https://www.vdi.de/richtlinien/unsere-richtlinien-highlights/vdi-6022 (Accessed October 14, 2023).
73. Rengasamy, S, Miller, A, Eimer, BC, and Shaffer, RE. Filtration performance of FDA-cleared surgical masks. J Int Soc Respir Prot. (2009) 26:54–70.
74. Fernández-Arribas, J, Moreno, T, Bartrolí, R, and Eljarrat, E. COVID-19 face masks: a new source of human and environmental exposure to organophosphate esters. Environ Int. (2021) 154:106654. doi: 10.1016/j.envint.2021.106654
75. Ostrowski, P, Masiuk, H, Kulig, P, Skoryk, A, Wcisłek, A, Jursa-Kulesza, J, et al. Medical face masks do not affect acid-base balance yet might facilitate the transmission of Staphylococcus aureus in hospital settings during the COVID-19 pandemic. Int J Environ Res Public Health. (2023) 20:2474. doi: 10.3390/ijerph20032474
76. Sakr, A, Brégeon, F, Mège, J-L, Rolain, J-M, and Blin, O. Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front Microbiol. (2018) 9:2419. doi: 10.3389/fmicb.2018.02419
77. Dietert, RR, and Dietert, JM. The human superorganism: using microbes for freedom vs fear. Appl Microbiol. (2023) 3:883–905. doi: 10.3390/applmicrobiol3030061
78. Nowicka, D, and Grywalska, E. Staphylococcus aureus and host immunity in recurrent Furunculosis. Dermatology. (2019) 235:295–305. doi: 10.1159/000499184
79. Akhtar Danesh, L, Saiedi Nejad, Z, Sarmadian, H, Fooladvand, S, van Belkum, A, and Ghaznavi-Rad, E. Elimination of Staphylococcus aureus nasal carriage in intensive care patients lowers infection rates. Eur J Clin Microbiol Infect Dis. (2020) 39:333–8. doi: 10.1007/s10096-019-03729-2
80. Han, C, Shi, J, Chen, Y, and Zhang, Z. Increased flare of acne caused by long-time mask wearing during COVID-19 pandemic among general population. Dermatol Ther. (2020) 33:e13704. doi: 10.1111/dth.13704
81. Bortoluzzi, P, Boneschi, V, and Veraldi, S. “Mask” tinea: an increasing infection during COVID-19 pandemic. Mycopathologia. (2022) 187:141–2. doi: 10.1007/s11046-021-00612-7
82. Silkiss, RZ, Paap, MK, and Ugradar, S. Increased incidence of chalazion associated with face mask wear during the COVID-19 pandemic. Am J Ophthalmol Case Rep. (2021) 22:101032. doi: 10.1016/j.ajoc.2021.101032
83. Akioud, W, Sebbata, S, Mozarie, Y, and Oubaaz, A. Chalazion and face mask wear during COVID-19 pandemic: is there a link? Eur J Med Health Sci. (2023) 5:17–9. doi: 10.24018/ejmed.2023.5.2.1641
84. Molero-Senosiain, M, Tiew, S, Patel, A, Houben, I, and Dhillon, N. Impact of face mask wear on bacterial keratitis. J Fr Ophtalmol. (2023) 46:e37–9. doi: 10.1016/j.jfo.2022.04.028
85. Sakamoto, T, Terasaki, H, Yamashita, T, Shiihara, H, Funatsu, R, and Uemura, A. Increased incidence of endophthalmitis after vitrectomy relative to face mask wearing during COVID-19 pandemic. Br J Ophthalmol. (2023) 107:1472–7. doi: 10.1136/bjophthalmol-2022-321357
86. Goncheva, MI, Gibson, RM, Shouldice, AC, Dikeakos, JD, and Heinrichs, DE. The Staphylococcus aureus protein IsdA increases SARS CoV-2 replication by modulating JAK-STAT signaling. iScience. (2023) 26:105975. doi: 10.1016/j.isci.2023.105975
87. Otto, M. Staphylococcus epidermidis – the “accidental” pathogen. Nat Rev Microbiol. (2009) 7:555–67. doi: 10.1038/nrmicro2182
88. Schoenfelder, SMK, Lange, C, Eckart, M, Hennig, S, Kozytska, S, and Ziebuhr, W. Success through diversity – how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol. (2010) 300:380–6. doi: 10.1016/j.ijmm.2010.04.011
89. Heilmann, C, Ziebuhr, W, and Becker, K. Are coagulase-negative staphylococci virulent? Clin Microbiol Infect. (2019) 25:1071–80. doi: 10.1016/j.cmi.2018.11.012
90. Khorvash, F, Abdi, F, Kashani, HH, Naeini, FF, and Narimani, T. Staphylococcus aureus in acne pathogenesis: a case-control study. N Am J Med Sci. (2012) 4:573–6. doi: 10.4103/1947-2714.103317
91. Findley, K, and Grice, EA. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. (2014) 10:e1004436. doi: 10.1371/journal.ppat.1004436
92. Bjerre, RD, Bandier, J, Skov, L, Engstrand, L, and Johansen, JD. The role of the skin microbiome in atopic dermatitis: a systematic review. Br J Dermatol. (2017) 177:1272–8. doi: 10.1111/bjd.15390
93. Foo, CCI, Goon, ATJ, Leow, Y, and Goh, C. Adverse skin reactions to personal protective equipment against severe acute respiratory syndrome – a descriptive study in Singapore. Contact Derm. (2006) 55:291–4. doi: 10.1111/j.1600-0536.2006.00953.x
94. Rosner, E. Adverse effects of prolonged mask use among healthcare professionals during COVID-19. J Infect Dis Epidemiol. (2020) 6:130. doi: 10.23937/2474-3658/1510130
95. Techasatian, L, Lebsing, S, Uppala, R, Thaowandee, W, Chaiyarit, J, Supakunpinyo, C, et al. The effects of the face mask on the skin underneath: a prospective survey during the COVID-19 pandemic. J Prim Care Community Health. (2020) 11:2150132720966167. doi: 10.1177/2150132720966167
96. Abduljabbar, M, Kalthoum, DE, Bakarman, M, Wahby Salem, I, Alsulaimani, Z, Alharbi, W, et al. The correlation between wearing face masks and skin damage in adults during the COVID-19 pandemic: a cross-sectional study in Jeddah, Saudi Arabia. Cureus. (2022) 14:e31521. doi: 10.7759/cureus.31521
97. Villani, A, Fabbrocini, G, Annunziata, MC, and Potestio, L. Maskne prevalence and risk factors during the COVID-19 pandemic. J Eur Acad Dermatol Venereol. (2022) 36:e678–80. doi: 10.1111/jdv.18248
98. Bakhsh, RA, Saddeeg, SY, Basaqr, KM, Alshammrani, BM, and Zimmo, BS. Prevalence and associated factors of mask-induced acne (Maskne) in the general population of Jeddah during the COVID-19 pandemic. Cureus. (2022) 14:e26394. doi: 10.7759/cureus.26394
99. Dani, A, Eseonu, A, and Bibee, K. Risk factors for the development of acne in healthcare workers during the COVID-19 pandemic. Arch Dermatol Res. (2023) 315:1067–70. doi: 10.1007/s00403-022-02434-z
100. Falodun, O, Medugu, N, Sabir, L, Jibril, I, Oyakhire, N, and Adekeye, A. An epidemiological study on face masks and acne in a Nigerian population. PLoS One. (2022) 17:e0268224. doi: 10.1371/journal.pone.0268224
101. Cheng, Y-F, Zhao, H, Li, J, Lipa, KE, Xie, H-F, Wang, B, et al. Factors aggravating acne vulgaris during the COVID-19 pandemic in China: a web-based cross-sectional survey. Eur Rev Med Pharmacol Sci. (2022) 26:7305–12. doi: 10.26355/eurrev_202210_29925
102. Tallent, Sandra M., Knolhoff, Ann, Rhodehamel, E. Jeffery, Harmon, Stanley M., and Bennett, Reginald W., Bacteriological Analytical Manual (BAM); Chapter 14: Bacillus cereus., FDA. (1996). Available at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-14-bacillus-cereus (Accessed October 15, 2023).
103. Roberge, R, Benson, S, and Kim, J-H. Thermal burden of N95 filtering facepiece respirators. Ann Occup Hyg. (2012) 56:808–14. doi: 10.1093/annhyg/mes001
104. Roberge, RJ, Kim, J-H, and Benson, SM. Absence of consequential changes in physiological, thermal and subjective responses from wearing a surgical mask. Respir Physiol Neurobiol. (2012) 181:29–35. doi: 10.1016/j.resp.2012.01.010
105. Kim, J-H, Benson, SM, and Roberge, RJ. Pulmonary and heart rate responses to wearing N95 filtering facepiece respirators. Am J Infect Control. (2013) 41:24–7. doi: 10.1016/j.ajic.2012.02.037
106. Scarano, A, Inchingolo, F, and Lorusso, F. Facial skin temperature and discomfort when wearing protective face masks: thermal infrared imaging evaluation and hands moving the mask. Int J Environ Res Public Health. (2020) 17:4624. doi: 10.3390/ijerph17134624
107. Park, S-R, Han, J, Yeon, YM, Kang, NY, and Kim, E. Effect of face mask on skin characteristics changes during the COVID-19 pandemic. Skin Res Technol. (2021) 27:554–9. doi: 10.1111/srt.12983
108. Lee, Y-H, Kim, H, Heo, DW, Ahn, I-S, and Park, H-K. Oral microbiome of the inner surface of face masks and whole saliva during the COVID-19 pandemic. Front Oral Health. (2023) 4:1178020. doi: 10.3389/froh.2023.1178020
109. Szunerits, S, Dӧrfler, H, Pagneux, Q, Daniel, J, Wadekar, S, Woitrain, E, et al. Exhaled breath condensate as bioanalyte: from collection considerations to biomarker sensing. Anal Bioanal Chem. (2023) 415:27–34. doi: 10.1007/s00216-022-04433-5
110. Xiang, G, Xu, K, Jian, Y, He, L, Shen, Z, Li, M, et al. Prolonged mask wearing changed nasal microbial characterization of young adults during the COVID-19 pandemic in Shanghai, China. Front Immunol. (2023) 14:1266941. doi: 10.3389/fimmu.2023.1266941
111. Viola, IM, Peterson, B, Pisetta, G, Pavar, G, Akhtar, H, Menoloascina, F, et al. Face coverings, aerosol dispersion and mitigation of virus transmission risk, IEEE open J Eng. Med Biol. (2021) 2:26–35. doi: 10.1109/OJEMB.2021.3053215
112. Jia, Z, Ai, Z, Cao, S, and Bekö, G. Effectiveness of respiratory protective equipment on source control of exhaled pollutants. J Build Eng. (2024) 86:108742. doi: 10.1016/j.jobe.2024.108742
113. Barari, K, Si, X, and Xi, J. Impacts of mask wearing and leakages on cyclic respiratory flows and facial thermoregulation. Fluids. (2024) 9:9. doi: 10.3390/fluids9010009
114. Drewnick, F, Pikmann, J, Fachinger, F, Moormann, L, Sprang, F, and Borrmann, S. Aerosol filtration efficiency of household materials for homemade face masks: influence of material properties, particle size, particle electrical charge, face velocity, and leaks. Aerosol Sci Technol. (2021) 55:63–79. doi: 10.1080/02786826.2020.1817846
115. Shah, Y, Kurelek, JW, Peterson, SD, and Yarusevych, S. Experimental investigation of indoor aerosol dispersion and accumulation in the context of COVID-19: effects of masks and ventilation. Phys Fluids. (2021) 33:073315. doi: 10.1063/5.0057100
116. Datta, R. Use of surgical facemasks in the operation theatre: effective or habit? Med J Armed Forces India. (2010) 66:163–5. doi: 10.1016/S0377-1237(10)80133-9
117. Barbosa, MH, and Graziano, KU. Influence of wearing time on efficacy of disposable surgical masks as microbial barrier. Braz J Microbiol. (2006) 37:216–7. doi: 10.1590/S1517-83822006000300003
118. Kelkar, US, Gogate, B, Kurpad, S, Gogate, P, and Deshpande, M. How effective are face masks in operation theatre? A time frame analysis and recommendations. Int J Infect Control. (2013) 9:1–6. doi: 10.3396/ijic.v9i1.003.13
119. Tcharkhtchi, A, Abbasnezhad, N, Zarbini Seydani, M, Zirak, N, Farzaneh, S, and Shirinbayan, M. An overview of filtration efficiency through the masks: mechanisms of the aerosols penetration. Bioact Mater. (2021) 6:106–22. doi: 10.1016/j.bioactmat.2020.08.002
120. McCullough, NV, Brosseau, LM, and Vesley, D. Collection of three bacterial aerosols by respirator and surgical mask filters under varying conditions of flow and relative humidity. Ann Occup Hyg. (1997) 41:677–90. doi: 10.1016/S0003-4878(97)00022-7
121. Hadayer, A, Zahavi, A, Livny, E, Gal-Or, O, Gershoni, A, Mimouni, K, et al. Patients wearing face masks during intravitreal injections may be at a higher risk of endophthalmitis. Retina. (2020) 40:1651–6. doi: 10.1097/IAE.0000000000002919
122. Huber, C. Masks, false safety and real dangers, part 4: proposed mechanisms by which masks increase risk of COVID-19. Primary Doctor Med J. (2020) 1:1–9. doi: 10.6084/m9.figshare.14021057
123. Borovoy, B, Huber, C, and Crisler, M. Masks, false safety and real dangers, part 2: microbial challenges from masks. PDMJ. (2020) 1:1–19. Available at: https://pdmj.org/masks2/Mask_Risks_Part2.pdf
124. Wyszyńska, M, Czelakowska, A, Rosak, P, Białożyt-Bujak, E, Gruca, O, Rosak-Szyrocka, J, et al. Changes in the oral cavity mucosal surface under the influence of wearing protective face masks—nitric oxide concentration analysis—preliminary report. Coatings. (2022) 12:1164. doi: 10.3390/coatings12081164
125. ICRP: Human Respiratory Tract Model for Radiological Protection. A report of a task Group of the International Commission on radiological protection. Ann ICRP. (1994) 24:1–482.
126. Everard, ML, Hardy, JG, and Milner, AD. Comparison of nebulised aerosol deposition in the lungs of healthy adults following oral and nasal inhalation. Thorax. (1993) 48:1045–6. doi: 10.1136/thx.48.10.1045
127. Cengiz, C, and Can, İH. The effect of N95 and surgical masks on mucociliary clearance function and sinonasal complaints. Eur Arch Otorrinolaringol. (2022) 279:759–64. doi: 10.1007/s00405-021-06838-x
128. Sangkham, S, Faikhaw, O, Munkong, N, Sakunkoo, P, Arunlertaree, C, Chavali, M, et al. A review on microplastics and nanoplastics in the environment: their occurrence, exposure routes, toxic studies, and potential effects on human health. Mar Pollut Bull. (2022) 181:113832. doi: 10.1016/j.marpolbul.2022.113832
129. Kisielinski, K, Hockertz, S, Hirsch, O, Korupp, S, Klosterhalfen, B, Schnepf, A, et al. Wearing face masks as a potential source for inhalation and oral uptake of inanimate toxins – a scoping review. Ecotoxicol Environ Saf. (2024) 275:115858. doi: 10.1016/j.ecoenv.2023.115858
130. Khan, A, and Jia, Z. Recent insights into uptake, toxicity, and molecular targets of microplastics and nanoplastics relevant to human health impacts. iScience. (2023) 26:106061. doi: 10.1016/j.isci.2023.106061
131. Liang, H, Ji, Y, Ge, W, Wu, J, Song, N, Yin, Z, et al. Release kinetics of microplastics from disposable face masks into the aqueous environment. Sci Total Environ. (2022) 816:151650. doi: 10.1016/j.scitotenv.2021.151650
132. Ma, J, Chen, F, Xu, H, Jiang, H, Liu, J, Li, P, et al. Face masks as a source of nanoplastics and microplastics in the environment: quantification, characterization, and potential for bioaccumulation. Environ Pollut. (2021) 288:117748. doi: 10.1016/j.envpol.2021.117748
133. Zhang, M, Liu, T, Zhang, L, Hua, Z, Guo, Z, Dong, J, et al. Assessment of microplastic exposure in nasal lavage fluid and the influence of face masks. J Hazard Mater. (2024) 480:136069. doi: 10.1016/j.jhazmat.2024.136069
134. Wieland, S, Balmes, A, Bender, J, Kitzinger, J, Meyer, F, Ramsperger, AF, et al. From properties to toxicity: comparing microplastics to other airborne microparticles. J Hazard Mater. (2022) 428:128151. doi: 10.1016/j.jhazmat.2021.128151
135. Kasloff, SB, Leung, A, Strong, JE, Funk, D, and Cutts, T. Stability of SARS-CoV-2 on critical personal protective equipment. Sci Rep. (2021) 11:984. doi: 10.1038/s41598-020-80098-3
136. Teo, W-L. The “Maskne” microbiome – pathophysiology and therapeutics. Int J Dermatol. (2021) 60:799–809. doi: 10.1111/ijd.15425
137. Sawada, Y. Occupational skin dermatitis among healthcare workers associated with the COVID-19 pandemic: a review of the literature. Int J Mol Sci. (2023) 24:2989. doi: 10.3390/ijms24032989
138. Tunçer Vural, A. The development of acne vulgaris due to face masks during the pandemic, risk awareness and attitudes of a group of university students. J Cosmet Dermatol. (2022) 21:5306–13. doi: 10.1111/jocd.15120
139. Liu, Y, Zhao, H, Chen, H, Li, X, Ran, C, Sun, H, et al. Does mask wearing affect skin health? An untargeted skin metabolomics study. Environ Int. (2023) 178:108073. doi: 10.1016/j.envint.2023.108073
140. Brooks, JK, Sultan, AS, and Jabra-Rizk, MA. Prolonged facial mask wear is a concern for the development of dysbiotic microbiome. Respirat Med Res. (2022) 81:100877. doi: 10.1016/j.resmer.2021.100877
141. Koshevarova, VA, Westenhaver, ZK, Schmitz-Brown, M, McKinnon, BJ, Merkley, KH, and Gupta, PK. Blepharoconjunctivitis and otolaryngological disease trends in the context of mask wearing during the COVID-19 pandemic. Clin Pract. (2022) 12:619–27. doi: 10.3390/clinpract12040065
142. Schultheis, WG, Sharpe, JE, Zhang, Q, Patel, SN, Kuriyan, AE, Chiang, A, et al. Effect of taping face masks on quantitative particle counts near the eye: implications for intravitreal injections in the COVID-19 era. Am J Ophthalmol. (2021) 225:166–71. doi: 10.1016/j.ajo.2021.01.021
143. Burgos-Blasco, B, Arriola-Villalobos, P, Fernandez-Vigo, JI, Oribio-Quinto, C, Ariño-Gutierrez, M, Diaz-Valle, D, et al. Face mask use and effects on the ocular surface health: a comprehensive review. Ocul Surf. (2023) 27:56–66. doi: 10.1016/j.jtos.2022.12.006
144. Moshirfar, M, West, WB, and Marx, DP. Face mask-associated ocular irritation and dryness. Ophthalmol Ther. (2020) 9:397–400. doi: 10.1007/s40123-020-00282-6
145. Boccardo, L. Self-reported symptoms of mask-associated dry eye: a survey study of 3,605 people. Contact Lens Anterior Eye. (2022) 45:101408. doi: 10.1016/j.clae.2021.01.003
146. Islam, SR, Prusty, D, Maiti, S, Dutta, R, Chattopadhyay, P, and Manna, SKK. Effect of short-term use of FFP2 (N95) mask on salivary metabolome of young healthy volunteers: a pilot study. Mol Omics. (2023) 19:383–94. doi: 10.1039/D2MO00232A
147. Arora, U, Priyadarshi, M, Katiyar, V, Soneja, M, Garg, P, Gupta, I, et al. Risk factors for coronavirus disease-associated mucormycosis. J Infect. (2022) 84:383–90. doi: 10.1016/j.jinf.2021.12.039
148. Kisielinski, K, Steigleder-Schweiger, C, Wagner, S, Korupp, S, Hockertz, S, and Hirsch, O. Risks and benefits of face masks in children. Preprints. (2024) 1–51. doi: 10.20944/preprints202409.1508.v1
149. Belkin, NL. The evolution of the surgical mask: filtering efficiency versus effectiveness. Infect Control Hosp Epidemiol. (1997) 18:49–57. doi: 10.1086/647501
150. Matuschek, C, Moll, F, Fangerau, H, Fischer, JC, Zänker, K, van Griensven, M, et al. The history and value of face masks. Eur J Med Res. (2020) 25:23. doi: 10.1186/s40001-020-00423-4
151. Lee, S-A, Grinshpun, SA, and Reponen, T. Respiratory performance offered by N95 respirators and surgical masks: human subject evaluation with NaCl aerosol representing bacterial and viral particle size range. Ann Occup Hyg. (2008) 52:177–85. doi: 10.1093/annhyg/men005
152. Ntlailane, MGL, and Wichmann, J. Effectiveness of N95 respirators for nanoparticle exposure control (2000–2016): a systematic review and meta-analysis. J Nanopart Res. (2019) 21:170. doi: 10.1007/s11051-019-4596-0
153. Samaranayake, LP, Fakhruddin, KS, Ngo, HC, Chang, JWW, and Panduwawala, C. The effectiveness and efficacy of respiratory protective equipment (RPE) in dentistry and other health care settings: a systematic review. Acta Odontol Scand. (2020) 78:626–39. doi: 10.1080/00016357.2020.1810769
154. Willeke, K, Qian, Y, Donnelly, J, Grinshpun, S, and Ulevicius, V. Penetration of airborne microorganisms through a surgical mask and a dust/mist respirator. Am Ind Hyg Assoc J. (1996) 57:348–55. doi: 10.1080/15428119691014882
155. Hodous, TK, and Coffey, CC. The role of respiratory protective devices in the control of tuberculosis. Occup Med. (1994) 9:631–57.
156. Qian, Y, Willeke, K, Grinshpun, SA, Donnelly, J, and Coffey, CC. Performance of N95 respirators: filtration efficiency for airborne microbial and inert particles. Am Ind Hyg Assoc J. (1998) 59:128–32. doi: 10.1080/15428119891010389
157. Loeb, M, Dafoe, N, Mahony, J, John, M, Sarabia, A, Glavin, V, et al. Surgical mask vs N95 respirator for preventing influenza among health care workers: a randomized trial. JAMA. (2009) 302:1865–71. doi: 10.1001/jama.2009.1466
158. Smith, JD, MacDougall, CC, Johnstone, J, Copes, RA, Schwartz, B, and Garber, GE. Effectiveness of N95 respirators versus surgical masks in protecting health care workers from acute respiratory infection: a systematic review and meta-analysis. CMAJ. (2016) 188:567–74. doi: 10.1503/cmaj.150835
159. Liu, I, Prasad, V, and Darrow, J. Evidence for community face masking to limit the spread of SARS-CoV-2: a critical review. Health Matrix J Law Med. (2023) 33:1. Available at: https://scholarlycommons.law.case.edu/healthmatrix/vol33/iss1/1/
160. Vincent, M, and Edwards, P. Disposable surgical face masks for preventing surgical wound infection in clean surgery. Cochrane Database Syst Rev. (2016) 2016:CD002929. doi: 10.1002/14651858.CD002929.pub3
161. Burdick, HN, and Maibach, H. Clinical relevance of masks in the operating room? A systematic review. Clin Infect Pract. (2021) 12:100087. doi: 10.1016/j.clinpr.2021.100087
162. Carbon, C-C. Wearing face masks strongly confuses counterparts in reading emotions. Front Psychol. (2020) 11:566886. doi: 10.3389/fpsyg.2020.566886
163. World Medical Association, WMA - The World Medical Association-Declaration of Helsinki. (2013). Available at: https://www.wma.net/what-we-do/medical-ethics/declaration-of-helsinki/ (Accessed November 8, 2021).
164. WHO, World Medical Association (WMA): Declaration of Helsinki. Ethical principles for medical research involving human subjects. Bull World Health Organ. (2001) 79:373–4.
165. Elgersma, IH, Fretheim, A, Elstrøm, P, and Aavitsland, P. Association between face mask use and risk of SARS-CoV-2 infection: cross-sectional study. Epidemiol Infect. (2023) 151:e194. doi: 10.1017/S0950268823001826
166. Boretti, A. Efficacy of generalized face masking mandates, health services research and managerial. Epidemiology. (2021) 8:23333928211058023. doi: 10.1177/23333928211058023
167. Galanis, P, Vraka, I, Fragkou, D, Bilali, A, and Kaitelidou, D. Impact of personal protective equipment use on health care workers’ physical health during the COVID-19 pandemic: a systematic review and meta-analysis. Am J Infect Control. (2021) 49:1305–15. doi: 10.1016/j.ajic.2021.04.084
168. Unoki, T, Sakuramoto, H, Sato, R, Ouchi, A, Kuribara, T, Furumaya, T, et al. Adverse effects of personal protective equipment among intensive care unit healthcare professionals during the COVID-19 pandemic: a scoping review. SAGE Open Nurs. (2021) 7:23779608211026164. doi: 10.1177/23779608211026164
169. Dirol, H, Alkan, E, Sindel, M, Ozdemir, T, and Erbas, D. The physiological and disturbing effects of surgical face masks in the COVID-19 era. BLL. (2021) 122:821–5. doi: 10.4149/BLL_2021_131
170. Gaikwad, RP, Banodkar, AB, and Nandgaonkar, VP. Respiratory consequences of N95 mask during Covid-19 pandemic- an observational study. Int J Health Sci Res. (2021) 11:55–61. doi: 10.52403/ijhsr.20210407
171. Walach, H, Traindl, H, Prentice, J, Weikl, R, Diemer, A, Kappes, A, et al. Carbon dioxide rises beyond acceptable safety levels in children under nose and mouth covering: results of an experimental measurement study in healthy children. Environ Res. (2022) 212:113564. doi: 10.1016/j.envres.2022.113564
172. Acuti Martellucci, C, Flacco, ME, Martellucci, M, Violante, FS, and Manzoli, L. Inhaled CO2 concentration while wearing face masks: a pilot study using capnography. Environ Health Insights. (2022) 16:11786302221123573. doi: 10.1177/11786302221123573
173. Ahmad, MDF, Wahab, S, Ali Ahmad, F, Intakhab Alam, M, Ather, H, Siddiqua, A, et al. A novel perspective approach to explore pros and cons of face mask in prevention the spread of SARS-CoV-2 and other pathogens. Saudi Pharm J. (2021) 29:121–33. doi: 10.1016/j.jsps.2020.12.014
174. Shobako, N. Lessons from the health policies for children during the pandemic in Japan. Front Public Health. (2022) 10:1015955. doi: 10.3389/fpubh.2022.1015955
175. Kampf, G. Effect of face masking on transmission of SARS-CoV-2 In: N Rezaei, editor. The COVID-19 aftermath: volume II: lessons learned. Switzerland, Cham: Springer Nature (2024). 175–99. doi: 10.1007/978-3-031-61943-4_12
176. Beauchamp, JD, and Mayhew, CA. Revisiting the rationale of mandatory masking. J Breath Res. (2023) 17:042001. doi: 10.1088/1752-7163/acdf12
177. Sandlund, J, Duriseti, R, Ladhani, SN, Stuart, K, Noble, J, and Beth Høeg, T. Face masks and protection against COVID-19 and other viral respiratory infections: assessment of benefits and harms in children. Paediatr Respir Rev. (2024). doi: 10.1016/j.prrv.2024.08.003
178. Mastropasqua, L, Lanzini, M, Brescia, L, D’Aloisio, R, Nubile, M, Ciancaglini, M, et al. Face mask-related ocular surface modifications during COVID-19 pandemic: a clinical, in vivo confocal microscopy, and immune-cytology study. Transl Vis Sci Technol. (2021) 10:22. doi: 10.1167/tvst.10.3.22
179. D’Souza, S, Vaidya, T, Nair, AP, Shetty, R, Kumar, NR, Bisht, A, et al. Altered ocular surface health status and tear film immune profile due to prolonged daily mask wear in health care workers. Biomedicines. (2022) 10:1160. doi: 10.3390/biomedicines10051160
180. Jin, S, Wetzel, D, and Schirmer, M. Deciphering mechanisms and implications of bacterial translocation in human health and disease. Curr Opin Microbiol. (2022) 67:102147. doi: 10.1016/j.mib.2022.102147
181. Asadi, S, Cappa, CD, Barreda, S, Wexler, AS, Bouvier, NM, and Ristenpart, WD. Efficacy of masks and face coverings in controlling outward aerosol particle emission from expiratory activities. Sci Rep. (2020) 10:15665. doi: 10.1038/s41598-020-72798-7
182. Bagchi, S, Basu, S, Chaudhuri, S, and Saha, A. Penetration and secondary atomization of droplets impacted on wet facemasks. Phys Rev Fluids. (2021) 6:110510. doi: 10.1103/PhysRevFluids.6.110510
183. Rebmann, T, Carrico, R, and Wang, J. Physiologic and other effects and compliance with long-term respirator use among medical intensive care unit nurses. Am J Infect Control. (2013) 41:1218–23. doi: 10.1016/j.ajic.2013.02.017
184. Matusiak, Ł, Szepietowska, M, Krajewski, P, Białynicki-Birula, R, and Szepietowski, JC. Inconveniences due to the use of face masks during the COVID-19 pandemic: a survey study of 876 young people. Dermatol Ther. (2020) 33:e13567. doi: 10.1111/dth.13567
185. Naylor, G, Burke, LA, and Holman, JA. Covid-19 lockdown affects hearing disability and handicap in diverse ways: a rapid online survey study. Ear Hear. (2020) 41:1442–9. doi: 10.1097/aud.0000000000000948
186. Thomas, F, Allen, C, Butts, W, Rhoades, C, Brandon, C, and Handrahan, DL. Does wearing a surgical facemask or N95-respirator impair radio communication? Air Med J. (2011) 30:97–102. doi: 10.1016/j.amj.2010.12.007
187. Heider, CA, Álvarez, ML, Fuentes-López, E, González, CA, León, NI, Verástegui, DC, et al. Prevalence of voice disorders in healthcare workers in the universal masking COVID-19 era. Laryngoscope. (2020) 131:E1227–33. doi: 10.1002/lary.29172
188. Zarei, N, Negarandeh, R, and Neshat, H. Communication challenges caused by wearing masks and strategies used by pediatric nurses during the COVID-19 pandemic: a qualitative study. J Pediatr Nurs. (2024) 77:e54–61. doi: 10.1016/j.pedn.2024.03.020
189. Sezer, H, Çınar, D, and Kılıç Akça, N. The effect of prolonged use of surgical masks during face-to-face teaching on cognitive and physiological parameters of nursing students: a cross-sectional and descriptive study. Nurse Educ Pract. (2023) 72:103779. doi: 10.1016/j.nepr.2023.103779
190. Elbl, C, Brunner, JX, Schier, D, Junge, A, and Junge, H. Protective face masks add significant dead space. Eur Respir J. (2021) 58:2101131. doi: 10.1183/13993003.01131-2021
191. Chandra, A, and Høeg, TB. Lack of correlation between school mask mandates and paediatric COVID-19 cases in a large cohort. J Infect. (2022) 85:671–5. doi: 10.1016/j.jinf.2022.09.019
192. Xiao, J, Shiu, EYC, Gao, H, Wong, JY, Fong, MW, Ryu, S, et al. Nonpharmaceutical measures for pandemic influenza in nonhealthcare settings—personal protective and environmental measures. Emerg Infect Dis J CDC. (2020) 26:967–75. doi: 10.3201/eid2605.190994
193. Wang, MX, Gwee, SXW, Chua, PEY, and Pang, J. Effectiveness of surgical face masks in reducing acute respiratory infections in non-healthcare settings: a systematic review and Meta-analysis. Front Med. (2020) 7:564280. doi: 10.3389/fmed.2020.564280
194. Fønhus, M.S., Dalsbø, T.K., and Brurberg, K.G., Facemasks to prevent transmission of respiratory illness, such as COVID-19, Norwegian Institute of Public Health. (2021). Available at: https://hdl.handle.net/11250/2756758
195. Aiello, AE, Perez, V, Coulborn, RM, Davis, BM, Uddin, M, and Monto, AS. Facemasks, hand hygiene, and influenza among young adults: a randomized intervention trial. PLoS One. (2012) 7:e29744. doi: 10.1371/journal.pone.0029744
196. Zain Alabdeen, E, Choudhry, A, and Al-Naji, D. Effect of use of face mask on hajj related acute respiratory infection among hajjis from Riyadh-a health promotion intervention study. FETP Saudi Epidemiol Bull. (2005) 12:27–8. Available at: https://saudifetp.org/seb/effect-use-face-mask-hajj-related-acute-respiratory-infection-among-hajjis-riyadh-health
197. Alfelali, M, Haworth, EA, Barasheed, O, Badahdah, A-M, Bokhary, H, Tashani, M, et al. Hajj research team, facemask against viral respiratory infections among hajj pilgrims: a challenging cluster-randomized trial. PLoS One. (2020) 15:e0240287. doi: 10.1371/journal.pone.0240287
198. Canini, L, Andréoletti, L, Ferrari, P, D’Angelo, R, Blanchon, T, Lemaitre, M, et al. Surgical mask to prevent influenza transmission in households: a cluster randomized trial. PLoS One. (2010) 5:e13998. doi: 10.1371/journal.pone.0013998
199. MacIntyre, CR, Cauchemez, S, Dwyer, DE, Seale, H, Cheung, P, Browne, G, et al. Face mask use and control of respiratory virus transmission in households. Emerg Infect Dis. (2009) 15:233–41. doi: 10.3201/eid1502.081166
200. MacIntyre, CR, Seale, H, Dung, TC, Hien, NT, Nga, PT, Chughtai, AA, et al. A cluster randomised trial of cloth masks compared with medical masks in healthcare workers. BMJ Open. (2015) 5:e006577. doi: 10.1136/bmjopen-2014-006577
201. Simmerman, JM, Suntarattiwong, P, Levy, J, Jarman, RG, Kaewchana, S, Gibbons, RV, et al. Findings from a household randomized controlled trial of hand washing and face masks to reduce influenza transmission in Bangkok, Thailand. Influenza Other Respir Viruses. (2011) 5:256–67. doi: 10.1111/j.1750-2659.2011.00205.x
202. Cowling, BJ, Fung, ROP, Cheng, CKY, Fang, VJ, Chan, KH, Seto, WH, et al. Preliminary findings of a randomized trial of non-pharmaceutical interventions to prevent influenza transmission in households. PLoS One. (2008) 3:e2101. doi: 10.1371/journal.pone.0002101
203. Cowling, BJ, Chan, K-H, Fang, VJ, Cheng, CKY, Fung, ROP, Wai, W, et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med. (2009) 151:437–46. doi: 10.7326/0003-4819-151-7-200910060-00142
204. Suess, T, Remschmidt, C, Schink, SB, Schweiger, B, Nitsche, A, Schroeder, K, et al. The role of facemasks and hand hygiene in the prevention of influenza transmission in households: results from a cluster randomised trial; Berlin, Germany, 2009-2011. BMC Infect Dis. (2012) 12:26. doi: 10.1186/1471-2334-12-26
205. Larson, EL, Ferng, Y, Wong-McLoughlin, J, Wang, S, Haber, M, and Morse, SS. Impact of non-pharmaceutical interventions on URIs and influenza in crowded, urban households. Public Health Rep. (2010) 125:178–91. doi: 10.1177/003335491012500206
206. Jacobs, JL, Ohde, S, Takahashi, O, Tokuda, Y, Omata, F, and Fukui, T. Use of surgical face masks to reduce the incidence of the common cold among health care workers in Japan: a randomized controlled trial. Am J Infect Control. (2009) 37:417–9. doi: 10.1016/j.ajic.2008.11.002
207. Bundgaard, H, Bundgaard, JS, Raaschou-Pedersen, DET, von Buchwald, C, Todsen, T, Norsk, JB, et al. Effectiveness of adding a mask recommendation to other public health measures to prevent SARS-CoV-2 infection in Danish mask wearers. Ann Intern Med. (2020). doi: 10.7326/M20-6817
208. Juutinen, A, Sarvikivi, E, Laukkanen-Nevala, P, and Helve, O. Face mask recommendations in schools did not impact COVID-19 incidence among 10–12-year-olds in Finland – joinpoint regression analysis. BMC Public Health. (2023) 23:730. doi: 10.1186/s12889-023-15624-9
Keywords: 16S rRNA gene amplicon sequencing, adverse effects, bacterial contamination, Rose Bengal staining, surgical mask, N95, personal protective equipment, risk
Citation: Kisielinski K, Wojtasik B, Zalewska A, Livermore DM and Jurczak-Kurek A (2024) The bacterial burden of worn face masks—observational research and literature review. Front. Public Health. 12:1460981. doi: 10.3389/fpubh.2024.1460981
Edited by:
Adwoa Asante-Poku, University of Ghana, GhanaReviewed by:
Gerald Mboowa, Makerere University, UgandaRachid Ait Addi, Cadi Ayyad University, Morocco
Copyright © 2024 Kisielinski, Wojtasik, Zalewska, Livermore and Jurczak-Kurek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Kisielinski, a2Fpa2lzaWVsaW5za2lAeWFob28uZGU=; Agata Jurczak-Kurek, YWdhdGEuanVyY3phay1rdXJla0B1Zy5lZHUucGw=
 Kai Kisielinski
Kai Kisielinski Barbara Wojtasik2
Barbara Wojtasik2 Aleksandra Zalewska
Aleksandra Zalewska Agata Jurczak-Kurek
Agata Jurczak-Kurek