- Environment and Life Sciences Research Center, Kuwait Institute for Scientific Research, Safat, Kuwait
Even though death due to COVID-19 is no longer a public health emergency, less virulent but highly transmissible forms of SARS-CoV-2 continue to spread in many countries leading to outbreaks and rise in hospitalizations in the affected regions. Lessons learned during the pandemic must be put into action to protect the world's population from another catastrophe like COVID-19. Novel approaches that were developed for tracking the spread of SARS-CoV-2 included analysis of wastewater, air samples, and various environmental surfaces. We conducted a study in Kuwait during the peak of COVID-19 pandemic to examine if SARS-CoV-2 could be detected in swabs taken from frequently touched environmental surfaces. We selected 12 Cooperative Society Stores—two from each governorate of Kuwait—for collection of surface samples. The Cooperative Society Stores are widely distributed across the whole country and cater to daily household needs including groceries and other essential items. These stores operated even during the “lockdown” imposed at the height of the pandemic. We collected swabs from high-touch surfaces including the handles of the shopping carts and freezers, the elevators, the keypads of the point-of-service terminals of cash counters, and the automated teller machines. All the surfaces tested showed a variable presence of SARS-CoV-2 by reverse transcriptase quantitative PCR, showing the validity of the proof-of-concept study. Monitoring of the presence of SARS-CoV-2 by surface sampling thus offers a cheap but effective means of environmental surveillance for coronaviruses. We therefore strongly recommend the addition of surface environmental sampling as a strategy for pandemic preparedness everywhere.
Introduction
More than seven million lives were lost to COVID-19 since SARS-CoV-2 was first detected in December 2019 (1). Despite the availability of highly advanced technologies for detection of pathogens of pandemic potentials, the most developed countries of the world could not protect their population from the devastating consequences of the pandemic. While the virus continues to spread in a less virulent form, the governments face the challenge of producing the best possible strategy to prevent the next outbreak of the so-called “Disease X” (2). One Health approach has been proposed to integrate many diverse levels of infectious disease surveillance for early detection and efficient management of threats posed by the emerging and re-emerging pathogens of pandemic potential (3–6). The role of an Intergovernmental Panel for One Health for strengthening pandemic preparedness has also been critically evaluated by health policy experts (7). This paper presents the findings of a proof-of-concept study carried out in Kuwait during the COVID-19 pandemic for detection of SARS-CoV-2 on the environmental surfaces and discusses the implications of this approach for surveillance of emerging and re-emerging viral outbreaks.
Coronaviruses are important human and animal pathogens that may have originated in bats (8, 9). The major symptoms of COVID-19 were dry cough, sore-throat, fever, and various degrees of respiratory difficulties (10). The alarming rate of the spread of COVID-19 across the globe prompted many governments to take stringent isolation strategies including complete lock down of affected regions with closure of schools and offices. Even curfews have been implemented in many countries including Kuwait to restrict the mobility of the people and limit person-to-person spread. Human-to-human transmission of SARS-CoV-2 has already been confirmed (11). The major route of transmission of the virus is thought to be via respiratory droplets of infected humans, while transmission from contaminated inanimate surfaces to humans has also been documented (12–15). The virus has been detected in throat swab, blood, urine, and stool of infected patients by reverse-transcriptase quantitative PCR (RT-qPCR), and the viral RNA was detectable up to 25 days after the onset of symptom (12). The possibility of asymptomatic healthy people carrying the virus raises concerns about transmission in the community although the significance of this mode of transmission is unknown. WHO recommends frequent hand washing with soap and water and the use of face mask and personal protective equipment (PPE) to protect individuals who have the risk of exposure.
Previous studies on the persistence of human and veterinary coronaviruses on inanimate surfaces revealed that human coronaviruses such as SARS coronavirus, MERS coronavirus or endemic human coronaviruses can persist on inanimate surfaces like metal, glass, or plastic for up to 9 days (14). A controlled experimental study showed that SARS-CoV-2 may survive in the air for < 4 h while it may remain viable for 2 days on plastic and 3 days on metallic surface (15). It is unknown how long the virus remains viable in the different environments that exist in different countries with diverse climatic conditions where the virus continues to spread. The present study was conducted to detect the virus in the environment of Kuwait when the disease activity was already established in the community.
Materials and methods
Ethics statement
All the samples collected and analyzed in this study were obtained from environmental surfaces only. Although the viral RNA that were isolated from the surface samples most likely originated from humans, we did not collect any samples directly from any identifiable human subject. The study was, therefore, exempted from obtaining ethical approval or informed consent. However, the investigators considered the significant health risks posed to the research team while conducting this study, and obtained approval from the Ministry of Health of Kuwait to conduct the study although it was not a human subject research that required ethical approval and informed consent.
Sample collection
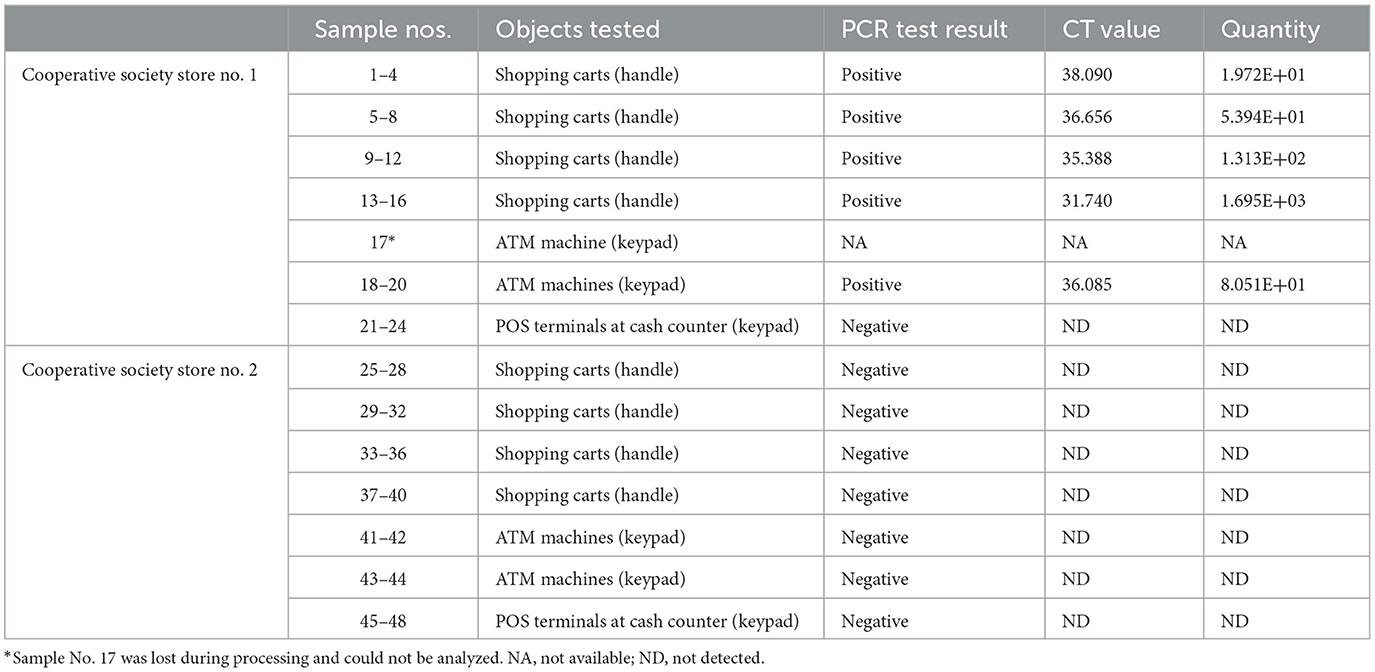
Samples were collected from high-touch surfaces from twelve main Cooperative Society Stores, also known as Co-ops—two from each governorate of Kuwait—between June 6 and 23 of 2021 (Figure 1). The types of surfaces included building entrance doors outside the store and shopping cart handles, freezer door handles, keypads of automatic teller machines (ATMs), keypads of the point-of-service (POS) terminals of cash counters and elevator buttons inside the store. Sterile cotton-tipped swabs saturated with 1X PBS were used for sampling. Samples were collected from 25 cm2 of the surface by gently swabbing the entire area horizontally and vertically with uniform movement and occasionally rotating the swab for uniform surface coverage (16, 17). The surfaces used for collection of samples from the cooperative stores included the handles of the shopping carts (n = 159) and fridges (n = 32), the buttons of the elevators (n = 12), the keypads of the POS terminals of cash counters (n = 44), and the ATM machines (n = 22) located inside or adjacent to the same store. A few stores had an outside door with handles that were frequently used by the clients—therefore swab samples were collected from these door handles (n = 3) as well. In total, 272 swab samples were collected from the selected cooperative stores. One sample was lost in transport and 271 were analyzed by reverse transcriptase-quantitative PCR (RT-qPCR) as described below. In order to maximize the chances of detection of the virus, most of the samples (95.6%) collected from similar types of surfaces were pooled by combining 2–4 samples while < 5% of the samples were analyzed without pooling as shown in Tables 2–7. The total number of DNA samples actually analyzed by RT-qPCR was 83, denoted by “np,” whereas the actual number of samples collected and processed was 271, denoted by “n.”
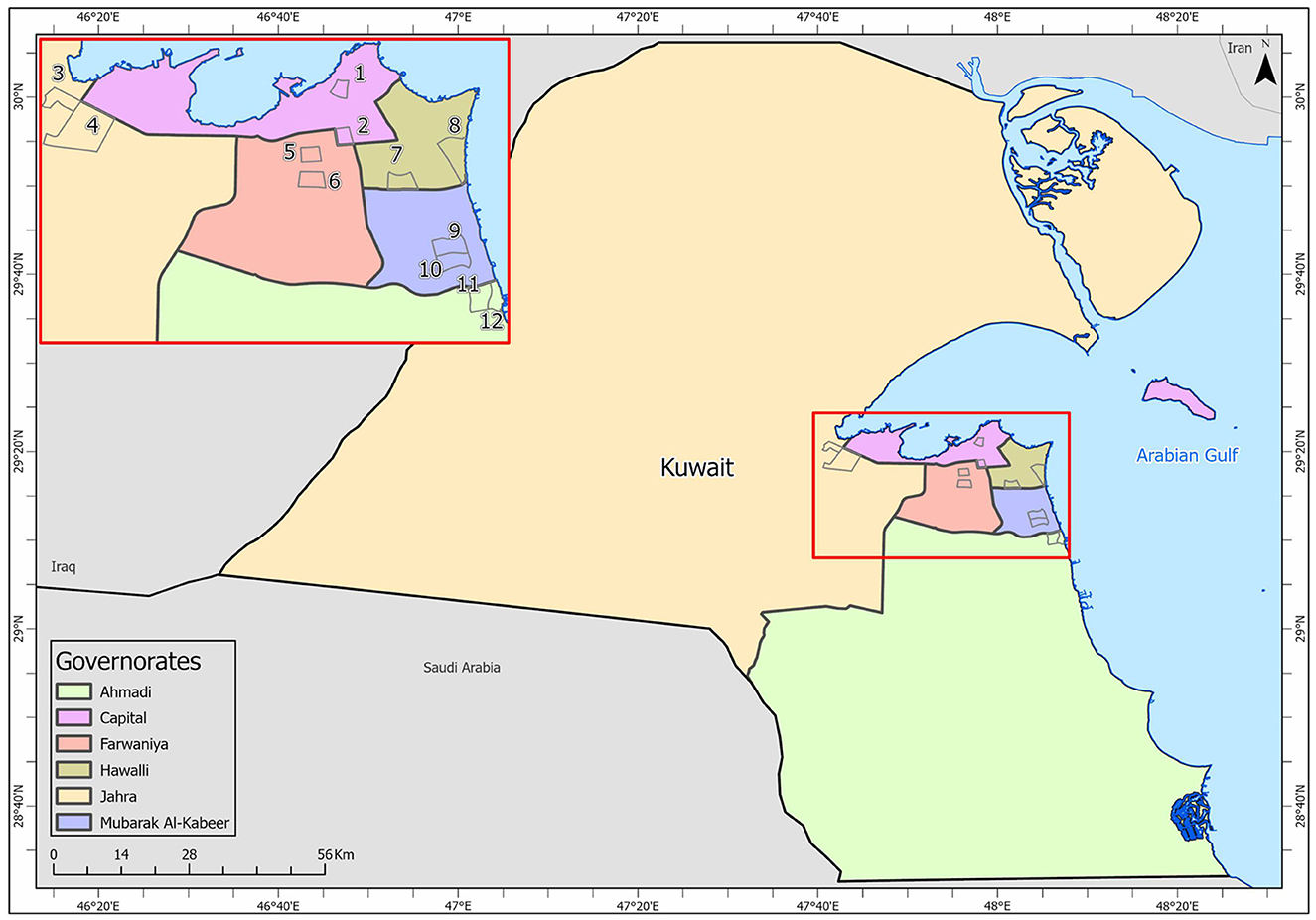
Figure 1. Kuwait map showing the sampling sites (Nos. 1–12) in the six governorates: Shamiya (1) & Yarmouk (2) in Capital Governorate; Qasr (3) & Saad Al-Abdullah (4) in Jahra Governorate; Rabiyah (5) & Ishbilia (6) in Farwaniya Governorate; Shuhada (7) & Salwa (8) in Hawalli Governorate; Al Qusour (9) & Qurain (10) in Mubarak Al Kabeer Governorate; Egaila (11) & Fintas (12) in Al Ahmadi Governorate.
Extraction of nucleic acids and RT-qPCR analysis
The swabs were dipped in 1 ml of TRIzol LS solution (Invitrogen, USA), and the RNA was isolated using the standard procedure described in the instruction manual (18). The total RNA was eluted in 10 μl of RNAse free water. All the steps were performed in BSL2 cabinet (Telstar BioII Advance Class II cabinet, Spain). Pooled samples were prepared by combining 5 μl aliquots of eluted RNA from 4 independent replicates of the same type of solid surface. A previously validated SARS-CoV-2 RT-qPCR protocol following the CDC guidelines and primers was used (16, 17, 19). RNA samples (2 μl) were converted to complementary DNA (cDNA) using iScript Reverse Transcription Supermix (BioRad, USA). The reverse transcription was carried out using random primers for 20 min at 46°C after priming for 5 min at 20°C. The reverse transcriptase enzyme was inactivated by incubating the reaction mix at 95°C for 1 min. The qPCR assays were performed on RNA samples to detect and quantify the SARS-CoV-2. A total of 6 serial dilutions ranging from 101 to 106 copies was prepared with a non-infectious synthetic lyophilized cDNA target encoding N gene and ORF1ab gene (Viasure SARS-CoV-2 positive control, CerTest Biotec S.L., Spain). This standard curve was used to determine the abundance of viral particles in RNA samples isolated from the surface swabs. The data was analyzed using the QuantStudio 5 software (Applied Biosystems, USA) by interpolating the absolute quantity of target in the test samples using the standard curve.
The cDNA (2 μl) was added to the reaction mixture with the forward primer 5′-GGGGAACTTCTCCTGCTAGAAT-3′ and reverse primer 5′-CAGACATTTTGCTCTCAAGCTG-3′ to amplify the N gene target of SARS-CoV-2 virus (15, 16). The total reaction volume was made up to 18μl with iTaq Universal SYBR Green Supermix, Forward and reverse primers, nuclease-free water, and 2 μl of sample cDNA was added. The PCR 20 μl reaction mix was loaded on the QuantStudio 5 Real-Time PCR System (Applied Biosystems, Singapore). Thermal cycling was performed at 95°C for 2 min and 40 cycles of 95°C for 5 s, 59.9°C for 1 min in QuantStudio® 3 Applied Biosystems™ (Applied Biosystems, USA). The above-mentioned positive control from VIASURE SARS-CoV- Real-Time PCR Detection Kit-CE- IVD (Certest-Biotec, Spain) was used to validate the qPCR reactions. In addition, a known concentration of non-infectious synthetic construct of cDNA (2,000 copies/μl) comprising the N gene and ORF1ab genes of SARS-CoV- 2 were used as a positive control. The Ct values of the positive samples were quantified using the QuantStudio Design and Analysis software V1.5.2. (Applied Biosystems, USA).
Results
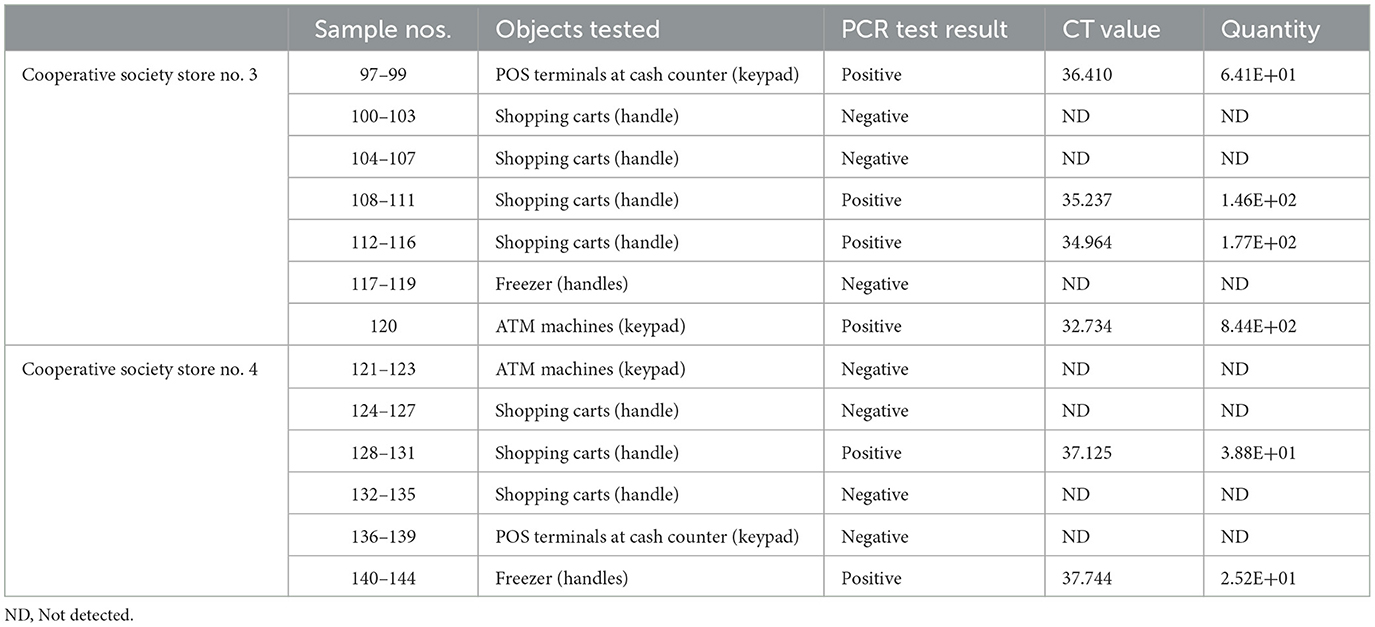
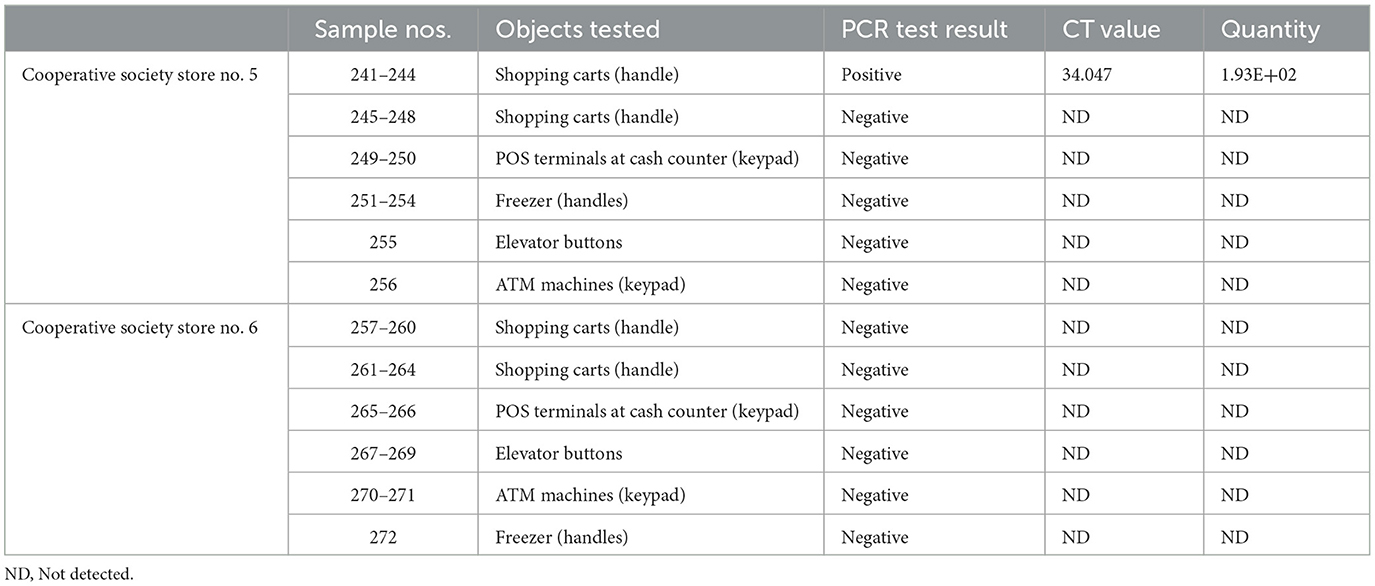
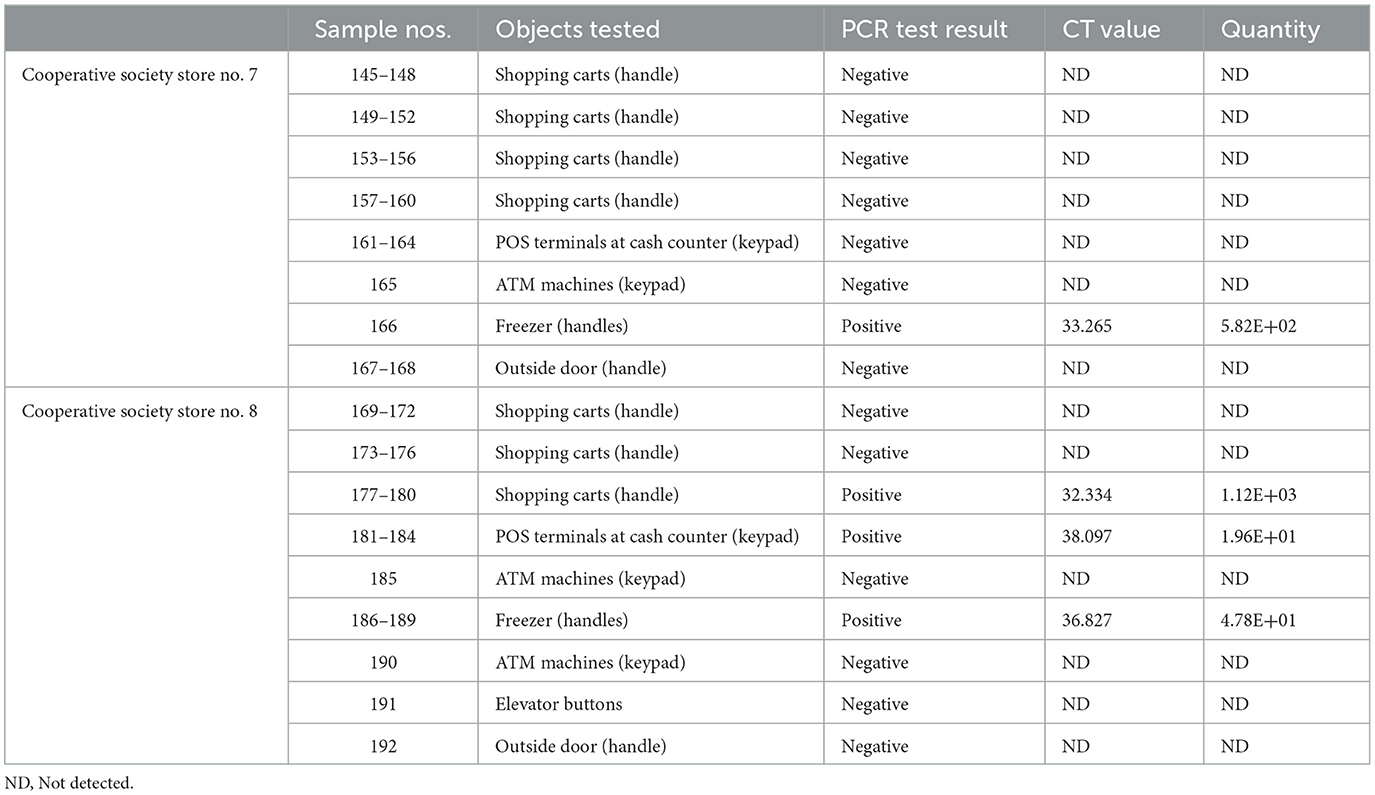
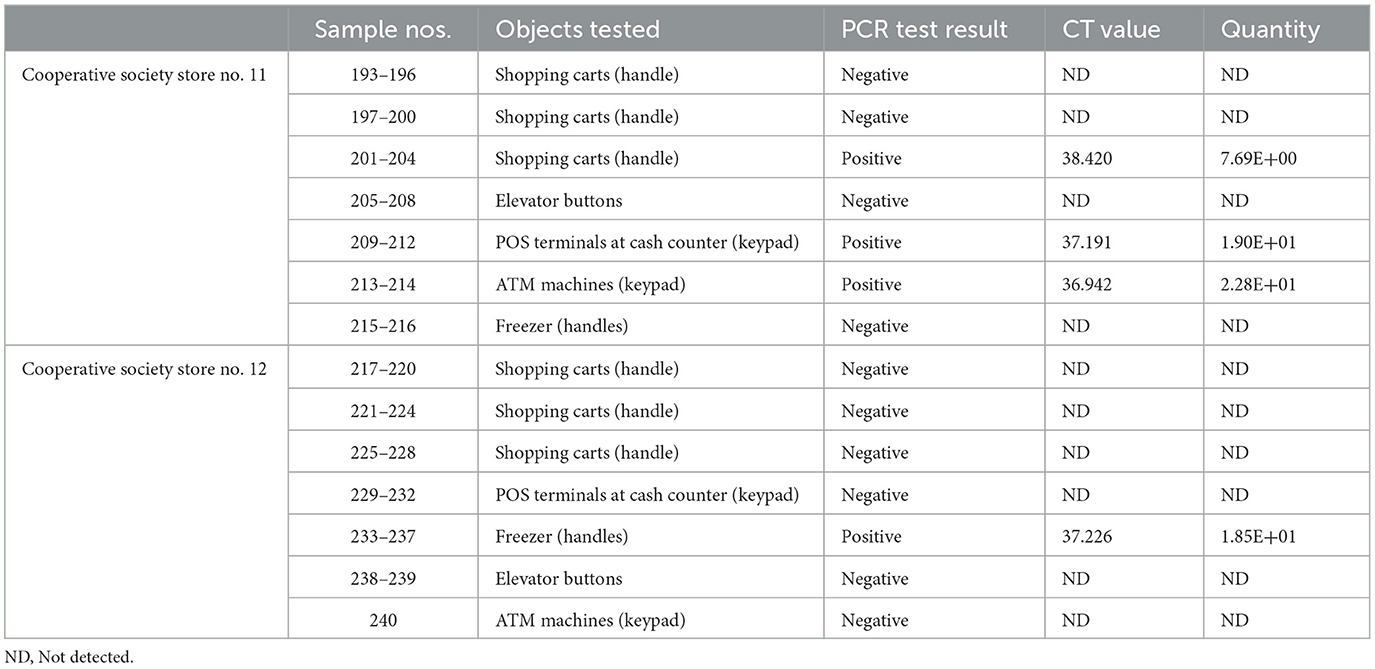
In pooled samples analyzed by RT-qPCR (np = 83), the rate of positive test was highest with the freezer handles (50%) followed by the shopping carts (32.5%), the POS terminals (25%), ATMs (23%), and the elevators (16.66%) (Table 1, Figure 2). Tables 2–7 show the results of the RT-qPCR performed in the surface samples collected from all the cooperative societies. The samples were numbered according to the sequence of collection which was scheduled in coordination with the respective Cooperative Society Stores. Thus, the two stores of the same governorate were not always sampled in the same week as indicated by the differences in the sample numbers.
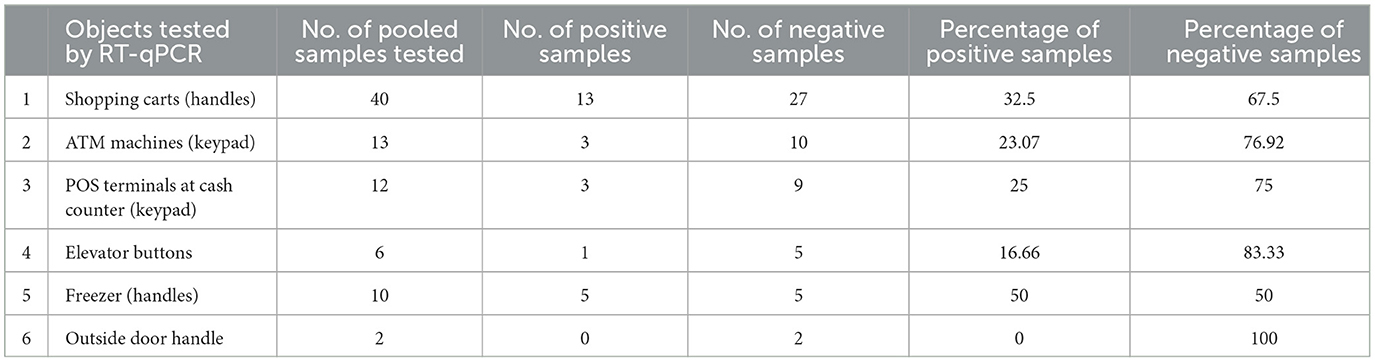
Table 1. Results of RT-qPCR performed on pooled samples (nP = 83) collected from the surfaces of various objects at the study sites.
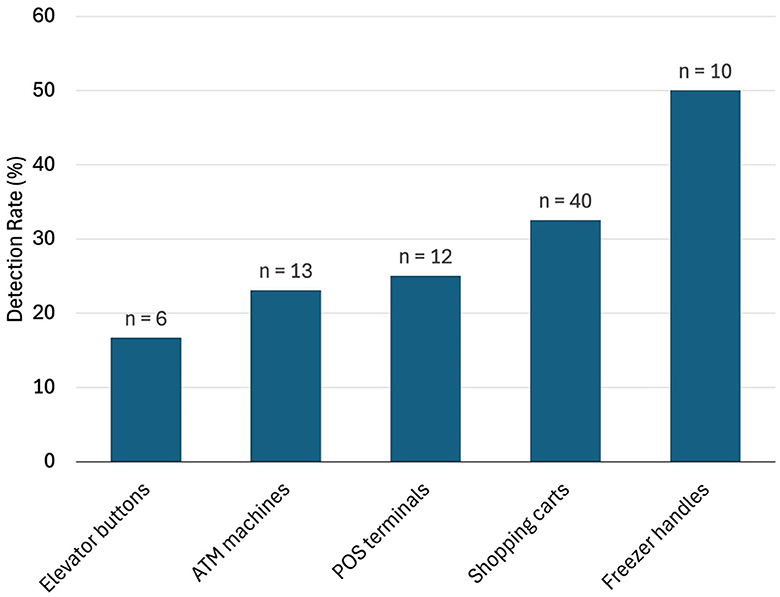
Figure 2. Rate of detection (percent) of SARS-Co-V-2 in pooled samples (nP = 83) collected from the different types of surfaces (data from the outside door handles were all negative and not shown here).
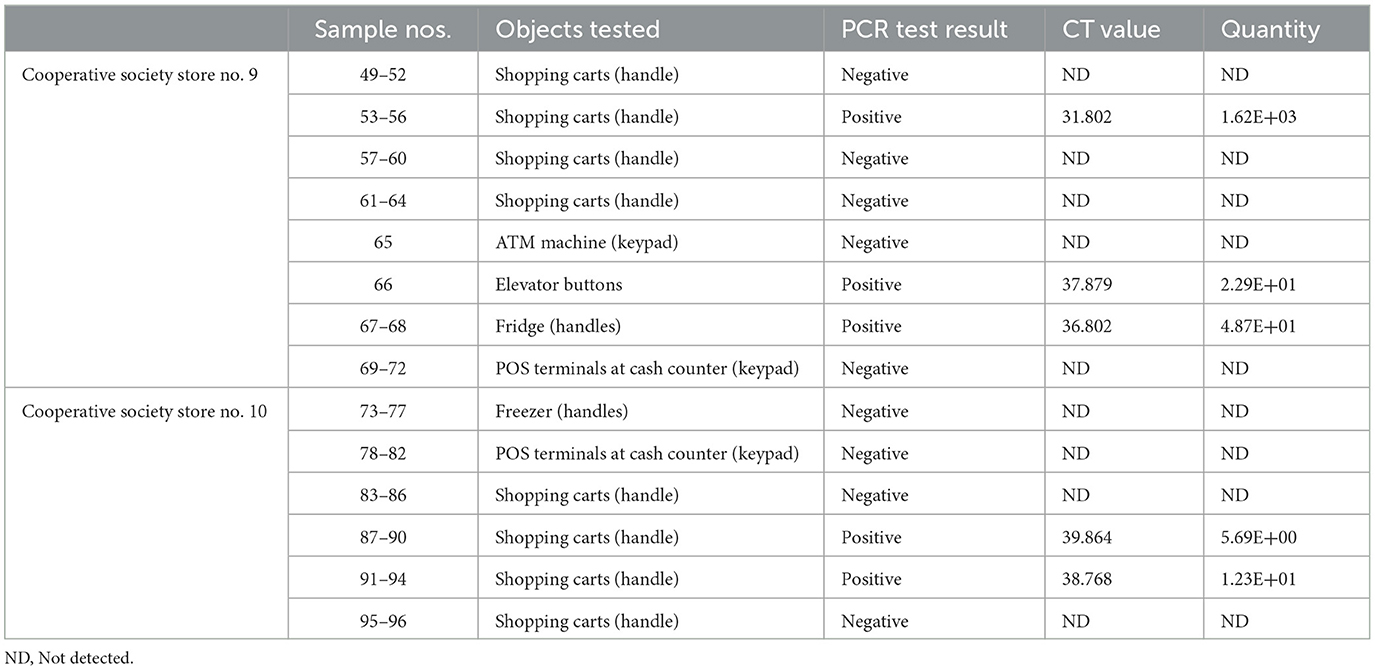
Table 6. Detection of SARS-CoV-2 in the surface samples collected from Mubarak Al-Kabeer governorate.
Discussion
Kuwait has a population of 4.8 million people of which one-third are Kuwaiti citizens and the remaining are foreign nationals (20). According to the World Bank development indicators, 100 per cent of Kuwait's population have an urban life (21). Kuwait is divided into six governorates or provinces namely, Al-Asimah, Jahra, Hawalli, Farwaniya, Mubarak Al-Kabeer, and Al-Ahmadi. In Kuwait, there are around 60 Cooperative Society Stores, also known as Co-Ops, which are public entities distributed across the six governorates. The Co-ops are the grocery and convenience stores that account for most of the retail trade in Kuwait. In order to save lives, Kuwait Government took stringent measures including complete lockdown, curfews, and international travel bans during the COVID-19 pandemic. The Co-Ops, however, remained in service during the whole period of the pandemic but required online appointments for access. Since most of the institutions including offices, schools, and most shopping malls have been shut down during the pandemic, the Co-Ops were chosen for collection of swab samples from high-touch surfaces with all precautions recommended by the Ministry of Health of Kuwait.
Active surveillance of any infectious disease is traditionally conducted by collecting appropriate body fluids (blood, urine, stool, saliva, throat swab, etc.) from an adequate number of exposed people. Conducting active surveillance for SARS-CoV-2 in the community was extremely risky because of high virulence of the coronavirus. On the other hand, collection of swab samples from the high-touch surfaces in the Co-ops for detection of SARS-CoV-2 was a much safer approach as compared with collection of nasopharyngeal swabs from the exposed community dwellers. Although previous studies have confirmed the presence and viability of coronaviruses on some environmental surfaces in hospitals and elsewhere, the abundance and detectability of SARS-CoV-2 on environmental surfaces within the convenience stores were unknown. The unique distribution of the Co-ops across the whole country offered the study team the opportunity for collecting surface samples from these locations that were visited by many people daily even during the height of the pandemic.
The surface samples collected from the Co-Ops represented all the governorates of Kuwait. Almost every type of high touch surfaces tested in this study showed the presence of SARS-CoV-2 in variable proportions. While the handles of the shopping carts and the freezer handles showed high rate of positivity, the handles of the outside door were negative although they were used by many people visiting the same store. A possible explanation of this could be that the lower temperature inside the air-conditioned store had facilitated the prolonged viability of the virus on those surfaces whereas the outside high temperature reaching up to 125°F in June might have resulted in rapid degradation of the viral RNA on the entrance door handles. The longer contact time with the shopping carts as compared with the other objects tested could have accounted for the higher rate of viral detection in the former. The rate of detection of the virus on the keypads of the POS terminals was lower than that on the ATM machines probably because the former were frequently cleaned with disinfectants. Also, many clients made contactless payment using their cards without making any physical contact with the keypads with the keypads of the POS terminals. Thus, avoidance of physical contact with contaminated surfaces by the customers and frequent sanitization of the keypads could have resulted in lesser frequency of occurrence of virus particles on these surfaces.
Several research groups have examined the occurrence of SARS-CoV-2 on various types of touch surfaces during the COVID-19 outbreak (14–16, 22–27). A meta-analysis of 22 studies by Kampf et al. (14) revealed that human coronaviruses such as SARS coronavirus, MERS coronavirus, or endemic human coronaviruses can persist on inanimate surfaces like metal, glass, or plastic for up to 9 days, but can be efficiently inactivated by surface disinfection procedures. Also, a high frequency of occurrence of SARS-CoV-2 RNA was found on stainless steel and plastic surfaces (22). The results of the study we report here clearly show that high-touch surfaces are prone to accumulate viral particles that are detectable by RT-qPCR which has a 100% specificity for detection of the infection in humans. The application of environmental sampling in infectious disease epidemiology has a long history. Wastewater-based epidemiology (WBE) has been used by the WHO for poliovirus monitoring since 2003 (28). However, its application for tracking community-level infection trends gained widespread popularity during the COVID-19 pandemic. The worldwide interest in the use of WBE underscores the potential of environmental surveillance to complement traditional epidemiological methods (29). The detection of viral RNA in wastewater, regardless of viability of the virus, provided crucial information for health regulators to implement life-saving measures and mitigate the viral spread within the community. The surface sampling method employed in our study fundamentally shared the same objective as wastewater-based epidemiology (WBE)—to assess the presence and prevalence of SARS-CoV-2 in the environment. The relative ease of swab sample collection and testing offers a powerful tool for environmental surveillance of SARS-CoV-2. While the role of surface transmission remains uncertain, detecting the virus on surfaces can effectively monitor human cases and carriers within a community, allowing governments to optimize resource allocation and manage the situation effectively. Moreover, environmental surface testing provides a unique opportunity to detect potential SARS-CoV-2 resurgences, enhancing preparedness for future outbreaks or pandemics. We strongly advocate incorporating this approach into the arsenal of preventative measures against a future pandemic.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
KJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft. NA: Formal analysis, Methodology, Writing – original draft. SF: Formal analysis, Methodology, Writing – review & editing. HK: Conceptualization, Methodology, Project administration, Writing – review & editing. AA-M: Investigation, Project administration, Writing – review & editing. BA-F: Investigation, Writing – review & editing. IA: Writing – original draft, Methodology, Project administration. VK: Writing – original draft, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors received funding from the Kuwait Foundation for Advancement of Sciences (KFAS grant CORONA PROP 39) to conduct this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. WHO COVID-19 Dashboard. COVID-19 epidemiological update. (2024). Available at: https://www.who.int/publications/m/item/covid-19-epidemiological-update-16-february-2024 (accessed February 21, 2024).
2. Simpson S, Kaufmann MC, Glozman V, Chakrabarti A. Disease X: accelerating the development of medical countermeasures for the next pandemic. Lancet Infect Dis. (2020) 20:e108–15. doi: 10.1016/S1473-3099(20)30123-7
3. Aarestrup FM, Bonten M, Koopmans M. Pandemics—One Health preparedness for the next. Lancet Reg Health Eur. (2021) 9:100210. doi: 10.1016/j.lanepe.2021.100210
4. Keusch GT, Amuasi JH, Anderson DE, Daszak P, Eckerle I, Field H, et al. Pandemic origins and a One Health approach to preparedness and prevention: solutions based on SARS-CoV-2 and other RNA viruses. Proc Natl Acad Sci USA. (2022) 119:e2202871119. doi: 10.1073/pnas.2202871119
5. WHO. Quadripartite One Health intelligence scoping study: actions to develop an effective global One Health intelligence system. (2022). Available at: https://www.who.int/publications/m/item/quadripartite-one-health-intelligence-scoping-study (accessed February 22, 2024).
6. Amuasi JH, Walzer C, Heymann D, Carabin H, Haines A, Winkler AS. Calling for a COVID-19 one health research coalition. Lancet. (2020) 395:1543–44. doi: 10.1016/S0140-6736(20)31028-X
7. Hobeika A, Stauffer MHT, Dub T, van Bortel W, Beniston M, Bukachi S, et al. The values and risks of an Intergovernmental Panel for One Health to strengthen pandemic prevention, preparedness, and response. Lancet Glob Health. (2023) 11:e1301–7. doi: 10.1016/S2214-109X(23)00246-2
8. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–192. doi: 10.1038/s41579-018-0118-9
9. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
10. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, china: a descriptive study. Lancet. (2020) 395:507–13. doi: 10.1016/S0140-6736(20)30211-7
11. Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. New Engl J Med. (2020) 382:872–4. doi: 10.1056/NEJMc2001272
12. To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior xropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. (2020) 20:565–574. doi: 10.1016/S1473-3099(20)30196-1
13. Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. Jama. (2020) 323:1610–2. doi: 10.1001/jama.2020.3227
14. Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hospital Infect. (2020) 104:246–51. doi: 10.1016/j.jhin.2020.01.022
15. Van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New Engl J Med. (2020) 382:1564–7. doi: 10.1056/NEJMc2004973
16. WHO. Surface Sampling WHO of Coronavirus Disease (COVID-19): A Practical “How to” Protocol for Health Care and Public Health Professionals. Geneva: World Health Organization (2020).
17. Bloise I, Gómez-Arroyo B, García-Rodríguez J. Detection of SARS-CoV-2 on high-touch surfaces in a clinical microbiology laboratory. J Hospital Infect. (2020) 105:784–6. doi: 10.1016/j.jhin.2020.05.017
18. Rio DC, Ares M, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Prot. (2010) 2010:prot5439. doi: 10.1101/pdb.prot5439
19. Kuchinski KS, Jassem AN, Prystajecky NA. Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_Strainer pipeline. J Clin Virol. (2020) 131:104581. doi: 10.1016/j.jcv.2020.104581
20. Available at: https://kuwaittimes.com/article/11439/kuwait/expats-still-make-up-two-thirds-of-population-as-some-communities-grow/ (accessed April 11, 2024).
21. Available at: https://data.worldbank.org/indicator/SP.URB.TOTL.IN.ZS?locations=KW (accessed April 11, 2024).
22. Falahi S, Bastani E, Pakzad I, Rashidi A, Abdoli A, Kenarkoohi A. Environmental surface contamination with SARS-CoV-2: toilets as the most contaminated surfaces in COVID-19 referral hospital. Hosp Top. (2021) 101:65–72. doi: 10.1080/00185868.2021.1969870
23. Gholipour S, Nikaeen M, Manesh RM, Aboutalebian S, Shamsizadeh Z, Nasri E, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) contamination of high-touch surfaces in field settings. Biomed Environ Sci. (2020) 33:925–9. doi: 10.3967/bes2020.126
24. Lee SE, Lee DY, Lee WG, Kang B, Jang YS, Ryu B, et al. Detection of novel coronavirus on the surface of environmental materials contaminated by COVID-19 patients in the Republic of Korea. Osong Public Health Res Perspect. (2020) 11:128–32. doi: 10.24171/j.phrp.2020.11.3.03
25. Li YH, Fan YZ, Jiang L, Wang HB. Aerosol and environmental surface monitoring for SARS-CoV-2 RNA in a designated hospital for severe COVID-19 patients. Epidemiol Infect. (2020) 148:e154. doi: 10.1017/S0950268820001570
26. Luo L, Liu D, Zhang H, Li Z, Zhen R, Zhang X, et al. Air and surface contamination in non-health care settings among 641 environmental specimens of 39 COVID-19 cases. PLoS Negl Trop Dis. (2020) 14:e0008570. doi: 10.1371/journal.pntd.0008570
27. Zhou J, Otter JA, Price JR, Cimpeanu C, Garcia DM, Kinross J, et al. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin Infect Dis. (2020) 2020:ciaa905. doi: 10.1101/2020.05.24.20110346
28. Singer AC, Thompson JR, Filho CRM, Street R, Li X, Castiglioni S, et al. A world of wastewater-based epidemiology. Nat Water. (2023) 1:408–15. doi: 10.1038/s44221-023-00083-8
Keywords: SARS-CoV-2, coronavirus, pandemic preparedness, environmental monitoring, surface sampling, RT-qPCR
Citation: Jamil K, Abdulrazack N, Fakhraldeen S, Kamal H, Al-Mutairi A, Al-Feili B, Ahmed I and Kumar V (2024) Detection of SARS-CoV-2 on the environmental surfaces and its implications for pandemic preparedness. Front. Public Health 12:1396334. doi: 10.3389/fpubh.2024.1396334
Received: 05 March 2024; Accepted: 20 August 2024;
Published: 10 September 2024.
Edited by:
Amira Adel Taha Abdel Aleem AL-Hosary, Assiut University, EgyptReviewed by:
Hosnia Abdel-Mohsein, Assiut University, EgyptOtun Saha, Washington State University, United States
Copyright © 2024 Jamil, Abdulrazack, Fakhraldeen, Kamal, Al-Mutairi, Al-Feili, Ahmed and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazi Jamil, a2phbWlsQGtpc3IuZWR1Lmt3; Vinod Kumar, dmlub2RrQGtpc3IuZWR1Lmt3
 Kazi Jamil
Kazi Jamil Nasreem Abdulrazack
Nasreem Abdulrazack Saja Fakhraldeen
Saja Fakhraldeen Heba Kamal
Heba Kamal Anwar Al-Mutairi
Anwar Al-Mutairi Batool Al-Feili
Batool Al-Feili Imtiaz Ahmed
Imtiaz Ahmed Vinod Kumar
Vinod Kumar