- 1Clinical Medical College of Chengdu Medical College, Chengdu, Sichuan, China
- 2Department of Pulmonary and Critical Care Medicine, The First Affiliated Hospital of Chengdu Medical College, Chengdu, Sichuan, China
- 3Emergency Department, The First Affiliated Hospital of Chengdu Medical College, Chengdu, Sichuan, China
Background: The timely diagnosis of tuberculosis through innovative biomarkers that do not rely on sputum samples is a primary focus for strategies aimed at eradicating tuberculosis. miR-29 is an important regulator of tuberculosis pathogenesis. Its differential expression pattern in healthy, latent, and active people who develop tuberculosis has revealed its potential as a biomarker in recent studies. Therefore, a systematic review and meta-analysis were performed for the role of miR-29 in the diagnosis of tuberculosis.
Methods: EMBASE, PubMed, CNKI, Web of Science, and Cochrane Library databases were searched utilizing predefined keywords for literature published from 2000 to February 2024.Included in the analysis were studies reporting on the accuracy of miR-29 in the diagnosis of tuberculosis, while articles assessing other small RNAs were not considered. All types of study designs, including case–control, cross-sectional, and cohort studies, were included, whether prospectively or retrospectively sampled, and the quality of included studies was determined utilizing the QUADAS-2 tool. Publication bias was analyzed via the construction of funnel plots. Heterogeneity among studies and summary results for specificity, sensitivity, and diagnostic odds ratio (DOR) are depicted in forest plots.
Results: A total of 227 studies were acquired from the various databases, and 18 articles were selected for quantitative analysis. These articles encompassed a total of 2,825 subjects, primarily sourced from the Asian region. Patient specimens, including sputum, peripheral blood mononuclear cells, cerebrospinal fluid and serum/plasma samples, were collected upon admission and during hospitalization for tuberculosis testing. miR-29a had an overall sensitivity of 82% (95% CI 77, 85%) and an overall specificity of 82% (95% CI 78, 86%) for detecting tuberculosis. DOR was 21 (95% CI 16–28), and the area under the curve was 0.89 (95% CI 0.86, 0.91). miR-29a had slightly different diagnostic efficacy in different specimens. miR-29a showed good performance in both the diagnosis of pulmonary tuberculosis and extrapulmonary tuberculosis. miR-29b and miR-29c also had a good performance in diagnosis of tuberculosis.
Conclusion: As can be seen from the diagnostic performance of miR-29, miR-29 can be used as a potential biomarker for the rapid detection of tuberculosis.
Systematic Review Registration: https://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=461107.
1 Introduction
Tuberculosis (TB) diagnosis continues to pose a worldwide challenge, and misdiagnoses have contributed to heightened morbidity and mortality rates (1). Hence, the imperative lies in the identification of TB infection biomarkers to facilitate the diagnosis of active TB. Among the assessed TB biomarkers, microRNA (miRNA) is one such type under evaluation (2). miRNAs have been proposed as a novel biological diagnostic indicator in various settings encompassing diabetes, heart disease, infectious diseases, pregnancy, cancer, and psoriasis. Mycobacterium tuberculosis is among the numerous microorganisms that, upon infection, lead to modifications in epigenetic patterns within the host. Epigenetic processes have the capacity to modify gene expression without making changes to the underlying DNA sequence. These processes encompass DNA methylation, histone modifications, and the action of miRNAs (3). MiRNA stands as a promising avenue for application as a molecular diagnostic marker in diverse infectious diseases, serving as biomarkers for disease diagnosis, treatment outcome, and prognosis.
In addition, miRNA has been implicated in various diseases such as immunity, heart, infectious diseases, and cancer (4). As the disease progresses, the affected organs have the capacity to release specific miRNAs into the bloodstream. Compared to healthy individuals, miRNA levels in blood significantly differ in individuals with multiple diseases. Up until now, miRNAs have been utilized as molecular diagnostic indicators for a range of conditions, including cancer, mental disorders, diabetes, heart disease, and infectious diseases (5).
An essential element of the World Health Organization’s 2035 milestone strategy to eradicate TB involves the early diagnosis of the disease through systematic screening of high-risk populations. This call was made to identify and validate clear biomarkers capable of discriminating between active and latent TB, as well as predicting disease progression. Circulating miRNA has emerged as a potent biomarker in TB diagnosis (6). Furthermore, miRNA has been employed as a biological indicator to differentiate between latent and active TB (7) and as a biomarker for TB coinfection in individuals with HIV (8). Recent studies have also showcased the significant role of these biomarkers in predicting the progression of the disease (9). In prior investigations, microarray technology has been employed in multiple studies to pinpoint miRNAs as potential biomarkers for TB in multiple sample sources, such as serum, blood, sputum macrophages, and peripheral blood mononuclear cells of TB patients (10–12). For individuals with active pulmonary TB, previous studies identified 59 kinds of miRNAs upregulated and 33 kinds of miRNAs downregulated compared with healthy controls (10). Among the upregulated miRNAs, miRNA-29 family (miR-29a, miR-29b, and miR-29c), particularly miR-29a, stands out, as it inhibits immune responses by post-transcriptionally inhibiting interferon (INF)-γ expression in T cells. This mechanism potentially enhances susceptibility to TB (13). In the context of Mycobacterium tuberculosis infection, the miR-29 family assumes a crucial role in influencing innate and adaptive immunity by exerting an impact on gene expression within macrophages, dendritic cells, B cells, and T cells (14). Multiple in vitro and in vivo studies have highlighted the function of the miR-29 family in host immune regulatory mechanisms during TB pathogenesis. These mechanisms encompass the modulation of apoptotic pathways, induction of autophagy, stimulation of IFN-γ, and secretion of TNF-α. Mycobacterium tuberculosis uses a variety of strategies to counteract these regulatory mechanisms and enhance its survival capabilities (15–17).
In this research, the available literature on the potential of the miR-29 family as a diagnostic biomarker for tuberculosis was reviewed, and the reported diagnostic accuracy of miR-29 was evaluated. The candidate gene miR-29 was identified, which could be employed as a reliable diagnostic marker for TB.
2 Methods
The protocol has been registered in the PROSPERO registry (ID: CRD42023461107).1
2.1 Search strategy
The systematic assessment and subsequent meta-analysis followed the guidelines established by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (18), and a corresponding report was generated. Extensive literature searches in the PubMed and CNKI databases were finished, and articles from the Web of Science and Embase databases were also retrieved. The search platform encompassed the inclusion of the Cochrane Library as well. MeSH vocabulary and keywords pertaining to TB and miR-29 were employed for the search, as outlined in the Supplementary Table 1. To identify potential additional eligible studies, both forward and backward citation checks were conducted based on relevant review articles.
2.2 Criteria for eligibility
2.2.1 Time frame
Articles published in Chinese and English from January 2000 to February 2024 were encompassed in the study.
2.2.2 Study type
All types of study designs, including case–control, cross-sectional, and cohort studies, were included, whether prospectively or retrospectively sampled. Non-original articles, such as narrative and systematic reviews, meta-analyses, and conference abstracts, were excluded. Animal studies were also not included.
2.2.3 Biomarker criteria
Included in the analysis were studies reporting on miR-29, while articles assessing other small RNAs were not taken into account. In light of the defined scope of this systematic review, studies that had assessed miR-29 as a biological marker for diseases other than respiratory diseases and TB were excluded. Studies that lacked specificity, sensitivity, and area under the curve (AUC) data were not taken into consideration.
2.2.4 Study population
No exclusion criteria were applied based on patient characteristics, encompassing studies involving both adult and pediatric populations.
2.3 Ethical review
Meta-analyses are a form of research that analyses previous research data without requiring ethical approval.
2.4 Screening and data extraction
All research articles identified through the search were imported into Endnote X9(EndNote X9, Clarivate Analytics, Philadelphia, PA, United States), and duplicate records were eliminated. The management of the included studies was conducted using Rayyan (19). Initially, two researchers conducted the screening of titles and abstracts from electronically retrieved publications to determine potentially eligible studies. Then, they subsequently conducted independent full-text screening to identify pertinent studies. In cases where specific articles lacked sufficient or complete data for a definitive inclusion or exclusion decision, the authors of the respective papers were contacted to obtain any absent data. The following data points were collected: publication year, sample size (cases and controls recorded separately), country, mean or median age, male-to-female ratio, study population (active TB/latent tuberculosis infection (LTBI)/healthy), sample source, reference test, screening and validation methods, identified miRNA (miR-29a, miR-29b, and miR-29c), and diagnostic accuracy indicators (specificity, sensitivity, and AUC). A data sheet was designed, and extracted data was entered into an Excel database. The values for True Positive (TP), False Positive (FP), True Negative (TN), and False Negative (FN) derived from the index test outcomes were documented. In cases where these values were not explicitly reported, they were computed based on the reported specificity, sensitivity, and sample size. For studies that did not contain adequate information to create a 2 × 2 table, the study authors were contacted and requested to provide the necessary details. If the study author could not provide this information, the study was not included in the meta-analysis, but the qualitative analysis within the narrative analysis section was retained. Two reviewers validated the extracted data, resolving any disagreements or discrepancies by discussion until a consensus was reached.
2.5 Evaluation of the quality of included studies
Methodological quality was evaluated for all-encompassed studies utilizing QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies) (20). The QUADAS-2 instrument includes four domains—patient selection, indicator testing, reference standards, and processes and timing, aimed at assessing the risk of bias and clinical applicability of the study. Within each domain, specific questions are posed to determine whether bias and applicability are rated as “low,” “high,” or “uncertain.” Studies were deemed to have good quality if they provided critical information, including details about biomarker identification methods and accurate classification of the study cohort (active or latent TB and control groups).
2.6 Data synthesis and statistical analysis
Meta-Disc 1.4(version 1.4; Universidad Complutense, Barcelona, Spain) and Stata 11.0(version 11.0; StataCorp LLC, College Station, TX, United States) software were utilized for all calculations. Since miR-29a is different from miR-29b and miR-2c, we separated them in our analyses. Spearman correlation coefficient was utilized to explore the presence of a threshold effect. Heterogeneity between studies was assessed utilizing the I2 test with a critical point of ≥50% and a p value of <0.10. In cases where heterogeneity was observed, a random-effects model was applied. Conversely, a fixed-effects model was utilized when no significant heterogeneity was detected. Pooled statistics were presented by specificity, sensitivity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and 95% confidence interval. A summary receiver operating characteristic curve (SROC curve) was plotted, and the area under the curve (AUC) was computed. Subgroup analysis was used to further investigate the diagnostic efficacy of miR-29(miR-29a, miR-29b, and miR-29c) in different subgroups. Furthermore, publication bias was determined by generating the funnel chart (Deeks’ funnel plot). In full-text statistics, p < 0.05 was deemed as a statistically significant value. Besides, the results of individual studies will be summarized descriptively if fewer studies are unable to carry out a meta-analysis.
3 Results
3.1 Attributes of included studies
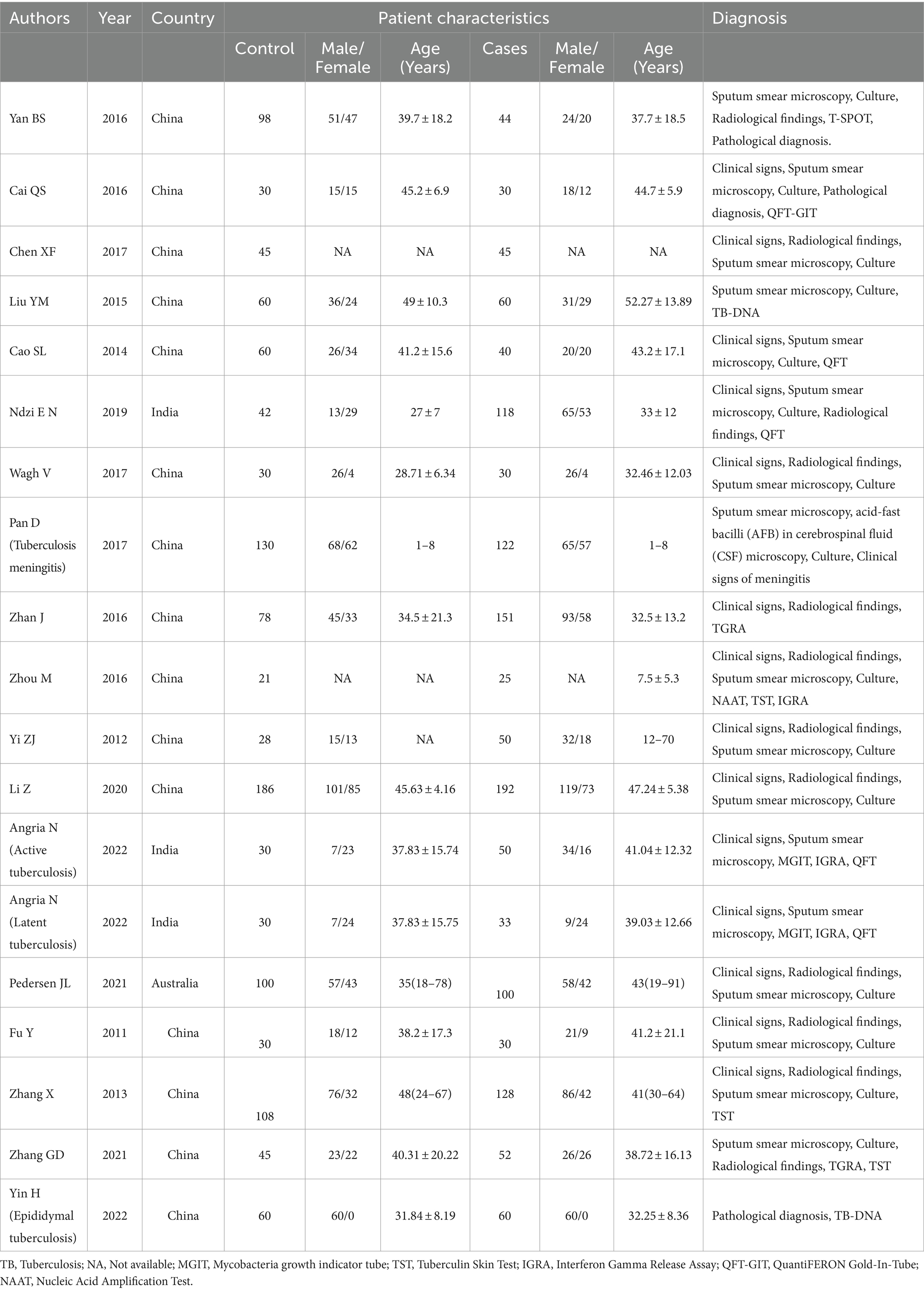
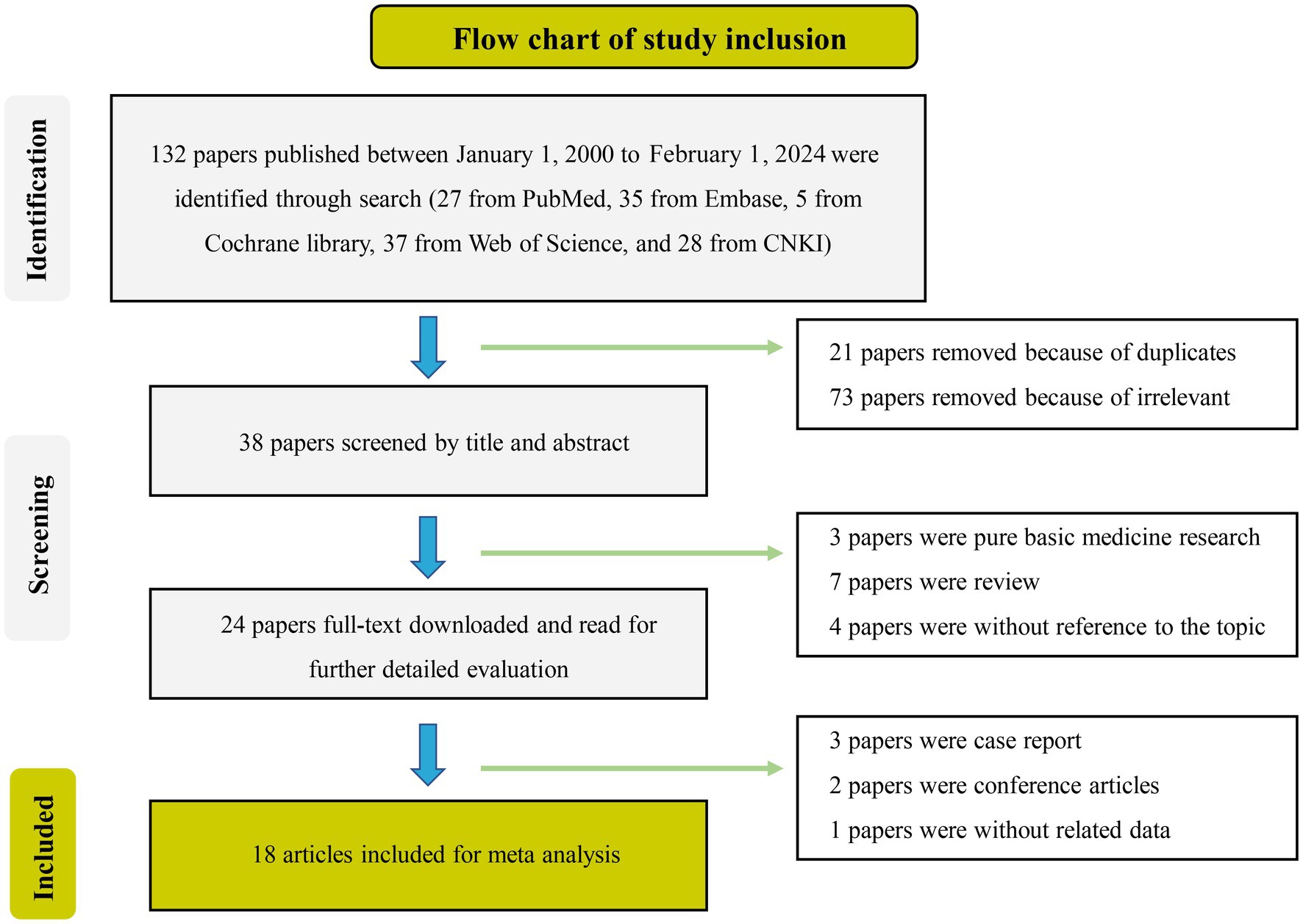
Out of the initial pool of 132 papers identified through the search formula, a total of 18 papers (3, 10, 11, 21–35) encompassing 20 studies were ultimately included in the analyses. This selection process involved the exclusion of irrelevant articles, duplicate studies, case reports, reviews, and other ineligible publications. These studies were mainly from Asian regions, involving 19 studies of active TB, 1 latent TB, 17 pulmonary TB, 3 extrapulmonary TB, 14 adult TB, and 3 pediatric TB. Eighteen studies on miR-29a, one on miR-29b, and one on miR-29c met inclusion criteria. Details of each included study are depicted in Tables 1, 2. The specific process of inclusion and exclusion is shown in Figure 1. Next, the quality assessment of the studies included in the analysis was performed using the QUADAS-2 tool. Studies that did not meet the inclusion criteria were assessed to have a high risk of bias (Figure 2).
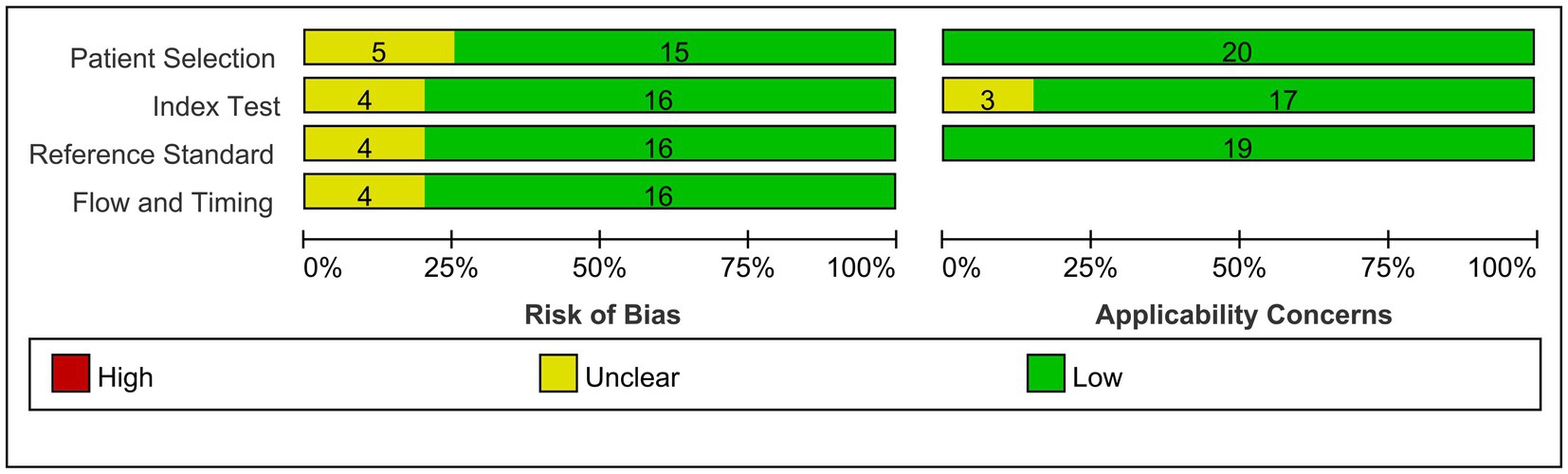
Figure 2. Overview of applicability concerns and the risk of bias of included studies as per the QUADAS-2 tool.
3.2 Literature data extraction
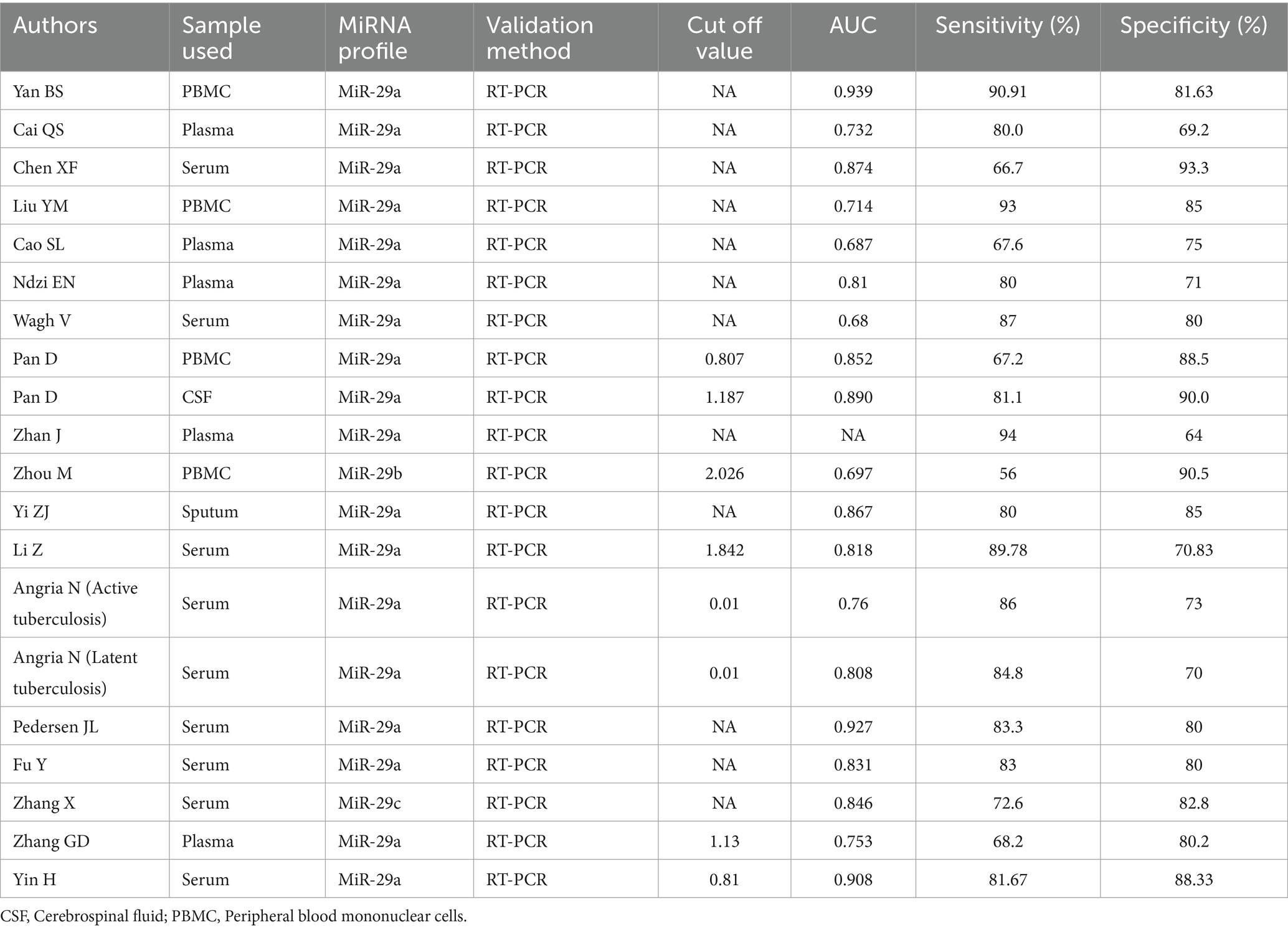
These 20 studies involved 2,543 participants. Each study clearly described the sensitivity and specificity of miR-29 for TB diagnosis. miR-29 was analyzed by quantitative PCR in all studies, but the sources of specimens varied, with sputum in one study, peripheral blood mononuclear cells in 4 studies, cerebrospinal fluid in one study, serum in nine studies, and plasma in 5 studies. Eight studies clearly provided the data of best cut-off values.
3.3 Meta-analysis
Of the included studies, 18 studies were on miR-29a, one reported miR-29b, and one reported miR-29c, thus, diagnostic meta-analysis was performed on miR-29a.The remaining data (miR-29b and miR-29c) were analyzed descriptively.
3.3.1 Threshold effect analysis
A threshold effect emerges due to variations in sensitivity, specificity, and DOR of the diagnostic test resulting from different thresholds. Therefore, it is essential to initially examine whether the diagnostic method exhibits a threshold effect. To assess this, Spearman correlation analysis was conducted utilizing Meta-Disc 1.4 software, yielding a correlation coefficient of r = 0.396 (p = 0.104). The absence of a significant threshold effect was indicated, allowing for the subsequent pooled analysis to proceed.
3.3.2 Pooled analysis
The pooled sensitivity and specificity across the 18 studies were 82% (95% CI, 77, 85%, I2 = 75.1%) and 82% (95% CI, 78, 86%, I2 = 69.5%), respectively (Figure 3A). The PLR was 4.7 (95% CI, 3.8, 5.7), and the NLR was 0.22 (95% CI, 0.18, 0.28). DOR was 21 (95% CI, 16, 28), and the AUC was 0.89 (95% CI, 0.86, 0.90; Table 3). The SROC curve of miR-29a with its confidence and prediction regions is shown in Figure 3B. In addition, the data on active TB was pooled for analysis to investigate the diagnostic ability of the miR-29a test for active TB. The diagnostic sensitivity and specificity for active TB were 81% (95% CI, 77, 85%) and 83% (95% CI, 79, 86%).
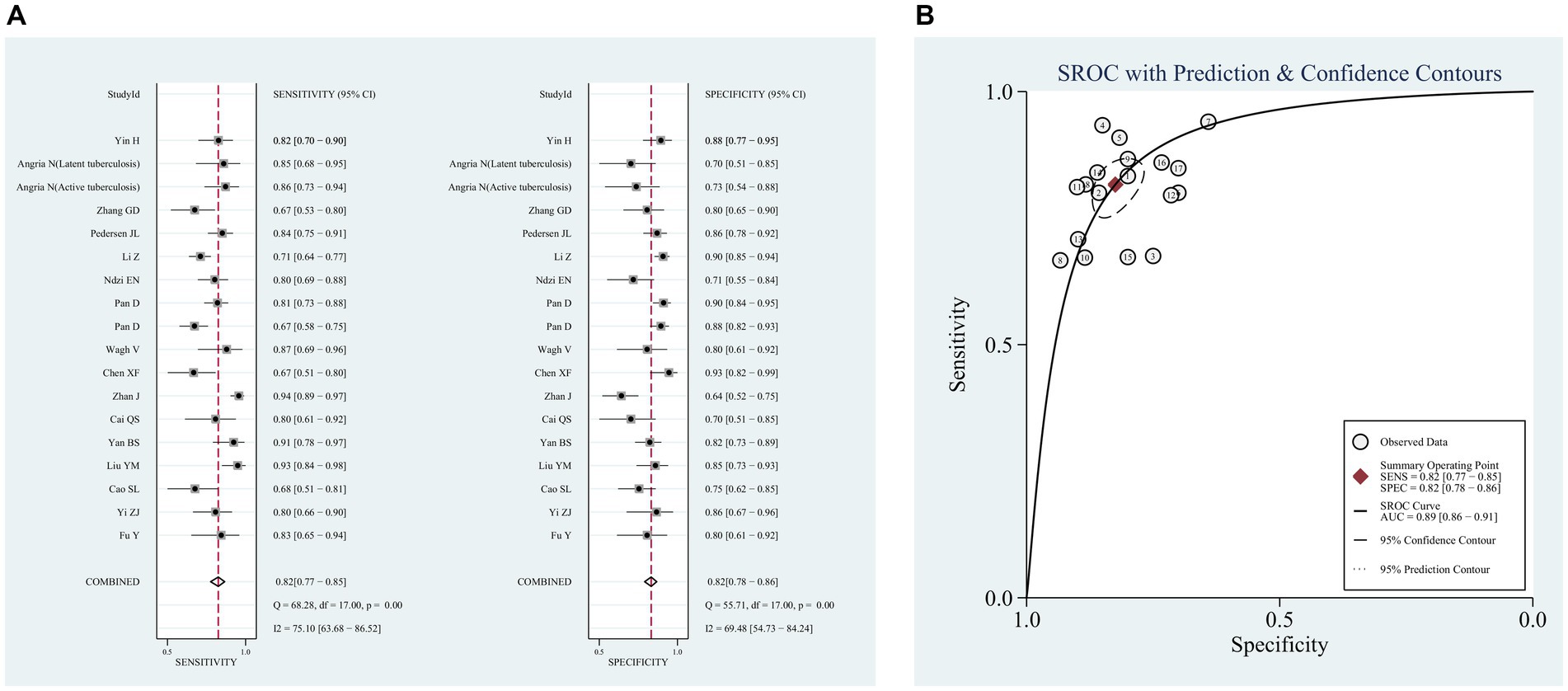
Figure 3. Pooled analysis of the diagnostic accuracy of miR-29a for patients with tuberculosis. (A) Sensitivity and Specificity, (B) SROC curve.
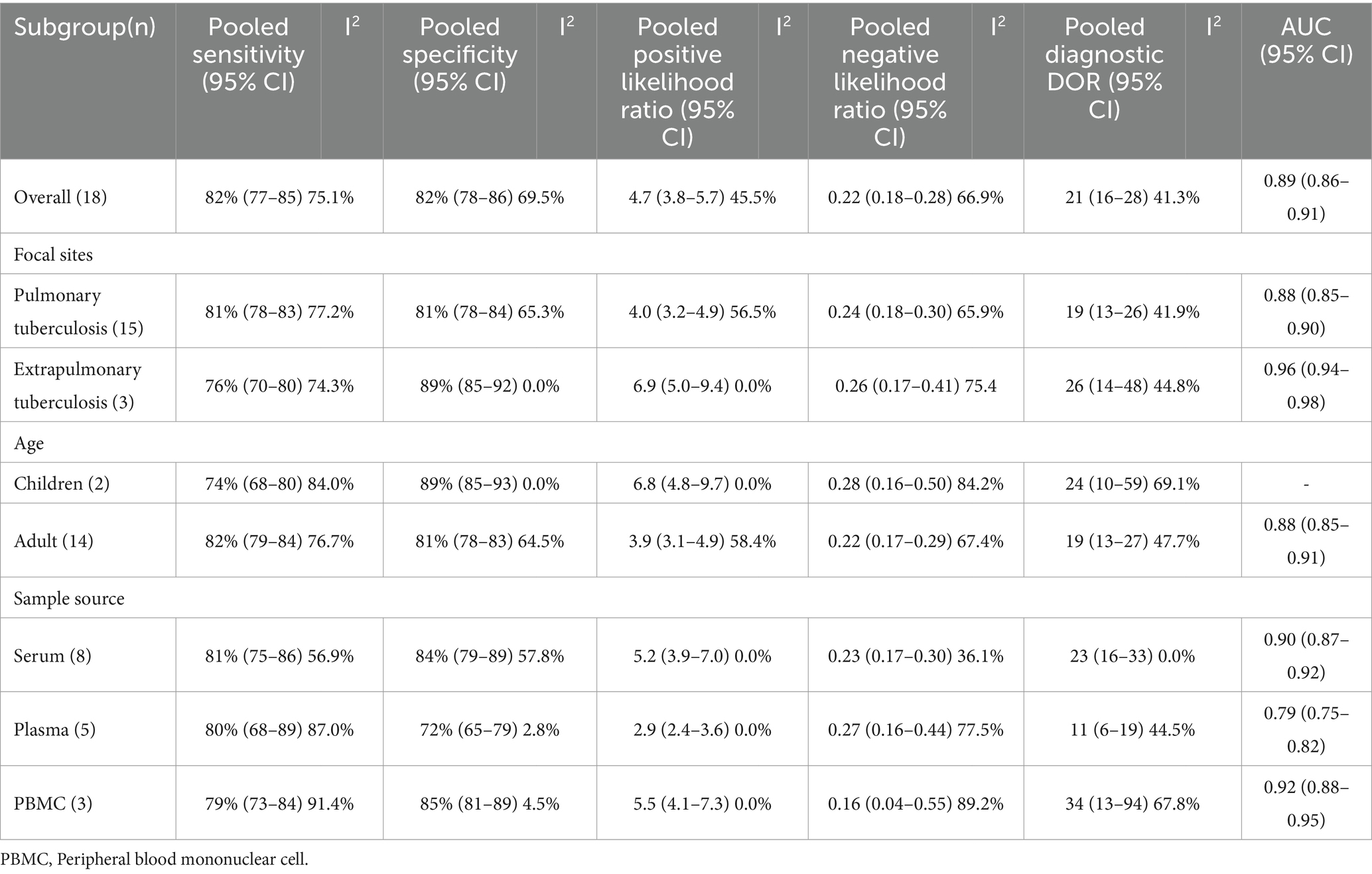
Table 3. Subgroup analysis of the included studies based on the differences of the purpose of testing and the positive result definition (miR-29a).
3.3.3 Subgroup analyses
3.3.3.1 Lesion location of TB
The subgroup analysis was conducted according to the site of TB occurrence, aiming to further explore the origin of heterogeneity. Subgroup analysis showed that miR-29a was used for pulmonary TB screening with a sensitivity of 81% (95% CI: 78, 83%), specificity of 81% (95% CI: 78, 84%; Figure 4A), PLR of 4.0 (95% CI: 3.2, 4.9), NLR of 0.24 (95% CI: 0.18, 0.30), DOR of 19 (95% CI: 13, 26), and AUC of 0.88 (95% CI: 0.85, 0.90). When miR-29a was used for screening extrapulmonary TB, it had a sensitivity of 76% (95% CI: 70, 80%), specificity of 89% (95% CI: 85, 92%; Figure 4B), PLR of 6.9 (95% CI: 5.0, 9.4), NLR of 0.26 (95% CI: 0.17, 0.41), DOR of 26 (95% CI: 14, 48), and AUC of 0.96 (95% CI: 0.94, 0.98; Table 3).
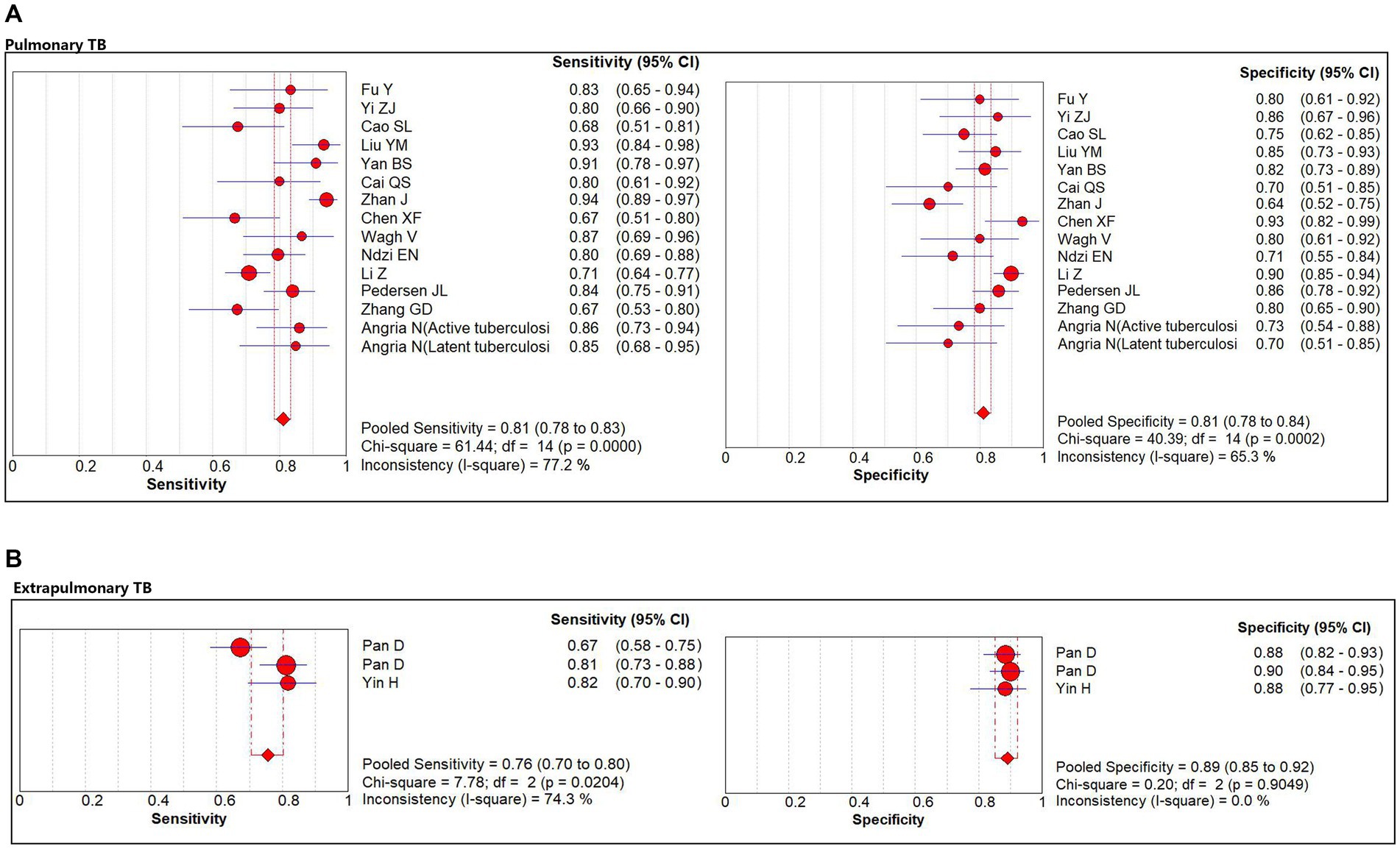
Figure 4. The sensitivity and specificity of miR-29a for patients with pulmonary TB and extrapulmonary TB. (A) Pulmonary TB. (B) Extrapulmonary TB. TB, tuberculosis.
3.3.3.2 Age
The subgroup analysis was performed according to age. In the adult group, miR-29a had a sensitivity of 82% (95% CI: 79, 84%), specificity of 81% (95% CI: 78, 83%), PLR of 3.9 (95% CI: 3.1, 4.9), NLR of 0.22 (95% CI: 0.17, 0.29), DOR of 19 (95% CI: 13, 27), and AUC of 0.88 (95% CI: 0.85, 0.91) for the diagnosis of TB. In the pediatric group, miR-29a had a sensitivity of 74% (95% CI: 68, 80%), specificity of 89% (95% CI: 85, 93%), PLR of 6.8 (95% CI: 4.8, 9.7), NLR of 0.28 (95% CI: 0.25, 0.37), DOR of 24 (95% CI: 10, 59) for the diagnosis of TB (Table 3).
3.3.3.3 Specimens
The subgroup analysis was executed according to the origin of the specimens. miR-29a in serum had a sensitivity of 81% (95% CI: 75, 86%), specificity of 84% (95% CI: 79, 89%), PLR of 5.2 (95% CI: 3.9, 7.0%), NLR of 0.23 (95% CI: 0.17, 0.30), DOR of 23 (95% CI: 16, 33), and AUC of 0.90 (95% CI: 0.87, 0.92) for the diagnosis of TB. miR-29a in plasma had a sensitivity of 80% (95% CI: 68, 89%), specificity of 72% (95% CI: 65, 79%), PLR of 2.9 (95% CI: 2.4, 3.6), NLR of 0.27 (95% CI: 0.16, 0.44), DOR of 11 (95% CI: 6, 19), and AUC of 0.79 (95% CI: 0.75, 0.82) for the diagnosis of TB. Furthermore, miR-29a in peripheral blood mononuclear cell plasma had a sensitivity of 79% (95% CI: 73, 84%), specificity of 85% (95% CI: 81, 89%), PLR of 5.5 (95% CI: 4.1, 7.3), NLR of 0.16 (95% CI: 0.04, 0.55), DOR of 34 (95% CI: 13, 94), and AUC of 0.92 (95% CI: 0.88, 0.95) for the diagnosis of TB (Table 3).
3.3.4 Sensitivity analysis and publication bias analysis
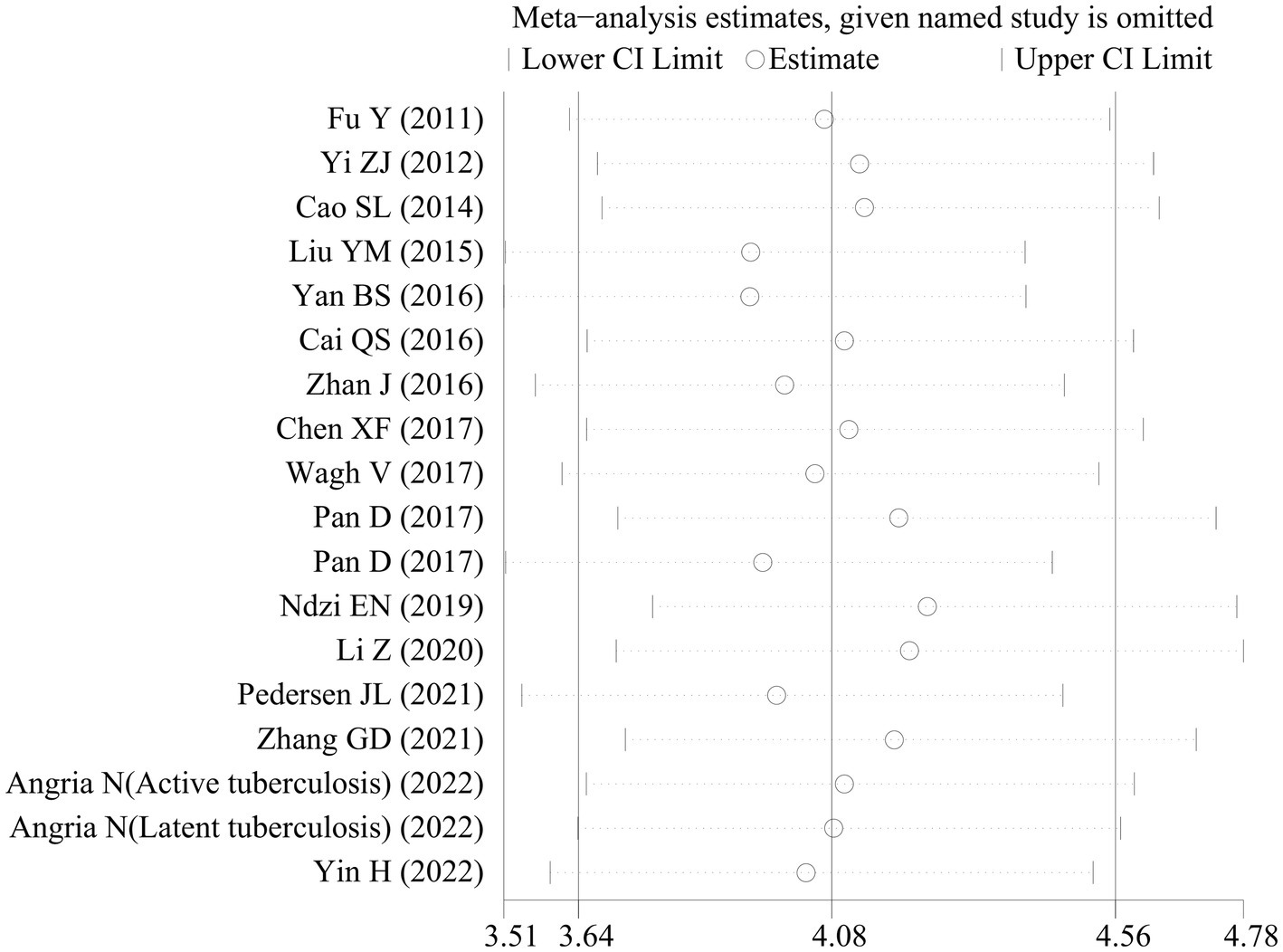
Sensitivity analyses were executed to examine the reliability of the findings. These analyses demonstrated that the outcomes of the meta-analysis remained largely consistent even when individual studies were removed one by one. This consistency suggested the absence of significant bias in the included studies and confirmed the reliability and stability of the results (Figure 5). Furthermore, Deeks’ funnel plot was employed to assess publication bias, revealing no notable publication bias (p = 0.32; Figure 6).
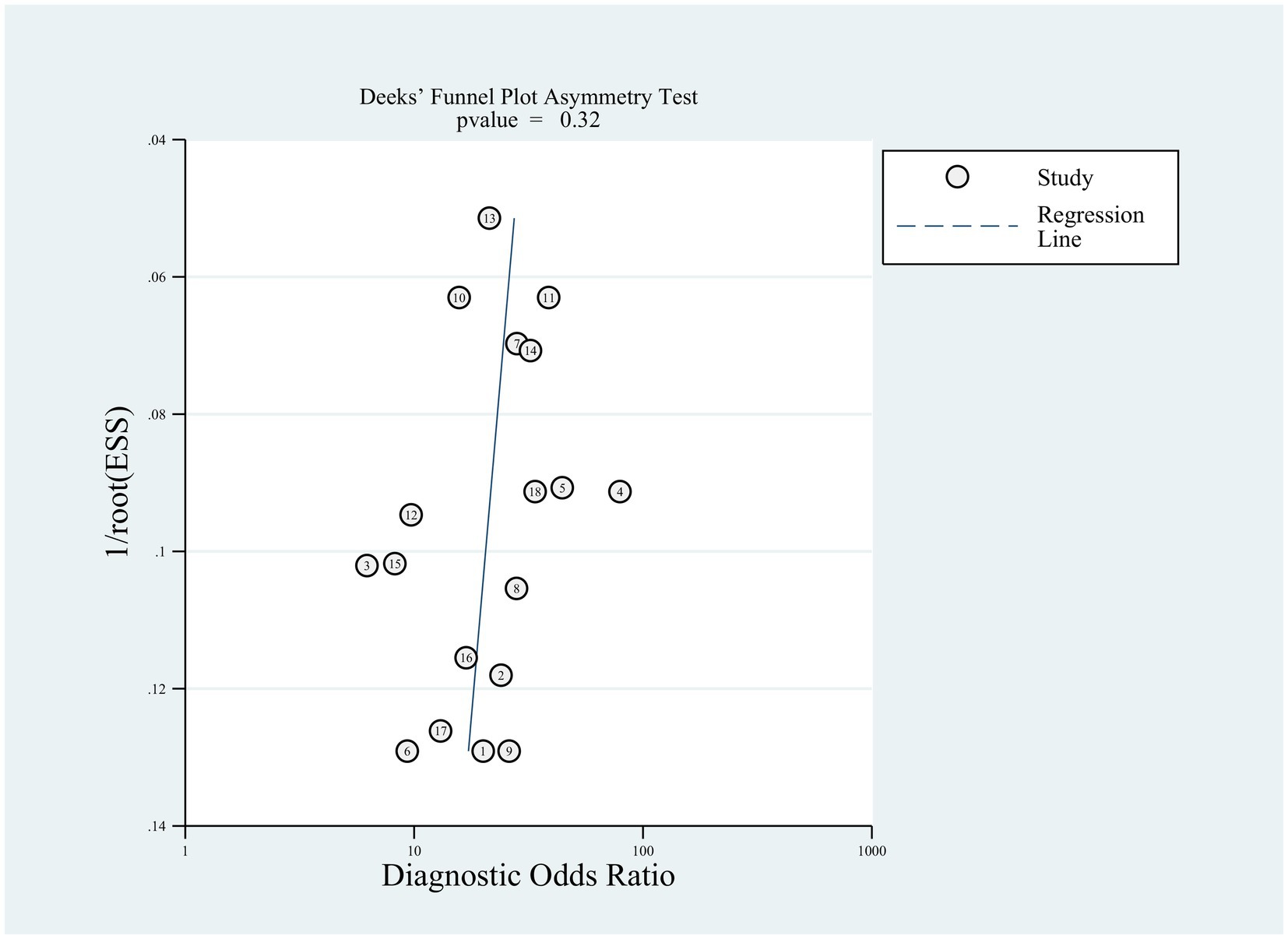
Figure 6. Publication bias analysis of the included studies by Deeks’ funnel plot asymmetric linear regression test.
3.4 Descriptive analysis
Zhou et al. (11) showed that PBMC miR-29b as a diagnostic for tuberculosis was specific (90.5%), but not sensitive (56%) in children. When compared with healthy controls, PBMC miR-29 was upregulated in the TB cases. Zhang et al. (35) found that the performance values of miR-29c are 72.6, 82.8% and 0.846 for sensitivity, specificity, and AUC value, respectively. Meanwhile, they found that the expression levels of miR-29c in TB serums were higher than those in normal control serums.
Yi et al. (26) detected miR-29a expression in the sputum of patients with pulmonary TB, and their results showed that miR-29a exhibited a sensitivity of 80%, a specificity of 85%, and an AUC value of 0.867 for the diagnosis of TB. Pan et al. (33) detected miR-29a expression in cerebrospinal fluid of children with tuberculosis meningitis. Their results showed that miR-29a expression was considerably elevated in the cerebrospinal fluid of children with tuberculosis meningitis. The sensitivity, specificity, and AUC of miR-29a in cerebrospinal fluid for the diagnosis of tuberculosis meningitis were 81.1, 90.0%, and 0.890, respectively.
4 Discussion
In this systematic review and meta-analysis, the diagnostic accuracy of the miR-29 family as a TB biomarker was assessed. This study showed that miR-29 family has high diagnostic efficacy accuracy in detecting TB, and the pooled sensitivity and specificity of miR-29a were 82 and 82%, respectively, which demonstrates that the missed diagnosis and misdiagnosis rates of TB using miR-29 have been as low as 18 and 18%, respectively. Results of our subgroup analysis in sensitivity and specificity also supported the above assumption. These findings were similar to Yao et al.’s study (36). As the sensitivity and specificity were higher than 80%, the diagnostic accuracy of miR-29a for tuberculosis was quite high. In addition, higher than 5 of positive likelihood ratio and lower than 0.3 of negative likelihood ratio are considered good diagnostic significance (37). In our meta-analysis, the positive likelihood ratio was 4.7, indicating that the probability of a miR-29a-positive diagnosis of TB was 4.7-fold higher than that of non-TB controls. DOR is a measure of diagnostic test efficiency that combines sensitivity and specificity; the higher the value of DOR, the better the performance of the discriminatory test (38). In this study, the DOR was 21, indicating that miR-29a is a good marker to distinguish TB from non-TB groups. The SROC curve also suggested that miR-29a had an excellent performance in diagnosing TB; the AUC value reached 0.89, which represents a high over accuracy with high values of sensitivity and specificity. Therefore, miR-29a could be an accurate molecular marker to support the diagnosis of TB. Nevertheless, it cannot be used as the only diagnostic marker of TB as it has a misdiagnosis rate of 18%. In addition, miR-29b and miR-29c also had a good performance in diagnosis of tuberculosis.
Notably, miR-29 exhibited reduced expression in IFN-γ-producing cells such as (natural killer) NK cells and CD4 + and CD8 + T cells following Mycobacterium tuberculosis BCG infection in mice (14). Its expression significantly increased in both latent and active TB cases, leading to reduced IFN-γ levels and modifications in signaling pathways (39). Ndzi EN et al. (31) also indicated that miR-29a demonstrated strong performance in distinguishing active pulmonary TB from healthy subjects (AUC = 81.37%) and exhibited good diagnostic performance in distinguishing active pulmonary TB from latent pulmonary TB (AUC = 84.35%). The performance of miR-29a-3p present in blood in distinguishing active TB from healthy control groups suggested that it may be a useful biomarker for TB diagnosis. The presence of this miRNA in readily obtainable blood (plasma) samples suggests its potential utility in diagnosing TB, particularly in children and cases of extrapulmonary TB, as compared to sputum-based methods. The findings also substantiated the aforementioned viewpoints. miR-29a exhibited a sensitivity of 82% and a specificity of 81% for TB diagnosis in the adult population and a sensitivity of 74% and a specificity of 89% for pediatric TB diagnosis. Moreover, this study also indicated that miR-29a, sourced from various specimen types, demonstrated strong performance in effectively distinguishing TB cases from healthy control groups.
miRNA is a single-stranded RNA molecule typically spanning 18 to 25 nucleotides in length. Its primary function is to restrict the activity of target genes during the post-transcriptional phase of gene expression. miRNA is kind of RNA that does not encode proteins. Instead, its final transcripts engage with the mRNA of target genes. miRNA serves to regulate the translation of mRNA, along with other regulatory components, including transcription factors (40). The majority of miRNAs are deemed to be situated within genomic regions classified as non-coding. An estimated 2 to 5% of human genes across the entire genome are believed to encode miRNAs (41). In many cases, Multi-gene transcripts often contain the genetic code for miRNAs. It is estimated that miRNAs possess the ability to regulate more than one-third of human genes, primarily because a single miRNA can target multiple mRNA molecules (42). Furthermore, various studies have demonstrated differences in gene expression profiles of macrophages and NK cells among participants with active TB, latent TB, and healthy controls. As reported by Fu et al. (10) and Sharbati et al. (43), miR-29 was initially identified as an mRNA repressor with a specific target: the HIV-13’UTR region in viral infections. Notably, during systemic infections caused by Mycobacterium bovis, miR-29 exhibited upregulation in NK cells, leading to the inhibition of the target gene IFN-γ. The miR-29a-mediated pathway enhanced host resistance to Listeria infection. Conversely, miR-29a demonstrated upregulation in both serum and sputum samples acquired from individuals with active pulmonary TB in comparison to healthy control groups. During Mycobacterium avium infection in human macrophages, the expression of miR-29a was also induced. This induction led to the targeting of genes involved in the apoptotic pathway, specifically caspase 7 and 3. Consequently, miR-29a exerted control over alterations in the composition of immune cells and the gene expression of associated target genes under TB infection (44).
miR-29a is the miRNA related to Mycobacterium tuberculosis infection (45). Mycobacterium tuberculosis infection leads to overexpression of miR-29a in host cells. miR-29a suppresses immune responses to Mycobacterium tuberculosis by reducing IFN-γ levels. Apart from its targeting of the 3′ untranslated region of IFN-γ mRNA, miR-29a also facilitates the binding of IFN-γ mRNA to the protein Argonaute 2. This interaction results in the formation of an RNA-silencing complex, which then leads to the post-transcriptional reduction of IFN-γ expression (46). Furthermore, multiple studies have provided evidence that miR-29a also targets genes such as anti-apoptotic B-cell lymphoma 22, myeloid leukemia-1, GTP-bound cell division cycle 42, and the p85 kinase gene, underscoring its involvement in the regulation of apoptotic pathways. In the anti-TB response, overexpression of miR-29a in TB infection hinders macrophages from engulfing tubercle bacilli by suppressing IFN-γ and promoting apoptosis (10).
Despite some promising results, significant heterogeneity differences between studies were observed. Numerous factors encompassing sample type, sample storage and processing, miRNA profiling techniques, validation techniques, and data normalization procedures may bring about potential variations, potentially resulting in inconsistent findings. Because there was a scarcity of available studies for analysis, it was not feasible to conduct additional subgroup analyses on the basis of sample sources or profiling methods. Furthermore, variations in reporting, with some studies including-3p or -5p variants while others did not, may have contributed to a potential reduction in the precision of the analysis. An additional significant limitation pertained to the relatively small sample sizes in each of the included studies. In biomarker discovery studies, the cohort size assumes critical importance and should ideally be representative of the target population. Furthermore, most of the studies were from China, which might lead to the patient selection bias and might represent the clinical characteristics of Chinese patients with tuberculosis. In addition, most of the subjects in the study population were from China and India with the highest burden of TB. miR-29 sensitivity might be lower in low- countries such as European and American countries. As a screening marker for tuberculosis, the cut-off value of miR-29 is supposed to be reduced in these low-burden countries to increase its sensitivity. Moreover, it is essential to diagnose active tuberculosis and latent tuberculosis infection to control the spread of Mycobacterium tuberculosis (Mtb), a complex pathogen increasingly resistant to antibiotics. Unfortunately, due to the small amount of available literature concerning miR-29 diagnostic efficacy between latent pulmonary TB and active pulmonary TB, the authors were unable to confirm these findings with literature sources. Nonetheless, despite these acknowledged limitations, a comprehensive pooled analysis of available data from the miR-29 family was carried out to examine the involvement of miR-29 in the field of TB.
Progress in science and technology has opened avenues for the development of diverse innovative miRNA detection platforms. These include Bead Array-based profiling, microfluidic technology, miRNA activity reporting assays, and amplification assays (cycle-mediated isothermal amplification, exponential isothermal amplification, rolling circle amplification). These platforms hold the potential to facilitate the creation of rapid and cost-effective assays for the sensitive diagnosis of TB. Additionally, these platforms allow for the identification of candidate miRNAs that are poised to become the most promising in this context (47). Using the miR-29 family as a diagnostic reagent will aid in developing rapid, cost-effective assays for TB, avoiding the shortcomings of traditional diagnostic tests.
5 Conclusion
The meta-analysis conducted in this study has demonstrated that miR-29 exhibits the potential to distinguish active TB patients from healthy control groups. It has high performance in diagnosing both active TB and extrapulmonary TB across both pediatric and adult patient populations. Consequently, the most studied circulating miRNA, especially circulating miR-29a, may be a highly accurate diagnostic tool for tuberculosis. Nevertheless, it is imperative to underscore that further large-scale prospective studies are necessary to validate the findings of this investigation before considering the clinical implementation of miR-29 as a diagnostic marker.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JX: Methodology, Software, Writing – original draft. YH: Formal analysis, Investigation, Methodology, Software, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Priming Scientific Research Foundation for the Introduced Talents of The First Affiliated Hospital of Chengdu Medical College (CYFY-GQ59).
Acknowledgments
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript. Moreover, we thank Na Huang and Jiaqing Jiang for critical revision of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1384510/full#supplementary-material
Abbreviations
TB, Tuberculosis; QUADAS-2, Cochrane Collaboration’s Diagnostic Accuracy Studies Quality Assessment 2; SROC, summary receiver operating characteristic; AUC, area under the curve; DOR, diagnostic odds ratio; 95%CI, 95% confidence intervals; NA, Not Available.
Footnotes
References
1. Mahatha, AC, Mal, S, Majumder, D, Saha, S, Ghosh, A, Basu, J, et al. RegX3 activates whiB3 under acid stress and subverts lysosomal trafficking of Mycobacterium tuberculosis in a WhiB3-dependent manner. Front Microbiol. (2020) 11:572433. doi: 10.3389/fmicb.2020.572433
2. Sun, Z, Pang, X, Wang, X, and Zeng, H. Differential expression analysis of miRNAs in macrophage-derived exosomes in the tuberculosis-infected bone microenvironment. Front Microbiol. (2023) 14:1236012. doi: 10.3389/fmicb.2023.1236012
3. Angria, N, Massi, MN, Bukhari, A, Djaharuddin, I, Jumadi, O, Ahmad, A, et al. Expression of miRNA-29a-3p and IFN-γ as biomarkers in active and latent pulmonary tuberculosis. Ann Med Surg (Lond). (2022) 83:104786. doi: 10.1016/j.amsu.2022.104786
4. Ullah, H, Tian, Y, Arbab, S, Li, K, Khan, MIU, Rahman, SU, et al. Circulatory microRNAs in helminthiases: potent as diagnostics biomarker, its potential role and limitations. Front Vet Sci. (2022) 9:1018872. doi: 10.3389/fvets.2022.1018872
5. Mazurek, M, Litak, J, Kamieniak, P, Osuchowska, I, Maciejewski, R, Roliński, J, et al. Micro RNA molecules as modulators of treatment resistance, immune checkpoints controllers and sensitive biomarkers in glioblastoma Multiforme. Int J Mol Sci. (2020) 21:21. doi: 10.3390/ijms21041507
6. Pattnaik, B, Patnaik, N, Mittal, S, Mohan, A, Agrawal, A, Guleria, R, et al. Micro RNAs as potential biomarkers in tuberculosis: a systematic review. Noncoding RNA Res. (2022) 7:16–26. doi: 10.1016/j.ncrna.2021.12.005
7. Ulger, M, Serin, MS, Tezcan Ulger, S, Aslan, G, Ilvan, A, Tiftik, EN, et al. Possible relation between expression of circulating microRNA and plasma cytokine levels in cases of pulmonary tuberculosis. J Infect Dev Ctries. (2022) 16:1166–73. doi: 10.3855/jidc.15831
8. Olsson, O, Tesfaye, F, Søkilde, R, Mazurek, J, Abebe, M, Yeba, H, et al. Expression of MicroRNAs is dysregulated by HIV while Mycobacterium tuberculosis drives alterations of small nucleolar RNAs in HIV positive adults with active tuberculosis. Front Microbiol. (2021) 12:808250. doi: 10.3389/fmicb.2021.808250
9. Fu, Y, Yang, X, Chen, H, and Lu, Y. Diagnostic value of miR-145 and its regulatory role in macrophage immune response in tuberculosis. Genet Mol Biol. (2020) 43:e20190238. doi: 10.1590/1678-4685-gmb-2019-0238
10. Fu, Y, Yi, Z, Wu, X, Li, J, and Xu, F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. (2011) 49:4246–51. doi: 10.1128/JCM.05459-11
11. Zhou, M, Yu, G, Yang, X, Zhu, C, Zhang, Z, and Zhan, X. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol Med Rep. (2016) 13:4620–6. doi: 10.3892/mmr.2016.5097
12. Lu, Y, Wang, X, Dong, H, Wang, X, Yang, P, Han, L, et al. Bioinformatics analysis of microRNA expression between patients with and without latent tuberculosis infections. Exp Ther Med. (2019) 17:3977–88. doi: 10.3892/etm.2019.7424
13. Daniel, EA, Sathiyamani, B, Thiruvengadam, K, Vivekanandan, S, Vembuli, H, and Hanna, LE. MicroRNAs as diagnostic biomarkers for tuberculosis: a systematic review and meta- analysis. Front Immunol. (2022) 13:954396. doi: 10.3389/fimmu.2022.954396
14. Ma, F, Xu, S, Liu, X, Zhang, Q, Xu, X, Liu, M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. (2011) 12:861–9. doi: 10.1038/ni.2073
15. Yareta, J, Galarza, M, Capristano, S, Pellón, O, Sánchez, C, Ballon, J, et al. Differential expression of circulating micro-RNAs in patients with active and latent tuberculosis. Rev Peru Med Exp Salud Publica. (2020) 37:51–6. doi: 10.17843/rpmesp.2020.371.4468
16. Kathirvel, M, Saranya, S, and Mahadevan, S. Expression levels of candidate circulating microRNAs in pediatric tuberculosis. Pathog Glob Health. (2020) 114:262–70. doi: 10.1080/20477724.2020.1761140
17. Corral-Fernández, NE, Cortes-García, JD, Bruno, RS, Romano-Moreno, S, Medellín-Garibay, SE, Magaña-Aquino, M, et al. Analysis of transcription factors, microRNAs and cytokines involved in T lymphocyte differentiation in patients with tuberculosis after directly observed treatment short-course. Tuberculosis (Edinb). (2017) 105:1–8. doi: 10.1016/j.tube.2017.03.007
18. Moher, D, Liberati, A, Tetzlaff, J, and Altman, DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
19. Ouzzani, M, Hammady, H, Fedorowicz, Z, and Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst Rev. (2016) 5:210. doi: 10.1186/s13643-016-0384-4
20. Huang, QX, and Huang, XW. QUADAS-2 tool for quality assessment in diagnostic meta-analysis. Ann Palliat Med. (2022) 11:1844–5. doi: 10.21037/apm-22-204
21. Cai, QS, Chen, YY, and Xia, Q. Study on the expression and clinical significance of miRNA in peripheral blood of patients with pulmonary tuberculosis. Chinese J General Prac. (2016) 4:546–88. doi: 10.16766/j.cnki.issn.1674-4152.2016.04.010
22. Cao, SL, Shen, AP, and Zhu, QF. A study on the possibility of using circulating microRNAs as biomarkers for the diagnosis of tubercu losis. Chin J Microbiol Immunol. (2014) 10:787–92. doi: 10.3760/cma.j.issn.0254-5101.2014.10.012
23. Chen, XF, and Xu, WF. The expression and clinical significance of miR-29a in peripheral blood of patients with active pulmonary tuberculosis. Chin J Microecol. (2017) 12:1416–23. doi: 10.13381/j.cnki.cjm.201712012
24. Liu, YM, Li, DD, and Zhu, SY. Expression of miR-29a in peripheral blood leucocyte as biomarker of pulmonary tuberculosis and its diagnostic potential. J Prev Med Inf. (2015) 10:755–8.
25. Yan, BS, Wang, J, and Luo, J. The diagnostic value of miR-101, miR-223 and miR-424 as potential biomarkers on pulmonary tuberculosis. ChongQing Med. (2016) 14:1902–5. doi: 10.3969/j.issn.1671-8348.2016.14.010
26. Yi, ZJ, Fu, YR, and Li, RF. Expression of miR-29a in sputum of active patients with pulmonary tuberculosis. Chin J Microbiol Immunol. (2012) 4:333–4. doi: 10.3760/cma.j.issn.0254-5101.2012.04.011
27. Yin, H, Fan, ZC, and Huang, TH. Diagnostic value of mycobacterium tuberculosis DNA in epididymal nodule puncture specimens combined with serum miR-29a-3p and in the diagnosis of epididymal tuberculosis. Chinese J Human Sexuality. (2022) 11:5–8.
28. Zhan, J, Liu, SY, and Wu, TW. Changes of the expression of miR-29a and IFN-γ in peripheral blood patients with active pulmonary tuberculosis and latent tuberculosis. Chinese J Microcirculation. (2016) 4:24–7. doi: 10.3969/j.issn.1005-1740.2016.04.007
29. Zhang, GD, Ding, L, and Zhu, YB. Expression and significance of the plasma miR-29 family and interferon-gamma in serum and peripheral blood mononuclear cells of patients with active pulmonary tuberculosis. Chin J Diffic and Compl Cas. (2021) 11:1131–5.
30. Li, Z, Li, C, Bao, R, and Liu, Z. Expressions of miR-29a, TNF-alpha and vascular endothelial growth factor in peripheral blood of pulmonary tuberculosis patients and their clinical significance. Iran J Public Health. (2020) 49:1683–91. doi: 10.18502/ijph.v49i9.4085
31. Ndzi, EN, Nkenfou, CN, Mekue, LM, Zentilin, L, Tamgue, O, Pefura, EWY, et al. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis (Edinb). (2019) 114:69–76. doi: 10.1016/j.tube.2018.12.001
32. Pan, D, Pan, M, and Xu, YM. Mir-29a expressions in peripheral blood mononuclear cell and cerebrospinal fluid: diagnostic value in patients with pediatric tuberculous meningitis. Brain Res Bull. (2017) 130:231–5. doi: 10.1016/j.brainresbull.2017.01.013
33. Pedersen, JL, Barry, SE, Bokil, NJ, Ellis, M, Yang, Y, Guan, G, et al. High sensitivity and specificity of a 5-analyte protein and microRNA biosignature for identification of active tuberculosis. Clin Transl Immunol. (2021) 10:e1298. doi: 10.1002/cti2.1298
34. Wagh, V, Urhekar, A, and Modi, D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis (Edinb). (2017) 102:24–30. doi: 10.1016/j.tube.2016.10.007
35. Zhang, X, Guo, J, Fan, S, Li, Y, Wei, L, Yang, X, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One. (2013) 8:e81076. doi: 10.1371/journal.pone.0081076
36. Yao, S, Liu, B, Hu, X, Tan, Y, Liu, K, He, M, et al. Diagnostic value of microRNAs in active tuberculosis based on quantitative and enrichment analyses. Diagn Microbiol Infect Dis. (2024) 108:116172. doi: 10.1016/j.diagmicrobio.2024.116172
37. Akaishi, T, and Ishii, T. Coronavirus disease 2019 transmission and symptoms in young children during the severe acute respiratory syndrome coronavirus 2 Delta variant and omicron variant outbreaks. J Int Med Res. (2022) 50:3000605221102079. doi: 10.1177/03000605221102079
38. Gao, L, Xu, L, and Wang, X. A systematic review of tRNA-derived small non-coding RNAs as diagnostic and prognostic markers in Cancer. Technol Cancer Res Treat. (2022) 21:15330338211072989. doi: 10.1177/15330338211072989
39. Yi, ZJ, Fu, YR, Li, JH, and Zhang, B. Expression and bioinformatic analysis of miR-29 family, target gene IFN-γ in CD4(+) T cells from subjects with latent tuberculosis infection. Zhonghua Yu Fang Yi Xue Za Zhi. (2013) 47:632–6. doi: 10.3760/cma.j.issn.0253-9624.2013.07.012
40. Guo, X, Cheng, M, Ke, W, Wang, Y, and Ji, X. MicroRNA-214 suppresses propofol-induced neuroapoptosis through activation of phosphoinositide 3-kinase/protein kinase B signaling by targeting phosphatase and tensin homolog expression. Int J Mol Med. (2018) 42:2527–37. doi: 10.3892/ijmm.2018.3814
41. Persano, S, Guevara, ML, Wolfram, J, Blanco, E, Shen, H, Ferrari, M, et al. Label-free isothermal amplification assay for specific and highly sensitive colorimetric miRNA detection. ACS Omega. (2016) 1:448–55. doi: 10.1021/acsomega.6b00109
42. Lu, WJ, Liang, HB, Li, YF, Tu, XQ, He, JR, Ding, KQ, et al. MicroRNA-210-3p targets RGMA to enhance the Angiogenic functions of endothelial progenitor cells under hypoxic conditions. Front Cell Neurosci. (2019) 13:223. doi: 10.3389/fncel.2019.00223
43. Sharbati, J, Lewin, A, Kutz-Lohroff, B, Kamal, E, Einspanier, R, and Sharbati, S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. (2011) 6:e20258. doi: 10.1371/journal.pone.0020258
44. Bartel, DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/s0092-8674(04)00045-5
45. Javadi, A, Shamaei, M, Tabarsi, P, Ainy, E, and Kazemi, B. Extracellular vesicles from serum of mycobacteria patients accelerate expression of apoptosis miRNAs and facilitate THP-1 monocyte cell death. Tanaffos. (2022) 21:434–47. doi: 10.1186/s12890-022-01839-w
46. Sun, L, Li, H, Fu, Q, Hu, S, and Zhao, W. Significance of detecting the levels of miR-29a, survivin and interferon gamma release assay in patients with lung cancer and tuberculosis. Adv Clin Exp Med. (2022) 31:1073–80. doi: 10.17219/acem/150306
Keywords: tuberculosis, miR-29, diagnosis, biomarkers, meta-analysis
Citation: He J, Xiong J and Huang Y (2024) miR-29 as diagnostic biomarkers for tuberculosis: a systematic review and meta-analysis. Front. Public Health. 12:1384510. doi: 10.3389/fpubh.2024.1384510
Edited by:
Daniel Diaz, National Autonomous University of Mexico, MexicoReviewed by:
Dongdong Li, Sichuan University, ChinaPavel Eduardo Hernández Carreño, National Autonomous University of Mexico, Mexico
Copyright © 2024 He, Xiong and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie He, MTM1NDAyNDY5NzRAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Jie He
Jie He Juan Xiong
Juan Xiong Yuanyuan Huang
Yuanyuan Huang