- 1Department of Epidemiology, School of Public Health, Southern Medical University, Guangzhou, Guangdong, China
- 2Department of Emergency, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
- 3Dermatology Hospital of Southern Medical University, Guangzhou, China
- 4Chenzhou No.1 People's Hospital, Chenzhou, China
Introduction: Among the different antigens used in the detection of anti-Chlamydia trachomatis antibodies, significant differences in sensitivity and specificity have been observed. Further evaluation of C. trachomatis antigens in antibody detection is urgently needed for the development and application of C. trachomatis serologic assays.
Methods: Chlamydia trachomatis antigens Pgp3, TmeA, InaC, and HSP60 were selected and used in luciferase immunosorbent assay (LISA). The detection results obtained from well-defined C. trachomatis positive and negative samples were compared with the commercial C. trachomatis ELISA (Mikrogen) for performance evaluation.
Results: Pgp3, TmeA, InaC, and HSP60-based LISA showed sensitivity of 92.8, 88.8, 90.4, and 94.4%, and specificity of 99.2, 99.2, 99.2, and 92%, respectively. ROC analysis indicated that Pgp3-based LISA showed similar performance to Mikrogen ELISA (AUC 0.986 vs. 0.993, p = 0.207). Furthermore, four C. trachomatis antigens achieved strong diagnostic efficiency, i.e., positive likelihood ratios [+LR] ≥ 10 in C. trachomatis-infected women and negative likelihood ratios [−LR] ≤ 0.1 in C. trachomatis negative low exposure risk children, but only Pgp3 and TmeA showed strong diagnostic value in general adults. In addition, Pgp3, TmeA, and InaC, but not HSP60, achieved high performance, i.e., both positive predictive value (PPV) and negative predictive value (NPV) ≥ 90.9%, and showed no significant cross-reactivity with anti-Chlamydiapneumoniae.
Conclusion: Three C. trachomatis species-specific antigens Pgp3, TmeA, and InaC show superior performance in the detection of anti-C. trachomatis antibody, indicating the potential to be used in developing C. trachomatis serologic tests.
1 Introduction
Chlamydia trachomatis, an obligate intracellular bacterium, widely spreads and can cause both urogenital and ocular infections (1). Chlamydia trachomatis can be classified into 19 genotypes according to the diversity of its OmpA gene sequences (2, 3). Genotypes D-K mainly cause genital infection, the most common bacterial sexually transmitted infection, with an annual 131 million new cases worldwide (4). Unfortunately, most genital chlamydial infections are asymptomatic and undetected, resulting in continued transmission and delayed or no treatment (5). Untreated C. trachomatis infection is able to cause more serious complications, especially pelvic inflammatory disease, ectopic pregnancy, and tubal factor infertility in women (1, 6, 7). Additionally, C. trachomatis infection may enhance human immunodeficiency virus (HIV) infection and help develop cervical cancer (8–10). However, screening and timely treatment of C. trachomatis infection are still an unmet goal. Currently, the diagnosis of C. trachomatis infection primarily relies on nucleic acid amplification tests (NAATs), which are resource and labor intensive, and difficult for routine diagnosis in resource-limiting settings (11). Furthermore, NAAT results mean current and active infection of C. trachomatis (12, 13), and cannot determine the prevalence of C. trachomatis exposure and disease burden unless serological assays are used in epidemiological or retrospective analyses (14, 15). Chlamydia trachomatis serology is also useful in understanding the natural history of C. trachomatis and providing support for the development and evaluation of C. trachomatis vaccines (1, 16).
However, a major issue for C. trachomatis serological detection is the cross-reactivity between C. trachomatis and other Chlamydia spp., especially Chlamydia pneumoniae. Microimmunofluorescence (MIF), which is the “gold standard” for serodiagnosis of chlamydial infection, is affected by the cross-reactivity of antibodies against C. trachomatis elementary body (EB) antigens with those against other Chlamydia species (17–19). The most commonly used C. trachomatis antigens, such as EBs, lipopolysaccharide (LPS), and major outer membrane protein (MOMP), also show high cross-reactivity in the simple enzyme-linked immunosorbent assay (ELISA) format due to genus-specific B cell epitopes (20–22). Although recombinant antigens of C. trachomatis-specific peptides can reduce cross-reactivity, the corresponding assays are usually not sensitive enough with a sensitivity from 45.7% (IgG pELISA plus medac assay) to 82.9% (Pgp3 double-antigen) in women, and from 40.0% (SeroCT) to 54.4% (Pgp3 double-antigen) in men when compared with NAAT (23–27). Although combined use of peptides of C. trachomatis-specific B cell epitopes could increase the sensitivity to 91.8%, testing IgG1 and IgG3 antibodies against 11 peptides is too complicated to apply in population screening (28). In general, the immunogenicity of C. trachomatis-specific peptides is much less than that of intact C. trachomatis antigens. Recently, several C. trachomatis species-specific antigens have been used in serological assays to improve their specificity. Among them, plasmid-encoded protein 3 (Pgp3) is a promising C. trachomatis species-specific and immunodominant antigen since it is highly conserved across C. trachomatis strains and rarely identified in C. pneumoniae (29–31). Pgp3-based ELISA methods show a sensitivity of 53.0–80.9% and a specificity of 80.0–98.0% (20, 23, 27, 32–39). In addition, translocated membrane-associated effector A (TmeA) encoded by CT694 gene and inclusion membrane protein for actin assembly (InaC) encoded by CT813 gene are species-specific antigens of C. trachomatis since there are no known homologous proteins in C. pneumoniae (40, 41). TmeA is an effector protein of the type III secretion system that facilitates C. trachomatis invasion (42–45). Antibody assays using TmeA as the antigen show a sensitivity of 30.8–91.0% and a specificity of 69.4–98.0% (41, 44). InaC is expressed on the membrane of C. trachomatis inclusion bodies and can polymerize the cytoskeleton of host cells to regulate C. trachomatis reproduction (46, 47). InaC-based ELISA also shows a sensitivity of 60.0% and a specificity of 100.0% (48). At present, an unresolved problem is the significant difference in detection sensitivity and specificity between the assays based on different C. trachomatis species-specific antigens due to multiple factors including lack of standardized evaluation and well-defined positive and negative controls, differences in assay format and antigens (1, 39).
We have previously reported a high-throughput luciferase immunosorbent assay (LISA) for the qualitative and semi-quantitative detection of antibodies, which is more convenient, straightforward and highly sensitive (49). In the current study, we introduced C. trachomatis species-specific and immunodominant antigens including Pgp3, TmeA, and InaC into our “in-house” LISA to evaluate its serodiagnositic value. In addition, heat shock protein 60 (HSP60), another antigen commonly used in C. trachomatis immunoassay, was selected to provide complementary information to better characterize the assays since it is one of the most conserved proteins in evolution. Previous studies indicated that anti-HSP60 antibody is associated with tubal factor infertility (TFI) (50, 51) although it has been found to have cross-reactivity with C. pneumoniae (32).
2 Materials and methods
2.1 Serum samples
A total of 450 serum samples collected from Chenzhou No.1 People’s Hospital, Guangzhou Dermatology Hospital and The Third Affiliated Hospital of Southern Medical University were used in this study. Group 1 contains 125 serum samples from C. trachomatis infected women who are C. trachomatis positive by NAAT, and 125 plasma specimens collected from healthy children aged 1–6 years old with low exposure risk to C. trachomatis and anti-C. trachomatis IgG negative. Samples of group 2 were collected from 200 general adults whose C. trachomatis infection was unknown. The characteristics of these subjects were shown in Supplementary Table S1. All the samples were stored at −80°C until they were processed. The study followed the ethical recommendations of the Declaration of Helsinki and obtained the informed consent of the participants and the parents or guardians of the children. Ethics Committee of Dermatology Hospital of Southern Medical University approved this study (GDDHLS-20181207).
Nucleic acid amplification test for screening C. trachomatis nucleic acid-positive women was performed by using cobas® 4800 CT/NG Amplification/Detection Kit (Roche Diagnostics, United States) in Guangzhou Dermatology Hospital or by using home-made reagents in Chenzhou No.1 People’s Hospital. The homemade NAAT assay used specific primers based on the highly conserved cryptic plasmid Pgp2 gene of C. trachomatis and included forward primer of 5′-TTCCCCTTGTAATTCGTTGC-3′ and reverse primer of 5′-TAGTAACTGCCACTTCATCA-3′. After DNA extraction from the patients’ cervical swabs by using QIAamp DNA Mini Kit (Qiagen, Germany), target nucleic acid was amplified by PCR. Briefly, 12.5 μL Taq Plus Master Mix (CoWin Biotechnology Co., Ltd., JiangSu, China), 0.5 μL of forward and reverse primers (10 μM), 9.5 μL of nuclease-free water, and 2 μL of target template were mixed. The reaction was run at 95°C for 5 min followed by 35 cycles of 95°C for 50 s, 55°C for 45 s, and 72°C for 45 s.
2.2 Chlamydia trachomatis antigen-based luciferase immunosorbent assay
Four C. trachomatis antigens Pgp3, TmeA, InaC, and HSP60 (Supplementary Table S2) were amplified and cloned into pNLF1-N luciferase expression vector (Promega, Madison, WI, United States) downstream of the Nluc luciferase gene as previously described (49). All materials and reactive used in this study were consistent with those of Wang. All recombinant plasmids were confirmed by DNA sequencing and transfected into HEK-293 T cells (ATCC CRL-3216). The expressed fusion proteins of Nluc-C. trachomatis in cell lysates were harvested for further confirmation using anti-luciferase antibody, and then stored at −80°C until use. Anti-C. trachomatis antibodies in sera were detected in LISA in which they were first captured by protein G-coated microtiter plate and detected by Nluc-C. trachomatis antigen lysate in the presence of luciferase substrate as described previously (49). Each sample was tested in triplicate and the relative light units (RLU) were calculated by dividing each sample’s average luciferase light units with the average LU of the control wells. The level of anti-C. trachomatis antibody was expressed as Log2RLU. The cut-off value of C. trachomatis LISA was determined according to the maximum value of Youden’s index obtained through the receiver operating characteristic (ROC) curve.
2.3 Commercial Chlamydia trachomatis/Chlamydia pneumoniae enzyme linked immunosorbent assay
A commercial recomWell C. trachomatis IgG ELISA kit that can detect antibodies against MOMP, translocated actin-recruiting phosphoprotein (TARP) and chlamydial protease-like activity factor (CPAF) was used to detect anti-C. trachomatis IgG in the sera. RecomWell Chlamydia pneumoniae IgG ELISA kit that detects antibodies against the outer membrane complex of elementary bodies and reticulate bodies of Chlamydia pneumoniae (COMC) was used for evaluating the specificity of C. trachomatis LISA. The ELISA kits were purchased from Mikrogen (Mikrogen GmbH, Neuried, Germany), and the cut-off values were determined according to the manufacturers’ instructions.
2.4 Determination of likelihood ratio and predictive value of Chlamydia trachomatis LISA
The positive likelihood ratio (+LR) and negative likelihood ratio (−LR) were calculated as sensitivity/(1 − specificity) and (1 − sensitivity)/specificity, respectively according to the sensitivity and specificity data of ROC curves. +LR ≥ 10 and −LR ≤ 0.1 indicate strong evidence to rule in or rule out the diagnosis, respectively (28). For different prevalence, positive and negative predictive values (PPV and NPV) were calculated as PPV = [sensitivity prevalence] [(sensitivity prevalence) + (1 − specificity) (1 − prevalence)] while NPV = [specificity (1 − prevalence)] [(1 − sensitivity) prevalence + specificity (1 − prevalence)] (52).
2.5 Statistical analysis
All data were analyzed using IBM SPSS 27.0 (SPSS Inc., Chicago, United States) and GraphPad Prism 8.0.2 (GraphPad Software, California, United States). Sensitivity, specificity and kappa coefficient were calculated by using two-by-two tables, and McNemar test were used to compare the difference of sensitivity and specificity between assays. The kappa coefficient was used to assess the agreement between different assays (53). Receiver operating characteristic (ROC) analysis was performed to determine the optimized cut-off values and calculate the area under ROC curves (AUCs). Delong test was used to compare ROC curves while Student’s t-tests were used to compare the average sensitivity between different assays. False positive rates were compared by Chi-square tests. p values ≤0.05 were considered statistically significant.
3 Results
3.1 Detection of anti-Chlamydia trachomatis IgG antibodies by ELISA and LISA
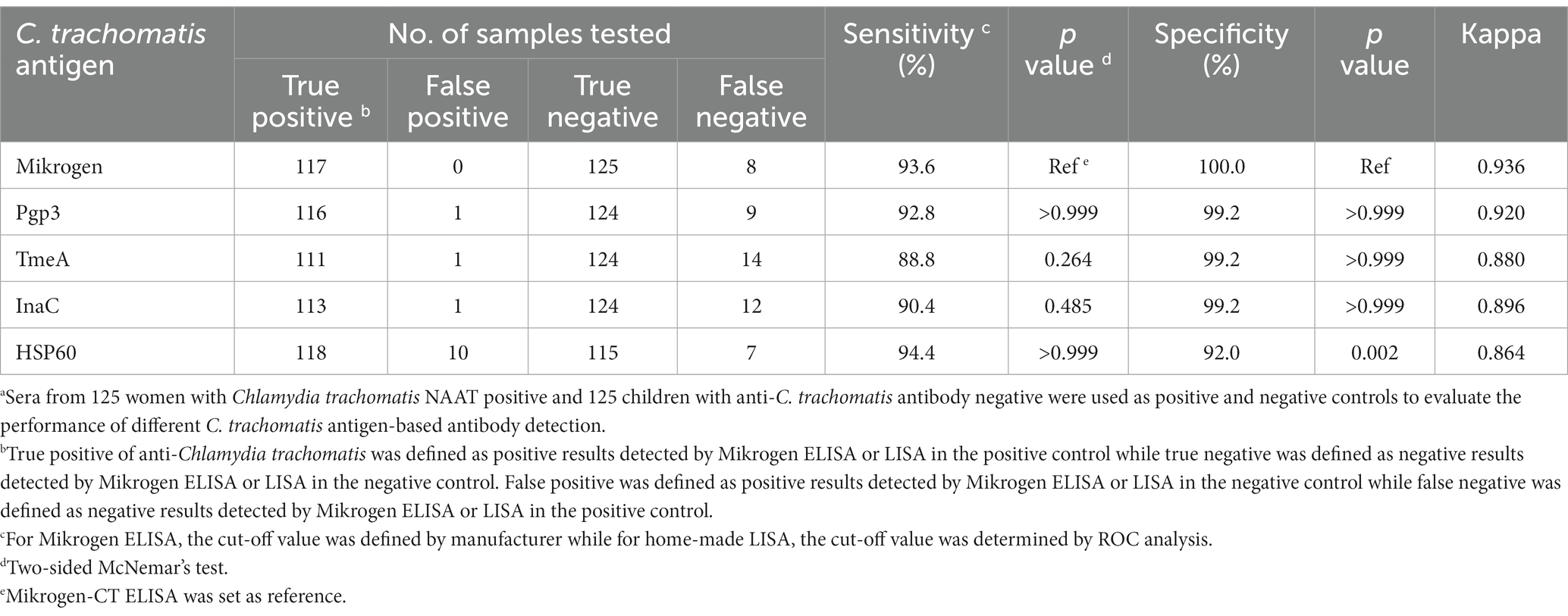
For the 125 serum samples from women with active C. trachomatis infection, 93.6% (117/125) were anti-C. trachomatis IgG positive by Mikrogen ELISA while none of the 125 children sera were positive. Then, ROC analysis was conducted to determine the optimal cut-off value for each C. trachomatis antigen-based LISA. Based on the cut-off values we selected, the detection sensitivity was 92.8, 88.8, 90.4, and 94.4% for Pgp3, TmeA, InaC, and HSP60-based LISA, respectively, and the corresponding specificities were 99.2, 99.2, 99.2, and 92.0%, respectively (Table 1). Pgp3, TmeA, and InaC-based LISA showed similar sensitivity and specificity to Mikrogen-CT ELISA whereas HSP60-based LISA showed significantly lower specificity (92.0%, p = 0.002). Pgp3-based LISA showed the best agreement with Mikrogen-CT ELISA (kappa = 0.920) among the four C. trachomatis antigens analyzed.
Of note, 5.6% (7/125) of the sera from actively C. trachomatis-infected women were anti-C. trachomatis antibody negative by both CT ELISA and LISA (Supplementary Figure S1), probably due to window period, i.e., NAAT positive but antibody negative during very early acute C. trachomatis infection. In addition, there were only one child’s serum positive by Pgp3-LISA, one child’s serum positive by TmeA-LISA, one child’s serum positive by InaC-LISA, and 10 children’s sera positive by HSP60-LISA, respectively. However, no children’s sera were positive for any two C. trachomatis antigen-LISAs simultaneously, suggesting a false positivity.
3.2 Performance of Chlamydia trachomatis antigen-based LISA
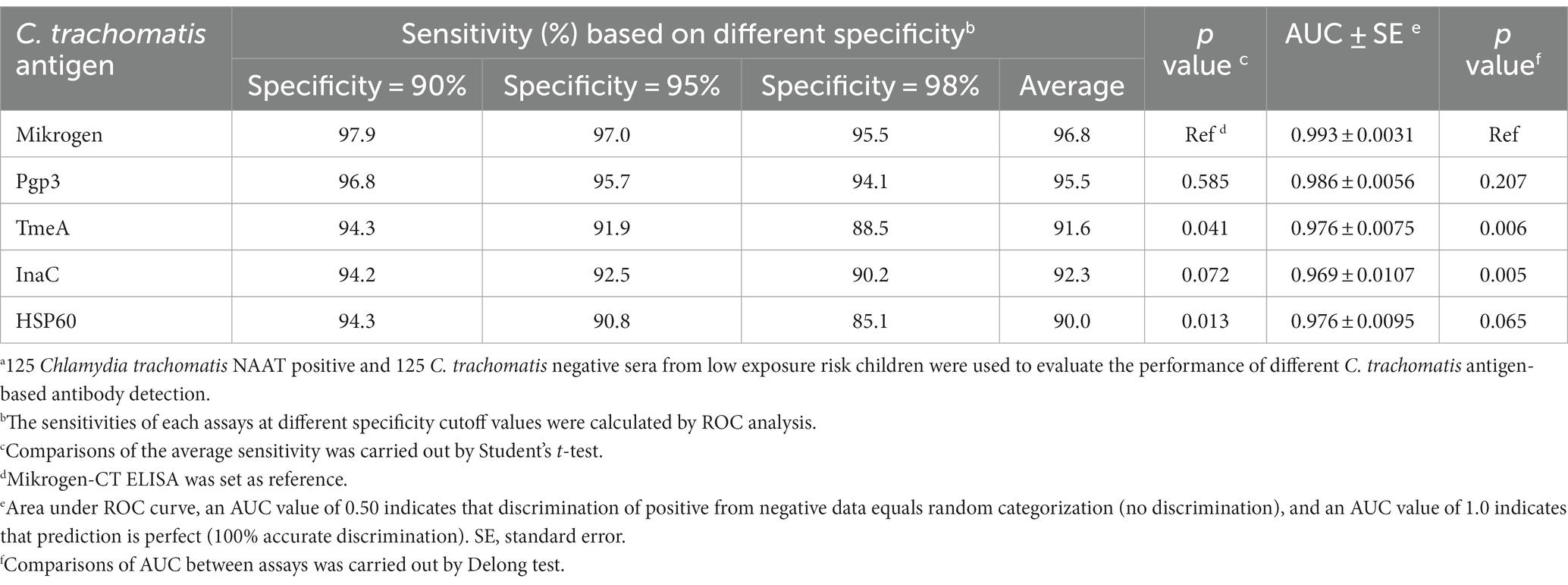
When the specificity was set as 90, 95, and 98%, Pgp3-based LISA achieved the highest sensitivity with an average sensitivity of 95.5%, followed by InaC (91.6%), TmeA (92.3%), and HSP60 (90.0%), respectively (Table 2). There was no significant difference for the detection sensitivity between Mikrogen ELISA and Pgp3 or InaC-based LISA (for Pgp3, 96.8 vs. 95.5%, p = 0.585; for InaC, 92.3 vs. 95.5%, p = 0.072). However, TmeA and HSP60-based LISA showed significantly lower average sensitivity (for TmeA, 91.6%, p = 0.041; for HSP60, 90.0%, p = 0.013). ROC-AUC analysis, a cutoff-independent global assay performance measurement, on the other hand, found no statistically significant difference in performance between Pgp3-based LISA and Mikrogen ELISA (AUC 0.986 vs. 0.993, p = 0.207, Table 2; Figure 1).
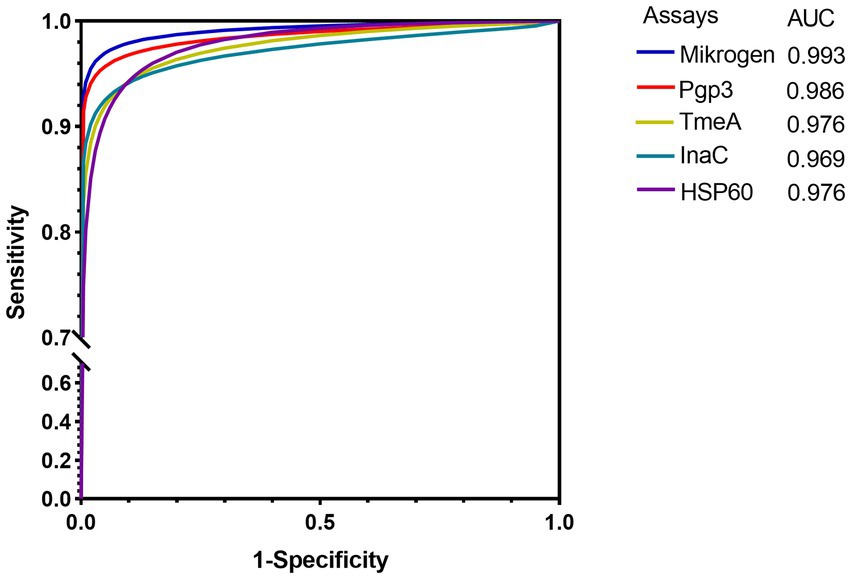
Figure 1. ROC curves of different Chlamydia trachomatis antigen-based LISA and commercial ELISA in detecting anti-C. trachomatis antibody. 125 C. trachomatis NAAT-positive and 125 C. trachomatis negative from low exposure risk children sera were used. The average Log2RLU signals of LISAs and the OD values of commercial ELISA were used as predictor variables. The solid color lines indicate the maximum likelihood-fitted ROC curves.
3.3 Diagnostic utility of Chlamydia trachomatis antigen-based LISA
When detecting anti-C. trachomatis antibody in active C. trachomatis infected women and anti-C. trachomatis negative low exposure risk children, at the specificity level of 91–99%, the corresponding sensitivity was 97.8–94.2% for Mikrogen ELISA and 96.6–92.8% for Pgp3-based LISA while the positive-likelihood ratios were ≥ 10 and the negative-likelihood ratios were ≤ 0.1 (Figure 2A; Supplementary Table S3). Similar results were obtained for TmeA, InaC, and HSP60-based LISA with the corresponding sensitivity of 93.9–91.1, 93.9–90.2, and 93.8–90.8%, respectively (Figure 2A; Supplementary Table S3), indicating strong diagnostic efficiency for both Mikrogen ELISA and C. trachomatis antigen-based LISA.
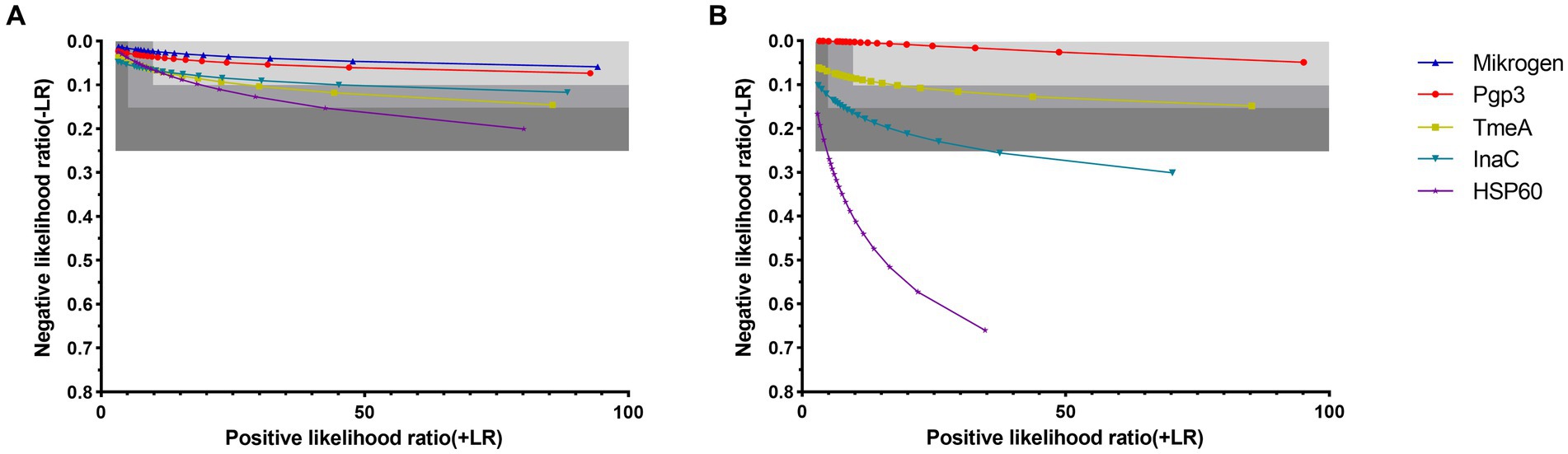
Figure 2. Diagnostic utility of anti-Chlamydia trachomatis antibody assays by likelihood ratios in control sera (A) and general adults (B). Sensitivities were calculated at specificities ranging from 70% (left) to 99% (right). Using sensitivity and specificity data of ROC curves which C. trachomatis exposure status was set as the gold standard for control sera and the determination of Mikrogen ELISA was set as the gold standard for general adults, positive-likelihood ratios (+LR) and negative-likelihood ratios (−LR) were calculated. The three-gray shaded areas indicate the zones of strong (+LR ≥ 10, −LR ≤ 0.1), moderate (+LR ≥ 5, −LR ≤ 0.15), and poor (+LR ≥ 2.5, −LR ≤ 0.25) performance of the assays, corresponding to the strong, moderate and poor diagnostic efficiencies.
When detecting anti-C. trachomatis antibody in general adults, Pgp3 and TmeA-based LISA showed strong diagnostic efficiency with specificity of 91–99 and 91–94%, respectively whereas InaC-based LISA showed only moderate diagnostic efficiency with specificity of 85–88% (Figure 2B; Supplementary Table S3). However, HSP60-based LISA showed poor diagnostic efficiency with +LR ≥ 2.5 and -LR ≤ 0.25, indicating a significantly lower diagnostic power in general adults.
3.4 Predictive value of Chlamydia trachomatis antigen-based LISA depends on the positive rate or prevalence of anti-Chlamydia trachomatis
Next, we adapted the predictive value model to compare and evaluate the diagnostic suitability of C. trachomatis antigen-based LISA at different levels of anti-C. trachomatis positivity or prevalence. All four C. trachomatis antigen-based LISAs and Mikrogen ELISA achieved high performance with both PPV and NPV ≥ 90.9% at the specificity level of 90–99%, in women with active C. trachomatis infection and anti-C. trachomatis negative low exposure risk children (Figures 3A–D). Furthermore, when the assay specificity increased, the optimal prevalence range changed. For example, the prevalence of anti-C. trachomatis that could be achieved by Mikrogen ELISA ranged from 51 to 81% at the specificity level of 90%, and from 10 to 63% at 99% specificity, indicating 23% increase of the optimal prevalence range when the specificity increased from 90 to 99% (Supplementary Table S4). At the specificity of 99%, the prevalence range of anti-C. trachomatis for the assays tested ranked as Mikrogen ELISA (10–63%) > Pgp3-based LISA (10–57%) > InaC-based LISA (11–46%) > TmeA-based LISA (11–40%) > HSP60-based LISA (12–33%) (Figure 3; Supplementary Table S4).
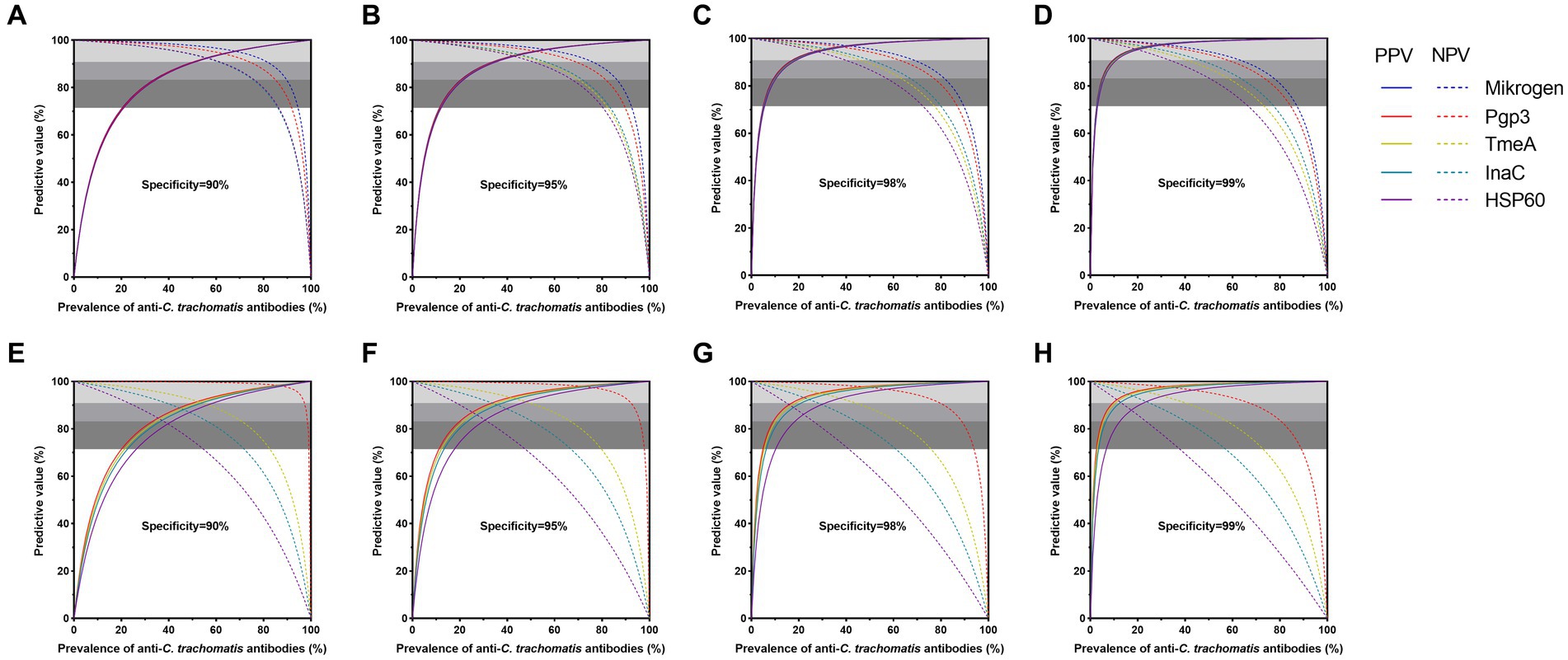
Figure 3. Diagnostic suitability of anti-Chlamydia trachomatis antibody assays by predictive values in control sera (A–D) and general adults (E–F). Positive and negative predictive values are dependent on the prevalence of populations of anti-C. trachomatis antibodies. Four specificity cut-off values and resultant sensitivities in ROC curve analysis were selected for panels (A,E; 90% specificity), (B,F; 95%), (C,G; 98%), and (D,H; 99%). The three-gray shaded areas indicate the zones of high (PPV and NPV ≥ 90.9%), moderate (PPV and NPV ≥ 83.3%), and poor (PPV and NPV ≥ 71.4%) diagnostic efficiency of assays.
Of note, further predictive value analysis of anti-C. trachomatis antibody detection assays was conducted in general adults to explore its feasibility in general population with relatively low prevalence of C. trachomatis infection. High performance was achieved for Pgp3-based LISA with a specificity of 90–99% while the specificity for TmeA-based LISA was 90–99% and InaC-based LISA was 98–99% at different positive rates or prevalence levels of anti-C. trachomatis antibody. However, HSP60-based LISA only performed moderately well (PPV and NPV ≥ 83.3%) at the specificity of 98–99% (Figures 3E–H; Supplementary Table S4). At the specificity level of 99%, Pgp3-based LISA showed the largest prevalence range of 10–67%, followed by TmeA-based LISA (11–40%) and InaC-based LISA (13–24%, Figures 3E–H; Supplementary Table S4).
3.5 Cross-reactivity of Chlamydia trachomatis antigen-based LISA with anti-Chlamydia pneumoniae ELISA
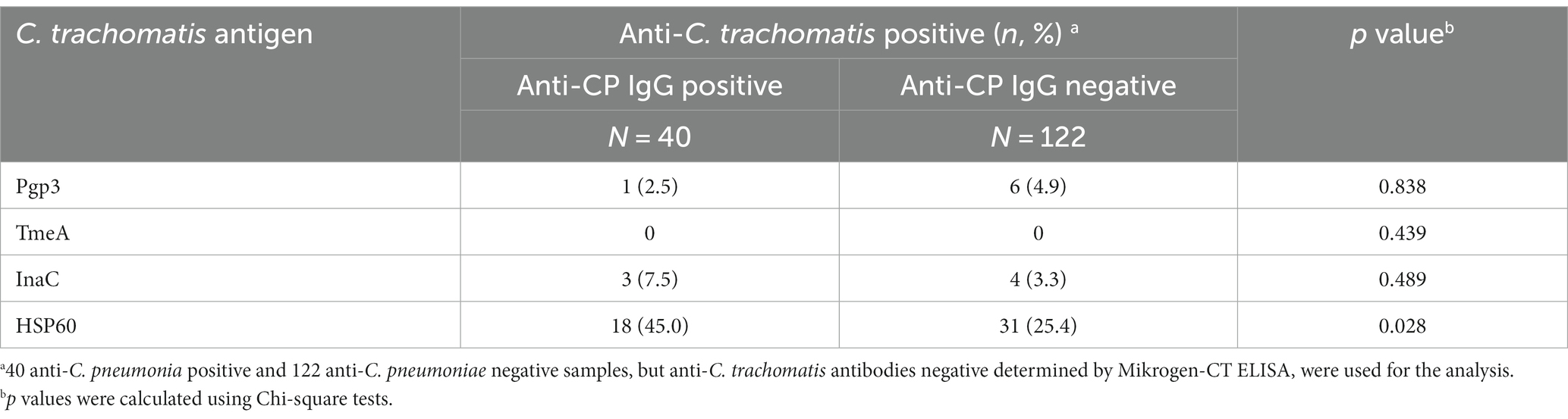
Of the 162 anti-C. trachomatis negative sera from general adults, 40 (24.7%) were positive for anti-C. pneumoniae antibody (Table 3). HSP60-based LISA showed high false positivity of anti-C. trachomatis antibody when testing anti-C. pneumoniae positive samples (p = 0.028; Table 3). No cross-reactivity with anti-C. pneumoniae positive samples were observed for Pgp3, TmeA, and InaC-based LISA (p > 0.4; Table 3), demonstrating the high specificity of the three C. trachomatis species-specific antigens.
4 Discussion
In the present study, we comprehensively evaluated the performance of C. trachomatis antigen-based LISAs in sera from current C. trachomatis-infected women, anti-C. trachomatis negative children with low-risk exposure to C. trachomatis, and general adults with unknown C. trachomatis infection status to analyze the diagnostic value of different C. trachomatis antigens including Pgp3, TmeA, InaC, and HSP60 in C. trachomatis serological testing. Our results demonstrated the great performance of all four C. trachomatis antigens in testing both C. trachomatis positive and negative sera while only Pgp3 and TmeA-based LISA performed well in detecting anti-C. trachomatis antibodies in general adults. Our data confirmed that C. trachomatis species specific antigen Pgp3 exhibited excellent diagnostic value in different populations according to its high sensitivity, specificity, PPV and NPV as well as the wide range of anti-C. trachomatis prevalence. Pgp3 has widely been used as a detecting antigen in serological assays including ELISA, multiplex bead array, and point-of-care testing, with sensitivity of 44.2–92.0% and specificity of 80.0–98.0% (20, 23, 27, 32–34, 39, 54, 55). When evaluating in a large population, a sensitivity of 82.9% and a specificity of 97.8% were reported for a double-antigen Pgp3 ELISA in 158 C. trachomatis-positive women and 494 C. trachomatis-negative pediatric sera (23). In our study, the Pgp3-based LISA showed a sensitivity of 92.8% and specificity of 99.2% in 125 C. trachomatis NAAT-positive women and 125 C. trachomatis negative children, indicating the potential and advantage of Pgp3 as a C. trachomatis species-specific antigen and immunodominant protein in the development of antibody detection assays including LISA.
Evaluation of anti-C. trachomatis serological assays relies on the selection of serum samples. In general, C. trachomatis NAAT-positive samples are commonly chosen as positive control in which more than 90% of NAAT-positive samples were positive for C. trachomatis-specific IgG antibodies (27, 28, 32). In the current research, we included 125 C. trachomatis NAAT-positive samples as positive controls and 125 anti-C. trachomatis antibody negative sera in children aged 1–6 years old as negative controls since children are considered at lower risk of exposure to C. trachomatis (27). In the current research, we included 125 C. trachomatis NAAT-positive samples as positive controls and 125 anti-C. trachomatis antibody negative sera in children aged 1–6 years old as negative controls. We used the commercial Mikrogen ELISA C. trachomatis kit instead of NAAT to screen negative children, since anti-C. trachomatis antibody can persist for many years when NAAT becomes negative (23) while anti-C. trachomatis antibody may be negative at the window period when NAAT becomes positive, and our study is mainly focused on the antibody detection of C. trachomatis rather than to determine the infection status of C. trachomatis. We found that 93.6% of NAAT positive sera determined positive for C. trachomatis IgG antibodies, which was consistent with previous study (56). Lack of anti-C. trachomatis specific antibody response has previously been reported in 5.5–10.2% of C. trachomatis-infected subjects (57–60) and can be explained by several possible reasons including early acute infection, delayed seroconversion, reduced infection duration, and antigenic burden caused by early treatment, or transient infection due to inadequate infection dose of C. trachomatis, or undiagnosed immunosuppressive diseases (58). Of note, of the 125 children sera, 0.8% were seropositive for by Pgp3, TmeA, or InaC-based LISA while 8.0% were positive by HSP60-based LISA. Wills et al. (27) also reported the C. trachomatis seropositive rate of 3.3% in pediatric sera by MIF, which may be due to the vertical transmission of C. trachomatis infection (61) or C. trachomatis eye disease (62). However, in our study, the positive results observed in children were more likely to be false positive as none was positive for any two C. trachomatis antigens simultaneously.
By using these well-defined samples, we found that TmeA-based LISA showed a sensitivity of 88.8% and a specificity of 99.2%, which is similar to earlier studies (41, 46). Wang et al. found that anti-TmeA antibody could be detected in approximately 80.8% (80/99) of C. trachomatis NAAT-positive sera. A TmeA-based multiplex bead assay showed a sensitivity of 90.9% and a specificity of 98.4% by using samples collected from 11 C. trachomatis PCR-positive children in Tanzania and 122 children from non-endemic areas in the United States. In addition, we found a sensitivity of 90.4% in our InaC-based LISA in this study, which is significantly higher than the sensitivity of 79% by the InaC-based ELISA in the study of Wang et al. (46), and 60% in the study of Gao et al. (48). The difference may be due to the relatively higher sensitivity of LISA than ELISA as observed in the previous study (49). The relative lower sensitivity and specificity of HSP60-based assays have also been reported (32).
Besides sensitivity, specificity, and AUC values, we also adopted likelihood ratio, predictive values, and range of antibody prevalence to evaluate the performance of C. trachomatis antigen-based LISAs. Likelihood ratio is a comprehensive index of both sensitivity and specificity whereas the predictive value can reflect the benefit gained from the actual application of diagnostic tests (63). The reliable and useful serological assays should be able to maintain a high predictive value across a wide range of antibody prevalence as well as a high positive likelihood ratio and a low negative likelihood ratio (28). Based on the likelihood ratios and predictive values we observed, the four C. trachomatis-antigen LISAs and the commercial Mikrogen ELISA showed moderate to strong diagnostic efficiency in detecting current C. trachomatis-infected women and children with low exposure risk to C. trachomatis. However, when detecting general adults, HSP60-based LISA showed poor diagnostic efficiency probably due to cross-reactivity with C. pneumoniae. Pgp3-based LISA, in contrast, maintained strong diagnostic efficiency even in general adults, suggesting its potential as a robust assay (28). TmeA-based LISA also achieved strong diagnostic efficiency with 91–94% specificity and 92.2–90.9% sensitivity while the antibody prevalence ranged from 36 to 54%, which is higher than the prevalence range of 20–30% in general population reported previously (23, 64–66), which suggests its feasibility in screening population with high prevalence of C. trachomatis infection (67).
The cross-reactivity between C. trachomatis and C. pneumoniae remains a basic problem in serological assays. However, except for HSP60, our results suggest that C. pneumoniae exposure had limited influence on the LISAs based on Pgp3, TmeA, and InaC, likely because these are C. trachomatis-specific antigens and have no equivalent orthologues in C. pneumoniae, which were consistent with previous studies (27, 40, 46). In contrast, HSP60, a highly-conserved protein that shares 89–95% identity with C. pneumoniae (28, 68, 69), showed strong cross-reactivity with C. pneumoniae. However, the cross-reactivity with C. pneumoniae cannot explain the high false positive rate of 25% observed in the C. pneumoniae negative group, suggesting that there may be other unrevealed factors, such as the cross-reactivity with human HSP60, which shares 48% identity with C. trachomatis (70, 71).
There are some limitations in our study. First, there is no recognized gold standard method for antibody detection of C. trachomatis. In our study, we only used one commercially available ELISA kit as a reference to evaluate the performance of 4 different C. trachomatis antigens in LISA. The results should be interpreted carefully. Second, the C. trachomatis NAAT positive sera were all from women, we did not evaluate the influence of gender on assay performance. Nevertheless, our previous longitudinal study on the seroepidemiology of C. trachomatis infection in the general population of China revealed no significant gender differences in terms of the prevalence of anti-C. trachomatis antibody (72). Third, the anti-C. pneumoniae antibody status was determined by only one commercial C. pneumoniae ELISA without additional assessment. Fourth, the serum IgG concentration may be different in adults and young children, which may affect the results of the analysis. Therefore, further studies are needed to assess the contribution of gender and C. pneumoniae to the performance of C. trachomatis LISA. In addition, the Pgp3 protein may highly conserve in plasmids of most other Chlamydia species that infect animals (73) and result in cross-reactive in the human sera that exposure to the Chlamydia from animals. However, the incidence of C. psittaci pneumonia may be very low. For example, a systematic review and meta-analysis showed that pneumonia caused by C. psittaci accounted for only 1% of community-acquired pneumonia cases (74). A multi-center observational study in China showed that, in a total of 4,545 patients with complicated or atypical pulmonary infection, the prevalence of C. psittaci was determined to be 2.1% (96/4545) by using metagenomic next generation sequencing, suggesting that prevalence of parrot fever remains low and sporadic in China (75). Finally, anti-Pgp3 may be absent in certain pgp3 plasmid-free C trachomatis strains (76, 77), which is a limitation of our manuscript. Fortunately, previous studies indicate that infection with plasmid-free C. trachomatis is rare in the general population (78, 79). We should still interpret the findings with caution.
In conclusion, we evaluated four C. trachomatis species-specific antigens Pgp3, TmeA, and InaC, and HSP60 in our LISA platform to detect anti-C. trachomatis and confirmed the great performance and diagnostic efficiency of Pgp3-based LISA. Our results indicate that Pgp3 can be used in C. trachomatis screening in both high-risk subjects and low-risk general population.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of Dermatology Hospital of Southern Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
YP: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. JS: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. CL: Investigation, Resources, Writing – review & editing. YL: Investigation, Resources, Writing – review & editing. HC: Investigation, Resources, Writing – review & editing. ST: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Bureau of Science and Information Technology of Guangzhou Municipality (Grant no. 201704020219) and the National Natural Science Foundation of China (Grant no. 31771007).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1333559/full#supplementary-material
References
1. Woodhall, SC, Gorwitz, RJ, Migchelsen, SJ, Gottlieb, SL, Horner, PJ, Geisler, WM, et al. Advancing the public health applications of Chlamydia trachomatis serology. Lancet Infect Dis. (2018) 18:e399–407. doi: 10.1016/s1473-3099(18)30159-2
2. Rawre, J, Juyal, D, and Dhawan, B. Molecular typing of Chlamydia trachomatis: an overview. Indian J Med Microbiol. (2017) 35:17–26. doi: 10.4103/ijmm.IJMM_16_341
3. Ward, ME. Chlamydial classification, development and structure. Br Med Bull. (1983) 39:109–15. doi: 10.1093/oxfordjournals.bmb.a071800
4. Newman, L, Rowley, J, Vander Hoorn, S, Wijesooriya, NS, Unemo, M, Low, N, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One. (2015) 10:e0143304. doi: 10.1371/journal.pone.0143304
5. Marcone, V, Recine, N, Gallinelli, C, Nicosia, R, Lichtner, M, Degener, AM, et al. Epidemiology of Chlamydia trachomatis endocervical infection in a previously unscreened population in Rome, Italy, 2000 to 2009. Euro Surveill. (2012) 17:20203. doi: 10.2807/ese.17.25.20203-en
6. Haggerty, CL, Gottlieb, SL, Taylor, BD, Low, N, Xu, F, and Ness, RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. (2010) 201:134–55. doi: 10.1086/652395
7. Manavi, K. A review on infection with Chlamydia trachomatis. Best Pract Res Clin Obstet Gynaecol. (2006) 20:941–51. doi: 10.1016/j.bpobgyn.2006.06.003
8. Anttila, T, Saikku, P, Koskela, P, Bloigu, A, Dillner, J, Ikäheimo, I, et al. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA. (2001) 285:47–51. doi: 10.1001/jama.285.1.47
9. Koskela, P, Anttila, T, Bjørge, T, Brunsvig, A, Dillner, J, Hakama, M, et al. Chlamydia trachomatis infection as a risk factor for invasive cervical cancer. Int J Cancer. (2000) 85:35–9. doi: 10.1002/(sici)1097-0215(20000101)85:1<35::aid-ijc6>3.0.co;2-a
10. Sangani, P, Rutherford, G, and Wilkinson, D. Population-based interventions for reducing sexually transmitted infections, including HIV infection. Cochrane Database Syst Rev. (2004) 2:Cd001220. doi: 10.1002/14651858.CD001220.pub2
11. Fine, D, Thomas, KK, Nakatsukasa-Ono, W, and Marrazzo, J. Chlamydia positivity in women screened in family planning clinics: racial/ethnic differences and trends in the northwest U.S., 1997-2006. Public Health Rep. (2012) 127:38–51. doi: 10.1177/003335491212700105
12. Jackson, LJ, Auguste, P, Low, N, and Roberts, TE. Valuing the health states associated with Chlamydia trachomatis infections and their sequelae: a systematic review of economic evaluations and primary studies. Value Health. (2014) 17:116–30. doi: 10.1016/j.jval.2013.10.005
13. Rahman, KS, and Kaltenboeck, B. Multipeptide assays for sensitive and differential detection of anti-Chlamydia Trachomatis antibodies. J Infect Dis. (2021) 224:S86–s95. doi: 10.1093/infdis/jiab016
14. Horner, P. Can Chlamydia serology be used to help inform a potential future Chlamydia vaccination strategy? Sex Transm Dis. (2017) 44:722–4. doi: 10.1097/olq.0000000000000732
15. Puolakkainen, M. Laboratory diagnosis of persistent human chlamydial infection. Front Cell Infect Microbiol. (2013) 3:99. doi: 10.3389/fcimb.2013.00099
16. Poston, TB, Gottlieb, SL, and Darville, T. Status of vaccine research and development of vaccines for Chlamydia trachomatis infection. Vaccine. (2019) 37:7289–94. doi: 10.1016/j.vaccine.2017.01.023
17. Kern, DG, Neill, MA, and Schachter, J. A seroepidemiologic study of Chlamydia pneumoniae in Rhode Island. Evidence of serologic cross-reactivity. Chest. (1993) 104:208–13. doi: 10.1378/chest.104.1.208
18. Ozanne, G, and Lefebvre, J. Specificity of the microimmunofluorescence assay for the serodiagnosis of Chlamydia pneumoniae infections. Can J Microbiol. (1992) 38:1185–9. doi: 10.1139/m92-194
19. Wong, YK, Sueur, JM, Fall, CH, Orfila, J, and Ward, ME. The species specificity of the microimmunofluorescence antibody test and comparisons with a time resolved fluoroscopic immunoassay for measuring IgG antibodies against Chlamydia pneumoniae. J Clin Pathol. (1999) 52:99–102. doi: 10.1136/jcp.52.2.99
20. Bas, S, Genevay, S, Schenkel, MC, and Vischer, TL. Importance of species-specific antigens in the serodiagnosis of Chlamydia trachomatis reactive arthritis. Rheumatology (Oxford). (2002) 41:1017–20. doi: 10.1093/rheumatology/41.9.1017
21. Campbell, LA, Kuo, CC, and Grayston, JT. Structural and antigenic analysis of Chlamydia pneumoniae. Infect Immun. (1990) 58:93–7. doi: 10.1128/iai.58.1.93-97.1990
22. Haralambieva, I, Iankov, I, Petrov, D, Ivanova, R, Kamarinchev, B, and Mitov, I. Cross-reaction between the genus-specific lipopolysaccharide antigen of Chlamydia spp. and the lipopolysaccharides of Porphyromonas gingivalis, Escherichia coli O119 and Salmonella Newington: implications for diagnosis. Diagn Microbiol Infect Dis. (2001) 41:99–106. doi: 10.1016/s0732-8893(01)00299-1
23. Horner, PJ, Wills, GS, Righarts, A, Vieira, S, Kounali, D, Samuel, D, et al. Chlamydia trachomatis Pgp3 antibody persists and correlates with self-reported infection and Behavioural risks in a blinded cohort study. PLoS One. (2016) 11:e0151497. doi: 10.1371/journal.pone.0151497
24. Land, JA, Gijsen, AP, Kessels, AG, Slobbe, ME, and Bruggeman, CA. Performance of five serological chlamydia antibody tests in subfertile women. Hum Reprod. (2003) 18:2621–7. doi: 10.1093/humrep/deg479
25. Morré, SA, Munk, C, Persson, K, Krüger-Kjaer, S, van Dijk, R, Meijer, CJ, et al. Comparison of three commercially available peptide-based immunoglobulin G (IgG) and IgA assays to microimmunofluorescence assay for detection of Chlamydia trachomatis antibodies. J Clin Microbiol. (2002) 40:584–7. doi: 10.1128/jcm.40.2.584-587.2002
26. Mygind, P, Christiansen, G, Persson, K, and Birkelund, S. Detection of Chlamydia trachomatis-specific antibodies in human sera by recombinant major outer-membrane protein polyantigens. J Med Microbiol. (2000) 49:457–65. doi: 10.1099/0022-1317-49-5-457
27. Wills, GS, Horner, PJ, Reynolds, R, Johnson, AM, Muir, DA, Brown, DW, et al. Pgp3 antibody enzyme-linked immunosorbent assay, a sensitive and specific assay for seroepidemiological analysis of Chlamydia trachomatis infection. Clin Vaccine Immunol. (2009) 16:835–43. doi: 10.1128/cvi.00021-09
28. Rahman, KS, Darville, T, Russell, AN, O'Connell, CM, Wiesenfeld, HC, Hillier, SL, et al. Comprehensive molecular serology of human Chlamydia trachomatis infections by peptide enzyme-linked immunosorbent assays. mSphere. (2018b) 3:e00253–18. doi: 10.1128/mSphere.00253-18
29. Comanducci, M, Cevenini, R, Moroni, A, Giuliani, MM, Ricci, S, Scarlato, V, et al. Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28 kDa antigen. J Gen Microbiol. (1993) 139:1083–92. doi: 10.1099/00221287-139-5-1083
30. Gwyn, S, Cooley, G, Goodhew, B, Kohlhoff, S, Banniettis, N, Wiegand, R, et al. Comparison of platforms for testing antibody responses against the Chlamydia trachomatis antigen Pgp3. Am J Trop Med Hyg. (2017) 97:1662–8. doi: 10.4269/ajtmh.17-0292
31. Kaur, H, Dize, L, Munoz, B, Gaydos, C, and West, SK. Evaluation of the reproducibility of a serological test for antibodies to Chlamydia trachomatis pgp3: a potential surveillance tool for trachoma programs. J Microbiol Methods. (2018) 147:56–8. doi: 10.1016/j.mimet.2018.02.017
32. Bas, S, Muzzin, P, Ninet, B, Bornand, JE, Scieux, C, and Vischer, TL. Chlamydial serology: comparative diagnostic value of immunoblotting, microimmunofluorescence test, and immunoassays using different recombinant proteins as antigens. J Clin Microbiol. (2001a) 39:1368–77. doi: 10.1128/jcm.39.4.1368-1377.2001
33. Bas, S, Muzzin, P, and Vischer, TL. Chlamydia trachomatis serology: diagnostic value of outer membrane protein 2 compared with that of other antigens. J Clin Microbiol. (2001b) 39:4082–5. doi: 10.1128/jcm.39.11.4082-4085.2001
34. Comanducci, M, Manetti, R, Bini, L, Santucci, A, Pallini, V, Cevenini, R, et al. Humoral immune response to plasmid protein pgp3 in patients with Chlamydia trachomatis infection. Infect Immun. (1994) 62:5491–7. doi: 10.1128/iai.62.12.5491-5497.1994
35. Danavall, DC, Gwyn, S, Anyalechi, GE, Bowden, KE, Hong, J, Kirkcaldy, RD, et al. Assessment and utility of 2 Chlamydia trachomatis Pgp3 serological assays for seroprevalence studies among women in the United States. Diagn Microbiol Infect Dis. (2021) 101:115480. doi: 10.1016/j.diagmicrobio.2021.115480
36. Dize, L, Martin, D, Gwyn, S, Perin, J, Gaydos, C, and Trent, M. Comparison of three serological assays to measure antibody response to Chlamydia antigen Pgp3 in adolescent and young adults with pelvic inflammatory disease. Int J STD AIDS. (2018) 29:1324–9. doi: 10.1177/0956462418785244
37. Donati, M, Laroucau, K, Storni, E, Mazzeo, C, Magnino, S, Di Francesco, A, et al. Serological response to pgp3 protein in animal and human chlamydial infections. Vet Microbiol. (2009) 135:181–5. doi: 10.1016/j.vetmic.2008.09.037
38. Horner, PJ, Wills, GS, Reynolds, R, Johnson, AM, Muir, DA, Winston, A, et al. Effect of time since exposure to Chlamydia trachomatis on chlamydia antibody detection in women: a cross-sectional study. Sex Transm Infect. (2013) 89:398–403. doi: 10.1136/sextrans-2011-050386
39. Winstanley, CE, Ramsey, KH, Marsh, P, and Clarke, IN. Development and evaluation of an enzyme-linked immunosorbent assay for the detection of antibodies to a common urogenital derivative of Chlamydia trachomatis plasmid-encoded PGP3. J Immunol Methods. (2017) 445:23–30. doi: 10.1016/j.jim.2017.03.002
40. Chen, C, Chen, D, Sharma, J, Cheng, W, Zhong, Y, Liu, K, et al. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect Immun. (2006) 74:4826–40. doi: 10.1128/iai.00081-06
41. Goodhew, EB, Priest, JW, Moss, DM, Zhong, G, Munoz, B, Mkocha, H, et al. CT694 and pgp3 as serological tools for monitoring trachoma programs. PLoS Negl Trop Dis. (2012) 6:e1873. doi: 10.1371/journal.pntd.0001873
42. Bullock, HD, Hower, S, and Fields, KA. Domain analyses reveal that Chlamydia trachomatis CT694 protein belongs to the membrane-localized family of type III effector proteins. J Biol Chem. (2012) 287:28078–86. doi: 10.1074/jbc.M112.386904
43. CostaPinto, L, Olavarria, VG, Grassi, MF, Lyrio, LD, Oliveira, RP, Santana, IU, et al. Prevalence of Chlamydia trachomatis endocervical infection in systemic lupus erythematosus patients and evaluation of the risk for HPV-induced lesions. Rheumatol Int. (2013) 33:631–6. doi: 10.1007/s00296-012-2419-z
44. Frikha-Gargouri, O, Gdoura, R, Znazen, A, Gargouri, J, Rebai, A, and Hammami, A. Diagnostic value of an enzyme-linked immunosorbent assay using the recombinant CT694 species-specific protein of Chlamydia trachomatis. J Appl Microbiol. (2009) 107:1875–82. doi: 10.1111/j.1365-2672.2009.04365.x
45. Pereira, SF, Henriques, AO, Pinho, MG, de Lencastre, H, and Tomasz, A. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol. (2009) 72:895–904. doi: 10.1111/j.1365-2958.2009.06687.x
46. Wang, J, Zhang, Y, Lu, C, Lei, L, Yu, P, and Zhong, G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol. (2010) 185:1670–80. doi: 10.4049/jimmunol.1001240
47. Wesolowski, J, Weber, MM, Nawrotek, A, Dooley, CA, Calderon, M, St Croix, CM, et al. Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and Golgi complex positioning. MBio. (2017) 8:e02280–16. doi: 10.1128/mBio.02280-16
48. Gao, XB, Xiao, M, Wang, J, Liu, YJ, Liu, QZ, and Qi, ML. Optimization of candidate proteins for serological screening of Chlamydia trachomatis infection. Genet Mol Res. (2015) 14:12240–6. doi: 10.4238/2015.October.9.12
49. Wang, H, Cai, Q, Liang, Y, Shui, J, and Tang, S. A simple and high-throughput luciferase immunosorbent assay for both qualitative and semi-quantitative detection of anti-HIV-1 antibodies. Virus Res. (2019) 263:9–15. doi: 10.1016/j.virusres.2018.12.017
50. Hjelholt, A, Christiansen, G, Johannesson, TG, Ingerslev, HJ, and Birkelund, S. Tubal factor infertility is associated with antibodies against Chlamydia trachomatis heat shock protein 60 (HSP60) but not human HSP60. Hum Reprod. (2011) 26:2069–76. doi: 10.1093/humrep/der167
51. Peeling, RW, Kimani, J, Plummer, F, Maclean, I, Cheang, M, Bwayo, J, et al. Antibody to chlamydial hsp60 predicts an increased risk for chlamydial pelvic inflammatory disease. J Infect Dis. (1997) 175:1153–8. doi: 10.1086/516454
52. Maxim, LD, Niebo, R, and Utell, MJ. Screening tests: a review with examples. Inhal Toxicol. (2014) 26:811–28. doi: 10.3109/08958378.2014.955932
53. Landis, JR, and Koch, GG. The measurement of observer agreement for categorical data. Biometrics. (1977) 33:159–74. doi: 10.2307/2529310
54. Ghaem-Maghami, S, Ratti, G, Ghaem-Maghami, M, Comanducci, M, Hay, PE, Bailey, RL, et al. Mucosal and systemic immune responses to plasmid protein pgp3 in patients with genital and ocular Chlamydia trachomatis infection. Clin Exp Immunol. (2003) 132:436–42. doi: 10.1046/j.1365-2249.2003.02163.x
55. Gwyn, S, Mitchell, A, Dean, D, Mkocha, H, Handali, S, and Martin, DL. Lateral flow-based antibody testing for Chlamydia trachomatis. J Immunol Methods. (2016) 435:27–31. doi: 10.1016/j.jim.2016.05.008
56. van Ess, EF, Ouburg, S, Spaargaren, J, Land, JA, and Morré, SA. Performance of the multitarget Mikrogen Chlamydia trachomatis IgG ELISA in the prediction of tubal factor infertility (TFI) in subfertile women: comparison with the Medac MOMP IgG ELISA plus. Pathog Dis. (2017) 75:ftx067. doi: 10.1093/femspd/ftx067
57. Bakshi, RK, Gupta, K, Jordan, SJ, Brown, LT, Press, CG, Gorwitz, RJ, et al. Immunoglobulin-based investigation of spontaneous resolution of Chlamydia trachomatis infection. J Infect Dis. (2017) 215:1653–6. doi: 10.1093/infdis/jix194
58. Geisler, WM, Morrison, SG, Doemland, ML, Iqbal, SM, Su, J, Mancevski, A, et al. Immunoglobulin-specific responses to Chlamydia elementary bodies in individuals with and at risk for genital chlamydial infection. J Infect Dis. (2012) 206:1836–43. doi: 10.1093/infdis/jis621
59. Komoda, T. Kinetic study of antibodies (IgG, IgA) to Chlamydia trachomatis: importance of IgA antibody in screening test for C. trachomatis infection by peptide-based enzyme immunosorbent assay. Jpn J Infect Dis. (2007) 60:347–51. doi: 10.7883/yoken.JJID.2007.347
60. Wang, SP, and Grayston, JT. Human serology in Chlamydia trachomatis infection with microimmunofluorescence. J Infect Dis. (1974) 130:388–97. doi: 10.1093/infdis/130.4.388
61. Schachter, J, Grossman, M, Sweet, RL, Holt, J, Jordan, C, and Bishop, E. Prospective study of perinatal transmission of Chlamydia trachomatis. JAMA. (1986) 255:3374–7. doi: 10.1001/jama.1986.03370240044034
62. Derrick, T, Roberts, C, Last, AR, Burr, SE, and Holland, MJ. Trachoma and ocular chlamydial infection in the era of genomics. Mediat Inflamm. (2015) 2015:791847. doi: 10.1155/2015/791847
63. van Ess, EF, Ouburg, S, Land, JA, and Morré, SA. Comparison of the Mikrogen multi-target ELISA with the Mikrogen recomLine immunoblot for the detection of Chlamydia trachomatis IgG antibodies in serum in infertile women. J Microbiol Methods. (2018) 150:5–8. doi: 10.1016/j.mimet.2018.05.006
64. Petersen, MR, Patel, EU, Grabowski, MK, Gaydos, CA, Quinn, TC, and Tobian, AAR. Seroprevalence of Chlamydia trachomatis among female adults in the United States: the National Health and nutrition examination surveys. Clin Infect Dis. (2021) 73:e629–37. doi: 10.1093/cid/ciaa1879
65. van Aar, F, de Moraes, M, Morré, SA, van Bergen, JE, van der Klis, FR, Land, JA, et al. Chlamydia trachomatis IgG seroprevalence in the general population of the Netherlands in 1996 and in 2007: differential changes by gender and age. Sex Transm Infect. (2014) 90:434–40. doi: 10.1136/sextrans-2013-051074
66. Woodhall, SC, Wills, GS, Horner, PJ, Craig, R, Mindell, JS, Murphy, G, et al. Chlamydia trachomatis Pgp3 antibody population Seroprevalence before and during an era of widespread opportunistic Chlamydia screening in England (1994-2012). PLoS One. (2017) 12:e0152810. doi: 10.1371/journal.pone.0152810
67. Parish, WL, Laumann, EO, Cohen, MS, Pan, S, Zheng, H, Hoffman, I, et al. Population-based study of chlamydial infection in China: a hidden epidemic. JAMA. (2003) 289:1265–73. doi: 10.1001/jama.289.10.1265
68. Kikuta, LC, Puolakkainen, M, Kuo, CC, and Campbell, LA. Isolation and sequence analysis of the Chlamydia pneumoniae GroE operon. Infect Immun. (1991) 59:4665–9. doi: 10.1128/iai.59.12.4665-4669.1991
69. Yuan, Y, Lyng, K, Zhang, YX, Rockey, DD, and Morrison, RP. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect Immun. (1992) 60:2288–96. doi: 10.1128/iai.60.6.2288-2296.1992
70. Rahman, KS, Darville, T, Russell, AN, O'Connell, CM, Wiesenfeld, HC, Hillier, SL, et al. Discovery of human-specific Immunodominant Chlamydia trachomatis B cell epitopes. mSphere. (2018a) 3:e00246–18. doi: 10.1128/mSphere.00246-18
71. Xiao, M, Wang, J, Liu, Q, and Qi, M. Detection of antibodies against immunodominant proteins of Chlamydia trachomatis in the sera of patients with urogenital Chlamydia trachomatis infection. Chin J Dermatol. (2013) 46:889–91. doi: 10.3760/cma.j.issn.0412-4030.2013.12.012
72. Shui, J, Xie, D, Zhao, J, Ao, C, Lin, H, Liang, Y, et al. Seroepidemiology of Chlamydia trachomatis infection in the general population of northern China: the Jidong community cohort study. Front Microbiol. (2021) 12:729016. doi: 10.3389/fmicb.2021.729016
73. Szabo, KV, O'Neill, CE, and Clarke, IN. Diversity in chlamydial plasmids. PLoS One. (2020) 15:e0233298. doi: 10.1371/journal.pone.0233298
74. Hogerwerf, L, Gier, DEB, Baan, B, and Van der Hoek, W. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. (2017) 145:3096–105. doi: 10.1017/s0950268817002060
75. Huang, W, Wang, F, Cai, Q, Xu, H, Hong, D, Wu, H, et al. Epidemiological and clinical characteristics of psittacosis among cases with complicated or atypical pulmonary infection using metagenomic next-generation sequencing: a multi-center observational study in China. Ann Clin Microbiol Antimicrob. (2023) 22:80. doi: 10.1186/s12941-023-00631-w
76. Escobedo-Guerra, MR, Katoku-Herrera, M, Lopez-Hurtado, M, Villagrana-Zesati, JR, de Haro-Cruz, MJ, and Guerra-Infante, FM. Identification of a new variant of Chlamydia trachomatis in Mexico. Enferm Infecc Microbiol Clin. (2019) 37:93–9. doi: 10.1016/j.eimc.2018.02.008
77. Yeow, TC, Wong, WF, Sabet, NS, Sulaiman, S, Shahhosseini, F, Tan, GM, et al. Prevalence of plasmid-bearing and plasmid-free Chlamydia trachomatis infection among women who visited obstetrics and gynecology clinics in Malaysia. BMC Microbiol. (2016) 16:45. doi: 10.1186/s12866-016-0671-1
78. Dahlberg, J, Hadad, R, Elfving, K, Larsson, I, Isaksson, J, Magnuson, A, et al. Ten years transmission of the new variant of Chlamydia trachomatis in Sweden: prevalence of infections and associated complications. Sex Transm Infect. (2018) 94:100–4. doi: 10.1136/sextrans-2016-052992
Keywords: Chlamydia trachomatis, luciferase immunosorbent assay (LISA), plasmid-encoded protein 3 (Pgp3), translocated membrane-associated effector A (TmeA), inclusion membrane protein for actin assembly (InaC), heat shock protein 60 (HSP60)
Citation: Pang Y, Shui J, Li C, Li Y, Chen H and Tang S (2024) The serodiagnositic value of Chlamydia trachomatis antigens in antibody detection using luciferase immunosorbent assay. Front. Public Health. 12:1333559. doi: 10.3389/fpubh.2024.1333559
Edited by:
Miguel Angel Sanchez-Aleman, National Institute of Public Health, MexicoReviewed by:
Heng Choon Cheong, University of Malaya, MalaysiaFernando Guerra-Infante, Instituto Nacional de Perinatología (INPER), Mexico
Copyright © 2024 Pang, Shui, Li, Li, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shixing Tang, dGFtZ3NoaXhpbmdAc211LmVkdS5jbg==
†These authors have contributed equally to this work
 Yulian Pang
Yulian Pang Jingwei Shui
Jingwei Shui Changchang Li3
Changchang Li3 Yongzhi Li
Yongzhi Li Hongliang Chen
Hongliang Chen Shixing Tang
Shixing Tang