- 1School of Environmental Science and Safety Engineering, Tianjin University of Technology, Tianjin, China
- 2Tianjin Fourth Central Hospital, Tianjin, China
- 3Tianjin Key Laboratory of Hazardous Waste Safety Disposal and Recycling Technology, Tianjin, China
- 4Institute of Disaster and Emergency Medicine, Tianjin University, Tianjin, China
- 5Tianjin Key Laboratory of Disaster Medicine Technology, Tianjin University, Tianjin, China
- 6Wenzhou Safety (Emergency) Institute, Tianjin University, Wenzhou, China
- 7Tianjin Institute of Medical and Pharmaceutical Sciences, Tianjin, China
- 8National Institute of Metrology, Beijing, China
- 9Hebei Research Center for Geoanalysis, Baoding, Hebei, China
Background: Phthalates (PAEs) are important synthetic substances in plastics, attracting much attention due to their potential effects on the cardiovascular system.
Methods: In this study, urine and blood samples from 39 individuals were collected in Tianjin, China. Phthalates and phthalate metabolites (mPAEs) were analyzed using gas chromatography-mass spectrometry (GC-MS) and high-performance liquid chromatography-mass spectrometry (HPLC-MS), respectively. The polymerase chain reaction (PCR) products from bisulfite-treated mitochondrial DNA (mtDNA) samples were analyzed using pyrosequencing technology.
Results: The detection frequencies for 9 PAEs varied from 2.56 to 92.31%, and those for 10 mPAEs varied from 30.77 to 100%. The estimated daily intakes (EDIs) and cumulative risk of PAEs were calculated based on the experimental statistics of urinary PAEs and mPAEs. For PAEs, the HIRfD (hazard index corresponding to reference doses) values of 10.26% of participants and the HITDI (hazard index corresponding to tolerable daily intake) values of 30.77% of participants were estimated to exceed 1, suggesting a relatively high exposure risk. The mtDNA methylation levels in the MT-ATP8 and MT-ND5 were observed to be lower than in the MT-ATP6. Mono-ethyl phthalate (MEP) and MT-ATP8 were positively correlated with triglyceride levels (p < 0.05). Based on the association of PAEs, mtDNA methylation, and triglycerides, the mediating role of mtDNA methylation between PAEs and cardiovascular diseases (CVDs) was analyzed in this study, but no mediated effect was observed.
Conclusion: The effects of PAE exposure on cardiovascular diseases (CVDs) should be investigated further.
1. Introduction
Phthalates (PAEs) are synthetic organic compounds that are widely used as additives in plastic products, such as food packing, personal care products, and medical devices (1). Since PAEs are not chemically bound to plastics, they are easily released into the environment (2). Therefore, PAEs have been widely detected in environmental matrices, such as soil, sediment, food, and air (3), and humans can be exposed to PAEs through various routes, including ingestion, inhalation, and dermal absorption from the environment (4, 5). Urine has been recognized as a suitable indicating matrix for exposure estimation due to rapid metabolization and short half-lives (<24 h) of PAEs in it (6, 7). Most recent studies on estimated daily intakes (EDIs) of PAEs focused on mPAEs (1, 5, 8), but one study detected the existence of PAEs in urine (9). Thus, the EDIs of PAEs may be underestimated.
The toxic effects of PAEs are causing increasing public concern, with several epidemiological studies revealing that PAEs can lead to oxidative stress (10–13) and cardiovascular diseases (CVDs) (14–17). Virani et al. reported that high levels of triglycerides, total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and lower levels of high-density lipoprotein cholesterol (HDL-C) are well-known CVD risk factors (18). In addition, Mínguez-Alarcón et al. found that certain PAE exposures were associated with lipid biomarkers (HDL-C, triglycerides, LDL-C, and TC) (19). However, studies about the relationships between PAE exposure and CVD were limited, and the potential biological mechanism remains under-recognized.
Increasing studies have confirmed that epigenetic patterns can be modified by some environmental exposures, and DNA methylation is the most studied and best-understood type of epigenetic modification (20, 21). A study has shown that DNA methylation is associated with benzene, dichlorodiphenyl trichloroethane, and polychlorinated biphenyls (22). Tian et al. (23) found that sperm LINE-1 DNA methylation levels were negatively correlated with bis(2-ethylhexyl) phthalate (DEHP) exposure, mediating the relationship between DEHP and sperm motility.
Over the years, methylation in mitochondrial DNA (mtDNA) has drawn attention. As mtDNA lacks a protector protein and effective repair mechanism, it is more vulnerable to reactive oxygen species (ROS) and mutations than nuclear DNA (24–26). An ENVIRONAGE birth cohort study suggested that mtDNA reacts to particulate matter that leads to ROS (27). mtDNA methylation could affect the normal function of mitochondria, causing mitochondrial damage and dysfunction, and has been recognized as a biomarker in many human diseases. Han et al. showed that mtDNA methylation may be a potential biomarker for predicting breast cancer risk (28). Stoccoro et al. (29) found mtDNA methylation differences in obese individuals and patients with colorectal cancer and CVD (30). Therefore, mtDNA methylation, as an epigenetic pattern, can be considered a potential mechanism of environmental exposures and CVDs.
Despite emerging interests and breakthroughs in mtDNA methylation in recent years, environmental exposures influencing CVDs through epigenetic mechanisms have yet to be thoroughly explored, and investigations on humans are still urgently needed. In this study, urine and blood samples from 39 participants were collected in Tianjin. The main objectives of this study are (1) the determination of PAEs and mPAEs in urine samples, (2) the investigation of the relationships of PAEs and mPAEs with lipid levels and mtDNA methylations, and (3) the assessment of the EDIs and potential health risks.
2. Materials and methods
2.1. Study population
In this study, we recruited 39 men (aged 27–43 years, referred by a physician) from Project ELEFANT (26, 31–35). At recruitment, all participants excluded autoimmune diseases, cardiovascular diseases, severe liver diseases, thyroid diseases, and blood diseases, and completed a physical examination under the guidance of nurses, signed an informed consent form, and filled in basic information (including age and BMI) listed in Table 1. This project complies with the Human Medical Ethic Review Law of the Ministry of Health and the Declaration of Helsinki. All procedures and study protocols were authorized by the ethics committee of Tianjin Medical University.
2.2. Measurement of urinary PAEs and mPAEs
In this study, we monitored 9 PAEs and 10 mPAEs, including di-methyl phthalate (DMP), di-ethyl phthalate (DEP), di-iso-butyl phthalate (DiBP), di-butyl phthalate (DBP), 1,2-benzenedicarboxylicacid (BMPP), di-pentyl phthalate (DPP), diethylhexyl phthalate (DEHP), di-n-octyl phthalate (DNOP), di-phenyl phthalate (DPHP), mono-methyl phthalate (MMP), mono-ethyl phthalate (MEP), mono-isobutyl phthalate (MiBP), mono-n-butyl phthalate (MBP), mono-ethyl phthalate (MEHP), mono-benzyl phthalate (MBzP), mono-octyl phthalate (MOP), mono-(2-ethyl-5-hydroxy-hexyl) phthalate (MEHHP), mono-(2-ethyl-5-oxo-hexyl) phthalate (MEOHP), and mono-(2-ethyl-5-carboxy-pentyl) phthalate (MECPP). To avoid the transfer of PAEs from PVC into urine samples, all materials were kept away from plastic vessels. Urine samples were collected, stored, and analyzed using glass tubes. Urine samples were gathered and stored at −20°C until analysis. To determine PAEs, 10 μl of internal standard (D4-DEHP, D4-DOP, and D4-DBP mixed solution, 10 μg/ml, Sigma) and 0.5 ml n-hexane was added to 0.5 ml of urine sample, and the sample mix solution was vortexed for 10 min. The sample mixture solution was centrifuged at 9,000 r/min for 10 min. The supernatant was collected and filtered through a 0.22-μm glass fiber. Finally, each sample was measured using gas chromatography-mass spectrometry (GC-MS, Thermo Fisher Scientific, Waltham, MA, USA) and calibrated and evaluated using the internal standard method. To determine mPAEs, 1.0 ml of urine sample was mixed with 0.25 ml ammonium acetate buffer solution (pH = 6.5, Sigma) and 10 μl β-glucuronidase standard solution (85,000 U/ml, Sigma), and placed in a 37°C water bath for 2 h. Then, the sample mix solution was filtered by a 0.22-μm glass fiber filter for further analysis.
2.3. mtDNA methylation analyses
After a medical examination, fasting blood samples were collected from 39 individuals by venipuncture using vacuum anticoagulant tubes with 2 ml EDTA, followed by centrifugation at 3,000 rpm for 10 min to separate plasma. Then 39 samples were stored at −80°C until mtDNA methylation analysis for platelets.
First, 300 μl of plasma was removed from each sample, and platelet mitochondria were extracted using a DNA methylation kit (Zymo Research, Orange, CA, USA) and treated with bisulfite. A 24-μl system was prepared from the bisulfite-treated DNA, and the DNA was amplified by polymerase chain reaction (PCR). Then the DNA was amplified 45 times under the conditions of 5 min at 95°C, 15 s at 95°C, 30 s at 52°C, 15 s at 72°C, and 5 min at 72°C. The PCR amplifications were conducted in a PCR amplifier (BIO-RAD, USA). The treated samples were run on 0.8% agarose gel and visualized under UV light. Finally, the rate of mtDNA methylation was tested by the PyroMark Q48 autoprep pyrophosphate sequencer (QIAGEN, Hilden, Germany) (26).
2.4. Instrumental analysis
Phthalates were determined by Agilent 8890-5977B GC-MS. The gas chromatography (GC) conditions were as follows: column, DB-5MS UI (30 m × 0.25 mm × 0.25 μm); carrier gas, helium. The injection volume was 1 mul splitless, with an injection port temperature of 250°C. The carrier gas was helium, with a flow rate of 1.0 ml/min (constant flow mode). The temperature program was as follows: the initial temperature was 80°C for 60 s, followed by 300°C (10°C/min) for 120 s, and then 340°C (5°C/min) for 60 s. MS conditions were as follows: scan mode: SIM, electron energy: 70 eV; interface temperature: 280°C; ion source temperature: 230°C.
High-performance liquid chromatography-mass spectrometry (HPLC-MS; Agilent 1290, USA) was used to detect mPAEs. HPLC Nexera LC-30A high-performance liquid chromatograph (Shimadzu). The parameters of HPLC were: chromatographic column: Kinetex XB-C18-100A LC column (100 × 3 mm, 2.6 μm; Phenomenex, USA); flow rate: 0.3 ml/min; injection volume: 10 μl; column temperature: 40°C. Mobile phase A contained an aqueous solution with 0.1% formic acid, and mobile phase B contained methanol (HPLC grade; Fisher Scientific, Houston, TX, USA). The mobile phase gradient program is listed in Supplementary Table 1.
mtDNA methylation was determined using a pyrosequencer (PyroMark Q48 Autoprep, QIAGEN, Hilden, Germany). The process was as follows: (1) 200 μl of enzyme-free water was added to the instrument's reagent clip for cleaning before sequencing each time, and the cleaning step was repeated once; (2) dNTP, denaturation buffer, enzyme mixture, mixed substrate, and annealing buffer were added to the instrument; (3) After the instrument was detected, 3 μl of magnetic beads (PyroMark Q48 Magnetic Beads) and 10 μl of the sample to be tested were added. The 48-well sample tray was added to the instrument, and then it was put on the instrument for mtDNA methylation sequencing; (4) The reagent clips were cleaned again after the test. mtDNA sequences used for targeted bisulfite sequencing are listed in Supplementary Table 2.
2.5. Quality assurance and quality control
Blank analysis was performed on each batch of samples. The recoveries of all the target analytes in the matrix-spiked samples were 85–112% (PAEs) and 89–115% (mPAEs). The relative standard deviations were below 10%. No obvious ionization restraint or enhancement was discovered, and the matrix effects of the target analyte were on the scale of 86–110% (PAEs) and 85–106% (mPAEs). The limit of quantification (LOQ) and limit of detection (LOD) were defined as 10 and three times the signal-to-noise ratio (S/N), respectively. The LOQ and LOD were estimated at 0.02–1.27 ng/ml and 0.006–0.4 ng/ml for PAEs, and 0.01–6.54 ng/ml and 0.003–2.1 ng/ml for mPAEs. Concentrations below the LOQ were specified as zero values for statistical analysis.
Strict QA/QC procedures were also carried out during the analysis of mtDNA methylation. Every equipment was sterilized, non-pyrogenic, and DNAse/RNAse-free during the analysis of mtDNA methylation, and then two quality controls were included. In each 96-well plate, two blank samples were set as negative controls in PCR. Two samples with known global DNA methylation levels, namely, low (1%) and high (99%), were added as positive controls in pyrophosphate sequencing. Finally, two duplicate pyrosequencing analyses were performed for each DNA sequence (26).
2.6. Statistical analysis
This study used R 4.1.0 software from Origin 2018 and SPSS software for data analysis. In this study, the concentrations of analytes below LOD were set to zero. Considering that PAE exposure may affect mtDNA methylation levels and lipid levels, we analyzed the relationships between PAE exposure, mtDNA methylation, and lipids by adjusting a generalized linear model of several covariates (including age, BMI, and seniority). Different PAEs and mPAEs were included in the generalized linear model together with age, BMI, and seniority covariates as independent variables. Since the levels of triglycerides and some mtDNA did not correspond to the normal distribution, the natural logarithm was used for statistical analysis (Shapiro-test P < 0.05). In addition, generalized linear models were used to analyze the relationships between mtDNA methylation, in vivo pollutants, and lipid levels after adjusting for age, BMI, and seniority. The p-value of < 0.05 was considered to indicate statistical significance and the p-value of < 0.01 was considered to indicate high significance.
2.7. Health calculations
The EDI (μg/kg bw/day) of PAE exposure was evaluated by the mPAEs values in urinary samples using the following equation (1, 5, 36):
Where UCm (ng/ml) is the concentration of urinary mPAE, UV (2.0 L/day) is the average adult volume of urine every day (37). Bw (kg) is the body weight of the participant. UCp (ng/ml) is the urinary PAE concentration. The EDI of DEHP is the sum of the calculated DEHP concentrations based on DEHP, MEHP, MEHHP, MECPP, and MEOHP (11). MWp is the molecular weight of PAE; MWm (g/mol) is the molecular weight of mPAE; and Fue is the molar fraction of mPAE. The values of MWp, MWm, and Fue are listed in Supplementary Table 3.
The hazard quotient (HQ) for exposure to PAEs was computed using equation (38):
EDI (μg/kg bw/day) is the estimated total daily intake of PAE; TDI (μg/kg bw/day) is recommended by the European Food Safety Authority (EFSA); the TDI values of DiBP and DEHP were 10 and 50 μg/kg bw/day, respectively. RfD (μg/kg bw/day) is recommended by the Environmental Protection Agency (EPA), the RfD values for DEP, DiBP, and DEHP were 800, 20, and 100 μg/kg bw/day. An HQ < 1 is considered safe, whereas an HQ > 1 indicates that the chemical may have potential adverse health effects on humans.
HI was estimated by summing the different values of HQ as follows:
3. Results
3.1. The level and relationship of PAEs and mPAEs
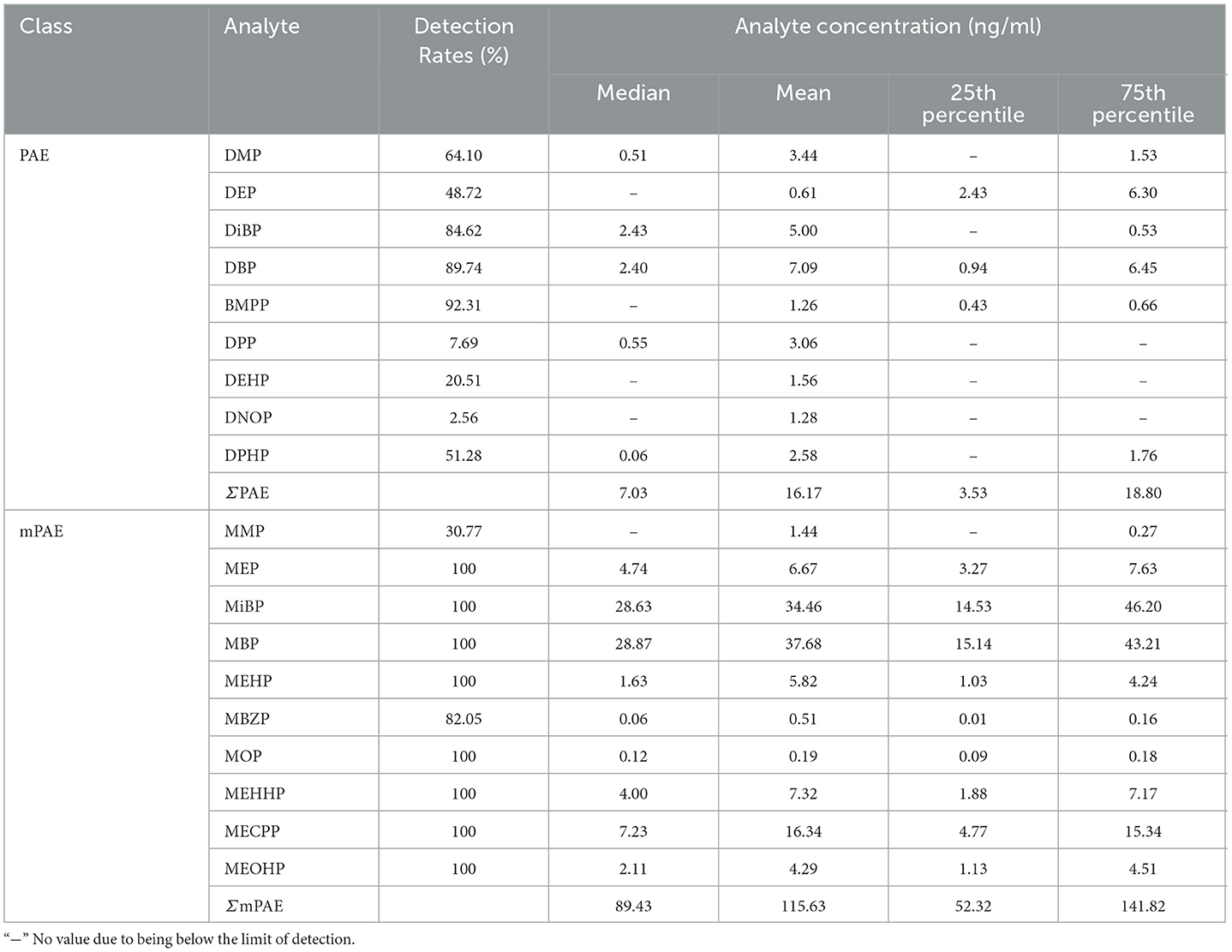
Out of 16 PAEs, nine were detected in urine samples in this study, including DMP, DEP, DiBP, DBP, BMPP, DPP, DEHP, DPHP, and DNOP. The detection rates ranged from 2.56 to 92.31%. The highest median concentration was DIBP (2.43 ng/ml), followed by DBP (2.40 ng/ml), DPP (0.55 ng/ml), DMP (0.51 ng/ml), and DPHP (0.06 ng/ml) as shown in Table 2.
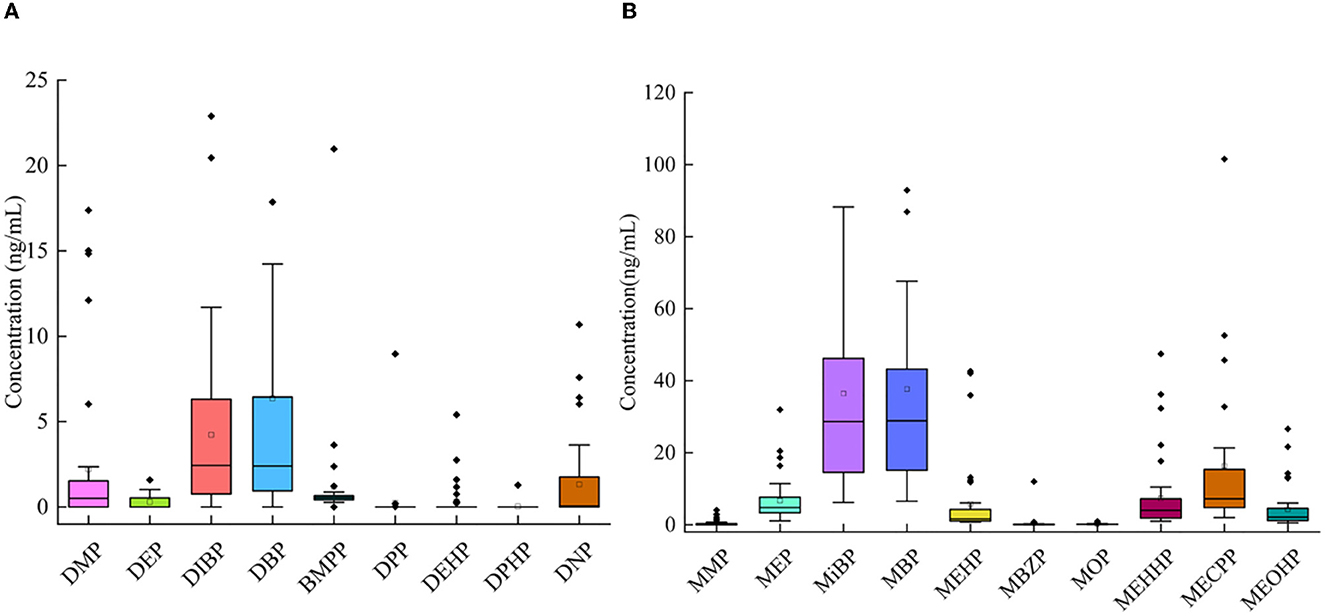
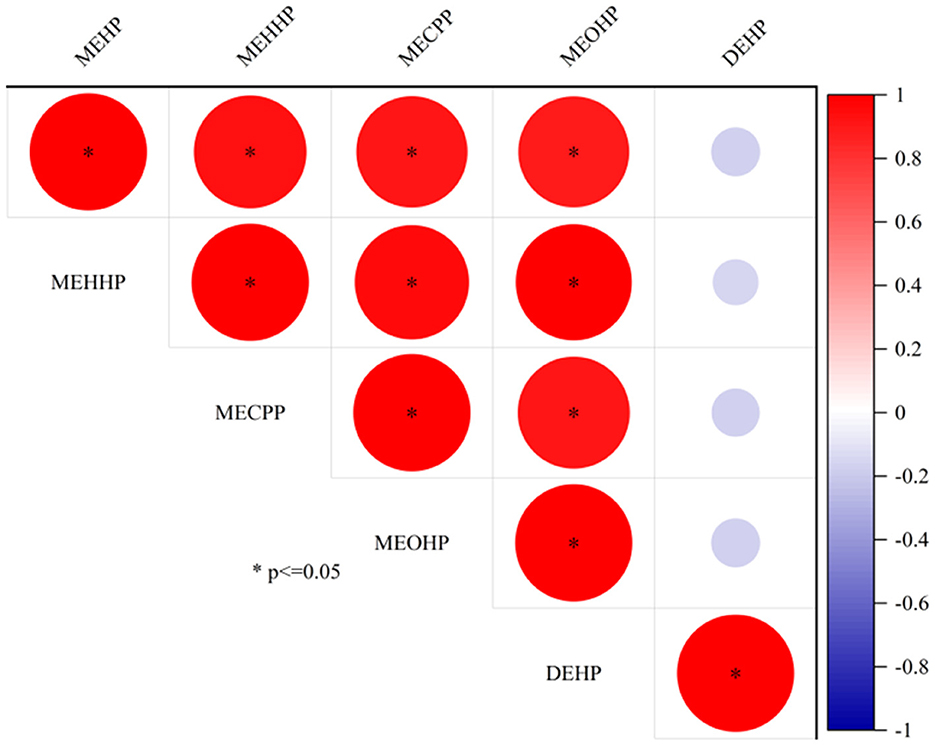
In the samples, 10 mPAEs, including MMP, MEP, MiBP, MBP, MBZP, and MOP, and four metabolites of DEHP (including MEHP, MEHHP, MECPP, and MEOHP) were detected. MiBP, MECPP, MEHHP, MEP, MEHP, MEOHP, MBP, and MOP were found in 100% of the samples, whereas MMP and MBZP were less frequently detected (30.77 and 82.05%, respectively). The highest median concentration was MBP (28.87 ng/ml), followed by MiBP (28.63 ng/ml), MECPP (7.23 ng/ml), MEP (4.74 ng/ml), MEHHP (4.00 ng/ml), MEOHP (2.11 ng/ml), MEHP (1.63 ng/ml), MOP (0.12 ng/ml), and MBZP (0.06 ng/ml). Figures 1A, B show that higher concentrations were found for MiBP and MBP, which correspond to the concentrations of their parent diesters (DIBP and DBP). The average contributions of nine PAEs (Σ9 PAEs) and 10 mPAEs (Σ10 mPAEs) were 12.3 and 87.7%, respectively. The results showed a significant positive correlation between the urinary metabolites of DEHP (MEHP, MEHHP, MEOHP, and MECPP) (p < 0.05), respectively (Figure 2).
3.2. The relationship between urinary concentrations of PAEs and mPAEs with lipid levels
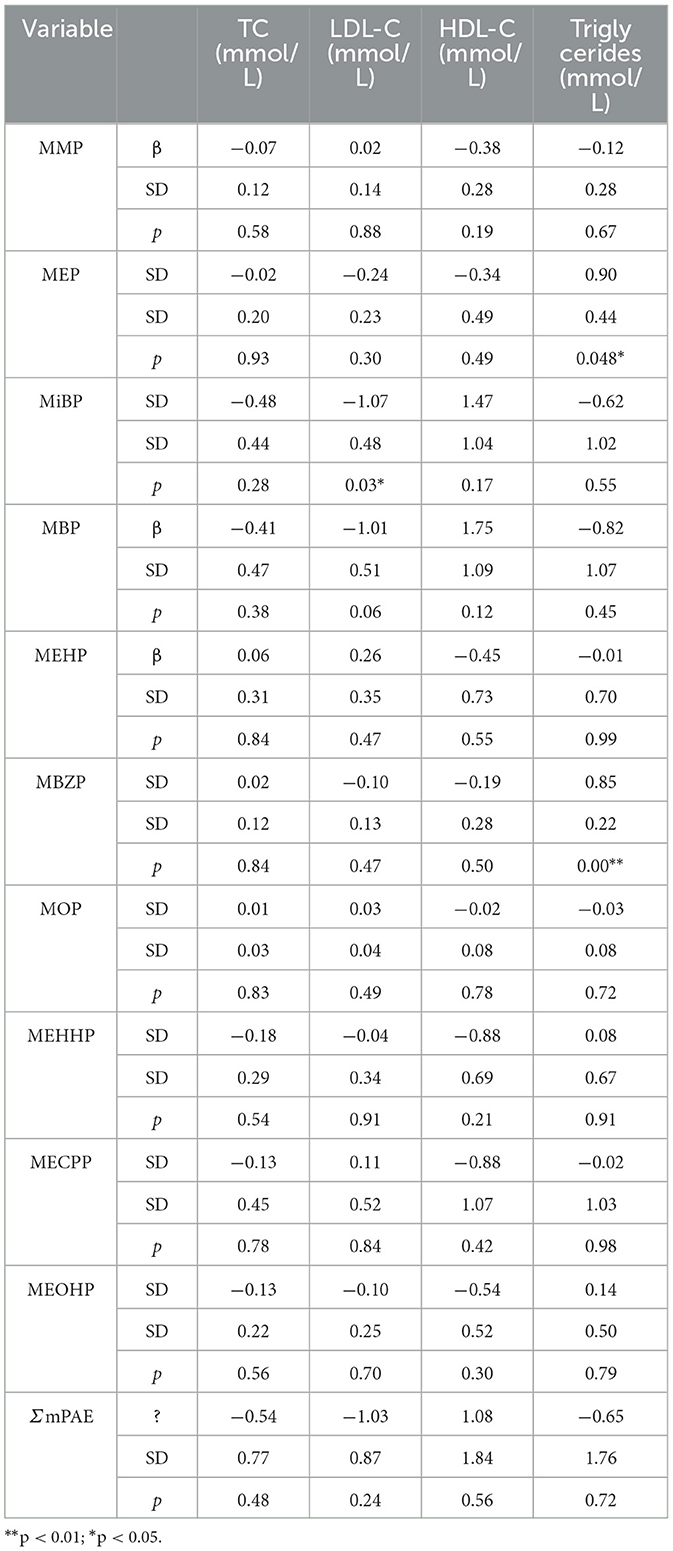
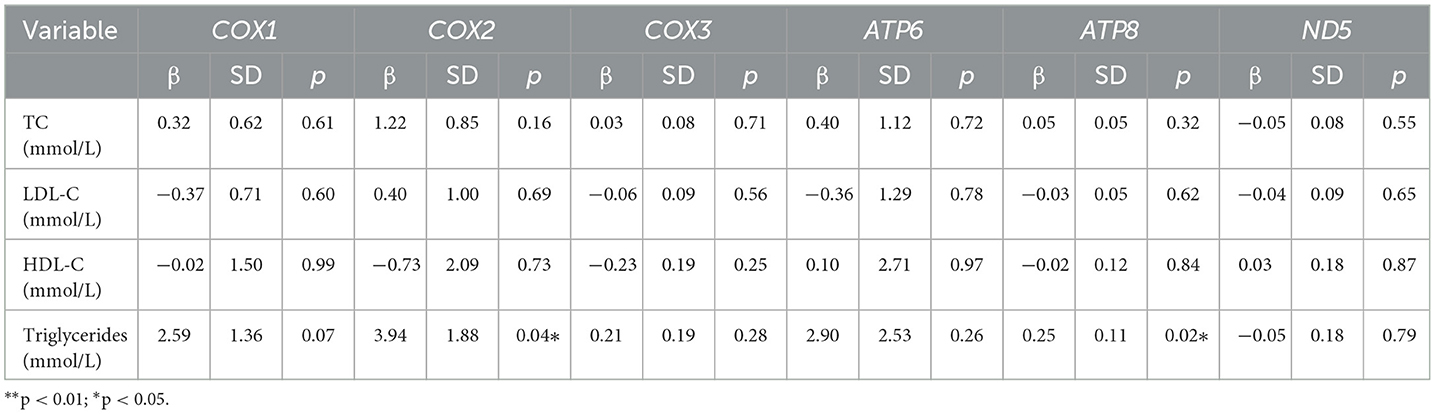
Using a generalized linear model, we tested associations between PAEs and mPAEs with lipid levels (including TC, triglycerides, HDL-C, and LDL-C). PAEs and lipid levels did not have a significant relationship (Supplementary Table 4). Both MEP and MBZP showed a positive influence on triglycerides (β = 0.90, p < 0.05; β f= 0.85, p < 0.001, respectively) (Table 3). Because a high level of triglycerides is one of the causes of CVDs (18), MEP and MBZP may increase the risk of CVDs.
3.3. The relationship between mtDNA methylation and lipid levels
For methylation analysis, six genes, including MT-COX1, MT-COX2, MT-COX3, MT-ATP6, MT-ATP8, and MT-ND5, were selected. Genetic mutations in cyclooxygenase (COX) genes are significantly associated with fatal metabolic disorders (39). ATPase subunit 6 (ATP6) and ATPase subunit 8 (ATP8) firsthand affect ATP synthesis and energy metabolism (40). NADH dehydrogenase subunit 5 (ND5) is the vital gene encoding NADH dehydrogenase. Hence, these genes mainly affect organs with high energy demands (41), such as the heart.
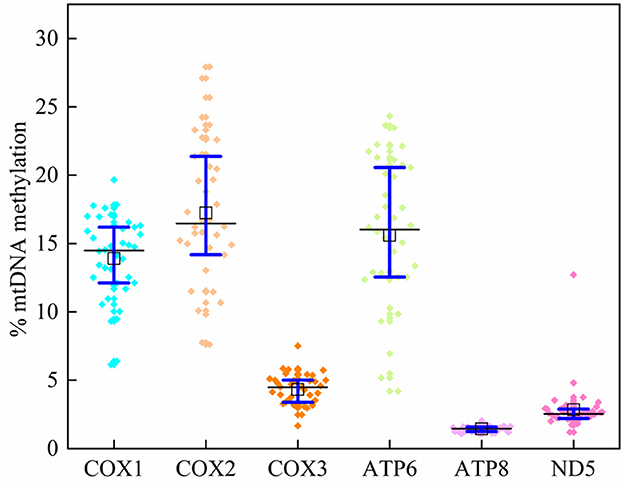
In 39 plasma samples, the mean methylation level [interquartile range (IQR) 25th−75th percentile] in the COX1 gene was 14.02 (12.23–16.17)%, 14.73 (11.12–17.35)% for COX2, 2.34 (2.12–2.62)% for COX3, 15.59 (12.55–20.45)% for ATP6, 1.43 (1.25–1.59)% for ATP8, and 2.84 (2.21–2.89)% for ND5 (Supplementary Table 5; Figure 3). The average methylation rates of MT-COX2 and MT-ATP8 were positively correlated with triglycerides (p < 0.05, respectively), indicating that the mtDNA methylation of specific genes could induce CVDs, which needs to be further explored (Table 4).
3.4. The relationships between PAEs and mPAEs concentrations with mtDNA methylation levels
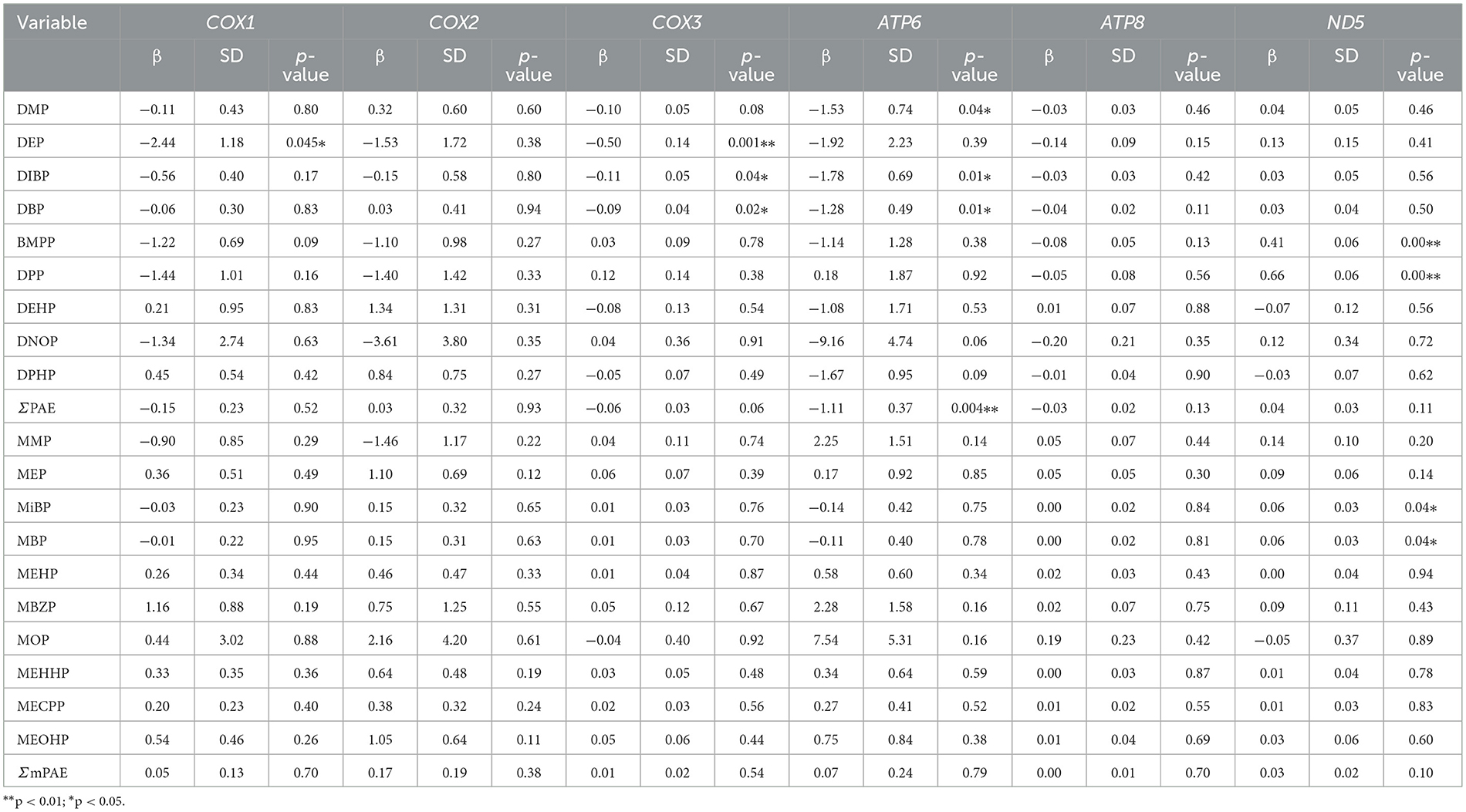
Relationships of PAEs and mPAEs concentrations with mtDNA methylation levels were analyzed by generalized linear models. A 1-ng/ml increment in DEP concentration was associated with a 2.44 and 0.50% decrease in MT-COX1 and MT-COX3 methylation rates (p < 0.05, p = 0.001, respectively) among all participants (Table 5). Each 1 ng/ml increase in DBP and DiBP concentrations was associated with a 1.28 and 1.78% decrease in the MT-ATP6 methylation rates (p < 0.05, respectively). The MT-ND5 methylation rate was positively associated with BMPP (β = 0.41, p < 0.05), DPP (β = 0.66, p < 0.05), MiBP (β = 0.06, p < 0.05), and MBP (β = 0.06, p < 0.05). These results suggested that the mtDNA methylation level was susceptible to changes in response to PAE exposure.
3.5. Mediation analysis on mtDNA methylation and lipid levels
The triglyceride levels were significantly associated with MEP concentration and MT-ATP8 methylation rate (p < 0.05, β = 0.90; p = 0.02, β = 0.25, respectively). To clarify the role of MEPs in the epigenetic regulation of the mitochondrial genome, we conducted a mediation analysis to examine the MT-ATP8 methylation rate with MEP and triglyceride levels (Table 6). However, we did not find the mediated effect of the above exposure levels on triglycerides through the mtDNA methylation pathway (p > 0.05).
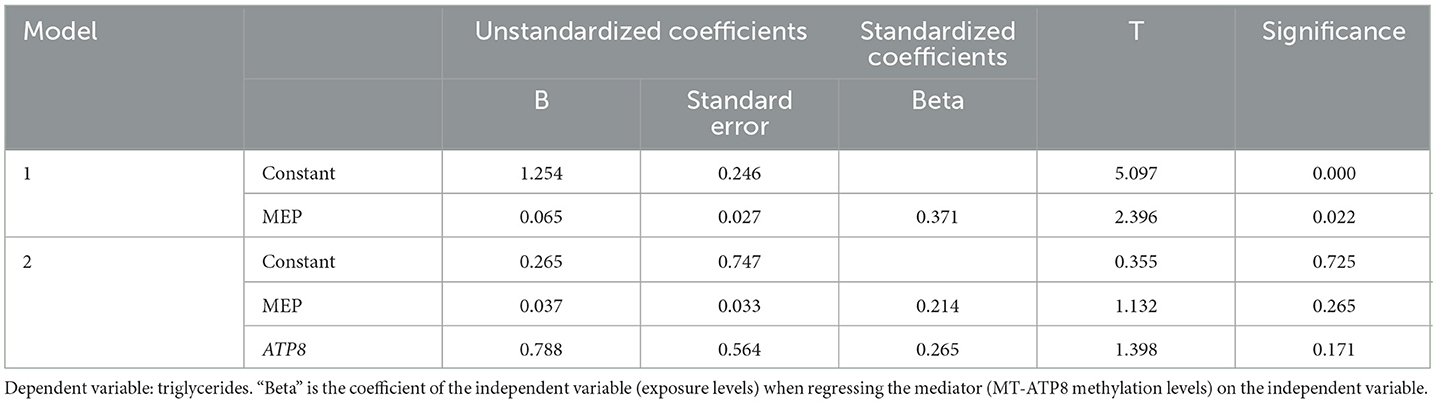
Table 6. The mediation effects of MT-ATP8 methylation on the association of MEP levels with triglycerides.
3.6. Human risk assessment
The EDIs, HQs, and HIs were calculated based on the levels of PAEs (DEP, DiBP, and DEHP). The median EDIs of DEP, DiBP, and DEHP were 0.438, 3.676, and 4.374 μg/kg bw/day, with details listed in Table 7. HQ showed a risk for a single PAE, while HI indicated a cumulative risk for several PAEs (calculated by the sum of DEP, DiBP, and DEHP). HQ<1 and HI<1 are considered significant daily intake doses and safe adverse effects caused by PAEs. In this study, 10% of participants showed HIRfD>1. Additionally, it should be noted that 12 of the 39 men in this study showed HITDI>1, indicating that ~31% of the population has potential health risks.
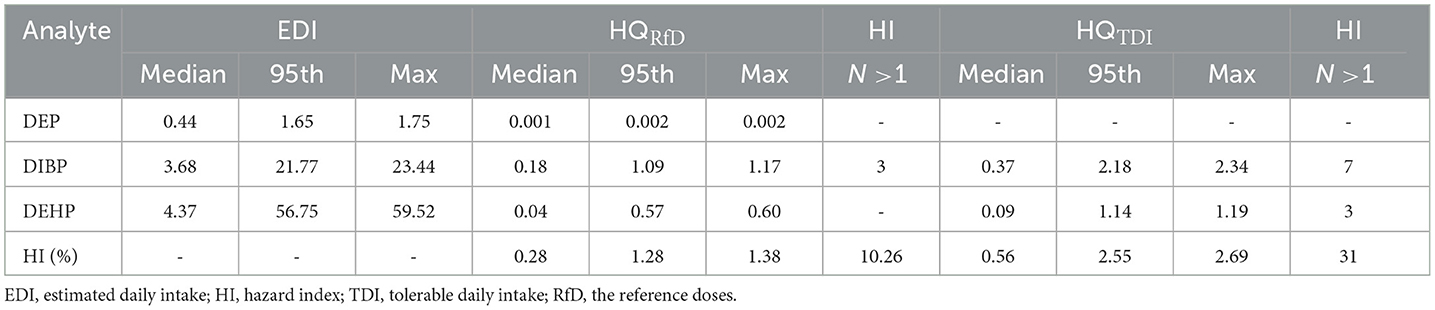
Table 7. Estimated daily intake (μg/kg bw/day), hazard quotient, and hazard index for 3 PAEs in 39 men.
4. Discussion
In this study, 9 PAEs and 10 mPAEs were detected in the urine of 39 men in Tianjin, with the average detection rate exceeding 30.77%. The detection rate in this study was similar to that of 88 male volunteers in Tianjin, China (31.8%) (42), but lower than that of 84 primiparas in Shenzhen, China (73%) (8) and 84 male adults in Fujian, China (64.3%) (43). In this study, the most widely detected were MBP, MiBP, and MEHP (100%, respectively), higher than the detection frequency of the general population in Guangzhou, China (84.2, 46.3, and 60.4%, respectively) (44), and higher than the prospective cohort of 2,298 children in Fujian, China (99.76, 97.23, and 54.52%, respectively) (45). This finding could be explained by the following factors: (1) PAE exposure characteristics could vary widely in different geographical areas. (2) Some factors, such as individual genetic susceptibility, could influence pollutant metabolism.
The primary metabolites of DEHP were MEHP, and the secondary metabolites were MECPP, MEHHP, and MEOHP. Strongly positive relationships were found between MEHP, MEHHP, MEOHP, MECPP, and DEHP (Figure 2), suggesting that these metabolites originated from the same exposure sources. We found that the median concentration of the three secondary metabolites of DEHP was 1.29–4.43 times higher than that of the primary metabolites, a result similar to that found in Canadian men (2.38–4.47 times) (46), which could be explained by the short half-life time of the MEHP.
Except for DEHP, the metabolism of other components is dominated by primary metabolism. MBP and MiBP were the most abundant compounds, with a median concentration of 28.87 and 28.63 ng/ml, respectively. The possible reason might be that their parent compounds (DBP and DiBP) are commonly used plasticizers. Compared to the results reported in the literature (Table 8), the concentrations of MBP in this study were much lower than that of primiparas in Shenzhen, China (median: 139 ng/ml), the general population in Guangzhou, China (median: 89.4 ng/ml), children in Fujian, China (median: 172.6 ng/ml), and the general population in Fujian, China (median: 75.3 ng/ml), but higher than that of pregnant women in Massachusetts, USA (median: 16.5 ng/ml) and men in Canada (median: 16.7 ng/ml) (8, 43–47). In addition, the median concentration of MiBP was close to that reported for children in Fujian, China (median: 23.16 ng/ml) (45). They were generally higher than those pregnant women in Massachusetts, USA (median: 7.57 ng/ml) and men in Canada (median: 12.6 ng/ml), but much lower than those men in Tianjin, China (median: 190 ng/ml) (42, 46, 47). This finding could be explained by the following factors: (1) Different countries have different levels of PAEs in the industry; and (2) different climate scenarios have different effects on the exposure levels (48).
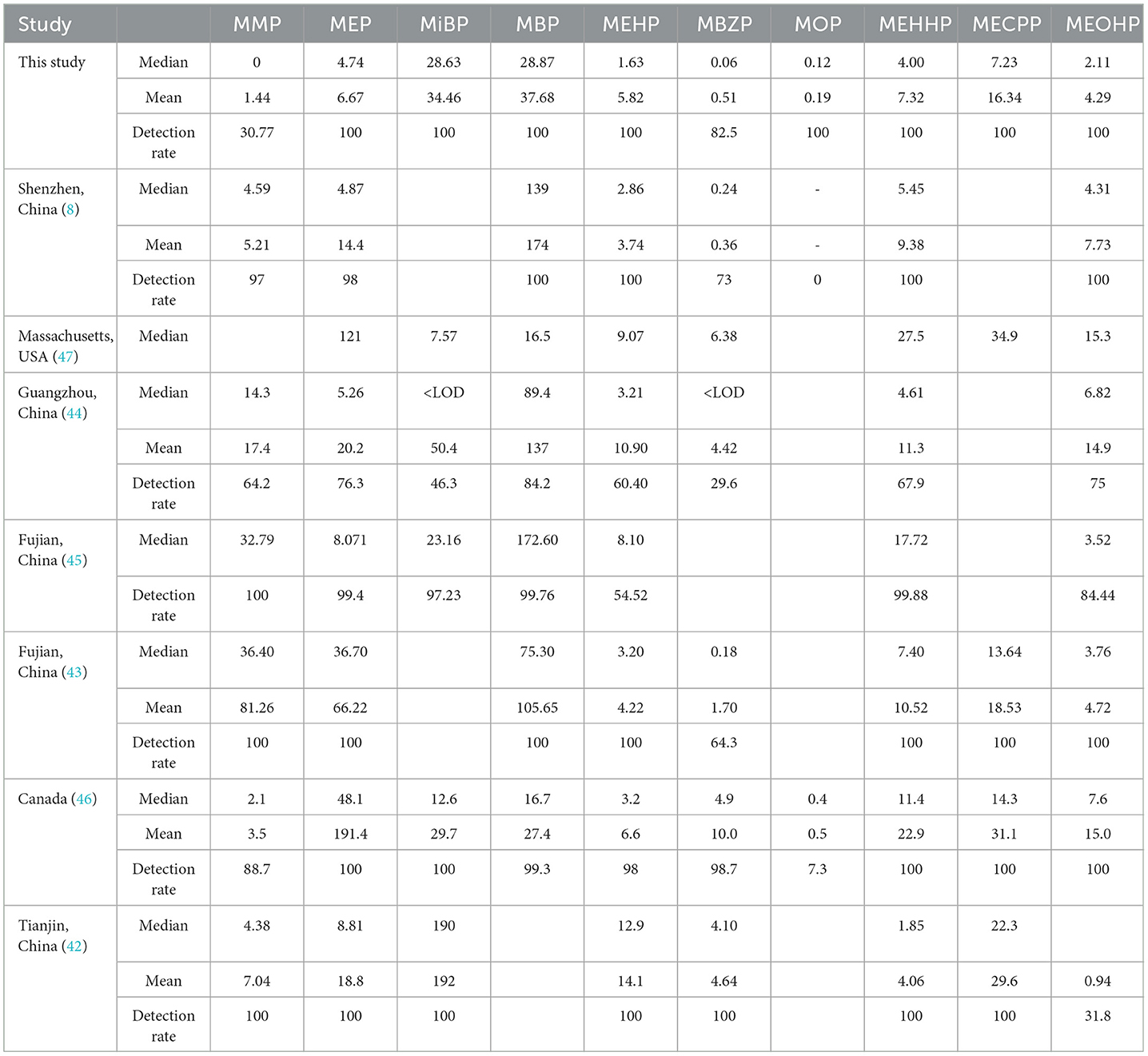
Table 8. Comparison of concentrations and levels of mPAEs in this study and previous studies (ng/ml).
In this study, the TDI value of DiBP exceeded 17.9% of men, and the TDI of DEHP exceeded 7.6%. For 39 men, 10 and 31% of HI values exceeded the RfD and TDI thresholds, respectively. Compared to our study, Chen et al. (8) found that 2 and 5% of EDI values in primiparas were greater than TDI and RfD values, and ~55% of HITDI values exceeded 1 in Shenzhen, China. Gao et al. (11) found that 40% of young adults in China showed HI values exceeding 1. These results demonstrated that adults probably have a health risk from PAE exposure in China. Given that PAE exposure has been associated with substantial adverse health effects, more attention should be paid to the cumulative risks of PAE exposure in Chinese adults.
This study speculates that mitochondrial epigenetic alterations can link PAE to adverse cardiovascular outcomes. We discovered that MEP and MBZP showed a significant relationship with triglycerides. Recently, most investigations have explored the PAEs associated with CVD risk (12, 14). PAE exposure levels have been related to oxidative stress, possibly through the activation of peroxisome proliferator-activated receptors or changes in mitochondrial membrane potential and permeability (49). Rosado-Berrios et al. (50) found that MEHP and DEHP affected mitochondrial membrane potential and promoted reactive oxygen species generation, then proved oxidative stress is a core underlying mechanism in the progression of CVDs (51). A meta-study showed that PAEs play a role similar to estrogen in the human body, which may bind to estrogen receptor α or receptors α and γ (pPARα and pPARγ) and damage β cell function to control carbohydrate metabolism and fat production (19). Given that triglycerides decompose into carbohydrates and water after liver metabolism, it is reasonable to assume that PAEs affect triglyceride metabolism, thereby leading to metabolic disorders and triglyceride accumulation (52, 53). As an essential subunit in ATP synthase, ATP8 directly affects ATP synthesis and energy metabolism (26). Therefore, the ATP8 gene plays a vital role in triglyceride metabolism. PAEs, mtDNA methylation, and triglyceride levels may have a potential association.
Through the statistical analysis, no mediator effect of PAE exposure on triglycerides through MT-ATP8 expression was observed. However, it is still worth noting that we found that 10.26–31.00% HI exceeded 1 and had a broad range of mtDNA methylation levels (1.09–24.31%, more than 22-fold differences), which is potentially risky. Baccarelli et al. found that CVDs are associated with the methylation of different segments of mtDNA (19, 54, 55). Liu et al. (20) carried out a study with 110 participants, which found that due to air pollutants, methylation levels in the MT-COX1, MT-ATP6, and MT-ATP8 regions were significantly lower, while COX2 and COX3 regions were obviously higher, providing a direction on the epigenetic regulation research of mitochondrial function.
This study was the first to relate urine PAE exposure and mtDNA methylation with lipid levels in men. PAE concentrations and health risks were assessed. In addition, the study evaluated the relationships between PAE levels and mtDNA methylation with lipid levels. The results provide a new direction to study PAEs and CVDs. However, some aspects could still be improved in this study. The small sample size in this study may only represent some of the population in Tianjin. The HI was only calculated with three PAEs due to the lack of parameters for other highly detectable PAEs. The relationships between PAEs, mPAEs concentrations, and mtDNA methylation levels with lipid levels might be influenced by other pollutants. Further studies are needed to explain the specific response relationship and the mechanisms of action on CVDs.
5. Conclusion
The risk assessments of the concentration accumulation of PAEs in men were analyzed in this study. MBP and MiBP were the dominant compounds in Chinese young men. We detected mtDNA methylation levels by using sequencing technology and assessed the associations between lipid levels with mtDNA methylation rates and PAEs. Significant associations between urine MEP concentrations and MT-ATP8 methylation with triglycerides were found. Those findings supported that PAE exposure is a risk factor for CVDs. We also explored the mediating effects of PAEs on the association between mtDNA methylation and lipid levels of CVDs, but the mediating effect failed to be found.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Tianjin Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
WL, JF, LZ, SS, TF, LG, CL, PL, and LW defined the research theme. WL and JF analyzed the data, and WL wrote the study. All authors have contributed to, read, and approved the manuscript.
Funding
This study was supported by the China Postdoctoral Science Foundation (2020M670667), Tianjin Technical Expert Project (21YDTPJC00280), Hebei Key Laboratory of Mineral Resources and Ecological Environment Monitoring (HBMREEM202104), Sino-Singapore Tianjin Eco-City Science and Technology Project of 2019, and Tianjin Science and Technology Project (22ZYCGCG00920).
Acknowledgments
The authors thank Dr. Hyang-Min Byun for research support for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2023.1108555/full#supplementary-material
References
1. Li X, Duan Y, Sun H, Zhang P, Xu J, Hua X, et al. Human exposure levels of PAEs in an e-waste recycling area: get insight into impacts of spatial variation and manipulation mode. Environ Int. (2019) 133:105143. doi: 10.1016/j.envint.2019.105143
2. Chen F, Chen Y, Chen C, Feng L, Dong Y, Chen J, et al. High-efficiency degradation of phthalic acid esters (PAEs) by Pseudarthrobacter defluvii E5: performance, degradative pathway, and key genes. Sci Total Environ. (2021) 794:148719. doi: 10.1016/j.scitotenv.2021.148719
3. Wu W, Wu P, Yang F, Sun DL, Zhang DX, Zhou YK. Association of phthalate exposure with anthropometric indices and blood pressure in first-grade children. Environ Sci Pollut Res Int. (2018) 25:23125–34. doi: 10.1007/s11356-018-2447-7
4. Gong M, Weschler CJ, Liu L, Shen H, Huang L, Sundell J, et al. Phthalate metabolites in urine samples from Beijing children and correlations with phthalate levels in their handwipes. Indoor Air. (2015) 25:572–81. doi: 10.1111/ina.12179
5. Guo Y, Wu Q, Kannan K. Phthalate metabolites in urine from China, and implications for human exposures. Environ Int. (2011) 37:893–8. doi: 10.1016/j.envint.2011.03.005
6. Pacyga DC, Sathyanarayana S, Strakovsky RS. Dietary predictors of phthalate and bisphenol exposures in pregnant women. Adv Nutr. (2019) 10:803–15. doi: 10.1093/advances/nmz029
7. Kessler W, Numtip W, Volkel W, Seckin E, Csanady GA, Putz C, et al. Kinetics of di(2-ethylhexyl) phthalate (DEHP) and mono(2-ethylhexyl) phthalate in blood and of DEHP metabolites in urine of male volunteers after single ingestion of ring-deuterated DEHP. Toxicol Appl Pharmacol. (2012) 264:284–91. doi: 10.1016/j.taap.2012.08.009
8. Chen Y, Jiang L, Lu S, Kang L, Luo X, Liu G, et al. Organophosphate ester and phthalate ester metabolites in urine from primiparas in Shenzhen, China: implications for health risks. Environ Pollut. (2019) 247:944–52. doi: 10.1016/j.envpol.2019.01.107
9. Ye Z, Ma M, Guo Y, Zong M, Ding M, Fan L. Determination of phthalate and metabolites in human urine by lithium bis(trifluoromethanesulfonyl)imide-enhanced dual microextraction method optimized by central composite design. Chromatographia. (2020) 83:397–408. doi: 10.1007/s10337-020-03858-3
10. Martino-Andrade AJ, Chahoud I. Reproductive toxicity of phthalate esters. Mol Nutr Food Res. (2010) 54:148–57. doi: 10.1002/mnfr.200800312
11. Gao CJ, Liu LY, Ma WL, Ren NQ, Guo Y, Zhu NZ, et al. Phthalate metabolites in urine of Chinese young adults: concentration, profile, exposure and cumulative risk assessment. Sci Total Environ. (2016) 543:19–27. doi: 10.1016/j.scitotenv.2015.11.005
12. Trasande L, Liu B, Bao W. Phthalates and attributable mortality: a population-based longitudinal cohort study and cost analysis. Environ Pollut. (2022) 292:118021. doi: 10.1016/j.envpol.2021.118021
13. Rattan S, Beers HK, Kannan A, Ramakrishnan A, Brehm E, Bagchi I, et al. Prenatal and ancestral exposure to di(2-ethylhexyl) phthalate alters gene expression and DNA methylation in mouse ovaries. Toxicol Appl Pharmacol. (2019) 379:114629. doi: 10.1016/j.taap.2019.114629
14. Fu X, Xu J, Zhang R, Yu J. The association between environmental endocrine disruptors and cardiovascular diseases: a systematic review and meta-analysis. Environ Res. (2020) 187:109464. doi: 10.1016/j.envres.2020.109464
15. Zhu X, Yin T, Yue X, Liao S, Cheang I, Zhu Q, et al. Association of urinary phthalate metabolites with cardiovascular disease among the general adult population. Environ Res. (2021) 202:111764. doi: 10.1016/j.envres.2021.111764
16. Bai PY, Wittert G, Taylor AW, Martin SA, Milne RW, Jenkins AJ, et al. The association between total phthalate concentration and non-communicable diseases and chronic inflammation in South Australian urban dwelling men. Environ Res. (2017) 158:366–72. doi: 10.1016/j.envres.2017.06.021
17. Wen ZJ, Wang ZY, Zhang YF. Adverse cardiovascular effects and potential molecular mechanisms of DEHP and its metabolites-A review. Sci Total Environ. (2022) 847:157443. doi: 10.1016/j.scitotenv.2022.157443
18. Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, et al. Heart disease and stroke statistics-2021 update: a report from the American Heart Association. Circulation. (2021) 143:e254–e743. doi: 10.1161/CIR.0000000000000950
19. Minguez-Alarcon L, Williams PL, James-Todd T, Souter I, Ford JB, Rexrode KM, et al. Association of urinary phthalate and phthalate replacement metabolite concentrations with serum lipid biomarker levels among pregnant women attending a fertility center. Toxics. (2022) 10:292. doi: 10.3390/toxics10060292
20. Liu Q, Li H, Guo L, Chen Q, Gao X, Li PH, et al. Effects of short-term personal exposure to air pollution on platelet mitochondrial DNA methylation levels and the potential mitigation by L-arginine supplementation. J Hazard Mater. (2021) 417:125963. doi: 10.1016/j.jhazmat.2021.125963
21. Morris MJ, Hesson LB, Poulos RC, Ward RL, Wong JWH, Youngson NA. Reduced nuclear DNA methylation and mitochondrial transcript changes in adenomas do not associate with mtDNA methylation. Biomark Res. (2018) 6:37. doi: 10.1186/s40364-018-0151-x
22. Huen K, Yousefi P, Bradman A, Yan L, Harley KG, Kogut K, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ Mol Mutagen. (2014) 55:209–22. doi: 10.1002/em.21845
23. Tian M, Liu L, Zhang J, Huang Q, Shen H. Positive association of low-level environmental phthalate exposure with sperm motility was mediated by DNA methylation: a pilot study. Chemosphere. (2019) 220:459–67. doi: 10.1016/j.chemosphere.2018.12.155
24. Quan Y, Xin Y, Tian G, Zhou J, Liu X. Mitochondrial ROS-Modulated mtDNA: a potential target for cardiac aging. Oxid Med Cell Longev. (2020) 2020:9423593. doi: 10.1155/2020/9423593
25. Nissanka N, Moraes CT. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. (2018) 592:728–42. doi: 10.1002/1873-3468.12956
26. Zhang J, Liu Z, Song S, Fang J, Wang L, Zhao L, et al. The exposure levels and health risk assessment of antibiotics in urine and its association with platelet mitochondrial DNA methylation in adults from Tianjin, China: a preliminary study. Ecotoxicol Environ Saf. (2022) 231:113204. doi: 10.1016/j.ecoenv.2022.113204
27. Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: an ENVIRONAGE birth cohort study. Epigenetics. (2015) 10:536–44. doi: 10.1080/15592294.2015.1048412
28. Han X, Zhao Z, Zhang M, Li G, Yang C, Du F, et al. Maternal trans-general analysis of the human mitochondrial DNA pattern. Biochem Biophys Res Commun. (2017) 493:643–9. doi: 10.1016/j.bbrc.2017.08.138
29. Stoccoro A, Coppede F. Mitochondrial DNA methylation and human diseases. Int J Mol Sci. (2021) 22:4594. doi: 10.3390/ijms22094594
30. Mposhi A, van der Wijst MG, Faber KN, Rots MG. Regulation of mitochondrial gene expression, the epigenetic enigma. Front Biosci Landmark Ed. (2017) 22:1099–113. doi: 10.2741/4535
31. Barrow TM, Peng C, Wilson A, Wang H, Liu H, Shen L, et al. Psychosocial stress is associated with benign breast disease in young Chinese women: results from Project ELEFANT. Breast Cancer Res Treat. (2019) 173:217–24. doi: 10.1007/s10549-018-4979-4
32. Guo L, Peng C, Xu H, Wilson A, Li PH, Wang H, et al. Age at menarche and prevention of hypertension through lifestyle in young Chinese adult women: result from project ELEFANT. BMC Womens Health. (2018) 18:182. doi: 10.1186/s12905-018-0677-y
33. Xu H, Li PH, Barrow TM, Colicino E, Li C, Song R, et al. Obesity as an effect modifier of the association between menstrual abnormalities and hypertension in young adult women: results from Project ELEFANT. PLoS ONE. (2018) 13:e0207929. doi: 10.1371/journal.pone.0207929
34. Cayir A, Barrow TM, Wang H, Liu H, Li C, Ding N, et al. Occupational noise exposure is associated with hypertension in China: results from project ELEFANT. PLoS ONE. (2018) 13:e0209041. doi: 10.1371/journal.pone.0209041
35. Guo L, Liu Z, Li P, Ji Y, Song S, Zheng N, et al. Association between mitochondrial DNA methylation and internal exposure to polycyclic aromatic hydrocarbons (PAHs), nitrated-PAHs (NPAHs) and oxygenated-PAHs (OPAHs) in young adults from Tianjin, China. Ecotoxicol Environ Saf. (2022) 241:113799. doi: 10.1016/j.ecoenv.2022.113799
36. Koch HM, Drexler H, Angerer J. An estimation of the daily intake of di(2-ethylhexyl)phthalate (DEHP) and other phthalates in the general population. Int J Hyg Environ Health. (2003) 206:77–83. doi: 10.1078/1438-4639-00205
37. Perucca J, Bouby N, Valeix P, Bankir L. Sex difference in urine concentration across differing ages, sodium intake, and level of kidney disease. Am J Physiol Regul Integr Comp Physiol. (2007) 292:R700–5. doi: 10.1152/ajpregu.00500.2006
38. Li J, Zhao H, Xia W, Zhou Y, Xu S, Cai Z. Nine phthalate metabolites in human urine for the comparison of health risk between population groups with different water consumptions. Science of The Total Environment. (2019) 649:1532–40. doi: 10.1016/j.scitotenv.2018.08.294
39. Pecina P, Houst kova H, Zeman J, Houstek J. Genetic defects of cytochrome C oxidase assembly. Physiol Res. (2004) 53:S213–S23.
40. Yi L, Ai Y, Ming L, Hai L, He J, Guo F-C, et al. Molecular diversity and phylogenetic analysis of domestic and wild Bactrian camel populations based on the mitochondrial ATP8 and ATP6 genes. Livest Sci. (2017) 199:95–100. doi: 10.1016/j.livsci.2017.03.015
41. Iannaccone M, Ianni A, Contaldi F, Esposito S, Martino C, Bennato F, et al. Whole blood transcriptome analysis in ewes fed with hemp seed supplemented diet. Sci Rep. (2019) 9:16192. doi: 10.1038/s41598-019-52712-6
42. Wang B, Qin X, Xiao N, Yao Y, Duan Y, Cui X, et al. Phthalate exposure and semen quality in infertile male population from Tianjin, China: associations and potential mediation by reproductive hormones. Sci Total Environ. (2020) 744:140673. doi: 10.1016/j.scitotenv.2020.140673
43. Tian M, Liu L, Wang H, Wang X, Martin FL, Zhang J, et al. Phthalates induce androgenic effects at exposure levels that can be environmentally relevant in humans. Environ Sci Technol. (2018) 5:232–6. doi: 10.1021/acs.estlett.8b00138
44. Li X, Zhong Y, He W, Huang S, Li Q, Guo C, et al. Co-exposure and health risks of parabens, bisphenols, triclosan, phthalate metabolites and hydroxyl polycyclic aromatic hydrocarbons based on simultaneous detection in urine samples from guangzhou, South China. Environ Pollut. (2021) 272:115990. doi: 10.1016/j.envpol.2020.115990
45. Dong Y, Gao D, Li Y, Yang Z, Wang X, Chen M, et al. Effect of childhood phthalates exposure on the risk of overweight and obesity: a nested case-control study in China. Environ Int. (2022) 158:106886. doi: 10.1016/j.envint.2021.106886
46. Feng Y-L, Liao X, Grenier G, Nguyen N, Chan P. Determination of 18 phthalate metabolites in human urine using a liquid chromatography-tandem mass spectrometer equipped with a core–shell column for rapid separation. Analytical Methods. (2015) 7:8048–59. doi: 10.1039/C5AY00107B
47. Ferguson KK, McElrath TF, Chen YH, Mukherjee B, Meeker JD. Urinary phthalate metabolites and biomarkers of oxidative stress in pregnant women: a repeated measures analysis. Environ Health Perspect. (2015) 123:210–6. doi: 10.1289/ehp.1307996
48. Huang L, Qiao Y, Deng S, Zhou M, Zhao W, Yue Y. Airborne phthalates in indoor environment: partition state and influential built environmental conditions. Chemosphere. (2020) 254:126782. doi: 10.1016/j.chemosphere.2020.126782
49. Hurst CH, Waxman DJ. Activation of PPARalpha and PPARgamma by environmental phthalate monoesters. Toxicol Sci. (2003) 74:297–308. doi: 10.1093/toxsci/kfg145
50. Rosado-Berrios CA, Velez C, Zayas B. Mitochondrial permeability and toxicity of diethylhexyl and monoethylhexyl phthalates on TK6 human lymphoblasts cells. Toxicol In Vitro. (2011) 25:2010–6. doi: 10.1016/j.tiv.2011.08.001
51. Callaghan MA, Alatorre-Hinojosa S, Connors LT, Singh RD, Thompson JA. Plasticizers and cardiovascular health: role of adipose tissue dysfunction. Front Pharmacol. (2020) 11:626448. doi: 10.3389/fphar.2020.626448
52. Latini G, Scoditti E, Verrotti A, De Felice C, Massaro M. Peroxisome proliferator-activated receptors as mediators of phthalate-induced effects in the male and female reproductive tract: epidemiological and experimental evidence. PPAR Res. (2008) 2008:359267. doi: 10.1155/2008/359267
53. Hao C, Cheng X, Xia H, Ma X. The endocrine disruptor mono-(2-ethylhexyl) phthalate promotes adipocyte differentiation and induces obesity in mice. Biosci Rep. (2012) 32:619–29. doi: 10.1042/BSR20120042
54. Baccarelli AA, Byun HM. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clin Epigenetics. (2015) 7:44. doi: 10.1186/s13148-015-0078-0
Keywords: phthalate, phthalate metabolites, health risk assessment, mtDNA methylation, cardiovascular diseases
Citation: Li W, Guo L, Fang J, Zhao L, Song S, Fang T, Li C, Wang L and Li P (2023) Phthalates and phthalate metabolites in urine from Tianjin and implications for platelet mitochondrial DNA methylation. Front. Public Health 11:1108555. doi: 10.3389/fpubh.2023.1108555
Received: 26 November 2022; Accepted: 27 March 2023;
Published: 26 April 2023.
Edited by:
Ciro Fernando Bustillo LeCompte, Toronto Metropolitan University, CanadaReviewed by:
Octavio Jiménez-Garza, University of Guanajuato, MexicoRadha Dutt Singh, University of California, San Francisco, United States
Copyright © 2023 Li, Guo, Fang, Zhao, Song, Fang, Li, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Wang, d2FuZ2xlaTg4MTJAMTI2LmNvbQ==; Penghui Li, bGlwZW5naHVpNDA2QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Weixia Li1,2,3†
Weixia Li1,2,3† Liqiong Guo
Liqiong Guo Penghui Li
Penghui Li