- 1Department of Clinical Laboratory, The First Affiliated Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China
- 2Xiamen Key Laboratory of Genetic Testing, School of Medicine, Xiamen University, Xiamen, China
- 3Centre of Clinical Laboratory, Zhongshan Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China
- 4Institute of Infectious Disease, School of Medicine, Xiamen University, Xiamen, China
Background: The global epidemiological situation of COVID-19 remains serious. The rapid hunting of SARS-CoV-2 infection is the key means for preventing transmission.
Methods: A total of 40,689 consecutive overseas arrivals were screened for SARS-CoV-2 infection based on PCR and serologic testing. The yield and efficiency of different screening algorithms were evaluated.
Result: Among the 40,689 consecutive overseas arrivals, 56 (0.14%) subjects were confirmed to have SARS-CoV-2 infection. The asymptomatic rate was 76.8%. When the algorithm based on PCR alone was used, the identification yield of a single round of PCR (PCR1) was only 39.3% (95% CI: 26.1–52.5%). It took at least four rounds of PCR to achieve a yield of 92.9% (95% CI: 85.9–99.8%). Fortunately, an algorithm based on a single round of PCR combined with a single round of serologic testing (PCR1+ Ab1) greatly improved the screening yield to 98.2% (95% CI: 94.6–100.0%) and required 42,299 PCR and 40,689 serologic tests that cost 6,052,855 yuan. By achieving a similar yield, the cost of PCR1+ Ab1 was 39.2% of that of four rounds of PCR. For hunting one case in PCR1+ Ab1, 769 PCR and 740 serologic tests were required, costing 110,052 yuan, which was 63.0% of that of the PCR1 algorithm.
Conclusion: Comparing an algorithm based on PCR alone, PCR combined with a serologic testing algorithm greatly improved the yield and efficiency of the identification of SARS-CoV-2 infection.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), and its rapid transmission and high virulence have resulted in the ongoing COVID-19 pandemic (1). The global epidemiological situation of COVID-19 remains serious. The rapid discovery of SARS-CoV-2 infection and the quick isolation of patients and tracing of their close contacts are currently the most effective means for preventing transmission. In low-prevalence areas in particular, the identification of SARS-CoV-2 infection is crucial. Currently, algorithms based on PCR alone are widely used to diagnose COVID-19 and for the detection and quantification of SARS-CoV-2 RNA (2, 3). There are several limitations to the use of PCR alone for diagnosing SARS-CoV-2 infection, including the potential for false-negative results, which may be linked to inadequate nasopharyngeal sampling, varying levels of the virus at different anatomical sites and different times during the disease course, and the inability to diagnose pre- or asymptomatic infections (4–6). Therefore, the yield of PCR is unsatisfactory. Due to rapid transmission and strong infectiousness of the disease, failure to detect a SARS-CoV-2 infection could greatly decrease the efficacy of prevention. To achieve higher yields, PCR must be performed repeatedly in all subjects and should consume considerable human and material resources during the initial COVID-19 pandemic. Compared with PCR, serologic testing is relatively easier to perform and faster (7). Unlike PCR, which can detect only acutely infected persons, serologic tests help determine whether the individual being tested was previously infected, even if that person never showed symptoms (8). However, the potential for false-positive results in serologic tests limits their use in low-prevalence populations (9, 10). Both PCR- and serology-based methods have obvious defects, but they possibly complement each other throughout the disease course. The efficiency of screening algorithms based on PCR combined with serologic testing for identifying SARS-CoV-2 infection in practice is unclear. In our region, the government used PCR combined with a serologic testing algorithm to hunt SARS-CoV-2 infection in consecutive overseas arrivals between July 2020 and September 2020. Therefore, we investigated different screening algorithms from an economic perspective to evaluate whether PCR combined with a serologic testing algorithm could improve the yield and efficiency of the identification of SARS-CoV-2 infection.
Methods
Study design and participants
A total of 40,689 consecutive overseas arrivals, which were screened for SARS-CoV-2 infection based on PCR and serologic testing in Xiamen city between July 2020 and September 2020 were retrospectively investigated. All individuals underwent the first round of serologic testing, total antibodies against SARS-CoV-2 (Ab), and PCR tests to identify SARS-CoV-2 infection on the first day of entry. The individuals who were Ab-positive (Ab+) were assigned to a key screening population, followed up for 14 days, and subjected to multiple rounds of PCR that were serially determined at 3, 5, 7, 10, and 14 days after entry. The individuals who were Ab-negative (Ab-) underwent serologic testing at 7 days after entry and underwent PCR at 7 and 14 days after entry. All individuals were followed up for 28 days. When the PCR result was positive (PCR+), the individual was escorted directly to the hospital for a comprehensive evaluation and epidemiological investigation. Based on the COVID-19 Diagnosis and Treatment Plan (eighth edition) of the National Health Commission of the People's Republic of China, the subjects were diagnosed according to their epidemiological history, clinical symptoms, imaging findings, and laboratory test results. Individuals with SARS-CoV-2 infection included asymptomatic and symptomatic individuals. According to the neutralization test and epidemiological history, Ab-positive (Ab+) was classified as true-positive or false-positive. Previously infected individuals were defined as those with true-positive antibody results and positive neutralization test results, but no symptoms or signs of COVID-19 and negative RT-PCR results during the study period. Finally, 40,498 individuals were non-infected. The age of the subjects ranged from 1 to 93 years, with a median age of 29 years, and 17,473 (43.1%) individuals were women. For the previously infected, the ages ranged from 21 to 65 years, with a median age of 34 years, and 65 (44.4%) individuals were women. To investigate different screening algorithms from an economic perspective, the testing algorithm for screening SARS-CoV-2 infection was set to nine algorithms, according to round and combination of PCR and serologic testing (Table 1). To determine the incremental yield of each SARS-CoV-2 test algorithm, we determined the number of additional SARS-CoV-2 infections detected relative to the number detected with the single round of PCR (PCR1). To determine the efficiency of each algorithm, we determined the number of serology and PCR tests used and the number needed to test (NNT) to detect one case of SARS-CoV-2 infection for each test.
PCR assays for SARS-CoV-2
Upper respiratory tract (URT) samples were collected from both nasopharyngeal and oropharyngeal swabs collected by trained medical staff (physicians and nurses). For lower respiratory tract (LRT) specimens, participants were given instructions the night before to collect the first morning sputum samples (after gargling) in a specimen cup. The Stream SP96 automatic nucleic acid extraction instrument (Da An Gene Co., Ltd. Guangzhou, China) was used for nucleic acid extraction. RT-qPCR was conducted using the SLAN-96P real-time PCR system (Shanghai Hongshi Biotechnology Co., Ltd., Shanghai, China). PCR assays for SARS-CoV-2 were performed using the Liferiver 2019-nCoV assay (Shanghai ZJ Bio-Tech Co., Ltd, Shanghai, China) to determine the presence of the virus through the identification of three genetic markers: the envelope (env) gene, the open reading frame (ORF) 1ab gene, and the nucleocapsid protein (N) gene. The cycle threshold (Ct) determined during RT-PCR testing refers to the cycle in which the detection of viral amplicons occurs, and it is inversely correlated with the amount of RNA present. A lower Ct value indicates larger quantities of viral RNA. The results were considered positive when the Ct values of all genes were <40 cycles. The assay had a sensitivity of 89.3% and a specificity of 100.0%, and no cross-reactivity was observed in clinical diagnostic efficacy (11). Two consecutive single-site positives or double-site positives are judged to indicate RT-PCR positivity according to the COVID-19 Prevention and Control Plan (eighth edition).
Serologic testing
Blood samples were centrifuged at 3,000 rpm, and the upper serum layer was used for testing. The total antibodies against SARS-CoV-2 were measured using a Wantai® Caris 200 system, based on a chemiluminescence microparticle immunoassay (CMIA) (Wantai Biological Pharmacy Enterprise Co., Ltd, Beijing, China). The detection experiments were performed according to the manufacturer's instructions. The kit was provided by Wantai Biological Pharmacy Enterprise Co., Ltd, Beijing, China. TAb detection was based on a double-antigen sandwich immunoassay using two types of mammalian cell-expressed recombinant antigens containing the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 as the immobilized and HRP-conjugated antigens. The antibody titer was calculated according to the cutoff and was recorded as the cutoff index (COI). COI<1.00 was considered negative, and COI≥1.00 was considered positive. The assay had a sensitivity of 96.7% and a specificity of 99.5%, and no cross-reactivity was observed (12).
Statistical analysis
Statistical analysis was carried out using IBM SPSS Statistics version 20 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism version 8.00 (GraphPad Software, San Diego, CA, USA). Continuous variables that did not follow a normal distribution are reported as medians with interquartile ranges (IQRs). The statistical analysis for group comparisons was conducted using the Kruskal–Wallis and Mann–Whitney U-tests. To determine the diagnostic yields of different algorithms, the clinical diagnostic results were used as the gold standard according to their epidemiological history, clinical symptoms, imaging findings, and laboratory test results. We compared the differences in the proportions of infection detected by each algorithm relative to the single round of PCR (PCR1) algorithm using the McNemar chi-square test of paired proportions. Statistical significance was indicated by a p-value of <0.05 (<0.05).
Results
Outcomes of SARS-CoV-2 screening
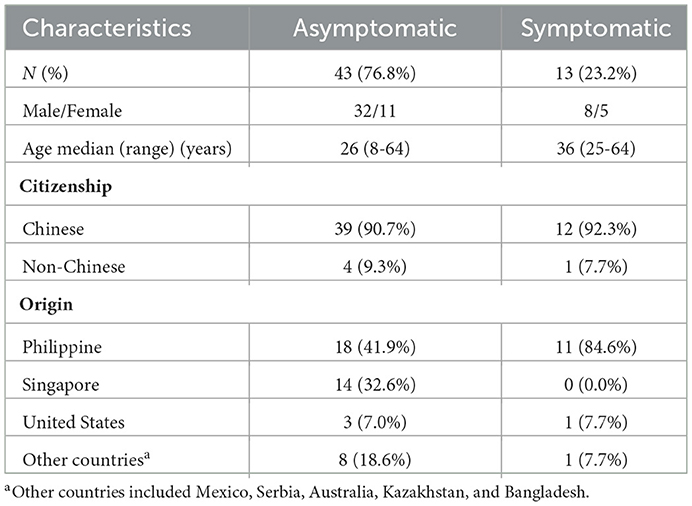
We performed PCR and serologic tests on samples collected from 40,689 subjects between July 2020 and September 2020. A total of 56 (0.14%) subjects with SARS-CoV-2 infection were identified. They came from eight foreign countries (Table 2). The asymptomatic rate was 76.8%. There were no new cases of SARS-CoV-2 infection found among the remaining subjects during the 28-day follow-up (Figure 1). The first round of serology tests revealed 359 subjects to be Ab-positive. Among Ab-positive subjects, 54 (96.4%) subjects with SARS-CoV-2 infections were identified (Figure 1). Notably, 14 (25.0%) subjects with SARS-CoV-2 infections were discovered after more than three rounds of PCR were performed because Ab positivity was classified as the key subject, which was screened by multiple rounds of PCR. In addition, four (7.14%) subjects with SARS-CoV-2 infections were found after four rounds of PCR (Figure 1). On the other hand, among the 40,330 Ab-negative subjects, two subjects showed seroconversion within a week, both of whom were confirmed SARS-CoV-2 infection, one was confirmed on the first round of PCR, and the other was confirmed on the third round of PCR.
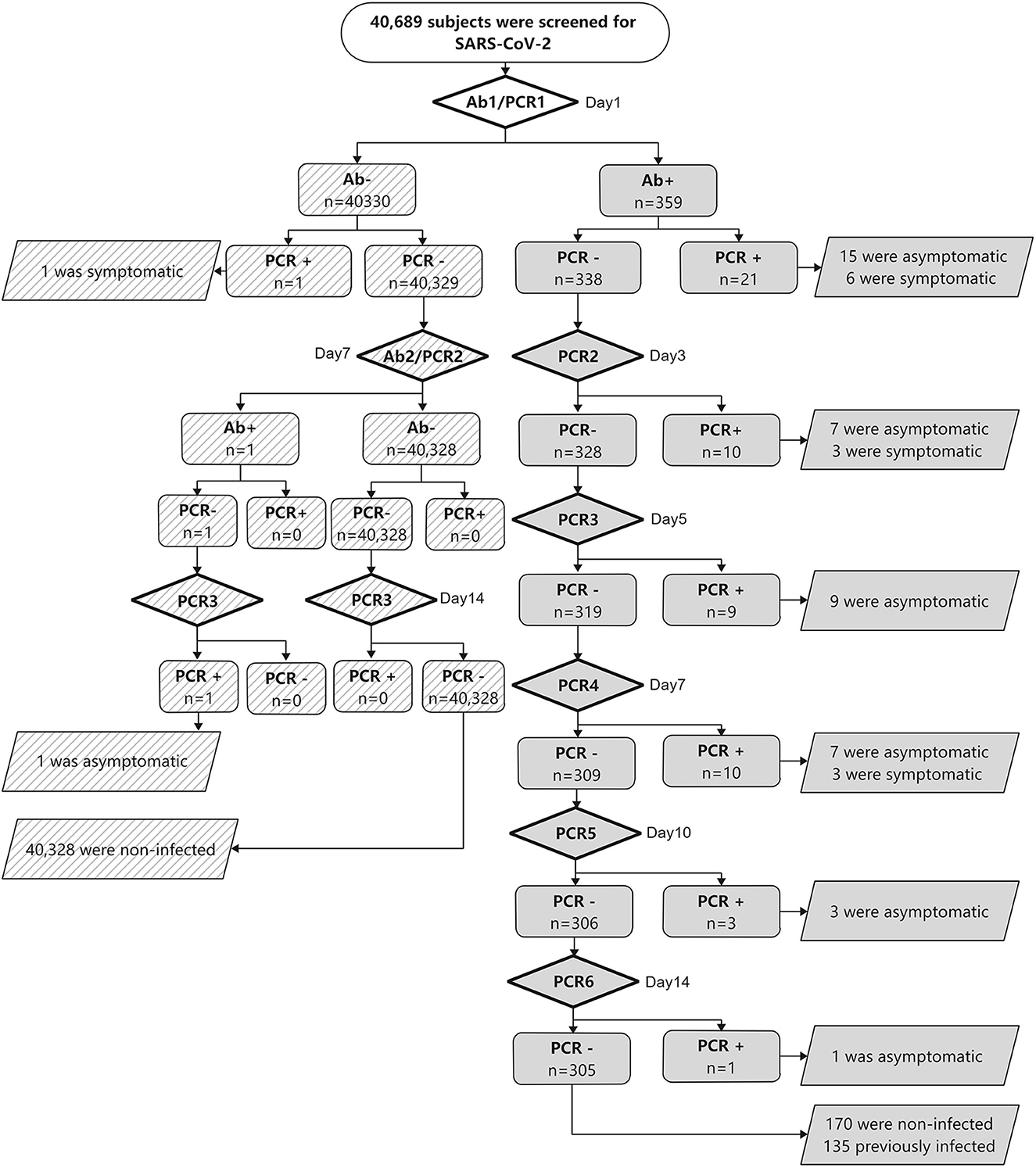
Figure 1. Results of SARS-CoV-2 screening. Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; PCR, polymerase chain reaction to detect SARS-CoV-2; Ab, total anti-SARS-CoV-2 antibody level.
The yield of the SARS-CoV-2 screening algorithm
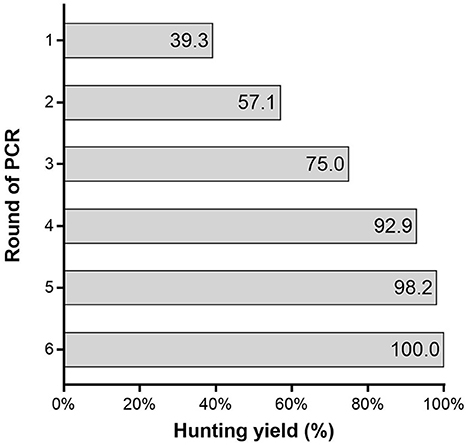
The identification yield of a single round of PCR was only 39.3% (95% CI: 26.1–52.5%). It took at least four rounds of PCR to achieve a yield of 92.9% (95% CI: 85.9–99.8%) (Figure 2). The diagnostic yield of the algorithm based on PCR alone increased as the number of PCR rounds increased. It was noted that the application of an algorithm based on PCR combined with serologic testing greatly improved the screening yield to 98.2% (95% CI: 94.6–100.0%). However, with the addition of serologic testing, the number of false positives increased to 305, which was 5.4 times greater than the number of SARS-CoV-2 infections (Table 3).
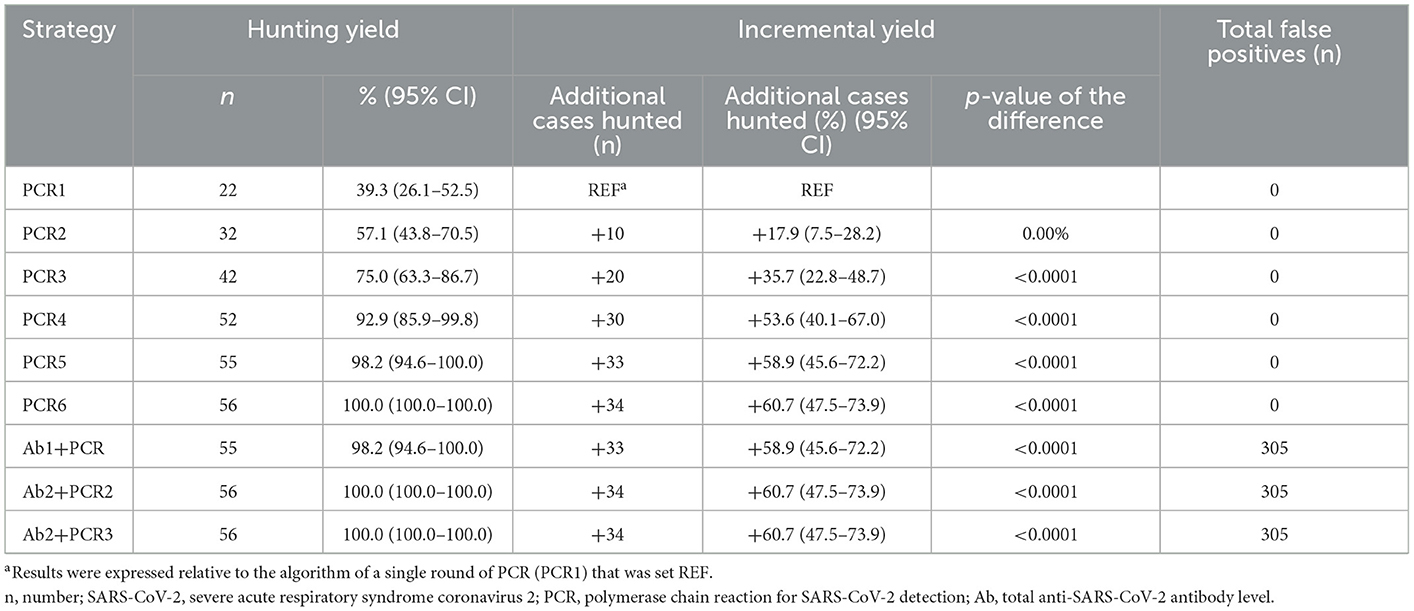
Table 3. Hunting yield, incremental yield, and number of false positives obtained with different algorithms.
The cost of different SARS-CoV-2 test algorithms
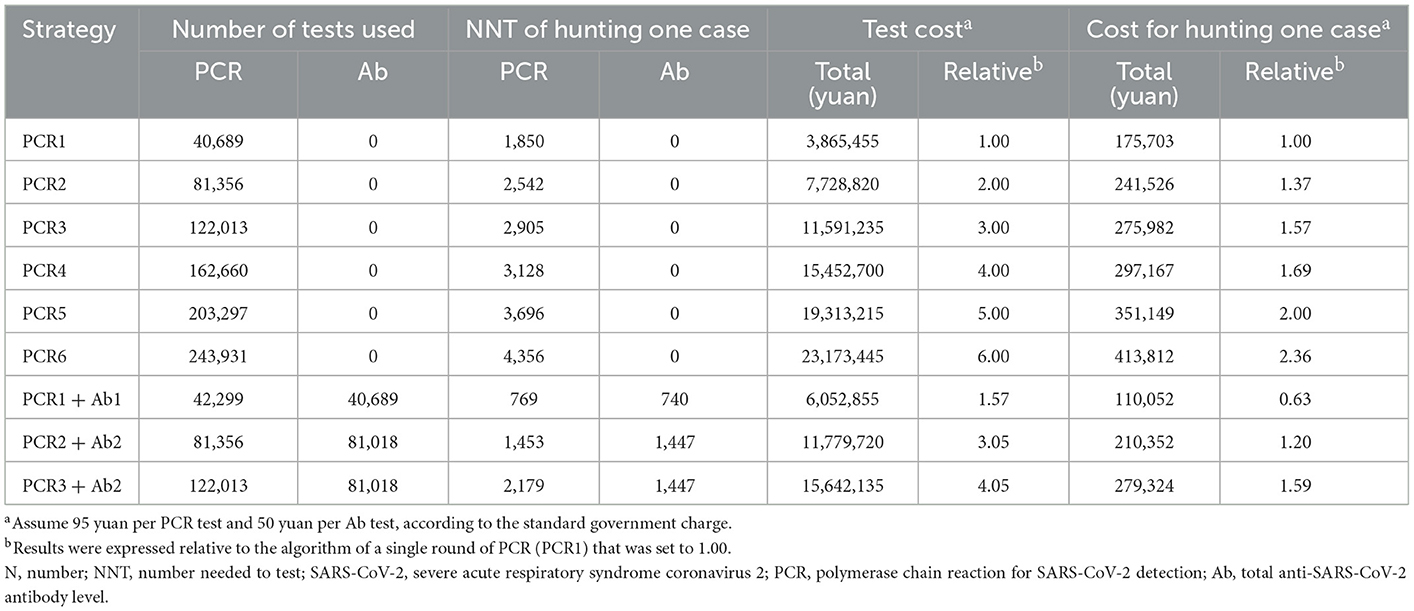
For the algorithm based on a single round of PCR, 40,689 tests were conducted at a cost of 3,865,455 yuan, and 1,850 tests costing 175,703 yuan were required to detect each case. However, this approach missed 34 (60.7%) SARS-CoV-2 infection cases. To achieve higher yields, the number of tests and the cost associated with the algorithm based on PCR alone rapidly increased as the number of rounds of PCR increased (Table 4). In four rounds of PCR (PCR4) achieving a yield of 92.9%, 162,660 tests were conducted at a cost of 15,452,700 yuan, and 3,128 tests costing 297,167 yuan were requiredfor hunting each case. Relative to the PCR1 algorithm, the test number and cost were increased 4.00-fold and 1.69-fold, respectively. Fortunately, the algorithm based on serologic testing combined with PCR was more effective. With a 98.2% yield, the algorithm based on a single round of PCR combined with a single round of serologic testing (PCR1+ Ab1) required 42,299 PCR and 40,689 serologic tests that cost 6,052,855 yuan. With a similar yield, the cost of PCR1+ Ab1 was 39.2% of that of four rounds of PCR. For hunting one case, 769 PCR and 740 serologic tests were required and cost 110,052 yuan, which was 63.0% of that of the PCR1 algorithm. For the algorithm based on PCR combined with serologic testing, the number of tests required and the cost of discovering one case increased as the round of testing increased, but the yield did not increase significantly (Table 4).
Serologic testing in the population
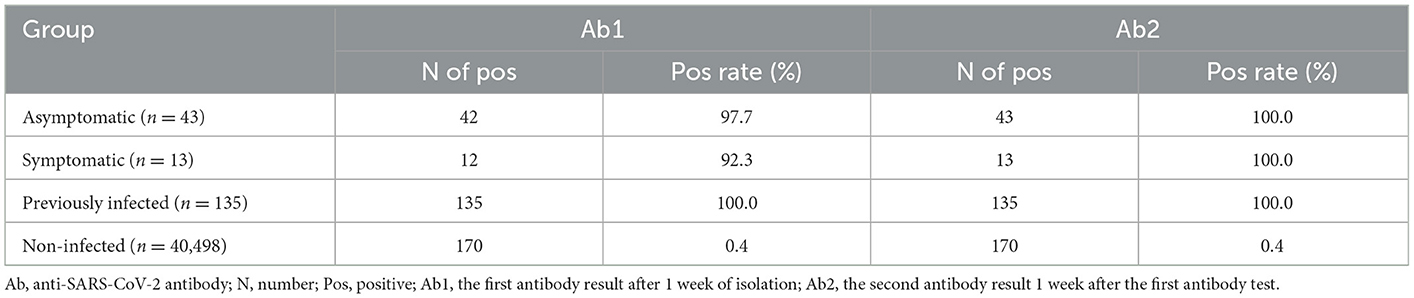
A total of 170 subjects showed false-positive results, and the false-positive rate was 0.42%. The Ab titers of the asymptomatic, symptomatic, previously infected, and false-positive groups were 43.00 (20.34–109.00) COI, 92.92 (18.63–215.20) COI, 49.98 (10.37–156.8) COI, and 1.99 (1.25–4.28) COI, respectively. A significantly lower titer was found in the false-positive group than in the other groups (Figure 3), with 94.70% of the subjects in the false-positive group showing a value below 20 COI. Within 1 week, seroconversion occurred in one case in the symptomatic group and one case in the asymptomatic group, and the positive rate of SARS-CoV-2 infection reached 100% (Table 5).

Figure 3. The titer of anti-SARS-CoV-2 antibody. Statistically significant differences correspond to the differences in the antibody titer between the false-positive group and the other groups. Abbreviations: COI, cut-off index. ***p < 0.001.
Discussion
The rapid hunting of SARS-CoV-2 infection and the quick isolation of patients are currently the most effective means for preventing the transmission of COVID-19 (13, 14). Different countries and regions adopted different SARS-CoV-2 screening algorithms and had different effects (15, 16). The successful hunting of SARS-CoV-2 infection depends heavily on the use of accurate tests performed at the appropriate time. In our study, the diagnostic yield of the algorithm based on one round of PCR was only 39.3%, indicating poor efficiency. At least four rounds of PCR (PCR4) were required to achieve a yield of over 90.0%. The diagnostic yield of the algorithm based on PCR alone increased as the number of PCR cycles increased. Furthermore, relative to the PCR1 algorithm, the test number and cost were increased 4.00 and 1.69-fold, respectively. This is a huge burden for any institution or country. Thus, the feasibility of this approach is highly questionable. Furthermore, 7.14% of infected subjects were missed, representing a huge hidden danger in epidemic control. The highest sensitivity of PCR testing based on nasopharyngeal sampling is observed from 0 to 4 days post-symptom onset, at 89%, dropping to 54% after 10 to 14 days (17). In the later stage of SARS-CoV-2 infection, the sensitivity of PCR was low and might not facilitate the identification. It suggested that the algorithm based on PCR alone could not meet the screening requirements.
Fortunately, it was noted that the use of an algorithm based on serologic testing combined with PCR could easily improve the screening yield to over 90.0%. The diagnostic yield of the algorithm based on a single serologic test combined with a single round of PCR was 98.2%. The success of diagnosis depends heavily on test accuracy and the use of accurate tests at the appropriate time. Serological tests show the lowest sensitivity at 0–7 days after symptom onset and the highest at >14 days (2, 3, 18). The sensitivity of IgG-, IgM-, and TAb-based tests is 25, 34, and 36%, respectively, during the first 7 days after symptom onset but increases to 62, 65, and 80% at 8–14 days post-symptom onset and 90, 85, and 93% after 14 days post-symptom onset (7, 18). In the early stage of SARS-CoV-2 infection, the sensitivity of serological testing was low and might not facilitate the identification. Fortunately, the advantages and disadvantages of PCR and serologic tests complement each other, covering all stages of the disease. The other reasons for the improvement achieved by the combined strategy were as follows. First, the amount of anti-SARS-CoV-2 antibody produced by asymptomatic individuals is similar to that produced by symptomatic individuals. Second, a highly sensitive detection method was used in this procedure. Third, the test time was several days after travel from the site of origin to the destination.
Furthermore, the algorithm based on serologic testing combined with PCR was more effective. With a 98.2% yield, the algorithm based on a single round of PCR combined with a single round of serologic tests (PCR1+ Ab1) required 42,299 PCR and 40,689 serologic tests that cost 6,052,855 yuan. With a similar yield, the cost of PCR1+ Ab1 was 39.2% of that of four rounds of PCR. For hunting one case, 769 PCR and 740 serologic tests were required at a cost of 110,052 yuan, which was 63.0% of that of the PCR1 algorithm. It is worth mentioning that by adding serologic testing, the size of the focus population was reduced from 40,689 to 361. Relative to the algorithm based on PCR alone, the combined approach substantially decreased the number of PCR tests required to obtain a similar yield. Compared with PCR, serologic testing is easier to perform and faster. In addition, blood samples are stable, easy to store, and less likely to contain infectious SARS-CoV-2 virus than respiratory specimens, decreasing the potential risk of infection for laboratory staff (7). Thus, the use of an algorithm based on serologic testing combined with PCR is more practical and inexpensive in the initial epidemic.
With the addition of serologic testing, the number of false positives was increased to 305, which was 5.4-fold more than that of SARS-CoV-2 infections. Serology is another important method for the investigation of SARS-CoV-2 infections (19). Nevertheless, the value of SARS-CoV-2 antibody testing for diagnosis, prevention, and control remains controversial (20). In the present study, total anti-SARS-CoV-2 antibody levels were measured, and the sensitivity for asymptomatic and symptomatic patients was 97.7 and 92.3%, respectively, which is higher than those reported previously (18, 21, 22). However, 170 subjects showed false positives, corresponding to a false-positive rate of 0.42%. Considering the 0.14% prevalence identified in this study, false positives were a great confounding factor. This suggests that serologic testing is not recommended as the primary approach for the diagnosis of SARS-CoV-2 infection (23, 24). A low titer was one of the characteristics of false positivity in the study. If a risk assessment dictates an overriding concern, the cutoff value can be set accordingly (10, 25). To exclude false positives, the cutoff value should be evaluated according to the specific objective and population.
This study has some limitations. First, because the time of subjects was in the early stage of the COVID-19 pandemic, a vaccinated population was not included in this study, and the variants were not involved. Second, the subjects came from all over the world and were highly mobile; thus, it was not possible to obtain a detailed disease course. Third, before boarding their plane, some subjects had been screened for SARS-CoV-2, which caused the true prevalence to be underestimated. Finally, due to the low number of SARS-CoV-2 infections, the relationship between the diagnostic efficacy of serologic testing, symptoms, and the cycle threshold was not determined.
Conclusion
PCR combined with the serologic testing algorithm greatly improved the yield and efficiency of the hunting of SARS-CoV-2 infection. The study provides a reference for a high-performance screening algorithm of PCR combined with serologic testing for the early detection of virus epidemics.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by (#xmzsyyky2021196) Institutional Ethics Committee of Zhongshan Hospital of Xiamen University, School of Medicine, Xiamen University. Written informed consent from the participants' legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
X-ML had full access to all the data in the study, took responsibility for the integrity of the data and the accuracy of the data analysis, and supported funding. L-LF and X-ML contributed to the protocol and design of the manuscript. L-LF, J-HZ, and X-ML drafted the manuscript. All authors designed and conducted the statistical analysis, and accessed and verified the underlying data. All authors critically revised the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Fujian Province, China [grant numbers 2020J011208 and 2020J011233] and the project for Xiamen Medical and Health Guidance [grant number 3502Z20214ZD1037].
Acknowledgments
The authors thank all the contributors to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
COVID-19, coronavirus disease 2019; SARS-CoV-2; severe acute respiratory syndrome coronavirus; URT, upper respiratory tract; LRT, lower respiratory tract; CMIA, chemiluminescence microparticle immunoassay; RBD, receptor-binding domain; COI, cut-off index; IQR, interquartile range; NNT, the number needed to test; PCR, polymerase chain reaction for SARS-CoV-2; N, number; Ab, total anti-SARS-CoV-2 antibody.
References
1. Van Elslande J, Houben E, Depypere M, Brackenier A, Desmet S, Andre E, et al. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. (2020) 26:1082–7. doi: 10.1016/j.cmi.2020.05.023
2. To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. (2020) 20:565–74. doi: 10.1016/S1473-3099(20)30196-1
3. Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. (2020) 581:465–9. doi: 10.1038/s41586-020-2196-x
4. Pray IW, Gibbons-Burgener SN, Rosenberg AZ, Cole D, Borenstein S, Bateman A, et al. COVID-19 outbreak at an overnight summer school retreat—wisconsin, July–August 2020. MMWR Morb Mortal Wkly Rep. (2020) 69:1600–4. doi: 10.15585/mmwr.mm6943a4
5. Paradiso AV, De Summa S, Loconsole D, Procacci V, Sallustio A, Centrone F, et al. Rapid serological assays and SARS-CoV-2 real-time polymerase chain reaction assays for the detection of SARS-CoV-2: comparative study. J Med Internet Res. (2020) 22:e19152. doi: 10.2196/19152
6. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. (2020) 296:E32–40. doi: 10.1148/radiol.2020200642
7. Zainol Rashid Z, Othman SN, Abdul Samat MN, Ali UK, Wong KK. Diagnostic performance of COVID-19 serology assays. Malays J Pathol. (2020) 42:13–21.
8. Prevntion, CfDCa. Overview of Testing for SARS-CoV-2 (COVID-19). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html (accessed December 29, 2020).
9. Shibata S, Ishiguro T, Kobayashi Y, Koike M, Numano T, Shimizu Y, et al. High incidence of false-positive results of IgG antibody against SARS-CoV-2 with rapid immunochromatographic antibody test due to human common cold coronavirus infection. Respiratory Med Case Rep. (2020) 31:101180. doi: 10.1016/j.rmcr.2020.101180
10. Zhang YV, Wiencek J, Meng QH, Theel ES, Babic N, Sepiashvili L, et al. AACC practical recommendations for implementing and interpreting SARS-CoV-2 emergency use authorization and laboratory-developed test serologic testing in clinical laboratories. Clin Chem. (2021) 67:1188–200. doi: 10.1093/clinchem/hvab051
11. Yang M, Cao S, Liu Y, Zhang Z, Zheng R, Li Y, et al. Performance verification of five commercial RT-qPCR diagnostic kits for SARS-CoV-2. Clin Chim Acta. (2022) 525:46–53. doi: 10.1016/j.cca.2021.12.004
12. Harritshoj LH, Gybel-Brask M, Afzal S, Kamstrup PR, Jorgensen CS, Thomsen MK, et al. Comparison of 16 serological SARS-CoV-2 immunoassays in 16 clinical laboratories. J Clin Microbiol. (2021) 59:e02596–20. doi: 10.1128/JCM.02596-20
13. Hassan H, Abo ElSood H, Abd ElGawad B, Kamel R, Fahim M, El Shourbagy S, et al. The value of contact tracing and isolation in mitigation of COVID-19 epidemic: findings from outbreak investigation of COVID-19 onboard Nile Cruise Ship, Egypt, March 2020. BMJ Global Health. (2022) 7:e008681. doi: 10.1136/bmjgh-2022-008681
14. Borro M, Salerno G, Montori A, Petrucca A, Anibaldi P, Marcolongo A, et al. SARS-CoV-2 transmission control measures in the emergency department: the role of rapid antigenic testing in asymptomatic subjects. Healthcare. (2022) 10:790. doi: 10.3390/healthcare10050790
15. Tay MZ, Ang LW, Wei WE, Lee VJM, Leo YS, Toh M. Health-seeking behavior of COVID-19 cases during the first eight weeks of the outbreak in Singapore: differences between local community and imported cases and having visits to single or multiple healthcare providers. BMC Public Health. (2022) 22:239. doi: 10.1186/s12889-022-12637-8
16. Li G, Lin J, Xu D. Epidemiological characteristics of COVID-19 and effective public health interventions in Shenzhen, China. Front Public Health. (2022) 10:923175. doi: 10.3389/fpubh.2022.923175
17. Mallett S, Allen AJ, Graziadio S, Taylor SA, Sakai NS, Green K, et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med. (2020) 18:346. doi: 10.1186/s12916-020-01810-8
18. Wang H, Ai J, Loeffelholz MJ, Tang YW, Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. (2020) 9:2200–11. doi: 10.1080/22221751.2020.1826362
19. Nakagama Y, Komase Y, Candray K, Nakagama S, Sano F, Tsuchida T, et al. Serological testing reveals the hidden COVID-19 burden among health care workers experiencing a SARS-CoV-2 nosocomial outbreak. Microbiol Spectr. (2021) 9:e0108221. doi: 10.1128/Spectrum.01082-21
20. Goldberg SA, Lennerz J, Klompas M, Mark E, Pierce VM, Thompson RW, et al. Presymptomatic transmission of severe acute respiratory syndrome coronavirus 2 among residents and staff at a skilled nursing facility: results of real-time polymerase chain reaction and serologic testing. Clin Infect Dis. (2021) 72:686–9. doi: 10.1093/cid/ciaa991
21. Ng OT, Marimuthu K, Koh V, Pang J, Linn KZ, Sun J, et al. SARS-CoV-2 seroprevalence and transmission risk factors among high-risk close contacts: a retrospective cohort study. Lancet Infect Dis. (2021) 21:333–43. doi: 10.1016/S1473-3099(20)30833-1
22. Perico L, Tomasoni S, Peracchi T, Perna A, Pezzotta A, Remuzzi G, et al. COVID-19 and lombardy: TESTing the impact of the first wave of the pandemic. EBioMedicine. (2020) 61:103069. doi: 10.1016/j.ebiom.2020.103069
23. Mitani A, Horie T, Yokoyama R, Nakano Y, Hamada K, Inoue Y, et al. Interpretations of SARS-CoV-2 IgM and IgG antibody titers in the seroepidemiological study of asymptomatic healthy volunteers. J Infect Chemother. (2022) 28:266–72. doi: 10.1016/j.jiac.2021.11.020
24. Escribano P, Álvarez-Uría A, Alonso R, Catalán P, Alcalá L, Muñoz P, et al. Detection of SARS-CoV-2 antibodies is insufficient for the diagnosis of active or cured COVID-19. Sci Rep. (2020) 10:19893. doi: 10.1038/s41598-020-76914-5
Keywords: SARS-CoV-2, serologic test, PCR, screening algorithm, yield, efficiency
Citation: Fang L-L, Zhu J-H, Cai M-J, Zhang J-W, Jiang L-C, Dai Z, Lin Y and Liang X-M (2023) PCR combined with serologic testing improves the yield and efficiency of SARS-CoV-2 infection hunting: A study in 40,689 consecutive overseas arrivals. Front. Public Health 11:1077075. doi: 10.3389/fpubh.2023.1077075
Received: 22 October 2022; Accepted: 17 January 2023;
Published: 13 February 2023.
Edited by:
Reza Lashgari, Shahid Beheshti University, IranReviewed by:
Yasutoshi Kido, Osaka City University, JapanClaudio Acuña-Castillo, University of Santiago of Chile, Chile
Copyright © 2023 Fang, Zhu, Cai, Zhang, Jiang, Dai, Lin and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Ming Liang,  MTc4MjkyOTU4QHFxLmNvbQ==
MTc4MjkyOTU4QHFxLmNvbQ==
 Li-Li Fang
Li-Li Fang Jian-Hui Zhu3,4
Jian-Hui Zhu3,4 Yu Lin
Yu Lin Xian-Ming Liang
Xian-Ming Liang