- 1Department of Family Medicine, Zhongxing Branch, Taipei City Hospital, Taipei, Taiwan
- 2Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei, Taiwan
- 3General Education Center, University of Taipei, Taipei, Taiwan
- 4Institute of Computer Science and Information Engineering, National Taiwan University, Taipei, Taiwan
- 5Department of Orthopedics, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 6Department of Orthopedics, Shuang Ho Hospital, Taipei Medical University, New Taipei, Taiwan
- 7Graduate Institute of Biomedical Optomechatronics, College of Biomedical Engineering, Research Center of Biomedical Device, Taipei Medical University, Taipei, Taiwan
- 8International Ph.D. Program in Biomedical Engineering, College of Biomedical Engineering, Taipei Medical University, Taipei, Taiwan
- 9Department of Orthopedics, Cathay General Hospital, Taipei, Taiwan
- 10Department of Orthopedics, Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan
- 11School of Medicine, College of Medicine, Fu Jen Catholic University, Taipei, Taiwan
- 12Department of Biomedical Engineering, National Taiwan University, Taipei, Taiwan
- 13Department of Orthopaedics, Ditmanson Medical Foundation Chia-Yi Christian Hospital, Chiayi, Taiwan
- 14Department of Long-Term Care and Management, WuFeng University, Chiayi, Taiwan
- 15Department of Orthopaedic Surgery, Taoyuan General Hospital, Ministry of Health and Welfare, Taoyuan, Taiwan
- 16Department of Orthopaedic Surgery, National Taiwan University Hospital, Taipei, Taiwan
- 17Department of Nursing, Yuanpei University of Medical Technology, Hsinchu, Taiwan
- 18Department of Orthopedics, Postal Hospital, Taipei, Taiwan
- 19Department of Physical Medicine and Rehabilitation, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 20Division of Nephrology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 21TMU Biodesign Center, Taipei Medical University, Taipei, Taiwan
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic can be effectively controlled by rapid and accurate identification of SARS-CoV-2-infected cases through large-scale screening. Hypercube pooling polymerase chain reaction (PCR) is frequently used as a pooling technique because of its high speed and efficiency. We attempted to implement the hypercube pooling strategy and found it had a large quantization effect. This raised two questions: is hypercube pooling with edge = 3 actually the optimal strategy? If not, what is the best edge and dimension? We used a C++ program to calculate the expected number of PCR tests per patient for different values of prevalence, edge, and dimension. The results showed that every edge had a best performance range. Then, using C++ again, we created a program to calculate the optimal edge and dimension required for pooling samples when entering prevalence into our program. Our program will be provided as freeware in the hope that it can help governments fight the SARS-CoV-2 pandemic.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has drastically changed the life of people worldwide. Currently, there are two strategies that are being used to contain the spread of the virus. One method is to impose strict restrictions on normal activities, such as China and many other countries imposing severe lockdowns. The other is to achieve high vaccination coverage. For example, the vaccination coverages in the United Kingdom and Unites States are both relatively high.
Effective control of SARS-CoV-2 infections requires rapid and precise identification of infected people through large-scale polymerase chain reaction (PCR) tests. However, no country has enough time and resources to test each individual sample separately. One efficient alternative, which many countries have adopted, is to pool samples and test them together. Many pooling strategies and techniques have been proposed (1–35).
Mutesa et al. proposed hypercube pooling, which is both efficient and fast (36, 37). The authors proposed edge three to be the most suitable edge, and the dimension can be calculated by ln (0.35/p) where p is the prevalence. Then, pooling is illustrated in Figure 1.
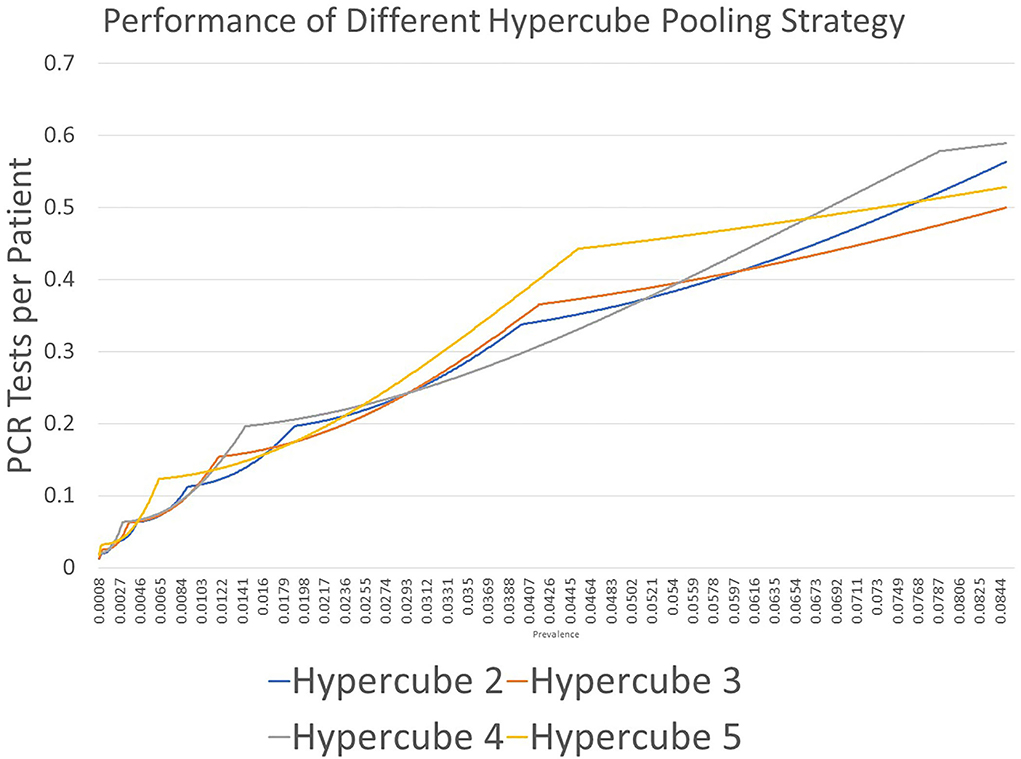
Figure 1. Results of PCR tests for each patient using the hypercube pooling strategy with edge values of 2 (Hypercube 2), 3 (Hypercube 3), 4 (Hypercube 4), and 5 (Hypercube 5). The quantization effects are displayed. Every strategy has an area in which its performance is strong.
We observed a significant quantization effect during the use of hypercube pooling, which dramatically increases the efficiency despite a minor prevalence change. This raised our interest to explore the underlying mechanism of this method. Mutesa et al. (36, 37) suggested that using edge three in hypercubic pooling should be the most efficient strategy. In that case, we wondered the following: (1) if the quantization effect does influence efficiency, is hypercube pooling with edge three really the best strategy? And (2), if the answer is no, what is the best edge and dimension?
Methods
The approach to selecting the estimated number of PCR tests per patient was first constructed in accordance with the concept of the hypercubic method and its variables: prevalence, edge, and dimension.
By using probability mathematics, the formula for the estimated number of PCR tests per patient was established as follows:
where n is the edge, d is the dimension, and p is the prevalence.
We used n = 2, 3, 4, and 5 in Formula (1) to determine which strategy works best under various values for the disease prevalence p.
C++ was used for implementation.
By leveraging a computer's computational ability, we simply computed every test possibility by using the brute force method. We then assigned dimension = 2 and edge = 2 in the formula to yield the maximal edge and dimension, respectively. Subsequently, we used the formula to calculate the estimates from edge = 2 to the maximal edge and from dimension = 2 to the maximal dimension, and then we chose the smallest number from all the estimated numbers of PCR tests per patient (1).
Results
Figure 2 shows the result of the first part of the study. We demonstrated that every proposed strategy had an area in which its performance was best. To obtain the best edge and dimension for the highest performance under a specific prevalence, we needed to conduct the second part of the investigation. For example, using edge = 2 (Hypercube 2 in Figure 2) resulted in the lowest estimate for the number of PCR tests per patient at prevalence of 0.0123–0.0537; at prevalence between 0.0261 and 0.0744, edge = 3 (Hypercube 3 in Figure 2) resulted in the lowest estimate.
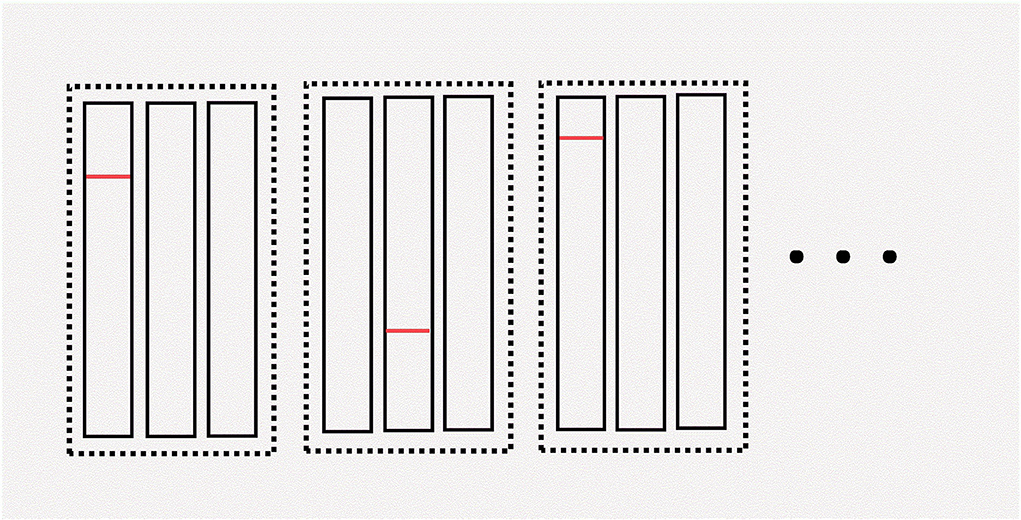
Figure 2. Illustration of hypercube pooling. The “dimension” is the total number of dotted rectangles. The “edge” is the number of rectangles within each dotted rectangle. The red line in this figure denotes a positive sample. In this example, the first pool in the first group, the second pool in the second group, and the first pool in the third group are all positive. We can determine which patient is positive during this round of pooling by using the hypercube pooling strategy. For more details about hypercube pooling, the readers can refer to previous literature (36, 37).
Therefore, we wrote a program for the second part of the study. The program uses the brute force method that examines every possible dimension number and edge number. Then, we calculated its performance by Formula (1). Finally, we chose the one which performs the best. When we entered the prevalence, the program calculated the best performance edge, dimension, and estimated number of PCR tests per patient (Figure 3).
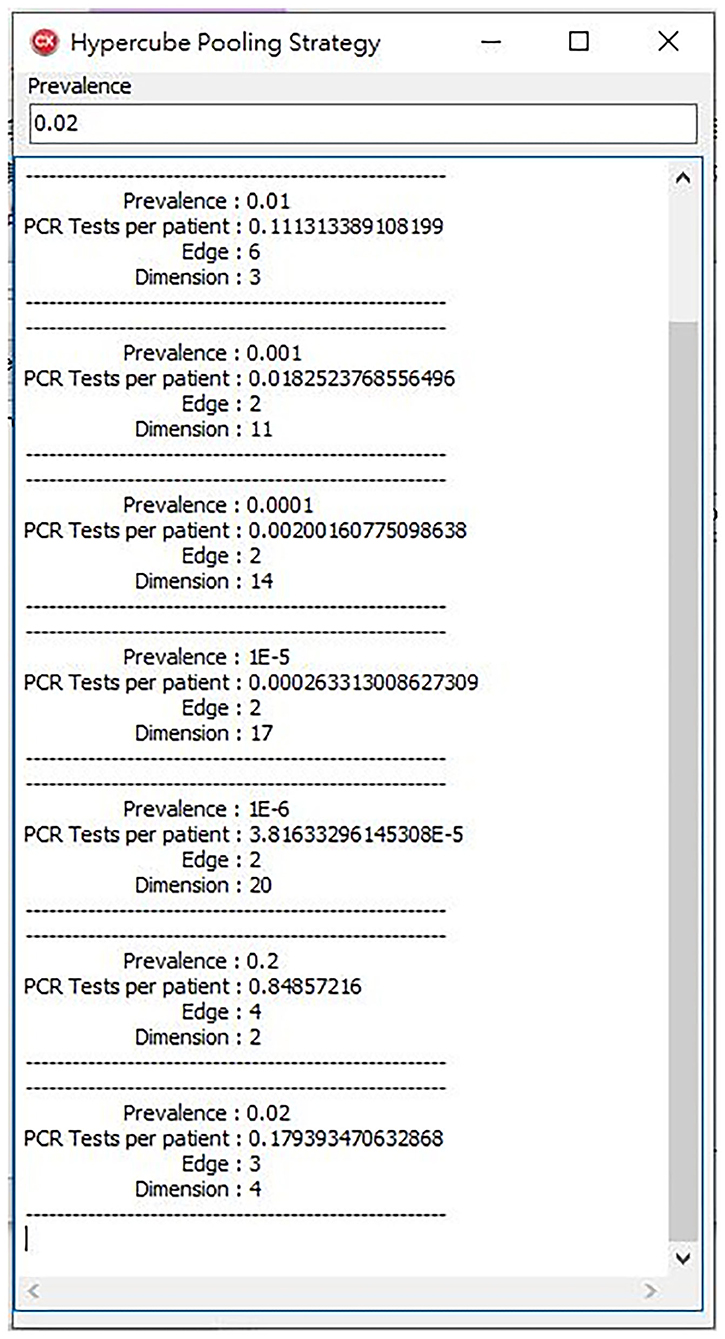
Figure 3. Our program offers a high-performance hypercube pooling strategy. The dimension, edge, and expected number of PCR tests per patient are the output variables.
Discussion
Various pooling techniques have been described, such as single pooling, n n double pooling, n2 n double pooling, and array pooling. In our opinion, one-dimensional hypercube pooling is single pooling, and array pooling is two-dimensional pooling.
Some pooling strategies—such as no pooling, array pooling, and hypercube pooling—can identify infected patients with a single round of PCR tests. According to a study by Mutesa et al., we can be 96% certain of positive cases in just the first round of pooled PCR tests (36, 37). Only 4% require second-round PCR tests. Hypercube pooling is a fast and efficient pooling method. Furthermore, a similar pooling approach was proposed almost at the same time (1). However, the authors did not explicitly mention the method or how it calculated the optimal pooling size. In this study, we made the complex probability mathematics more calculable by switching it and writing it into a program. Our method is much simpler and clearer than other methods.
Every PCR machine that uses a specific primer has a different maximal allowable dilution. However, the cutoff cycle threshold (Ct) can also change the maximum dilution. Therefore, we should conduct a pilot study to understand the limits of our machine. Additionally, we cannot pool more samples during hypercube pooling than the maximal allowable dilution.
Another challenge in the use of this method is the high number of dimensions. Two dimensions or even three dimensions are easy to imagine and visualize, but, in higher dimensions, we may need to code every sample with a specific number to identify individuals with infection accurately and continue the hypercube pooling process.
The prevalence of infection is unknown when conducting large-scale screening. Therefore, we recommend to begin pooling with edge = 2, which produces the maximum dimension that satisfies Equation (2). Subsequently, the prevalence can be calculated, and the strategy for the rest of the tests can be adjusted.
However, pooling may not yield significant benefits in areas with high disease prevalence. If a prevalence value is entered into the software and the expected number of PCR tests per patient is greater than one, pooling is not necessary. Instead, pooling will waste time and resources.
Conclusion
This pooling approach for SARS-CoV-2 detection is hypercube pooling PCR with an optimal edge and a dimension. This type of pooling can be completed in only one round of testing. We offer a tool to calculate the edge and the dimension required for pooling. We hope that it can be widely used, especially in large-scale screening, to better detect SARS-CoV-2 cases.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
CK-L, T-YW, and Y-CL conceptualized the study and wrote the paper. CK-L, P-WW, J-YW, C-YC, and Y-MH helped acquire the funding. Y-CL deduced the mathematics. C-SF, C-PC, Y-LC, C-KC, and K-LY wrote an algorithm. C-HY, H-KW, W-PL, T-HL, M-SW, and Y-MH simulated data and tested. All authors edited the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grants from the Taiwan Ministry of Science and Technology (Grant No. 109-2314-B-038-029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Shental N, Levy S, Wuvshet V, Skorniakov S, Shalem B, Ottolenghi A, et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci Adv. (2020) 6:eabc5961. doi: 10.1126/sciadv.abc5961
2. Aubry M, Teiti I, Teissier A, Richard V, Mariteragi-Helle T, Chung K, et al. Self-collection and pooling of samples as resources-saving strategies for RT-PCR-based SARS-CoV-2 surveillance, the example of travelers in French Polynesia. PLoS ONE. (2021) 16:e0256877. doi: 10.1371/journal.pone.0256877
3. Barak N, Ben-Ami R, Sido T, Perri A, Shtoyer A, Rivkin M, et al. Lessons from applied large-scale pooling of 133, 816 SARS-CoV-2 RT-PCR tests. Sci Transl Med. (2021) 13:eabf223. doi: 10.1101/2020.10.16.20213405
4. Bateman AC, Mueller S, Guenther K, Shult P. Assessing the dilution effect of specimen pooling on the sensitivity of SARS-CoV-2 PCR tests. J Med Virol. (2021) 93:1568–72. doi: 10.1002/jmv.26519
5. Chhikara K, Kanta P, Ghosh A, Prakash RC, Goyal K, Singh MP. Validation of SARS CoV-2 detection by real-time PCR in matched pooled and deconvoluted clinical samples before and after nucleic acid extraction: a study in tertiary care hospital of North India. Diagn Microbiol Infect Dis. (2021) 99:115206. doi: 10.1016/j.diagmicrobio.2020.115206
6. Chu AWH, Yip CCY, Chan WM, Ng ACK, Chan DLS, Siu RHP, et al. Evaluation of an automated high-throughput liquid-based RNA extraction platform on pooled nasopharyngeal or saliva specimens for SARS-CoV-2 RT-PCR. Viruses. (2021) 13:615. doi: 10.3390/v13040615
7. Cruceriu D, Baldasici O, Balacescu L, Gligor-Popa S, Flonta M, Man MA., et al. Critical aspects concerning the development of a pooling approach for SARS-CoV-2 diagnosis using large-scale PCR testing. Viruses. (2021) 13:902. doi: 10.3390/v13050902
8. Daon Y, Huppert A, Obolski U. An accurate model for SARS-CoV-2 pooled RT-PCR test errors. R Soc Open Sci. (2021) 8:210704. doi: 10.1098/rsos.210704
9. Estevez A, Catalan P, Alonso R, Marin M, Bouza E, Munoz P, et al. Sample pooling is efficient in PCR testing of SARS-CoV-2: a study in 7400 healthcare professionals. Diagn Microbiol Infect Dis. (2021) 100:115330. doi: 10.1016/j.diagmicrobio.2021.115330
10. Gangwar M, Shukla A, Patel VK, Prakash P, Nath G. Assessment of successful qRT-PCR of SARS-CoV-2 assay in pool screening using isopropyl alcohol purification step in RNA extraction. Biomed Res Int. (2021) 2021:6653950. doi: 10.1155/2021/6653950
11. Garg A, Ghoshal U, Patel SS, Singh DV, Arya AK, Vasanth S, et al. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. (2021) 93:2281–6. doi: 10.1002/jmv.26691
12. Garg J, Garg A. Evaluation of SARS CoV-2 RT-PCR in a multiple sample pool. Indian J Med Res. (2021) 153:699–700. doi: 10.4103/ijmr.IJMR_4282_20
13. Garg J, Singh V, Pandey P, Verma A, Sen M, Das A, et al. Evaluation of sample pooling for diagnosis of COVID-19 by real time-PCR: a resource-saving combat strategy. J Med Virol. (2021) 93:1526–31. doi: 10.1002/jmv.26475
14. Handous I, Hannachi N, Marzouk M, Hazgui O, Ben Alaya NBE, Boukadida J. Pooling nasopharyngeal swab specimens to improve testing capacity for SARS-CoV-2 by real-time RT-PCR. Biol Proced Online. (2021) 23:19. doi: 10.1186/s12575-021-00156-6
15. Klenske ED. Optimal test pooling for efficient PCR testing of SARS-CoV2. Ir J Med Sci. (2021) 190:481–2. doi: 10.1007/s11845-020-02278-4
16. Laverack M, Tallmadge RL, Venugopalan R, Cronk B, Zhang XL, Rauh R, et al. Clinical evaluation of a multiplex real-time RT-PCR assay for detection of SARS-CoV-2 in individual and pooled upper respiratory tract samples. Arch Virol. (2021) 166:2551–61. doi: 10.1007/s00705-021-05148-1
17. Lu XY, Sakthivel SK, Wang LJ, Lynch B, Dollard SM. Enhanced throughput of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) real-time RT-PCR panel by assay multiplexing and specimen pooling. J Virol Methods. (2021) 293:114149. doi: 10.1016/j.jviromet.2021.114149
18. More S, Narayanan S, Patil G, Ghosh P, Pushparaj S, Cooper E, et al. Pooling of nasopharyngeal swab samples to overcome a global shortage of real-time reverse transcription-PCR COVID-19 test kits. J Clin Microbiol. (2021) 59:e01295-20. doi: 10.1128/JCM.01295-20
19. Mungmunpuntipantip R, Wiwanitkit V. Pooled Sample RT-PCR Test for COVID-19 screening: cost reduction analysis. Clin Lab. (2021) 67:1963–4. doi: 10.7754/Clin.Lab.2021.201232
20. Musumeci A, Vinci M, L'episcopo F, Ragalmuto A, Neri V, Roccella M, et al. Implementation of sample pooling procedure using a rapid SARS-CoV-2 diagnostic real-time PCR test performed prior to hospital admission of people with intellectual disabilities. Int J Environ Res Public Health. (2021) 18:9317. doi: 10.3390/ijerph18179317
21. Praharaj I, Jain A, Singh M, Balakrishnan A, Dhodapkar R, Borkakoty B, et al. Evaluation of SARS CoV-2 RT-PCR in a multiple sample pool reply. Indian J Med Res. (2021) 153:700–1.
22. Sahajpal NS, Mondal AK, Ananth S, Njau A, Ahluwalia P, Newnam G, et al. SalivaSTAT: direct-PCR and pooling of saliva samples collected in healthcare and community setting for SARS-CoV-2 mass surveillance. Diagnostics. (2021) 11:904. doi: 10.3390/diagnostics11050904
23. Sanghani HR, Nawrot DA, Marmolejo-Cossio F, Taylor JM, Craft J, Kalimeris E, et al. Concentrating pooled COVID-19 patient lysates to improve reverse transcription quantitative PCR sensitivity and efficiency. Clin Chem. (2021) 67:797–8. doi: 10.1093/clinchem/hvab035
24. Shukla S, Upadhyay V, Maurya VK. Evaluating the efficiency of specimen (sample) pooling for real-time PCR based diagnosis of COVID-19. Indian J Med Microbiol. (2021) 39:339–42. doi: 10.1016/j.ijmmb.2021.03.011
25. Singh AK, Nema RK, Joshi A, Shankar P, Gupta S, Yadav AK, et al. Testing of four-sample pools offers resource optimization without compromising diagnostic performance of real time reverse transcriptase-PCR assay for COVID-19. PLoS ONE. (2021) 16:e0251891. doi: 10.1371/journal.pone.0251891
26. Sun Q, Li J, Ren H, Pastor L, Loginova Y, Madej R, et al. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLoS ONE. (2021) 16:e0243183. doi: 10.1371/journal.pone.0243183
27. Yip CCY, Leung KH, Ng ACK, Chan KH, To KKW, Chan JFW, et al. Comparative evaluation of a dual-target real-time RT-PCR assay for COVID-19 diagnosis and assessment of performance in pooled saliva and nasopharyngeal swab samples. Expert Rev Mol Diagn. (2021) 21:741–7. doi: 10.1080/14737159.2021.1933445
28. Yu JL, Huang YD, Shen ZJ. Optimizing and evaluating PCR-based pooled screening during COVID-19 pandemics. Sci Rep. (2021) 11:21460. doi: 10.1038/s41598-021-01065-0
29. Barathidasan R, Sharmila FM, Raj RV, Dhanalakshmi G, Anitha G, Dhodapkar R. Pooled sample testing for COVID-19 diagnosis: evaluation of bi-directional matrix pooling strategies. J Virol Methods. (2022) 304:114524. doi: 10.1016/j.jviromet.2022.114524
30. Esteves E, Mendes AK, Barros M, Figueiredo C, Andrade J, Capelo J, et al. Population wide testing pooling strategy for SARS-CoV-2 detection using saliva. PLoS ONE. (2022) 17:e0263033. doi: 10.1371/journal.pone.0263033
31. Hueda-Zavaleta M, Copaja-Corzo C, Benites-Zapata VA, Cardenas-Rueda P, Maguina JL, Rodriguez-Morales AJ. Diagnostic performance of RT-PCR-based sample pooling strategy for the detection of SARS-CoV-2. Ann Clin Microbiol Antimicrob. (2022) 21:11. doi: 10.1186/s12941-022-00501-x
32. Jayakody H, Rowland D, Pereira C, Blackwell R, Lasota T, Laverick M, et al. Development of a high sensitivity RT-PCR assay for detection of SARS-CoV-2 in individual and pooled nasopharyngeal samples. Sci Rep. (2022) 12:5369. doi: 10.1038/s41598-022-09254-1
33. Lee HR, Wang FY, Li JL, Chou TY, Ho HL. Real-world evaluation of a sample pooling strategy for large-scale rapid COVID-19 testing. J Clin Virol. (2022) 149:105133. doi: 10.1016/j.jcv.2022.105133
34. Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, Cantu-Cuevas MA, Madrid-Gonzalez DA, Baron-Olivares H, et al. Pooling saliva samples as an excellent option to increase the surveillance for SARS-CoV-2 when re-opening community settings. PLoS ONE. (2022) 17:e0263114. doi: 10.1371/journal.pone.0263114
35. Perea S, Tretina K, O'donnell KN, Love R, Bethlendy G, Wirtz M, et al. Saliva-based, COVID-19 RT-PCR pooled screening strategy to keep schools open. Disaster Med Public Health Prep. (2022) 589:1–6. doi: 10.1017/dmp.2021.337
36. Mutesa L, Ndishimye P, Butera Y, Souopgui J, Uwineza A, Rutayisire R, et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. (2021) 589:276–80. doi: 10.1038/s41586-020-2885-5
Keywords: pooling PCR, hypercube pooling, SARS-CoV-2, COVID-19, one round pooling PCR
Citation: Wu T-Y, Liao Y-C, Fuh C-S, Weng P-W, Wang J-Y, Chen C-Y, Huang Y-M, Chen C-P, Chu Y-L, Chen C-K, Yeh K-L, Yu C-H, Wu H-K, Lin W-P, Liou T-H, Wu M-S and Liaw C-K (2022) An improvement of current hypercube pooling PCR tests for SARS-CoV-2 detection. Front. Public Health 10:994712. doi: 10.3389/fpubh.2022.994712
Received: 15 July 2022; Accepted: 20 September 2022;
Published: 19 October 2022.
Edited by:
Reza Lashgari, Shahid Beheshti University, IranReviewed by:
Linda R. Lara-Jacobo, San Diego State University, United StatesAnand Maurya, Banaras Hindu University, India
Copyright © 2022 Wu, Liao, Fuh, Weng, Wang, Chen, Huang, Chen, Chu, Chen, Yeh, Yu, Wu, Lin, Liou, Wu and Liaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen-Kun Liaw, Y2hlbmt1bmxpYXdAdG11LmVkdS50dw==; Y2hlbmt1bmxpYXdAZ21haWwuY29t
 Tai-Yin Wu1,2,3
Tai-Yin Wu1,2,3 Kuei-Lin Yeh
Kuei-Lin Yeh Wei-Peng Lin
Wei-Peng Lin Tsan-Hon Liou
Tsan-Hon Liou Chen-Kun Liaw
Chen-Kun Liaw