- 1Department of Paediatrics I, “George Emil Palade” University of Medicine, Pharmacy, Sciences, and Technology of Târgu Mureş, Târgu Mureş, Romania
- 2Department of Paediatrics III, “George Emil Palade” University of Medicine, Pharmacy, Sciences, and Technology of Târgu Mureş, Târgu Mureş, Romania
- 3Center for Advanced Medical and Pharmaceutical Research, “George Emil Palade” University of Medicine, Pharmacy, Science, and Technology of Târgu Mureş, Târgu Mureş, Romania
Pediatric obesity presents a multifactorial etiology, which involves genetic traits as well, including single nucleotide polymorphisms. The aim of the study is to investigate the contribution of PPARG gene polymorphisms (namely Pro12Ala rs1801282, His447His rs3856806, and Pro115Gln rs1800571) and PPARGC1A rs8192678 SNP on the anthropometric and metabolic parameters in a population of Romanian children. We conducted a cross-sectional study of 295 Caucasian children, divided according to the body mass index (BMI) z-score into the study (obese and overweight) group of 130 children and the control (normoponderal) group of 165 children. Anthropometric parameters were greater in the obese and overweight population as opposed to controls, with significant differences (p < 0.01) found for the weight (2.77 ± 1.54 SD vs. −0.04 ± 1.15 SD), body mass index (BMI) (2.28 ± 0.97 SD vs. −0.18 ± 1.19 SD), mid-upper arm circumference (MUAC) (4.59 ± 2.28 SD vs. 0.28 ± 3.45 SD), tricipital skin-fold (TSF) (3.31 ± 3.09 SD vs. 0.62 ± 7.28 SD) and waist-to-height ratio (WHtR) (0.61 ± 1.51 SD vs. −0.35 ± 1.35 SD) z-scores. Moreover, triglyceride values were higher in the study group (118.70 ± 71.99 SD vs. 77.09 ± 37.39 SD). No significant difference in the allele and genotype distribution of investigates gene polymorphisms was observed between the studied groups (p > 0.05). PPARG (rs1801282, rs3856806, and rs1800571) were not associated with demographic, anthropometric, and laboratory parameters. However, PPARGC1A rs8192678 CC genotype was associated with TSF z-score (p = 0.03), whereas total and LDL cholesterol levels were significantly higher among TT homozygotes (p < 0.01). Our data suggest that PPARG (rs1801282, rs3856806, and rs1800571) and PPARGC1A (rs8192678) gene polymorphisms were not associated with childhood and adolescence overweight and obesity. The present study identified a significant increase in fasting glucose levels, triglyceride, albumin, and ALT levels in children with excess weight, as well as expected important upward variation of anthropometric parameters (BMI, MUAC, TSF z-scores).
Introduction
In the last four decades, we faced a dramatic, ten-fold increase in obesity incidence. The World Health Organization (WHO) estimates that nearly 1 in 5 children and adolescents are overweight or obese (1). Nutritional status across Europe is miscellaneous, but a high prevalence of childhood obesity has been reported throughout the entire continent, especially in Southern Europe (2, 3). Overweight and obesity prevalence numbers in children from Romania are comparable to other European countries, with one recent study showing that every 1 in 4 individuals aged 6–19 years are overweight or obese (2, 4).
A multifactorial etiology stands behind excessive weight gain, ranging from high caloric intake to sedentary lifestyle, hormonal imbalance, gut microbiota imbalance, or hereditary factors (5). A lot of attention has been given in the past years to the implications of genetic backgrounds in the development of obesity, with studies evolving around various genes and different populations (6–8). Contradictory theories have emerged regarding the role of gene polymorphisms in the onset of childhood obesity, given that some single nucleotide polymorphisms (SNPs) constitute protecting factors in the development of multiple chronic diseases. Thus, associations between polymorphisms of genes such as leptin receptor (LEPR), fat mass obesity-associated receptors (FTO), melanocortin-4 receptors (MC4R), peroxisome proliferator-activated receptor-gamma (PPARG), and weight gain in school-aged children have been intensely studied (9–11).
Peroxisome proliferator-activated receptors, in particular, are abundantly expressed in adipose tissue, are one of the candidate genes in terms of metabolic regulation, by modulating adipocyte differentiation, lipid oxidation as well as glucose homeostasis, and insulin sensitivity (12, 13). There are multiple isoforms of PPAR gamma, namely γ1, γ2, and γ3. PPARγ1 and 3 are highly expressed in adipose tissue and at a lower level in other tissues as well (liver, colon, heart, etc) while PPARγ2 (which presents an additional 28 amino acids sequence on the N-terminal of PPARγ1) is expressed almost only in adipose tissue (14).
Mapped to chromosome 3p25.2 (Online Mendelian Inheritance in Man—OMIM number 601487), PPARG contains 9 exons and spans over more than 100 kb (12). Its transcription activity is reduced by a missense mutation, namely a proline12-to-alanine (Pro12Ala) substitution in the PPARG gene (rs1801282), close to the NH2-terminal region of the protein (15). Two of the most common variants in the PPARG gene are the ones of Pro12Ala (rs1801282 C > G), as well as of His447His (rs3856806 C>T) polymorphisms (16). Several studies conducted on the adult population showed a positive association between PPARG Pro12Ala polymorphism (rs1801282) and obesity in adults, ultimately proving the involvement of the variant G allele (17, 18). Furthermore, a systematic review also pointed out a relationship between the Pro12Ala polymorphism and an increase in body mass index (BMI) in healthy adults (19). His447His (rs3856806), also known as C161T, the other frequently encountered polymorphism of PPARG, seems to be involved in the maintenance of lipid metabolism equilibrium, with a possible indirect link to an increased risk of coronary heart disease (CHD), especially in patients with type 2 diabetes mellitus (T2DM) (20–22). Furthermore, Ristow et al. showed that the PPARG Pro115Gln (rs1800571) missense variant is associated with severe obesity, as it determines an increased activity of the gene, thus leading to enhanced adipocyte differentiation (23). Contradictorily, another study conducted by Bluher et al. concluded that rs1800571 SNP did not have an important role in the development of insulin resistance and obesity (24).
A transcriptional coactivator of PPARG, the peroxisome proliferator-activated receptor-G coactivator 1-alpha (PPARGC1A) also plays an essential role in regulating energy metabolism. PPARGC1A acts on the nuclear receptor PPARG, thus interacting with transcription factors (25). The GLy482Ser (rs8192678) SNP is a variant in the protein coding sequence of PPARGC1A, whose deleterious effect has been related to obesity, as well as its associated complications, which include CHD, T2DM, non-alcoholic fatty liver disease (NAFLD), and arterial hypertension (26).
To date, there are little data on the involvement of PPARG SNPs in the development of obesity in children, and reports on the implications of gene polymorphisms in the process and consequences of excessive weight gain are even scarcer in Romanian populations. Thus, the current study aims to investigate the contribution of three PPARG polymorphisms (namely Pro12Ala rs1801282, His447His rs3856806, and Pro115Gln rs1800571) and PPARGC1A rs8192678 SNP on the anthropometric and metabolic parameters in a population of Romanian children.
Materials and methods
We conducted a prospective study on 295 Caucasian children aged between 3 months and 17 years, recruited within a timeframe of 36 months in a tertiary pediatric hospital from Tîrgu Mureş, Romania. Anthropometric parameters, biochemical traits, and SNPs of PPARG2 and PPARGC1A were cross-sectionally assessed in each child, during the first visit. The division of the population sample among the two study groups (study group- obese, plus overweight and controls- weight within normal ranges) was dependent upon the values of the body mass index (BMI) z score, as recommended by the WHO. Thus, in children aged under 5 years, a z score between 1 and 2 standard deviations (SDs) defined risk of overweight, a z score of at least 2 SD was suggestive of overweight and a z score of at least 3 SD was interpreted as suggestive of obesity, whereas in children older than 5 years, a z score between 1 and 2 SDs was indicative of overweight and one exceeding 2 SDs translated into obesity (27, 28). Z score was applied instead of percentiles due to recent studies supporting its use over percentiles in classifying childhood overweight and obesity (29). Comparison of anthropometric parameters, biochemical traits, and PPARG2 and PPARGC1A SNPs was therefore conducted between the two study groups.
Children included in the study were presented for regular check-ups or digestive complaints such as constipation, regurgitations, heartburn, abdominal pain, bloating. Patients with previously known chronic conditions or those who were diagnosed with food intolerances or chronic disorders of the digestive tract as a result of paraclinical investigations were excluded from the study.
Anthropometric parameters
Anthropometric measurements were done by a single qualified person. Weight was measured using a properly calibrated scale, while height was quantified with the help of a calibrated length board/stadiometer, depending on the subject's age. BMI was calculated by dividing weight (kg) by squared height (in meters). Weight, height, waist-to-height ratio (WHtR), BMI, mid-upper arm circumference (MUAC), and tricipital skin-fold (TSF) z-scores were calculated in accordance with WHO growth charts. All anthropometric measurements (weight, height, waist circumference (WC), hip circumference (HC), MUAC, and TSF were obtained from shoeless children wearing lightweight clothing by applying a method that had been described by the authors in their previous research (9, 10). A novel parameter, which can assess excess weight and central obesity status, namely waist-to-height ratio (WHtR) was also calculated and interpreted according to recent recommendations: normal values < 0.45, overweight for values ≥0.45, and obesity for values exceeding 0.5 (30).
Biochemical variables
Serum biochemical and metabolic traits from venous blood sample (glucose, total cholesterol (TC), high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c), triglycerides (TG), liver enzymes, total protein levels, fasting glucose levels, serum albumin) were assessed using standard laboratory methods (with the help of a Cobas Integra 6000 plus automated analyzer, Roche Diagnostics GmbH, Mannheim, Germany).
DNA extraction and genotyping
For DNA extraction 2 mm of venous blood were collected on tubes with Ethylenediamine tetraacetic acid (EDTA) as anticoagulated from each child included in the study. PureLink Genomic DNA Mini Kit (Invitrogen, Thermo Fischer Scientific, USA) was used to obtain genomic DNA. Genotyping of PPARG rs1801282, rs3856806, and rs180057 and PPARGC1A rs8192678 polymorphism were determined through PCR (polymerase chain reaction) followed by digestion with the restriction enzyme (RFLP restriction fragment length polymorphism method). The PCR conditions, primers, and restriction enzymes used were according to previously described protocols (31–34).
Statistical analysis
Statistical analysis was done using GraphPad Prism T software, version 9.0. Descriptive statistics were applied to quantitative variables and expressed as mean ± standard deviation (SD). The frequency of categorical variables was expressed in percentages (%). The number and frequency distributions of genetic polymorphisms were assessed using the Poisson random field (PRF) model (35). Shapiro-Wilk test was used to assess distribution characteristics of analyzed data. Consequently, mean comparison was conducted using the Mann-Whitney test for non-Gaussian distributed variables and Student's unpaired t-test for variables complying with a Gaussian distribution pattern. Comparison of multiple data sets was performed with the help of analysis of variance (ANOVA), namely after applying the non-parametric Kruskal-Wallis test. Categorical variables such as sex, genotype, allele frequency, as well as the Hardy-Weinberg equilibrium, were analyzed using the Chi-square test. Mul-tiple linear regressions were applied to assess the associ-ation between genotype frequency and children's BMI, on which the division of the study groups was conducted, while also taking different confounders into consideration. Adjustment for confounding factors consisted of two models, namely model A, in which maternal age, parity, weight (devised according to BMI), educational level and ethnicity was taken into account, and model B, in which paternal age, weight (devised according to BMI) and ethnicity were considered. The level of significance was set at p < 0.05, corresponding to a confidence interval of 95%.
Results
The study cohort consisted of 295 Caucasian children, who had a mean age of 10.24 ± 4.38 years and were divided in accordance with the BMI z score into two groups, a study (obese and overweight) group of 130 children (85 obese and 45 overweight children) and a control (normoponderal) group of 165 children. The comparison of demographic, anthropometric, and metabolic data of all subjects are presented in Table 1. There was no significant difference between the two groups in terms of mean age, but the male sex was significantly more prevalent among the study group (p < 0.01, OR = 2.10). Anthropometric parameters were expectably greater in the study population as opposed to controls, with significant differences (p < 0.01) found for weight (2.77 ± 1.54 SD vs. −0.04 ± 1.15 SD), BMI (2.28 ± 0.97 SD vs. −0.18 ± 1.19 SD), MUAC (4.59 ± 2.28 SD vs. 0.28 ± 3.45 SD), TSF (3.31 ± 3.09 SD vs. 0.62 ± 7.28 SD) and I/L (0.61 ± 1.51 SD vs. −0.35 ± 1.35 SD) z scores. The lipid profile comparison showed insignificant variation between the two groups of total HDL and LDL cholesterol levels, but triglyceride values were higher in the study group (118.70 ± 71.99 SD vs. 77.09 ± 37.39 SD). An ascending trend was also found for basal glucose (91.66 ± 19.33 SD vs. 83.13 ± 10.75 SD), ALT (31.18 ± 36.49 SD vs. 22.30 ± 15.86 SD), and total protein levels (4.64 ± 0.50 SD vs. 4.35 ± 0.55 SD) in obese and overweight children.
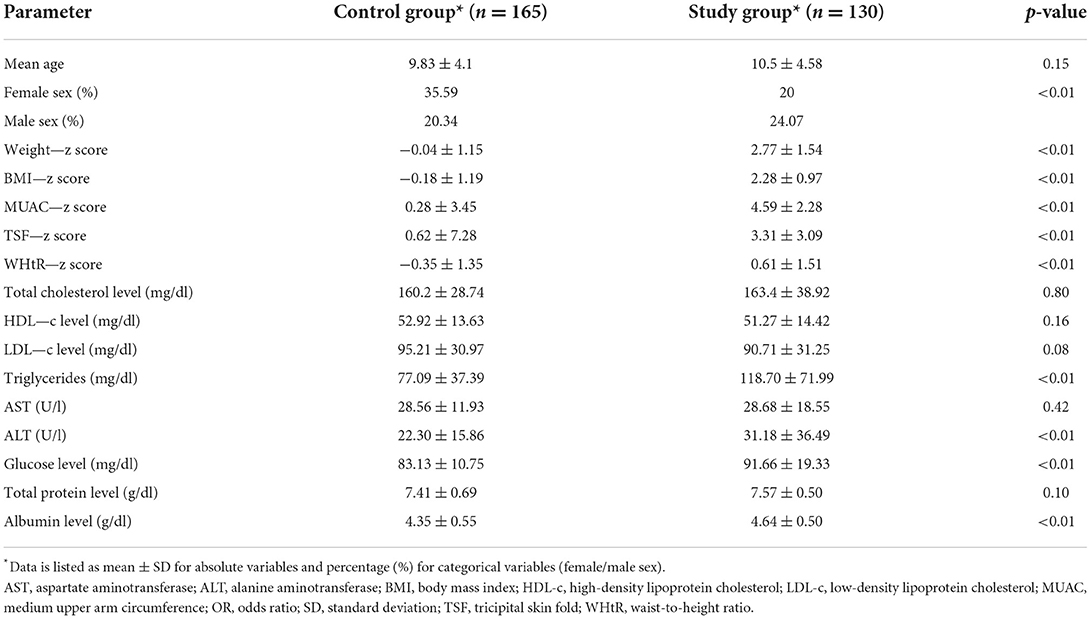
Table 1. Comparison of demographic, anthropometric, metabolic, and paraclinical data of the study population.
The genotype frequencies of the investigated PPARG (rs1801282, rs3856806, rs1800571) and PPARGC1A rs8192678 polymorphisms were in agreement with the Hardy–Weinberg equilibrium (p < 0.05). The comparative assessment between the two study groups of genotype and allele frequency for the four studied polymorphisms can be visualized in Table 2. Comparison of PPARG genotype and allele frequency for the three studied polymorphisms, namely rs1801282, rs3856806, rs1800571, revealed no significant dissimilarities. The rs1800571 (Pro115Gln) SNP was distinguished among others through an identification of the wild type homozygous genotype or the wild type allele among nearly every subject included in the study (Table 2). Assessment of the rs8192678 SNP of PPARGC1A, revealed the dominant frequency of the C allele within both the study and the control group but without a significant difference.
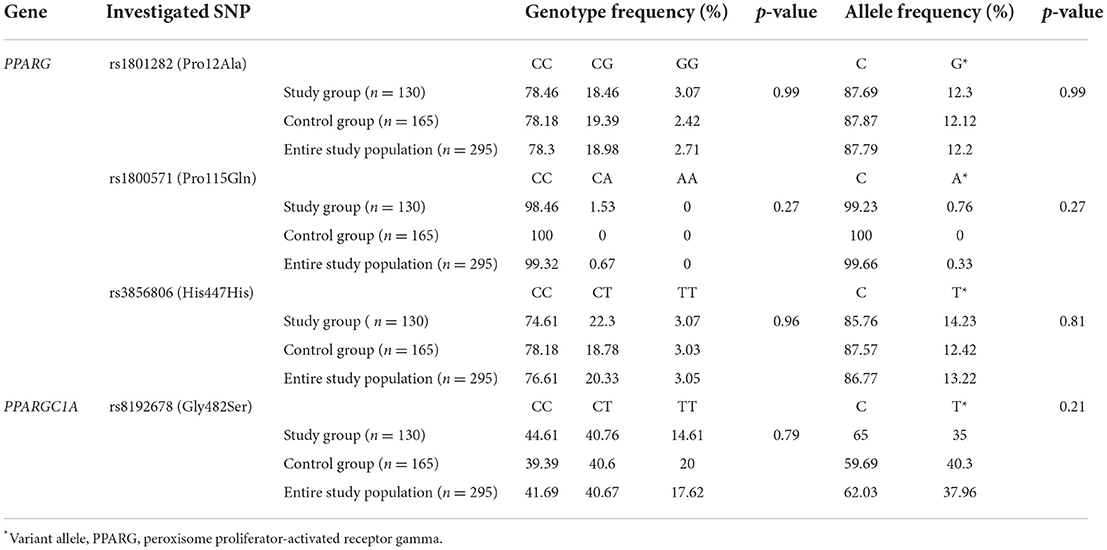
Table 2. Comparison of the genotype and allele frequencies of investigated SNPs within the entire study cohort, between the study (overweight/obese) group and control group.
Ethnicity, maternal and paternal characteristics, including age, weight (devised according to BMI), parity and educational level were compared between the two study groups (Table 3). There were no significant differences between the two groups in terms of parental age. However, parents of children belonging to the study group presented significantly higher prevalence of overweight and obesity. High maternal educational level and low parity represented protective factors against abnormally high child BMI (p < 0.01), whereas Roma ethnicity were more prone to overweight and obesity (p < 0.01, OR = 3.04). Race, maternal and parental characteristics were afterwards considered as confounding factors for assessment of correlation between genotype of the studied SNPs and patient's BMI. Multiple linear regression was applied using two different models, as further detailed in Table 4, but no significant results were obtained.
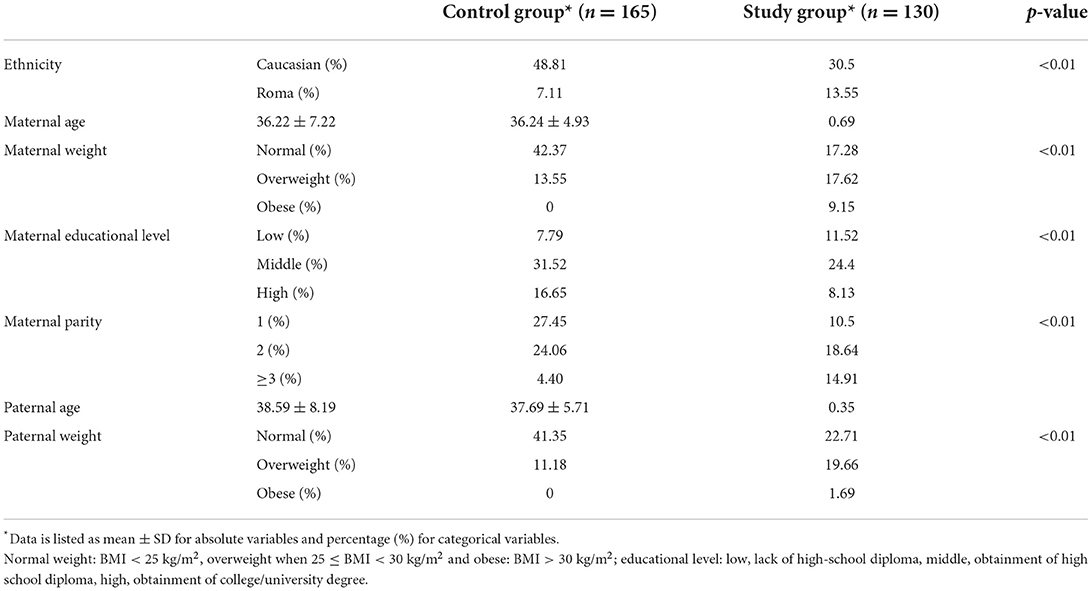
Table 3. Comparison of ethnicity, parental weight, age, maternal, parity and educational level between the two study groups.
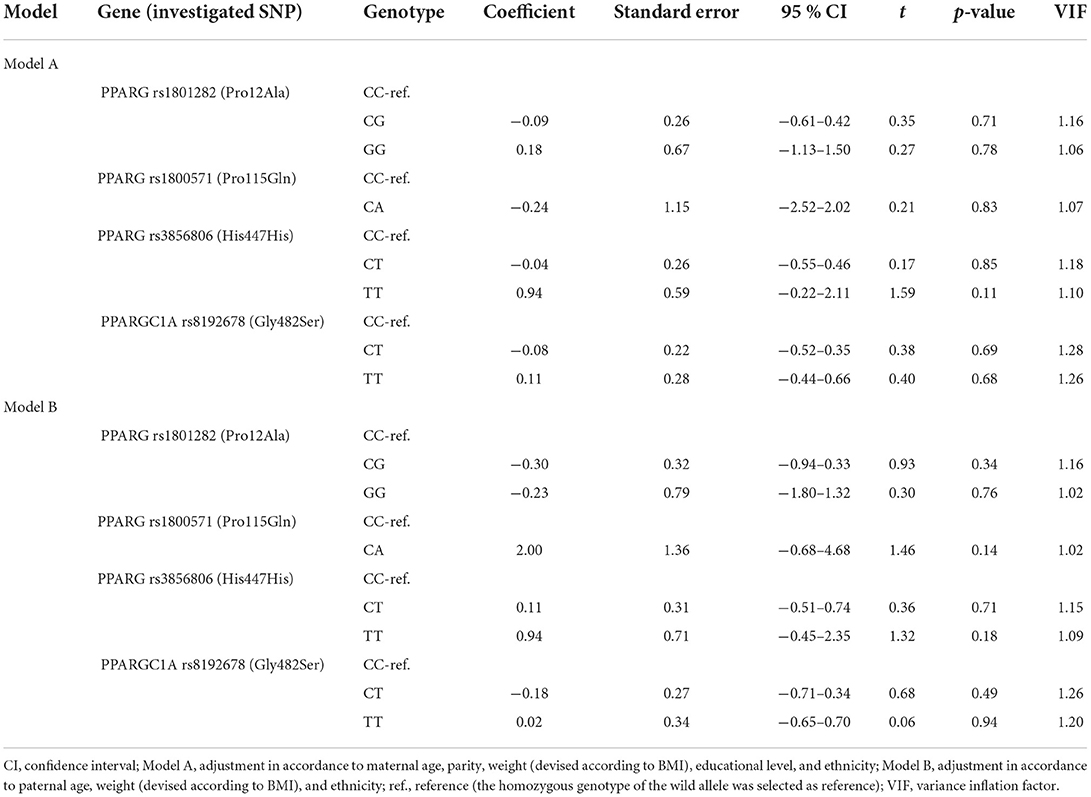
Table 4. Multiple linear regression for association between genotype frequency and patients' BMI, after adjustment for parental confounders using two models.
Comparison of demographic and laboratory parameters presented in Table 1 was also conducted by dividing the entire study population, as well as the obese/overweight group by genotype distribution. This analysis did not include rs1800571, due to the very limited number of subjects with variant heterozygous genotypes and the absence of subjects with variant homozygous genotypes. Analyzed parameters presented similar mean values, regardless of genotype in the case of PPARG rs1801282, rs3856806 SNPs. Genotype-based separation of demographic and paraclinical data showed no statistical significance for any of the studied parameters (Table 5). For PPARGC1A, the same analysis was performed by dividing the study subjects between CC homozygous genotype and both variant homozygous and heterozygous genotypes. Total and LDL cholesterol levels were significantly higher among TT homozygotes (p < 0.01).
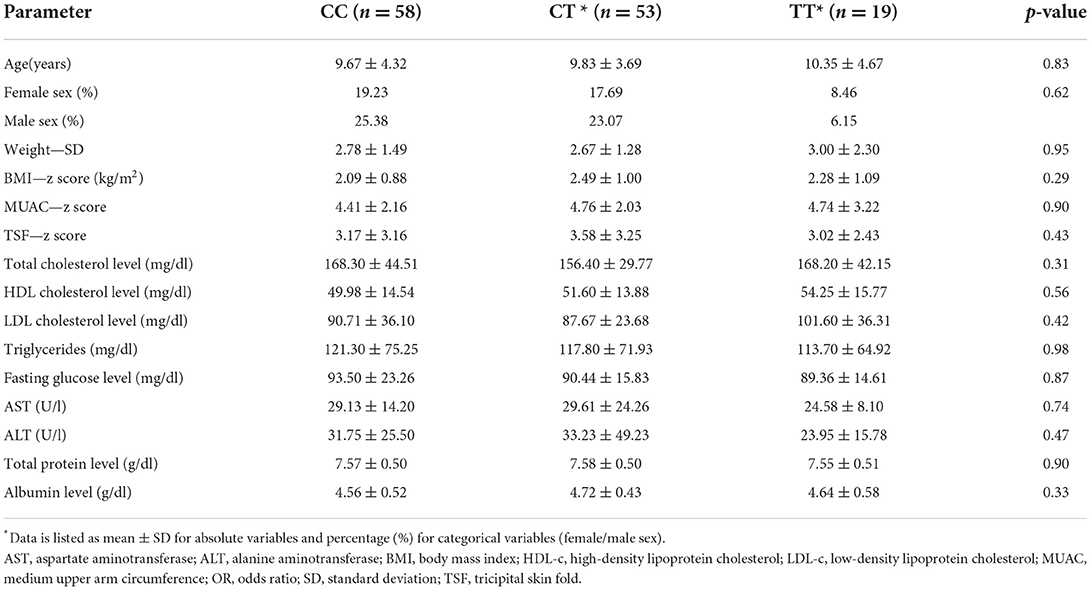
Table 5. Comparison of demographic, anthropometric, metabolic, and paraclinical data of the overweight/obese group divided in accordance with genotype distribution of PPARGC1A rs8192678 SNP.
The other parameters showed no significant allele-dependent variation, although an ascending trend can be seen for mean anthropometric constants, as well as for liver enzyme, total protein, albumin, triglyceride values, and fasting glucose levels. Furthermore, homozygotes for C allele presented higher mean values of the TSF (p = 0.03). The same type of analysis was carried out in the obese/overweight population group without obtaining statistically significant results.
Allele-based comparisons were also carried out both for PPARG rs1801282 (CC vs. CG + GG) and rs3856806 (CC vs. CT + TT) SNPs within the entire study population and within the obese/overweight group but analyzed parameter variation was insignificant with only one exception. An important increase in mean HDL levels was associated with homozygous CC genotype in the case of PPARG rs3856806 SNP (53.47 ± 14.96 SD vs. 44.77 ± 10.42 SD, p < 0.01), as illustrated in Figure 1.
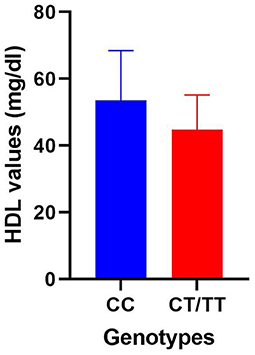
Figure 1. Comparison of HDL mean values within the obese/overweight group divided between homozygous CC genotype and a group consisting of variant heterozygous and homozygous genotypes for the PPARGC1A rs3856806 polymorphism.
A comparison of obesity/anthropometry parameters between subjects with concomitant presence of variant alleles and those with wild-type alleles was conducted for those three genotypes included in the study (PPARG rs1801282, PPARG rs3856806, and PPARGC1A rs8192678) in which an important prevalence of the variant allele was encountered. No significant discrepancies were found. Therefore, separate, similar, individual comparisons of anthropometric and obesity parameters were carried out for two polymorphisms of those three aforementioned. An important decrease in HDL levels was linked to the concomitant presence of variant alleles of PPARG rs3856806 and PPARGC1A rs8192678 (44.79 ± 10.23 SD vs. 51.97 ± 15.34 SD, p = 0.04). Variant allele carriers of both PPARG rs1801282 and PPARGC1A rs8192678 exhibited higher mean TSF values (3.96 ± 2.29 SD vs. 3.23 ± 3.35 SD, p = 0.03). Other similar comparisons for other parameters or the other two combinations of genotypes (PPARG rs1801282 and PPARG rs3856806) yielded insignificant results.
Discussion
Childhood and adolescence obesity can determine metabolic complications that persist through adulthood and translate into cardiovascular risk factors (36). Dyslipidemia, a predictor of atherosclerosis, is greatly influenced by physical activity level, dietary or genetic factors (37). It has been associated with anthropometric parameters, with many studies confirming a relationship between measured constants which are suggestive of an excessively represented adipose tissue, such as BMI, MUA, TSF, WC or HC (38). Still, the term “metabolically healthy obesity” (MHO) has recently emerged, and defines obese subjects whose triglycerides, high-density lipoprotein cholesterol, blood pressure, and fasting glucose levels are within normal ranges. Its prevalence has been reported at 31% in one pediatric study, but varies in accordance to definitions used and types of parameters evaluated (39, 40). In our study, we found no significant differences in terms of total cholesterol, HDL- or LDL-cholesterol values, but triglycerides presented an ascending trend in children with obesity as opposed to the normoponderal ones. The same pattern was observed for fasting glucose levels, in resonance with other studies, which also sustain the proven effect of weight loss on basal glucose values (41, 42). Moreover, a significant increase in ALT values was found in children with obesity, which might constitute an important, yet not single marker of childhood NAFLD (43).
Research regarding the involvement of SNPs in pediatric metabolic imbalances is still ongoing. SNPs seem to have an impact on glucose hemostasis, lipid metabolism, anthropometric parameters, leptin and adiponectin levels in subjects with obesity, starting from an early age (44–47). Several recent studies have highlighted the influence of SNPs in childhood obesity in Romania, with particular focus on FTO rs9939609, LEPR 223, and 1019 gene polymorphisms, interleukin 6 (IL-6) 572 C/G, 190 C/T and angiotensin-converting enzyme gene insertion/deletion (ACE I/D) gene polymorphisms (9, 10, 48, 49). However, the impact of PPARG SNPs on the obesity risk in children has been scarcely studied, literature data reporting mostly associations with adulthood obesity and its related complications (26, 50). PPARG gene expression has been negatively correlated with IL-6 and TNF-α, which are known to play an important role in regulating inflammatory status related to pediatric obesity (51). Furthermore, PPARG rs1801282 polymorphism has been controversially linked to a decrease in BMI, according to a study conducted in Portugal (11). Other research data supported an association between PPARG Pro12Ala (rs1801282) polymorphism and insulin resistance in Mexican children with dyslipidemia (52). Hence, the role of PPARG polymorphism in pediatric weight gain and its related complications still remains unclear.
Through this study we tried to investigate a possible association between three polymorphisms of the PPARG gene, as well as one polymorphism of the PPARGC1A gene and pediatric obesity and overweight.
Pediatric studies have focused on PPARG mostly. A very recent study in teenagers from the North of Mexico revealed that carriers of the PPARG G allele (Pro12Ala or rs1801282) present an increased risk for high WHtR or LDL-c (53). These results expectingly contradict our conclusions, in light of a scarcity of homozygous GG genotype within our study. The very low prevalence of homozygous genotype Ala/Ala (GG) frequency in obese and overweight children is in resonance with the one reported by Aiman et al. in their study that did not identify a single subject with a GG genotype for the PPARG Pro12Ala polymorphism. Moreover, the authors found a significant association between Pro/Pro (CC), heterozygous Pro/Ala (CG) carriers of Pro12Ala PPARG gene variant and a higher mean BMI, WC, and HC in overweight and obese Malay children population (54). On the other hand, in our research, the C allele (Pro12Ala or rs1801282 polymorphism) (although more prevalent as well within our study population) did not exhibit an influence upon anthropometric or biochemical parameters.
In addition, we investigated the presence of at least 2 variant genotype for PPARG (rs1801282, rs3856806, and rs1800571) polymorphisms in investigated children, in an attempt to assess their contribution in the determinism of excess weight but no relevant results were obtained, even after adjusting for potential parental confounders.
Polymorphism variation is dependent upon race, as proven by a nationwide study involving pre-school children from the United States of America which reported a higher prevalence of obesity among Hispanic populations as in Caucasians (55). This might explain miscellaneous results obtained by various authors which prove a lack of association between genetic SNPs of multiple genes, including PPARG and obesity. Queiroz et al. reported no association for PPARG rs1801282 and several anthropometric and metabolically-related parameters (BMI, WC, total cholesterol, LDL/HDL cholesterol, insulin, and HOMA-IR, body fat percentage, birth weight, systolic blood pressure), within a research article conducted on Brazilian children and adolescents (56). Two other reports described an insignificant impact of PPARG rs3856806 polymorphism on BMI value variations among children and adolescents (57, 58). In our study we failed to identify an association between three polymorphisms of PPARG and children with overweight and obesity. Our results are similar to those reported by Clement et al., whose study highlighted the lack of correlation between PPARG rs1800571 (Pro115Gln), rs1801282 (Pro12Ala) variants and obesity in French Caucasians. Furthermore, the study underlined the absence of the Pro115Gln mutation in the entire study population (59). Considering the low number of patients with rs1800571 variant (only two heterozygotes) within our study groups as well, the significance of this mutation within Caucasian populations is raised into question. Nevertheless, no association was found in another Portuguese study between PPARG polymorphisms and obesity in premenopausal women, which concludes this SNP may differ between populations and probably should not be considered as a strong genetic marker to evaluate risk for obesity as FTO rs9939609 has been reported (60).
Relevant associations between metabolic parameters, lipid profile, and PPARG polymorphisms have though been found, even among studies that could not conclude that the analyzed gene variants represent an obesity/overweight risk. Particularly, Vidovic et al. found lower mean values of HDL-c in the CT and TT genotype of PPARG rs3856806, despite of a lack of association between the aforementioned polymorphism and overweight/obesity among Serbian adolescents (57). These findings closely resemble results obtained in our study, in which CC genotype (PPARG rs3856806) was significantly associated with higher mean HDL-c values and carriers of variant alleles of both PPARG rs3856806 and PPARGC1A rs8192678 presented significantly lower serum HDL-c levels. Still, the authors also report higher fasting blood glucose levels within the same CT and TT genotype group (57), which we could not trace in our study. Contradictorily, Fan et al. reported a correlation between minor allele variants of rs3856806 and elevated LDL-c levels in Chinese adults (61). Once again, these miscellaneous results prove the race variation of genetic polymorphisms.
A previous study on unrelated German subjects demonstrated that the PPARG Pro115Gln (rs1800571) missense variant is possibly pathogenic for severe obesity (23). On the other hand, Hamer et al. claimed that the Pro115Gln polymorphism does not epidemiologically influence the prevalence of morbid obesity (62). Similarly, the same polymorphism did not seem to be related to obesity in Malaysian teenagers (63). Furthermore, the rs1800571 SNP did not contribute considerably to insulin resistance and obesity occurrence, according to Blüher et al. (24). In our research, variant allele of the PPARG rs1800571 was found in a very small number of subjects, in accordance with literature data performed on Caucasian populations. The presence of the PPARG rs1800571 variant allele in only 2 subjects supports results obtained by Ek et al. and Bidzinska et al. in studies conducted on Caucasian, European populations, which failed to identify the Pro115Gln mutation in any of the enrolled subjects (64, 65). Furthermore, this mutation has been classified as part of the rare sequence variants by a study conducted in South Africa (66). Thus, all these data sustain the theory that this variant is highly unlikely to be involved in the pathogenesis of obesity.
In our geographical area, research on PPARG and obesity has been limited so far to two studies. One of them is a cross-sectional study which proved that the wild-type C allele of the PPARG rs1801282 gene polymorphism is associated with an increased risk of childhood obesity in the offspring of obese mothers (67). Moreover, a second study conducted on Romanian and Moldavian children underlined the lack of clinical impact of PPARG gene polymorphism on insulin resistance among an overweight population sample (4). Hence, research on PPARG polymorphisms in Romania is scarce and few information is available regarding variant allele carriers in healthy populations.
We identified no significant association between the PPARGC1A Gly482Ser polymorphism and a study group consisting of overweight and obese children, as suggested by the significantly higher prevalence of the C allele. Still, for the same polymorphism, significantly higher mean TSF values were correlated with the CC homozygotes, as opposed to the CT and TT genotypes. On the other hand, higher mean TSF values were also encountered in those patients which were variant carriers of both G allele and T allele of PPARG rs1801282 and PPARGC1A rs8192678, respectively. Although anthropometric and biochemical parameters were not associated with any of the rs8192678 polymorphism genotypes, total and LDL cholesterol levels were higher among homozygous variant (T) allele carriers, similarly with the results obtained in another Romanian study, which enrolled an adult population affected by metabolic syndrome (34). Still, this polymorphism has been linked to variations in HDL-c concentrations among obese subjects, as well as with insulin resistance within an obese population (68, 69). Other studies have reported a lack of association between PPARGC1A Gly482Ser polymorphism and obesity or a decreased metabolic risk for those with 482Gly/Gly genotype (70, 71). It is important though to mention that these studies have involved only adults, pediatric research articles reporting limited data regarding PPARGC1A polymorphisms, mainly a relationship between this gene's A risk allele and NAFLD in obese children (72).
A recent study found that PPRAG rs1801282 modified the association between n-3 long-chain PUFA (LCPUFA) and serum insulin with an inverse association in minor allele (Ala12 = G) carriers only and no interaction between fatty acid desaturase FADS or n-3 LCPUFA and plasma glucose (73). This is in agreement with the results of previous research, where fish oil reduced plasma glucose only in carriers of the rs1801282 G allele in the PPRG2 gene (74). Damsgaard et al. (73) found an inverse association between seric n-3 LCPUFA and triglycerides only in the PPARG2 rs1801282 G allele carriers, which concurs with the results of a previous study in infants (74). Blood n-3 LCPUFA was positively associated with LDL-c in children who were C allele homozygous of the rs1801282 SNP in the PPARG2 gene (73).
Another study found that T allele carriers of PPARGC1A rs8192678 had a higher decrease in TC and LDL-c levels following a low-fat diet compared to CC homozygotes (75) in overweight and obese Spanish subjects. A similar trend was noticed in the A allele carriers vs. CC homozygotes in those with a moderately high-protein diet group. A significant decrease in the rate/percentage of hypercholesterolemia was observed just in CC homozygotes who underwent the moderately high-protein diet and not in the low-fat diet group (75). Furthermore, this study conducted by Ramos-Lopez et al. indicated that an energy-restricted moderately high-protein diet yielded better results in decreasing serum cholesterol among PPARGC1A CC homozygotes than a low-fat diet (75).
PPARG polymorphisms have also been studied in relation to aerobic training response. In a previous review that studied the effectiveness of PPARG and their coactivators' polymorphism genes' influence on human training response, the cases with TT PPARGC1A rs8192678 genotype were non-responders for aerobic training while those with CC PPARGC1A rs8192678 genotype were the best responders to aerobic training (76). Moreover, the presence of C allele for PPARG rs1801282 carriers has been associated with a better response to exercise as well as to nutritional intervention (76). The same meta-analysis found that subjects with PPARG rs1801282 CC genotype are the negative responders to aerobic training (decreased fasting insulin response and glucose tolerance after aerobic training). In their study, Zarebska et al. found that the PPARG genotype can modulate training response and induced/produced body mass and composition parameters changes (77). They found that young Polish women with CC PPARG rs1801282 genotype had a better response for body fat mass variables compared to G allele carriers after a 12-week training program (77). However, data on this previously debated matter is scarce in the pediatric population and has been so far limited to adolescents, in whom a physical exercise intervention might imply a better compliance as opposed to younger ages (78).
The main strength of our study was the assessment of three different polymorphisms of PPARG, which were not simultaneously analyzed before in Romanian children, as well as the evaluation of PPARGC1A in the same subjects, only briefly discussed in pediatric literature. The relatively small sample size for a study of this type, as well as the unicentric enrollment character of the study constitute limitations of our research which cannot be neglected and might have influenced our results. A low statistical power has been noted through a post ad-hoc analysis particularly within the combined anthropometric parameter comparisons of three of the studied polymorphisms. Moreover, we did not investigate a possible influence of PPARG polymorphisms on nutritional intervention and exercise benefits in the obese/overweight study group, as follow-up of subjects included in the study would have implied a longitudinal design, particularly hard to achieve at young ages. Still, a clearer image of PPARG and PPARGC1A polymorphisms involved in childhood obesity most likely requires further studies performed in various geographical regions.
Conclusion
Genetic traits of obesity represent an attractive research topic in children, given the challenges faced by future metabolic-related complications and the limited literature data on the subject. Our findings showed no association of variant genotypes of PPARG (rs1801282, rs3856806, and rs1800571) and PPARGC1A (rs8192678) gene polymorphisms with childhood and adolescence overweight and obesity. Our study identified a significant increase in fasting glucose levels, triglyceride, albumin, ALT levels in children with excess weight, as well as expected important upward variation of anthropometric parameters (BMI, MUAC, and TSF z scores).
The limited number of patients, enrolled from a single referral center is a limitation of our study which once again highlights the need of expanding future research on PPARG, PPARGC1A SNPs and other polymorphisms on pediatric populations from various parts of the world. Therefore, the importance of race and geographical region cannot be overseen in studies evolving around clinical impact of polymorphism in weight gain.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Ethics Committee of the George Emil Palade University of Medicine, Pharmacy, Sciences, and Technology of Târgu Mureş (Institutional Review Board no. 13/18 July 2011). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
CM and CB contributed to the design of the study. MOS performed the statistical analysis. AC and CB carried out genetic analyses. CM, MOS, AC, and CB were involved to interpret the data. CM, MOS, and CB wrote and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was partly supported by a project financed by the Romanian Ministry of Education and Research, CNCS-UEFISCDI, Project No PN-III-P4-ID-PCE-2020-1928, within the PNCDI III, Contract No. PCE 72/2021.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Report of the Comission on Ending Childhood Obesity. WHO, Geneva (2016).
2. Brug J, van Stralen MM, Te Velde SJ, Chinapaw MJM, De Bourdeaudhuij I, Lien N, et al. Differences in weight status and energy-balance related behaviors among schoolchildren across Europe: the ENERGY-project. PLoS ONE. (2012) 7:e34742. doi: 10.1371/journal.pone.0034742
3. Ahrens W, Branca F. From data to action: combatting childhood obesity in Europe and beyond. Obes Rev. (2021) 22:e13302. doi: 10.1111/obr.13302
4. Chirita-Emandi A, Munteanu D, Andreescu N, Tutac P, Paul C, Velea IP, et al. No clinical utility of common polymorphisms in IGF1, IRS1, GCKR, PPARG, GCK1 and KCTD1 genes previously associated with insulin resistance in overweight children from Romania and Moldova. J Pediatr Endocrinol Metab. (2019) 32:33–9. doi: 10.1515/jpem-2018-0288
5. Guerra JVS, Dias MMG, Brilhante AJVC, Terra MF, García-Arévalo M, Figueira ACM. multifactorial basis and therapeutic strategies in metabolism-related diseases. Nutrients. (2021) 13:2830. doi: 10.3390/nu13082830
6. Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al. The human obesity gene map: the 2005 update. Obesity. (2006) 14:529–644. doi: 10.1038/oby.2006.71
7. Sözen MA, Özcan MU, Çildir M, Dogru IH, Aygök AG, Balkan KÜ. association of the human PPARγ2 Pro12Ala polymorphism with obesity in a population from Turkey. Acta Endocrinol. (2018) 14:459–65. doi: 10.4183/aeb.2018.459
8. Mǎrginean CO, Mǎrginean C, Melit LE. New insights regarding genetic aspects of childhood obesity: a minireview. Front Pediatr. (2018) 6:271. doi: 10.3389/fped.2018.00271
9. Duicu C, Mărginean CO, Voidăzan S, Tripon F, Bănescu C. FTO rs 9939609 SNP is associated with adiponectin and leptin levels and the risk of obesity in a cohort of romanian children population. Medicine. (2016) 95:e3709. doi: 10.1097/MD.0000000000003709
10. Mărginean C, Mărginean CO, Iancu M, Melit LE, Tripon F, Bănescu C. The FTO rs9939609 and LEPR rs1137101 mothers-newborns gene polymorphisms and maternal fat mass index effects on anthropometric characteristics in newborns: a cross-sectional study on mothers-newborns gene polymorphisms—the FTO-LEPR study (STROBE-compliant article). Medicine. (2016) 95:e5551. doi: 10.1097/MD.0000000000005551
11. Almeida SM, Furtado JM, Mascarenhas P, Ferraz ME, Ferreira JC, Monteiro MP, et al. Association between LEPR, FTO, MC4R, and PPARG-2 polymorphisms with obesity traits and metabolic phenotypes in school-aged children. Endocrine. (2018) 60:466–78. doi: 10.1007/s12020-018-1587-3
12. García-Ricobaraza M, García-Bermúdez M, Torres-Espinola FJ, Segura Moreno MT, Bleyere MN, Díaz-Prieto LE, et al. Association study of rs1801282 PPARG gene polymorphism and immune cells and cytokine levels in a Spanish pregnant women cohort and their offspring. J Biomed Sci. (2020) 27:101. doi: 10.1186/s12929-020-00694-3
13. Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. (2003) 65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528
14. Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol. (2021) 12:624112. doi: 10.3389/fendo.2021.624112
15. Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, et al. Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. (1998) 20:284–7. doi: 10.1038/3099
16. Lin J, Chen Y, Tang W-F, Liu C, Zhang S, Guo Z-Q, et al. rs3856806 C >T Polymorphism Increased the risk of colorectal cancer: a case-control study in Eastern Chinese Han Population. Front Oncol. (2019) 9:63. doi: 10.3389/fonc.2019.00063
17. Dedoussis GV, Vidra N, Butler J, Papoutsakis C, Yannakoulia M, Hirschhorn JN, et al. Peroxisome proliferator-activated receptor-gamma (PPARgamma) Pro12Ala polymorphism and risk for pediatric obesity. Clin Chem Lab Med. (2009) 47:1047–50. doi: 10.1515/CCLM.2009.242
18. Masud S, Ye S. SAS Group. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J Med Genet. (2003) 40:773–80. doi: 10.1136/jmg.40.10.773
19. Mansoori A, Amini M, Kolahdooz F, Seyedrezazadeh E. Obesity and Pro12Ala polymorphism of peroxisome proliferator-activated receptor-gamma gene in healthy adults: a systematic review and meta-analysis. Ann Nutr Metab. (2015) 67:104–18. doi: 10.1159/000439285
20. Wei W-M, Wu X-Y, Li S-T, Shen Q. PPARG gene C161T CT/TT associated with lower blood lipid levels and ischemic stroke from large-artery atherosclerosis in a Han population in Guangdong. Neurol Res. (2016) 38:620–4. doi: 10.1080/01616412.2016.1189056
21. Li Q, Chen R, Bie L, Zhao D, Huang C, Hong J. Association of the variants in the PPARG gene and serum lipid levels: a meta-analysis of 74 studies. J Cell Mol Med. (2015) 19:198–209. doi: 10.1111/jcmm.12417
22. Yilmaz-Aydogan H, Kurnaz O, Kurt O, Akadam-Teker B, Kucukhuseyin O, Tekeli A, et al. Effects of the PPARG P12A and C161T gene variants on serum lipids in coronary heart disease patients with and without Type 2 diabetes. Mol Cell Biochem. (2011) 358:355–63. doi: 10.1007/s11010-011-0987-y
23. Ristow M, Müller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N Engl J Med. (1998) 339:953–9. doi: 10.1056/NEJM199810013391403
24. Blüher M, Lübben G, Paschke R. Analysis of the relationship between the Pro12Ala variant in the PPAR-gamma2 gene and the response rate to therapy with pioglitazone in patients with type 2 diabetes. Diabetes Care. (2003) 26:825–31. doi: 10.2337/diacare.26.3.825
25. Sanchis-Gomar F, García-Giménez JL, Gómez-Cabrera MC, Pallardó FV. Mitochondrial biogenesis in health and disease. Molecular and therapeutic approaches. Curr Pharm Des. (2014) 20:5619–33. doi: 10.2174/1381612820666140306095106
26. Taghvaei S, Saremi L, Babaniamansour S. Computational analysis of gly482ser single-nucleotide polymorphism in PPARGC1A gene associated with CAD, NAFLD, T2DM, obesity, hypertension, and metabolic diseases. PPAR Res. (2021) 2021:5544233. doi: 10.1155/2021/5544233
27. de Onis M, Lobstein T. Defining obesity risk status in the general childhood population: which cut-offs should we use? Int J Pediatr Obes. (2010) 5:458–60. doi: 10.3109/17477161003615583
28. Obesity and Overweight. Available online at: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed September 4, 2022).
29. Anderson LN, Carsley S, Lebovic G, Borkhoff CM, Maguire JL, Parkin PC, et al. Misclassification of child body mass index from cut-points defined by rounded percentiles instead of Z-scores. BMC Res Notes. (2017) 10:639. doi: 10.1186/s13104-017-2983-0
30. Mokha JS, Srinivasan SR, Dasmahapatra P, Fernandez C, Chen W, Xu J, et al. Utility of waist-to-height ratio in assessing the status of central obesity and related cardiometabolic risk profile among normal weight and overweight/obese children: the Bogalusa Heart Study. BMC Pediatr. (2010) 10:73. doi: 10.1186/1471-2431-10-73
31. Oh EY, Min KM, Chung JH, Min YK, Lee MS, Kim KW, et al. Significance of Pro12Ala mutation in peroxisome proliferator-activated receptor-gamma2 in Korean diabetic and obese subjects. J Clin Endocrinol Metab. (2000) 85:1801–4. doi: 10.1210/jcem.85.5.6499
32. Krasznai M, Szaniszlo K, Kraxner H, Vargha E, Kovacs M, Kaszas E, et al. Association of PPAR polymorphisms with cytokine levels in allergic rhinitis. Eur Arch Otorhinolaryngol. (2012) 269:513–21. doi: 10.1007/s00405-011-1710-z
33. Shabana SS. The Pro115Gln missense mutation of peroxisome proliferator activated receptor γ (PPARγ) gene in diabetes in the Pakistani population Avicenna. J Clin Microbiol Infect. (2016) 3:35148–35148. doi: 10.17795/ajcmi-35148
34. Csép K, Szigeti E, Vitai M, Korányi L. The PPARGC1A—GLY482SER polymorphism (RS8192678) and the metabolic syndrome in a central Romanian population. Acta Endocrinol. (2017) 13:161–7. doi: 10.4183/aeb.2017.161
35. Desai MM, Plotkin JB. The polymorphism frequency spectrum of finitely many sites under selection. Genetics. (2008) 180:2175–91. doi: 10.1534/genetics.108.087361
36. Després J-P, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. (2006) 444:881–7. doi: 10.1038/nature05488
37. Lartey A, Marquis GS, Aryeetey R, Nti H. Lipid profile and dyslipidemia among school-age children in urban Ghana. BMC Public Health. (2018) 18:320. doi: 10.1186/s12889-018-5196-0
38. Furtado JM, Almeida SM, Mascarenhas P, Ferraz ME, Ferreira JC, Vilanova M, et al. Anthropometric features as predictors of atherogenic dyslipidemia and cardiovascular risk in a large population of school-aged children. PLoS ONE. (2018) 13:e0197922. doi: 10.1371/journal.pone.0197922
39. Damanhoury S, Morrison KM, Mian R, McPhee PG, Kozyrskyj AL, Newton AS, et al. Metabolically healthy obesity in children enrolled in the Canadian pediatric weight management registry (CANPWR): an exploratory secondary analysis of baseline data. Clin Obes. (2021) 12:e12490. doi: 10.1111/cob.12490
40. Vukovic R, Dos Santos TJ, Ybarra M, Atar M. Children with metabolically healthy obesity: a review. Front Endocrinol. (2019) 10:865. doi: 10.3389/fendo.2019.00865
41. Vermeiren E, Bruyndonckx L, De Winter B, Verhulst S, Van Eyck A, Van Hoorenbeeck K. The effect of weight regain on cardiometabolic health in children with obesity: a systematic review of clinical studies. Nutr Metab Cardiovasc Dis. (2021) 31:2575–86. doi: 10.1016/j.numecd.2021.05.020
42. Munusamy J, Yadav J, Kumar R, Bhalla A, Dayal D. Metabolic complications of childhood obesity. J Family Med Prim Care. (2021) 10:2325–30. doi: 10.4103/jfmpc.jfmpc_975_20
43. Putri RR, Casswall T, Hagman E. Prevalence of increased transaminases and its association with sex, age, and metabolic parameters in children and adolescents with obesity - a nationwide cross-sectional cohort study. BMC Pediatr. (2021) 21:271. doi: 10.1186/s12887-021-02747-4
44. Balkhiyarova Z, Luciano R, Kaakinen M, Ulrich A, Shmeliov A, Bianchi M, et al. Relationship between glucose homeostasis and obesity in early life—a study of Italian children and adolescents. Hum Mol Genet. (2021) 31:ddab287. doi: 10.1093/hmg/ddab287
45. Raskiliene A, Smalinskiene A, Kriaucioniene V, Lesauskaite V, Petkeviciene J. Associations of MC4R, LEP, and LEPR polymorphisms with obesity-related parameters in childhood and adulthood. Genes. (2021) 12:949. doi: 10.3390/genes12060949
46. Metelcová T, Vanková M, Zamrazilová H, Hovhannisyan M, Stanková B, Tvrzická E, et al. FADS1 gene polymorphism(s) and fatty acid composition of serum lipids in adolescents. Lipids. (2021) 56:499–508. doi: 10.1002/lipd.12317
47. Kulaeva ED, Volchik VV, Bocharova OV, Teplakova ED, Shkurat MA, Derevyanchuk EG, et al. Association of SNPs in lipid metabolism gene single nucleotide polymorphism with the risk of obesity in children. Genet Test Mol Biomarkers. (2021) 25:419–25. doi: 10.1089/gtmb.2020.0343
48. Marginean CO, Banescu C, Voidazan S, Duicu C. IL-6 572 C/G, 190 C/T, and 174 G/C gene polymorphisms in children's malnutrition. J Pediatr Gastroenterol Nutr. (2014) 59:666–73. doi: 10.1097/MPG.0000000000000492
49. Mărginean CO, Bănescu C, Duicu C, Voidăzan S, Mărginean C. Angiotensin-converting enzyme gene insertion/deletion polymorphism in nutritional disorders in children. Eur J Nutr. (2015) 54:1245–54. doi: 10.1007/s00394-014-0802-0
50. Velazquez-Roman J, Angulo-Zamudio UA, León-Sicairos N, Medina-Serrano J, DeLira-Bustillos N, Villamil-Ramírez H, et al. Association of FTO, ABCA1, ADRB3, and PPARG variants with obesity, type 2 diabetes, and metabolic syndrome in a Northwest Mexican adult population. J Diabetes Compl. (2021) 35:108025. doi: 10.1016/j.jdiacomp.2021.108025
51. Vargas-Sánchez K, Vargas L, Urrutia Y, Beltrán I, Rossi AB, Lozano HY, et al. PPARα and PPARβ/δ are negatively correlated with proinflammatory markers in leukocytes of an obese pediatric population. J Inflamm. (2020) 17:35. doi: 10.1186/s12950-020-00264-2
52. Stryjecki C, Peralta-Romero J, Alyass A, Karam-Araujo R, Suarez F, Gomez-Zamudio J, et al. Association between PPAR-γ2 Pro12Ala genotype and insulin resistance is modified by circulating lipids in Mexican children. Sci Rep. (2016) 6:24472. doi: 10.1038/srep24472
53. Carrillo-Venzor MA, Erives-Anchondo NR, Moreno-González JG, Moreno-Brito V, Licón-Trillo A, González-Rodríguez E, et al. Pro12Ala PPAR-γ2 and +294T/C PPAR-δ Polymorphisms and association with metabolic traits in teenagers from Northern Mexico. Genes (Basel). (2020) 11:E776. doi: 10.3390/genes11070776
54. Rahman ANA, Jawis MN, Yusoff S. Peroxisome proliferator activated receptor gamma (PPARγ) gene variant in relation to physical activity and obesity among malay children. MJHG. (2020) 1:45–52. Available online at: http://www.kk.usm.my/journal/index.php/mjhg/article/view/37
55. Anderson SE, Whitaker RC. Prevalence of obesity among US preschool children in different racial and ethnic groups. Arch Pediatr Adolesc Med. (2009) 163:344–8. doi: 10.1001/archpediatrics.2009.18
56. Queiroz EM, Cândido APC, Castro IM, Bastos AQA, Machado-Coelho GLL, Freitas RN. IGF2, LEPR, POMC, PPARG, and PPARGC1 gene variants are associated with obesity-related risk phenotypes in Brazilian children and adolescents. Braz J Med Biol Res. (2015) 48:595–602. doi: 10.1590/1414-431X20154155
57. Vidović V, Maksimović N, Vidović S, Damnjanović T, Novaković I. Association of PPARG rs3856806 C>T polymorphism with body mass index, glycaemia, and lipid parameters in Serbian adolescents. Scr Med. (2021) 52:15–21. doi: 10.5937/scriptamed52-29376
58. León-Mimila P, Villamil-Ramírez H, Villalobos-Comparán M, Villarreal-Molina T, Romero-Hidalgo S, López-Contreras B, et al. Contribution of common genetic variants to obesity and obesity-related traits in Mexican children and adults. PLoS ONE. (2013) 8:e70640. doi: 10.1371/journal.pone.0070640
59. Clement K, Hercberg S, Passinge B, Galan P, Varroud-Vial M, Shuldiner AR, et al. The Pro115Gln and Pro12Ala PPAR gamma gene mutations in obesity and type 2 diabetes. Int J Obes Relat Metab Disord. (2000) 24:391–3. doi: 10.1038/sj.ijo.0801191
60. Carlos FF, Silva-Nunes J, Flores O, Brito M, Doria G, Veiga L, et al. Association of FTO and PPARG polymorphisms with obesity in Portuguese women. Diabetes Metab Syndr Obes. (2013) 6:241–5. doi: 10.2147/DMSO.S45779
61. Fan W, Shen C, Wu M, Zhou Z-Y, Guo Z-R. Association and interaction of PPARα, δ, and γ gene polymorphisms with low-density lipoprotein-cholesterol in a Chinese Han population. Genet Test Mol Biomarkers. (2015) 19:379–86. doi: 10.1089/gtmb.2015.0002
62. Hamer OW, Forstner D, Ottinger I, Ristow M, Bollheimer LC, Schölmerich J, et al. The Pro115Gln polymorphism within the PPAR gamma2 gene has no epidemiological impact on morbid obesity. Exp Clin Endocrinol Diabetes. (2002) 110:230–4. doi: 10.1055/s-2002-33072
63. Zaharan NL, Muhamad NH, Jalaludin MY, Su TT, Mohamed Z, Mohamed MNA. A Majid H. Non-synonymous single-nucleotide polymorphisms and physical activity interactions on adiposity parameters in Malaysian adolescents. Front Endocrinol. (2018) 9:209. doi: 10.3389/fendo.2018.00209
64. Ek J, Urhammer SA, Sørensen TI, Andersen T, Auwerx J, Pedersen O. Homozygosity of the Pro12Ala variant of the peroxisome proliferation-activated receptor-gamma2 (PPAR-gamma2): divergent modulating effects on body mass index in obese and lean Caucasian men. Diabetologia. (1999) 42:892–5. doi: 10.1007/s001250051243
65. Bidzinska B, Demissie M, Tworowska U, Slezak R, Dobosz T. Milewicz A. Is obesity associated with a polymorphism Pro12Ala and Pro115Gln in PPARgamma gene? Pol Merkur Lekarski. (2004) 16:434–7.
66. Vergotine Z, Kengne AP, Erasmus RT, Yako YY, Matsha TE. Rare mutations of peroxisome proliferator-activated receptor gamma: frequencies and relationship with insulin resistance and diabetes risk in the mixed ancestry population from South Africa. Int J Endocrinol. (2014) 2014:187985. doi: 10.1155/2014/187985
67. Mărginean C, Mărginean CO, Iancu M, Szabo B, Cucerea M, Melit LE, et al. The role of TGF-β1 869 T > C and PPAR γ2 34 C > G polymorphisms, fat mass, and anthropometric characteristics in predicting childhood obesity at birth: a cross-sectional study according the parental characteristics and newborn's risk for child obesity (the newborns obesity's risk) NOR study. Medicine. (2016) 95:e4265. doi: 10.1097/MD.0000000000004265
68. Vohl M-C, Houde A, Lebel S, Hould F-S, Marceau P. Effects of the peroxisome proliferator-activated receptor-gamma co-activator-1 Gly482Ser variant on features of the metabolic syndrome. Mol Genet Metab. (2005) 86:300–6. doi: 10.1016/j.ymgme.2005.07.002
69. Fanelli M, Filippi E, Sentinelli F, Romeo S, Fallarino M, Buzzetti R, et al. The Gly482Ser missense mutation of the peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1 alpha) gene associates with reduced insulin sensitivity in normal and glucose-intolerant obese subjects. Dis Markers. (2005) 21:175–80. doi: 10.1155/2005/576748
70. Castro GV, Latorre AFS, Korndorfer FP, de Carlos Back LK, Lofgren SE. The impact of variants in four genes: MC4R, FTO, PPARG and PPARGC1A in overweight and obesity in a large sample of the Brazilian population. Biochem Genet. (2021) 59:1666–79. doi: 10.1007/s10528-021-10079-2
71. Vázquez-Del Mercado M, Guzmán-Ornelas M-O, Corona Meraz F-I, Ríos-Ibarra C-P, Reyes-Serratos E-A, Castro-Albarran J, et al. The 482Ser of PPARGC1A and 12Pro of PPARG2 alleles are associated with reduction of metabolic risk factors even obesity in a Mexican–Mestizo population. Biomed Res Int. (2015) 2015:285491. doi: 10.1155/2015/285491
72. Lin Y-C, Chang P-F, Chang M-H, Ni Y-H, A. common variant in the peroxisome proliferator-activated receptor-γ coactivator-1α gene is associated with nonalcoholic fatty liver disease in obese children. Am J Clin Nutr. (2013) 97:326–31. doi: 10.3945/ajcn.112.046417
73. Damsgaard CT, Vuholm S, Teisen MN, Stark KD, Lauritzen L. Does polymorphisms in PPAR and APOE genes modify associations between fatty acid desaturase (FADS), n-3 long-chain PUFA and cardiometabolic markers in 8-11-year-old Danish children? Br J Nutr. (2021) 125:369–76. doi: 10.1017/S0007114520002822
74. Harsløf LBS, Damsgaard CT, Hellgren LI, Andersen AD, Vogel U, Lauritzen L. Effects on metabolic markers are modified by PPARG2 and COX2 polymorphisms in infants randomized to fish oil. Genes Nutr. (2014) 9:396. doi: 10.1007/s12263-014-0396-4
75. Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Goni L, Cuervo M, Martinez JA. Association of the Gly482Ser PPARGC1A gene variant with different cholesterol outcomes in response to two energy-restricted diets in subjects with excessive weight. Nutrition. (2018) 47:83–9. doi: 10.1016/j.nut.2017.10.008
76. Petr M, Stastny P, Zajac A, Tufano JJ, Maciejewska-Skrendo A. The role of peroxisome proliferator-activated receptors and their transcriptional coactivators gene variations in human trainability: a systematic review. Int J Mol Sci. (2018) 19:E1472. doi: 10.3390/ijms19051472
77. Zarebska A, Jastrzebski Z, Cieszczyk P, Leonska-Duniec A, Kotarska K, Kaczmarczyk M, et al. The Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma gene modifies the association of physical activity and body mass changes in polish women. PPAR Res. (2014) 2014:373782. doi: 10.1155/2014/373782
Keywords: PPARG, obesity, anthropometric parameters, metabolic parameters, child
Citation: Muntean C, Sasaran MO, Crisan A and Banescu C (2022) Effects of PPARG and PPARGC1A gene polymorphisms on obesity markers. Front. Public Health 10:962852. doi: 10.3389/fpubh.2022.962852
Received: 06 June 2022; Accepted: 24 October 2022;
Published: 16 November 2022.
Edited by:
Qingzhong Wang, Shanghai University of Traditional Chinese Medicine, ChinaReviewed by:
Petr Stastny, Charles University, CzechiaMalgorzata Wojcik, Jagiellonian University Medical College, Poland
Copyright © 2022 Muntean, Sasaran, Crisan and Banescu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Muntean, ZHVpY3VjYXJtZW5AeWFob28uY29t
†These authors have contributed equally to this work and share first authorship
 Carmen Muntean
Carmen Muntean Maria Oana Sasaran
Maria Oana Sasaran Adriana Crisan3
Adriana Crisan3 Claudia Banescu
Claudia Banescu