- 1National Clinical Laboratory on Tuberculosis, Beijing Key Laboratory for Drug-Resistant Tuberculosis Research, Beijing Chest Hospital, Capital Medical University, Beijing Tuberculosis and Thoracic Tumor Institute, Beijing, China
- 2Beijing Chest Hospital, Capital Medical University, Beijing Tuberculosis and Thoracic Tumor Institute, Beijing, China
Background: Pulmonary non-tuberculous mycobacteria (NTM) infection has become a public health concern in China and around the world. The objective of this study was to describe the longitudinal changes in the frequency and diversity of NTM in northern China.
Methods: We retrospectively analyzed data on mycobacterium species in Beijing Chest Hospital from January 2014 to December 2021. The isolates were identified to species level by targeted DNA sequencing.
Results: After excluding duplicates, 1,755 NTM strains were analyzed, which were from 27 provinces in China over 8 years. Among all mycobacteria, the proportion of NTM increased each year, from 4.24% in 2014 to 12.68% in 2021. Overall, 39 different NTM species were identified, including 23 slow growing mycobacteria (SGM) and 16 rapid growing mycobacteria (RGM). The most common species were M. intracellulare (51.62%), M. abscessus (22.22%), M. kansasii (8.32%), M. avium (7.75%) and M. fortuitum (2.05%). The number of NTM species identified also increased each year from 9 in 2014 to 26 in 2021. Most species showed stable isolation rates over the years; however, the proportion of M. avium increased from 3.85 to 10.42% during the study period. Besides, 81 non-mycobacteria strains, including Gordonia (21 isolates), Nocardia (19 isolates) and Tsukamurella (17 isolates), etc., were also discovered.
Conclusion: The proportion of NTM and species diversity increased considerably in northern China from 2014 to 2021. M. intracellulare was the most common NTM isolated among respiratory specimens, followed by M. abscessus and M. kansasii. Rare NTM species and non-mycobacteria pathogens also need attention.
Introduction
Non-tuberculous mycobacteria (NTM) can cause opportunistic infection and present a threat to public health (1). To date, more than 270 species/subspecies of NTM have been identified (http://www.bacterio.net/mycobacterium.html), and up to 60 NTM species have been proved to be human pathogens (2). The distribution of NTM species isolated from human clinical samples is geographically specific (3). As NTM differ strongly in their drug susceptibility profiles (4), understanding this diversity has significant reference value for treating and managing of these infections.
China ranked second among 30 high tuberculosis (TB) burden countries in 2020. NTM infections are prone to be misdiagnosed as multidrug-resistant TB in China, when only acid-fast staining and mycobacterial culture are used for the diagnosis of TB (5). Increased awareness of NTM identification is very important. However, precise incidence and prevalence data about NTM infection are lacking in China. Although three national TB epidemiological sampling surveys carried out in China in 1990, 2000 and 2010 reported the proportion of NTM among mycobacterial isolates, these NTM isolates were not identified to species. A few studies have reported NTM epidemiology mainly based on regional or local data (6–11). Liu et al. (12) reported the incidence of NTM in China from a national survey conducted in 2013. However, most of the reports were cross-sectional studies. The aim of this study was to determine the longitudinal changes in the frequency and diversity of NTM related to pulmonary disease over 8 years at the National Tuberculosis Clinical Laboratory of the Beijing Chest Hospital in China.
Materials and Methods
Data Collection
From January 2014 to December 2021, the basic patient demographic information and species identification results of positive mycobacterial cultures from respiratory samples were collected through the laboratory information system at the National Tuberculosis Clinical Laboratory of the Beijing Chest Hospital (Beijing, China).
Smear and Culture
Direct smears were prepared and stained with auramine and examined by light-emitting diode microscopy. After processing with NALC/NaOH and centrifugation, 500 μl suspensions were inoculated into a 7 ml MGIT tube (Becton, Dickinson and Company, USA), and/or 100 μl suspensions were inoculated onto LJ medium (Encode Medical Engineering Co., Ltd, China). LJ tubes were incubated at 37 °C and examined weekly for growth for a maximum of 8 weeks and MGIT tubes were incubated in the BACTEC MGIT 960 system for 6 weeks. All of the culture-positive isolates were primarily identified as M. tuberculosis complex (MTBC) by MPT64 antigen testing. Isolates that were initially identified as not MTBC by the MPT64 antigen testing were further identification to the species level using target DNA sequencing.
Species Identification
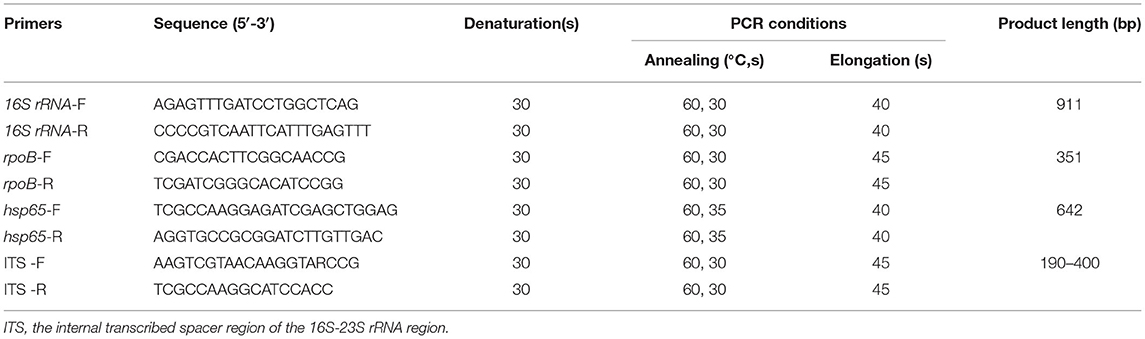
We identified the isolates to species level by target DNA sequencing, including 16S rRNA, rpoB, hsp65, and the internal transcribed spacer region of the 16S−23S rRNA region (ITS) (13, 14). Genomic DNA was isolated from isolates by boiling method. The primer sets and conditions for amplification are shown in Table 1. There were 70 mycobacterial reference strains stored in the Bio-bank in Beijing Chest Hospital (Beijing, China), which were obtained either from the American Type Culture Collection (ATCC) or from the German Collection of Microorganisms (DSM). Multigene sequence similarity for the clinical isolates was determined in comparison with the reference sequences in our Bio-bank or the multigene database using the basic local alignment search tool (BLAST). Values above 99% sequence similarity for 16S rRNA, and above 97% similarity for hsp65, rpoB and ITS genes were used for species distinction.
Statistical Analyses
The χ2 test was used to compare differences in proportions. Statistical analysis was performed using SPSS version 22.0. Differences were considered statistically significant at P < 0.05.
Results
Changes of NTM Proportion
A total of 20,719 mycobacterial isolates, including 1,755 NTM and 18,964 MTBC strains, were identified. The proportion of NTM among all mycobacteria increased each year, from 4.24% (78/1,839) in 2014 to 12.68% (451/3,558) in 2021 (Figure 1). The trend was statistically significant (χ2 = 278, P < 0.001); the greatest increase was from 2017 (6.48%) to 2018 (9.93%). Fewer patients were tested in 2020, due to the SARS-CoV2 pandemic started in 2019.
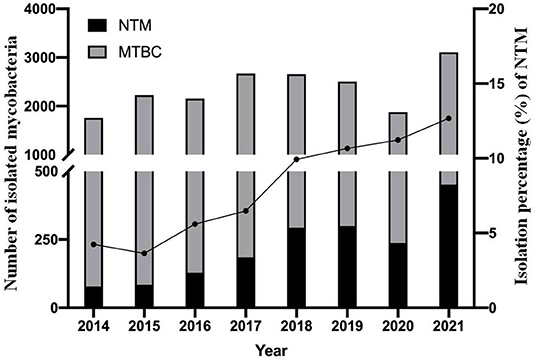
Figure 1. Continued upward trend in the proportion of NTM isolates over the 8-year study period. Filled circles (•) denote the percentage of NTM isolates each year. MTBC, mycobacterium tuberculosis complex; NTM, non-tuberculous mycobacteria.
The Spectrum of NTM Species
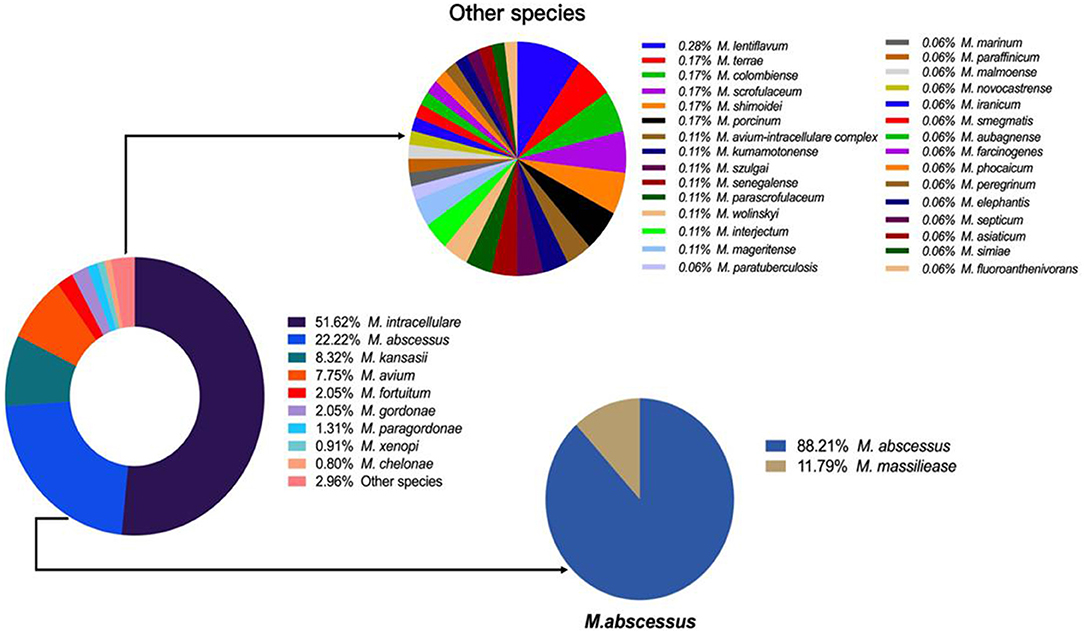
Overall, 39 different NTM species were identified, including 23 slow growing mycobacteria (SGM) and 16 rapid growing mycobacteria (RGM). M. intracellulare (51.62%), M. abscessus (22.22%), M. kansasii (8.32%), M. avium (7.75%) and M. fortuitum (2.05%) were the five most common isolated NTM, accounting for 91.96% of all NTM species (Figure 2).
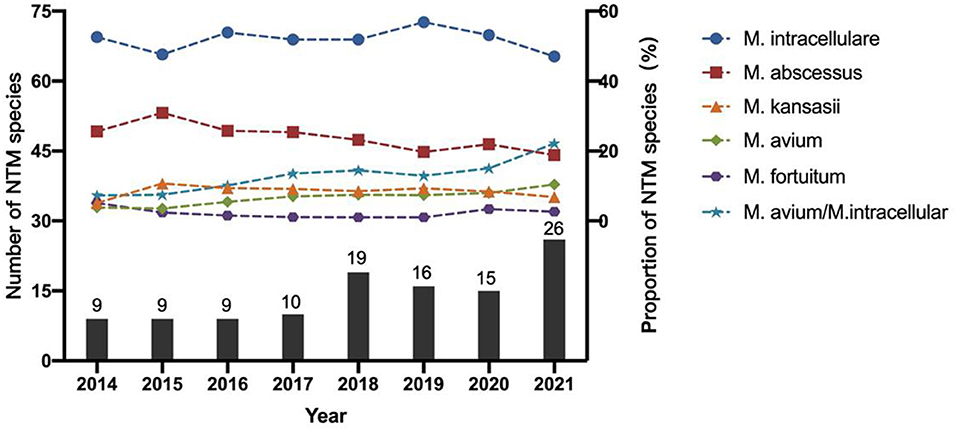
The number of NTM species identified also increased each year from 9 in 2014 to 26 in 2021 (Figure 3). M. intracellulare predominated in the Mycobacterium avium complex (MAC); however, the relative ratio of M. avium to M. intracellulare increased each year (Table 2; Figure 3). M. abscessus samples included two subspecies: M. abscessus. abscessus (88.21%, 344/390) and M. abscessus. massiliense (11.79%, 46/390). Most species showed stable isolation rates over the years; however, the proportion of M. avium increased from 3.85% in 2014 to 10.42% in 2021 (Figure 3).
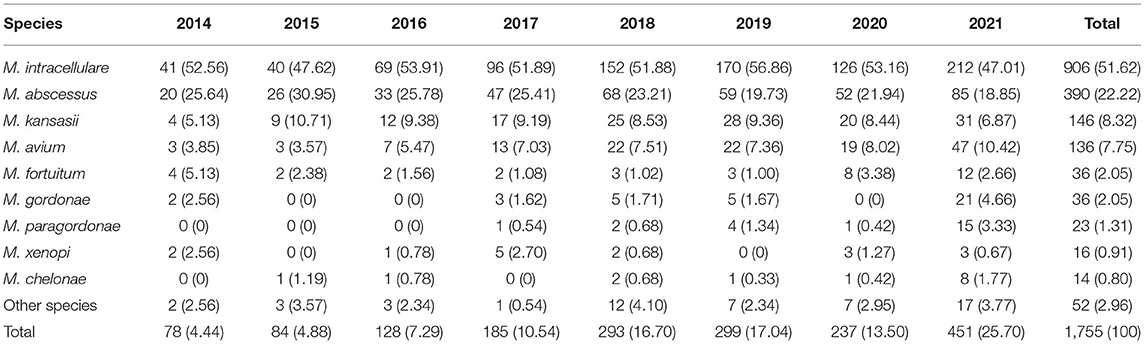
Table 2. Non-tuberculous mycobacteria species isolated from respiratory specimens in China, 2014–2021.
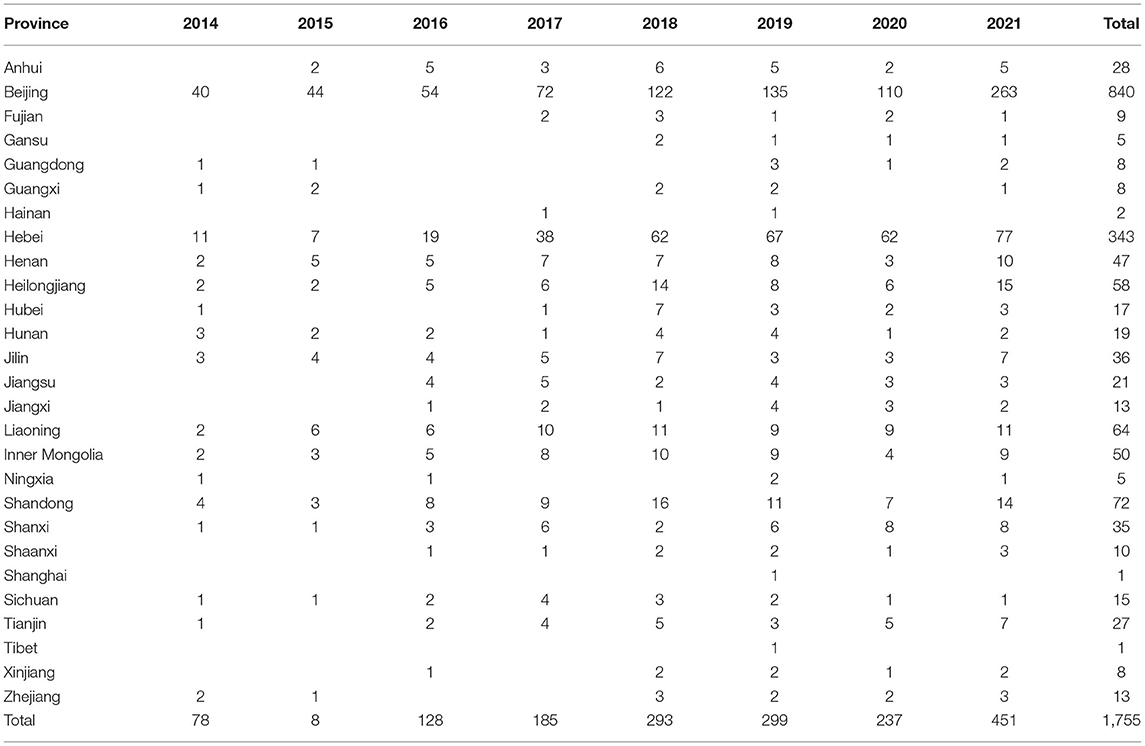
The 1,755 NTM isolates were from 27 provinces in China (Figure 4; Table 3). A large proportion of patients came from the north of China, with Beijing, Hebei, Liaoning, Heilongjiang, and Inner Mongolia accounting for 47.86% (840/1,755), 19.54% (343/1,755), 3.65% (64/1,755), 3.30% (58/1,755) and 2.85% (50/1,755) of all NTM identified, respectively.
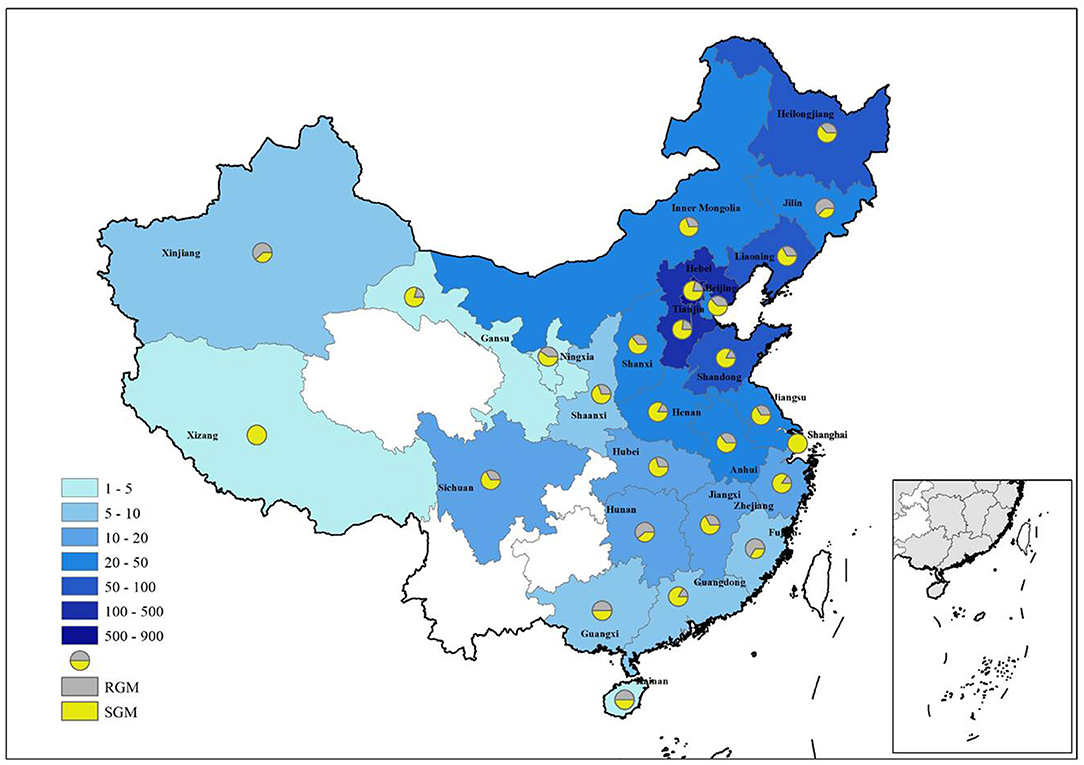
Figure 4. Distribution of NTM in different provinces of China. RGM rapid growing mycobacteria, SGM slow growing mycobacteria.
Non-mycobacteria Pathogens Identification
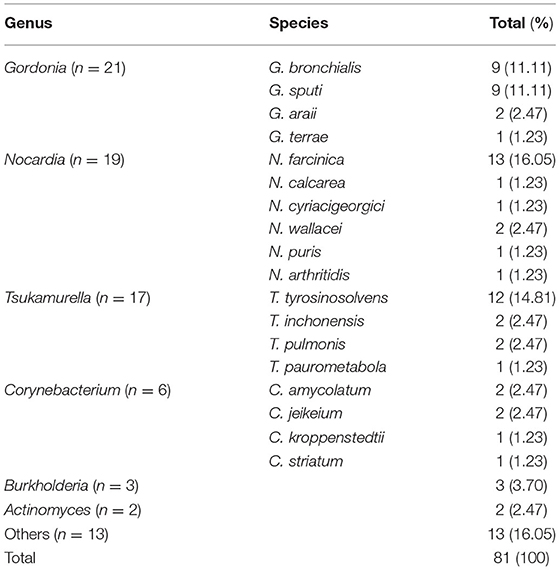
In addition to Mycobacteria spp., we also identified 81 non-mycobacteria strains, which were acid-fast-staining-positive or resistant to NALC-NaOH decontamination. Totally, 21 Gordonia, 19 Nocardia, 17 Tsukamurella, 6 Corynebacterium, 3 Burkholderia, 2 Actinomyces and 13 others were identified (Table 4). The similarity of the primers used to the equivalent regions in 16S rRNA, rpoB, and hsp65 in these species with mycobacterium was about 90, 80, and 85%, respectively.
Demographic Data of NTM Positive Patients
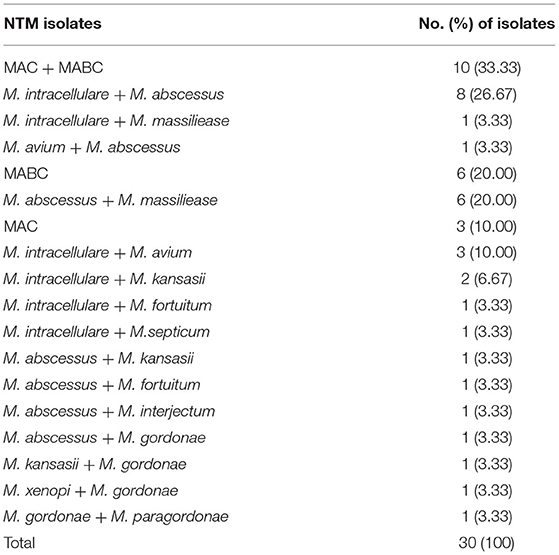
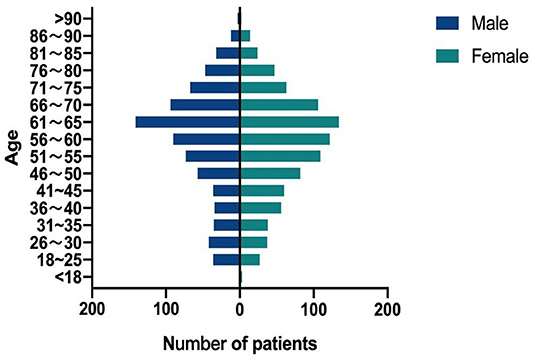
Thirty patients had two NTM species isolated (Table 5), among which MAC (M. avium or M. intracellulare) and MABC (M. abscessus or M. massiliense) accounting for 33.33% (10/30). The proportion of female patients in this study was 53.57% (924/1,725). The ages of the NTM patients ranged from 4 to 94 years. Patients aged 51–70 years accounted for 50.38% (869/1,725) of the total patients. In the 41–50 and 51–60 years groups, female patients had a higher proportion of NTM than male patients (P = 0.022) (Figure 5).
Discussion
This study provided the NTM epidemiology in China from 2014 to 2021 when the same protocols were used for samples. Our group previously reported that the isolation rates of NTM was 2.6% (95/3,714) in respiratory samples from persons in northern China, 2008–2011 (15). The overall NTM pulmonary infection rate was 6.4% in China from a national survey conducted in 2013 (12). During our study period, the proportion of NTM almost tripled from 4.24% in 2014 to 12.68% in 2021, suggesting a considerable increase in the NTM epidemic in China.
Total 39 different NTM species were identified, indicating the diversity of NTM species circulating among patients in China. M. intracellulare was the most common NTM pathogen, which was similar to previous findings in Australia, Korea, India, Uruguay and so on (3, 16–19). Furthermore, we found the incidence of M. avium grew faster than others from 2014 to 2021, and the ratio of M. avium to M. intracellulare increased each year, suggesting a more prominent role for M. avium infections in China. MAC and M. abscessus accounted for 81.59% of the NTM strains during this study period. Beyond these common NTM species, many rare NTM species were also identified, for example M. lentiflavum, M. terrae, M. colombiensee, M. scrofulaceum, M.shimoridei, M. porcinum, M. kumamotonense, etc. Studies had also reported some of these species isolated from pulmonary samples (20). Precisely species identification should be conducted to facilitate pulmonary NTM treatment.
Interestingly, 81 non-mycobacteria strains including Gordonia, Nocardia, Tsukamurella, etc., were discovered. Actinomycetes with mycolic acid had been classified under genera such as Corynebacterium, Gordonia, Mycobacterium, Nocardia, Rhodococcus, Tsukamurella, Skermania and Williamsia (21, 22). The genus Gordonia is a gram-positive, partially acid-fast organism. Gordonial infections may be misdiagnosed as NTM, Nocardia or other actinomycetes infections due to the similar clinical manifestations (23). Nocardia species are gram-positive, slightly acid-fast, opportunistic pathogens. The lung is the most common infection site and symptoms of nocardiosis are similar to TB or NTM infection, such as fever, cough and chest pain (24). The genus Tsukamurella is an aerobic actinomycete and was a cause of opportunistic infection. Burkholderia are Gram-negative bacilli and opportunistic human pathogens (25–29). Several studies reported that Gordonia, Nocardia and Tsukamurella had also been identified in suspected Mycobacterium species isolates in China (8, 12, 30). In addition to NTM and MTBC infection, non-mycobacteria pathogens should also be tested when using acid-fast staining or mycobacterial culture to diagnose pulmonary disease.
Non-mycobacteria isolates could not be differentiated from NTM using traditional methods such as PNB differential media, acid-fast stains, and MPB64 protein assay. Commonly used commercial kits could only detect ~20 NTM species, and could not identify the newly discovered NTM species and non-mycobacteria pathogens (31, 32). Target DNA sequencing, such as 16S rRNA, rpoB, hsp65, and ITS, not only could identify the NTM strains to the species level, but also could distinguish NTM from non-mycobacterial but related genera, such as Gordonia, Nocardia and Tsukamurella.
More NTM strains were isolated from female (53.57%) than from male (46.43%) in this study. The NTM patients were most common in the 51–70 (50.38%) years age group. Several reports also indicated that older women were more susceptible to NTM infection (33–35).
The large sample size, resolution power of our species identification method, and avoiding duplication were important strengths of our study, but its limitations should also be noted. First, this is a retrospective study and there may be an underestimation of the proportion of NTM. Some NTM species require specific medium requirements and low or high temperatures for growth, so some strains may be not cultivable on routine condition. Second, all of the strains were isolated from respiratory specimens. In fact respiratory samples could best reflect the distribution of NTM species in local environments. However, there may be a selection bias, as the low isolation rate of some NTM species may reflect an inability to persist in human airways. Third, the data were collected from the National Tuberculosis Clinical Laboratory of the Beijing Chest Hospital. Due to the location of Beijing Chest Hospital, most patients were from the north of China. Since the southern region had higher NTM prevalence rate than the northern region in China. Our study showed that the proportion of NTM increased considerably in northern China from 2014 to 2021, and more attention need to be taken to combat NTM.
In conclusion, the proportion of NTM and species diversity increased considerably in China from 2014 to 2021. M. intracellulare was the most common NTM isolated among respiratory samples, followed by M. abscessus and M. kansasii. Rare NTM species and non-mycobacteria pathogens also need attention.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
JP, GW, and QS: substantial contributions to the conception of the work. JY, XL, ChaW, CheW, LD, and FW: acquisition and analysis of data. QS, GJ, and HH: interpretation of data. QS and GW: writing the first draft. JP and GW: revision of manuscript. QS, JY, XL, ChaW, CheW, GJ, LD, FW, HH, GW, and JP: final approval of the version to be published. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Capital's Funds for Health Improvement and Research (2022-1G-2162), Beijing Public Health Experts Project (2022-3-040), Beijing Tongzhou Municipal Science & Technology commission (KJ2022CX044), Tongzhou Yunhe Project under Grant (YH201917), and Beijing Municipal Administration of Hospitals' Ascent Plan (DFL20181602).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dean SG, Ricotta EE, Fintzi J, Lai YL, Kadri SS, Olivier KN, et al. Mycobacterial testing trends, United States, 2009-2015. Emerg Infect Dis. (2020) 26:2243–6. doi: 10.3201/eid2609.200749
2. Mortaz E, Moloudizargari M, Varahram M, Movassaghi M, Garssen J, Kazempour Dizagie M, et al. What immunological defects predispose to non-tuberculosis mycobacterial infections? Iran J Allergy Asthma Immunol. (2018) 17:100–9.
3. Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. (2013) 42:1604–13. doi: 10.1183/09031936.00149212
4. Li G, Pang H, Guo Q, Huang M, Tan Y, Li C, et al. Antimicrobial susceptibility and MIC distribution of 41 drugs against clinical isolates from China and reference strains of nontuberculous mycobacteria. Int J Antimicrob Agents. (2017) 49:364–74. doi: 10.1016/j.ijantimicag.2016.10.024
5. Jing H, Tan W, Deng Y, Gao D, Li L, Lu Z, et al. Diagnostic delay of pulmonary nontuberculous mycobacterial infection in China. Multidiscip Respir Med. (2014) 9:48. doi: 10.1186/2049-6958-9-48
6. Zhu Y, Hua W, Liu Z, Zhang M, Wang X, Wu B, et al. Identification and characterization of nontuberculous mycobacteria isolated from suspected pulmonary tuberculosis patients in eastern china from 2009 to 2019 using an identification array system. Braz J Infect Dis. (2022) 26:102346. doi: 10.1016/j.bjid.2022.102346
7. Huang JJ, Li YX, Zhao Y, Yang WH, Xiao M, Kudinha T, et al. Prevalence of nontuberculous mycobacteria in a tertiary hospital in Beijing, China, January 2013 to December 2018. BMC Microbiol. (2020) 20:158. doi: 10.1186/s12866-020-01840-5
8. Yu XL, Lu L, Chen GZ, Liu ZG, Lei H, Song YZ, et al. Identification and characterization of non-tuberculous mycobacteria isolated from tuberculosis suspects in Southern-central China. PLoS ONE. (2014) 9:e114353. doi: 10.1371/journal.pone.0114353
9. Ji LC, Chen S, Piao W, Hong CY, Li JL, Jiang Q. Increasing trends and species diversity of nontuberculous mycobacteria in a coastal Migrant City-Shenzhen, China. Biomed Environ Sci. (2022) 35:146–50. doi: 10.3967/bes2022.020
10. Pang Y, Tan Y, Chen J, Li Y, Zheng H, Song Y, et al. Diversity of nontuberculous mycobacteria in eastern and southern China: a cross-sectional study. Eur Respir J. (2017) 49:1601429. doi: 10.1183/13993003.01429-2016
11. Shao Y, Chen C, Song H, Li G, Liu Q, Li Y, et al. The epidemiology and geographic distribution of nontuberculous mycobacteria clinical isolates from sputum samples in the eastern region of China. PLoS Negl Trop Dis. (2015) 9:e0003623. doi: 10.1371/journal.pntd.0003623
12. Liu CF, Song YM, He WC, Liu DX, He P, Bao JJ, et al. Nontuberculous mycobacteria in China: incidence and antimicrobial resistance spectrum from a nationwide survey. Infect Dis Poverty. (2021) 10:59. doi: 10.1186/s40249-021-00844-1
13. Kim MJ, Kim KM, Shin JI, Ha JH, Lee DH, Choi JG, et al. Identification of nontuberculous mycobacteria in patients with pulmonary diseases in gyeongnam, Korea, using multiplex PCR and multigene sequence-based analysis. Can J Infect Dis Med Microbiol. (2021) 2021:8844306. doi: 10.1155/2021/8844306
14. Adekambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. (2003) 41:5699–708. doi: 10.1128/JCM.41.12.5699-5708.2003
15. Wang X, Li H, Jiang G, Zhao L, Ma Y, Javid B, et al. Prevalence and drug resistance of nontuberculous mycobacteria, northern China, 2008-2011. Emerg Infect Dis. (2014) 20:1252–3. doi: 10.3201/eid2007.131801
16. Thangavelu K, Krishnakumariamma K, Pallam G, Dharm Prakash D, Chandrashekar L, Kalaiarasan E, et al. Prevalence and speciation of non-tuberculous mycobacteria among pulmonary and extrapulmonary tuberculosis suspects in South India. J Infect Public Health. (2021) 14:320–3. doi: 10.1016/j.jiph.2020.12.027
17. Lee YM, Kim MJ, Kim YJ. Increasing trend of nontuberculous mycobacteria isolation in a referral clinical laboratory in South Korea. Medicina. (2021) 57:720. doi: 10.3390/medicina57070720
18. Chou MP, Clements AC, Thomson RM. A spatial epidemiological analysis of nontuberculous mycobacterial infections in Queensland, Australia. BMC Infect Dis. (2014) 14:279. doi: 10.1186/1471-2334-14-279
19. Greif G, Coitinho C, van Ingen J, Robello C. Species distribution and isolation frequency of nontuberculous mycobacteria, uruguay. Emerg Infect Dis. (2020) 26:1014–8. doi: 10.3201/eid2605.191631
20. Okoi C, Anderson STB, Antonio M, Mulwa SN, Gehre F, Adetifa IMO. Non-tuberculous mycobacteria isolated from pulmonary samples in sub-saharan africa - a systematic review and meta analyses. Sci Rep. (2017) 7:12002. doi: 10.1038/s41598-017-12175-z
21. Drzyzga O. The strengths and weaknesses of Gordonia: a review of an emerging genus with increasing biotechnological potential. Crit Rev Microbiol. (2012) 38:300–16. doi: 10.3109/1040841X.2012.668134
22. Andalibi F, Fatahi-Bafghi M. Gordonia: isolation and identification in clinical samples and role in biotechnology. Folia Microbiol. (2017) 62:245–52. doi: 10.1007/s12223-017-0491-1
23. Gil-Sande E, Brun-Otero M, Campo-Cerecedo F, Esteban E, Aguilar L, Garcia-de-Lomas J. Etiological misidentification by routine biochemical tests of bacteremia caused by Gordonia terrae infection in the course of an episode of acute cholecystitis. J Clin Microbiol. (2006) 44:2645–7. doi: 10.1128/JCM.00444-06
24. Khadka P, Basnet RB, Rijal BP, Sherchand JB. Pulmonary nocardiosis masquerading renascence of tuberculosis in an immunocompetent host: a case report from Nepal. BMC Res Notes. (2018) 11:488. doi: 10.1186/s13104-018-3604-2
25. Scoffone VC, Chiarelli LR, Trespidi G, Mentasti M, Riccardi G, Buroni S. Burkholderia cenocepacia infections in cystic fibrosis patients: drug resistance and therapeutic approaches. Front Microbiol. (2017) 8:1592. doi: 10.3389/fmicb.2017.01592
26. Gee JE, Elrod MG, Gulvik CA, Haselow DT, Waters C, Liu L, et al. Burkholderia thailandensis isolated from infected wound, Arkansas, USA. Emerg Infect Dis. (2018) 24:2091–4. doi: 10.3201/eid2411.180821
27. Fomda B, Velayudhan A, Siromany VA, Bashir G, Nazir S, Ali A, et al. An outbreak of Burkholderia cepacia bloodstream infections in a tertiary-care facility in northern India detected by a healthcare-associated infection surveillance network. Infect Control Hosp Epidemiol. (2022) 2022:1–7. doi: 10.1017/ice.2022.111
28. Dobrovic K, Marekovic I, Payerl-Pal M, Andrijasevic N, Skrobo T, Koscak V, et al. Outbreak of healthcare-associated bacteremia caused by Burkholderia gladioli due to contaminated multidose vials with saline solutions in three Croatian hospitals. Int J Infect Dis. (2022) 121:152–6. doi: 10.1016/j.ijid.2022.05.012
29. Huse HK, Lee MJ, Wootton M, Sharp SE, Traczewski M, LiPuma JJ, et al. Evaluation of antimicrobial susceptibility testing methods for burkholderia cenocepacia and burkholderia multivorans isolates from cystic fibrosis patients. J Clin Microbiol. (2021) 59:e0144721. doi: 10.1128/JCM.01447-21
30. Liu H, Lian L, Jiang Y, Huang M, Tan Y, Zhao X, et al. Identification of species of nontuberculous mycobacteria clinical isolates from 8 provinces of China. Biomed Res Int. (2016) 2016:2153910. doi: 10.1155/2016/2153910
31. Singh AK, Maurya AK, Umrao J, Kant S, Kushwaha RA, Nag VL, et al. Role of genotype((R)) mycobacterium common mycobacteria/additional species assay for rapid differentiation between mycobacterium tuberculosis complex and different species of non-tuberculous mycobacteria. J Lab Phys. (2013) 5:83–9. doi: 10.4103/0974-2727.119847
32. Xu Y, Liang B, Du C, Tian X, Cai X, Hou Y, et al. Rapid identification of clinically relevant mycobacterium species by multicolor melting curve analysis. J Clin Microbiol. (2019) 57. doi: 10.1128/JCM.01096-18
33. Lee H, Myung W, Koh WJ, Moon SM, Jhun BW. Epidemiology of nontuberculous mycobacterial infection, South Korea, 2007-2016. Emerg Infect Dis. (2019) 25:569–72. doi: 10.3201/eid2503.181597
34. Russell CD, Claxton P, Doig C, Seagar AL, Rayner A, Laurenson IF. Non-tuberculous mycobacteria: a retrospective review of Scottish isolates from 2000 to 2010. Thorax. (2014) 69:593–5. doi: 10.1136/thoraxjnl-2013-204260
Keywords: mycobacterium, non-tuberculous mycobacteria, species, identification, M. intracellulare
Citation: Sun Q, Yan J, Liao X, Wang C, Wang C, Jiang G, Dong L, Wang F, Huang H, Wang G and Pan J (2022) Trends and Species Diversity of Non-tuberculous Mycobacteria Isolated From Respiratory Samples in Northern China, 2014–2021. Front. Public Health 10:923968. doi: 10.3389/fpubh.2022.923968
Received: 20 April 2022; Accepted: 23 June 2022;
Published: 13 July 2022.
Edited by:
Jun Chen, Fudan University, ChinaReviewed by:
Teca C. Galvao, Oswaldo Cruz Foundation (Fiocruz), BrazilJulius Mugweru, University of Embu, Kenya
Copyright © 2022 Sun, Yan, Liao, Wang, Wang, Jiang, Dong, Wang, Huang, Wang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhua Pan, cHhtMTk2MEBzb2h1LmNvbQ==; Guirong Wang, d2FuZ2d1aXJvbmcxMjMwQGNjbXUuZWR1LmNu
†These authors have contributed equally to this work
 Qing Sun1†
Qing Sun1† Guirong Wang
Guirong Wang