
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 26 July 2022
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.876949
The use of machine learning (ML) for diagnosis support has advanced in the field of health. In the present paper, the results of studying ML techniques in a tuberculosis diagnosis loop in a scenario of limited resources are presented. Data are analyzed using a tuberculosis (TB) therapy program at a health institution in a main city of a developing country using five ML models. Logistic regression, classification trees, random forest, support vector machines, and artificial neural networks are trained under physician supervision following physicians' typical daily work. The models are trained on seven main variables collected when patients arrive at the facility. Additionally, the variables applied to train the models are analyzed, and the models' advantages and limitations are discussed in the context of the automated ML techniques. The results show that artificial neural networks obtain the best results in terms of accuracy, sensitivity, and area under the receiver operating curve. These results represent an improvement over smear microscopy, which is commonly used techniques to detect TB for special cases. Findings demonstrate that ML in the TB diagnosis loop can be reinforced with available data to serve as an alternative diagnosis tool based on data processing in places where the health infrastructure is limited.
Artificial intelligence (AI) is a set of bioinspired algorithms that are used to solve problems in different applications. Within this wide area, machine learning (ML) is a common subfield in which models learn from examples of data, taking advantage of the idea of adjusting parameters in classification or regression tasks (1). There are several different ML models according to the fundamental concepts for adapting the parameters, with diverse examples including naive Bayes, decision or classification trees, support vector machines (SVM), and artificial neural networks (ANNs), which emulate the behavior of the brain through connectionist models. Besides these and other ML models, new models are continuously being proposed (2).
Tuberculosis (TB) is a disease caused by the Mycobacterium tuberculosis bacillus, and the World Health Organization still considers it a global emergency because of its high estimate of more than 1.4 million fatalities in the last 3 years (3). In developing countries, TB incidence is as high as 282,000 new cases in recent years with a mortality rate of 2.4 per 100,000 populations. In one specific place, Colombia, the reported TB incidence was 33, the prevalence was 48, and the mortality was 1.6 per 100,000 populations. Given these numbers, any contribution to decreasing TB fatalities is welcomed. M. tuberculosis is slow-growing and replicates itself every 24 h, an important fact that determines subacute symptoms. Additionally, the main organ affected by TB is the lung, and because of this, the main signs of the disease are respiratory-related (3). Coughing and expectoration allow for assessing the probability of TB by studying sputum; however, because TB is an infectious disease, the accurate diagnosis is microbiological (4).
In the health area, AI has been applied to solve problems in public health, medical images analysis, and diagnosis support systems (5–8). For TB, different approaches have been proposed since 1999 with the work of El-Solh et al. (9), for whom medical images were the main source of information. Advances in this field have allowed for better detecting thoracic diseases including TB, pneumonia, asthma, and cancer (10, 11). Investigators have widely used specific ML models in health systems to contribute to improving TB diagnosis by taking advantage of available meaningful data (12, 13), such as data from clinical information (14–16), or molecular biology (17, 18).
ANNs have been particularly valuable in incorporating ML into TB diagnosis through different architectures such as multilayer perceptrons (MLP), self-organizing maps, and adaptive resonance theory (ART) joined to fuzzy models in the Fuzzy-ART approach to support detection and clustering in risk groups for pulmonary TB (19–21) and pleural TB (22–24). Researchers have used different data sources to support health professionals in daily tasks such as collecting breathing acoustic signals (25) and other clinical variables (20, 26).
Finally, TB researchers have used deep learning (DL) architecture using vast data sets to provide scenarios based on images (27–29). For instance, one important task was establishing the ImageCLEF data set, which allowed users to determine TB type and treatment resistance using coaxial tomography images (28, 30); researchers have also used images from radiography to support health professionals' decision making (31–33). Generally, DL has been widely applied in assisting with medical diagnosis, utilizing radiography images, and obtaining highlight results (34, 35). Additionally, one DL subfield, transfer learning, entails refining large pretrained models with new data, and several researchers have applied transfer learning to the same kinds of medical images (27, 36).
Nevertheless, despite its demonstrable benefits, ML's effectiveness can be limited by data availability constraints related to inadequate information technology infrastructure. Precarious health systems that cannot or do not collect radiographic information or conduct specialized testing significantly complicate the implementation of ML models. Researchers have analyzed these characteristics and proposed infrastructure for developing regions that can accommodate few variables and poor information systems have been treated for developing regions (19, 21).
The present work proposes ML techniques as a tool in the loop of TB diagnosis, where health professionals make decisions but with extra help based on limited available data. This scenario is studied for using ML in situations with limited infrastructure for application within the complete TB diagnosis protocol.
The concept of the “algorithm-in-the-loop” is related to the use of ML models to support decision making and improve both human–computer interactions and human performance (37). Interaction between the model and users in a loop is not limited to simple representations of performance such as numbers but extends to a global idea that articulates ethics, policies, and standards (38). Including AI and ML stages in the clinical decision making support workflow can ultimately improve patient experiences and outcomes and optimize health system performance (8). Interactive ML is another term for when algorithms and humans work together to improve the results in terms of metrics, understandability, and outcomes (39).
For the case of TB, diagnosis was long based on respiratory symptoms followed by testing suspicious patients with a serial sputum smear; however, although this test is simple, it is necessary to consider some aspects in determining its usefulness. Smear microscopy is performed using sputum smear and staining that allows direct microscopic visualization of the bacillus. However, diagnostic sensitivity is low, around 60%, because a high number of microorganisms per cubic millimeter of a sample is required to obtain results (40). Indeed, a high percentage of people with the disease cannot be diagnosed using this method, and furthermore, detected bacillus could be a non-TB mycobacterium. A more sensitive assay is a culture in either solid or liquid medium, which needs at least 2 weeks to obtain results (41). Following more recent advances, molecular testing is now available: Polymerase chain reaction (PCR) identifies the TB bacillus with high sensitivity and in approximately 2 h (42). However, the infrastructure for this technology is limited in developing countries such as Colombia.
From the ML point of view, different applications have particular characteristics such as requiring biomedical data that have high uncertainty and incompleteness (43), and strategies beyond straightforward ML are sometimes demanded. For the present study, ML in the loop (MLL) is investigated; this strategy depends on how the ML tool will be used. Researchers have analyzed the necessary workflows to improve results (44), but in medicine, where health professionals play an indispensable role, other investigators have studied the doctor-in-the-loop in terms of system performance (45, 46). Today, how ML models perform is no longer the sole concern; models' generalizability and functionality during human interaction are also important. Assessing these broader aspects of performance allows for understanding important aspects of decision making and operation that must be considered in system designs (47).
Figure 1 depicts the MLL process for TB diagnosis support that was studied for the present work. First, a subject with respiratory disease symptoms arrives at the medical center for either a consultation or an emergency. There, a member of the medical staff examines the possible patient and then sends the patient to internal medicine for a more detailed examination. After this deeper analysis, if the patient's respiratory symptoms continue, medical staff request three main exams to detect pulmonary TB: sputum smear microscopy, sputum culture, and molecular assay (GenXpert®). If results from these three exams indicate infection, the patient begins antituberculosis therapy. Meanwhile the results are definitive, there is no positive diagnosis. However, the patient initialize the antituberculosis treatment. It is at this point where ML was applied to assist the medical staff members in diagnosis.
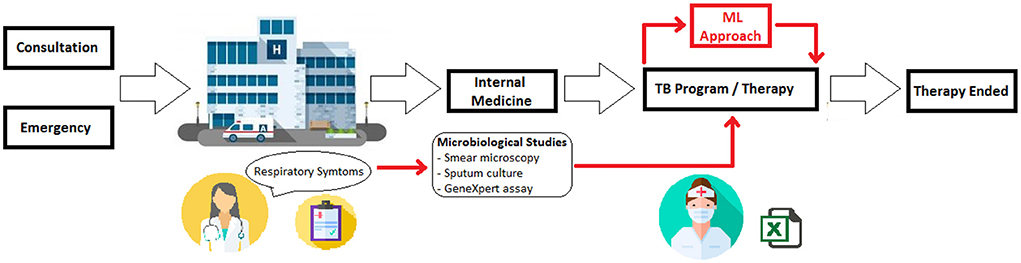
Figure 1. Schematic of using ML in TB diagnosis. During the TB diagnosis, ML tools are employed to support the decision about the antituberculosis therapy beginning.
At the study facility, the health care workers are responsible for acquiring basic patient information equivalent to the medical records obtained in other stages. This information is input into a registry for the use of the institution's TB program; the protocol to detect TB can be time-consuming, and using ML with this registry could expedite diagnosis. This study proposed to apply MLL searches to support health care workers during the time the test results take. This allows staff to efficiently manage patient treatment according to the need for isolation, hospital capacity, and necessary medications.
Data were acquired through the TB program at Hospital Santa Clara (HSC) in Bogotá D.C., Colombia. The HSC is an important public institution associated with the Subred Integrada de Servicios de Salud Centro Oriente (SCO, Middle East Subnetwork of Health Services) that treats vulnerable populations with low socioeconomic status or high risk of sexually transmitted infections as well as persons who live in overcrowded conditions.
As explained earlier, the data were collected within the hospital's traditional TB diagnosis process. Information was considered from 233 clinical suspected pulmonary TB subjects whose data had been acquired in the period from January 2017 to December 2019. From this set, 184 subjects (79%) had TB confirmed and 36 subjects (15%) were determined to be disease-free based on smear microscopy, culture, and molecular examination following the national protocol to diagnose TB (48). Thirteen subjects were not considered because they had no available information on their TB status. The Ethics and Research Committee of the SCO approved this study on the basis of the use of anonymous data with only population-related variables that posed no risks to subjects. Informed consent was not required because all data were retrospective and anonymous.
At the HSC, electronic health records are used, but they are not standardized across the country; records can include diagnoses and symptoms of medical conditions such as diabetes, chronic kidney disease, and immunosuppression such as by the human immunodeficiency virus (HIV). Sociodemographic variables are also important for TB diagnosis (49), and the SCO commonly treats vulnerable populations such as persons who are indigenous, homeless, migrants, or refugees for TB. Although some of the data are available, the different information systems do not always communicate with each other. For this reason, only the variables that were available at the beginning of the TB program were applied for this study, as specified above. Using only these data allowed for simulating a scenario with limited information.
Health care workers at this point of TB diagnosis collect only seven variables, which were the ones considered in the present work: sex, age, type of population, city location, HIV/AIDS (acquired immunodeficiency syndrome) status, antiretroviral treatment status, and the number of days since treatment onset (see Table 1). Age and number of days were discrete numeric variables that were normalized by maximum of 100 and 15, respectively. Sex was a binary variable where a patient was either male or female, and this variable was set at 00 when no data were available. HIV and antiretroviral treatment status could take either of three possible values: positive, negative, or unknown. Finally, the type of population and city location were, respectively, coded with zeros and ones to reflect if a clinic visitor was a member of a specific vulnerable group and where in Bogotá City the client resided based on established geographic divisions.
ML models are a set of algorithms that learn from data (50). For the present study, four MLL models were compared for their usefulness to health professionals and for the interactions between available features in the TB decision making process. In health sciences, logistic regression (LR) algorithms are widely applied to associate predictors or input variables to an output that represents a detection or estimation of the illness (41, 51). To evaluate the present scenario, LR was the fifth model considered to determine the possible contribution of traditional tools. The optimization algorithm was based on a quasi-Newton method, the Broden–Fletcher–Goldfarb–Shanno (lbfgs) approximation; additionally, penalization was used with a maximum of 100 iterations.
Classification or decision tree (DT) algorithms are trained through supervised learning and are considered a non-parametric method for classification or regression (52). DT structure is based on nodes and leaves, where each node is represented by a function that divides the information flow into two or more classes according to the function's output. For the present case, this function was based on the Gini coefficient. A notable advantage of this ML model is that it allows for visually determining the conditions for the input variables and the leaves. Random forest (RF) is a special DT model, in which more tree structures are analyzed and tested (53, 54). Then, the best configuration of trees is selected for the classification or regression, according to a sample from the data set and avoiding model overfitting.
SVMs deal with the boundary between hyperplanes that divides the data classes from input variables represented in a features space (55, 56). The hyperplanes are built from support vectors obtained from the training data and optimized according to the support vectors with the best performance. This model is widely applied with kernelling, modifying the initial non-linear separable space into a linear separation through a non-linear kernel that for the present case was Gaussian.
Finally, an MLP was applied as a model to detect the TB cases because the results were known in this specific problem (57). For this case, an architecture with one hidden layer was trained to detect TB. The number of input nodes was equal to the number of variables, and there was one output node. Resilient backpropagation was applied for training and stop criteria with a maximum of 500 epochs, zero gradients, and early stopping, the first time early stopping was considered.
Cross-validation was conducted to assess the performance and generalization of the models (58). Based on the special scenario under study, the mode of data acquisition, and the possibility of a system application in the future, the data were divided into three sets. This allowed for establishing the models based on 2 years of data that were validated and tested for generalizability in the third year. Through this process, the tool can be used using previous information with similar properties. Table 2 shows these sets, the year of acquisition, and the number of instances per set.
A process to balance the classes was implemented, searching to adjust the inequality between positive and negative TB for the classes. In this case, a weighted training process of internal parameters for each model was regulated according to the frequency of the instances by class (59).
Study variables were analyzed through the performance computation for each ML model under study. The variables in Table 1 were converted to zero and then applied to the best trained of the DT, LR, RF, SVM, and MLP models. Subsequently, model performance metrics such as accuracy, sensitivity, and specificity were compared.
Automated ML (aML) was also tested to find the best models (60), and the Tree-based Pipeline Optimization Tool (TPOT) was applied to obtain the best detectors (61). This was carried out because of differences in the ML models' performance. Here aML and TPOT were used to compare the individual models' performance and to determine the influences of the ML model parameters in the search results.
Table 3 shows the findings for the training process and the test scores with data from the year left out in the cross-validation described before; accuracy (ACC), sensitivity (SE), and specificity (SP) were collected to determine the differences due to the balance between positive and negative TB for each year (see Table 2). Additionally, the area under the receiver operating curve (AUC) allowed for considering SE and SP simultaneously.
The LR, RF, and MLP models achieved the best results, obtaining the highest AUC, 0.84, in the test set (see Table 3). This value can be compared with the maximum AUC of 0.96 in the DT model for the training set, demonstrating that it was difficult to generalize the findings from the present application.
Table 4 presents the ACC, SE, SP, and AUC means and standard deviations for the three test data subsets. The table shows that MLP obtained the best results for ACC, SE, and AUC and that SP was the best with the LR model. These findings suggest that combining models might give better results for these metrics. Nevertheless, although SP was the best with the LR, that model had the worst results for ACC and SE, which suggests this model's suitability for the objective task of finding negative TB cases. Finally, the SVM model gave the worst results for most metrics.
Table 5 presents the best results for each metric for all the studied models and the full data set, showing that the LR model had the best accuracy, SVM had the best sensitivity, and MLP had the best specificity. Additionally, following subsection 3.3, all models were checked for relevance. Specifically, for each model, the input variables (see Table 1) were set at 0, and then, ACC, SE, and SP were computed. Figure 2 shows the effect of this processing, notably that type of population was not important in the LR, RF, and MLP models; when the zero values were eliminated, the models' performance improved. Figure 2D shows that age caused significant differences in the SVM model. Finally, all variables were relevant in the MLP model.

Figure 2. Sensitivity, accuracy, and specificity for all five ML models: (A) Logistic regression; (B) Classification tree; (C) Random forest; (D) Support vector machine; (E) Multilayer perceptron neural network. For all ML models is visualized the effect of using or not each one of the considered variables in terms of sensitivity (blue), specificity (green) and accuracy (orange). There it is possible to see how the metrics change, according to the inclusion or exclusion of the seven variables.
Table 6 presents the findings from testing aML and TPOT, which require less intensive user exploration of the hyperparameters. The table shows that the automated ML was more successful than manual exploration (see Table 3), although the results were similar. The first model, for the year 2019, applied six ML models: two passive-aggressive, two MLPs, one extra tree, and one gradient boosting. The second model, for 2018, had 28 models that included a number of the different strategies presented here (e.g., MLP, RF, and logistic regressors). For the 2017 case, aML produced a combination of five models (two random forests, one mlp, one passive-aggressive, and one stochastic gradient descent). Table 7 presents the aML and TPOT results for all 3 years. Specificity is considerably affected in this automatic generation of models, which is ineffective and not appropriate in the context of diagnosis support.
TB detection in earlier stages is important to prevent transmission of the disease. However, irrespective of when a patient is diagnosed, patients in the populations studied in this work must be kept in isolation because these patients tend not to maintain safe distances as they are being treated.
Because of the lack of specific clinical symptoms, it is difficult for physicians to diagnose tuberculosis, but meanwhile, patients require rapid isolation to prevent spreading the disease to others. Presumptive TB cases require further analysis, and tools for completing specific tasks could reduce the workloads of health professionals. ML and AI could be effective in this context while keeping decisions under the purview of the medical staff. Furthermore, in developing or low-income countries such as Colombia, ML and AI can extend the availability of health care to remote regions with limited infrastructure and few if any health care personnel.
There remain many challenges to applying ML and AI in the health informatics field, but doing so can contribute to easing burdens for clinical personnel; further testing of these applications in real-world settings will be highly beneficial. Furthermore, the coworking between health professionals and health care AI is a challenge. The American Medical Association calls for considering AI an augmentation to human intelligence rather than a replacement (62). Recent authors have reported on developing this kind of articulation with health professionals as the center of the entire strategy (12).
In this study, the high incidence rate in the analyzed data set was related to the stage of the diagnosis process, although despite this, it is possible to see that not all presumptive TB cases were ultimately diagnosed as positive TB. This indicates that the ML tool identified variables that were imperceptible to humans, which could help improve therapy management as well as increase the efficient allocation of clinical resources (time, professional staff, medicaments, space, etc.). However, it was determined in this study that the unbalance between positive and negative TB cases could be offer a difficulty of the ML models training (59). However, the RF, LR, and MLP models achieved similar results for SE and SP, consistent with earlier findings for MLP models (19, 21, 33, 55); these findings support RF, LR, and MLP as appropriate models for diagnosis support. In the present study, MLP had the best AUC metric, which exhibits best balance between SE and SP. Additionally, the proposed models can decrease the number of cases for which treatment begins without a confirmed diagnosis, which should decrease health system costs in time and other resources. Regarding aML and TPOT, finding the hyperparameters was not a dilemma, but the SP results were not as good as they were with other models. Furthermore, it is common for health informatics applications to have access to only small data sets or represent only rare events, and these conditions significantly reduce the accuracy of the results from aML approaches (60, 61).
Diagnostic algorithms have been incorporated into several national and international recommendations and guidelines for optimizing patient approaches. In the case of Colombia, health entities must notify the alert surveillance system of public health diseases, to epidemiologically monitor and clinically control TB to verify the success of the treatment. National TB registries allow for acquiring adequate global information on all the current clinical and sociodemographic aspects of TB as well as the success of the treatment strategies used.
In terms of limitations of the present study, there was a high incidence of TB in the data set, which could have induced bias in the analyzed data; addressing this will require more specific scenarios that involve clinical observation. Additionally, TB culture is considered the gold standard for diagnosis in some cases, especially when the infrastructure of GenExpert is not available. In this study, although the hospital database can only hold a limited number of patients, the HSC is an important center for TB treatment in Bogotá City; future researchers could incorporate data from more institutions that treat TB. Finally, researchers could incorporate more technical aspects such as including ensemble methods, combining different ML models, and considering more sophisticated models as the next steps.
The findings of this study make it possible to conclude that sensitive ML algorithms can support TB diagnosis by considering the clinical features of the cases as well as medical and sociodemographic risk factors of the patients. TB continues to be a global leading cause of death, and challenges remain in identifying, treating, and containing the disease in several communities. The mycobacteria–host relationship can delay diagnosis for a host of reasons, as can limited clinical resources for diagnosis. Computational tools such as those studied here can support timely TB diagnosis and treatment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Conceptualization: AO-C and CA. Methodology, supervision, and resources: AO-C and AJ. Software, writing—original draft preparation, funding acquisition, and visualization: AO-C. Validation: AO-C, CA, EV, and AP. Formal analysis, investigation, and writing—review and editing: AO-C, AJ, CA, EV, and AP. Data curation: AO-C, CA, and AJ. Project administration: AJ. All authors have read and agreed to the published version of the manuscript.
This research was funded by Ministerio de Ciencia, Tecnologia e Innovación of Colombia—Minciencias, grant number 123380762899.
The authors acknowledge the support of the Ministerio de Ciencia y Tecnología–Minciencias of Colombia, funded through project 123380762899. Additionally, Universidad Antonio Nariño, the Subred Integrada de Servicios de Salud Centro Oriente, and Universidad del Rosario were relevant for the development of this work, according to the availability of computational resources and staff time dedicated to the authors team.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Panch T, Szolovits P, Atun R. Artificial intelligence, machine learning and health systems. J Glob Health. (2018) 8:020303. doi: 10.7189/jogh.08.020303
2. Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical Machine Learning Tools and Techniques. New York, NY, USA: Morgan Kaufmann (2016).
4. Fogel N. Tuberculosis: a disease without boundaries. Tuberculosis. (2015) 95:527–31. doi: 10.1016/j.tube.2015.05.017
5. Wahl B, Cossy-Gantner A, Germann S, Schwalbe NR. Artificial intelligence (AI) and global health: how can AI contribute to health in resource-poor settings? BMJ Glob Heal. (2018) 3:e000798. doi: 10.1136/bmjgh-2018-000798
6. Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. (2017) 2:230–43. doi: 10.1136/svn-2017-000101
7. For International Development, U.S.A. Artificial Intelligence in Global Health (2019). Available online at: https://www.usaid.gov/sites/default/files/documents/1864/AI-in-Global-Health_webFinal_508.pdf
8. Chen M, Decary M. Artificial intelligence in healthcare: an essential guide for health leaders. Healthc Manage Forum. (2020) 33:10–8. doi: 10.1177/0840470419873123
9. El-Solh AA, Hsiao C-B, Goodnough S, Serghani J, Grant BJB. Predicting active pulmonary tuberculosis using an artificial neural network. Chest J. (1999) 116:968–73. doi: 10.1378/chest.116.4.968
10. Er O, Yumusak N, Temurtas F. Chest diseases diagnosis using artificial neural networks. Expert Syst Appl. (2010) 37:7648–55. doi: 10.1016/j.eswa.2010.04.078
11. Meraj SS, Yaakob R, Azman A, Rum SNM, Nazri ASA. Artificial intelligence in diagnosing tuberculosis: a review. Int J Adv Sci Eng Inf Technol. (2019) 9:81–91. doi: 10.18517/ijaseit.9.1.7567
12. Awaysheh A, Wilcke J, Elvinger F, Rees L, Fan W, Zimmerman KL. Review of medical decision support and machine-learning methods. Vet Pathol. (2019) 56:512–25. doi: 10.1177/0300985819829524
13. Michael KY, Ma J, Fisher J, Kreisberg JF, Raphael BJ, Ideker T. Visible machine learning for biomedicine. Cell. (2018) 173:1562–5. doi: 10.1016/j.cell.2018.05.056
14. Whang J, Wang C, Wenyu Z. Data analysis and forecasting of tuberculosis prevalence rates for smart healthcare based on a novel combination model. Appl Sci. (2018) 8:1–24. doi: 10.3390/app8091693
15. Nagabhushanam D, Naresh N, Raghunath A, Praveen Kumar K. Prediction of tuberculosis using data mining techniques on indian patients data. IJCST. (2013) 4:262–5.
16. dos Santos Alves E, Souza Filho JBO, Galliez RM, Kritski A. Specialized MLP classifiers to support the isolation of patients suspected of pulmonary tuberculosis. In Proceedings of the Computational Intelligence and 11th Brazilian Congress on Computational Intelligence (BRICS-CCI & CBIC). 2013 BRICS Congress on (2013). p. 40–5.
17. Deelder W, Christakoudi S, Phelan J, Benavente ED, Campino S, McNerney R, et al. Machine learning predicts accurately Mycobacterium tuberculosis drug resistance from whole genome sequencing data. Front Genet. (2019) 10:922. doi: 10.3389/fgene.2019.00922
18. Bobak CA, Titus AJ, Hill JE. Comparison of common machine learning models for classification of tuberculosis using transcriptional biomarkers from integrated datasets. Appl Soft Comput. (2019) 74:264–73. doi: 10.1016/j.asoc.2018.10.005
19. Orjuela-Cañón AD, Mendoza JEC, García CEA, Vela EPV. Tuberculosis diagnosis support analysis for precarious health information systems. Comput Methods Programs Biomed. (2018) 157:11-7. doi: 10.1016/j.cmpb.2018.01.009
20. E Souza JBdO, Sanchez M, de Seixas JM, Maidantchik C, Galliez R, Moreira A da SR, et al. Screening for active pulmonary tuberculosis: development and applicability of artificial neural network models. Tuberculosis. (2018) 111:94–101. doi: 10.1016/j.tube.2018.05.012
21. Aguiar FS, Torres RC, Pinto JVF, Kritski AL, Seixas JM, Mello FCQ. Development of two artificial neural network models to support the diagnosis of pulmonary tuberculosis in hospitalized patients in Rio de Janeiro, Brazil. Med Biol Eng Comput. (2016) 54:1751–9. doi: 10.1007/s11517-016-1465-1
22. Orjuela-Cañón AD, de Seixas JM, Trajman A. SOM Neural Networks as a Tool in Pleural Tuberculosis Diagnostic. In: Braga AdeP, Bastos Filho CJA, Editors. Proceedings of the Annals of the 11th Brazilian Congress on Computational Intelligence. Porto de Galinhas, PE: SBIC (2013). p. 1–5.
23. Orjuela-Canon AD, De Seixas J. Fuzzy-ART neural networks for triage in pleural tuberculosis. In Proceedings of the Pan American Health Care Exchanges, PAHCE. (Medellin, Colombia) (2013). doi: 10.1109/PAHCE.2013.6568342
24. Seixas JM, Faria J, Souza F, Vieira AFM, Kritski A, Trajman A. Artificial neural network models to support the diagnosis of pleural tuberculosis in adult patients. Int J Tuberc Lung Dis. (2013) 17:682–6. doi: 10.5588/ijtld.12.0829
25. Becker KW, Scheffer C, Blanckenberg MM, Diacon AH. Analysis of adventitious lung sounds originating from pulmonary tuberculosis. In: Proceedings of the Engineering in Medicine and Biology Society (EMBC), 2013 35th Annual International Conference of the IEEE (2013). p. 4334–7. doi: 10.1109/EMBC.2013.6610505
27. Rajaraman S, Antani SK. Modality-specific deep learning model ensembles toward improving TB detection in chest radiographs. IEEE Access. (2020) 8:27318–26. doi: 10.1109/ACCESS.2020.2971257
28. Gao XW, Qian Y. Prediction of multidrug-resistant TB from CT pulmonary images based on deep learning techniques. Mol Pharm. (2017) 15:4326–35. doi: 10.1021/acs.molpharmaceut.7b00875
29. Nash M, Kadavigere R, Andrade J, Sukumar CA, Chawla K, Shenoy VP, et al. Deep learning, computer-aided radiography reading for tuberculosis: a diagnostic accuracy study from a tertiary hospital in India. Sci Rep. (2020) 10:1–10. doi: 10.1038/s41598-019-56589-3
30. Cid YD, Kalinovsky A, Liauchuk V, Kovalev V, Müller H. Overview of the imageclef 2017 tuberculosis task-predicting tuberculosis type and drug resistances. In: Proceedings of the CLEF (Working Notes) (Dublin, Ireland) (2017).
31. Jaeger S, Karargyris A, Candemir S, Folio L, Siegelman J, Callaghan F, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging. (2014) 33:233–45. doi: 10.1109/TMI.2013.2284099
32. Ding M, Antani S, Jaeger S, Xue Z, Candemir S, Kohli M, et al. Local-global classifier fusion for screening chest radiographs. in proceedings of the medical imaging 2017. Imag Inform Healthcare Res Appl. (2017) 10138:101380A. doi: 10.1117/12.2252459
33. Hwang S, Kim H-E, Jeong J, Kim H-J. A novel approach for tuberculosis screening based on deep convolutional neural networks. Proc Med Imag 2016 Comput Aided Diagn. (2016) 9785:97852W. doi: 10.1117/12.2216198
34. Hwang EJ, Park S, Jin K-N, Kim JI, Choi SY, Lee JH, et al. Development and validation of a deep learning–based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin Infect Dis. (2019) 69:739–47. doi: 10.1093/cid/ciy967
35. Qin ZZ, Sander MS, Rai B, Titahong CN, Sudrungrot S, Laah SN, et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: a multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci Rep. (2019) 9:1–10. doi: 10.1038/s41598-019-51503-3
36. Paul HY, Kim TK, Lin CT. Generalizability of deep learning tuberculosis classifier to COVID-19 chest radiographs: new tricks for an old algorithm? J Thorac Imaging. (2020) 35:W102-4. doi: 10.1097/RTI.0000000000000532
37. Green B, Chen Y. Disparate interactions: an algorithm-in-the-loop analysis of fairness in risk assessments. In: Proceedings of the Proceedings of the Conference on Fairness, Accountability, and Transparency (New York, NY, USA) (2019). p. 90–9. doi: 10.1145/3287560.3287563
38. Green B, Chen Y. The principles and limits of algorithm-in-the-loop decision making. Proc ACM Human Comput Interact. (2019) 3:1–24. doi: 10.1145/3359152
39. Holzinger A. Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inform. (2016) 3:119–31. doi: 10.1007/s40708-016-0042-6
40. Lewinsohn DM, Leonard MK, LoBue PA, Cohn DL, Daley CL, Desmond E, et al. Official american thoracic society/infectious diseases society of america/centers for disease control and prevention clinical practice guidelines: diagnosis of tuberculosis in adults and children. Clin Infect Dis. (2017) 64:e1–33. doi: 10.1093/cid/ciw694
41. Ghazvini K, Yousefi M, Firoozeh F, Mansouri S. Predictors of tuberculosis: Application of a logistic regression model. Gene Rep. (2019) 17:100527. doi: 10.1016/j.genrep.2019.100527
42. Berra TZ, Gomes D, Ramos ACV, Alves YM, Bruce ATI, Arroyo LH, et al. Effectiveness and trend forecasting of tuberculosis diagnosis after the introduction of GeneXpert in a city in south-eastern Brazil. PLoS ONE. (2021) 16:e0252375. doi: 10.1371/journal.pone.0252375
43. Holzinger A. Biomedical Informatics: Discovering Knowledge in Big Data. Graz, Austria: Springer (2014). doi: 10.1007/978-3-319-04528-3
44. Xin D, Ma L, Liu J, Macke S, Song S, Parameswaran A. Accelerating human-in-the-loop machine learning: challenges and opportunities. In: Proceedings of the Proceedings of the Second Workshop on Data Management for End-To-End Machine Learning (New York, NY, USA) (2018). p. 1–4. doi: 10.1145/3209889.3209897
45. Holzinger A. Trends in Interactive Knowledge Discovery For Personalized Medicine: Cognitive Science Meets Machine Learning (2014).
46. Robert S, Büttner S, Röcker C, Holzinger A. Reasoning under uncertainty: Towards collaborative interactive machine learning. In: Machine Learning for Health Informatics. Springer (2016). p. 357–76. doi: 10.1007/978-3-319-50478-0_18
47. Nay J, Strandburg KJ. Generalizability: Machine Learning and Humans-in-the-Loop. In: Res. Handb. BIG DATA LAW (rol. Vogl, ed., Edward Elgar, 2020 Forthcoming) (2019). p. 20–7. doi: 10.2139/ssrn.3417436
48. de Salud IN. Tuberculosis: Protocolo de Vigilancia en Salud Pública. Colombia: Instituto acional de Salud (2020).
49. Parsons LM, Somoskövi Á, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. (2011) 24:314–50. doi: 10.1128/CMR.00059-10
50. Calamuneri A, Donato L, Scimone C, Costa A, D'Angelo R, Sidoti A. On Machine Learning in Biomedicine. Life Saf Secur. (2017) 5:96–9. doi: 10.12882/2283-7604.2017.5.12
51. Ohene S-A, Fordah S, Boni P. Dela Childhood tuberculosis and treatment outcomes in Accra: a retrospective analysis. BMC Infect Dis. (2019) 19:1–9. doi: 10.1186/s12879-019-4392-6
52. Cruz APD, Tumibay GM. Predicting tuberculosis treatment relapse: a decision tree analysis of J48 for data mining. J Comput Commun. (2019) 7:243–51. doi: 10.4236/jcc.2019.77020
53. Wu Y, Wang H, Wu F. Automatic classification of pulmonary tuberculosis and sarcoidosis based on random forest. In: Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) (Piscataway, New Jersey) (2017). p. 1–5. doi: 10.1109/CISP-BMEI.2017.8302280
54. Sugirtha GE, Murugesan G. Detection of tuberculosis bacilli from microscopic sputum smear images. In: Proceedings of the 2017 Third International Conference on Biosignals, Images and Instrumentation (ICBSII) (Red Hook, NY, USA) (2017). p. 1–6. doi: 10.1109/ICBSII.2017.8082271
55. Yahiaoui A, Er O, Yumusak N. A new method of automatic recognition for tuberculosis disease diagnosis using support vector machines. Biomed Res. (2017) 28:4208-12.
56. Zulvia FE, Kuo RJ, Roflin E. An Initial Screening Method for Tuberculosis Diseases Using a Multi-objective Gradient Evolution-Based Support Vector Machine and C5. 0 Decision Tree. In: Proceedings of the 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC) (2017). p. 204–9. doi: 10.1109/COMPSAC.2017.57
57. Khan MT, Kaushik AC, Ji L, Malik SI, Ali S, Wei D-Q. Artificial neural networks for prediction of tuberculosis disease. Front Microbiol. (2019) 10:395. doi: 10.3389/fmicb.2019.00395
58. Haykin S. Neural Networks and Learning Machines. Neural networks and learning machines. Prentice Hall (2009). ISBN 978-0-13-147139-9.
59. Han W, Huang Z, Li S, Jia Y. Distribution-sensitive unbalanced data oversampling method for medical diagnosis. J Med Syst. (2019) 43:39. doi: 10.1007/s10916-018-1154-8
60. Feurer M, Klein A, Eggensperger K, Springenberg J, Blum M, Hutter F. Efficient and Robust Automated Machine Learning. In: Cortes C, Lawrence ND, Lee DD, Sugiyama M, Garnett R, Editors. Advances in Neural Information Processing Systems. Curran Associates Inc. (2015). p. 2962–70.
61. Olson RS, Bartley N, Urbanowicz RJ, Moore JH. Evaluation of a tree-based pipeline optimization tool for automating data science. In: Proceedings of the Proceedings of the Genetic and Evolutionary Computation Conference 2016. New York, NY: Association for Computing Machinery (2016). p. 485–92. doi: 10.1145/2908812.2908918
Keywords: tuberculosis diagnosis, machine learning, relevance analysis, machine learning in the loop, diagnosis support systems
Citation: Orjuela-Cañón AD, Jutinico AL, Awad C, Vergara E and Palencia A (2022) Machine learning in the loop for tuberculosis diagnosis support. Front. Public Health 10:876949. doi: 10.3389/fpubh.2022.876949
Received: 16 February 2022; Accepted: 30 June 2022;
Published: 26 July 2022.
Edited by:
ZhiMin Xiao, University of Essex, United KingdomReviewed by:
Ivan Miguel Pires, Universidade da Beira Interior, PortugalCopyright © 2022 Orjuela-Cañón, Jutinico, Awad, Vergara and Palencia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alvaro D. Orjuela-Cañón, YWx2YXJvLm9yanVlbGFAdXJvc2FyaW8uZWR1LmNv
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.