
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 10 March 2022
Sec. Environmental Health and Exposome
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.794513
This article is part of the Research TopicEnvironmental Reservoirs of Antibiotic Resistance Determinants: A Ticking Time Bomb for the Future Emergence of Super-Bugs of Environmental and Public Health ImportanceView all 8 articles
Aquatic environments, under frequent anthropogenic pressure, could serve as reservoirs that provide an ideal condition for the acquisition and dissemination of antibiotic resistance genetic determinants. We investigated the prevalence and diversity of antibiotic-resistant Escherichia coli by focusing on their genetic diversity, virulence, and resistance genes in anthropogenic-impacted Larut River. The abundance of E. coli ranged from (estimated count) Est 1 to 4.7 × 105 (colony-forming units per 100 ml) CFU 100 ml−1 to Est 1 to 4.1 × 105 CFU 100 ml−1 with phylogenetic group B1 (46.72%), and A (34.39%) being the most predominant. The prevalence of multiple antibiotic resistance phenotypes of E. coli, with the presence of tet and sul resistance genes, was higher in wastewater effluents than in the river waters. These findings suggested that E. coli could be an important carrier of the resistance genes in freshwater river environments. The phylogenetic composition of E. coli and resistance genes was associated with physicochemical properties and antibiotic residues. These findings indicated that the anthropogenic inputs exerted an effect on the E. coli phylogroup composition, diversification of multiple antibiotic resistance phenotypes, and the distribution of resistance genes in the Larut River.
Escherichia coli (E. coli) of the family Enterobacteriaceae, is a Gram-negative, facultative anaerobe, non-spore-forming, rod-shaped, commensal, and potentially pathogenic bacterium that resides largely in the gastrointestinal tracts of warm-blooded vertebrate animals (1–3). Most E. coli strains are harmless, and only some are pathogenic. The pathogenic E. coli can be classified as either intestinal pathogenic E. coli (IPEC) or extraintestinal pathogenic E. coli (ExPEC). The IPECs are major diarrhoeagenic pathogens that cause gastroenteritis with six intestinal pathotype subgroups: enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and diffusely adherent E. coli (DAEC). Meanwhile, ExPEC consists of three human pathotype subgroups: neonatal-meningitis E. coli (NMEC), uropathogenic E. coli (UPEC), sepsis-associated pathogenic E. coli (SePEC), and the avian pathogenic E. coli (APEC) (4).
Studies have shown that E. coli is a highly adaptable bacterium that can survive and grow in hostile external environments-niches (5, 6). Environmental E. coli, from aquatic riverine environments, are genetically diverse (7) and with the current application of a PCR-based diagnostic method, seven major phylogenetic groups (A, B1, B2, C, D, E, and F) and five cryptic clades are categorized (8, 9). These different phylogroups have different associations with phenotypic and genotypic traits, metabolic properties, ecotype, lifestyle, and pathogenicity (10–13).
Studies have found that antibiotic-resistant (AR) E. coli are ubiquitous in the aquatic environment and the rising emergence of multiple AR (MAR) E. coli (14, 15), coupled with the potential presence of virulent strains, is a great concern for public health (16). The antibiotic residues, AR E. coli, and their resistome concomitantly enter into the aquatic environment through wastewater discharge from anthropogenic activities (17, 18). Thus, aquatic ecosystems may serve as reservoirs that provide an ideal setting for the acquisition and dissemination of AR resistome between the environmental bacterial communities and non-pathogenic and pathogenic bacteria via horizontal gene transfer (19–21), which is also known as a lateral gene transfer (22).
Despite the different studies on E. coli from different aquatic ecosystems, phenotypic and genotypic studies of antibiotic resistance of these bacteria, and their distribution remain scarce, particularly in tropical aquatic ecosystems. The rivers of Asia are amongst the most polluted in the world and contain up to three times as much bacteria from human waste, where the reported fecal count is 50 times that of the WHO guidelines (23). In Malaysia, the major causes of pollution in rivers are related to anthropogenic activities and the sources of contamination are mainly attributed to industrial areas, sewages, workshops, residential areas, animal, and agricultural farming activities according to the Department of Environment (DOE), Malaysia (24, 25). These anthropogenic influences subsequently lead to the deterioration of water quality with elevated concentrations of heavy metals, mercury, coliforms, and nutrient loads (26).
Ghaderpour et al. (27) reported the prevalence of diverse AR E. coli in the Larut River estuarine waters, with one of the largest mangrove forests in Malaysia, and suggested anthropogenic sources as the major contributor of antibiotic resistance. Larut River is the only river flowing through the town of Taiping, Perak. This study aimed to determine the impact of anthropogenic wastewaters from a hospital, a zoo, and a poultry slaughterhouse in the town of Taiping to the occurrence, genetic diversity, and virulence of AR E. coli, as well as their resistance genes in the riverine estuarine waters of Larut River.
Sampling was conducted at six sampling sites located in the upstream, middle, and downstream of the Larut River. Larut River is 20.9 km long, serving a population of approximately 217,647 (28). Water samples were collected from upstream (S1a, 04°51.158'N, 100°45.737'E), at the reserve forest Larut Hill (elevation: 1,250 m) followed by the middle downstream where the river water received wastewater discharges from a zoo (04°51.101'N, 100°45.045'E), a public hospital (04°51.149'N, 100°44.018'E), and a slaughterhouse (04°50.238'N, 100°44.709'E) before passing through downstream Larut (S1b, 04°50.535'N, 100°43.925'E) and finally reaching the Larut Estuary (S1c, 04°50.140'N, 100°37.583'E) (Figure 1).
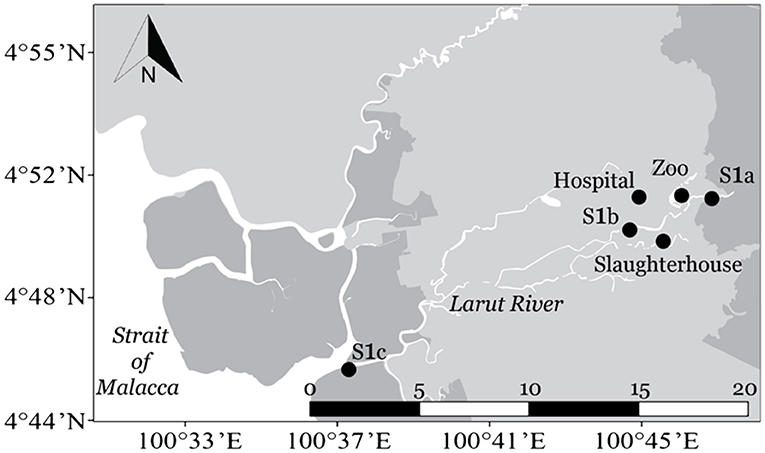
Figure 1. Map of sampling sites along Larut River. Geolocation: S1a (4°51.158'N, 100°45.737'E), Zoo (4°51.101'N, 100°45.045'E), Hospital (4°51.149'N, 100°44.018'E), Slaughterhouse (4°50.238'N, 100°44.709'E), S1b (4°50.535'N, 100°43.925'E), and S1c (4°50.140'N, 100°37.583'E).
The greywater effluents, with possible fecal contamination, were collected at the main outlet of a hospital, a slaughterhouse, and a zoo while the river surface waters were collected from S1a, S1b, and S1c of the Larut River. The water samples were collected in triplicates from each sampling site using sterilized 2 L amber glass bottles and were kept on ice until further analysis in the laboratory. A simultaneous study was conducted to measure the in situ physicochemical parameters [temperature, salinity, pH, and dissolved oxygen (DO)] and the dissolved inorganic nutrients [nitrate (NO3), nitrite (NO2), ammonium (NH4), phosphate (PO4), and silicate (SiO4)]. The physicochemical results were published earlier in Lye et al. (29).
For the isolation and enumeration of coliform and E. coli, the membrane filtration technique with the CHROMagarTM ECC media (CHROMagar Inc., Paris, France) was used. After filtering a suitable volume of sample onto a pre-sterilized 0.45 μm nitrocellulose membrane, the membrane filter was transferred onto the CHROMagarTM ECC media and incubated at 37 ± 0.5°C overnight. Blue colonies were enumerated as E. coli, whereas mauve colonies were enumerated as total coliform. Presumptive E. coli isolates were further purified with Luria Bertani medium and were preserved in stab and glycerol solution before further tests. The abundance of total coliform and E. coli were reported as colony-forming units per 100 ml (CFU 100 ml−1).
The identity of presumptive E. coli isolates was confirmed by a monoplex PCR targeting the E. coli housekeeping gene [alkaline phosphatase (phoA) gene] using primers and amplification conditions as described by Kong et al. (30). Detection of virulence genes was performed by two multiplex PCR assays, in which nine virulent genes were for the differentiation of six E. coli pathotypes. The two multiplex PCR was conducted in a 25 μl of reaction mixture consisting of 5 μl DNA, 1× Green GoTaq buffer (pH 8.5), 0.5 U of Taq DNA polymerase (Promega, USA), 1.65 mM of MgCl2, 220 μM dNTP, and 0.24 μM of each primer. The primers used and the amplification conditions were adopted from Nguyen et al. (31), Vidal et al. (32), and Aranda et al. (33) (Supplementary Table 1). The E. coli 2060-004, E2348/69, JM221, E9034A, C1845, and EC-12 were used as positive controls for EHEC, EPEC, EAEC, ETEC, DAEC, and EIEC, respectively, whereas sterile water was used as the negative control.
A quadruplex PCR assay was performed to determine the phylogroups (A, B1, B2, C, D, E, and F) that belong to E. coli sensu stricto, or one of the five cryptic clades (I-V) using primer concentrations and PCR conditions according to Clermont et al. (9) (Supplementary Table 1). For each quadruplex reaction, 20 μl reaction volume was carried out containing 3 μl DNA template, 2 μM each dNTP, 1X PCR buffer, 2 U Taq polymerase, 1 mM MgCl2, 1 μM for primers except for Acek-F (2 μM), ArpA1-R (2 μM), trpBA-F (0.6 μM), and trpBA-R (0.6 μM). The combination of presence or absence of the genes arpA, chuA, yjaA, and TspE4.C2 was used to determine the phylogenetic groups according to Clermont et al. (9).
The disc diffusion method on Mueller-Hinton agar (Difco, USA), as described by the Clinical Laboratory Standards Institute (34), was used to determine the antibiotic resistance profiles of the E. coli isolates. A total of 20 commonly used antibiotics from 11 classes were tested: tetracyclines [tetracycline (TE) 30 μg], quinolones [nalidixic acid (NA) 30 μg, oxolinic acid (OA) 2 μg, and ofloxacin (OFX) 5 μg], penicillins [ampicillin (AMP) 10 μg, amoxicillin/clavulanic acid (AMC) 30 μg], sulfonamides [sulfafurazole (SF) 300 μg and sulfamethoxazole/trimethoprim (SXT) 25 μg], fluoroquinolones [ciprofloxacin (CIP) 5 μg, ofloxacin 5 μg, and enrofloxacin (ENR) 5 μg], phenicols [chloramphenicol (C) 30 μg and florfenicol (FFC) 30 μg], aminoglycoside [streptomycin (S) 10 μg, gentamicin (CN) 10 μg, and neomycin (N) 30 μg], macrolide [azithromycin (AZM) 15 μg], cephalosporins [cephazolin (KZ) 30 μg and ceftriaxone (CRO) 30 μg], carbapenems [imipenem (IPM) 10 μg], and nitrofurans [nitrofurantoin (F) 100 μg]. The antibiotic discs were applied to the inoculated plates and were incubated at 37 ± 0.5°C overnight. The zone of inhibition for each E. coli isolate was analyzed according to the standards and the interpretive criteria of CLSI (34). The E. coli ATCC 25922, which is a recommended reference strain for antimicrobial susceptibility testing, was used as a control.
The average MAR index of all E. coli isolates was calculated using the following formula:
where “a” is the aggregate antimicrobial resistance score of all the E. coli isolates from one site, “b” is the number of E. coli isolates tested, and “c” is the number of antibiotics used in the study. A MAR index value of ≥0.2 indicates that the antibiotic resistance at the study area is rendered from contamination by antibiotics, whereas the value of <0.2 indicates that the antibiotic resistance at the study area is indigenous (35).
In this study, we assessed tetracycline and sulfonamide resistance genes as both tetracycline and sulfonamide are old antibiotics that were among the most widely used antibiotics in humans and animals particularly in Southeast Asia, either as therapeutic and/or prophylactic agents (36). For tet gene detection, multiplex PCR was used to screen 14 tet genes (37). All reaction mixtures were performed in 50 μl volume consisting of 2 μl DNA template, 1× PCR buffer, 2.5 U Taq polymerase, 300 μM of each dNTPs, 3 mM MgCl2, and selected primers with 0.2 μM for tet(D), 0.4 μM for tet(M) and tet(S), 0.6 μM for tet(A), tet(B), tet(E), tet(G), tet(K), tet(O), 0.8 μM for tet(C), tet(L), tet(Q), tet(X), and 1.6 μM for tetA(P). While for sul genes, duplex and monoplex PCR were performed according to Kozak et al. (38) and Pei et al. (39) by using gene-specific primers of sul1, sul2, and sul3, respectively. The PCR amplifications were carried out in 25 μl volume containing 1× PCR buffer, 1 mM MgCl2, 0.2 μM each primer, 2 μl DNA template, 1.5 μM of each dNTPs, and 2 U Taq polymerase for sul1 and sul2; while the reaction mixture for sul3 contained 1× PCR buffer, 1 mM MgCl2, 0.2 μM each primer, 1 μL DNA template, 0.2 μM of each dNTPs, and 1.75 U Taq polymerase.
The genetic diversity of the E. coli isolates was analyzed by a Repetitive Extragenic Palindromic-PCR (REP-PCR) using REP oligonucleotides as previously reported by Lim et al. (40). The E. coli isolates were prepared, and the PCR amplification was essentially conducted as described by Ghaderpour et al. (27). Amplifications were carried out in 25 μl volume including 4 μL DNA template, 200 μM of each dNTPs, 1× PCR buffer, 1 U Taq polymerase 2.5 mM MgCl2, and 0.5 μM primer. The fingerprint patterns were analyzed by BioNumerics, version 7.6.3 (Applied Maths, Kortrijk, Belgium). The similarity between profiles was calculated with the dice coefficient, while cluster analysis was performed using the unweighted pair group method using arithmetic averages (UPGMA) with Shannon diversity index (H'). Shannon diversity index (H') was calculated using the following equation:
where ni is the number of strains having each band pattern, N is the total number of isolates applied for rep-PCR.
Statistical analysis was performed with the SPSS version 21.0 (IBM, Chicago, USA). The criterion for statistical significance for all the following analyses was at p ≤ 0.05. Pearson's chi-square (goodness of fit) test was performed to determine the significant difference among phylogenetic groups, sul resistance genes, and tet resistance mechanism types according to frequency data, whereas the chi-square test for independence was applied to determine any association between phylogroups and antibiotic resistance. Prevalence of antibiotic resistance was defined as the proportion of resistant E. coli isolates over the total tested isolates. Correlation and linear regression analyses were performed to establish any association between water quality (29) toward E. coli and total coliform abundance. Cluster analysis for sampling sites was performed based on the antimicrobial susceptibility profile through the Bray-Curtis similarity index using PAST version 3.22 (41). Besides that, canonical correlation analysis (CCA) was performed using PAST to analyze the distribution of E. coli phylogenetic groups among sampling sites relative to the resistance genes sul and tet, water quality parameters, and antibiotic residue concentrations (42).
Coliform and E. coli were detected at all sampling sites; their concentrations at Larut River are up to 4.7 × 105 CFU 100 ml−1; up to 4.1 × 105 CFU 100 ml−1, respectively (Figure 2). The results clearly showed that Larut River was affected by anthropogenic wastewaters that harbored more coliform and E. coli (1–5 log CFU 100 ml−1 difference) compared to river waters (p ≤ 0.05). The highest total coliform and E. coli counts were observed in wastewater effluent from the slaughterhouse. In addition to lower anthropogenic influence, the comparatively low counts of coliform and E. coli detected at S1c were associated with the inhibitory effect of salinity (>16 ppt) on their survival and growth rates (29, 43), as coliform counts decreased significantly with increasing salinity in Larut River (R2 = 0.26, df = 16, and n = 18). The coliform and E. coli concentrations detected in this study were within the range that is previously reported for polluted Malaysian rivers and other locations (18, 44–47), which exceeded the standard maximum 100 CFU 100 ml−1 limit in the National Water Quality Standards (NWQS) class II for rivers set by the DOE Malaysia and the Malaysia Interim Marine Water Quality Standards (25). The important sources of fecal pollution in the Larut River are poultry manure, agriculture runoff, poorly treated or untreated sewage, and anthropogenic input from human and industrial activities along the river basins that contributed to the deterioration of water quality.
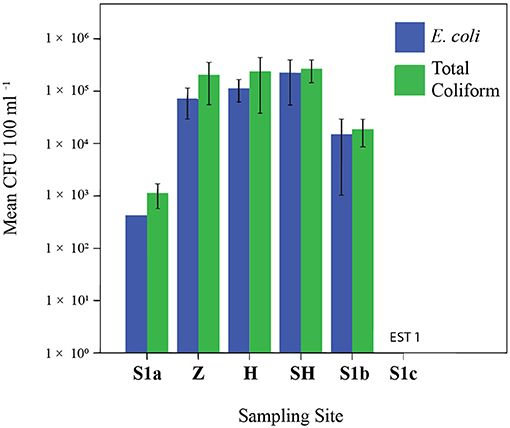
Figure 2. Abundance of Escherichia coli and total coliform. EST, estimation. Sampling sites: Z, Zoo; H, Hospital; SH, Slaughterhouse.
A total of 354 E. coli were isolated, and all the seven phylogenetic groups were observed with no cryptic clades. The majority of the E. coli isolated (77.97%) from Larut River were commensal strains, with phylogroup B1 (39.55%) and phylogroup A (38.42%) among the most prevalent. The predominance of groups A and B1 were consistent with other studies (27, 48). These two groups are sister groups that are consistently more predominant and prevalent in aquatic environments than virulent extraintestinal strains from groups B2 and D (27, 48). The phylogroup B1 was found higher at the wastewaters from zoo, slaughterhouse, and S1c of the Larut River, thus, receiving anthropogenic discharges from upstream. This may be associated with the influence of effluents discharge that contained more animal waste as the phylogroup B1 tend to be isolated from animal feces, particularly from herbivorous animals (49). Furthermore, this group of bacteria has already been shown to benefit from longer persistence in water than other phylogroups albeit for estuarine and coastal waters (6, 44, 50). In this study, it was expected to find a higher frequency of human-associated phylogroup A in wastewater effluent from the hospital, followed by slaughterhouse and S1b, which received the wastewater from the same point source. The least abundant and rarely detected phylogroups B2, C, D, E, and F isolated from Larut River were consistent with other studies (51, 52). The phylogroups B2 and D are known to be related to virulence factors that cause extraintestinal infections. Their higher detection frequencies at upstream (S1a) (B2: χ2 = 74.799, df = 5, p = 0.000; D: χ2 = 11.094, df = 5, and p = 0.050) probably resulted from commensal isolates from birds (53) and wild mammals (54, 55), as Larut Hill Forest Reserve is rich in biodiversity as it shelters 227 bird and 27 mammal species (56). This is in agreement with Petit et al. (57) and Ghaderpour et al. (27) who reported a higher abundance of B2 and D phylogroup at surface water collected near a forest with lesser human activities. Thus, fecal pollution from wildlife could present pathogenic E. coli infection risks. Hence determining the sources of fecal pollution in environmental waters is essential for pollution control and sustainable water quality management. The distribution of phylogenetic groups was not homogenous among the sites, and this may be attributed to the land use, sources of pollution, selective pressures in the waters, availability of nutrients, in situ physicochemical parameters (dissolved oxygen, pH, salinity, etc), protozoan and bacterial predators, and hydrological conditions (46, 58). The different survival abilities of E. coli and their abilities to overcome stresses will structure their community distribution and diversity in aquatic environments (6).
We examined the susceptibility of all the isolated E. coli in this study with 20 antibiotics that represented 11 different antibiotic classes. Of them, the highest frequency of resistance was detected for tetracycline class (34.32–86.84%), followed by quinolones (13.46–80.33%), penicillins (0–75%), sulfonamides (14.29–65.79%), amphenicols (5.71–60.53%), fluoroquinolones (8.57–57.38%), and aminoglycosides (0–47.37%) (Figure 3). The resistance rates against macrolides (1.92–19.74%), cephalosporins (0–18.57%), and nitrofurans (0–1.92%) were low. All the isolates were susceptible to imipenem and none of the isolates were resistant to all the antibiotics tested. Among these isolates, 265 (74.86%) were confirmed resistant to at least one or more antibiotics, two MAR E. coli isolates from phylogroups F and B1, each isolated from the slaughterhouse and hospital effluents, respectively, were resistant against 16 types of antibiotics. Only 25.14% of the isolates were susceptible to all antibiotics tested, indicating the prevalence of AR E. coli in the Larut River. Our results are consistent with other studies that reported the resistance of E. coli against old and widely used antibiotics in aquatic environments (27, 59–61). Furthermore, these antibiotic classes, except aminoglycosides, were the frequently detected antibiotics in Larut River (antibiotic concentration: LOD−1,092.49 ng/L) (29, 42). Our results showed that the frequency of MAR was higher in wastewater effluents (zoo > slaughterhouse > hospital) than in river waters (S1b > S1c > S1a). The prevalence of environmental MAR E. coli detected in this study was comparable to Kat River in South Africa (62) and Cochin Estuary in India (63), but higher than Matang Estuary in Malaysia (27), Tagus Estuary in Spain (18), Kshipra River in India (64), San Pedro River in Mexico (65), and generally lower than DongJiang River in China (88.00%) (66).
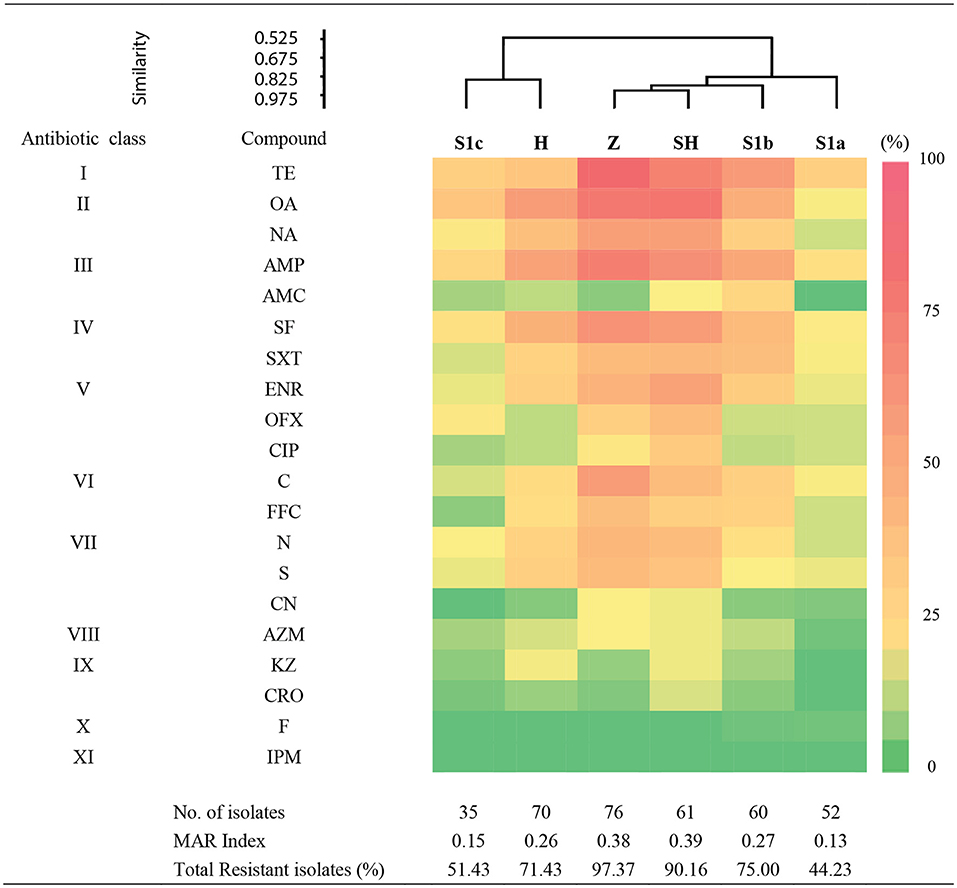
Figure 3. Antibiogram of antibiotic resistance phenotype detected in surface water among sampling sites in Larut River organied by similarity. Abbr, abbreviation. Antibiotic class: I, Tetracycline; II, Quinolone; III, Penicilin; IV, Sulfonamide; V, Fluoroquinolone; VI, Amphenicol; VI, Aminoglycoside; VIII, Macrolide; XI, Cephalosporin; X, Nitrofuran; XI, Carbapenem. Antibiotic type: TE, Tetracycline; OA, Oxolinic acid; NA, Nalidixic acid; AMP, Ampicilin; AMC, Amoxycillin/Clavulanic acid; SF, Sulfafurazole; SXT, Sulfamethoxazole/trimethoprim; ENR, Enrofloxacin; OFX, Ofloxacin; CIP, Ciprofloxacin; C, Chloramphenicol; FFC, Florfenicol; N, Neomycin; S, Streptomycin; CN, Gentamicin; AZM; Azithromycin; KZ, Cephazolin; CRO, Ceftriaxone; F, Nitrofurantion; IPM, Imipenem. Sampling sites: Z, Zoo; H, Hospital; SH, Slaughterhouse.
Cluster analysis revealed that the distribution of antibiotic resistance phenotype of E. coli isolates from the zoo and the slaughterhouse effluents were more similar than the hospital effluent and S1c. This could be due to the influence of wastewater effluent discharge that contained animal waste that received similar and commonly used veterinary antibiotics as previously reported by Low et al. (42). In contrast, the almost similar distribution of phylogenetic groups, together with the wastewater effluents containing human waste, could be a contributing factor to the similarity of antibiotic resistance phenotypes from the zoo and slaughterhouse effluent compared to S1b, which received the anthropogenic inputs from upstream. In this study, the phylogroup with the most resistant strains was observed in B1 followed by A, B2, D, E, F, and C. Our findings were consistent with other studies that MAR of E. coli mostly belonged to phylogroups B1 and A (27, 67). Despite these groups generally being considered harmless, their prevalence as intestinal pathogenic strains (68) has increased the risk on public health. The lower percentage of antibiotic resistance in phylogenetic groups B2 and D were also consistent with Ghaderpour et al. (27). These phylogroups were detected more upstream (S1a), which was associated with an animal origin. Phylogroup B2 was found to have higher resistance against penicillin, quinolones/fluoroquinolones, sulphonamides, and aminoglycosides, whereas Phylogroup D was detected to have higher resistance against tetracycline, penicillin, and sulfonamides. Studies have revealed their occurrence in wild, companion, and food animals, suggesting diet may be the main factor introducing MAR bacteria to animals (55, 69–72). Besides, rapid urbanization causes habitat loss and has simultaneously exposed animals to various environmental pollutants, which subsequently favors the transfer and emergence of MAR bacteria in animals through the food chain (72–74). The MAR index of 0.2 was observed at all the sampling sites, except for S1a and S1c, indicating that the midstream Larut River is contaminated with different anthropogenic inputs from the hospital, zoo, and slaughterhouse effluents that contained high risks sources, which were able to influence the prevalence and phylotype diversity of MDR E. coli in the Larut River.
Recent studies began using virulence factors as risk indicators in the environment, due to their close relation to pathogenicity among E. coli isolates (75). We found that 9 environmental E. coli isolates harbored an intestinal pathogenic E. coli (IPEC)-associated virulence gene. The relatively low abundance of presumptive IPEC in this study concurred with other aquatic environments with non-point source contamination (27, 44, 76), but contrasted with waters impacted by a sewage treatment plant (77). The virulence factor aggR, which indicates a positive EAEC pathotype, was the most prevalent gene (n = 5) detected in effluents from the zoo and the hospital. Runoff from sewage overflows likely transported these E. coli isolates, with virulent factors from feces of animals and humans, into environmental waters. Additionally, bfpA (n = 1), ST (n = 2), and eae (n = 1) virulence genes were also detected along the river continuum with high susceptibility to antibiotics except for a single phylogroup F isolate from zoo effluent. Nevertheless, these detected virulence genes still play their respective roles in lesion attachment, adherence, ion outflow induction, etc. (78). Although these E. coli isolates each carried a virulence gene, the observation of virulence factors itself does not always demonstrate pathogenicity (79). The present detection of single virulence gene patterns in E. coli isolates could likely be explained by a horizontal gene transfer among cells, which mediate the exchange of virulence factors located on mobile genetic elements (80). We identified five E. coli isolates (2.19%) from the zoo (n = 3) and hospital (n = 2) effluents to be EAEC strains. These are emerging diarrheal pathogens that cause acute diarrhea in developing countries (81). These EAEC isolates possessed MAR profiles of 5–16 antibiotics, with combinations of sul2 and sul3 genes, and tet(B), tet(A), tet(L), tet(M), and tet(X) genes. Studies have revealed similar findings on EAEC strains to be associated with water surfaces (82) and MAR EAEC belonging to phylogroup F from a stream with poor water quality (83). Our finding suggested that the dissemination of both virulence and resistance determinants could occur in the same anthropogenic site and pose a health risk. Either resistant or pathogenic isolates, when in contact with autochthonous bacteria, may then disseminate resistance and virulence determinants among natural ecosystems via a horizontal gene transfer (84). Interestingly, the phylogroup F strains isolated (n = 4/9) were found to carry virulent genes. El-shaer et al. (85) found that although environmental isolates which harbored virulence genes were located in phylogroup B1, the newly described phylogroup, such as phylogroup F, has also a virulence potential. The impact of this observation is not well-elucidated and further study is needed. Members of these phylogenetic groups are of particular interest because there is a relationship between the genetic background of a strain and its virulence factors (86).
Overall, 277 (78.25%) E. coli isolates harbored at least one of the tested tet genes except for tet(S). The majority of the resistance genes were found in the zoo (93.42%) effluents, slaughterhouse (85.25%) effluents, and S1b (78.33%). Seventy-seven E. coli isolates did not harbor any of the tested resistance genes. The three predominant tet genes were tet(A) (52.64%), followed by tet(L) (27.12%), and tet(X) (12.15%) (Figure 4A). Among sites, tet(A) was the dominant gene in zoo effluent, tet(L) was abundant in S1c, and tet(X) was in hospital effluent. We found that efflux genes were prevalent among the sites indicating that the active efflux via membrane-associated proteins were the main mechanisms for resistance in E. coli that resided in the Larut River. Both tet(A) and tet(L) belonged to the active efflux resistant mechanism. The predominance of these genes could be due to the low concentrations of tetracycline [mean: 64.4 ng L−1 (42)] in the environment as the expression of these genes are mainly induced at low tetracycline level (87). Both resistance mechanisms of active efflux via membrane-associated proteins, along with ribosomal protection proteins (RPP), were found in zoo wastewater effluent. The expression of the RPP tet(M) is mainly induced only at the high tetracycline level (88). This fact could explain the overall low abundance of tet(M) detected in this study. Certain E. coli (37.85%) in this study were found to carry multiple tet resistance genes with a high variation of tet gene combination. Our findings concurred with previous studies that tetracycline-resistant genes are ubiquitous in aquatic environments (89, 90). Among the 57 different tet combinations harbored by E. coli isolates, the most prevalent multiple tet resistance genes [tet(A)(L) (8.19%), tet(A)(M) (5.65%), and tet(A)(L)(M) (4.24%)] were significantly dominant (p ≤ 0.05) in wastewater effluents from the hospital. Notably, we had detected the presence of all three tet resistance mechanisms that include enzyme inactivation [tet(X)] in E. coli isolates isolated from hospital effluent. In contrast, E. coli isolates that harbored significantly fewer resistance genes were observed at less polluted sites. Our results indicated that tetracycline-resistant E. coli acquired multiple mechanisms to confer resistance.
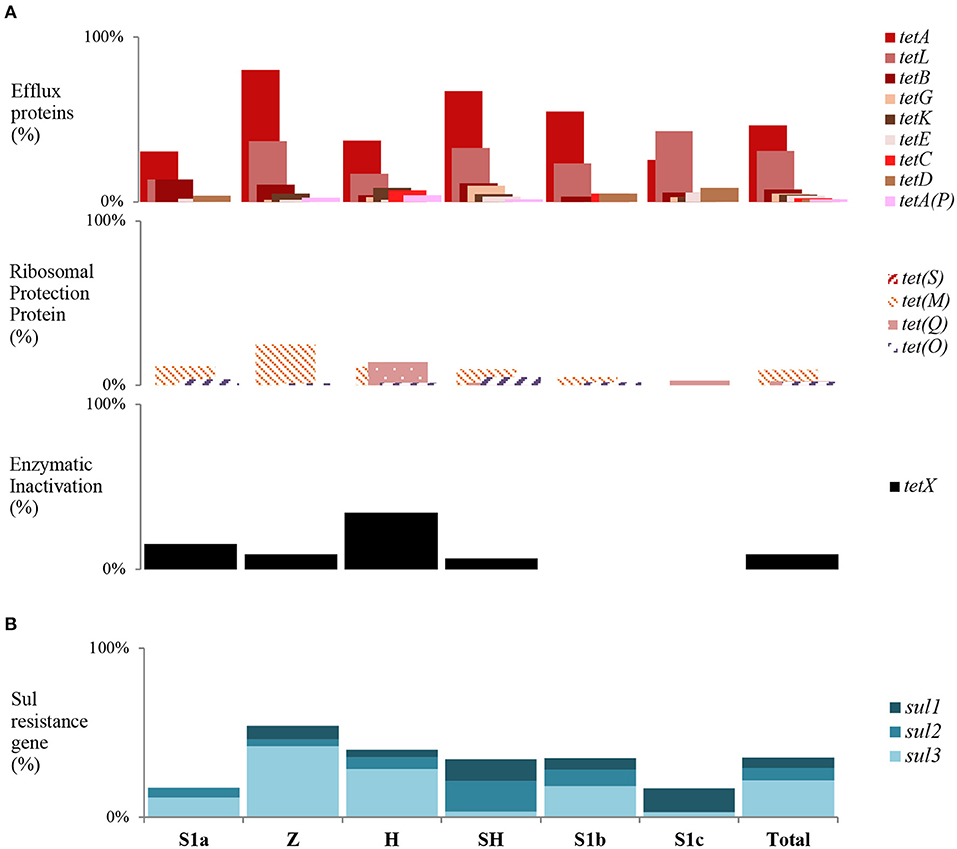
Figure 4. Trends of tet and sul gene among sampling sites in Larut River. (A) tet resistance gene type detected by site (B) Cumulative sul resistance gene type detected by site. Z, Zoo; H, Hospital; SH, Slaughterhouse.
For the sulfonamide resistance gene, there were 125 (35.31%) E. coli isolates that harbored at least one among all sites, with the gene-frequency distribution of sul3 (20.90%) > sul2 (5.08%) > sul1 (3.11%) (Figure 4B). Among the multiple sul gene combinations, the sul1 and sul3 combination was the most frequently detected in this study. The pattern of gene frequency distribution observed concurred with Lai et al. (91), where sul3 was prevalent in urban freshwater aquatic recipients of Sweden. The sul1 gene has been widely documented in receiving aquatic environments that can be regarded as a marker for anthropogenic pollutants (92, 93). Meanwhile, sul3 is initially suspected to be of human origin (94), but numerous studies have reported their prevalence in E. coli isolates from animals and livestock (95–97). Thus, the widespread of sul3 among E. coli isolates in this study was most likely due to the consumption of sulfonamide by humans and veterinary use (29, 36, 42). Sulfonamides are widely used in Asian countries, and in Malaysia, the usage in veterinary medicine was ~18,000 kg per year (98). While for the healthcare industry, the usage ranged from 0.0982 to 5.9900 daily defined dose (DDD)/1,000 population/day (99). In the absence of selective pressure from sulfonamide, sulfonamide-resistant bacteria may remain stable in the environment for at least 5–10 years longer than sulfonamide itself (100). The sulfonamide-resistant phenotypes that carried no resistant determinants was most likely due to the acquisition of other mechanisms (e.g., mutations in the chromosomal DHPS gene flop), as environmental E. coli strains may acquire genetically unrecognized resistance mechanisms more frequently compared to clinical E. coli strains (101, 102).
Relationship between physicochemical and antibiotic residues on E. coli phylogenetic distribution studies have shown that E. coli phylogenetic groups are adaptable and genotypically affected by environmental changes (103). In our study, the correlation coefficient has shown that salinity explained 26% of the variation in coliform abundance. Thus, other factors in combination may also contribute to the distribution of E. coli and their activity in the environment. Our CCA analysis revealed that the E. coli phylogroups A, B1, and C were greater in deteriorating water quality with SiO4, NH4, and PO4 (Figure 5A). The phylogroup B1 seemed to grow well in turbid water, with NO2, and have better tolerance to salinity, and pH, while the other lower frequency phylogroups (B2, D, E, and F) were associated with DO, temperature, and NO3. Our findings were consistent with Jang et al. (103) and Bong et al. (44), who reported that the occurrence and distribution of E. coli phylogenetic distribution can be affected by environmental variables. Collectively, a better adaptation to environmental drivers, paired with a high turnover of gene repertoires, made the phylogroup B1 exquisitely versatile in the environment compared to other phylogenetic groups (117). Our findings were in line with previous studies that nutrient concentrations (C, N, and P) are one of the important factors influencing the growth and survival of E. coli in the environment (104, 105). The addition of nutrient concentrations could also enhance the horizontal transfer of genetic resistance materials (106), which further enhanced the adaptive ability and plasticity of E. coli in a variety of environments. Lye et al. (29) detected a positive correlation between PO4 with sulfonamide-resistant heterotrophic bacteria and sulfonamide-enteric bacteria in Larut River. They had found that on four occasions, an exceptionally high PO4 concentration but low nitrogen concentration was observed in wastewater effluents from both the zoo and the hospital. However, further studies are still required to understand the nature of these anthropogenic stressors and the exact mechanisms in shaping the E. coli prevalence, diversity, and dissemination of antibiotic resistance in this river.
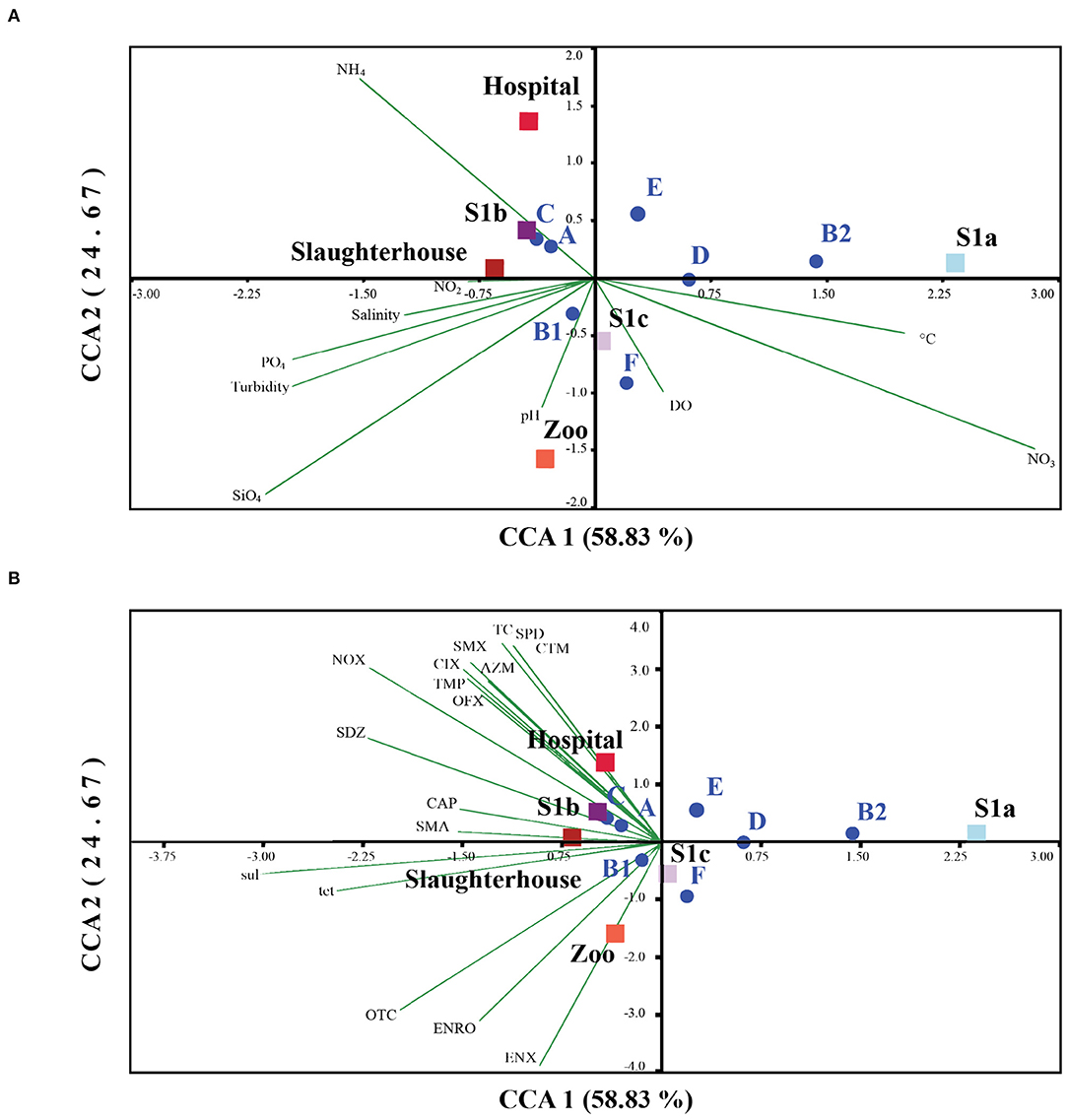
Figure 5. Square root normalized CCA ordination biplot showing the distribution of E. coli phylogenetic groups by sampling sites in Larut River with respect to their phylogenetic group and resistance genes tet and sul. CCA 1 on axis 1 explained 52.60% whereas CCA 2 on axis 2 explained 25.81% of all explanatory variables. (A) Relationship between E. coli and water physicochemical parameters of Larut River collected by Lye et al. (29). (B) Relationship between E. coli and water antibiotic residue detected in Larut River collected by Low et al. (42). Water physicochemical factors abbreviation: Temp (°C), salinity (ppt), pH, dissolved Oxygen (DO), turbidity and concentrations (μM) of Silicate (SiO4), Ammonium (NH4), Nitrite (NO2), Nitrate (NO3), Phosphate (PO4). Antibiotic resistance gene: sul, sulfonamide resistance gene; tet, tetracycline resistance gene. Antibiotic residue: RTM, Roxithromycin; CTM, clarithromycin, AZM, azithromycin; ETM-H2O, erythromycin-H2O; ENX, enoxacin; ENRO, enrofloxacin; NOX, norfloxacin; OFX, ofloxacin, CIX, ciprofloxacin; CTC, chlortetracycline; OTC, oxytetracycline; TC, tetracycline; FF, florfenicol; CAP, chloramphenicol; TMP, trimethoprim; SAAM, sulfacetamide; STZ, sulfathiazole; SDM, sulfadimethoxine; SMA, sulfadimidine; SPD, sulfapyridine; SDZ, sulfadiazine; SMX, sulfamethoxazole.
Our results also showed that the distribution of different phylogroups and antibiotic resistance genes were also affected by antibiotic use. The antibiotics detected in hospital, slaughterhouse, and S1b exhibited a significant correlation with the phylogenetic groups A, C, sul, and tet genes, while oxytetracycline (OTC) and fluoroquinolones (ENRO and ENX) detected in the zoo showed positive associations with phylogroup B1 (Figure 5B). However, no correlation was observed in the lower frequency phylogroups (B2, D, E, and F). This observation of correlations concurred with Varela et al. (107), Lye et al. (29), and Low et al. (42), who have revealed that the wastewater effluents from the zoo, slaughterhouse, and hospital are important antibiotic pollutant sources to the Larut River. Thus, the antibiotic residues in these effluents are expected to have a strong impact of selective pressure on antibiotic resistance in environmental E. coli compared to river waters. The variation in the response of different phylogenetic groups of E. coli, including resistant genes to different antibiotic residues, might be attributed to the types of antibiotics detected, physiochemical properties and persistence of antibiotics in water (64), water quality (6), acquired resistance mechanisms (108, 109), environmental fitness of E. coli, and indigenous microflora (58).
A total of 354 band patterns, with amplicon sizes ranging from 100 to 2,000 bp, were generated by REP-PCR. The genetic diversity of the E. coli population was high in effluents from the slaughterhouse (H', 3.53) and the zoo (H', 3.38) compared to river waters. Our observation concurred with Jang et al. (110) where diversity of E. coli genotypes tends to be greater with increasing proximity to anthropogenic urban sites. However, genetic heterogeneity between isolates from the natural environment may be caused by differences between the sampling sites (e.g., sampling sites were probably subjected to high pollution from various sources) (111). In contrast, hospital effluent had a low E. coli genetic diversity (H': 2.91) among other effluent sites, even though Low et al. (42) observed elevated antibiotic concentrations. A similar observation was found by McLellan (112) who reported lower diversity of E. coli in contaminated surface waters, in which environmental survival may be the factor that influences the recovery of the composition of strains from contaminated waters.
Due to the large population size, only the non-repeating E. coli genotypes (<85% similarity coefficient, n = 74) were selected to have their genetic relationships characterized. Seven major clusters were generated, with the highest number of isolates grouped in cluster I (n = 40), while the lowest number was shared among clusters II, IV, and VII (n = 4) (Figure 6). Cluster I isolates were found in a roughly equal distribution at all sites at Larut River, except for the hospital effluent and S1c with isolates (72%) that are characterized under the generalist phylogroups A and B1 with phenotypic resistance (77.50%). This cluster constituted the prevalence of background antibiotic resistance in this study (113–115). Similar to cluster I, cluster III (n = 10) were phylogroup A and B1 strains, with high antibiotic resistance, in which S1b has a higher distribution among the sites. In cluster VII, isolates from downstream site S1c co-clustered with an isolate from midstream hospital, thus, suggesting that the isolates along these sites shared a lineage. In contrast, low AR phylogroup B2 isolates were abundant in clusters IV and VI. These clusters collectively contained isolates (n = 14) that were located at non-anthropogenic sites, except a strain from the zoo. Similar to Liang et al. (116), there was no clear pattern of E. coli clustering according to sites in this study. It is important to note that the main aim of this study is not to trace the exact host sources of commensal E. coli in a complex aquatic environment. The diversity of E. coli genotypic and phenotypic is very large. According to literature, a collection of more than 20,000 isolates had only captured 27% of the predicted genotypes as estimated by a rarefaction analysis (46). This was evidenced by the hospital and the zoo isolates consisting of a diverse population with five and four different clades, respectively, which were also observed by Ghaderpour et al. (27). Given the limitation, this study did, however, demonstrate that AR E. coli population diversity in riverine estuarine water was, instead, related to the proximity of source contamination, stream order, and land use as observed by other studies (46, 76).
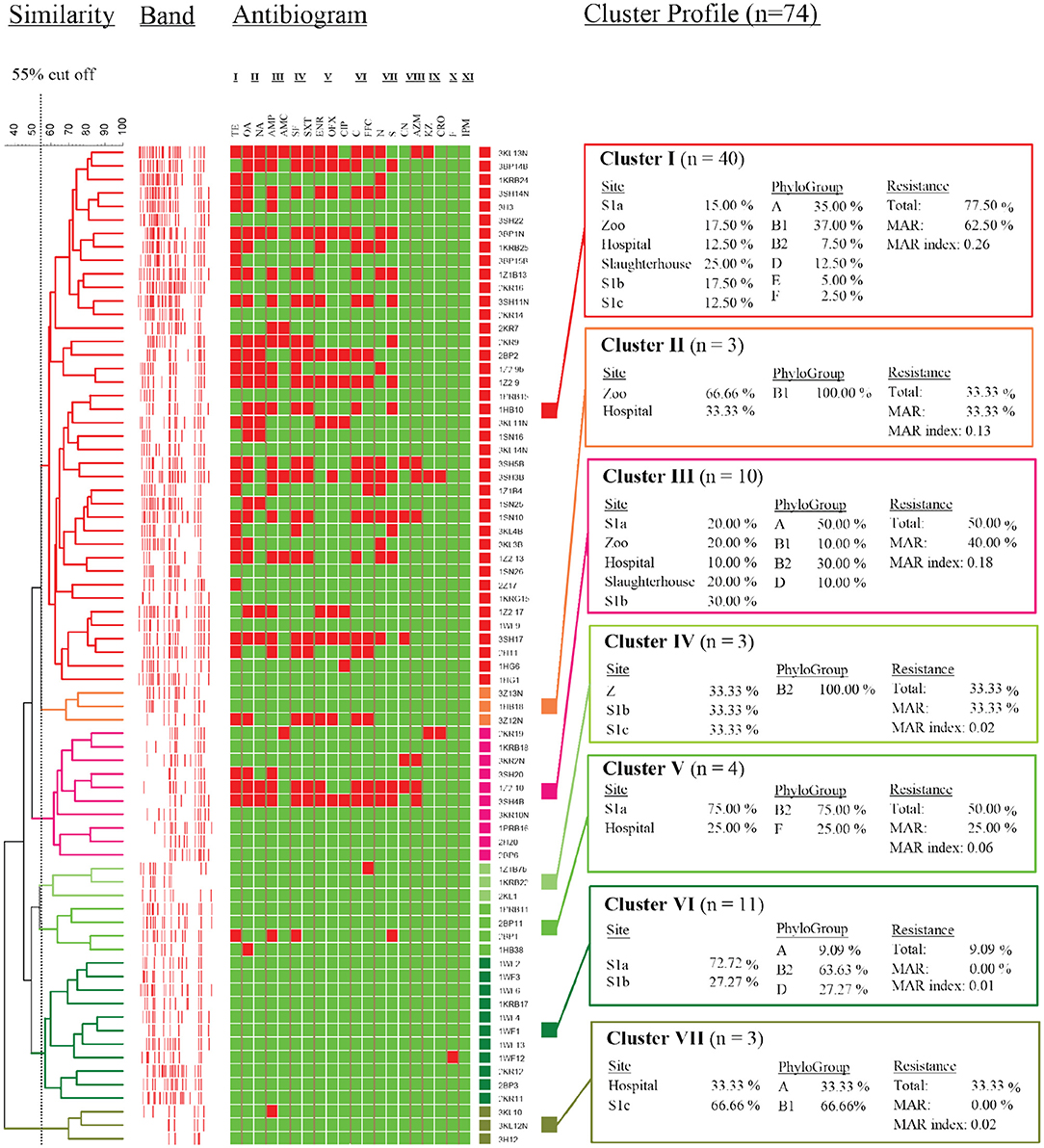
Figure 6. Dendrogram showing similarity of E. coli strains isolated from different sampling sites as determined by rep-PCR fingerprint analysis using REP primer. I, Tetracycline; II, Quinolone; III, Penicilin; IV, Sulfonamide; V, Fluoroquinolone; VI, Amphenicol; VII, Aminoglycoside; VIII, Macrolide; IX, Cephalosporin; X, Nitrofuran; XI, Carbapenem.
The present study affirmed that the prevalence and the diversification of antibiotic-resistant E. coli in Larut River were intensified by wastewater effluent from zoo, hospital, and slaughterhouse as sources of antibiotic residues. Our findings showed that phylogroups B1 and A were predominant with the presence of resistance genes. The cluster analysis revealed that the antibiotic resistance phenotype distribution of E. coli isolates from the zoo and the slaughterhouse effluents were more similar than the hospital effluent and downstream site (S1b). The tet efflux genes were detected in the majority of the E. coli isolates, thus, suggesting that E. coli may be an important carrier and/or reservoir of tetracycline resistance genes conferring resistance. The prevalence of the sul3 gene in E. coli isolates might be attributed to the consumption of sulfonamide for humans and veterinary use. The CCA analysis revealed a significant association between phylogroup and resistance genes with physicochemical properties and antibiotic residues on the environmental persistence of antibiotic-resistant E. coli. All these findings are important to provide information on the global comparison of persistence of antibiotic-resistant E. coli in different aquatic ecosystems, and the need to have surveillance and monitoring of virulence and antibiotic resistance in fresh river water to mitigate the emerging resistance and dissemination through water and environment.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
CB: contributed to conceptualization, methodology, writing, and editing. KL: contributed to methodology and editing. LC: contributed to conceptualization and editing. CL: contributed to conceptualization, editing, and funding. All authors contributed to the article and approved the submitted version.
This research was supported by the Ministry of Higher Education Malaysia under the Higher Institution Centre of Excellence (HICoE) Programme (IOES-2014D), University of Malaya (RU009D-2015), and private funding (PV009-2019) awarded to LC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.794513/full#supplementary-material
1. Leclerc H, Mossel DAA, Edberg SC, Struijk CB. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol. (2001) 55:201–34. doi: 10.1146/annurev.micro.55.1.201
2. Kaper JP, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. (2004) 2:123–40. doi: 10.1038/nrmicro818
3. Huang WC, Hsu BM, Kao PM, Tao CW, Ho YN, Kuo CW, et al. Seasonal distribution and prevalence of diarrheagenic Escherichia coli in different aquatic environments in Taiwan. Ecotox Environ Saf. (2016) 124:37–41. doi: 10.1016/j.ecoenv.2015.09.040
4. Lindstedt BA, Finton MD, Porcellato D, Brandal LT. High frequency of hybrid Escherichia coli strains with combined intestinal pathogenic Escherichia coli (IPEC) and extraintestinal pathogenic Escherichia coli (ExPEC) virulence factors isolated from human faecal samples. BMC Infect Dis. (2018) 18:544. doi: 10.1186/s12879-018-3449-2
5. Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. Presence and growth of naturalized Escherichia coli in temperate soils from lake superior watersheds. Appl Environ Microb. (2006) 72:612–21. doi: 10.1128/AEM.72.1.612-621.2006
6. Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microb. (2013) 79:4684–93. doi: 10.1128/AEM.00698-13
7. Chandrasekaran R, Hamilton MJ, Wang P, Staley C, Matteson S, Birr A, et al. Geographic isolation of Escherichia coli genotypes in sediments and water of the seven mile creek — a constructed riverine watershed. Sci Total Environ. (2015) 538:78–85. doi: 10.1016/j.scitotenv.2015.08.013
8. Gordon DM, Clermont O, Tolley H, Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol. (2008) 10:2484–96. doi: 10.1111/j.1462-2920.2008.01669.x
9. Clermont O, Christenson JK, Denamur E, Gordon DM. The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Env Microbial Rep. (2013) 5:58–65. doi: 10.1111/1758-2229.12019
10. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microb. (2000) 66:4555–8. doi: 10.1128/AEM.66.10.4555-4558.2000
11. Gordon DM. The influence of ecological factors on the distribution and the genetic structure of Escherichia coli. EcoSal Plus. (2004) 1:5. doi: 10.1128/ecosalplus.6.4.1
12. Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. (2010) 8:207–217. doi: 10.1038/nrmicro2298
13. Méric G, Kemsley EK, Falush D, Saggers EJ, Lucchini S. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ Microbiol. (2012) 152:487–501. doi: 10.1111/j.1462-2920.2012.02852.x
14. Djordjevic SP, Stokes HW, Chowdhury PR. Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front Microbiol. (2013) 4:86. doi: 10.3389/fmicb.2013.00086
15. Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev. (2014) 38:1–40. doi: 10.1111/1574-6976.12031
16. de Luca Rebello RC, Regua-Mangia AH. Potential enterovirulence and antimicrobial resistance in Escherichia coli isolates from aquatic environments in Rio de Janeiro, Brazil. Sci Total Environ. (2014) 490:19–27. doi: 10.1016/j.scitotenv.2014.04.040
17. Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. (2006) 40:7445–50. doi: 10.1021/es060413l
18. Pereira A, Santos A, Tacão M, Alves A, Henriques I, Correia A. Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci Total Environ. (2013) 461–2:65–71. doi: 10.1016/j.scitotenv.2013.04.067
19. Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. (2009) 157:2893–902. doi: 10.1016/j.envpol.2009.05.051
20. Partridge SR. Analysis of antibiotic resistance regions in gram-negative bacteria. FEMS Microbiol Rev. (2011) 35:820–55. doi: 10.1111/j.1574-6976.2011.00277.x
21. Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. (2015) 13:310–7. doi: 10.1038/nrmicro3439
22. Garcillán-Barcia MP, Alvarado A, De la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. (2011) 35:936–56. doi: 10.1111/j.1574-6976.2011.00291.x
23. ESCAP. State of the Environment in Asia and the Pacific 2000. New York, NY: United Nations (2000).
24. Department of Environment Malaysia. “Pollution Sources Inventory” in Environmental Quality Report 2018. Department of Environment Malaysia (2018). Available online at: https://enviro2.doe.gov.my/ekmc/digital-content/laporan-kualiti-alam-sekeliling-2019/ (accessed September 29, 2021).
25. Department of Environment Malaysia. Malaysia Marine Water Quality Standards and Index. Department of Environment Malaysia (2019). Available online at: https://www.doe.gov.my/portalv1/en/standard-dan-indeks-kualiti-jabatan-alam-sekitar (accessed September 22, 2021).
26. Khatri N, Tyagi S. Influences of natural and anthropogenic factors on surface and groundwater quality in rural and urban areas. Front Life Sci. (2015) 8:23–39. doi: 10.1080/21553769.2014.933716
27. Ghaderpour A, Ho WS, Chew LL, Bong CW, Chong VC, Thong KL, et al. Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front Microbiol. (2015) 6:977. doi: 10.3389/fmicb.2015.00977
28. Worldometers. Population of Cities in Malaysia. Worldometers (2017). Available online at: https://www.worldometers.info/world-population/malaysia-population/ (accessed September 15, 2021).
29. Lye YL, Bong CW, Lee CW, Zhang RJ, Zhang G, Suzuki S, et al. Anthropogenic impacts on sulfonamide residues and sulfonamide resistant bacteria and genes in Larut and Sangga Besar River, Perak. Sci Total Environ. (2019) 688:1335–47. doi: 10.1016/j.scitotenv.2019.06.304
30. Kong RYC, So CL, Law WF, Wu RSS. A sensitive and versatile multiplex PCR system for the rapid detection of enterotoxigenic (ETEC), enterohaemorrhagic (EHEC) and enteropathogenic (EPEC) strains of Escherichia coli. Mar Pollut Bull. (1999) 38:1207–15. doi: 10.1016/S0025-326X(99)00164-2
31. Nguyen TV, Van PL, Huy CL, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. (2005) 43:755–60. doi: 10.1128/JCM.43.2.755-760.2005
32. Vidal M, Kruger E, Durán C, Lagos R, Levine M, Prado V, et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol. (2005) 43:5362–5. doi: 10.1128/JCM.43.10.5362-5365.2005
33. Aranda KRS, Fabbricotti SH, Fagundes-Neto U, Scaletsky ICA. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol Lett. (2007) 267:145–50. doi: 10.1111/j.1574-6968.2006.00580.x
34. CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 30th edn. Wayne, PA: CLSI (2020).
35. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Micro. (1983) 46:165–70. doi: 10.1128/aem.46.1.165-170.1983
36. Thiang EL, Lee CW, Takada H, Seki K, Takei A, Suzuki S, et al. Antibiotic residues from aquaculture farms and their ecological risks in Southeast Asia: a case study from Malaysia. Ecosyst Health Sustain. (2021) 7:1926337. doi: 10.1080/20964129.2021.1926337
37. Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probe. (2001) 15:209–15. doi: 10.1006/mcpr.2001.0363
38. Kozak GK, Pearl DL, Parkman J, Reid-Smith RJ, Deckert A, Boerlin P. Distribution of sulfonamide resistance genes in Escherichia coli and salmonella isolates from swine and chickens at abattoirs in Ontario and Québec, Canada. Appl Environ Micro. (2009) 75:5999–6001. doi: 10.1128/AEM.02844-08
39. Pei R, Kim SC, Carlson KH, Pruden A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. (2006) 40:2427–35. doi: 10.1016/j.watres.2006.04.017
40. Lim KT, Yasin R, Yeo CC, Puthucheary S, Thong KL. Characterization of multidrug resistant ESBL-producing Escherichia coli isolates from hospitals in Malaysia. J Biomed Biotechnol. (2009) 2009:1–10. doi: 10.1155/2009/165637
41. Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. (2001) 4:1–9. Available online at: https://palaeo-electronica.org/2001_1/past/issue1_01.htm
42. Low K, Chai L, Lee C, Zhang G, Zhang R, Vahab V, et al. Prevalence and risk assessment of antibiotics in riverine estuarine waters of Larut and Sangga Besar River, Perak. J Ocean Limnol. (2021) 39:122–34. doi: 10.1007/s00343-020-9246-y
43. Rozen Y, Belkin S. Survival of enteric bacteria in seawater: molecular aspects. In: Oceans and Health: Pathogens in the Marine Environment. Boston, MA: Springer (2005). p. 93–107. doi: 10.1007/0-387-23709-7_4
44. Bong CW, Chai SK, Chai LC, Wang AJ, Lee CW. Prevalence and characterization of Escherichia coli in Kelantan River and its adjacent coastal waters. Water Supp. (2020) 20:930–42. doi: 10.2166/ws.2020.018
45. Chatterjee SK, Bhattacharjee I, Chandra G. Water quality assessment near an industrial site of Damodar River, India. Environ Monit Assess. (2010) 161:177–89. doi: 10.1007/s10661-008-0736-1
46. Lyautey E, Lu Z, Lapen DR, Wilkes G, Scott A, Berkers T, et al. Distribution and diversity of Escherichia coli populations in the South Nation River drainage basin, eastern Ontario, Canada. Appl Environ Microb. (2010) 76:1486–96. doi: 10.1128/AEM.02288-09
47. Al-Badaii F, Shuhaimi-Othman M. Water pollution and its impact on the prevalence of antibiotic-resistant E. coli and total coliform bacteria: a study of the Semenyih River, Peninsular Malaysia. Water Qual Expos Hea. (2015) 7:319–30. doi: 10.1007/s12403-014-0151-5
48. Figueira V, Serra E, Manaia CM. Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci Total Environ. (2011) 409:1017–23. doi: 10.1016/j.scitotenv.2010.12.011
49. Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MIZ, Gomes TAT, et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. (2010) 10:161. doi: 10.1186/1471-2180-10-161
50. Ratajczak M, Laroche E, Berthe T, Clermont O, Pawlak B, Denamur E, et al. Influence of hydrological conditions on the Escherichia coli population structure in the water of a creek on a rural watershed. BMC Microbiol. (2010) 10:222. doi: 10.1186/1471-2180-10-222
51. Sen K, Berglund T, Soares MA, Taheri B, Ma Y, Khalil L, et al. Antibiotic resistance of E. coli isolated from a constructed wetland dominated by a crow roost, with emphasis on ESBL and AmpC containing E. coli. Front Microbiol. (2019) 10:1034. doi: 10.3389/fmicb.2019.01034
52. Giacometti F, Pezzi A, Galletti G, Tamba M, Merialdi G, Piva S, et al. Antimicrobial resistance patterns in Salmonella enterica subsp. enterica and Escherichia coli isolated from bivalve molluscs and marine environment. Food Control. (2021) 121:107590. doi: 10.1016/j.foodcont.2020.107590
53. Nakhaee P, Peighambari SM, Razmyar J. Phylogenetic group determination of Escherichia coli isolated from broilers and layers with colibacillosis. Iran J Vet Sci Technol. (2015) 7:12–21. doi: 10.22067/veterinary.v7i1.43940
54. Lu S, Jin D, Wu S, Yang J, Lan R, Bai X, et al. Insights into the evolution of pathogenicity of Escherichia coli from genomic analysis of intestinal E. coli of Marmota himalayana in Qinghai-Tibet plateau of China. Emerg Microbes Infec. (2016) 5:1–9. doi: 10.1038/emi.2016.122
55. Alonso CA, Zarazaga M, Ben Sallem R, Jouini A, Ben Slama K, Torres C. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. (2017) 64:318–34. doi: 10.1111/lam.12724
56. World Wildlife Fund Malaysia. Study on the Development of Hill Stations. Vol. 2. Selangor: World Wildlife Fund (2001).
57. Petit F, Clermont O, Delannoy S, Servais P, Gourmelon M, Fach P, et al. Change in the structure of Escherichia coli population and the pattern of virulence genes along a rural aquatic continuum. Front Microbiol. (2017) 8:609. doi: 10.3389/fmicb.2017.00609
58. van Elsas JD, Semenov AV, Costa R, Trevors JT. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. (2011) 5:173–83. doi: 10.1038/ismej.2010.80
59. Watkinson AJ, Micalizzi GB, Graham GM, Bates JB, Costanzo SD. Antibiotic-resistant Escherichia coli in wastewaters, surface waters, and oysters from an urban riverine system. Appl Environ Microb. (2007) 73:5667–70. doi: 10.1128/AEM.00763-07
60. Alves MS, Pereira A, Araújo SM, Castro BB, Correia ACM, Henriques I. Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front Microbiol. (2014) 5:426. doi: 10.3389/fmicb.2014.00426
61. Divya SP, Hatha AAM. Screening of tropical estuarine water in south-west coast of India reveals emergence of ARGs-harboring hypervirulent Escherichia coli of global significance. Int J Hyg Envir Health. (2019) 222:235–48. doi: 10.1016/j.ijheh.2018.11.002
62. Nontongana N, Sibanda T, Ngwenya E, Okoh AI. Prevalence and antibiogram profiling of Escherichia coli pathotypes isolated from the Kat River and the Fort Beaufort abstraction water. Int J Env Res Pub Health. (2014) 11:8213–27. doi: 10.3390/ijerph110808213
63. Sukumaran DP, Durairaj S, Abdulla MH. Antibiotic resistance of Escherichia coli serotypes from Cochin Estuary. Interdisc Perspect Infec Dis. (2012) 2012:1–7. doi: 10.1155/2012/124879
64. Diwan V, Hanna N, Purohit M, Chandran S, Riggi E, Parashar V, et al. Seasonal variations in water-quality, antibiotic residues, resistant bacteria and antibiotic resistance genes of Escherichia coli isolates from water and sediments of the Kshipra River in Central India. Int J Env Res Pub Health. (2018) 15:1281. doi: 10.3390/ijerph15061281
65. Ramírez Castillo FY, Avelar González FJ, Garneau P, Marquez Díaz F, Guerrero Barrera AL, Harel J. Presence of multi-drug resistant pathogenic Escherichia coli in the San Pedro River located in the State of Aguascalientes, Mexico. Front Microbiol. (2013) 4:147. doi: 10.3389/fmicb.2013.00147
66. Su HC, Ying GG, Tao R, Zhang RQ, Zhao JL, Liu YS. Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ Pollut. (2012) 169:42–9. doi: 10.1016/j.envpol.2012.05.007
67. Vignaroli C, Di Sante L, Leoni F, Chierichetti S, Ottaviani D, Citterio B, et al. Multidrug-resistant and epidemic clones of Escherichia coli from natural beds of venus clam. Food Microbiol. (2016) 59:1–6. doi: 10.1016/j.fm.2016.05.003
68. Escobar-Páramo P, Grenet K, Le Menac'h A, Rode L, Salgado E, Amorin C, et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microb. (2004) 70:5698–700. doi: 10.1128/AEM.70.9.5698-5700.2004
69. Vredenburg J, Varela AR, Hasan B, Bertilsson S, Olsen B, Narciso-da-Rocha C, et al. Quinolone-resistant Escherichia coli isolated from birds of prey in Portugal are genetically distinct from those isolated from water environments and gulls in Portugal, Spain and Sweden. Environ Microbiol. (2014) 16:995–1004. doi: 10.1111/1462-2920.12231
70. Manges AR, Harel J, Masson L, Edens TJ, Portt A, Reid-Smith RJ, et al. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog Dis. (2015) 12:302–10. doi: 10.1089/fpd.2014.1860
71. Hertz FB, Nielsen JB, Schønning K, Littauer P, Knudsen JD, Løbner-Olesen A, et al. Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia coli from community-acquired urinary tract infections. BMC Microbiol. (2016) 16:63. doi: 10.1186/s12866-016-0681-z
72. Borges CA, Beraldo LG, Maluta RP, Cardozo MV, Barboza KB, Guastalli EAL, et al. Multidrug-resistant pathogenic Escherichia coli isolated from wild birds in a veterinary hospital. Avian Pathol. (2017) 46:76–83. doi: 10.1080/03079457.2016.1209298
73. Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, et al. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microb. (2010) 76:8126–34. doi: 10.1128/AEM.01446-10
74. Smith S, Wang J, Fanning S, McMahon BJ. Antimicrobial resistant bacteria in wild mammals and birds: a coincidence or cause for concern? Irish Vet J. 67:1–3. doi: 10.1186/2046-0481-67-8
75. Pereira AL, Ferraz LR, Silva RSN, Giugliano LG. Enteroaggretive Escherichia coli virulence markers: positive association with distinct clinical characteristics and segregation into 3 enteropathogenic E. coli serogroups. J Infect Dis. (2007) 195:366–74. doi: 10.1086/510538
76. Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Fairbrother J, et al. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl Environ Microb. (2007) 73:477–84. doi: 10.1128/AEM.01445-06
77. Anastasi EM, Matthews B, Stratton HM, Katouli M. Pathogenic Escherichia coli found in sewage treatment plants and environmental waters. Appl Environ Microb. (2012) 78:5536–41. doi: 10.1128/AEM.00657-12
78. Osińska A, Korzeniewska E, Harnisz M, Niestepski S. The prevalence and characterization of antibiotic-resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci Total Environ. (2017) 577:367–75. doi: 10.1016/j.scitotenv.2016.10.203
79. Rehman MU, Zhang H, Iqbal MK, Mehmood K, Huang S, Nabi F, et al. Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of Escherichia coli isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. (2017) 9:1–11. doi: 10.1186/s13099-017-0174-0
80. Sidhu JPS, Ahmed W, Hodgers L, Toze S. Occurrence of virulence genes associated with diarrheagenic pathotypes in Escherichia coli isolates from surface water. Appl Environ Microb. (2013) 79:328–35. doi: 10.1128/AEM.02888-12
81. Savarino SJ, Fasano A, Watson J, Martin BM, Levine MM, Guandalini S, et al. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA. (1993) 90:3093–7. doi: 10.1073/pnas.90.7.3093
82. Müller A, Stephan R, Nüesch-Inderbinen M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ. (2016) 541:667–72. doi: 10.1016/j.scitotenv.2015.09.135
83. Furlan JPR, Savazzi EA, Stehling EG. Widespread high-risk clones of multidrug-resistant extended-spectrum β-lactamase-producing Escherichia coli B2-ST131 and F-ST648 in public aquatic environments. Int J Antimicrob AG. (2020) 56:106040. doi: 10.1016/j.ijantimicag.2020.106040
84. Calhau V, Mendes C, Pena A, Mendonça N, Da Silva GJ. Virulence and plasmidic resistance determinants of Escherichia coli isolated from municipal and hospital wastewater treatment plants. J Water Health. (2015) 13:311–8. doi: 10.2166/wh.2014.327
85. El-shaer S, Abdel-Rhman SH, Barwa R, Hassan R. Virulence characteristics, serotyping and phylogenetic typing of clinical and environmental Escherichia coli isolates. Jundishapur J Microb. (2018) 11:12. doi: 10.5812/jjm.82835
86. Escobar-Páramo P, Clermont O, Blanc-Potard AB, Bui H, Le Bouguénec C, Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol Biol Evol. (2004) 21:1085–94. doi: 10.1093/molbev/msh118
87. Wang Q, Li X, Yang Q, Chen Y, Du B. Evolution of microbial community and drug resistance during enrichment of tetracycline-degrading bacteria. Ecotox Environ Saf. (2019) 171:746–52. doi: 10.1016/j.ecoenv.2019.01.047
88. Ammor MS, Gueimonde M, Danielsen M, Zagorec M, Van Hoek A. H. A. M., et al. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in Lactobacillus sakei rits 9. Appl Environ Microb. (2008) 74:1394–401. doi: 10.1128/AEM.01463-07
89. Jia S, He X, Bu Y, Shi P, Miao Y, Zhou H, et al. Environmental fate of tetracycline resistance genes originating from swine feedlots in river water. J Environ Sci Health B. (2014) 49:624–31. doi: 10.1080/03601234.2014.911594
90. Chen B, Hao L, Guo X, Wang N, Ye B. Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ Sci Pollut R. (2015) 22:13950–9. doi: 10.1007/s11356-015-4636-y
91. Lai FY, Muziasari W, Virta M, Wiberg K, Ahrens L. Profiles of environmental antibiotic resistomes in the urban aquatic recipients of Sweden using high-throughput quantitative PCR analysis. Environ Pollut. (2021) 287:117651. doi: 10.1016/j.envpol.2021.117651
92. Cacace D, Fatta-Kassinos D, Manaia CM, Cytryn E, Kreuzinger N, Rizzo L, et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: a pan-European survey of urban settings. Water Res. (2019) 162:320–30. doi: 10.1016/j.watres.2019.06.039
93. Sabri NA, Schmitt H, Van Der Zaan B, Gerritsen HW, Zuidema T, Rijnaarts HHM, et al. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J Environ Chem Eng. (2020) 8:102245. doi: 10.1016/j.jece.2018.03.004
94. Grape M, Sundström L, Kronvall G. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J Antimicrob Chemoth. (2003) 52:1022–4. doi: 10.1093/jac/dkg473
95. Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, Helmuth R. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemoth. (2003) 52:489–92. doi: 10.1093/jac/dkg362
96. Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Ch. (2003) 47:1169–72. doi: 10.1128/AAC.47.3.1169-1172.2003
97. Hammerum AM, Sandvang D, Andersen SR, Seyfarth AM, Porsbo LJ, Frimodt-Møller N, et al. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int J Food Microbiol. (2006) 106:235–7. doi: 10.1016/j.ijfoodmicro.2005.06.023
98. Marzuki Z. Use of Antimicrobial Agents in Veterinary Medicine in Malaysia. 2nd OIE Information Seminar for Practicing Veterinatians: Combating AMR (2017). Available online at: http://vam.org.my/home/wp-content/uploads/2017/11/Mazuki_AMU-in-Vet-Med.pdf (accessed August 29, 2019).
99. Pharmaceutical Services Division Ministry of Health Malaysia. Malaysian Statistics on Medicine 2011–2014. Kuala Lumpur: Ministry of Health (2017).
100. Gao P, Mao D, Luo Y, Wang L, Xu B, Xu L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. (2012) 46, 2355–64. doi: 10.1016/j.watres.2012.02.004
101. Changkaew K, Utrarachkij F, Siripanichgon K, Nakajima C, Suthienkul O, Suzuki Y. Characterization of antibiotic resistance in Escherichia coli isolated from shrimps and their environment. J. Food Protect. (2014) 77:1394–401. doi: 10.4315/0362-028X.JFP-13-510
102. Ogura Y, Ueda T, Nukazawa K, Hiroki H, Xie H, Arimizu Y, et al. The level of antimicrobial resistance of sewage isolates is higher than that of river isolates in different Escherichia coli lineages. Sci Rep. (2020) 10:17880. doi: 10.1038/s41598-020-75065-x
103. Jang J, Di DYW, Lee A, Unno T, Sadowsky MJ, Hur HG. Seasonal and genotypic changes in Escherichia coli phylogenetic groups in the Yeongsan River basin of South Korea. PLoS ONE. (2014) 9:e100585. doi: 10.1371/journal.pone.0100585
104. Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. Environmental Escherichia coli: ecology and public health implications—a review. J Appl Microbiol. (2017) 123:570–81. doi: 10.1111/jam.13468
105. Taabodi M, Hashem FM, Oscar TP, Parveen S, May EB. The possible roles of Escherichia coli in the nitrogen cycle. Int J Environ Res. (2019) 13:597–602. doi: 10.1007/s41742-019-00191-y
106. Blanco G, Lemus JA, Grande J. Microbial pollution in wildlife: linking agricultural manuring and bacterial antibiotic resistance in red-billed choughs. Environ Res. (2009) 109:405–12. doi: 10.1016/j.envres.2009.01.007
107. Varela AR, Andr é S, Nunes OC, Manaia CM. Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system. Water Res. (2014) 54:327–36. doi: 10.1016/j.watres.2014.02.003
108. Kawamura-Sato K, Yoshida R, Shibayama K, Ohta M. Virulence genes, quinolone and fluoroquinolone resistance, and phylogenetic background of uropathogenic Escherichia coli strains isolated in Japan. Jpn J Infect Dis. (2010) 63:113–5.
109. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Claeys G, et al. Characterization of extended-spectrum β-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb Drug Resist. (2010) 16:129–34. doi: 10.1089/mdr.2009.0132
110. Jang J, Unno T, Lee SW, Cho KH, Sadowsky MJ, Ko G, et al. Prevalence of season-specific Escherichia coli strains in the Yeongsan River Basin of South Korea. Environ Microbiol. (2011) 13:3103–13. doi: 10.1111/j.1462-2920.2011.02541.x
111. dos Anjos Borges LG, Dalla Vechia V, Corção G. Characterisation and genetic diversity via REP-PCR of Escherichia coli isolates from polluted waters in southern Brazil. FEMS Microbiol Ecol. (2003) 45:173–80. doi: 10.1016/S0168-6496(03)00147-8
112. McLellan SL. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl Environ Microb. (2004) 70:4658–65. doi: 10.1128/AEM.70.8.4658-4665.2004
113. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol R. (2010) 74:417–33. doi: 10.1128/MMBR.00016-10
114. D'Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, et al. Antibiotic resistance is ancient. Nature. (2011) 477:457–61. doi: 10.1038/nature10388
115. Tamminen M, Karkman A, Lõhmus A, Muziasari WI, Takasu H, Wada S, et al. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ Sci Tech. (2011) 45:386–391. doi: 10.1021/es102725n
116. Liang X, Liao C, Thompson ML, Soupir ML, Jarboe LR, Dixon PM. E. coli surface properties differ between stream water and sediment environments. Front Microbiol. (2016) 7:1732. doi: 10.3389/fmicb.2016.01732
Keywords: Escherichia coli, anthropogenic activities, antibiotic, resistant, resistance genes
Citation: Bong CW, Low KY, Chai LC and Lee CW (2022) Prevalence and Diversity of Antibiotic Resistant Escherichia coli From Anthropogenic-Impacted Larut River. Front. Public Health 10:794513. doi: 10.3389/fpubh.2022.794513
Received: 13 October 2021; Accepted: 20 January 2022;
Published: 10 March 2022.
Edited by:
Luther King Abia Akebe, University of KwaZulu-Natal, South AfricaReviewed by:
Daniel Gyamfi Amoako, National Institute for Communicable Diseases (NICD), South AfricaCopyright © 2022 Bong, Low, Chai and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chui Wei Bong, Y3dib25nQHVtLmVkdS5teQ==; bGVlQHVtLmVkdS5teQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.