
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 18 March 2022
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.787299
 Jingheng Zhang1
Jingheng Zhang1 Fang Yu1
Fang Yu1 Keyun Fu1
Keyun Fu1 Xinyu Ma1
Xinyu Ma1 Yi Han1
Yi Han1 Chi Ching Ali1
Chi Ching Ali1 Haonan Zhou2
Haonan Zhou2 Yantao Xu1,3
Yantao Xu1,3 Tingyue Zhang1
Tingyue Zhang1 Shuntong Kang1
Shuntong Kang1 Yiming Xu1
Yiming Xu1 Zhuolin Li2
Zhuolin Li2 Jiaqi Shi2
Jiaqi Shi2 Shuai Gao2
Shuai Gao2 Yongyi Chen4,5
Yongyi Chen4,5 Liyu Chen6*
Liyu Chen6* Jianglin Zhang7,8,9*
Jianglin Zhang7,8,9* Feizhou Zhu2,5,10*
Feizhou Zhu2,5,10*Background: Macrolides have been widely used to treat moderate-to-severe acne for more than 50 years. However, the prevalent antibiotic resistance of Propionibacterium acnes, along with the absence of clinically available resistance tests, has made macrolide misuse a frequent occurrence around the globe, with serious consequences.
Objective: We developed Cutibacterium acnes quantitative PCR (qPCR)-based antibiotics resistance assay (ACQUIRE) to enable fast and accurate detection of C. acnes macrolide resistance in clinical settings, representing an opportunity to administer antibiotics more wisely and improve the quality of care.
Methods: A cross-sectional observational study (n = 915) was conducted to probe into the macrolide resistance of C. acnes in patients with acne.
Results: The high sensitivity of ACQUIRE enabled us to reveal a much higher C. acnes 23S recombinant DNA (rDNA) point mutation rate (52%) and thus a higher macrolide resistance (75.5%) compared to previous reports. Carriage of ermX gene was discovered on 472 (53%) subjects, which concurs with previous studies.
Conclusion: The macrolide resistance of C. acnes is much higher than previously reported. Integrating ACQUIRE into acne treatment modalities may eliminate macrolide misuse and achieve better clinical improvements.
Acne vulgaris, one of the most common skin disorders worldwide, is inflicting billions of patients, significantly impairs their quality of life. The pathogenesis of acne involves the release of inflammatory mediators, skin hyper-keratinization, increased sebum secretion, and colonization of Propionibacterium acnes (recently reclassified as Cutibacterium acnes) (1–5). As the dominant commensal of the human pilosebaceous unit, C. acnes is intimately involved in the development of acne. It not only disturbs the proliferation of keratinocytes, but also carries lipase, protease, and hyaluronidase activity which can damage the pilosebaceous unit and induce inflammation (6). C. acnes can also activate toll-like receptors (7) and protease-activated receptors expressed by keratinocytes, which in turn induces the production of interleukins and matrix metalloproteinases (8).
Antibiotics have been used in acne treatment for more than 50 years, representing an essential part of the first-line treatment for moderate-to-severe acne (9). Macrolides (erythromycin, clarithromycin, and azithromycin) and tetracyclines (minocycline and doxycycline) are two classes of antibiotics that are frequently applied (1, 10). However, the extensive use of macrolides has caused escalating C. acnes resistance worldwide. Increases in C. acnes resistance have now been reported in all major regions within China and around the globe (11–20), with many countries reporting more than 50% of macrolide-resistant C. acnes (21). If macrolide is prescribed, the carriage of macrolide-resistant C. acnes strains may cause reduced treatment response, relapse, or prolonged course of disease for acne patients, increasing the disease burden of acne.
Although C. acnes exhibit far less resistance to tetracyclines (21), compared with macrolide, tetracyclines have several shortcomings, including more common adverse effects, potential liver and kidney toxicity, and the incompatibility with oral isotretinoin. Hence macrolide remains essential in acne management in China and many other countries. Also, simply abolishing macrolide and initiating the extensive use of tetracyclines will result in the rapid escalation of tetracycline resistance and cause waste of antibiotic resources that are insufficient already. Therefore, the most desirable practice for dermatologists would be prescribing tetracyclines to patients who carry macrolide-resistant C. acnes, and administering macrolide to those whose C. acnes is susceptible to macrolide.
However, the current antimicrobial susceptibility test (AST) of C. acnes employs a series of time-, labor-, and cost-intensive procedures due to the nature of C. acnes. As an anaerobe, C. acnes require a special culture environment and propagate slowly, so the test result could only be available after a week post sampling. The sensitivity of the current culture-based method is also unsatisfactory because the resistant C. acnes would be hard or even impossible to isolate if existing only in a low-percentage, which is often the case when the mutation occurred naturally but had not undergone antibiotic selection. Those drawbacks not only prevent any chance of applying the C. acnes resistance test clinically, but also impose huge restrictions on related scientific research.
Toward those ends, we developed C. acnes quantitative PCR (qPCR)-based antibiotics resistance assay (ACQUIRE), a method that utilizes the comedones (whitehead or blackhead) extracted from patients' follicles based on ARMS-qPCR (Amplification refractory mutation system qPCR), and is capable of determining the presence of macrolide-resistant C. acnes within 3 h, showing great potential to serve as a routine test for patients with clinical moderate-to-severe acne. ARMS-qPCR is widely used as a convenient and cost-saving tool to detect the point mutation and SNPs in nucleic acids (22). However, it has not been utilized to determine the resistance determinants in C. acnes before this study.
With ACQUIRE, we conducted a large-scale cross-sectional observational study to examine the macrolide resistance level of C. acnes.
A total of 915 moderate-to-severe acne patients were enrolled from December 2017 to January 2020 at both the Department of Dermatology, Xiangya Hospital, Central South University, and a skincare institution (Miao'miao-qingfu) to ensure a wide spectrum of enrolled patients. The inclusion criteria were: (1) 12–50 years of age; (2) moderate-to-severe acne, classified by the global acne grading system (GAGS) with a score ≥ 19. The exclusion criteria were: (1) history of systemic acne treatment within 6 months; (2) mild acne (GAGS score < 19). All patients provided informed consent and the study was approved by the ethics committee of Xiangya Hospital. Enrolled patients were all sampled and randomly assigned into two non-intersecting sets (Figure 1).
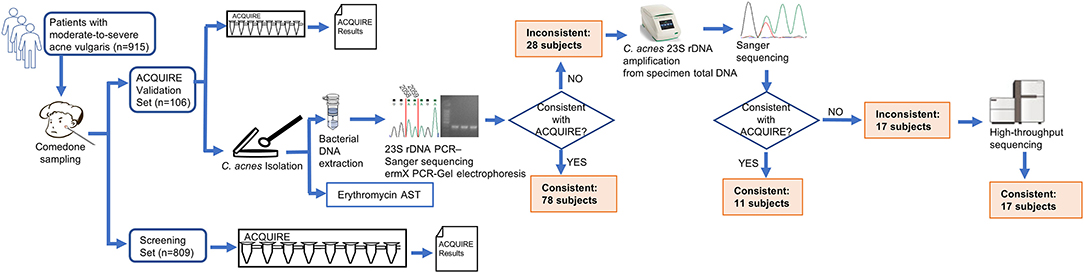
Figure 1. Schematic diagram of study design. The patients enrolled were sampled and randomly assigned into two sets. For patients in the Cutibacterium acnes quantitative PCR (qPCR)-based antibiotics resistance assay (ACQUIRE) Validation Set whose ACQUIRE results and result of culture method were inconsistent, the C. acnes 23S recombinant DNA (rDNA) was amplified from specimen total DNA and subject to Sanger sequencing. If the ACQUIRE results and the result of Sanger sequencing were still inconsistent, the amplification product was subject to high-throughput sequencing (Illumina Miseq), sequence filtration (sequences with <95% similarity to C. acnes were discarded), and mutation rate analysis.
To the ACQUIRE Validation Set (n = 106), acne lesions from multiple sites of the subjects' faces were squeezed using a sterile comedone extractor to obtain the comedone specimens (whitehead or blackhead). The specimens were then tested by ACQUIRE. A small portion of the specimens was reserved for C. acnes isolation in order to compare the results of ACQUIRE and the current method. To the Screening Set (n = 809), the specimens were also obtained and tested.
Specimens were obtained from 915 subjects (525 women and 390 men) and were tested by ACQUIRE. Subjects were 12–49 years of age (mean ± SD, 24.1 ± 5.7 years). Among 915 subjects, age and sex distributions among 12 genotypes were not significantly different.
Isolation, identification, 23S rDNA mutation, ermX detection, and AST of C. acnes were adopted from previous studies (23). Briefly, C. acnes were isolated on CDC anaerobic blood agar under an anaerobic atmosphere and at 37°C. The 16S recombinant RNA (rRNA) gene of isolated bacteria was amplified and sequenced to identify C. acnes from isolated strains. The ermX carriage status and the genotype of 23S rDNA of isolated C. acnes were determined by PCR -agarose gel electrophoresis and PCR-sanger sequencing, respectively. The minimum inhibitory concentration (MIC) of C. acnes to erythromycin was measured by broth microdilution. Brucella Broth with serial erythromycin concentration (0, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, and 256 ug/mL) was dispensed into 96-well plate, and strain suspension with a density of 0.5 Mcfarland Standard was inoculated. The plate was anaerobically incubated for 72 h and OD600 was measured with a plate reader (EnSpire 2300, Perkin Elmer). An MIC90 above or equal to 1 μg/ml was considered resistant. The anaerobic atmosphere in this study was generated by putting an Anaeropack sachet (C-1, Mitsubishi Gas Chemical) into an air-tight culture box, and was inspected by observing the color of an oxygen indicator (C-22, Mitsubishi Gas Chemical).
Mechanistically, C. acnes gains macrolide resistance through the presence of the ermX gene in its genome and 23S rDNA point mutation. The ermX gene, which often is carried by a corynebacterial-origin transposon Tn5432, makes C. acnes exhibit constitutive MLSB (macrolide-lincosamine-type B streptogramin) resistance (24). Point mutations in 23S rDNA at Escherichia coli-equivalent bases 2,058(A>T, A>G) and 2,059(A>G), which affect the peptidyl transferase region of the ribosome, makes C. acnes exhibit constitutive MLSB resistance and macrolide-only resistance, respectively (25).
As the dominant organism of skin follicle microbiota, C. acnes takes up ~50–90% of organisms in skin follicles (26, 27), which is a huge advantage of utilizing comedone as the test specimen. To eliminate the interference caused by the DNA of other organisms, primers were designed against the sites that showed the greatest inter-species variation. Sequences of ACQUIRE primers and probes were:
C.acnes-23S-F: GGGGACCTGAGCTGTC;
ARMS-58A-R: CTATAGTAAAGGTCCCGGGGTCGYT;
ARMS-58T-R: CTATAGTAAAGGTCCCGGGGTCGYA;
ARMS-58G-F: CTATAGTAAAGGTCCCGGGGTCGYC;
ARMS-59A-R: CTATAGTAAAGGTCCCGGGGTCAT;
ARMS-59G-R: CTATAGTAAAGGTCCCGGGGTCAC;
23S-probe: FAM-CATCTTTACTCGTAATGCAATTTCACC-BHQ1;
ermX-F: GATGATGACGGCTCAGTGGT;
ermX-R: TTTGCGCTGCTCTATCGGAA;
ermX-probe: FAM-TTCACATTTCACCTGGGTTCTCGGGTACCAA-BHQ1; C.acnes-16S-F: ACCGCTTTCGCCTGTGAC;
C.acnes-16S-R: AGCCCC AAGATTACACTTCCGAC;
16S-probe: VIC-AAGCGTGAGTGACGGTAATGGGTAAAGAAGCACC-BHQ1.
Once the specimen was obtained, it was transferred into a 1.5 ml microcentrifuge tube prefilled with 175 μl lysis buffer (Cat. 939016, Qiagen) and glass beads (40 mesh, 0.425 mm in diameter). Then, the microcentrifuge tube was fixed on a tissue disruptor (4,000 rpm, 10 min) to release the C. acnes that resided inside the specimen. Then, the total bacterial DNA was extracted using a universal Gram-positive bacteria DNA extraction kit (Cat. FP211, Tiangen Biotech, China), and dissolved in 100 μl sterile TE buffer (Cat. 17000-10, Qiagen). The eluted DNA was loaded into an ACQUIRE strip tube (0.5 μL eluted DNA per tube) prefilled with primers, probes, water, and a qPCR reaction mix. Then, the qPCR protocol was run and the raw results were analyzed and the assay result was determined accordingly (Figure 2).
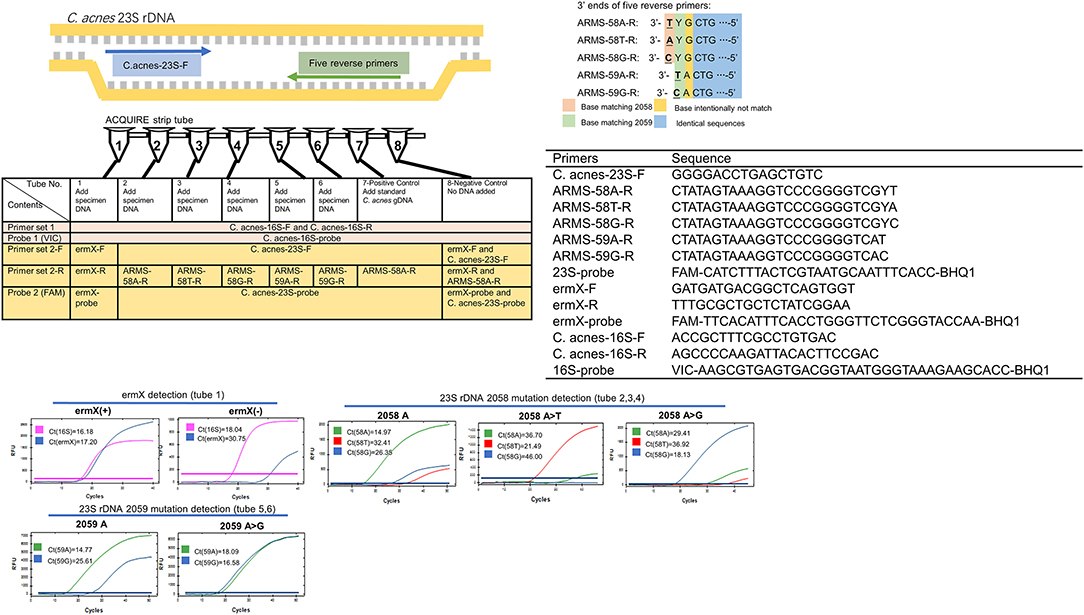
Figure 2. Working principle, composition, and typical results of ACQUIRE. Composed of 10 PCR primers and three fluorescence probes, ACQUIRE is integrated into one 8-strip tube. Tube 1 interrogates ermX presence, tubes 2, 3, and 4 discriminate 2,058 A>T and 2,058 A>G from 2,058A. Tubes 5 and 6 discriminate 2,059 A>G from 2,059A. Tube 7 and 8 serve as a positive and negative control, respectively. The discrimination of 23S rDNA mutations was achieved by the Ct value difference caused by the match or mismatch of 3' ends of five reverse primers. Additional mutations (base intentionally not match) were introduced into primers to strengthen this effect.
Statistical analysis was performed on SPSS 22.0 software (IBM, Armonk, NY, USA). Continuous variables were expressed as mean ± SD and compared using the Mann-Whitney U-test for two groups. Categorical variables were expressed as frequencies and percentages and compared using the chi-squared test (n > 5) or Fisher's exact test (n < 5). For all statistical analyses, a 2-sided P < 0.05 was accepted as statistically significant.
Among 106 subjects, the results of ACQUIRE and validation were not significantly different concerning the macrolide susceptibility phenotype (P = 0.377) (Table 1). In the ACQUIRE results, 31 (29.2%) subjects were macrolide-susceptible genotype and 75 (70.8%) were macrolide-resistant genotype. In the validation result, consistent with the ACQUIRE results, 37 (34.9%) subjects were macrolide-susceptible genotype and 69 (65.1%) were macrolide-resistant genotype (Table 1). Compared with ACQUIRE, the PCR+Sequencing method, which is culture-dependent, costs much more time (Figure 3). The detailed results of the ACQUIRE validation process were recorded in Supplementary Table 1.
A total of 123 C. acnes strains were isolated from 106 subjects, and their genotype and susceptibility to erythromycin were determined. Consistent with previous studies, the genotype of C. acnes well-correlated with their phenotype of macrolide susceptibility (P = 0.702), with only three inconsistent strains (Table 1).
Among 915 subjects whose specimen was tested by ACQUIRE, ermX was the most frequently detected macrolide-resistance determinant and was discovered on 472 (53%) subjects, which concurs with previous studies (21). However, the prevalence of 23S rDNA mutation is much higher than previous reports, with 468 (52%) subjects carrying at least one mutation. The proportion of macrolide-resistant subjects was also higher, with 691 (75.5%) being resistant to macrolide (Table 1).
The increasing C. acnes resistance asks for dermatologists to utilize those antibiotics that are still effective, which is a limited and diminishing clinical asset, more prudently and smartly. However, due to the inability of the current culture-based method, the status quo of C. acnes resistance is underestimated, and the related clinical and microbiological studies are insufficient.
Of note, simply switching the oral antibiotic to tetracyclines is not the once-and-for-all solution for C. acnes resistance and associated treatment failure, since resistant mutations for tetracyclines can also emerge in C. acnes and accumulate in the population.
Over the last 20 years, multiple studies (12, 13, 18, 23–25) have well-established and confirmed the correlation between the C. acnes genotype (23S rDNA 2058_2059 mutation and ermX carriage status) and its macrolide susceptibility phenotype, which laid the foundation for developing a macrolide resistance assay for C. acnes based on its genotype. However, the molecular basis of C. acnes resistance to tetracyclines has not been well-elucidated, which regrettably made it impossible to integrate the tetracycline resistance detection of C. acnes into ACQUIRE.
As a supplement to the current acne treatment algorithm, ACQUIRE can be used whenever a macrolide agent is intended to be prescribed, including topical and oral agents, and leave the rest of treatment options uninfluenced, which made it easy to integrate ACQUIRE into current treatment modalities and guidelines. ACQUIRE also provides an easy, scalable, and economical way to probe into the C. acnes resistance status in a large population.
By eliminating antibiotic misuse, ACQUIRE can make acne treatment more effective with less cost and shortened course of the disease, representing an opportunity to improve the quality of care. In this proof-of-concept study, we demonstrated the possibility and efficacy of integrating ACQUIRE into acne treatment modalities and achieved a better clinical outcome. However, large-scale multi-center randomized controlled trials are urgently needed to provide more solid evidence.
In the future, with advancements in the field, functions of ACQUIRE can be further extended, for example, by adding more resistance determinants into the current assay panel to enable the antibiotic resistance detection of tetracyclines and other antibiotics, or enabling the discrimination and quantification of different C. acnes subgroups, or probing into the carriage status of acne susceptible genes in the genome of the patient.
Meanwhile, general principles to slow the development of antibiotic resistance should always be encouraged, like using Benzoyl Peroxide (BPO), strictly limiting long-term (more than 2 months) antibiotic usage (28), and adopting alternative therapies like laser and light-based treatments (29).
The macrolide resistance of C. acnes is higher than previously reported due to more prevalent 23S rDNA mutations. Future studies should focus on the efficacy of treatment guided by ACQUIRE in large-scale cohorts.
The high-throughput sequencing data involved in this article has been deposited in NCBI SRA database under the accession number PRJNA768297 and is made publically accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA768297.
The studies involving human participants were reviewed and approved by Ethics Committee of Xiangya Hosiptal, Central South University. The patients/participants provided their written informed consent to participate in this study.
JHZ designed the study, conducted the experiments, and drafted the manuscript. FY conducted half of the qPCR experiment and collected patient demographic data. KYF, XYM, YH, CCA, HNZ, and YMX processed clinical specimen and isolated bacterial strains. TYZ, YTX, and STK provided valuable opinion, analyzed data, and revised the manuscript. ZLL, JQS, SG, and YYC provided conceptual insights and conducted microbiology experiments. LYC, JLZ, and FZZ participated in the study design, manuscript revision, acquired funding, and supervised the research. All authors contributed to this manuscript by drafting or revising the manuscript and approved the submitted version.
This study was supported by China Hunan Provincial Science and Technology Plan (2018SK7005 and 2017SK2092), Innovative Education Reform Program of Central South University (2019CG045), National Undergraduate Innovation Training Program of Central South University (201810533233 and GS201910533150X), and Hunan Science and Technology Innovation Talent Program (to YMX).
FZZ, JLZ, LYC, JHZ, and FY reported China Patent 2020101633011, a method that promotes the detection accuracy and efficiency of C. acnes antibiotic resistance.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.787299/full#supplementary-material
2. Williams HC, Dellavalle RP, Garner S. Acne vulgaris. Lancet. (2012) 379:361–72. doi: 10.1016/S0140-6736(11)60321-8
3. Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov, Cutibacterium gen nov, and Pseudopropionibacterium gen nov. Int J Systemat. Evol. Microbiol. (2016) 66:4422–32. doi: 10.1099/ijsem.0.001367
4. Thiboutot D, Gollnick H, Bettoli V, Dreno B, Kang S, Leyden JJ, et al. New insights into the management of acne: an update from the Global Alliance to Improve Outcomes in Acne Group. J Am Acad Dermatol. (2009) 60:S1–50. doi: 10.1016/j.jaad.2009.01.019
5. Tuchayi SM, Makrantonaki E, Ganceviciene R, Dessinioti C, Feldman SR, Zouboulis CC. Acne vulgaris. Nat Rev Dis Primer. (2015) 1:29. doi: 10.1038/nrdp.2015.29
6. Kwon HH, Suh DH. Recent progress in the research about Propionibacterium acnes strain diversity and acne: pathogen or bystander? Int J Dermatol. (2016) 55:1196–204. doi: 10.1111/ijd.13282
7. Heymann WR. Toll-like receptors in acne vulgaris. J Am Acad Dermatol. (2006) 55:691–92. doi: 10.1016/j.jaad.2006.05.049
8. Dreno B. What is new in the pathophysiology of acne, an overview. J Eur Acad Dermatol Venereol. (2017) 31:8–12. doi: 10.1111/jdv.14374
9. Thiboutot D, Dreno B, Sanders V, Rueda MJ, Gollnick H. Changes in the management of acne: 2009-2019. J Am Acad Dermatol. (2020) 82:1268–69. doi: 10.1016/j.jaad.2019.04.012
10. Zaenglein AL, Pathy AL, Schlosser BJ, Alikhan A, Baldwin HE, Berson DE, et al. Guidelines of care for the management of acne vulgaris. J Am Acad Dermatol. (2016) 74:945. doi: 10.1016/j.jaad.2015.12.037
11. Yang SS, Long V, Liau MM, Lee SH, Toh M, Teo J, et al. A profile of Propionibacterium acnes resistance and sensitivity at a tertiary dermatological centre in Singapore. Br J Dermatol. (2018) 179:200–1. doi: 10.1111/bjd.16380
12. Zhu T, Zhu W, Wang Q, He L, Wu W, Liu J, et al. Antibiotic susceptibility of Propionibacterium acnes isolated from patients with acne in a public hospital in Southwest China: prospective cross-sectional study. BMJ Open. (2019) 9:22938. doi: 10.1136/bmjopen-2018-022938
13. Schafer F, Fich F, Lam M, Garate C, Wozniak A, Garcia P. Antimicrobial susceptibility and genetic characteristics of Propionibacterium acnes isolated from patients with acne. Int J Dermatol. (2013) 52:418–25. doi: 10.1111/j.1365-4632.2011.05371.x
14. Fan Y, Hao F, Wang W, Lu Y, He L, Wag Gm, et al. Multicenter cross-sectional observational study of antibiotic resistance and the genotypes of Propionibacterium acnes isolated from Chinese patients with acne vulgaris. J Dermatol. (2016) 43:406–13. doi: 10.1111/1346-8138.13149
15. Toyne H, Webber C, Collignon P, Dwan K, Kljakovic M. Propionibacterium acnes (P. acnes) resistance and antibiotic use in patients attending Australian general practice. Austral J Dermatol. (2012) 53:106–11. doi: 10.1111/j.1440-0960.2011.00867.x
16. Zandi S, Vares B, Abdollahi H. Determination of microbial agents of acne vulgaris and Propionibacterium acnes antibiotic resistance in patients referred to dermatology clinics in Kerman, Iran. Jundishapur J Microbiol. (2011) 4:17–22.
17. Mendoza N, Hernandez PO, Tyring SK, Haitz KA, Motta A. Antimicrobial susceptibility of Propionibacterium acnes isolates from acne patients in Colombia. Int J Dermatol. (2013) 52:688–92. doi: 10.1111/j.1365-4632.2011.05403.x
18. Ross JI, Snelling AM, Carnegie E, Coates P, Cunliffe WJ, Bettoli V, et al. Antibiotic-resistant acne: lessons from Europe. Br J Dermatol. (2003) 148:467–78. doi: 10.1046/j.1365-2133.2003.05067.x
19. Nakase K, Hayashi N, Akiyama Y, Aoki S, Noguchi N. Antimicrobial susceptibility and phylogenetic analysis of Propionibacterium acnes isolated from acne patients in Japan between 2013 and 2015. J Dermatol. (2017) 44:1248–54. doi: 10.1111/1346-8138.13913
20. Fattah NSAA, Darwish YW. In vitro antibiotic susceptibility patterns of Propionibacterium acnes isolated from acne patients: an Egyptian university hospital-based study. J Eur Acad Dermatol Venereol. (2013) 27:1546–51. doi: 10.1111/jdv.12057
21. Walsh TR, Efthimiou J, Dreno B. Systematic review of antibiotic resistance in acne: an increasing topical and oral threat. Lancet Infect Dis. (2016) 16:E23–33. doi: 10.1016/S1473-3099(15)00527-7
22. Zhu F, Peng Y, Qi L, Bi G, He Y, Jie C, et al. Nested ARMS-qPCR is a fast and cost-saving method for single nucleotide polymorphism genotyping in clinical service. Int J Clin Exp Med. (2016) 9:16292–300.
23. Ross JI, Snelling AM, Eady EA, Cove JH, Cunliffe WJ, Leyden JJ, et al. Phenotypic and genotypic characterization of antibiotic-resistant Propionibacterium acnes isolated from acne patients attending dermatology clinics in Europe, the USA, Japan and Australia. Br J Dermatol. (2001) 144:339–46. doi: 10.1046/j.1365-2133.2001.03956.x
24. Ross JI, Eady EA, Carnegie E, Cove JH. Detection of transposon Tn5432-mediated macrolide-lincosamide-streptogramin B (MLSB) resistance in cutaneous propionibacteria from six European cities. J Antimicrob Chemother. (2002) 49:165–68. doi: 10.1093/jac/49.1.165
25. Ross JI, Eady EA, Cove JH, Ratyal AH, Cunliffe WJ. Resistance to erythromycin and clindamycin in cutaneous propionibacteria is associated with mutations in 23S rRNA. Dermatology. (1998) 196:69–70. doi: 10.1159/000017871
26. Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Investig Dermatol. (2013) 133:2152–60. doi: 10.1038/jid.2013.21
27. Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. (2016) 6:srep39491. doi: 10.1038/srep39491
28. Nagler AR, Milam EC, Orlow SJ. The use of oral antibiotics before isotretinoin therapy in patients with acne. J Am Acad Dermatol. (2016) 74:273–79. doi: 10.1016/j.jaad.2015.09.046
Keywords: acne (acne vulgaris), antimicrobial resistance, quantitative PCR (qPCR), antimicrobial, macrolide-resistant gene
Citation: Zhang J, Yu F, Fu K, Ma X, Han Y, Ali CC, Zhou H, Xu Y, Zhang T, Kang S, Xu Y, Li Z, Shi J, Gao S, Chen Y, Chen L, Zhang J and Zhu F (2022) C. acnes qPCR-Based Antibiotics Resistance Assay (ACQUIRE) Reveals Widespread Macrolide Resistance in Acne Patients and Can Eliminate Macrolide Misuse in Acne Treatment. Front. Public Health 10:787299. doi: 10.3389/fpubh.2022.787299
Received: 02 October 2021; Accepted: 14 February 2022;
Published: 18 March 2022.
Edited by:
Monica Catarina Botelho, Instituto Nacional de Saúde Doutor Ricardo Jorge (INSA), PortugalReviewed by:
Mehmet Demirci, Kırklareli University, TurkeyCopyright © 2022 Zhang, Yu, Fu, Ma, Han, Ali, Zhou, Xu, Zhang, Kang, Xu, Li, Shi, Gao, Chen, Chen, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feizhou Zhu, emh1ZmVpemhvdUBjc3UuZWR1LmNu; Jianglin Zhang, bGVvempsMTAxMEAxMjYuY29t; Liyu Chen, Y2hlbmxpeXVAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.