
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 12 January 2023
Sec. Occupational Health and Safety
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.1083826
 Luca Ferrari1,2*†
Luca Ferrari1,2*† Paola Monti1†
Paola Monti1† Chiara Favero1
Chiara Favero1 Michele Carugno1,2
Michele Carugno1,2 Letizia Tarantini1
Letizia Tarantini1 Cristina Maggioni1
Cristina Maggioni1 Matteo Bonzini1,2
Matteo Bonzini1,2 Angela Cecilia Pesatori1,2
Angela Cecilia Pesatori1,2 Valentina Bollati1,2
Valentina Bollati1,2Introduction: Night shift (NS) work has been associated with an increased risk of different conditions characterized by altered inflammatory and immune responses, such as cardio-metabolic and infectious diseases, cancer, and obesity. Epigenetic modifications, such as DNA methylation, might mirror alterations in biological processes that are influenced by NS work.
Methods: The present study was conducted on 94 healthy female workers with different working schedules and aimed at identifying whether NS was associated with plasmatic concentrations of the inflammatory proteins NLRP3 and TNF-alpha, as well as with DNA methylation levels of ten human endogenous retroviral (HERV) sequences, and nine genes selected for their role in immune and inflammatory processes. We also explored the possible role of the body mass index (BMI) as an additional susceptibility factor that might influence the effects of NS work on the tested epigenetic modifications.
Results and discussion: We observed a positive association between NS and NLRP3 levels (p-value 0.0379). Moreover, NS workers retained different methylation levels for ERVFRD-1 (p-value = 0.0274), HERV-L (p-value = 0.0377), and HERV-P (p-value = 0.0140) elements, and for BIRC2 (p-value = 0.0460), FLRT3 (p-value = 0.0422), MIG6 (p-value = 0.0085), and SIRT1 (p-value = 0.0497) genes. We also observed that the BMI modified the relationship between NS and the methylation of ERVE, HERV-L, and ERVW-1 elements. Overall, our results suggest that HERV methylation could pose as a promising biomolecular sensor to monitor not only the effect of NS work but also the cumulative effect of multiple stressors.
In modern “24-h society,” flexible working schedules are becoming increasingly common, with the healthcare industry having one of the highest percentages of night shift (NS) workers to guarantee continuous assistance to patients (1, 2). However, irregular sleep/wake patterns and chronic sleep deprivation could result in disruption of circadian rhythms, i.e., biochemical and behavioral fluctuations synchronized with external 24 h light/dark cycles, with detrimental consequences on both physical and mental health; indeed, circadian rhythms control a wide array of cellular processes, including metabolic homeostasis and innate immunity (3). Therefore, it is not surprising that NS work has been associated with an increased risk of several pathological conditions characterized by altered immune and inflammatory responses, such as infectious diseases, cancer, obesity, and cardiometabolic diseases (4, 5). In this scenario, the identification of molecular modifications mirroring such a complex pattern of biological processes affected by NS work could be of great interest. Besides, this could also help monitor the efficacy of health promotion strategies aimed at reducing NS detrimental effects. Epigenetic modifications such as DNA methylation (i.e., the covalent addition of a methyl group to the C5 position of cytosines, generally occurring at CpG sites) may be suitable candidates since they are known to modulate gene expression in response to a wide array of environmental and lifestyle stressors (6–8). A growing body of literature has reported that NS workers retain an altered methylation profile of clock genes, i.e., the set of genes controlling circadian rhythmicity (9–12). Also, genes involved in the inflammatory process (13, 14) and cancer development (3, 14, 15) are differentially methylated in subjects working at night. Such methylation modifications could also be exacerbated by additional stressful conditions associated with systemic low-grade inflammation, such as being overweight (16).
In this framework, human endogenous retroviruses (HERVs) are one of the most abundant classes of repetitive elements (REs), accounting for 8% of the human genome (17). These sequences originated from ancestral retroviruses that infected the primates' germ line and integrated into the host genome, thus being vertically transmitted across generations; however, the progressive accumulation of sequence changes within HERVs has re-shaped them into novel elements at the service of the host cell innate immune system, modulating the inflammatory response after different environmental triggers (18, 19).
Based on this evidence, HERV sequences may likely respond to NS work by changing their methylation status, with potential implications in the innate immune/inflammatory processes. Being NS work a potential risk factor for infectious and cancer diseases, the immune escape mechanisms might also be involved. Indeed, environmental stressors can affect the capacity of pathogen and tumor cells to evade the immunogenic response by altering the host defense mechanisms (20, 21).
To test this hypothesis, we designed a cross-sectional study to investigate the effects of NS work on inflammation and the methylation of HERV elements and a subset of immune escape-related genes. The analyses were conducted on blood samples of healthy nurses with different NS working schedules, who have been recruited in the context of previous research (15, 22). First, we evaluated whether NS work is associated with the plasmatic concentration of the NLR family pyrin domain containing 3 (NLRP3) and the tumor necrosis factor-alpha (TNF-alpha) inflammatory markers. Second, we tested the association between NS work and the methylation of a panel of 10 HERV elements and 10 genes involved in immune evasion. Finally, we explored the possible role of the body mass index (BMI) as an additional susceptibility factor, potentially influencing the association of NS work with gene methylation.
The study population consisted of 97 female nurses working at the Fondazione IRCCS Ca' Granda Policlinico Hospital in Milan, Italy, who were enrolled voluntarily in the context of previous research works, as extensively described in previous studies (15, 22). The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board Comitato Etico Milano Area 2 of the Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico (approval number 702_2015). As previously reported (15), eligibility criteria included: being a female of Caucasian ethnicity, age 30–45 years, and with a length of service of at least 1 year. Subjects were excluded if, at the time of the enrolment were affected by cancer, neurological diseases (e.g., multiple sclerosis, Alzheimer's or Parkinson's disease, epilepsy), or acute relapses of systemic diseases (e.g., cardiovascular diseases, diabetes). Other exclusion criteria were antihypertensive or steroid drug assumption, pregnancy or menopausal status, and body mass index (BMI) >30. These criteria were adopted to avoid any confounding due to reverse causation (23).
After giving signed informed consent, all participants were administered a semi-structured interview to collect personal data about demographics, lifestyle, and medical conditions/treatments, as well as information about their shift work schedules. Current night shift (NS) workers (N = 46), i.e., nurses working NS for at least 2 years, were matched by age and length of service to day shifts (DS) workers. NS workers include former NS workers, i.e., nurses who have worked NS for at least 2 years and who have quit it for <1 year up to 20 years before recruitment, as well as nurses who have never worked NS.
Each subject agreed to donate a 12 ml blood sample, which was drawn in the morning (9.00–10.30 a.m.) at the end of the night shift (for NS nurses) or the beginning of the working day (for day shifters). Body parameters collected in the questionnaire were used to calculate the body mass index (BMI), i.e., weight divided by square height (kg/m2), taken as an indicator of metabolic health. Complete outcome data (see below) could not be assessed for three subjects, and we thus performed our analyses on 94 nurses.
Blood samples were collected in EDTA tubes and centrifuged at 1,200 g for 15 min to separate plasma, buffy coat, and red blood cell fractions within 4 h after withdrawal. Plasma aliquots were stored at −80°C until use, making sure to avoid repeated freeze-thaw cycles.
The quantification of plasma NLRP3 was carried out using a commercial enzyme-linked immunosorbent assay (ELISA) kit (cat. MBS3802246, MyBioSource), according to the manufacturer's instructions. The assay was performed using 10 μl of plasma for each subject. The quantification of plasma TNF-alpha was carried out using the Human TNF-alpha Quantikine HS ELISA kit (cat. HSTA00E, R&D Systems) according to the manufacturer's instruction, using 50 μl plasma samples for each subject. A Synergy HT-BioTek spectrophotometer was used to read the optical density at 450 nm. A blank correction was applied for both assays. For TNF-alpha measurement, we applied an additional wavelength correction at 540 nm, as suggested in the assay protocol. Protein concentration (pg/ml) was determined from the standard curve (R2 > 0.99). As each sample and standard were tested in duplicate, the mean value of the two runs was used in the statistical analysis.
DNA extraction from Buffy coat and sodium bisulfite treatment, followed by amplification of the DNA sequences of interest and pyrosequencing, were performed according to the procedure described in Monti et al. (16). PCR cycling condition and primer sequences used for PCR amplification and pyrosequencing are reported in Supplementary Table S1. CpG sites were queried within regulatory regions (promoter and enhancer regions) of the following genes: ERVE, ERVFRD-1, ERVH, ERVK, HERV-L, HERV-P, ERV3-1, ERV9-1, ERVW-1, HRES1, BIRC2, FLRT3, GAL9, IDO1, LPHN1, MIG6, NLRC5, SIRT1, and XIAP. Every sample was measured twice for each gene to test the reproducibility of the experimental setting.
For each gene, methylation levels were calculated as the percentage of methylated cytosines out of the total number of cytosines (5-methyl-cytosine + unmethylated cytosines) at each CpG site of interest. The designed assays allow the assessment of a variable number of CpG sites (1–5). Coefficients of variation for each assay are as follows: ERVE = 0.02, ERVFRD-1 = 0.01, ERVH = 0.01, ERVK = 0.03, ERVL < 0.01, HERV-P = 0.02, ERV3-1 = 0.01, ERV9-1 = 0.03, ERVW-1 = 0.01, HRES1 = 0.07, BIRC2 = 0.18, FLRT3 = 0.09, GAL9 = 0.12, IDO1 = 0.03, LPHN1 = 0.06, MIG6 = 0.03, NLRC5 = 0.04, SIRT1 = 0.14, and XIAP = 0.01.
Collected data were summarized by standard descriptive statistics, and a graphical inspection of the main variables of interest was performed to examine their distribution. Continuous variables were expressed as mean ± standard deviation (SD), whereas categorical data were reported as frequencies (%). The study population was divided into different groups: current NS workers vs. colleagues who are currently not working DS (“Yes” vs. “No”), current + former NS workers (“ever”) vs. colleagues who have never worked NS (“never”), and “current” vs. “former” vs. “never” shifters.
Baseline characteristics by current NS work (yes vs. no) were reported and compared with a t-test for continuous variables and the chi-square test or Fisher's exact test as appropriate for categorical variables.
To test the association of NS work with the concentration of plasmatic NLRP3 and TNF-alpha, we applied ANCOVA models adjusted for age, BMI, smoking habits, use of oral contraceptives, general health medications, and ELISA plate.
To estimate the effect of NS work on DNA methylation for the genes of interest, we fitted linear mixed-effect models to consider intra-individual correlation due to repeated-measure data structure. Models were adjusted for age, BMI, smoking habits, years of NS work, as well as run, CpG position, and their interaction (run*position). DNA methylation measurements for each subject were run in duplicate. The pyrosequencing-based DNA methylation analysis tested a variable number of CpG positions according to CpG density in the promoter assay. Linear mixed-effect models were used to account for each CpG dinucleotide position (as a random effect) measured in the two runs. An unstructured covariance structure was used to model within-subject errors. Methylation mean levels were calculated as marginal means.
To examine the potential modifying effect of BMI on the association between NS and gene methylation, we added an interaction term between BMI and NS work in each multivariable linear mixed-effect model. Differences between study groups were indicated as β mean differences, standard error (SE), and 95% confidence intervals (CI). We evaluated whether the effect of NS work on methylation levels differs depending on BMI levels. The cut-offs selected for BMI were: 25th percentile (19.8 kg/m2), 50th (22.0 kg/m2), 75th percentile (24.2 kg/m2), and 95th percentile (29.1 kg/m2).
Statistical analyses were performed with SAS software (version 9.4; SAS, Cary, NC, USA). A two-sided p-value of 0.05 was considered statistically significant.
Out of 94 eligible subjects, 44 were working NS at the enrollment (i.e., current NS), and were matched by age and length of service with 50 female colleagues who worked day shifts (DS; Table 1). Among the latter, 28 had never worked NS, and 22 were former NS workers. The majority of study participants had a healthy BMI [i.e., 18.5 ≤ BMI ≤ 24.9 kg/m2 (24)]. No significant differences were observed between DS and NS workers regarding age, length of service, BMI, smoking habits, and pharmacological treatments (oral contraceptive use, general health medications, anti-depressive medications, and sleeping pill). The only exception was the family situation, regarding the marital status (p-value < 0.0001) and the number of children (p-value = 0.0026).
The average NLRP3 concentration calculated on the whole study population was 15.38 ± 2.44 pg/ml and the TNF-alpha concentration was 0.8 ± 0.4 pg/ml. Then, we tested the possible association between NS work and the plasmatic mean concentration of NLRP3 and TNF-alpha (Table 2). We observed a positive association with NLRP3 levels (p-value = 0.0379). We further tested whether having ever been a shift worker (i.e., ever vs. never) and being currently a NS worker (i.e., current vs. former vs. never) was associated with NLRP3 and TNF alpha concentrations (Supplementary Table S2).
We tested whether DNA methylation levels of HERV elements (ERVE, ERVFRD-1, ERVH, ERVK, ERVL, HERV-P, ERV3-1, ERV9-1, ERVW-1, and HRES1) and immune-related genes (BIRC2, FLRT3, GAL9, IDO1, LPHN1, MIG6, NLRC5, SIRT1, and XIAP) were different in night and day shifters (Table 3). Statistically significant differences were observed for ERVFRD-1, ERVL, HERV-P, BIRC2, FLRT3, MIG6, and SIRT1. In particular, we observed a lower percentage of methylated CpGs for ERVFRD-1, FLRT3, and MIG6 in night- compared to day-shifters, whereas ERVL, HERV-P, BIRC2, and SIRT1 methylation was higher in NS nurses.
As overweight could represent an additional susceptibility factor for NS workers, we evaluated the modification effect of BMI on the association between current NS work and DNA methylation of the genes. Interestingly, the only significant interactions between current NS work and BMI were observed for HERV genes, in particular for ERVE, ERVL, and ERVW-1 (p-values = 0.007, =0.029, and =0.032, respectively) (Figure 1). The strength of these associations was evaluated at four fixed levels of BMI (25th percentile: 19.8 kg/m2, 50th percentile: 22.0 kg/m2, 75th percentile: 24.2 kg/m2, and 95th percentile: 29.1 kg/m2).
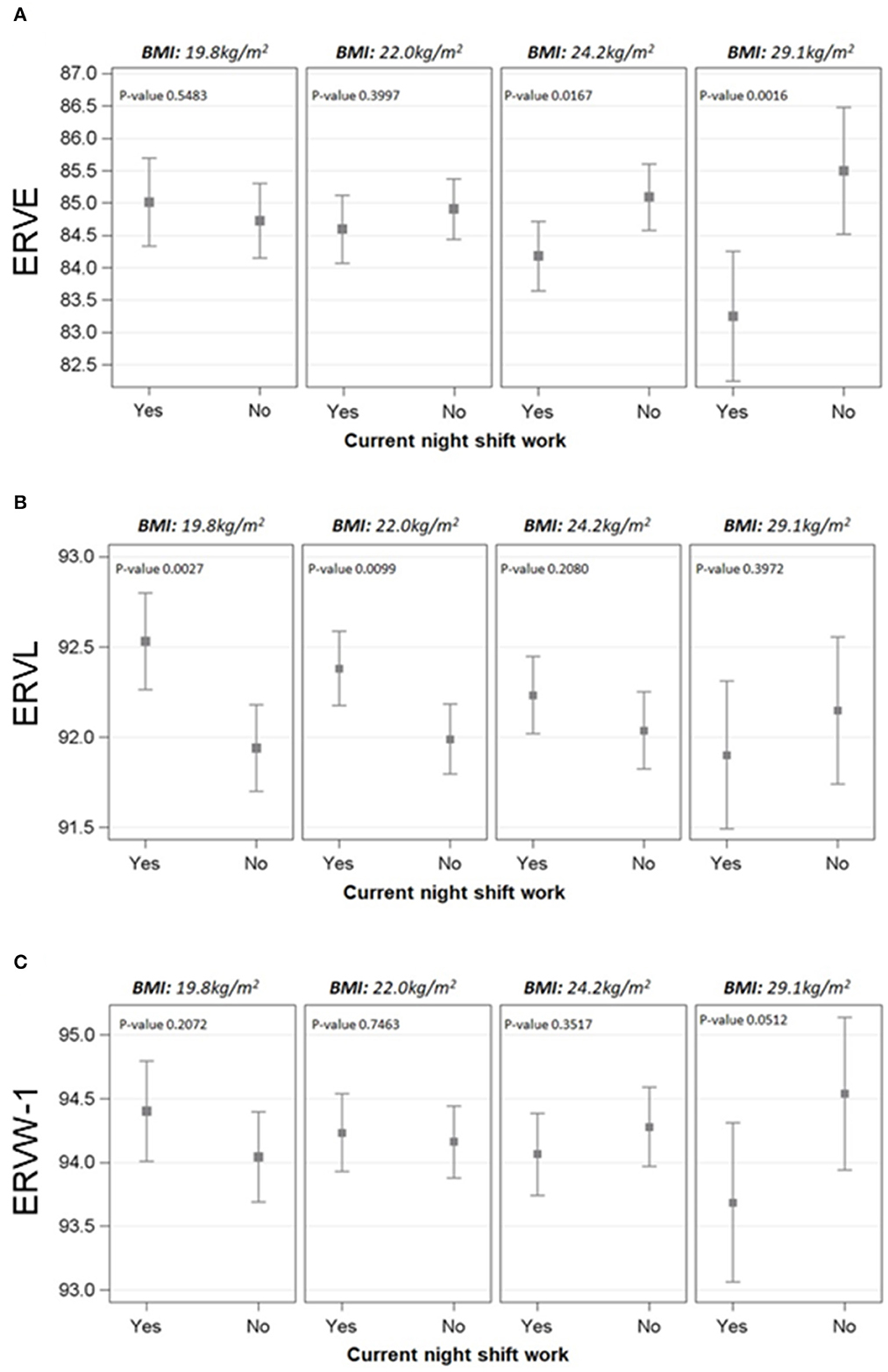
Figure 1. Effect modification of BMI on the association between NS work and methylation of ERVE (A), ERVL (B), and ERVW-1 (C). The cut-offs selected for BMI were 25th, 50th, 75th, and 95th percentile (19.8, 22.0, 24.2, and 29.1 kg/m2, respectively). Linear mixed regression models were adjusted for age, BMI, smoking habits, length of service with NS work, the interaction term BMI*current NS work, run, position, and their interaction. Methylation levels were calculated as marginal means.
For ERVE, the percentage of mCpG sites estimated at BMI = 24.2 kg/m2 was found to be lower in NS workers if compared to DS (β = −0.91; p-value = 0.0167). The difference becomes even wider at BMI = 29.1 kg/m2 (β = −2.25; p-value = 0.002) and it was not significant for lower levels of BMI. On the contrary, the association between NS and ERVL methylation was different for lower-mid BMI values, with higher methylation levels in NS if compared to DS at BMI = 19.8 kg/m2 (β = 0.59; p-value =0.003) and BMI = 22.0 kg/m2 (β = 0.39; p-value = 0.010). Regarding ERVW-1, a slight difference was observed at BMI = 29.1 kg/m2, with NS workers having lower methylation levels (β = −0.85; p-value = 0.052). The associations between current NS work and methylation levels at selected BMI values are reported in Supplementary Table S3.
In the present study conducted on 94 female workers with different working schedules, we first evaluated the association of NS work with plasmatic concentrations of NLRP3 and TNF-alpha.
NLRP3 and TNF-alpha were chosen as paradigmatic indicators of innate immune and inflammatory alterations (25). We found that plasmatic levels of NLRP3 were increased in NS nurses compared to colleagues with daily schedules. NLRP3 is a crucial player in host immune defenses by mediating the activation of caspase-1 and the secretion of proinflammatory cytokines IL-1β and IL-18 (26). NLRP3 expression is controlled by many endogenous and exogenous factors, including circadian rhythms (27). A recent study reported that the core clock component NR1D1 negatively regulates NLRP3 expression and activation, as well as the downstream release of IL-1β and IL-18 (28). Of note, circadian rhythm disruption results in impaired NLRP3 levels, as supported by a study reporting NLRP3 overexpression in mice subjected to sleep deprivation (29). On the contrary, we found no difference in plasma TNF-alpha concentrations between the two study groups. TNF-alpha alterations could be expected among NS workers, as the secretion of this proinflammatory cytokine is under circadian control (30). However, data available in the literature are inconsistent, with studies reporting either positive (31) or negative (30) associations between NS and plasmatic TNF-alpha levels, while others observed non-significant differences (32–34). The recent finding that TNF-alpha knockout and wild-type mice do not respond differently to sleep deprivation (35) led to speculation that other pathways (such as the NLRP3 one) might have a greater impact on the response to circadian disruption (25, 35).
Being DNA methylation affected by environmental triggers, including NS (22, 36), we also examined the methylation levels of HERV elements, which are implied in immune and inflammatory responses (18).
Three HERV genes were differentially methylated between NS and DS workers. In particular, ERVFRD-1 methylation was lower, while ERVL and HERV-P methylation levels were higher in nurses working at night. HERVs derive from ancestral retroviruses that infected the primates' germ line and subsequently integrated into the human genome, thus becoming endogenous elements stably inherited across generations (18). Indeed, the structural organization of HERVs resembles that of retroviral DNA, with the gag, pol, and env genes sided by two long terminal repeats (LTRs) with regulatory functions. However, the progressive accumulation of sequence changes within ancestral retroviral elements has led to the “domestication” of HERVs, leading in most cases to the loss of protein-coding capacity and the acquisition of novel functions at the interface between the “self” and “non-self.” HERVs are emerging as important players in the host's innate immune system by modulating the inflammatory response to pathogens and other external triggers (18). Being non-unique sequences, HERVs are generally subjected to transcriptional silencing to prevent genomic instability (37). Nevertheless, changes in HERV methylation levels have been reported in response to many proinflammatory stressors (e.g., environmental toxicants and lifestyle stressors) with possible pathological outcomes (19). Given their numerous and largely unexplored physiological functions, their alterations have been implicated in cancer, as well as in several autoimmune and inflammatory diseases (38, 39). HERVs can act as pathogen-associated molecular patterns, triggering both innate and adaptive immune responses (18, 40–42) by providing antigenic epitopes recognized by lymphocytes and stimulating the onset of specific T- and B- cells (43, 44). These mechanisms might be crucial for the involvement of HERVs in autoimmune diseases, such as multiple sclerosis (ERVW-1), systemic lupus erythematosus (ERVE and HRES), rheumatoid arthritis (ERVK), and amyotrophic lateral sclerosis (ERVK) (45, 46).
On the other hand, HERVs have also been involved in immune response downregulation, starting from their crucial role in maternal immune tolerance to their suggested protective action against undue immune activation (41). The immune response downregulation underlies HERV involvement in tumorigenesis. Interestingly, aberrant methylation and expression of HERV elements have been reported in many cancer types, including breast, colorectal, and prostate cancer (39, 46), for which an increased risk has been reported also in association with NS work (47, 48).
We observed that ERVE, ERVW-1, and ERVL were differentially methylated depending on the BMI. In particular, ERVE and ERVW-1 were differentially methylated between NS and DS at high BMI values, while ERVL methylation was different for lower-mid BMI values. These results suggest that the overweight condition might be an additional susceptibility factor interacting with NS in fostering an altered immune/inflammatory response modulation. We recently reported an increase in age acceleration per year in NS work in subjects with overweight/obesity (22). Thus, HERV sequences might be considered sensitive indicators of immune alterations caused by multiple environmental stress factors.
We further investigated the association between NS work and the methylation of a subset of immune-related genes. We found that BIRC2 and SIRT1 methylation was higher in NS nurses. BIRC2 (Baculoviral IAP Repeat Containing 2) encodes a member of the inhibitors of apoptosis (IAP) family of proteins. BIRC2 expression is downregulated by promoter hypermethylation (49, 50), which was also associated with increased IL-1β production (49, 51). Similarly, hypermethylation of the SIRT1 gene (encoding the Sirtuin 1, a deacetylase whose downregulation has been found in many disease conditions) correlates with lower transcript levels and with an increased risk of inflammatory and metabolic diseases (52, 53). SIRT1 affects multiple biological processes by de-acetylating a variety of proteins including histones and non-histone proteins. Alterations to SIRT1 methylation, expression, and activity were linked to inflammatory diseases (52). It is therefore possible that NS-induced hypermethylation of such genes might determine an increased inflammation and susceptibility to related pathological conditions.
On the contrary, FLRT3 and MIG6 methylation levels were reduced in NS workers. FLRT3 (Fibronectin leucine-rich transmembrane protein 3) is a protein involved in cell adhesion and receptor signaling. Although to our knowledge methylation levels of the FLRT3 gene have never been investigated before, FLRT3 overexpression was found to be protective toward tumorigenesis as it promotes apoptosis and suppresses epithelial-mesenchymal transition, as well as colorectal cancer cell proliferation, migration, and invasion (54). Besides, FLRT3 is overexpressed in breast cancer cells and participates in immune escape processes by regulating the immune receptor Tim-3 pathway (55). Also, MIG6 (Mitogen-inducible gene-6, also known as ERRFI1) has been principally studied in the context of cancer biology, where it poses as a tumor suppressor that inhibits EGFR signaling. MIG6 hypermethylation results in transcriptional downregulation (56), which has been observed in many cancer types; however, the impact of DNA methylation on MIG6 expression could depend on cancer type, as treatment with DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine did not always affect MIG6 expression (57).
In the present study, we selectively included workers without major pathological conditions at the time of enrollment to specifically focus on the effects of NS work on DNA methylation and exclude possible confounding due to ongoing disease states. The modifications observed might mirror alterations in biological processes affected by NS work. Future functional studies are needed to evaluate the effects of DNA methylation differences reported in the present associative study. Our findings suggest that DNA methylation of HERV elements and immune-related genes could be a promising marker to highlight the complex pattern of inflammatory alterations found in NS workers (supported by plasmatic NLRP3 increase). However, prospective studies are needed to explore the causal relationship between NS exposure and the observed biological modifications, and eventually evaluate their potential as biomarkers of NS-related diseases.
We acknowledge some limitations of the present study. First, the study population is quite small, especially when it comes to considering BMI classes (the number of study participants with BMI>29.1 kg/m2 is very small). Second, we selected the genes of interest based on current evidence in the literature, and not based on unbiased genome-wide methylation analysis; therefore, we could have missed additional methylation alterations occurring in uninvestigated regions. Third, the observed methylation differences between night and day shifters are slight. These results are expected upon exposure of a healthy population to environmental/occupational factors and have emerged thanks to the high precision technology employed and the analytical method applied. However, it is possible that the reduced variations detected might have been underestimated due to the analysis of bulk samples (i.e., buffy coat cell populations). Future studies should be conducted to clarify this aspect.
In the present study, although explorative and based on a small sample, we observed that exposure to NS work is associated with altered modulation of immune and inflammatory-related factors. Moreover, our findings indicate DNA and in particular HERV methylation as a potential biomolecular sensor to monitor the cumulative effect of multiple stressors (NS and metabolic alterations).
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board Comitato Etico Milano Area 2 of the Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico (approval number 702_2015). The patients/participants provided their written informed consent to participate in this study.
LF, ACP, and VB contributed to the conception and design of the study. LF, PM, and LT performed experimental analyses. CF and MC organized the database and performed the statistical analysis. CM and MB enrolled the subjects. LF and PM wrote the first draft of the manuscript. LF and VB supervised the whole study. All authors contributed to the manuscript revision, read, and approved the submitted version.
This study was supported by Programma di Finanziamento alla Ricerca 2020 dell'Università degli Studi di Milano PSR2020 to LF and by Istituto Nazionale Assicurazione Infortuni sul Lavoro INAIL BRIC 2019 (ID 8192866) PROAGEING to MB and VB.
The authors thank all the nurses of the Fondazione IRCCS Ca' Granda Policlinico Hospital who gave their consent to participate in the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1083826/full#supplementary-material
1. Costa G. Shift work and health: current problems and preventive actions. Saf Health Work. (2010) 1:112–23. doi: 10.5491/SHAW.2010.1.2.112
2. Wilson JL. The impact of shift patterns on healthcare professionals. J Nurs Manag. (2002) 10:211–9. doi: 10.1046/j.1365-2834.2002.00308.x
3. Labrecque N, Cermakian N. Circadian clocks in the immune system. J Biol Rhythms [Internet]. (2015) 30:277–90. doi: 10.1177/0748730415577723
4. Torquati L, Mielke GI, Brown WJ, Burton NW, Kolbe-Alexander TL. Shift work and poor mental health: a meta-analysis of longitudinal studies. Am J Public Health. (2019) 109:E13–20. doi: 10.2105/AJPH.2019.305278
5. Kecklund G, Axelsson J. Health consequences of shift work and insufficient sleep. BMJ. (2016) 355:5210. doi: 10.1136/bmj.i5210
6. Rider CF, Carlsten C. Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenetics. (2019) 11:7132. doi: 10.1186/s13148-019-0713-2
7. Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. (2011 ) 3:267. doi: 10.2217/epi.11.22
8. Martin EM, Fry RC. Environmental influences on the epigenome: exposure- associated dna methylation in human populations. Annu Rev Public Health. (2018) 39:309–33. doi: 10.1146/annurev-publhealth-040617-014629
9. Ritonja JA, Aronson KJ, Flaten L, Topouza DG, Duan QL, Durocher F, et al. Exploring the impact of night shift work on methylation of circadian genes. Epigenetics. (2021) 3:141. doi: 10.1136/OEM-2021-EPI.141
10. Reszka E, Wieczorek E, Przybek M, Jabłońska E, Kałuzny P, Bukowska-Damska A, et al. Circadian gene methylation in rotating-shift nurses: a cross-sectional study. Chronobiol Int. (2018) 35:111–21. doi: 10.1080/07420528.2017.1388252
11. Bukowska-Damska A, Reszka E, Kaluzny P, Wieczorek E, Przybek M, Zienolddiny S, et al. Sleep quality and methylation status of core circadian rhythm genes among nurses and midwives. Chronobiol Int. (2017) 34:1211–23. doi: 10.1080/07420528.2017.1358176
12. Ritonja JA, Aronson KJ, Leung M, Flaten L, Topouza DG, Duan QL, et al. Investigating the relationship between melatonin patterns and methylation in circadian genes among day shift and night shift workers. Occup Environ Med. (2022) 3:8111 doi: 10.1136/oemed-2021-108111
13. Bollati V, Baccarelli A, Sartori S, Tarantini L, Motta V, Rota F, et al. Epigenetic effects of shiftwork on blood dna methylation. Chronobiol Int. (2010) 27:1093–104. doi: 10.3109/07420528.2010.490065
14. Adams CD, Jordahl KM, Copeland W, Mirick DK, Song X, Sather CL, et al. Nightshift work, chronotype, and genome-wide DNA methylation in blood. Epigenetics. (2017) 12:833–40. doi: 10.1080/15592294.2017.1366407
15. Carugno M, Maggioni C, Crespi E, Bonzini M, Cuocina S, Dioni L, et al. Night shift work, DNA methylation and telomere length: an investigation on hospital female nurses. Int J Environ Res Public Health. (2019) 16:2292. doi: 10.3390/ijerph16132292
16. Monti P, Iodice S, Tarantini L, Sacchi F, Ferrari L, Ruscica M, et al. Effects of PM exposure on the methylation of clock genes in a population of subjects with overweight or obesity. Int J Environ Res Public Health. (2021) 18:1–15. doi: 10.3390/ijerph18031122
17. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. (2001) 409:860–921. doi: 10.1038/35057062
18. Grandi N, Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol. (2018) 9:2039. doi: 10.3389/fimmu.2018.02039
19. Cho K, Lee YK, Greenhalgh DG. Endogenous retroviruses in systemic response to stress signals. Shock. (2008) 30:105–16. doi: 10.1097/SHK.0b013e31816a363f
20. Franchini AM, Lawrence BP. Environmental exposures are hidden modifiers of anti-viral immunity. Curr Opin Toxicol. (2018) 10:54–9. doi: 10.1016/j.cotox.2018.01.004
21. Pozniak J, Nsengimana J, Laye JP, O'Shea SJ, Diaz JMS, Droop AP, et al. Genetic and environmental determinants of immune response to cutaneous melanoma. Cancer Res]. (2019) 79:2684–96. doi: 10.1158/0008-5472.CAN-18-2864
22. Carugno M, Maggioni C, Ruggiero V, Crespi E, Monti P, Ferrari L, et al. can night shift work affect biological age? Hints from a cross-sectional study on hospital female nurses. Int J Environ Res Public Health. (2021) 18:10639. doi: 10.3390/ijerph182010639
23. Richardson TG, Richmond RC, North TL, Hemani G, Davey Smith G, Sharp GC, et al. An integrative approach to detect epigenetic mechanisms that putatively mediate the influence of lifestyle exposures on disease susceptibility. Int J Epidemiol. (2019) 48:887–98. doi: 10.1093/ije/dyz119
24. Weir CB, Jan A. BMI Classification Ion Percentile and Cut Off Points. Treasure Island, FL: StatPearls Publishing (2022).
25. Zielinski MR, Gibbons AJ. Neuroinflammation, Sleep, and Circadian Rhythms. Front Cell Infect Microbiol. (2022) 12:3096. doi: 10.3389/fcimb.2022.853096
26. Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci MDPI AG. (2019) 59:3328. doi: 10.3390/ijms20133328
27. Pourcet B, Duez H. Circadian control of inflammasome pathways: implications for circadian medicine. Front Immunol. (2020) 11:1630. doi: 10.3389/fimmu.2020.01630
28. Pourcet B, Zecchin M, Ferri L, Beauchamp J, Sitaula S, Billon C, et al. Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology. (2018) 154:1449–64. doi: 10.1053/j.gastro.2017.12.019
29. Zielinski MR, Gerashchenko D, Karpova SA, Konanki V, McCarley RW, Sutterwala FS, et al. The NLRP3 inflammasome modulates sleep and NREM sleep delta power induced by spontaneous wakefulness, sleep deprivation and lipopolysaccharide. Brain Behav Immun. (2017) 62:137–50. doi: 10.1016/j.bbi.2017.01.012
30. Liu PY, Irwin MR, Krueger JM, Gaddameedhi S, Van Dongen HPA. Night shift schedule alters endogenous regulation of circulating cytokines. Neurobiol Sleep Circadian Rhythm. (2021) 10:63. doi: 10.1016/j.nbscr.2021.100063
31. Cakan P, Yildiz S. Effects of half- or whole-night shifts on physiological and cognitive parameters in women. Am J Med Sci. (2020) 360:525–36. doi: 10.1016/j.amjms.2019.12.002
32. Van Mark A, Weiler SW, Schröder M, Otto A, Jauch-Chara K, Groneberg DA, et al. The impact of shift work induced chronic circadian disruption on IL-6 and TNF-alpha immune responses. J Occup Med Toxicol. (2010) 5:18. doi: 10.1186/1745-6673-5-18
33. Copertaro A, Bracci M, Gesuita R, Carle F, Amati M, Baldassari M, et al. Influence of shift-work on selected immune variables in nurses. Ind Health. (2011) 49:597–604. doi: 10.2486/indhealth.MS1210
34. Loef B, Nanlohy NM, Jacobi RHJ, van de Ven C, Mariman R, van der Beek AJ, et al. Immunological effects of shift work in healthcare workers. Sci Rep. (2019) 9:4816. doi: 10.1038/s41598-019-54816-5
35. Szentirmai É, Kapás L. Sleep and body temperature in TNFα knockout mice: the effects of sleep deprivation, β3-AR stimulation and exogenous TNFα. Brain Behav Immun. (2019) 81:260. doi: 10.1016/j.bbi.2019.06.022
36. Ferrari L, Carugno M, Bollati V. Particulate matter exposure shapes DNA methylation through the lifespan. Clin Epigenetics. (2019) 11:726. doi: 10.1186/s13148-019-0726-x
37. Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci. (2017) 74:2055–65. doi: 10.1007/s00018-017-2454-8
38. Mustelin T, Ukadike KC. How retroviruses and retrotransposons in our genome may contribute to autoimmunity in rheumatological conditions. Front Immunol. (2020) 11:3891. doi: 10.3389/fimmu.2020.593891
39. Alcazer V, Bonaventura P, Depil S. Human endogenous retroviruses (HERVs): shaping the innate immune response in cancers. Cancers. (2020) 12:610. doi: 10.3390/cancers12030610
40. Wolff F, Leisch M, Greil R, Risch A, Pleyer L. The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun Signal. (2017) 15:0168. doi: 10.1186/s12964-017-0168-z
41. Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. (2012) 33:663–71. doi: 10.1016/j.placenta.2012.05.005
42. Hurst TP, Magiorkinis G. Activation of the innate immune response by endogenous retroviruses. J Gen Virol. (2015) 96(Pt 6):1207–18. doi: 10.1099/vir.0.000017
43. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. (2015) 162:961–73. doi: 10.1016/j.cell.2015.07.056
44. Trela M, Nelson PN, Rylance PB. The role of molecular mimicry and other factors in the association of human endogenous retroviruses and autoimmunity. APMIS. (2016) 124:88–104. doi: 10.1111/apm.12487
45. Buzdin AA, Prassolov V, Garazha A V. Friends-enemies: endogenous retroviruses are major transcriptional regulators of human DNA. Front Chem. (2017) 5:35. doi: 10.3389/fchem.2017.00035
46. Gröger V, Cynis H. Human endogenous retroviruses and their putative role in the development of autoimmune disorders such as multiple sclerosis. Front Microbiol. (2018) 9:265. doi: 10.3389/fmicb.2018.00265
47. Lie JAS, Kjuus H, Zienolddiny S, Haugen A, Stevens RG, Kjærheim K. Night work and breast cancer risk among Norwegian nurses: assessment by different exposure metrics. Am J Epidemiol. (2011) 173:1272–9. doi: 10.1093/aje/kwr014
48. Wegrzyn LR, Tamimi RM, Rosner BA, Brown SB, Stevens RG, Eliassen AH, et al. Rotating night-shift work and the risk of breast cancer in the nurses' health studies. Am J Epidemiol. (2017) 186:532–40. doi: 10.1093/aje/kwx140
49. Tseng CC, Liao WT, Wong MC, Chen CJ, Lee SC, Yen JH, et al. Cell lineage-specific methylome and genome alterations in gout. Aging. (2021) 13:3843. doi: 10.18632/aging.202353
50. Zhou XH, Lin W, Ren YM, Liu S, Fan BY, Wei ZJ, et al. Comparison of DNA Methylation in schwann cells before and after peripheral nerve injury in rats. Biomed Res Int. (2017) 2017:3268. doi: 10.1155/2017/5393268
51. Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T. Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des. (2013) 82:243–51. doi: 10.1111/cbdd.12176
52. Yang Y, Liu Y, Wang Y, Chao Y, Zhang J, Jia Y, et al. Regulation of SIRT1 and its roles in inflammation. Front Immunol. (2022) 13:872. doi: 10.3389/fimmu.2022.831168
53. Heidari L, Ghaderian SMH, Bastami M, Hosseini S, Alipour Parsa S, Heidari S, et al. Reverse expression pattern of sirtuin-1 and histone deacetylase-9 in coronary artery disease. Arch Physiol Biochem. (2020) 3:7100. doi: 10.1080/13813455.2020.1797100
54. Yang M, Li D, Jiang Z, Li C, Ji S, Sun J, et al. TGF-β-induced FLRT3 attenuation is essential for cancer-associated fibroblast-mediated epithelial-mesenchymal transition in colorectal cancer. Mol Cancer Res. (2022) 3:OF1–13. doi: 10.1158/1541-7786.MCR-21-0924
55. Yasinska IM, Sakhnevych SS, Pavlova L, Selnø ATH, Abeleira AMT, Benlaouer O, et al. The Tim-3-galectin-9 pathway and its regulatory mechanisms in human breast cancer. Front Immunol. (2019) 10:1594. doi: 10.3389/fimmu.2019.01594
56. Milewska M, Romano D, Herrero A, Guerriero ML, Birtwistle M, Quehenberger F, et al. Mitogen-inducible gene-6 mediates feedback inhibition from mutated BRAF towards the epidermal growth factor receptor and thereby limits malignant transformation. PLoS ONE. (2015) 10:9859. doi: 10.1371/journal.pone.0129859
Keywords: night shift work, DNA methylation, inflammation, HERV, immune-related genes
Citation: Ferrari L, Monti P, Favero C, Carugno M, Tarantini L, Maggioni C, Bonzini M, Pesatori AC and Bollati V (2023) Association between night shift work and methylation of a subset of immune-related genes. Front. Public Health 10:1083826. doi: 10.3389/fpubh.2022.1083826
Received: 29 October 2022; Accepted: 22 December 2022;
Published: 12 January 2023.
Edited by:
Luigi Vimercati, University of Bari Aldo Moro, ItalyReviewed by:
Rene Cortese, University of Missouri, United StatesCopyright © 2023 Ferrari, Monti, Favero, Carugno, Tarantini, Maggioni, Bonzini, Pesatori and Bollati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Ferrari,  bHVjYS5mZXJyYXJpQHVuaW1pLml0
bHVjYS5mZXJyYXJpQHVuaW1pLml0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.