
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 05 January 2023
Sec. Planetary Health
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.1069685
This article is part of the Research TopicAir Pollution Exposure and Respiratory DiseasesView all 4 articles
Background: Fine particulate matter (PM2.5) is a crucial risk factor for chronic obstructive pulmonary disease (COPD). However, the mechanisms whereby PM2.5 contribute to COPD risk have not been fully elucidated. Accumulating evidence suggests that epigenetics, including DNA methylation, play an important role in this process; however, the association between PM2.5 exposure and genome-wide DNA methylation in patients with COPD has not been studied.
Objective: To evaluate the association of personal exposure to PM2.5 and genome-wide DNA methylation changes in the peripheral blood of patients with COPD.
Methods: A panel study was conducted in Beijing, China. We repeatedly measured and collected personal PM2.5 data for 72 h. Genome-wide DNA-methylation of peripheral blood was analyzed using the Illumina Infinium Human Methylation BeadChip (850 k). A linear-mixed effect model was used to identify the differentially methylated probe (DMP) associated with PM2.5. Finally, we performed a functional enrichment analysis of the DMPs that were significantly associated with PM2.5.
Results: A total of 24 COPD patients were enrolled and 48 repeated DNA methylation measurements were associated in this study. When the false discovery rate was < 0.05, 19 DMPs were significantly associated with PM2.5 and were annotated to corresponding genes. Functional enrichment analysis of these genes showed that they were related to the response to toxic substances, regulation of tumor necrosis factor superfamily cytokine production, regulation of photosensitivity 3-kinase signaling, and other pathways.
Conclusion: This study provided evidence for a significant relationship between personal PM2.5 exposure and DNA methylation in patients with COPD. Our research also revealed a new biological pathway explaining the adverse effects of PM2.5 exposure on COPD risk.
Chronic obstructive pulmonary disease (COPD) is a heterogeneous disease characterized by persistent respiratory symptoms and airflow limitations. COPD is caused by the interaction between environmental and genetic risk factors (1). Fine particulate matter (PM2.5) is an important environmental risk factor for COPD. Epidemiological and clinical studies have shown that PM2.5 can exacerbate symptoms and accelerate lung function decline in patients with COPD and is an important risk factor for COPD and illness-associated mortality (2–6). Toxicological studies have shown that inflammation and oxidative stress are the key mechanisms whereby PM2.5 contributes to disease risk (7, 8). However, the potential biological pathways underscoring these relationships are far from clear.
Epigenetics, as a bridge linking environment-gene interactions, may provide a new biological mechanism for the adverse health effects of PM2.5 (9). Epigenetics describes changes in gene expression through DNA modification without changing the DNA sequence, including DNA methylation, histone modification, and non-coding RNA modification (10). Among them, DNA methylation is the most extensively studied and fully understood epigenetic modification. Abnormal DNA methylation patterns lead to the destruction of DNA integrity and gene expression changes (11).
External environmental factors, including air pollutants, can affect the level of DNA methylation, which in turn may alter gene expression profiles (12). Early studies provided evidence for the impact of air pollution on global DNA methylation by using long interspersed nucleotide element-1 (LINE-1) and Alu repetitive elements or the level of 5-methylcytosine (5 mC) in the genome (13–16). In addition, candidate-gene methylation analyses have yielded evidence suggesting that PM2.5 exposure is associated with CpG site methylation of genes such as TNF-α, sICAM-1, sCD40L, interleukin 6, and inducible nitric oxide synthase gene (iNOS) (9, 17–19). These results indicate that PM2.5 exposure reduces DNA methylation of specific gene loci, which are associated with the regulation of related gene expression.
Recently, several cohort studies used DNA methylation chip technology to explore the impact of air pollution exposure on genome-wide DNA methylation patterns and found that several CpG sites were related to air pollution exposure. The Women's Health Initiative and The Atherosclerosis Risk in Communities cohort studies found that DNA methylation at three CpG sites was significantly associated with PM exposure, and these sites were located in genes related with neurological, pulmonary, endocrine, and cardiovascular diseases (20). The UK LifeLines cohort study found that differential DNA methylation at seven CpG sites was significantly associated with NO2 exposure, of which two CpGs were considered potential mediators of the association between NO2 exposure and lung function (cg14938677 and cg18379295). NO2 exposure may thus affect lung function by changing DNA methylation patterns. (21) Panel studies conducted in Europe and China found that short-term exposure to PM2.5 was associated with changes in the methylation of multiple CpG sites in healthy non-smokers. Mediation analysis revealed that DNA methylation may play an important role in mediating the impact of PM2.5 on inflammatory pathways (17, 22, 23).
However, the relationship between PM2.5 exposure and peripheral blood genome-wide DNA methylation changes in patients with COPD remains unclear. Therefore, this study examined the association of PM2.5 and DNA methylation using the Illumina Infinium Human Methylation BeadChip (850 K) technology. We anticipated that this study might provide evidence for further exploration of the potential pathogenic mechanisms whereby PM2.5 contributes to COPD risk.
This is a panel study conducted in Beijing, China, between July 2017 and August 2019, which is part of the research that has been reported (24). We collected repeated measurements of personal PM2.5 exposure and health outcomes in patients with COPD who were followed up every 3 months for a total of four visits in 1 year. Briefly, a personal PM2.5-monitoring device was worn for 3 days at each follow-up to obtain the PM2.5-exposure data, and peripheral blood samples were collected. Information about individual demographic and medical characteristics such as sex, age, body mass index (BMI), medication use, duration of COPD, and the number of acute exacerbations was collected at the baseline visit. Written informed consent was obtained from each participant before participation. The study protocol was approved by the Ethics Committee of the China-Japan Friendship Hospital (2017-19). This study was conducted following the principles of the Declaration of Helsinki.
Participants were recruited from the outpatient department of the China-Japan Friendship Hospital. The inclusion criteria were as follows: (1) age 45–75 years; (2) COPD definitively diagnosed by a professional doctor according to the Global Initiatives for Chronic Obstructive Pulmonary Disease guidelines (25); (3) patients who never smoked or quit smoking for more than 6 months.
The exclusion criteria, designed to reduce some factors that may have an obvious impact on DNA methylation, were as follows: (1) patients with a medical history of comorbidities such as malignant tumors, severe cardiovascular and cerebrovascular diseases, hepatic and renal insufficiency, active tuberculosis, and neuropsychiatric disorders; (2) pregnant or lactating women; and (3) alcoholics. Ultimately, 24 patients were enrolled in the study.
The MicroPEM Personal Exposure Monitor (Version 3.2, RTI International, USA) was used to obtain personal PM2.5 exposure data during each follow up. In addition to monitoring the concentration of PM2.5 in real time, this instrument can detect the temperature and humidity of the surrounding environment. Its principle has been introduced in many other studies as well as in our prior research (24, 26). This instrument is small enough to fit in a backpack, and patients need to keep this backpack on their body during follow-up to continuously measure the PM2.5 exposure level for 72 h. Finally, we took the 72 h average as the personal PM2.5 exposure level during each follow-up period.
After each PM2.5 exposure monitoring, 4 mL of fasting venous blood was collected from the patient, processed, and transferred to −80°C within 2 h. Genomic DNA was extracted from peripheral blood according to the manufacturer's protocol for methylation analysis. Briefly, DNA was isolated from peripheral blood using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The purity and concentration of the DNA were estimated using a Nanodrop 2000 (Thermo Fisher Scientific Inc., USA).
In this study, we performed DNA methylation analysis from two blood samples with the highest and lowest PM2.5 exposure in each patient during the four follow-up visits due to the budget constraints, which is a reference to previous panel study conducted by Mostafavi et al. (22) and a cross-over study conducted by Jiang et al. (27). The Illumina Infinium Human Methylation BeadChip (850 K) was used to analyze genome-wide DNA methylation. The 850 K microarrays can detect 866,895 cytosine methylation sites, covering CpG islands, enhancers, gene promoters, and coding regions in the human genome. Briefly, 500 ng of DNA from each sample was used for bisulfite conversion using an EZ DNA Methylation Kit (Zymo Research, USA). The converted products were then placed into 850 K BeadChips following the manufacturer's instructions and protocol. The data were mainly analyzed using the ChAMP package in R. For data quality control, we removed probes with a detection p-value >0.01, probes with less than three beads in at least 10% of samples, non-CpG probes, probes located on chromosomes X and Y, and (SNP-related probes) in turn. We used the β value to show the percentage of methylated cytosines at a given CpG site. The β-value is a continuous variable ranging from 0 to 1, representing the ratio of the intensity of the methylated probe signal to the total locus signal intensity. Then β-value matrix was normalized using BMIQ to adjust type I and type II probe bias (28). In addition, based on DNA methylation data, the Hausman algorithm was used to estimate the relative proportion of five different leukocyte types (i.e., CD8+T lymphocytes, CD4+T lymphocytes, natural killers cells, B cells, monocytes, and granulocytes) in each sample (29).
To further evaluate the biological function of PM2.5-related unique CpG methylation sites, we conducted Gene Ontology (GO) enrichment using GenoGo Metacore. In addition, pathway analysis was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. P-values < 0.05 was considered significant.
To replicate our DMPs with results from previous clinical studies, we performed a systematic review in Pubmed (https://www.ncbi.nlm.nih.gov/pubmed/) from its inception to November 2022. We used the following search terms strategy:(((PM2.5) OR (Fine particulate matter)) OR (PM)) AND ((genome-wide DNA methylation) OR (epigenome-wide)). As no clinical study have been found on PM2.5 exposure and genome-wide DNA methylation in COPD patients currently. In order to verify the correlation between the DMPs and COPD disease, we looked up the DMPs in genome-wide DNA methylation studies of lung tissue in COPD patients (30). We set the cutoff of false discovery rate (FDR) < 0.05 for statistical significance for the look-up.
We used a linear mixed effects model to evaluate the impact of short-term exposure to PM2.5 on genome-wide DNA methylation. In this model, the random intercept for each patient to account for intra-individual correlation between repeated measurements, PM2.5 was regarded as the fixed-effect term. To control for potential confounding, we adjusted some fixed-effect covariates, such as sex, age, BMI, smoking status, temperature, relative humidity, and estimated leukocyte cell-type proportions. In addition, we controlled pulmonary function (FEV1%) in another model to exclude the influence of different severity of COPD disease itself. Finally, we performed the two-pollutant model to assess whether the estimated effects of PM2.5 are dependent on other air pollutants including carbon monoxide(CO), sulfur dioxide(SO2), ozone(O3) and nitrogen dioxide(NO2). Air pollutant data were obtained from environmental monitoring stations closest to the patients' home address. The average daily pollutants in the 3 days before blood sample collection were included in the two-pollutant model for analysis. The LME model was conducted using R software (version 4.0.0) with the package of “lmertest.” To control for type I errors in the genome-wide association, we used the Benjamini–Hochberg false discovery rate method (BH-FDR) to adjust the obtained p-values, and FDR < 0.05 was considered significant.
A total of 18 males and 6 females with an average disease duration of 6.0 ± 4.7 years were enrolled in this study. Basic information on the participants included in this study is shown in Table 1. The mean age of the participants was 62.2 ± 8.0 years, and the mean BMI was 26.2 ± 6.7 kg/m2. Among them, 66.7% were former smokers, with an average smoking index of 25.6 packs/year. The average FEV1/FVC ratio and FEV1% after inhalation of the bronchodilator were 57.4 and 51.8%, respectively. Common comorbidities included hypertension, diabetes, and hyperlipidemia.
The personal PM2.5 exposure, and meteorological data are shown in Table 2. During the study period, the average 72 h mean concentrations of personal PM2.5 exposure varied from 8.5 to 150 μg/m3, with a mean of 62 μg/m3. We conducted a correlation analysis between PM2.5 and meteorological data, and found no significant correlation between PM2.5, temperature, or humidity (Supplementary Table 1).
After removing probes that did not meet the quality control criteria, 840,157 CpGs were included in the LME model. Analyzing PM2.5 as a continuous variable, 19 DMPs were significantly associated with PM2.5 exposure under the condition of FDR < 0.05 (Figure 1). Among the 19 DMPs, 15 DMPs were negatively correlated with PM2.5. In other words, the methylation level decreased after exposure to PM2.5; four DMPs were positively correlated with PM2.5. The names of these DMPs, their corresponding gene names, their distribution, and location on gene structural elements, their chromosomes, and their correlation coefficients with PM2.5 are shown in Table 3. When FEV1% was adjusted in the LME model, 10 of the 19 DMPS were still statistically significant, and the estimated effect coefficients did not change significantly (Table 4).
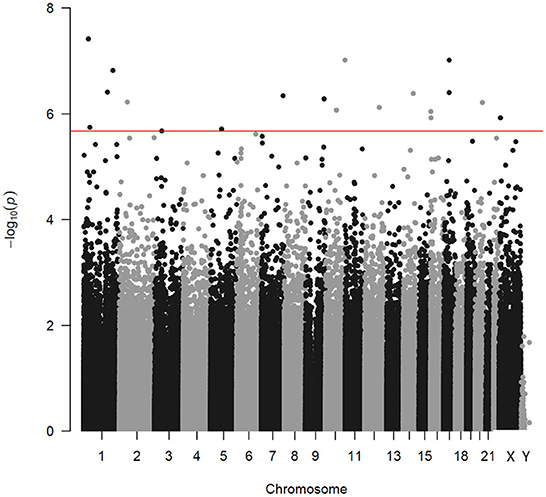
Figure 1. Manhattan plot indicating the associations between PM2.5 and DNA methylation. Every point corresponds to a CpG methylation site. The x-axis shows the chromosome of each CpG locus. The horizontal line corresponds to an adjusted p-value (FDR) of 0.05 level.
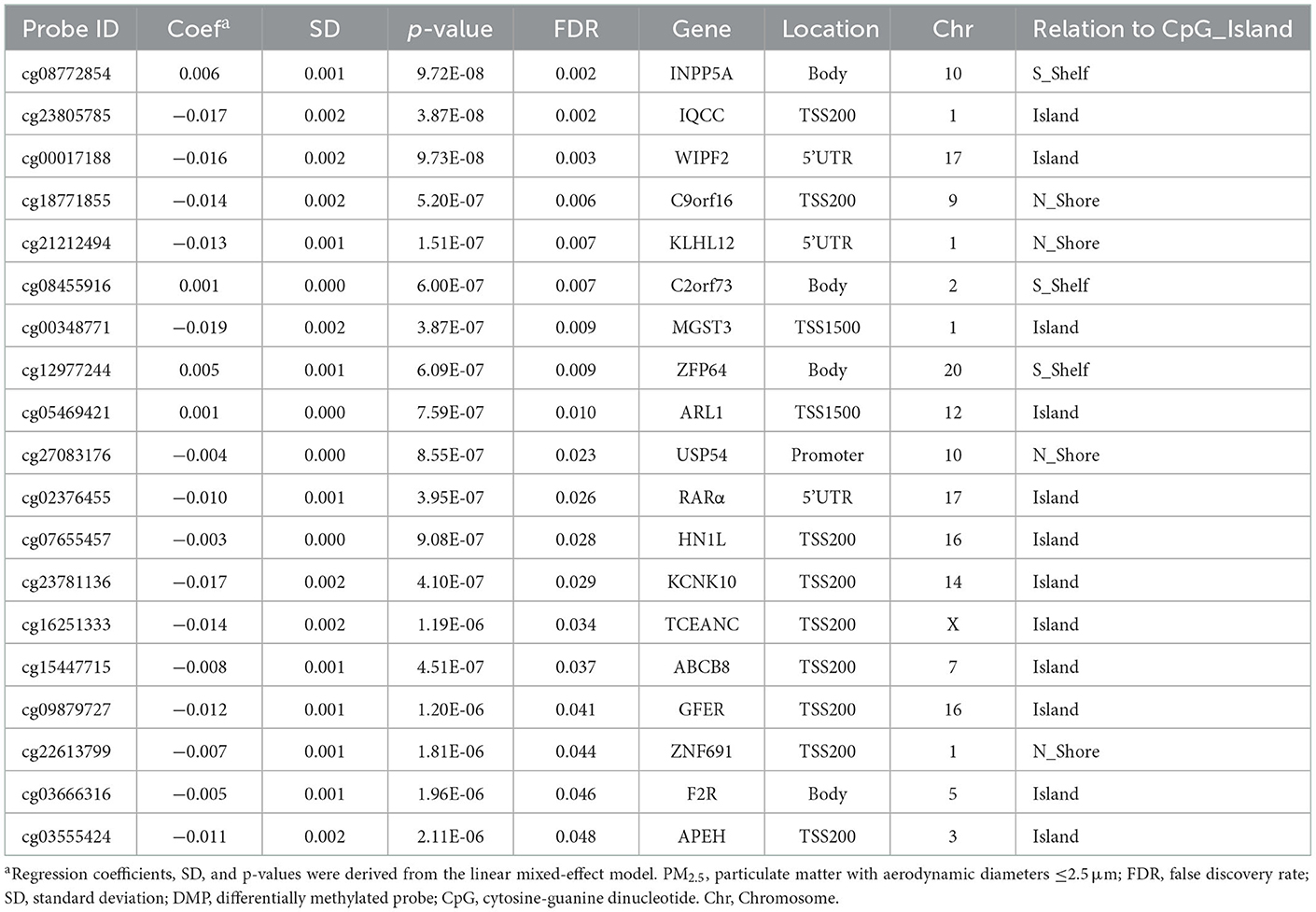
Table 3. Characteristics of DMPs associated with PM2.5 exposure in genome-wide methylation analysis, sorted by FDR.
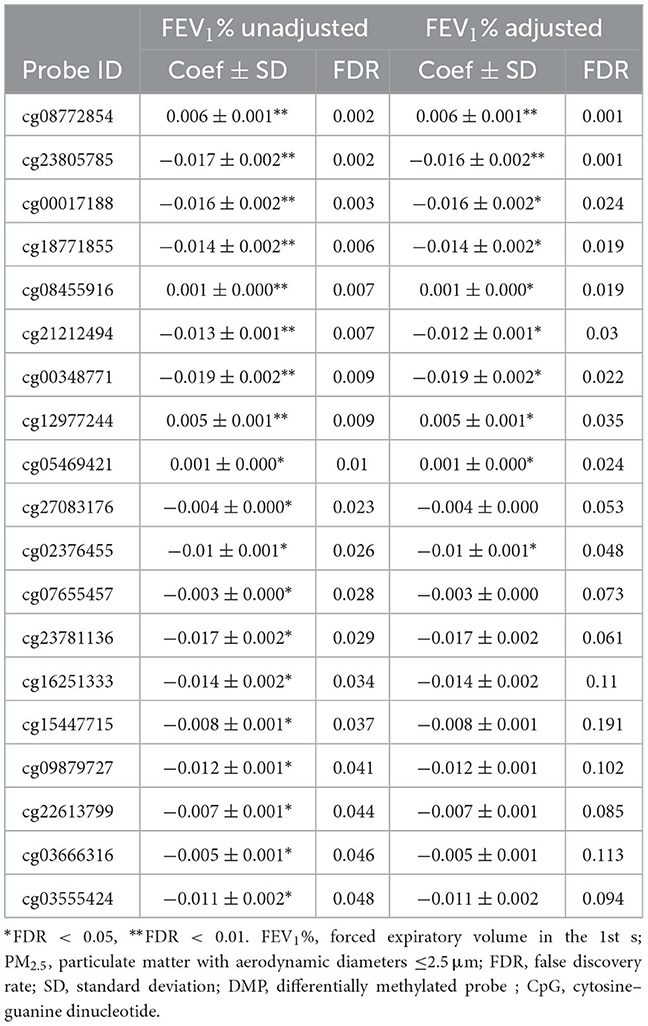
Table 4. Characteristics of DMPs associated with PM2.5 exposure in genome–wide methylation analysis after adjusted for lung function, sorted by FDR.
Table 5 shows the estimated effect of other air pollutants on PM2.5-related DMPs in two-pollutant models. After adjusting for O3, only 3 of the 19 DMPs remained statistically significant, suggesting that O3 may neutralize or weaken the estimated effect of PM2.5 on DNA methylation. Similarly, 6 DMPs were still statistically significant after adjusting for CO. However, when NO2 and SO2 were adjusted in the LME model, most DMPs remained statistically significant (14 and 17 DMPs, respectively), and the estimated effect coefficients did not change significantly, suggesting that the estimated effect of PM2.5 on DNA methylation was relatively independent of NO2 or SO2.
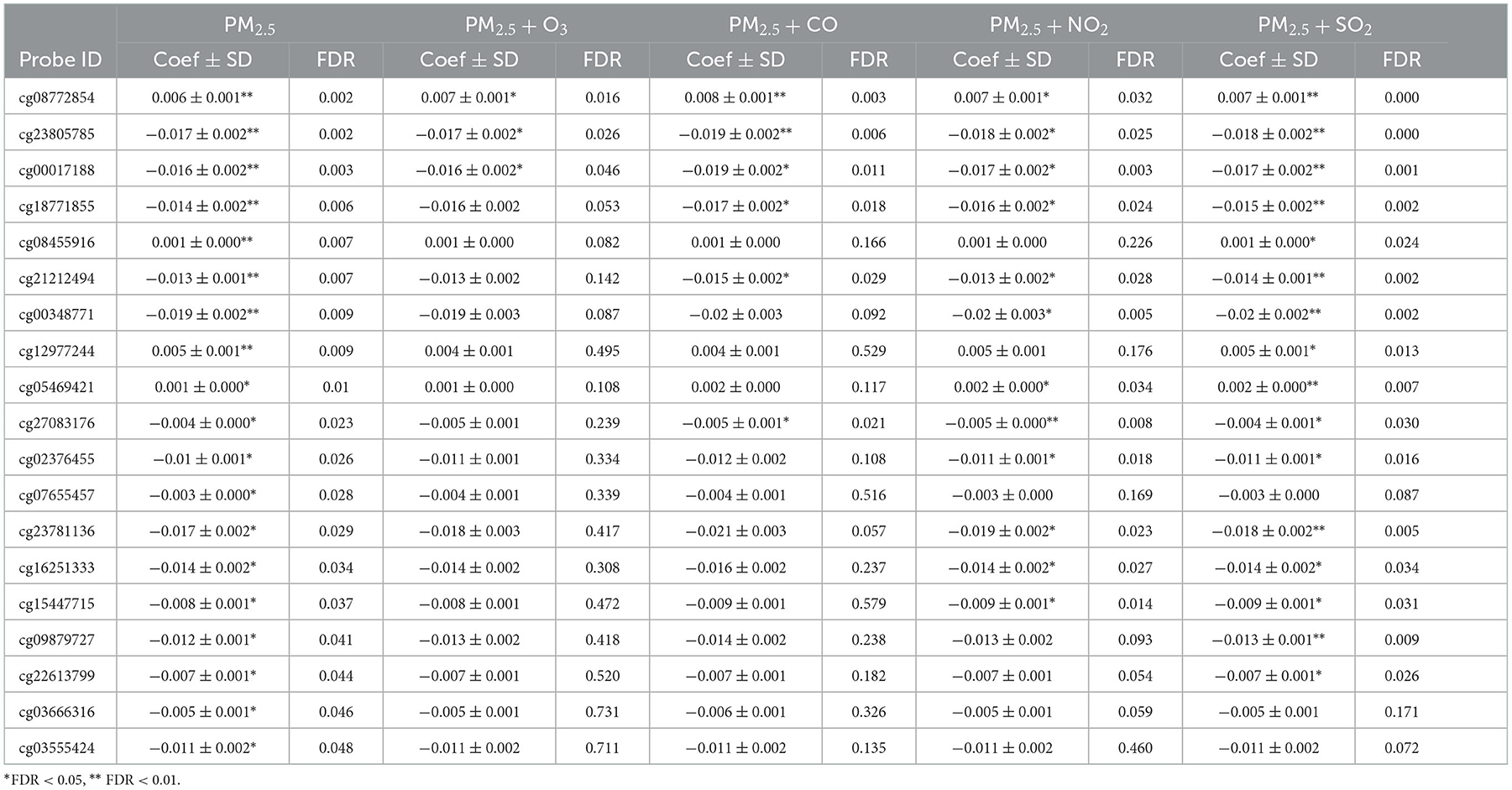
Table 5. Two-pollutant models show the estimated effects of other air pollutants on PM2.5-related DMPs, sorted by FDR of PM2.5 alone.
To evaluate the potential biological functions of the DMPs associated with PM2.5, we performed gene function enrichment analysis on these 19 DMPs. The ten most enriched GO pathways are shown in Figure 2, and the gene names involved in these GO pathways are shown in Supplementary Table 2. The GO enrichment mainly focused on cellular toxicity and metabolic responses to toxic substances, as well as the production of inflammatory factors, the response to toxic substances, regulation of tumor necrosis factor superfamily cytokine production, regulation of tumor necrosis factor production, secondary metabolic processes, organic hydroxy compound catabolic processes, and regulation of phosphatidylinositol 3-kinase signaling. In KEGG pathway enrichment analysis, a total of 30 pathways were involved in those DMPs, but only 3 of them reached statistical significance (P-values < 0.05). The three pathways are Pathogenic Escherichia coli infection, PI3K-Akt signaling pathway and Glutathione metabolism (Supplementary Table 3).
We initially identified 51 manuscripts based on the search terms. After reading the full manuscripts, 15 genome-wide DNA methylation clinical studies were selected according to our analysis requirements (20–22, 27, 29, 31–40). Of the 19 DMPs associated with PM2.5 in our study, only one DMPs (cg08772854, mapped to INPP5A gene) was validate in the 2 of 15 previous genome-wide DNA methylation studies above (23, 32). And our results have the same direction effects as those 2 studies, the estimated effect coefficients are 0.006, 0.15, and 0.005, respectively (Supplementary Table 4). Moreover, cg08772854 was verified in the COPD cohort, indicating that cg08772854 is associated with COPD. The 18 remaining DMPs related to PM2.5 in our study were novel, meaning not reported to be associated with PM2.5 in other earlier studies.
In this study, we used the latest genome-wide DNA methylation chip technology to explore the association between personal PM2.5 exposure and peripheral blood genome-wide DNA methylation in patients with COPD. We found that 19 DMPs were associated with PM2.5, and all 19 DMPs were annotated to specific genes. DNA methylation of these genes was reported for the first time to be related to exposure to PM2.5. Functional enrichment analysis showed that these genes were mainly involved in the metabolic response to external toxic substances and the production and regulation of inflammatory factors. To the best of our current knowledge, this is the first study to investigate the association of PM2.5 and epigenome-wide DNA methylation in COPD patients. The association of other air pollutants and DNA methylation in patients with COPD are also limited. Only a Korean cohort study (N = 100, which included 60 COPD patients) analyzed the effect of PM10 and NO2 on genome-wide DNA methylation (33). There was no duplication between our study and the latter. Therefore, this study provides new insights into the acute health effects of PM2.5 and COPD. We observed mostly negative estimated effect values between PM2.5 and genome-wide DNA methylation, in contrast to previous results that observed positive association between short-term PM exposure and DNA methylation levels at most DMPs (27, 32).
Inflammation and oxidative stress play important mediating roles in the effects of PM2.5 exposure on health. Some of the unique sites associated with PM2.5 exposure found in this study were genes associated with inflammation and oxidative stress. For example, we found that microsomal glutamate S-transfer 3 (mGST3) was hypo-methylation after PM2.5 exposure. mGST3 is a membrane-bound antioxidant enzyme with glutathione-dependent peroxidase activity, which is closely related to the antioxidant defense system and can protect mammals from oxidative stress damage caused by harmful substances in the environment (41). MGST3 plays an important role in protecting airway and alveolar epithelial cells by mitigating oxidative stress in the lung, which is also an important target in the treatment of COPD. Tang et al. (42) found that MGST3 was associated with FEV1/FVC in patients with COPD. In addition, mGST3 is involved in the catalysis of leukotriene C4 and reduced glutathione, thereby playing an important role in inflammation and oxidative stress. These findings suggest that PM2.5 can affect inflammation and oxidative stress in the lung by affecting the methylation status of genes related to inflammation and oxidative stress, thereby affecting the occurrence and development of COPD (43, 44).
Changes in gene methylation related to immune functions were also observed in this study. For example, Kelch-like family member 12 (KLHL12) and ZEP64 showed low methylation levels after exposure to PM2.5. KLHL12 is a nuclear protein that can specifically bind to the dopamine D4 receptor and participate in ubiquitination (45). KLHL12 is associated with a variety of autoimmune diseases. For example, KLHL12 has been identified as an autoantigen in Sjogren's syndrome and is a potential biomarker for primary biliary cirrhosis (46–48). Activation of Toll-like receptors (TLRs) is very important for initiating protective immune responses. Wang et al. found that overexpression of ZFP64 could significantly upregulate TLR-induced NF-κB activation and promote TNF-α, IL-6, and IFN-β production, while knockdown of ZFP64 expression significantly inhibited the production of these cytokines, indicating that ZFP64 can promote the activation of the TLR signaling pathway (49). The roles of these genes in COPD have not been studied, and further biological studies should be carried out to further explore the potential mechanisms of these genes in the pathogenesis of COPD.
We performed functional enrichment analysis of the genes where these 19 DMPs were located and provided biological implications for our findings. Significantly enriched pathways included responses to toxic substances, secondary metabolic processes, and other metabolic pathways to external toxic substances. PM2.5 not only contains harmful compounds such as polycyclic aromatic hydrocarbons, but also adsorbs other toxic substances on its surface. These toxic stimuli can cause changes in the state of cells, including gene expression. Our study showed that PM2.5 may stimulate the body's toxic and metabolic response by affecting the changes in DNA methylation patterns of genes involved in these pathways.
Functional enrichment analysis revealed that these 19 DMPs were also involved in inflammatory and oxidative stress-related pathways, e.g., the regulation of tumor necrosis factor production and phosphatidylinositol 3-kinase signaling (PI3K). Previous studies have shown that most members of the tumor necrosis factor superfamily can participate in inflammatory responses, regulate cell proliferation, differentiation, and apoptosis, and also play a role in various illnesses including cancer, cardiovascular disease, pulmonary conditions, and autoimmune disorders (50, 51). Moreover, members of the TNF superfamily can activate NF-κB, leading to a persistent inflammatory response in the lung. Multiple members of the tumor necrosis factor superfamily, such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (52), CD40L (53), and TNF-α (54), play an important role in the pathogenesis of COPD. The PI3K signaling pathway is an important signaling cascade involved in a variety of important pathophysiological processes, including oxidative stress and inflammatory responses. Several experimental studies have shown that PI3K signaling is significantly activated in COPD and plays a key role in chronic inflammation and oxidative stress in COPD (55, 56). In this study, functional enrichment analysis revealed the potential biological functions of aberrant DNA methylation genes related to PM2.5.
Our study had several strengths. First, we obtained personal PM2.5 exposure data using an individual sampler, allowing us to more accurately assess exposure to PM2.5. Additionally, we used a panel study approach, allowing everyone to act as their own control factor, avoiding confusion over time-invariant factors. Simultaneously, our study had some limitations. First, the expensive and cumbersome assessment of individual PM2.5 exposure limited the sample size. Second, only differentially methylated genes were analyzed, and the expression levels of related genes were not measured or verified. In the future, mRNA and protein expression of these genes will be analyzed to provide evidence for the role of PM2.5 exposure-related DNA methylation in the development of COPD.
In conclusion, our study provides strong evidence suggesting that short-term exposure to PM2.5, which is involved in response to toxic substances, tumor necrosis superfamily cytokine production, oxidative stress, and other biological pathways, may lead to changes in the methylation status of multiple CpG sites in the peripheral blood of patients with COPD. Further studies with larger sample sizes are necessary to confirm the findings of this study.
The studies involving human participants were reviewed and approved by China-Japan friendship hospital. The patients/participants provided their written informed consent to participate in this study.
TiY proposed this study and revised the manuscript. RD and HN completed participants' recruitment and follow-up. RD did the data analysis and completed the manuscript. TaY and XL collected samples and did quality control. All authors revised the report and approved the final version before submission.
This work was supported by the Scientific Research Project of China-Japan Friendship Hospital (Grant Number 2019-2-QN-74), the National Nature Science Foundation of China (Grant Number 92043000), the CAMS Innovation Fund for Medical Science (Grant Number 2020-I2M-2-009), and National Key Research and Development Program of China (Grant Number 2022YFC3702704).
The authors would like to thank the study participants for their dedicated participation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1069685/full#supplementary-material
1. Singh D, Agusti A, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the GOLD science committee report 2019. Eur Respir J. (2019) 53:1900164. doi: 10.1183/13993003.00164-2019
2. Sinharay R, Gong J, Barratt B, Ohman-Strickland P, Ernst S, Kelly FJ, et al. Respiratory and cardiovascular responses to walking down a traffic-polluted road compared with walking in a traffic-free area in participants aged 60 years and older with chronic lung or heart disease and age-matched healthy controls: a randomized, crossover study. Lancet. (2018) 391:339–49. doi: 10.1016/s0140-6736(17)32643-0
3. Doiron D, de Hoogh K, Probst-Hensch N, Fortier I, Cai Y, De Matteis S, et al. Air pollution, lung function and COPD: results from the population-based UK Biobank study. Eur Respir J. (2019) 54:1802140. doi: 10.1183/13993003.02140-2018
4. Liu S, Zhou Y, Liu S, Chen X, Zou W, Zhao D, et al. Association between exposure to ambient particulate matter and chronic obstructive pulmonary disease: results from a cross-sectional study in China. Thorax. (2017) 72:788–95. doi: 10.1136/thoraxjnl-2016-208910
5. Li MH, Fan LC, Mao B, Yang JW, Choi AMK, Cao WJ, et al. Short-term exposure to ambient fine particulate matter increases hospitalizations and mortality in COPD: a systematic review and meta-analysis. Chest. (2016) 149:447–58. doi: 10.1378/chest.15-0513
6. Kariisa M, Foraker R, Pennell M, Buckley T, Diaz P, Criner GJ, et al. Short- and long-term effects of ambient ozone and fine particulate matter on the respiratory health of chronic obstructive pulmonary disease subjects. Arch Environ Occup Health. (2015) 70:56–62. doi: 10.1080/19338244.2014.932753
7. Wang J, Li Y, Zhao P, Tian Y, Liu X, He H, et al. Exposure to air pollution exacerbates inflammation in rats with preexisting COPD. Mediators Inflamm. (2020) 8:4260204. doi: 10.1155/2020/4260204
8. Xu X, Wang H, Liu S, Xing C, Liu Y, Aodeng Q, et al. TP53-dependent autophagy links the ATR-CHEK1 axis activation to proinflammatory VEGFA production in human bronchial epithelial cells exposed to fine particulate matter. Autophagy. (2016) 12:1832–48. doi: 10.1080/15548627.2016.1204496
9. Chen R, Qiao L, Li H, Zhao Y, Zhang Y, Xu W, et al. Fine particulate matter constituents, nitric oxide synthase dna methylation and exhaled nitric oxide. Environ Sci Technol. (2015) 49:11859–65. doi: 10.1021/acs.est.5b02527
10. Weinhold B. Epigenetics: the science of change. Environ Health Perspect. (2006) 114:A160–167. doi: 10.1289/ehp.114-a160
11. de Hartog JJ, Ayres JG, Karakatsani A, Analitis A, Brink HT, Hameri K, et al. Lung function and indicators of exposure to indoor and outdoor particulate matter among asthma and COPD patients. Occup Environ Med. (2010) 67:2–10. doi: 10.1136/oem.2008.040857
12. Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb). (2010) 105:105–12. doi: 10.1038/hdy.2010.2
13. Wong JYY, Cawthon R, Dai Y, Vermeulen R, Bassig BA, Hu W, et al. Elevated Alu retroelement copy number among workers exposed to diesel engine exhaust. Occup Environ Med. (2021) 78:823–8. doi: 10.1136/oemed-2021-107462
14. Miao M, Zhou X, Li Y, Zhang O, Zhou Z, Li T, et al. LINE-1 hypomethylation in spermatozoa is associated with Bisphenol A exposure. Andrology. (2014) 2:138–44. doi: 10.1111/j.2047-2927.2013.00166.x
15. Wang Y, Wang T, Xu M, Yu H, Ding C, Wang Z, et al. Independent effect of main components in particulate matter on DNA methylation and DNA methyltransferase: a molecular epidemiology study. Environ Int. (2020) 134:105296. doi: 10.1016/j.envint.2019.105296
16. Bellavia A, Urch B, Speck M, Brook RD, Scott JA, Albetti B, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. (2013) 2:e000212. doi: 10.1161/JAHA.115.001981
17. Wang C, Chen R, Shi M, Cai J, Shi J, Yang C, et al. Possible mediation by methylation in acute inflammation following personal exposure to fine particulate air pollution. Am J Epidemiol. (2018) 187:484–93. doi: 10.1093/aje/kwx277
18. Kile ML, Fang S, Baccarelli AA, Tarantini L, Cavallari J, Christiani DC, et al. panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health. (2013) 12:47. doi: 10.1186/1476-069X-12-47
19. Wang C, O'Brien KM, Xu Z, Sandler DP, Taylor JA, Weinberg CR. Long-term ambient fine particulate matter and DNA methylation in inflammation pathways: results from the Sister study. Epigenetics. (2020) 15:524–35. doi: 10.1080/15592294.2019.1699894
20. Holliday KM, Justice AE, Mendez-Giraldez R, Stewart JD, et al. Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ Int. (2019) 132:104723. doi: 10.1016/j.envint.2019.03.071
21. de FC Lichtenfels AJ, Van Der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, et al. Long-term air pollution exposure, genome-wide dna methylation and lung function in the lifelines cohort study. Environ Health Perspect. (2018) 126:027004. doi: 10.1289/EHP2045
22. Mostafavi N, Vermeulen R, Ghantous A, Hoek G, Probst-Hensch N, Herceg Z, et al. Acute changes in DNA methylation in relation to 24 h personal air pollution exposure measurements: A panel study in four European countries. Environ Int. (2018) 120:11–21. doi: 10.1016/j.envint.2018.07.026
23. Li H, Chen R, Cai J, Cui X, Huang N, Kan H. Short-term exposure to fine particulate air pollution and genome-wide DNA methylation: a randomized, double-blind, crossover trial. Environ Int. (2018) 120:130–6. doi: 10.1016/j.envint.2018.07.041
24. Duan R, Niu H, Yu T, Huang K, Cui H, Chen C, et al. Adverse effects of short-term personal exposure to fine particulate matter on the lung function of patients with chronic obstructive pulmonary disease and asthma: a longitudinal panel study in Beijing, China. Environ Sci Pollut Res Int. (2021) 28:47463–73. doi: 10.1007/s11356-021-13811-y
25. Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. (2017) 195:557–82. doi: 10.1164/rccm.201701-0218PP
26. Lei X, Xiu G, Li B, Zhang K, Zhao M. Individual exposure of graduate students to PM25 and black carbon in Shanghai, China. Environ Sci Pollut Res Int. (2016) 23:12120–7. doi: 10.1007/s11356-016-6422-x
27. Jiang M, Wang X, Gao X, et al. Association of DNA methylation in circulating CD4(+) T cells with short-term PM(2.5) pollution waves: a quasi-experimental study of healthy young adults. Ecotoxicol Environ Safety. (2022) 239:113634. doi: 10.1016/j.ecoenv.2022.113634
28. Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. (2013) 29:189–96. doi: 10.1093/bioinformatics/bts680
29. Brunekreef B, Janssen NA, de Hartog J, Harssema H, Knape M, van Vliet P. Air pollution from truck traffic and lung function in children living near motorways. Epidemiology. (1997) 8:298–303. doi: 10.1097/00001648-199705000-00012
30. Eriksson Ström J, Kebede Merid S, Pourazar J, et al. Chronic obstructive pulmonary disease is associated with epigenome-wide differential methylation in BAL lung cells. Am J Respir Cell Mol Biol. (2022) 66:638–47. doi: 10.1165/rcmb.2021-0403OC
31. Dai L, Mehta A, Mordukhovich I, Just AC, Shen J, Hou L, et al. Differential DNA methylation and PM25 species in a 450K epigenome-wide association study. Epigenetics. (2017) 12:139–48. doi: 10.1080/15592294.2016.1271853
32. Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ Health Perspect. (2016) 124:983–90. doi: 10.1289/ehp.1509966
33. Lee MK, Xu CJ, Carnes MU, Nichols CE, Ward JM, Kwon SO, et al. Genome-wide DNA methylation and long-term ambient air pollution exposure in Korean adults. Clin Epigen. (2019) 11:37. doi: 10.1186/s13148-019-0635-z
34. White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, et al. Air pollution, particulate matter composition and methylation-based biologic age. Environ Int. (2019):132 105071. doi: 10.1016/j.envint.2019.105071
35. Wang C, Cardenas A, Hutchinson JN, Just A, Heiss J, Hou L, et al. Short- and intermediate-term exposure to ambient fine particulate elements and leukocyte epigenome-wide DNA methylation in older men: the normative aging study. Environ Int. (2022) 158:106955. doi: 10.1016/j.envint.2021.106955
36. Mu G, Nie X, Yang S, Ye Z, Cheng M, Fan L, et al. PM2.5-related DNA methylation and the association with lung function in non-smokers. Environ Pollut. (2022) 3:120700. doi: 10.1016/j.envpol.2022.120700
37. Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int. (2017) 108:127–36. doi: 10.1016/j.envint.2017.08.006
38. Honkova K, Rossnerova A, Chvojkova I, Milcova A, Margaryan H, Pastorkova A, et al. Genome-wide DNA methylation in policemen working in cities differing by major sources of air pollution. Int J Mol Sci. (2022) 23:1666. doi: 10.3390/ijms23031666
39. Sayols-Baixeras S, Fernández-Sanlés A, Prats-Uribe A, Subirana I, Plusquin M, Künzli N, et al. Association between long-term air pollution exposure and DNA methylation: the REGICOR study. Environ Res. (2019) 176:108550. doi: 10.1016/j.envres.2019.108550
40. Eze IC, Jeong A, Schaffner E, Rezwan FI, Ghantous A, Foraster M, et al. Genome-wide DNA methylation in peripheral blood and long-term exposure to source-specific transportation noise and air pollution: the SAPALDIA study. Environ Health Perspect. (2020) 128:67003. doi: 10.1289/ehp6174
41. Guo R, Ebenezer V, Ki JS. PmMGST3, a novel microsomal glutathione S-transferase gene in the dinoflagellate Prorocentrum minimum, is a potential biomarker of oxidative stress. Gene. (2014) 546:378–85. doi: 10.1016/j.gene.2014.05.046
42. Tang W, Bentley AR, Kritchevsky SB, Harris TB, Newman AB, Bauer DC, et al. Genetic variation in antioxidant enzymes, cigarette smoking, and longitudinal change in lung function. Free Radic Biol Med. (2013) 63:304–12. doi: 10.1016/j.freeradbiomed.2013.05.016
43. Gao N, Xu W, Ji J, Yang Y, Wang ST, Wang J, et al. Lung function and systemic inflammation associated with short-term air pollution exposure in chronic obstructive pulmonary disease patients in Beijing, China. Environ Health. (2020) 19:12. doi: 10.1186/s12940-020-0568-1
44. Yang SL, Chen LJ, Kong Y, Xu D, Lou YJ. Sodium nitroprusside regulates mRNA expressions of LTC4 synthesis enzymes in hepatic ischemia/reperfusion injury rats via NF-kappaB signaling pathway. Pharmacology. (2007) 80:11–20. doi: 10.1159/000102595
45. Rondou P, Haegeman G, Vanhoenacker P, Van Craenenbroeck K, BTB. Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. J Biol Chem. (2008) 283:11083–96. doi: 10.1074/jbc.M708473200
46. Uchida K, Akita Y, Matsuo K, Fujiwara S, Nakagawa A, Kazaoka Y, et al. Identification of specific autoantigens in Sjogren's syndrome by SEREX. Immunology. (2005) 116:53–63. doi: 10.1111/j.1365-2567.2005.02197.x
47. Norman GL, Yang CY, Ostendorff HP, Shums Z, Lim MJ, Wang J, et al. Anti-kelch-like 12 and anti-hexokinase 1: novel autoantibodies in primary biliary cirrhosis. Liver Int. (2015) 35:642–51. doi: 10.1111/liv.12690
48. Norman GL, Reig A, Vinas O, Mahler M, Wunsch E, Milkiewicz P, et al. The prevalence of anti-hexokinase-1 and anti-kelch-like 12 peptide antibodies in patients with primary biliary cholangitis is similar in Europe and North America: a large international, multi-center study. Front Immunol. (2019) 10:662. doi: 10.3389/fimmu.2019.00662
49. Wang C, Liu X, Liu Y, Zhang Q, Yao Z, Huang B, et al. Zinc finger protein 64 promotes Toll-like receptor-triggered proinflammatory and type I interferon production in macrophages by enhancing p65 subunit activation. J Biol Chem. (2013) 288:24600–8. doi: 10.1074/jbc.M113.473397
50. Figgett WA, Vincent FB, Saulep-Easton D, Mackay F. Roles of ligands from the TNF superfamily in B cell development, function, and regulation. Semin Immunol. (2014) 26:191–202. doi: 10.1016/j.smim.2014.06.001
51. Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. (2012) 119:651–65. doi: 10.1182/blood-2011-04-325225
52. Haw TJ, Starkey MR, Nair PM, Pavlidis S, Liu G, Nguyen DH, et al. A pathogenic role for tumor necrosis factor-related apoptosis-inducing ligand in chronic obstructive pulmonary disease. Mucosal Immunol. (2016) 9:859–72. doi: 10.1038/mi.2015.111
53. Liu Y, Liang WB, Gao LB, Wang YY, Zhang L. Association of CD40−1C/T polymorphism in the 5'-untranslated region and chronic obstructive pulmonary disease. Clin Chim Acta. (2009) 408:56–9. doi: 10.1016/j.cca.2009.07.005
54. Lundblad LK, Thompson-Figueroa J, Leclair T, Sullivan MJ, Poynter ME, Irvin CG, et al. Tumor necrosis factor-alpha overexpression in lung disease: a single cause behind a complex phenotype. Am J Respir Crit Care Med. (2005) 171:1363–70. doi: 10.1164/rccm.200410-1349OC
55. Stokes CA, Condliffe AM. Phosphoinositide 3-kinase δ (PI3Kδ) in respiratory disease. Biochem Soc Trans. (2018) 46:361–9. doi: 10.1042/BST20170467
Keywords: PM2.5, DNA-methylation, chronic obstructive pulmonary disease (COPD), health effect, inflammation
Citation: Duan R, Niu H, Dong F, Yu T, Li X, Wu H, Zhang Y and Yang T (2023) Short-term exposure to fine particulate matter and genome-wide DNA methylation in chronic obstructive pulmonary disease: A panel study conducted in Beijing, China. Front. Public Health 10:1069685. doi: 10.3389/fpubh.2022.1069685
Received: 14 October 2022; Accepted: 08 December 2022;
Published: 05 January 2023.
Edited by:
Chanjuan Sun, University of Shanghai for Science and Technology, ChinaReviewed by:
Shaowei Wu, Xi'an Jiaotong University Health Science Center, ChinaCopyright © 2023 Duan, Niu, Dong, Yu, Li, Wu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Yang,  enJ5eXlhbmd0aW5nQDE2My5jb20=
enJ5eXlhbmd0aW5nQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.