- 1Department of Clinical Laboratory, Hangzhou Women's Hospital (Hangzhou Maternity and Child Health Care Hospital), Zhejiang, China
- 2Department of Infectious Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, Zhejiang, China
- 4Regional Medical Center for National Institute of Respiratory Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 5Department of Clinical Laboratory, Tongxiang First People's Hospital, Tongxiang, Zhejiang, China
- 6Blood Center of Zhejiang Province, Hangzhou, Zhejiang, China
- 7Hospital Infection Control Office, Hospital of Zhejiang People's Armed Police, Zhejiang, China
- 8Department of Infectious Diseases, Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
Background: Surgical sites infections (SSIs) caused by Methicillin-resistant Staphylococcus aureus (MRSA) constitute a major clinical problem. Understanding the transmission mode of MRSA is important for its prevention and control.
Aim: We investigated the transmission mode of a MRSA outbreak in a trauma and orthopedic hospital ward.
Methods: Clinical data were collected from patients (n = 9) with MRSA infection in a trauma and orthopedic ward from January 1, 2015 to December 31, 2019. The wards (n = 18), patients (n = 48), medical staff (n = 23), and their households (n = 5) were screened for MRSA. The transmission mode of MRSA isolates was investigated using next-generation sequencing and phylogenetic analyses. The resistance genes, plasmids, and single-nucleotide variants of the isolates were analyzed to evaluate microevolution of MRSA isolates causing SSIs. The MRSA colonization-positive doctor was asked to suspend his medical activities to stop MRSA spread.
Findings: Nine MRSA infected patients were investigated, of which three patients were diagnosed with SSI and had prolonged hospitalization due to the persistent MRSA infection. After screening, MRSA isolates were not detected in environmental samples. The surgeon in charge of the patients with SSI caused by MRSA and his son were positive for MRSA colonization. The MRSA from the son was closely related to the isolates detected in MRSA-induced SSIs patients with 8–9 single-nucleotide variants, while ST88-MRSA isolates with three different spa types were detected in the surgeon's nasal cavity. Comparative genomic analysis showed that ST88-MRSA isolates acquired mutations in genes related to cell wall synthesis, colonization, metabolism, and virulence during their transmission. Suspending the medical activity of this surgeon interrupted the spread of MRSA infection in this ward.
Conclusion: Community-associated MRSA clones can invade hospitals and cause severe postoperative nosocomial infections. Further MRSA surveillance in the households of health workers may prevent the transition of MRSA from colonization to infection.
1. Introduction
Surgical site infection (SSI) after orthopedic surgery accounts for 14%−16% of all nosocomial infections and prolongs hospital stays by a median of 14 days per patient, increases hospitalization expenses by more than three-fold, and approximately doubles rehospitalization rates (1). Given the increasing infection rate of methicillin-resistant Staphylococcus aureus (MRSA), S. aureus is among the most important pathogens in SSIs (2). Staphylococcus aureus is a common pathogen; however, upon acquiring the staphylococcal cassette chromosome mec element, S. aureus can become resistant to almost all β-lactam antibiotics, giving rise to MRSA (3). Recently, because of increases in hypervirulent and low fitness-cost strains, community-acquired MRSA (CA-MRSA) clones are causing infections in hospitalized patients (4). Unlike hospital-acquired MRSA, CA-MRSA often causes infections in young adults (5). Acute CA-MRSA infection can develop rapidly and cause the death of the patient (6). Depending on the type of surgery performed and carrier status of individuals, the incidence of MRSA SSIs varies from 1 to 33% (7). Blocking the transmission of pathogens can effectively reduce the infection rate of hospitalized patients, the number of days in the hospital, and medical expenses.
Understanding the mechanism by which pathogens spread is a key factor in blocking their dissemination; however, the pattern of CA-MRSA infiltration into hospitals is poorly understood. Sequencing technology can be used to accurately identify and explore the characteristics of spreading pathogens (8, 9). Using genomic surveillance, Thiede et al. (10) revealed that the most prevalent CA-MRSA lineage, USA300, was circulating across both community and healthcare settings.
This study was conducted to investigate an S. aureus infection outbreak that occurred in a trauma and orthopedic ward of a secondary hospital. Medical records were reviewed, and the patients, medical staff, and households of the staff were screened for MRSA. Phylogenetic analysis was performed using whole-genome sequencing.
2. Methods
2.1. Clinical cases and isolates
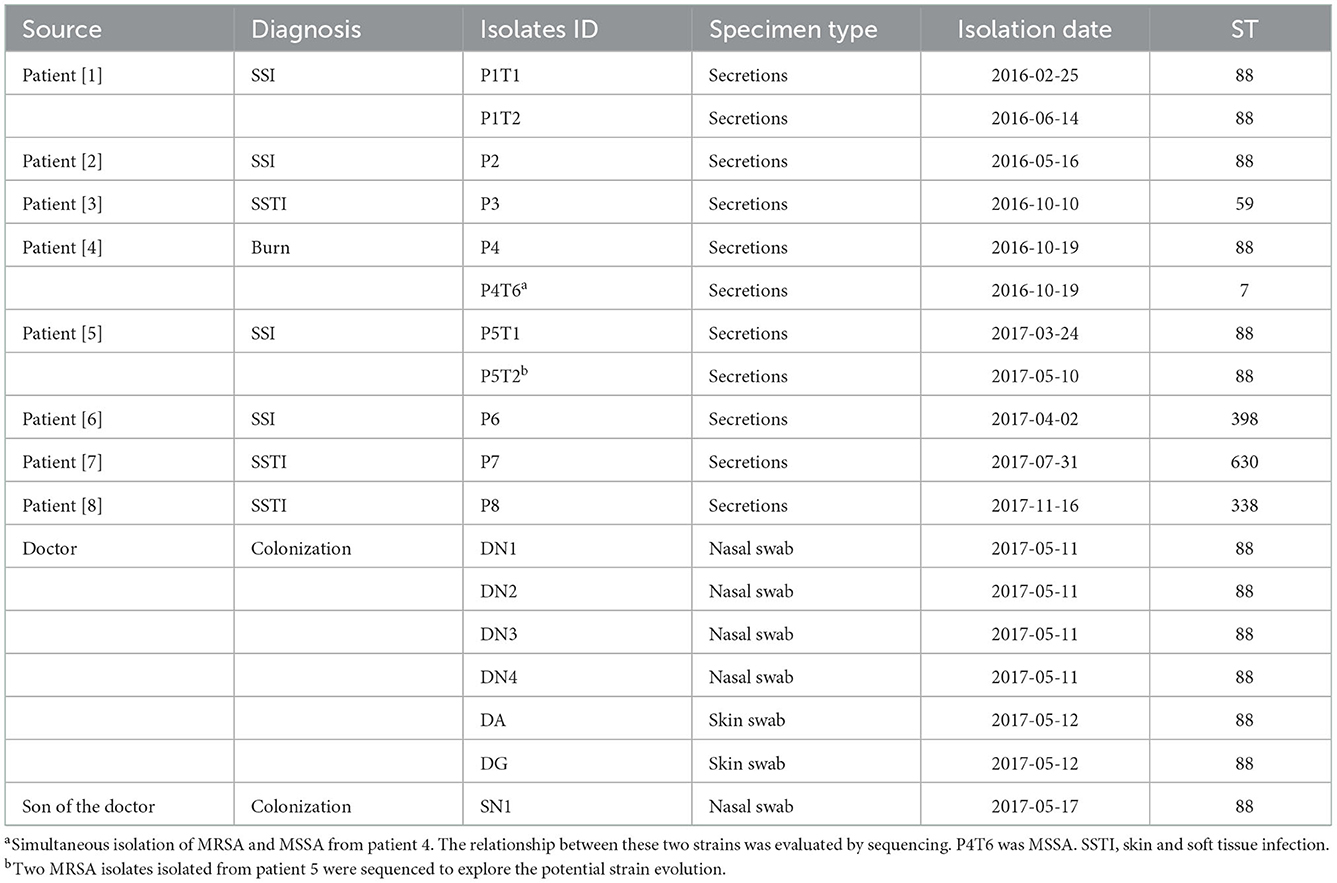
This study was conducted in a trauma and orthopedic ward with 45 beds in a hospital with 886 admissions per year. A total of nine cases of MRSA infection from January 1, 2015, to December 31, 2019, were included in this study. MRSA isolates were identified using a microTyper MS analyzer (Skyray, Jiangsu, China). Patient information was obtained from electronic medical records. All collected isolates were preserved at −80°C. The local ethics committee of Sir Run Run Shaw Hospital reviewed and approved this study (number 20190821-9). The patients/participants provided their consent to participate in this study.
2.2. MRSA screening
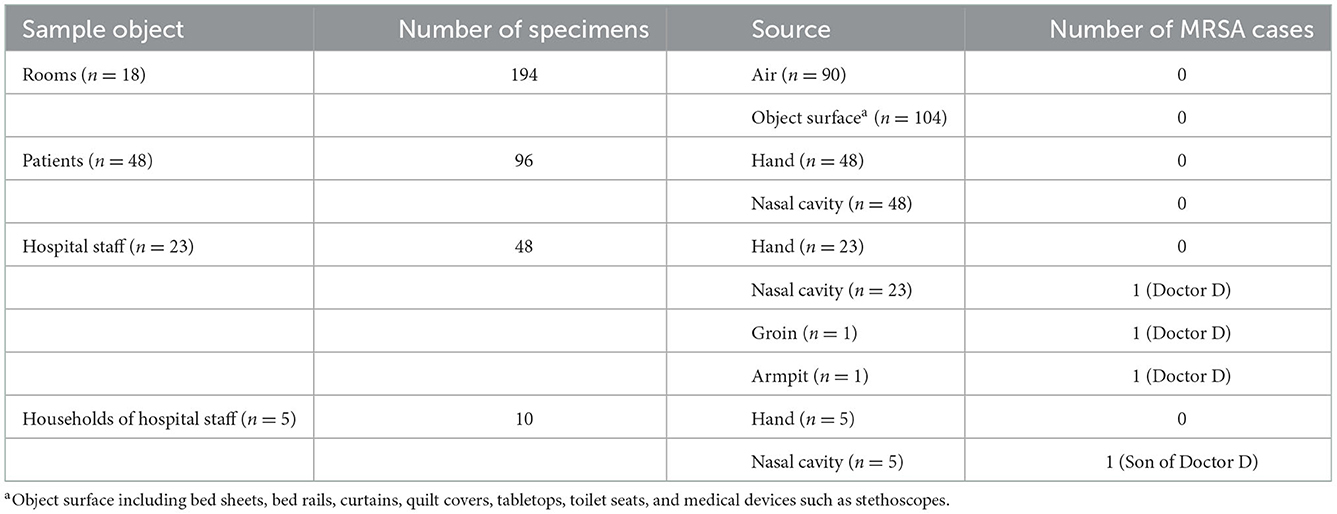
To trace the origin of the outbreak of MRSA infections in patients after bone and joint surgery, we conducted environmental MRSA screening in this trauma and orthopedic ward in May 2017. Several sample types were collected for MRSA screening. Air samples were collected from the ward using the sedimentation method. Opened Columbia blood agar plates were evenly placed in the east, south, west, north, and middle of the ward to allow microorganisms to settle as previously described (11). Samples from object surfaces were collected using saline-moistened cotton swabs, which were inoculated on CHROMagar Staph aureus plates (CHROMagar, Paris, France). Environmental samples were collected from bed sheets, bed rails, curtains, quilt covers, tabletops, toilet seats, and medical devices such as stethoscopes that were present in the ward. MRSA colonization screening were also conducted for the patients, healthcare staff and households of healthcare staff. Swab samples were taken from their nasal cavities and hands. For MRSA positive individual, the groins and armpits were also screened.
2.3. Antibiotic susceptibility testing
Antibiotic susceptibility testing of all isolates was performed using a BD Phoenix™-100 Automated Microbiology System (BD Biosciences, Franklin Lakes, NJ, USA). For the sequenced isolates, a drug susceptibility test was performed according to Clinical and Laboratory Standards Institute guidelines (12).
2.4. Genome sequencing
A primary MRSA isolate from each patient with MRSA infection was selected for sequencing. Since the MRSA isolates isolated from a patient in 2015 was not stocked, we sequenced MRSA isolates from eight patients as listed in Table 1. From the staff and the households, at least one representative isolate from each site was selected for sequencing (Table 1).
Total DNA was extracted from the MRSA isolates using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The extracted DNA was sequenced on an Illumina HiSeq Xten platform (San Diego, CA, USA) using the 2 × 150 base pair paired-end mode. The derived short reads were assembled into a FASTA file using the Shovill pipeline (version 4.4.5, https://github.com/tseemann/shovill). The minimum length and depth of Shovill were set to 200 and 10, respectively.
2.5. Molecular typing
The assemblies of sequencing data were entered into SeqSphere+ software (version 4.1.9; Ridom GmbH, Münster, Germany) for multilocus sequence typing, spa, and core genome multilocus sequence typing analyses using default parameters. The minimum spanning trees of these MRSA isolates were constructed based on differences in 1,788 alleles.
2.6. Plasmid and resistance gene detection
Plasmids and resistance genes were detected using the ABRIcate pipeline (version 1.0.0, https://github.com/tseemann/abricate). PlasmidFinder (13) and NCBI AMRFinderPlus (14) were used for plasmid and resistance gene screening, respectively. The thresholds for coverage and identity were 80 and 75%, respectively. Using the genome of an ST88-MRSA strain previously isolated from an SSI (15), the plasmid map was constructed by placing the FASTA file in BRIG (version 0.95).
2.7. Plasmid stability test
To verify that the plasmids carrying the erythromycin resistance gene (pSR02) in these strains are easily lost, passaging experiments were performed. Isolates P1, DN4, and SN1 were passaged in tryptic soy broth at 37 °C. The bacterial solution was collected every other day, cultivated on a tryptic soy agar (TSA) plate to select single clones, and simultaneously inoculated onto TSA and TSA containing 10 μg/ml erythromycin plates. The day on which clones grew on the TSA plates but not on the TSA plates containing erythromycin was recorded.
2.8. Phylogeny analysis
The FASTA file was annotated using the Prokka pipeline (version 1.14.6) (16). The annotated general feature format file was entered into the Panaroo pipeline (version 1.2.7) (17) in strict mode to generate a core-genome alignment file. The alignment file was placed in the IQ-TREE pipeline (version 2.1.2) (18) to generate a maximum-likelihood tree. The model for the tree was selected using ModelFinder (19). The number of bootstrap replicates was selected as 1,000 as recommended by the IQ-TREE Operation manual using UFBoot (20). Single-nucleotide variants (SNVs) were calculated using the Snippy pipeline (version 4.4.5, https://github.com/tseemann/snippy). A total of 2,516 core genes in the MRSA isolates were included in the phylogenetic analysis and SNV counting.
2.9. Screening of amino acid mutation sites
The P1T1 strain was the earliest MRSA strain isolated the index patient. Using this annotated strain as a reference, specific amino acid mutation sites were detected in our ST88-MRSA isolates using the Snippy pipeline (version 4.4.5, https://github.com/tseemann/snippy).
3. Results
3.1. MRSA outbreak in the trauma and orthopedic hospital ward
From February, 2016, to November, 2017, we observed that the MRSA infection rate was significantly higher than those recorded in 2014–2015 in this ward (Figure 2B), indicating a potential outbreak of MRSA, with eight MRSA-infected patients detected in 2016–2017, including four with SSI, three with skin and soft tissue infection, and one with burn wound infection (Table 1). A review of the medical data revealed that the three patients with SSI were treated by the same doctor (doctor D), indicating postoperative nosocomial infection.
Patient 1 (P1), a 21-year-old man, was admitted to the hospital with a right knee injury on September 17, 2015. On day six of admission, the patient underwent arthroscopic anterior cruciate ligament reconstruction surgery by doctor D. The patient was discharged from the hospital on January 29, 2016. One month after discharge, his right knee became swollen and ulcerated. On February 24, 2016, the patient was re-hospitalized for treatment. MRSA was isolated from wound secretions at that time, but the patient was not treated with anti-MRSA therapy and was discharged after 13 days. Because of rupture of the front of the right knee joint, the patient was admitted to the hospital for the third time on April 19, 2016. The patient continuously tested positive for MRSA during hospitalization and was administered teicoplanin. The patient subsequently underwent right knee surgery, during which the steel plates and screws were removed, and debridement was performed. However, MRSA was still continuously recovered in the secretions of the knee joint after surgery (Figure 1A).
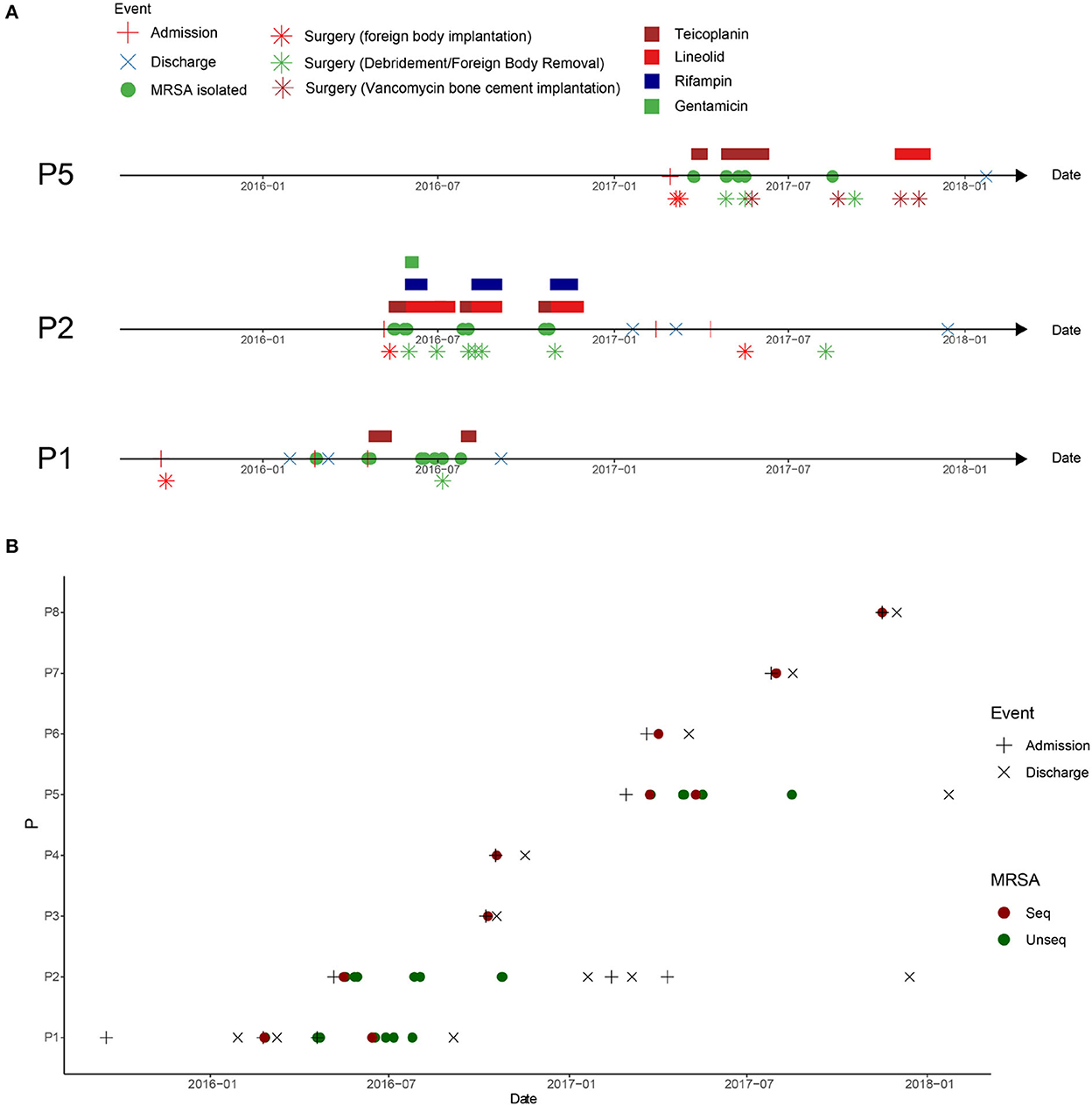
Figure 1. Clinical events in patients with methicillin-resistant Staphylococcus aureus (MRSA) infection in the orthopedic surgery ward. (A) Clinical events in patients with MRSA infection at the surgical site after osteoarthritis surgery. P1: patient 1, P2: patient 2, and P5: patient 5. (B) Timeline of MRSA isolation from MRSA-infected patients in this ward between admission of P1 and discharge of P5. The dotted line represents the screening time of the medical staff in the ward.
Patient 2 (P2), a 20-year-old man, was admitted to the hospital with an ankle fracture on May 6, 2016. Six days later, he underwent surgery on his right ankle performed by doctor D. After surgery, MRSA isolates were continuously isolated from the secretions at the surgical incision, and linezolid was used for treatment (Figure 1A, P2).
Patient 5 (P5), a 21-year-old man, was admitted to the hospital with ankle and fibular fractures on February 28, 2017. He underwent surgery for right ankle skeletal traction by doctor D on March 6, 2017. Four days later, the patient underwent right ankle fracture resection and internal fixation. Two weeks later, secretions from the surgical incision tested positive for MRSA. The doctors performed debridement and applied vancomycin bone cement (Figure 1A, P5).
To investigate the spread of MRSA isolates in the hospital, the medical records of all MRSA-infected patients in this ward during the MRSA outbreak period were reviewed. Eight cases of MRSA infection were reported from February, 2016 to May, 2017. The hospitalization period of these patients overlapped (Figure 1B).
3.2. MRSA screening statistics
To clarify the transmission mode of MRSA in the ward, medical staff (n = 23), households (n = 5) of hospital staff harboring MRSA, patients (n = 48) who stayed in the same ward as those with MRSA infection during the study period, and the ward (n = 18) environment were screened for MRSA. In total, 194 environmental samples and 156 samples from patients, hospital staffs, and households of hospital staffs were collected. No MRSA was isolated from the environmental samples. In addition, no MRSA isolates were found in samples obtained from the hands or nasal cavity of 48 patients (Table 2).
Among the 23 medical staffs who worked in this ward, MRSA was isolated only from the nasal cavity of Doctor D, who was the doctor in charge of patients P1, P2, and P5. Further screening revealed that this doctor also had MRSA colonization in the armpit and groin.
To further explore the transmission of MRSA, the households of Doctor D, including his wife and son, were screened for MRSA colonization. One MRSA isolate (SN1) was obtained from the nasal cavity of his 5-year-old son (Table 2). This result indicated that these MRSA strains may have spread among the patients, doctor D, and his son.
3.3. Molecular typing and phylogenetic analysis
Among the 18 MRSA isolates recovered from the patients (n = 11), Doctor D (n = 6), and the household of Doctor D (n = 1), six STs were identified, including ST88, ST59, ST630, ST338, ST398, and ST7. Based on the differences in the core gene alleles, the main epidemic MRSA clone in this department was ST88, which was isolated from P1, P2, P4, P5, Doctor D, and SN1 (Supplementary Figure 1).
To investigate the relationship among the ST88-MRSA isolates detected in this ward, phylogenetic analysis of the genomes of these strains was performed. The results showed that MRSA isolates from patients treated before (P1 and P2) and after (P4 and P5) October 2016 were divided into two clades: A and B. The MRSA isolates from SN1 belonged to clade A. Isolates DA, DN1, DN2, DN3, DN4, and DG, which were obtained from the different body parts of Doctor D, belonged to clade B (Figure 2A). Nine different SNVs were detected between SN1 and P1, whereas eight SNVs were detected between SN1 and P2. Single-nucleotide polymorphism analysis revealed a maximum of 21 (<40) nucleotide position differences between the ST88 isolates (Supplementary Figure 2). Using genomic information, we obtained strong evidence of the spread of these strains among patients, doctor D, and his son. Besides, transmission of these isolates between doctor D and his son may occur before P4 admission.
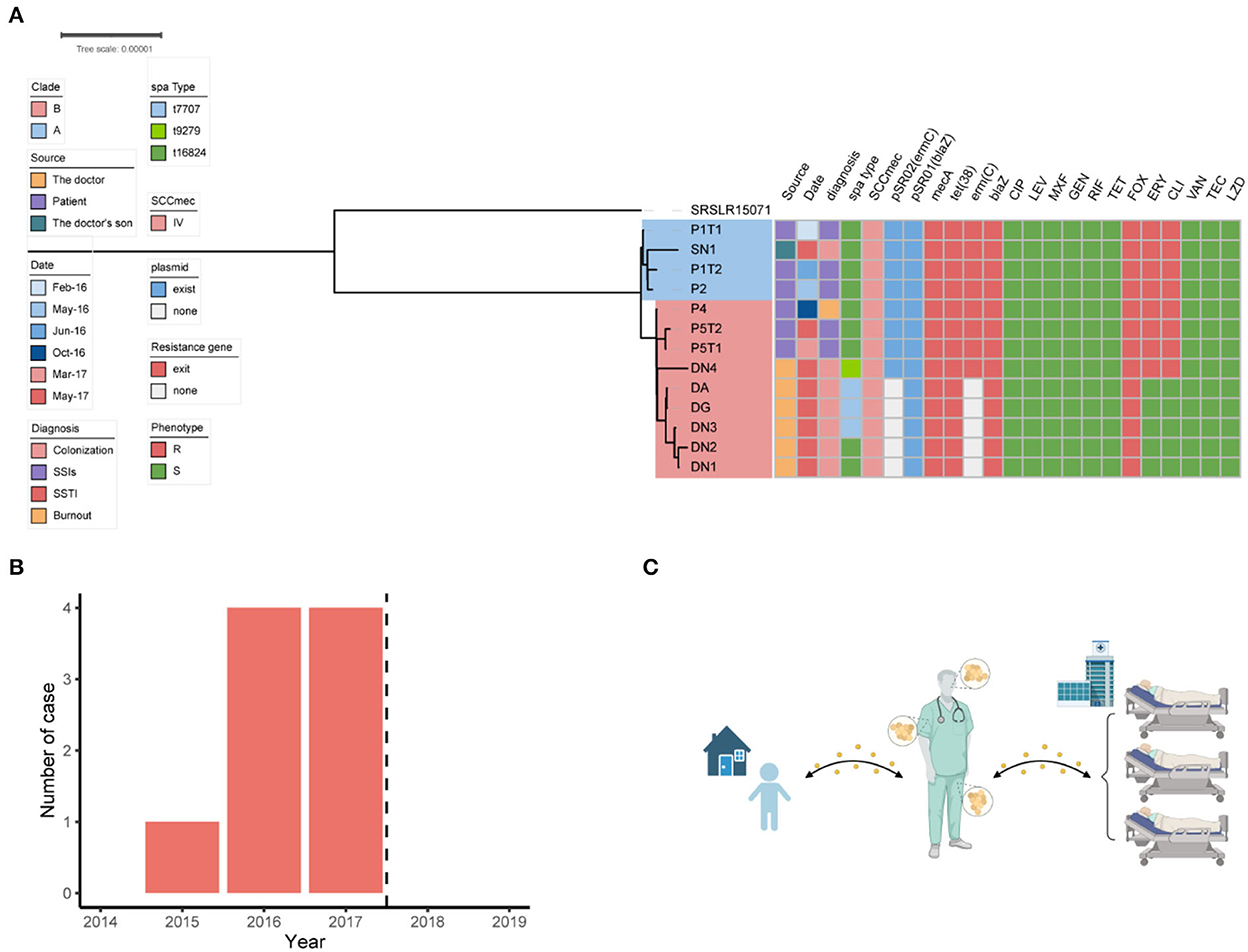
Figure 2. Dissemination analysis of the ST88 strain in this disease area. (A) Phylogenetic tree of the ST88-methicillin-resistant Staphylococcus aureus (MRSA) isolates disseminated in this ward. (B) Annual number of MRSA-infected patients in the ward. Dotted lines represent the dates when surgeons colonized with MRSA ceased their medical activity. (C) Transmission pattern of ST88-MRSA strains in this study.
3.4. Antimicrobial susceptibility and genotype of ST88-MRSA isolates
The antimicrobial susceptibility test results showed that the ST88-MRSA isolates from patients and the son of doctor D were resistant to erythromycin, clindamycin, and methicillin. Interestingly, both erythromycin-susceptible and erythromycin-resistant ST88-MRSA isolates were from Doctor D (Figure 2A).
The resistance genes of the ST88-MRSA isolates were also analyzed. Staphylococcal cassette chromosome mec IV was carried on the chromosomes in these isolates. ErmC and blaZ were carried on two plasmids (Figure 2A). BlaZ was carried on the 20,658-bp plasmid pSR01, whereas ermC was carried on the 2,473-bp plasmid pSR02 (Figure 3B). It is noteworthy that ermC-carrying plasmids were absent from several isolates from Doctor D (Figure 2A).
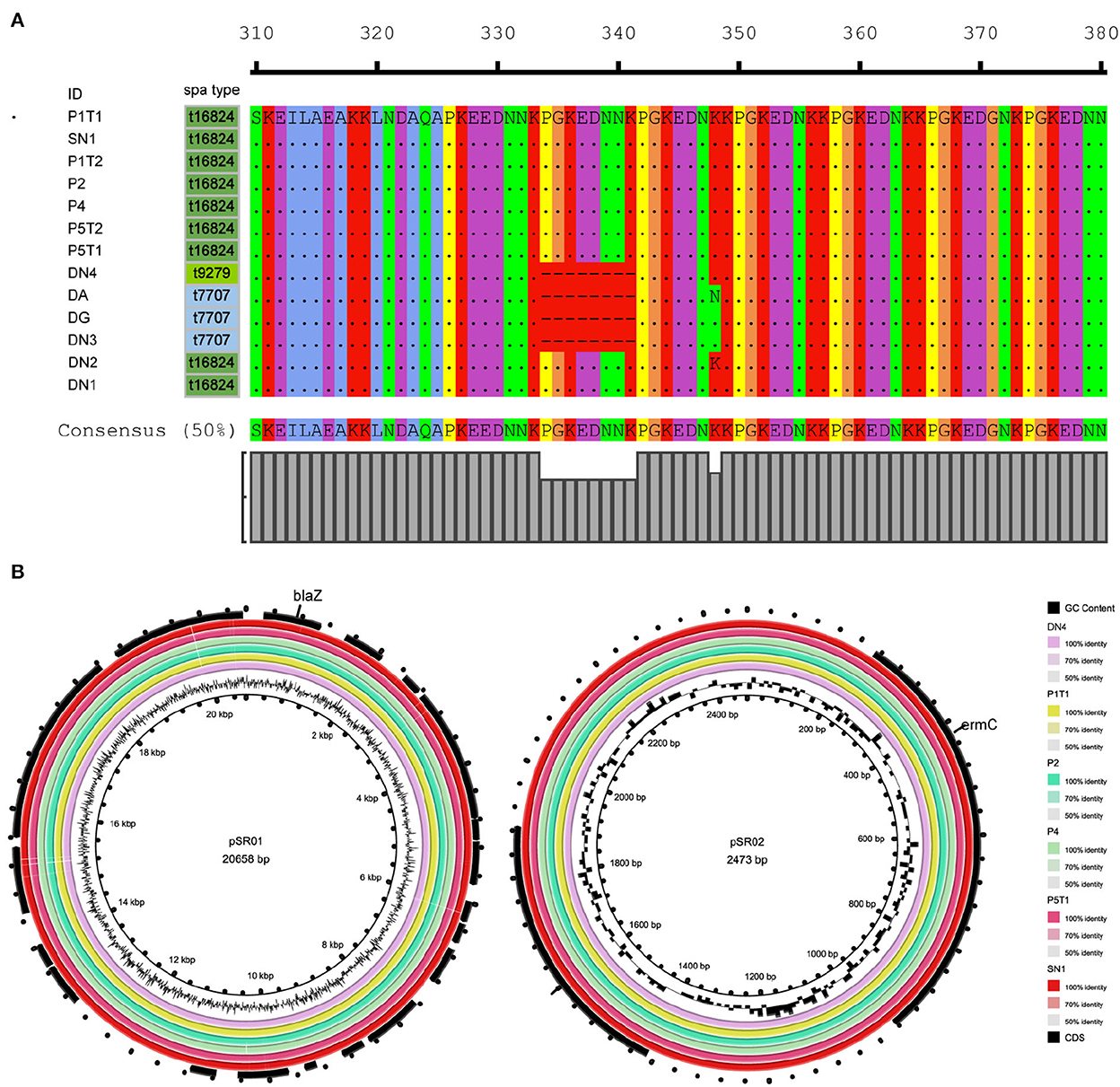
Figure 3. Molecular characteristics of ST88-methicillin-resistant Staphylococcus aureus (MRSA) isolates detected in this ward. (A) Amino acid sequence differences of protein A encoded by spa. (B) ST88-MRSA isolate plasmid features.
To analyze the stability of pSR02, the ST88-MRSA strains were evaluated in a passaging experiment. After subculture in tryptic soy broth without antibiotic pressure, a clone susceptible to erythromycin was isolated from the progeny of strain P1T1 on day 11. For the DN4 and SN1 isolates, erythromycin-susceptible clones were isolated after 8 days of subculture in tryptic soy broth without antibiotic pressure. PCR analysis confirmed that the ermC-carrying plasmids were lost from these clones (Supplementary Table 1).
3.5. In-host evolution of ST88-MRSA isolates
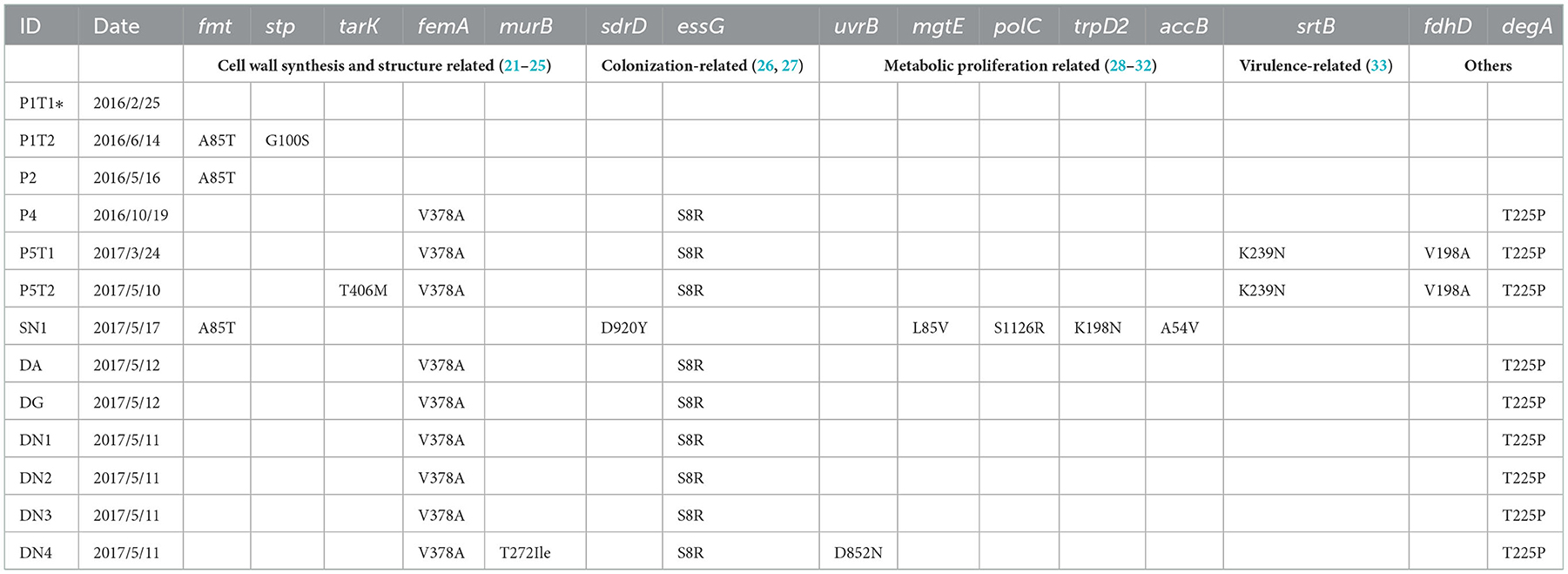
To further analyze the in-host evolution of ST88-MRSA isolates, the molecular characteristics of these isolates were analyzed. The MRSA strains isolated from the nasal cavity of Doctor D had three different spa types, t16824, t9279, and t7707 (Figure 2A), whereas the patient's strains were all carrying the same spa type, t16824. Further analysis based on whole-genome sequencing data confirmed the presence of multiple mutations in the spa genes that encode protein A, including the deletion of amino acid sites 334–341 of protein A (t16824–t9279) and an N348K mutation of protein A (t16824–t7707; Figure 3A). This means that these strains may have colonized the doctor's nasal cavity for a long time, having evolved under the pressure of long-term host immunity.
In addition to the spa mutations, other gene mutations among the ST88 isolates were analyzed using the P1T1 strain as a reference. We detected mutations in genes related to the synthesis and structure of the S. aureus cell wall, colonization ability, metabolic ability, and virulence. For strains P1T1 and P1T2, which were isolated from the same patients within 4 months, mutations were identified in fmt and stp which were related to cell wall synthesis. Similarly, most acquired mutation sites in the isolates from SN1 were in genes involved in pathways related to bacterial proliferation and metabolism, such as mgtE, polC, trpD2, and accB. For strains exposed to the hospital environment for a long time, the acquired mutation sites were mainly in cell wall synthesis genes and colonization ability-related genes, such as stp, tarK, femA, murB, sdrD, and essG. For P5T1 and P5T2, which were isolated from the most recently infected patient (P5), a mutation was detected in the virulence-related gene srtB (Table 3).
3.6. Intervetions to interrupt MRSA spreading
After identifying the MRSA isolates on the doctor's body, the doctor's medical activities were suspended, and decolonization was performed by rubbing disinfectant on the body surfaces. From the end of data collection (December 31, 2019), no additional cases of MRSA infection were detected in this department in the next 2 years (Figure 2B).
4. Discussion
Staphylococcus aureus is the most common pathogenic organism in infections, accounting for 26% of prosthetic joint infections and 42% of all bone and joint infections according to a previous study (34). Because of the increasing presence of antibiotic-resistant pathogens, there are no reliable treatment regimens for infections occurring after orthopedic joint surgery (35). Although vancomycin remains the first-line therapy for SSI caused by MRSA, the failure rate is as high as 35%−46% (36). Vonberg et al. (37) reported that among 191 outbreaks of S. aureus infection, there was a strong epidemiological evidence that S. aureus-colonized healthcare workers were the source in 14 outbreaks. Therefore, efforts to prevent postoperative infections in patients with joint injuries must be increased. However, the transmission mode of MRSA bone and joint infections in healthcare setting has not been widely evaluated, hindering the prevention and control of MRSA infections.
In this study, we investigated MRSA infection outbreaks in a trauma and orthopedic department. For three MRSA-induced SSI patients, although antimicrobial susceptibility tests showed that these MRSA isolates were susceptible to vancomycin and linezolid, the therapeutic effect was not satisfactory. The successive isolation of MRSA from the surgical sites of these three patients led us to suspect that there is an MRSA infection outbreak in this ward. Further investigation by MRSA screening showed that the environmental samples and hospitalized patients were negative for MRSA, but the surgeon in charge (doctor D) and his son were colonized by MRSA. Thus, we presumed that MRSA isolates could be transmitted between wards and the community via medical staff and their households. Using next-generation sequencing, we confirmed that MRSA isolated from patients with SSIs belonged to the ST88 clone, similar to the MRSA isolates from doctor D (and his son) who was in charge of these patients.
ST88-MRSA was previously reported as a prevalent CA-MRSA clone in Africa (38). While in Asia, previous studies showed that ST88-MRSA-IV was also the most colonized MRSA isolate from healthcare workers in Tehran, Iran (39). Furthermore, ST88-MRSA strains isolated from pigs and humans who come into contact with pigs have been reported (40). These studies suggest that ST88-MRSA is a widely distributed clone in animal husbandry and communities, with a strong colonization capacity in humans. Based on our genomic data, ST88-MRSA isolates causing nosocomial outbreaks might originated from the households of doctors. In a previous study, an SNV count of ≤ 40 was considered to indicate the same isolate (41). According to the thresholds previously reported, the ST88-MRSA isolates circulated among patients P1, P2, P4, P5, Doctor D, and Doctor D's household were closely related.
Further phylogenetic analysis showed that the ST88 isolates from the patients were divided into two clades that coincided with the timing of their acquisition. Interestingly, the MRSA isolates from SN1 had a similar genome as those detected in the early hospitalized patients (P1 and P2). This result indicates that these strains might have been transmitted between doctor D and SN1 before October 2016, which was before P4 was hospitalized. Because of the low selective pressure in the community environment, the colonized strain from SN1 evolved slowly. In contrast, the MRSA strains isolated from doctor D, which were exposed to the hospital environment, acquired more mutations than the strains from SN1. Based on these data, we presumed several possible propagation modes. First, the doctor's son spread the MRSA isolates to the doctor who transported them to the hospital, causing nosocomial transmission. Second, a patient brought the strain to the hospital and transmitted it to doctor D who brought it home and spread it to his son. At the same time, strains colonizing the nasal cavity of doctor D caused nosocomial MRSA dissemination. Third, mutual transmission between patients in a hospital is also a possible mode of transmission (Figure 2C).
Because limited MRSA strains preserved from this ward before the outbreak, it was difficult to determine the precise pattern of ST88-MRSA spread in the hospital among healthcare workers and their families. However, after the Doctor D stopping his medical activities, there were no additional cases of MRSA infection in this ward in the next 2 years. This evidenced that doctors and their households may act as reservoirs for the spread of MRSA in hospitals.
Molecular characterization of these strains supported that they have been colonizing Doctor D for a long time. The doctor's nasal cavity was colonized by three spa types of MRSA. As spa encodes protein A, which is associated with evasion of host immunity (42), the ST88-MRSA isolates may have evolved different spa types by interacting with the host immune system in the doctor's nasal cavity. Furthermore, monoclonal spa typing of isolates from the patient, the doctor's son, and other parts of the doctor was performed, but only spa type t16824 was detected (data not shown). Protein A encoded by spa is also associated with MRSA colonization, and spa-knockout strains showed increased IgG responses against staphylococcal colonization determinants (43). As amino acid changes in protein A may affect the colonization capacity of MRSA strains, our results revealed the evolutionary processes of the strain within a healthcare-associated worker. Moreover, MRSA strains with and without an ermC-carrying plasmid were present in the nasal cavity of Doctor D, and we confirmed that ST88 MRSA could lose their ermC-carrying plasmids in the absence of pressure, suggesting that gene loss during colonization contributed to heterogeneity in antimicrobial resistance among these closely related MRSA strains.
Further functional analysis of SNVs suggested that the evolution of ST88 strains is influenced by environmental selection. Most ST88-MRSA isolates in our study had mutations in cell wall synthesis and structure-related proteins, which may be attributed to the routine use of β-lactams to prevent infection before bone and joint surgeries in this hospital (data not shown). MRSA isolates from the doctor and his son acquired the mutation in colonization-related genes. MRSA isolated from the last patient carried mutations in the virulence-related gene srtB. Previous studies confirmed that srtB encoding Sortase B plays an important role in S. aureus infections (33). The backbone amide of Glu224 and the side chain of Arg233 of Sortase B are essential for enzymatic catalysis activity (44). In our isolates, the mutation Lys239Asn in Sortase B may have affected the virulence of the isolates by influencing the activity of Sortase B. Further studies are needed to determine the biological and clinical roles of these specific mutation sites.
After stopping doctor D's medical activities, there were no cases of MRSA infection in this ward during the study period. This result further validates that healthcare staff play an important role in MRSA outbreaks in healthcare settings and that expanding the screening and decolonization to the households of medical staff is also important.
There are some limitations in our research. Because of the small sample size, the origin of ST88 MRSA isolates was difficult to trace before the outbreak. Using whole gene sequencing, we obtained evidence of MRSA transmission between communities and hospitals, but the phenotypes related to the mutations detected in these ST88 MRSA are not fully known. However, our data provide valuable evidence for the invasion of CA-MRSA in healthcare settings.
5. Conclusion
We found that community-associated clones could invade the hospital via household transmission. Next-generation sequencing technology is a useful tool for tracing MRSA strains causing severe outbreaks. Furthermore, interventions such as MRSA screening and decolonization should not be limited to healthcare settings but should also be implemented in the household environment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: BioProject, PRJNA893823.
Ethics statement
The studies involving human participants were reviewed and approved by the local Ethics Committee of Sir Run Run Shaw Hospital reviewed and approved this study (number 20190821-9). The patients/participants provided their consent to participate in this study. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
YaC, YYu, and LoS designed this study. LoS, HZ, YYi, and LD collected the isolates and clinical data. HZ, XL, and ZW performed bioinformatics analysis. LoS, HZ, LD, XL, YYi, ZW, MC, SJia, YiC, FZ, and HW performed experiments. SJi, LuS, DW, YYu, and YaC supervised and directed this project. LoS, HZ, and YaC wrote the manuscript. All authors commented on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 81971977 and 82172308) and Zhejiang Provincial Natural Science Foundation of China (grant numbers LQ20H190005 and LY21H190002).
Acknowledgments
We thank Editage for English language editing. We also thank https://app.Biorender.com/ for drawing Figure 2C.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1053785/full#supplementary-material
References
1. Goyal N, Miller A, Tripathi M, Parvizi J. Methicillin-resistant Staphylococcus aureus (MRSA): colonisation and pre-operative screening. Bone Joint J. (2013) 95-b:4–9. doi: 10.1302/0301-620X.95B1.27973
2. Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect. (2008) 70:3–10. doi: 10.1016/S0195-6701(08)60017-1
3. Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. (2018) 31:e00020-18. doi: 10.1128/CMR.00020-18
4. Barcudi D, Sosa EJ, Lamberghini R, Garnero A, Tosoroni D, Decca L, et al. MRSA dynamic circulation between the community and the hospital setting: new insights from a cohort study. J Infect. (2020) 80:24–37. doi: 10.1016/j.jinf.2019.10.001
5. Chen Y, Sun L, Ba X, Jiang S, Zhuang H, Zhu F, et al. Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: a whole-genome-based survey. Clin Microbiol Infect. (2022) 28:85–92. doi: 10.1016/j.cmi.2021.05.024
6. Chen Y, Hong J, Chen Y, Wang H, Yu Y, Qu T, et al. Characterization of a community-acquired methicillin-resistant sequence type 338 Staphylococcus aureus strain containing a staphylococcal cassette chromosome mec type V(T). Int J Infect Dis. (2020) 90:181–7. doi: 10.1016/j.ijid.2019.10.034
7. Gurusamy KS, Koti R, Toon CD, Wilson P, Davidson BR. Antibiotic therapy for the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in surgical wounds. Cochrane Database Syst Rev. (2013) Cd009726. doi: 10.1002/14651858.CD009726.pub2
8. Chng KR, Li C, Bertrand D, Ng AHQ, Kwah JS, Low HM, et al. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat Med. (2020) 26:941–51. doi: 10.1101/644740
9. Raven KE, Blane B, Kumar N, Leek D, Bragin E, Coll F, et al. Defining metrics for whole-genome sequence analysis of MRSA in clinical practice. Microb Genom. (2020) 6:e000354. doi: 10.1099/mgen.0.000354
10. Thiede SN, Snitkin ES, Trick W, Payne D, Aroutcheva A, Weinstein RA, et al. Genomic epidemiology suggests community origins of healthcare-associated USA300 methicillin-resistant Staphylococcus aureus. J Infect Dis. (2022) 226:157–66. doi: 10.1093/infdis/jiac056
11. Hyvonen SM, Lohi JJ, Rasanen LA, Heinonen T, Mannerstrom M, Vaali K, et al. Association of toxic indoor air with multi-organ symptoms in pupils attending a moisture-damaged school in Finland. Am J Clin Exp Immunol. (2020) 9:101–13.
12. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
13. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
14. Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. (2019) 63:e00483-19. doi: 10.1128/AAC.00483-19
15. Sun L, Wu D, Chen Y, Wang Q, Wang H, Yu Y, et al. Characterization of a PVL-negative community-acquired methicillin-resistant Staphylococcus aureus strain of sequence type 88 in China. Int J Med Microbiol. (2017) 307:346–52. doi: 10.1016/j.ijmm.2017.07.002
16. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
17. Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. (2020) 21:180. doi: 10.1186/s13059-020-02090-4
18. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. (2020) 37:1530–4. doi: 10.1093/molbev/msaa015
19. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. (2017) 14:587–9. doi: 10.1038/nmeth.4285
20. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. (2018) 35:518–22. doi: 10.1093/molbev/msx281
21. Komatsuzawa H, Sugai M, Ohta K, Fujiwara T, Nakashima S, Suzuki J, et al. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (1997) 41:2355–61. doi: 10.1128/AAC.41.11.2355
22. Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun. (2009) 77:1406–16. doi: 10.1128/IAI.01499-08
23. Pereira MP, D'Elia MA, Troczynska J, Brown ED. Duplication of teichoic acid biosynthetic genes in Staphylococcus aureus leads to functionally redundant poly(ribitol phosphate) polymerases. J Bacteriol. (2008) 190:5642–9. doi: 10.1128/JB.00526-08
24. Maidhof H, Reinicke B, Blümel P, Berger-Bächi B, Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. (1991) 173:3507–13. doi: 10.1128/jb.173.11.3507-3513.1991
25. Yang Y, Severin A, Chopra R, Krishnamurthy G, Singh G, Hu W, et al. 3,5-dioxopyrazolidines, novel inhibitors of UDP-N- acetylenolpyruvylglucosamine reductase (MurB) with activity against gram-positive bacteria. Antimicrob Agents Chemother. (2006) 50:556–64. doi: 10.1128/AAC.50.2.556-564.2006
26. Ajayi C, Åberg E, Askarian F, Sollid JUE, Johannessen M, Hanssen AM, et al. Genetic variability in the sdrD gene in Staphylococcus aureus from healthy nasal carriers. BMC Microbiol. (2018) 18:34. doi: 10.1186/s12866-018-1179-7
27. Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. (2016) 2:16183. doi: 10.1038/nmicrobiol.2016.183
28. Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res. (2013) 12:547–58. doi: 10.1021/pr300394r
29. Trachsel E, Redder P, Linder P, Armitano J. Genetic screens reveal novel major and minor players in magnesium homeostasis of Staphylococcus aureus. PLoS Genet. (2019) 15:e1008336. doi: 10.1371/journal.pgen.1008336
30. Fagan SP, Mukherjee P, Jaremko WJ, Nelson-Rigg R, Wilson RC, Dangerfield TL, et al. Pyrophosphate release acts as a kinetic checkpoint during high-fidelity DNA replication by the Staphylococcus aureus replicative polymerase PolC. Nucleic Acids Res. (2021) 49:8324–38. doi: 10.1093/nar/gkab613
31. Caligiuri MG, Bauerle R. Subunit communication in the anthranilate synthase complex from Salmonella typhimurium. Science. (1991) 252:1845–8. doi: 10.1126/science.2063197
32. Mochalkin I, Miller JR, Evdokimov A, Lightle S, Yan C, Stover CK, et al. Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase. Protein Sci. (2008) 17:1706–18. doi: 10.1110/ps.035584.108
33. Wang G, Gao Y, Wang H, Niu X, Wang J. Baicalin weakens Staphylococcus aureus pathogenicity by targeting sortase B. Front Cell Infect Microbiol. (2018) 8:418. doi: 10.3389/fcimb.2018.00418
34. Lemaignen A, Bernard L, Marmor S, Ferry T, Grammatico-Guillon L, Astagneau P, et al. Epidemiology of complex bone and joint infections in France using a national registry: the CRIOAc network. J Infect. (2021) 82:199–206. doi: 10.1016/j.jinf.2020.12.010
35. Shirwaiker RA, Springer BD, Spangehl MJ, Garrigues GE, Lowenberg DW, Garras DN, et al. A clinical perspective on musculoskeletal infection treatment strategies and challenges. J Am Acad Orthop Surg. (2015) 23(Suppl)S44–54. doi: 10.5435/JAAOS-D-14-00379
36. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. (2019) 17:203–18. doi: 10.1038/s41579-018-0147-4
37. Vonberg RP, Stamm-Balderjahn S, Hansen S, Zuschneid I, Ruden H, Behnke M, et al. How often do asymptomatic healthcare workers cause methicillin-resistant Staphylococcus aureus outbreaks? A systematic evaluation. Infect Control Hosp Epidemiol. (2006) 27:1123–7. doi: 10.1086/507922
38. Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. (2015) 6:348. doi: 10.3389/fmicb.2015.00348
39. Ohadian Moghadam S, Modoodi Yaghooti M, Pourramezan N, Pourmand MR. Molecular characterization and antimicrobial susceptibility of the CA-MRSA isolated from healthcare workers, Tehran, Iran. Microb Pathog. (2017) 107:409–12. doi: 10.1016/j.micpath.2017.04.027
40. Otalu OJ, Kwaga JKP, Okolocha EC, Islam MZ, Moodley A. High genetic similarity of MRSA ST88 isolated from pigs and humans in Kogi State, Nigeria. Front Microbiol. (2018) 9:3098. doi: 10.3389/fmicb.2018.03098
41. Coll F, Raven KE, Knight GM, Blane B, Harrison EM, Leek D, et al. Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis. Lancet Microbe. (2020) 1:e328–35. doi: 10.1016/S2666-5247(20)30149-X
42. Kim HK, Falugi F, Missiakas DM, Schneewind O. Peptidoglycan-linked protein A promotes T cell-dependent antibody expansion during Staphylococcus aureus infection. Proc Natl Acad Sci U S A. (2016) 113:5718–23. doi: 10.1073/pnas.1524267113
43. Sun Y, Emolo C, Holtfreter S, Wiles S, Kreiswirth B, Missiakas D, et al. Staphylococcal protein A contributes to persistent colonization of mice with Staphylococcus aureus. J Bacteriol. (2018) 200:e00735-17. doi: 10.1128/JB.00735-17
Keywords: whole-genome sequencing, methicillin-resistant Staphylococcus aureus, surgical site infection, hospital, colonization
Citation: Sun L, Zhuang H, Di L, Ling X, Yin Y, Wang Z, Chen M, Jiang S, Chen Y, Zhu F, Wang H, Ji S, Sun L, Wu D, Yu Y and Chen Y (2023) Transmission and microevolution of methicillin-resistant Staphylococcus aureus ST88 strain among patients, healthcare workers, and household contacts at a trauma and orthopedic ward. Front. Public Health 10:1053785. doi: 10.3389/fpubh.2022.1053785
Received: 26 September 2022; Accepted: 19 December 2022;
Published: 09 January 2023.
Edited by:
Mel C. Melendrez, Anoka-Ramsey Community College, United StatesReviewed by:
Maryam Fazeli, Motamed Cancer Institute, IranTaj Azarian, University of Central Florida, United States
Copyright © 2023 Sun, Zhuang, Di, Ling, Yin, Wang, Chen, Jiang, Chen, Zhu, Wang, Ji, Sun, Wu, Yu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Chen,  Y2hlbnlhbkB6anUuZWR1LmNu; Yunsong Yu,
Y2hlbnlhbkB6anUuZWR1LmNu; Yunsong Yu,  eXZ5czExOUB6anUuZWR1LmNu
eXZ5czExOUB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Long Sun1†
Long Sun1† Hemu Zhuang
Hemu Zhuang Shengnan Jiang
Shengnan Jiang Yunsong Yu
Yunsong Yu Yan Chen
Yan Chen