- 1Department of Anesthesia, Hainan Hospital of Chinese People's Liberation Army General Hospital, Sanya, China
- 2Department of Geriatric Cardiology, Chinese People's Liberation Army General Hospital, Beijing, China
- 3Department of Cardiology, Hainan Hospital of Chinese People's Liberation Army General Hospital, Sanya, China
- 4School of Life Science, Beijing Institute of Technology, Beijing, China
- 5Department of Epidemiology, School of Public Health, Southern Medical University, Guangzhou, China
- 6Central Laboratory, Hainan Hospital of Chinese People's Liberation Army General Hospital, Sanya, China
- 7Center for Healthy Aging and Development Studies, National School of Development, Peking University, Beijing, China
- 8General Station for Drug and Instrument Supervision and Control, Joint Logistic Support Force of Chinese People's Liberation Army, Beijing, China
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen that causes coronavirus disease 2019 (COVID-19), infects humans through a strong interaction between the viral spike protein (S-protein) and angiotensin converting enzyme 2 (ACE2) receptors on the cell surface. The infection of host lung cells by SARS-CoV-2 leads to clinical symptoms in patients. However, ACE2 expression is not restricted to the lungs; altered receptors have been found in the nasal and oral mucosa, vessel, brain, pancreas, gastrointestinal tract, kidney, and heart. The future of COVID-19 is uncertain, however, new viral variants are likely to emerge. The SARS-CoV-2 Omicron variant has a total of 50 gene mutations compared with the original virus; 15 of which occur in the receptor binding domain (RBD). The RBD of the viral S-protein binds to the human ACE2 receptor for viral entry. Mutations of the ACE2–RBD interface enhance tight binding by increasing hydrogen bond interactions and expanding the accessible surface area. Extracorporeal membrane oxygenation, hyperbaric oxygen, and aggressive dialysis for the treatment of COVID-19 have shown various degrees of clinical success. The use of decoy receptors based on the ACE2 receptor as a broadly potent neutralizer of SARS-CoV-2 variants has potential as a therapeutic mechanism. Drugs such as 3E8 could block binding of the S1-subunit to ACE2 and restrict the infection of ACE2-expressing cells by a variety of coronaviruses. Here, we discuss the development of ACE2-targeted strategies for the treatment and prevention of COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—the causative pathogen of coronavirus disease 2019 (COVID-19)—is an enveloped non-segmented positive strand ribonucleic acid (RNA) virus belonging to the family Coronaviridae, order Nidovirales. SARS-CoV-2 infects upper respiratory tract cells and lung epithelial cells and can be detected in lower respiratory tract samples (1–3). Upper and lower respiratory symptoms are seen in individuals with COVID-19, and transmission occurs before symptoms develop. Typical clinical features are fever, sore throat, dry cough, rhinorrhea, sneezing, dyspnoea, headache, pneumonia, and increased cytokine levels (4). Severely infected individuals may succumb to severe sepsis and acute respiratory distress syndrome. In 2020, a global COVID-19 outbreak gave rise to public health concerns and warnings by the World Health Organization (5).
Coronaviruses are prevalent and widely distributed, in part attributable to their genetic diversity and genome reassortment, and cross-species infection and occasional spillover (6). Coronaviruses have an error prone RNA-dependent RNA polymerase, resulting in frequent mutation and reassortment events. For example, mutation of the SARS-CoV-2 virus has resulted in improved binding to its cellular receptors and optimized replication in human cells (7). Our knowledge of coronaviruses remains limited, however, and serious public health threats are likely to occur in the future (8). The current outbreak of COVID-19, and possible outbreaks of other coronaviruses in the future, indicate that exploration of innovative therapeutic strategies and methods is warranted (9). As one SARS-CoV-2 primary receptor, recognizing the role of ACE2 in different pathways would be key to evaluating the impact of SARS-CoV-2/ACE2 binding on the physiology of the organs and helping us find better diagnostic tools and therapeutic approaches of SARS-CoV-2 (10). It is currently documented that its mechanism of action may be related to the imbalance of renin-angiotensin-aldosterone system (RAAS) and kallikrein system (KKS) (11–13). Here, we discuss the development of angiotensin converting enzyme 2 (ACE2)-targeted strategies for the treatment and prevention of COVID-19 (Figure 1).
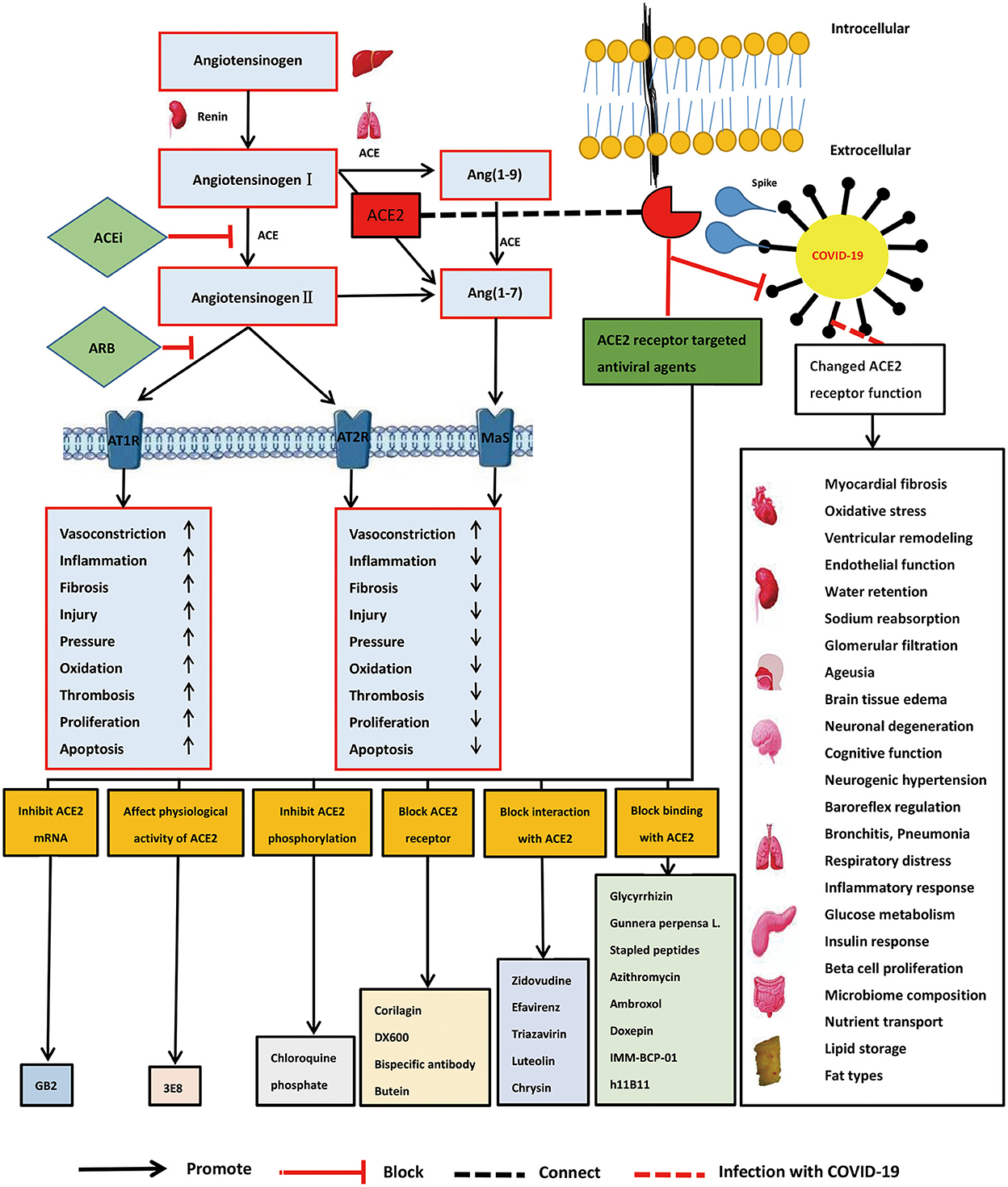
Figure 1. Role of angiotensin converting enzyme 2 (ACE2) receptor, ACE2 mechanism of viral action, and development of ACE2-targeted strategies in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Spike proteins and angiotensin converting enzyme 2 receptors
SARS-CoV-2 gains entry into sensitive cells and achieves human-to-human transmission via strong interactions between viral spike proteins (S-protein) and angiotensin converting enzyme 2 (ACE2) receptors on the cell surface (13). Wan et al. (14) discovered that ACE2 is the receptor for COVID-19. The S-protein–ACE2 binding pathway plays a significant role in human transmission and the pathogenic process of COVID-19 (15). The S-protein comprises two components: S1 with a receptor binding domain (RBD), and S2 with a fusion peptide (9). Sequence variation replacing Arg426 with Asn426 in the SARS-CoV-2 S-protein resulted in the loss of hydrogen bond interactions and an increase in binding free energy. In the S-protein, substituted residues at positions 442, 472, 479, and 487 did not alter the three-dimensional structure of the RBD domain and maintained certain van der Waals and electrostatic properties on the interaction interface (16). ACE2 is key for the entry of SARS-CoV-2 into HeLa cells and is concentrated in a small subset of type II alveolar cells (AT2).
ACE2 variants are strongly associated with susceptibility to COVID-19 infection (17–20). Thirty two variants of ACE2 have now been identified by studying Asian, American, African, and European populations (21), including seven hotspot variants (lys26arg, ile468val, ala627val, asn638ser, ser692pro, asn720asp, and leu731ile/leu731phe). Genetic variation among different populations affects ACE2 function (20). For example, analysis of ACE2 expression profiles in normal lung cells, revealed that ACE2 expression is higher in Asian men than in white or African populations (18). This result suggests that Asian men may be more susceptible to viral infection. Interference of viral transmission and pathogenicity by regulating the S-protein–ACE2 binding pathway in ACE2-expressing AT2 may be an effective strategy to prevent and treat COVID-19.
Omicron and ACE2 receptors
The emergence of the SARS-CoV-2 Omicron variant in Botswana and South Africa has influenced vaccine effectiveness and antibody capacity (22). Omicron has 50 gene mutations compared with the original strain, with 15 mutations in the RBD of the S-protein that binds to the ACE2 receptor. Thirty mutations of the Omicron S-protein are distributed over all domains of the trimeric protein, where the mutated residues are involved in intramolecular interactions, making it more stable. Seven mutations occur on the interaction interface between the RBD and the ACE2 receptor complex, including two ionic interactions, eight hydrogen bonds, and seven van der Waals interactions (23). Molecular features that have resulted in rapid diffusion of the Omicron variant include an increase in antibody evasion and the retention of strong interactions at the ACE2 interface (24). Unlike the Alpha, Beta, and Gamma variants, in Omicron the RBD binds to the human ACE2 receptor with a similar affinity to the prototypical RBD, likely owing to immune escape and compensation by multiple mutations that are transmissible (25). The structural basis for the binding of RBD-specific mutations to ACE2 receptors was revealed through the complex structures of Omicron RDB–ACE2 and Delta RDB–ACE2 (26). The Omicron RBD binds more strongly to the ACE2 protein, mainly through increased hydrogen bonding interactions and an enlarged accessible surface area (27).
ACE2 location and symptoms
Although infection of host lung cells by SARS-CoV-2 can result in severe symptoms in patients, ACE2 expression is not unique to the lungs and altered receptors have been found in other tissues (28–30). ACE2 is highly expressed in the oral cavity, which increases the risk of COVID-19 infection (30). In addition, the expression of ACE2 decreases as the virus replicates, which causes prolonged constriction of the arteries, leading to increased dysfunction and inflammation, thereby resulting in significant cardiovascular damage (31). Symptoms such as “COVID toes” (chilblain-like acral lesions), reported during the COVID-19 pandemic, are likely a result of impairment of the vasculature (32). High expression levels of ACE2 in the heart and kidneys makes them susceptible to infection (33). Among hospitalized patients severely infected with this virus, 58% had hypertension and 44% had arrhythmias (8). ACE2 is highly expressed at the site of insulin production in the pancreas—decreased insulin secretion and altered glucose tolerance are associated with ACE2 deletion (34)—and diabetes has been identified as a unique comorbidity of COVID-19 (28). ACE2 is also highly expressed in the brain, especially in the glial cells, neurons, and spinal fluid (35). Brain tissue edema and partial neuronal degeneration have been found during autopsy of patients with COVID-19.
ACE2 pathway and treatment
The renin–angiotensin II–aldosterone system plays a very important role in the body's regulation of circulatory and fluid homeostasis. Angiotensin II has immunomodulatory effects in the local pulmonary renin–angiotensin II system and the ACE/ACE2 balance is important for regulating angiotensin II levels. A homolog of ACE, ACE2 generates angiotensin 1–7 from angiotensin II. ACE2 plays an opposing role to ACE by counter balancing angiotensin II type 1 receptor (AT1R)-mediated actions and negatively regulates angiotensin II levels (10). Increased ACE and reduced ACE2 activities have been suggested to increase clinical susceptibility to acute and chronic pulmonary diseases. Loss of ACE2 expression increases vascular permeability, causing pulmonary edema and worsening lung function (36).
As a receptor for SARS-CoV-2, the downregulation of ACE2, and upregulation of ACE, may play causal roles in COVID-19 pathogenesis (37). ACE inhibitors have been confirmed to reduce ACE ability and increase ACE2 ability, and may prove beneficial in the treatment of COVID-19. Treatment of acute lung injury with active recombinant ACE2 protein can improve symptoms (37). ACE inhibitors can act as significant immunomodulators and decrease systemic cytokine levels (38). ACE inhibitors can also protect cardiopulmonary function and even improve the long-term prospects of patients with pulmonary disease (39). Current strategies for the treatment of diabetes and hypertension include ACE inhibitor drugs, angiotensin II receptor blockers, human recombinant ACE2, endogenous ACE2 activators, and ACE2 gene therapy (40). Soluble ACE2 (sACE2) can mediate viral entry into cells. Recombinant human ACE2 is an exogenous sACE2 that can complement endogenous ACE2, which may be an important option for the effective treatment of COVID-19 (41).
Vaccines are being used for active immunization against COVID-19 and drug repurposing and convalescent plasma may also be feasible treatment options (42). However, vaccinated persons have been infected with Omicron, and post-vaccination sera showed poor neutralization of the variant (43). The binding strength of the Omicron RBD to ACE2 is two-fold higher than that of prototype SARS-CoV-2. Spike mutations have promoted receptor binding to infect the respiratory system, and impaired antibody binding to evade the immune response (44). Novel treatments, including cocktail therapies, may be needed to treat Omicron infection.
Treatment potential using ACE2
Molecular detection and close surveillance are essential to identify potential COVID-19 cases and deliver timely treatment (45). During the first contact of a clinician with a suspected case, different diagnoses may be made based on clinical symptoms and rapid pathogen detection. Urgent measures include adopting the most effective control strategies to avoid viral transmission in the community. For critically ill patients, varying degrees of clinical success have been achieved using extracorporeal membrane oxygenation, aggressive dialysis, and hyperbaric oxygen. Remdesivir, an RNA polymerase inhibitor, is the first Food and Drug Administration-approved treatment (46). Broad-spectrum antivirals, such as lopinavir, ritonavir, remdesivir, and interferon beta, are being evaluated for activity against COVID-19 (47, 48). Corticosteroids are commonly used to treat severely symptomatic patients by reducing inflammation-induced lung injury. However, administration of corticosteroids showed no effect on mortality and can delay viral clearance (49). Therefore, corticosteroids should not be routinely administered as a systemic treatment for COVID-19. Further study is urgently needed to evaluate whether different antiviral drugs and systematic corticosteroid treatment are applicable for patients infected with COVID-19 (8).
The theoretical and clinical significance of the S-protein–ACE2 binding pathway in viral transmission and pathogenicity highlight its potential as a target for COVID-19 treatments. ACE/ACE2-targeted therapeutic strategies are a cornerstone of cardiovascular therapeutics, and the same methods may be valid for the treatment of pulmonary disease, promoting the concept of synchronous treatment of the heart and lungs. Further studies are needed to investigate the use of therapeutic drugs based on the S-protein–ACE2 binding pathway.
Spike-binding ACE2 decoys may be an effective treatment for this viral infection as a result of their enhanced affinity and neutralizing efficacy (44). The use of decoy receptors based on ACE2 as a widely effective neutralizer of SARS-CoV-2 variants could have a variety of therapeutic mechanisms (Table 1). Chen et al. (50) found that 3E8, an antibody against human ACE2, could block binding of the S1-subunit to ACE2 without affecting the physiological activity of ACE2 or causing severe toxicities in hACE2 “knock-in” mice. In addition, 3E8 may be a potent “broad-spectrum” blocker of multiple SARS-CoV-2 variants, such as Delta, Omicron, Alpha, Beta, Kappa, and Gamma, which utilize ACE2 as the entry receptor (51). Studies have explored the interaction of the SARS-CoV-2 S-protein RBD with the ACE2 receptor in three variants (Omicron, Delta, and WT). Despite the multiple mutations of Omicron and its relatively high viral spread, the calculated binding affinities of phytochemical limonoids and glycyrrhizic acid support that traditional medicines can be used to formulate adjunctive therapies to combat this variant (52, 53). The above treatment strategies may be potential antiviral agents for Omicron-infected patients.
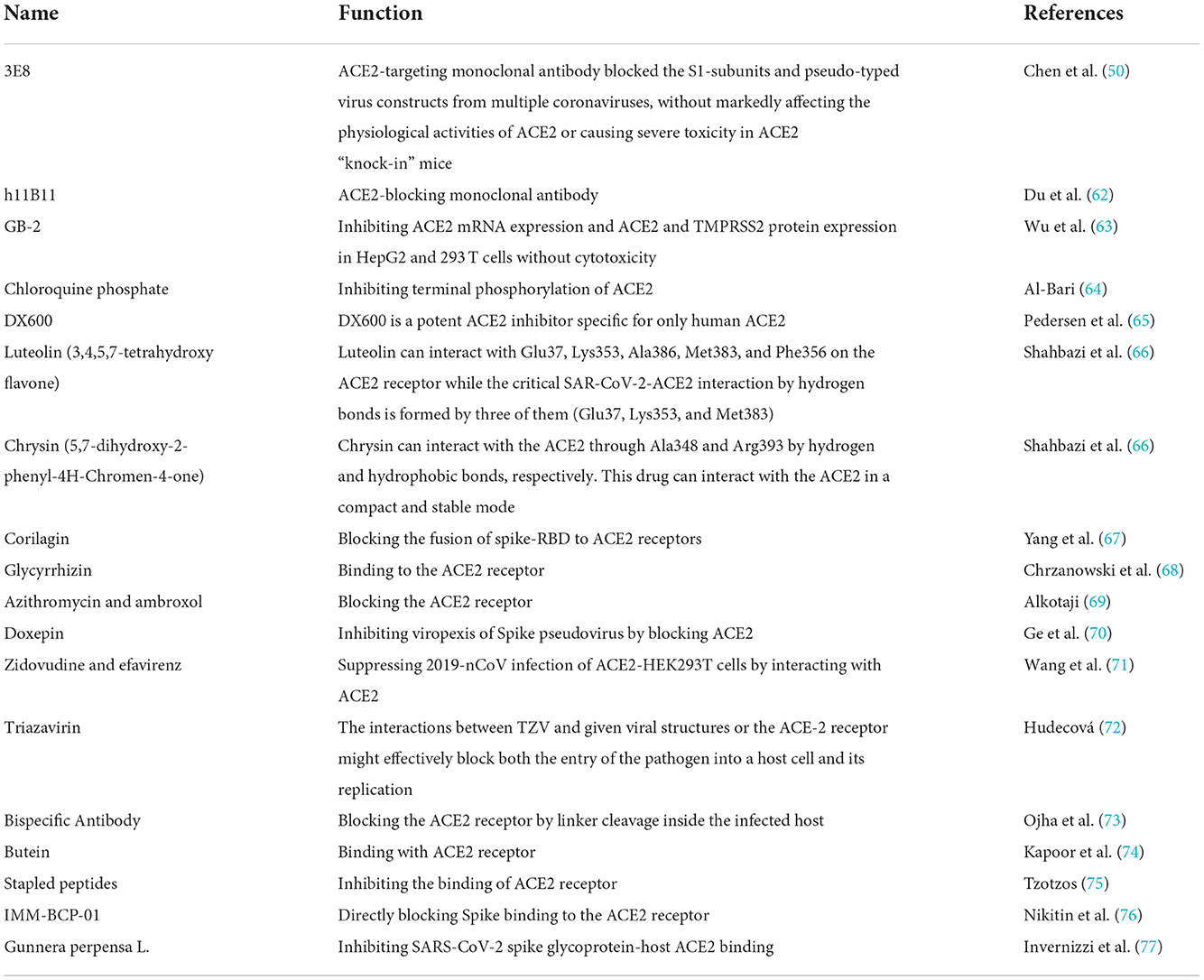
Table 1. Development of angiotensin converting enzyme 2 (ACE2) targeted strategies for the treatment and prevention of coronavirus disease 2019.
Omicron is characterized by high transmissibility and rapid spread, but its symptoms are less severe than those of previous variants. Early and prudent preventive measures, including vaccination, are key to inhibiting the Omicron variant (54). A recent study found that three doses of messenger RNA vaccine were more effective against the Omicron and Delta variants than not vaccinating or receiving two doses (55). Mutations in the S gene may generate novel variants with improved viral fitness through selective or survival advantages, such as increased ACE2 receptor affinity, replication, transmissibility, infectivity, immune escape, resistance to neutralizing antibodies, or disease severity (56). Quantitative analysis of the genetic transformation rate of the virus showed that the modified candidate drug catechin gallate can be repelled by ACE2, indicating that further modification of medical candidate drugs could produce effective docking inhibitors (57). Therefore, potential new solutions based on the ACE2 pathway could involve bait receptors based on ACE2, or mutations of the S gene.
Conclusion
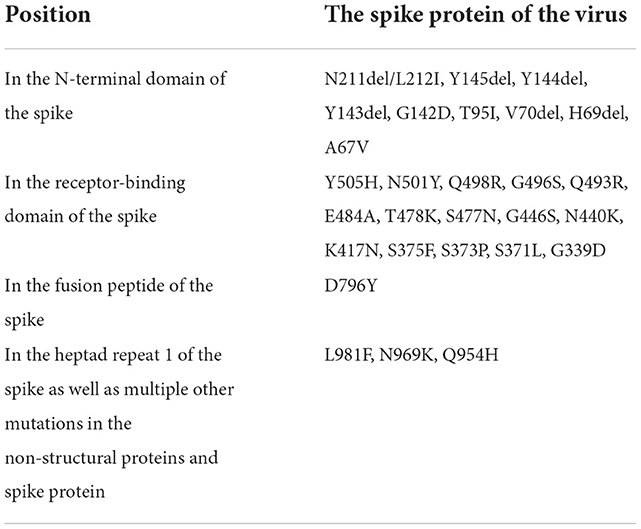
To date, there have been more than 620 million confirmed cases of COVID-19 worldwide and more than 6 million people have died (58). Since the outbreak of the pandemic, vaccines have been developed and administered, and the disease has been controlled to some extent. However, as the future of COVID-19 is uncertain, new viral variants may continue to emerge (59). Omicron showed 30 amino acid mutations in the S-protein, escaped the immunity of vaccinated individuals, and has shown increased infectivity and reinfection risk (Table 2) (60). Omicron has a lesser impact on the lower respiratory tract than previous variants and a reduced likelihood of hospitalization (61). Omicron remains infectious, however, and continues to influence work health policies and public health recommendations (54). New variants are likely to present new challenges for global control of COVID-19. Finding effective therapeutic drugs for COVID-19 is an urgent issue. In this context, therapeutic strategies that focus on the S-protein–ACE2 binding pathway are promising for treatment of COVID-19.
Author contributions
All authors contributed to study design, data collection and analyses, and drafted whole paper.
Funding
This work was supported by grants from the Military Medical Science and Technology Youth Incubation Program (20QNPY110 and 19QNP060), National Natural Science Foundation of China (81900357, 81941021, 81903392, 81901252, 82001476, 81802804, and 81801251), Excellent Youth Incubation Program of Chinese People's Liberation Army General Hospital (2020-YQPY-007), Natural Science Foundation of Hainan Province (821QN389 and 821MS112), Military Medicine Youth Program of Chinese People's Liberation Army General Hospital (QNF19069 and QNF19068), National Key R&D Program of China (2018YFC2000400), National S&T Resource Sharing Service Platform Project of China (YCZYPT[2018]07), Specific Research Fund of The Innovation Platform for Academicians of Hainan Province (YSPTZX202216), Hainan Major Scientific and Technological Cooperation Project (2016KJHZ0039), China Postdoctoral Science Foundation funded project (2019M650359, 2020M682816, and 2021T140298), Medical Big Data R&D Project of Chinese People's Liberation Army General Hospital (MBD2018030), National Geriatric Disease Clinical Medicine Research Center Project (NCRCG-PLAGH-2017-014), Central Health Care Scientific Research Project (W2017BJ12), Hainan Medical and Health Research Project (16A200057), Sanya Medical and Health Science and Technology Innovation Project (2016YW21, 2017YW22, 2018YW11, and 2018YW16), and Clinical Scientific Research Supporting Fund of Chinese People's Liberation Army General Hospital (2017FC-CXYY-3009). The sponsors had no role in the design, conduct, interpretation, review, approval, or control of this article.
Acknowledgments
We appreciate all the authors for their continued cooperation and contribution in field work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
2. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. (2020) 382:1199–207. doi: 10.1056/NEJMoa2001316
3. Booth CM, Matukas LM, Tomlinson GA, Rachlis AR, Rose DB, Dwosh HA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. (2003) 289:2801–9. doi: 10.1001/jama.289.21.JOC30885
4. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect Dis. (2013) 13:752–61. doi: 10.1016/S1473-3099(13)70204-4
5. Perlman S. Another decade, another coronavirus. N Engl J Med. (2020) 382:760–2. doi: 10.1056/NEJMe2001126
6. Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. (2004) 303:1666–9. doi: 10.1126/science.1092002
7. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
8. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
9. Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol. (2013) 87:8017–28. doi: 10.1128/JVI.00998-13
10. Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. (2005) 436:112–6. doi: 10.1038/nature03712
11. Schmaier AH. The plasma kallikrein-kinin system counterbalances the renin-angiotensin system. J Clin Invest. (2002) 109:1007–9. doi: 10.1172/JCI15490
12. Scialo F, Daniele A, Amato F, Pastore L, Matera MG, Cazzola M, et al. ACE2: the major cell entry receptor for SARS-CoV-2. Lung. (2020) 198:867–77. doi: 10.1007/s00408-020-00408-4
13. Ge XY Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. (2013) 503:535–8. doi: 10.1038/nature12711
14. Wan Y, Shang J, Graham R, Baric RS Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J Virol. (2020) 94:e00127–20. doi: 10.1128/JVI.00127-20
15. Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. (2020) 63:457–60. doi: 10.1007/s11427-020-1637-5
16. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. (2020). doi: 10.1101/2020.01.22.914952
17. Antony P, Vijayan R. Role of SARS-CoV-2 and ACE2 variations in COVID-19. Biomed J. (2021) 44:235–44. doi: 10.1016/j.bj.2021.04.006
18. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. (2020) 202:756–9. doi: 10.1164/rccm.202001-0179LE
19. Cai G. Bulk and single-cell transcriptomics identify tobacco-use disparity in lung gene expression of ACE2, the receptor of 2019-nCov. MedRxiv. (2020). doi: 10.20944/preprints202002.0051.v3
20. Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. (2020) 6:11. doi: 10.1038/s41421-020-0147-1
21. Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: Juxtaposing population groups, gender, and different tissues. Int J Environ Res Public Health. (2020) 17:3433. doi: 10.3390/ijerph17103433
22. Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. (2022) 376:eabn4947. doi: 10.1126/science.abn4947
23. Koley T, Kumar M, Goswami A, Ethayathulla AS, Hariprasad G. Structural modeling of Omicron spike protein and its complex with human ACE-2 receptor: Molecular basis for high transmissibility of the virus. Biochem Biophys Res Commun. (2022) 592:51–3. doi: 10.1016/j.bbrc.2021.12.082
24. Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS, et al. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. (2022) 375:760–4. doi: 10.1126/science.abn7760
25. Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. (2022) 185:630–40.e10. doi: 10.1016/j.cell.2022.01.001
26. Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. MedRxiv. (2021). doi: 10.1101/2021.12.08.21267417
27. Lupala CS, Ye Y, Chen H, Su XD, Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun. (2022) 590:34–41. doi: 10.1016/j.bbrc.2021.12.079
28. Ashraf UM, Abokor AA, Edwards JM, Waigi EW, Royfman RS, Hasan SA, et al. SARS-CoV-2, ACE2 expression, and systemic organ invasion. Physiol Genomics. (2021) 53:51–60. doi: 10.1152/physiolgenomics.00087.2020
29. Gembardt F, Sterner-Kock A, Imboden H, Spalteholz M, Reibitz F, Schultheiss HP, et al. Organ-specific distribution of ACE2 mRNA and correlating peptidase activity in rodents. Peptides. (2005) 26:1270–7. doi: 10.1016/j.peptides.2005.01.009
30. Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. (2020) 12:8. doi: 10.1038/s41368-020-0074-x
31. Vaduganathan M, Vardeny O, Michel T, McMurray JJV, Pfeffer MA, Solomon SD. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N Engl J Med. (2020) 382:1653–9. doi: 10.1056/NEJMsr2005760
32. Kanitakis J, Lesort C, Danset M, Jullien D. Chilblain-like acral lesions during the COVID-19 pandemic (‘COVID toes'): Histologic, immunofluorescence, and immunohistochemical study of 17 cases. J Am Acad Dermatol. (2020) 83:870–5. doi: 10.1016/j.jaad.2020.05.145
33. Simões e Silva AC, Teixeira MM. ACE inhibition, ACE2 and angiotensin-(1–7) axis in kidney and cardiac inflammation and fibrosis. Pharmacol Res. (2016) 107:154–62. doi: 10.1016/j.phrs.2016.03.018
34. Batlle D, Jose Soler M, Ye M. ACE2 and diabetes: ACE of ACEs? Diabetes. (2010) 59:2994–6. doi: 10.2337/db10-1205
35. Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. (2020) 8:420–2. doi: 10.1016/S2213-2600(20)30076-X
36. Ferreira AJ, Shenoy V, Yamazato Y, Sriramula S, Francis J, Yuan L, et al. Evidence for angiotensin-converting enzyme 2 as a therapeutic target for the prevention of pulmonary hypertension. Am J Respir Crit Care Med. (2009) 179:1048–54. doi: 10.1164/rccm.200811-1678OC
37. Kaparianos A, Argyropoulou E. Local renin-angiotensin II systems, angiotensin-converting enzyme and its homologue ACE2: Their potential role in the pathogenesis of chronic obstructive pulmonary diseases, pulmonary hypertension and acute respiratory distress syndrome. Curr Med Chem. (2011) 18:3506–15. doi: 10.2174/092986711796642562
38. Gullestad L, Aukrust P, Ueland T, Espevik T, Yee G, Vagelos R, et al. Effect of high- versus low-dose angiotensin converting enzyme inhibition on cytokine levels in chronic heart failure. J Am Coll Cardiol. (1999) 34:2061–7. doi: 10.1016/S0735-1097(99)00495-7
39. Mancini GB, Etminan M, Zhang B, Levesque LE, FitzGerald JM, Brophy JM. Reduction of morbidity and mortality by statins, angiotensin-converting enzyme inhibitors, and angiotensin receptor blockers in patients with chronic obstructive pulmonary disease. J Am Coll Cardiol. (2006) 47:2554–60. doi: 10.1016/j.jacc.2006.04.039
40. Shukla AK, Banerjee M. Angiotensin-converting-enzyme 2 and renin-angiotensin system inhibitors in COVID-19: An update. High Blood Press Cardiovasc Prev. (2021) 28:129–39. doi: 10.1007/s40292-021-00439-9
41. Wang J, Zhao H, An Y. ACE2 shedding and the role in COVID-19. Front Cell Infect Microbiol. (2021) 11:789180. doi: 10.3389/fcimb.2021.789180
42. Kumar P, Sah AK, Tripathi G, Kashyap A, Tripathi A, Rao R, et al. Role of ACE2 receptor and the landscape of treatment options from convalescent plasma therapy to the drug repurposing in COVID-19. Mol Cell Biochem. (2021) 476:553–74. doi: 10.1007/s11010-020-03924-2
43. Shah M, Woo HG. Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front Immunol. (2021) 12:830527. doi: 10.3389/fimmu.2021.830527
44. Zhang L, Narayanan KK, Cooper L, Chan KK, Devlin CA, Aguhob A, et al. An engineered ACE2 decoy receptor can be administered by inhalation and potently targets the BA.1 and BA.2 omicron variants of SARS-CoV-2 [Preprint]. bioRxiv. (2022). doi: 10.1101/2022.03.28.486075
45. Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E, A. novel coronavirus emerging in China - Key questions for impact assessment. N Engl J Med. (2020) 382:692–4. doi: 10.1056/NEJMp2000929
46. FDA news release,. Remdesivir EUA Letter of Authorization. (2020). Available online at: https://www.fda.gov/media/137564/download
47. Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. JAMA. (2020) 323:707–8. doi: 10.1001/jama.2020.0757
48. Graham BS, Mascola JR, Fauci AS. Novel vaccine technologies: Essential components of an adequate response to emerging viral diseases. JAMA. (2018) 319:1431–2. doi: 10.1001/jama.2018.0345
49. Lansbury L, Rodrigo C, Leonardi-Bee J, Nguyen-Van-Tam J, Lim WS. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst Rev. (2019) 2:CD010406. doi: 10.1002/14651858.CD010406.pub3
50. Chen Y, Zhang YN, Yan R, Wang G, Zhang Y, Zhang ZR, et al. ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker. Signal Transduct Target Ther. (2021) 6:315. doi: 10.1038/s41392-021-00740-y
51. Ou J, Zhang Y, Wang Y, Zhang Z, Wei H, Yu J, et al. ACE2-Targeting antibody suppresses SARS-CoV-2 Omicron and Delta variants. Signal Transduct Target Ther. (2022) 7:43. doi: 10.1038/s41392-022-00913-3
52. Vardhan S, Sahoo SK. Computational studies on the interaction of SARS-CoV-2 Omicron SGp RBD with human receptor ACE2, limonin and glycyrrhizic acid. Comput Biol Med. (2022) 144:105367. doi: 10.1016/j.compbiomed.2022.105367
53. Bharathi M, Sivamaruthi BS, Kesika P, Thangaleela S. Chaiyasut C. In silico screening of bioactive compounds of representative seaweeds to inhibit SARS-CoV-2 ACE2-bound omicron B11529 spike protein trimer. Mar Drugs. (2022) 20:148. doi: 10.3390/md20020148
54. Ren SY, Wang WB, Gao RD, Zhou AM. Omicron variant (B11529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. (2022) 10:1–11. doi: 10.12998/wjcc.v10.i1.1
55. Accorsi EK, Britton A, Fleming-Dutra KE, Smith ZR, Shang N, Derado G, et al. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 omicron and delta variants. JAMA. (2022) 327:639–51. doi: 10.1001/jama.2022.0470
56. Hirabara SM, Serdan TDA, Gorjao R, Masi LN, Pithon-Curi TC, Covas DT, et al. SARS-CoV-2 variants: differences and potential of immune evasion. Front Cell Infect Microbiol. (2021) 11:781429. doi: 10.3389/fcimb.2021.781429
57. Hanai T. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta. (2022) 240:123206. doi: 10.1016/j.talanta.2022.123206
58. Sharma A, Ahmad Farouk I, Lal SK. COVID-19: A review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses. (2021) 13:202. doi: 10.3390/v13020202
59. Papanikolaou V, Chrysovergis A, Ragos V, Tsiambas E, Katsinis S, Manoli A, et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene. (2022) 814:146134. doi: 10.1016/j.gene.2021.146134
60. Kannan S, Shaik Syed Ali P, Sheeza A. Omicron (B11529)—variant of concern—molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci. (2021) 25:8019–22. doi: 10.26355/eurrev_202112_27653
61. Menni C, Valdes AM, Polidori L, Antonelli M, Penamakuri S, Nogal A, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. (2022) 399:1618–24. doi: 10.1016/S0140-6736(22)00327-0
62. Du Y, Shi R, Zhang Y, Duan X, Li L, Zhang J et al. A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat Commun. (2021) 12:5000. doi: 10.1038/s41467-021-25331-x
63. Wu CY, Lin YS, Yang YH, Shu LH, Cheng YC, Liu HT. GB-2 inhibits ACE2 and TMPRSS2 expression: In vivo and in vitro studies. Biomed Pharmacother. (2020) 132:110816. doi: 10.1016/j.biopha.2020.110816
64. Al-Bari MAA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect. (2017) 5:e00293. doi: 10.1002/prp2.293
65. Pedersen KB, Sriramula S, Chhabra KH, Xia H, Lazartigues E. Species-specific inhibitor sensitivity of angiotensin-converting enzyme 2 (ACE2) and its implication for ACE2 activity assays. Am J Physiol Regul Integr Comp Physiol. (2011) 301:R1293–9. doi: 10.1152/ajpregu.00339.2011
66. Shahbazi B, Mafakher L, Teimoori-Toolabi L. Different compounds against Angiotensin-Converting Enzyme 2 (ACE2) receptor potentially containing the infectivity of SARS-CoV-2: an in silico study. J Mol Model. (2022) 28:82. doi: 10.1007/s00894-022-05059-1
67. Yang LJ, Chen RH, Hamdoun S, Coghi P, Ng JP, Zhang DW, et al. Corilagin prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding. Phytomed. (2021) 87:153591. doi: 10.1016/j.phymed.2021.153591
68. Chrzanowski J, Chrzanowska A, Graboń W. Glycyrrhizin: An old weapon against a novel coronavirus. Phytother Res. (2021) 35:629–36. doi: 10.1002/ptr.6852
69. Alkotaji M. Azithromycin and ambroxol as potential pharmacotherapy for SARS-CoV-2. Int J Antimicrob Agents. (2020) 56:106192. doi: 10.1016/j.ijantimicag.2020.106192
70. Ge S, Wang X, Hou Y, Lv Y, Wang C, He H. Repositioning of histamine H1 receptor antagonist: Doxepin inhibits viropexis of SARS-CoV-2 Spike pseudovirus by blocking ACE2. Eur J Pharmacol. (2021) 896:173897. doi: 10.1016/j.ejphar.2021.173897
71. Wang J, Zhang Y, Hu S, Bai H, Xue Z, Liu Y, et al. Antiviral drugs suppress infection of 2019-nCoV spike pseudotyped virus by interacting with ACE2 protein. J Biochem Mol Toxicol. (2022) 36:e22948. doi: 10.1002/jbt.22948
72. Hudecová IMJCGKMPL.Triazavirin might be the new hope to fight Severe Acute Respiratory Syndrome Coronavirus (2) (SARS-CoV-2). Ceska Slov Farm. (2021) 70:18–25. doi: 10.5817/CSF2021-1-18
73. Ojha R, Gurjar K, Ratnakar TS, Mishra A, Prajapati VK. Designing of a bispecific antibody against SARS-CoV-2 spike glycoprotein targeting human entry receptors DPP4 and ACE2. Hum Immunol. (2022) 83:346–55. doi: 10.1016/j.humimm.2022.01.004
74. Kapoor N, Ghorai SM, Khuswaha PK, Bandichhor R, Brogi S. Butein as a potential binder of human ACE2 receptor for interfering with SARS-CoV-2 entry: a computer-aided analysis. J Mol Model. (2022) 28:270. doi: 10.1007/s00894-022-05270-0
75. Tzotzos S. Stapled peptides as potential inhibitors of SARS-CoV-2 binding to the hACE2 receptor. J Pept Sci. (2022) 28:e3409. doi: 10.1002/psc.3409
76. Nikitin PA, DiMuzio JM, Dowling JP, Patel NB, Bingaman-Steele JL, Heimbach BC, et al. IMM-BCP-01, a patient-derived anti-SARS-CoV-2 antibody cocktail, is active across variants of concern including Omicron BA.1 and BA.2. Sci Immunol. (2022) 7:eabl9943. doi: 10.1126/sciimmunol.abl9943
77. Invernizzi L, Moyo P, Cassel J, Isaacs FJ, Salvino JM, Montaner LJ, et al. Use of hyphenated analytical techniques to identify the bioactive constituents of Gunnera perpensa L, a South African medicinal plant, which potently inhibit SARS-CoV-2 spike glycoprotein-host ACE2 binding. Anal Bioanal Chem. (2022) 414:3971–85. doi: 10.1007/s00216-022-04041-3
Keywords: angiotensin converting enzyme 2, coronavirus disease 2019, receptor binding domain, S-protein, 3E8
Citation: Feng L, Fu S, Zhang P, Zhang Y, Zhao Y, Yao Y, Luo L and Ping P (2022) Potential use of the S-protein–Angiotensin converting enzyme 2 binding pathway in the treatment of coronavirus disease 2019. Front. Public Health 10:1050034. doi: 10.3389/fpubh.2022.1050034
Received: 21 September 2022; Accepted: 15 November 2022;
Published: 28 November 2022.
Edited by:
Ahmed Mostafa, National Research Centre, EgyptReviewed by:
Mahmoud Shehata, University of Giessen, GermanyKonda Mani Saravanan, Bharath Institute of Higher Education and Research, India
Copyright © 2022 Feng, Fu, Zhang, Zhang, Zhao, Yao, Luo and Ping. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yali Zhao, emhhb3lsMzAxQDE2My5jb20=; Yao Yao, eWFveWFvQG5zZC5wa3UuZWR1LmNu; Leiming Luo, bHVvbGVpbWluZ0B2aXAuc2luYS5jb20=; Ping Ping, cGluZ3BpbmczMDFAMTI2LmNvbQ==
†These authors share first authorship
 Long Feng
Long Feng Shihui Fu
Shihui Fu Pei Zhang
Pei Zhang Yujie Zhang
Yujie Zhang Yali Zhao
Yali Zhao Yao Yao
Yao Yao Leiming Luo
Leiming Luo Ping Ping8*
Ping Ping8*