
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 30 November 2022
Sec. Radiation and Health
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.1031038
Objective: To reveal the potential targets and signaling pathways of dasatinib in the treatment of radiation ulcers through network pharmacology and molecular docking technology.
Methods: Pathological targets of radiation ulcers were screened using GeneCards database. At the same time, the pharmacological targets of dasatinib were obtained through SwissTargetPrediction (STP), Binding DB and Drugbank databases. Subsequently, the potential targets of dasatinib for anti-radiation ulcers were obtained after intersection by Venn diagram. Next, a protein-protein interaction (PPI) network was constructed through the STRING database and core targets were screened. Finally, the identified core targets were subjected to GO and KEGG enrichment analysis, co-expression network analysis, and molecular docking technology to verify the reliability of the core targets.
Results: A total of 76 potential targets for anti-radiation ulcer with dasatinib were obtained, and 6 core targets were screened, including EGFR, ERBB2, FYN, JAK2, KIT, and SRC. These genes were mainly enriched in Adherens junction, EGFR tyrosine kinase inhibitor resistance, Focal adhesion, Bladder cancer and PI3K-Akt signaling pathway. Molecular docking results showed that dasatinib binds well to the core target.
Conclusion: Dasatinib may play a role in the treatment of radiation ulcers by regulating EGFR, ERBB2, FYN, JAK2, KIT, and SRC. These core targets may provide new insights for follow-up studies of radiation ulcers.
Radiation ulcers are the result of skin damage in excess of the cumulative radiation dose during radiation therapy or accidental exposure to radiation sources (1, 2), and the histological features of the lesions include epidermal atrophy, dermal sclerosis, superficial vasodilation, adnexal structures loss, etc. (3, 4). However, so far, there is no clear clinical consensus on the treatment of radiation ulcers. Conservative treatment has a significant effect on radiation dermatitis (without ulcers), but severe cases, especially ulcers, require surgical intervention to promote wound healing (5, 6). Therefore, it is particularly important to minimize radiation dose to prevent radiation ulcers (7–9). In addition, patients should also receive appropriate skin care education and regular dermatological monitoring and evaluation when at therapeutic radiation dose levels (7, 8). Animal experiments showed that no skin changes were seen in the 10 Gy irradiation group; mild skin damage was seen in the 15 Gy irradiation group; skin ulcers appeared in some rats in the irradiation group exceeding 20 Gy; the rats irradiated with 35 to 55 Gy could successfully Induced radiation-induced skin ulcer model (10–12).
Tissue reconstruction of radiation skin ulcers presents a great challenge for plastic surgeons. Anti-inflammatory drugs, growth factors, and local anesthetics are commonly used clinically to relieve radiation skin ulcers, but the effect is not good (13). Hyperbaric oxygen therapy is also considered an effective way to reduce skin ulcers after radiation therapy, but the treatment time is very long (14, 15). The pathological mechanism of radiation skin ulcers is unclear, and there is no effective treatment (16). Therefore, there is an urgent need for a safe and effective drug that can relieve radiation skin ulcers. Previous studies have confirmed that Cordycepin can prevent and treat radiation ulcers by inhibiting NRF2 and AMPK-induced cellular senescence (17). Therefore, senescent cells may be a viable target for the treatment of radiation ulcers (18). Dasatinib is a tyrosine kinase inhibitor first approved for the treatment of chronic myeloid leukemia (19). Kinases inhibited by dasatinib mainly include Src family kinases (SFK), cKIT, platelet-derived growth factor receptor, TEC and SYK families (19, 20). Recent animal studies have shown that dasatinib and quercetin can promote the healing of radiation ulcers by removing senescent cells (21). Dasatinib has shown potential to treat radiation ulcers, but its specific molecular mechanism remains unclear. Therefore, this study explored the potential targets and signaling pathways of dasatinib in the treatment of radiation skin ulcers through network pharmacology and molecular docking. Our findings provide a rationale for the mechanism of action of dasatinib in the treatment of radiation skin ulcers, which can be validated before being used in clinical trials.
The SMILES (Simplified Molecular Linear Input Specification) is obtained from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database with the search term “Dasatinib,” which is an ASCII string that explicitly describes the three-dimensional chemistry of molecules structure. Import the retrieved SMILES into the SwissTargetPrediction (http://www.swisstargetprediction.ch/) database and the Binding DB (http://www.bindingdb.org/bind/index.jsp) database, with the respective filter conditions set as Probability > 0 and Similarity > 0.85. Subsequently, in the Drugbank (https://go.drugbank.com/) database, its potential targets were searched with the keyword “Dasatinib,” and all its drug target sources were supported by corresponding experimental verifications. The above search species are all set to “homo sapiens.” Finally, the final dasatinib pharmacological targets were obtained after screening and deduplication of the targets obtained from the three databases. The pathological targets were obtained by searching with “radiation skin ulcer” as the keyword, and setting the target with score > 1 form GeneCards database (https://www.genecards.org/). This is a database that provides detailed information on all currently annotated and predictable genes in humans.
The intersection targets of Dasatinib against radiation skin ulcer were obtained through the venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/index.html), and then the intersection targets were imported into the STRING (https://cn.string-db.org/) (Version 11.5) data analysis platform for PPI analysis (species set to “homo sapiens,” protein interaction score set with a high confidence level of 0.400). Subsequently, the PPI network was visualized by Cytoscape (version 3.9.0) software. KEGG and GO analyses were performed on the intersection targets by using the online analysis tool Kobas 3.0. Subsequently, its results were visualized by R software.
The six commonly used topological analysis methods (Closeness, Radiality, Degree, Betweenness, Stress, EPC) in Cytohubba, a plug-in of Cytoscape software, were used to identify the core targets of Dasatinib activity against radiation skin ulcer. Subsequently, the co-expression network and functional analysis of core targets were constructed through the GeneMANIA (http://www.genemania.org/) database, which is mainly used to generate hypotheses about gene functions and analyze gene lists. KEGG and GO analyses were performed on the core targets by Kobas 3.0. Subsequently, its results were visualized by R software. Finally, the Drug-target-disease network was drawn by Cytoscape software.
We enter the gene symbol of the core target in the PDB (https://www.rcsb.org/) database to obtain the protein structure crystal. In order to accurately obtain the PDB ID of the core target, the screening conditions are set as: Experimental method is set to X- ray diffraction, Refinement resolution is set to ≤ 2A, PH is selected to be 7.35–7.50, and chemical components are selected to have Ligand. We downloaded the 3D crystal structure of the core target from PDB database, selected to be saved in PDB format, served as the protein receptor. At the same time, download the 2D structure of Quercetin from the PubChem database, choose to save it as “SDF” format, and use OpenBabel (version 2.4.1) software to convert it to PDB format as a small molecule ligand. Using PyMOL software to remove water molecules and original ligands from protein molecules. Then, AutoDockTools (version 1.5.6) software was used to convert the PDB format files of proteins and small molecules into PDBQT format, including some operations: hydrogenation, charge calculation, atom type addition, determination of torque center (root), etc. Finally, the protein receptor and the small molecule ligand are docked by AutoDock vina (version 1.1.2). The size of the docking binding energy of the two indicates the strength of the binding activity. Small molecule ligands can bind spontaneously, and the smaller the binding energy, the better the binding activity and the stronger the binding stability. Pymol (version 2.2.0) software was used to visualize the docking results with minimal receptor-ligand binding energy.
In order to further improve the reliability of predicted core targets, we used Discovery Studio 2019 Client software (protein structure analysis software, widely used in molecular docking) to remove water molecules, hydrogenation, apply forcefield, clean protein and other operations on the core targets, and then obtain macromolecular receptors. Hydrogenation and apply forcefield are performed on the active ligand of the core target. The macromolecular receptor is molecularly docked with the treated active ligand, and the RMSD value is obtained (RMSD represents the structural difference between two molecules (or between two states of the same molecule), a smaller value indicates a more accurate docking method). After the reliability verification of the docking software, we began to do molecular docking between the core target and the drug. Before molecular docking, we performed operations such as removing water molecules, deleting ligand groups, hydrogenation, and cleaning proteins for the core targets, and preparing ligands for small molecule drugs.
Single cell sequencing data GSE193564 was downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193564), which has been previously uploaded by our team. Then we analyzed the expression changes of these core targets at different time points after irradiation in rats.
The bar chart, string chart and violin chart in this paper were analyzed and drawn in Sangerbox (http://sangerbox.com/home.html). Sangerbox is an online analysis tool developed based on R language, which has been widely recognized by peers (22).
The flow chart of this study design is shown in Figure 1. The chemical structures of Dasatinib are shown in Figure 2A. A total of 3,970 radiation skin ulcer-related pathological targets were obtained from the GeneCards database (Supplementary Table 1). A total of 216 Dasatinib targets were obtained from STP, Drugbank and Binding DB databases (Supplementary Tables 2–4), and a total of 137 targets were obtained after screening and deduplication.
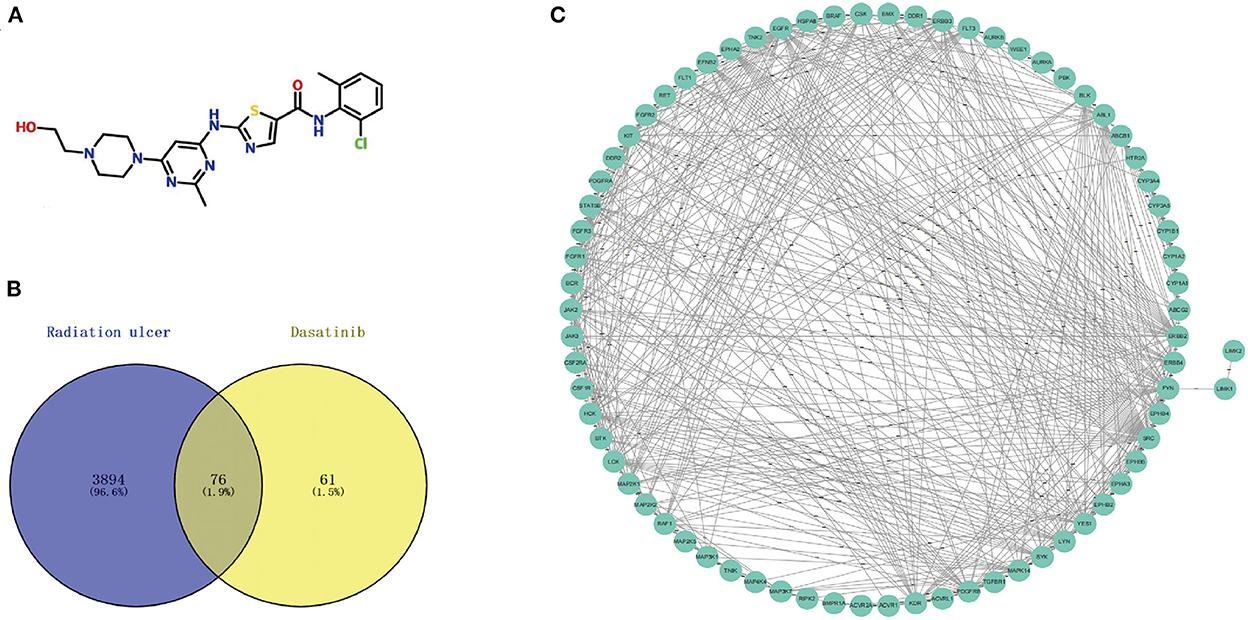
Figure 2. (A) Chemical structures of dasatinib. (B) A total of 76 cross targets were identified by Venn diagram. (C) PPI network of cross targets was constructed by using STRING database and Cytoscape software.
Using the online tool Draw venn diagram to draw a Venn diagram for the targets related to Dasatinib activity against radiation skin ulcer, a total of 76 intersection targets were obtained (Figure 2B; Supplementary Table 5). A PPI network with a protein interaction score > 0.4 was generated from the STRING database (Figure 2C), which contains 72 nodes and 503 interaction pairs. To determine the biological functions of the intersection targets, we performed GO and KEGG pathway enrichment analysis. Figure 3 presents the top 10 meaningful enrichment results, respectively. GO analysis results show that these genes were mainly enriched in ATP binding, peptidyl-tyrosine phosphorylation, protein tyrosine kinase activity, transmembrane receptor protein tyrosine kinase signaling pathway, protein autophosphorylation, receptor complex, transmembrane receptor protein tyrosine kinase activity, protein phosphorylation, positive regulation of kinase activity and protein serine/threonine kinase activity. In terms of KEGG Pathway, these genes were mainly enriched in MAPK signaling pathway, PI3K-Akt signaling pathway, Pathways in cancer, EGFR tyrosine kinase inhibitor resistance, Rap1 signaling pathway, Ras signaling pathway, Central carbon metabolism in cancer, ErbB signaling pathway, Signaling pathways regulating pluripotency of stem cells and Regulation of actin cytoskeleton.
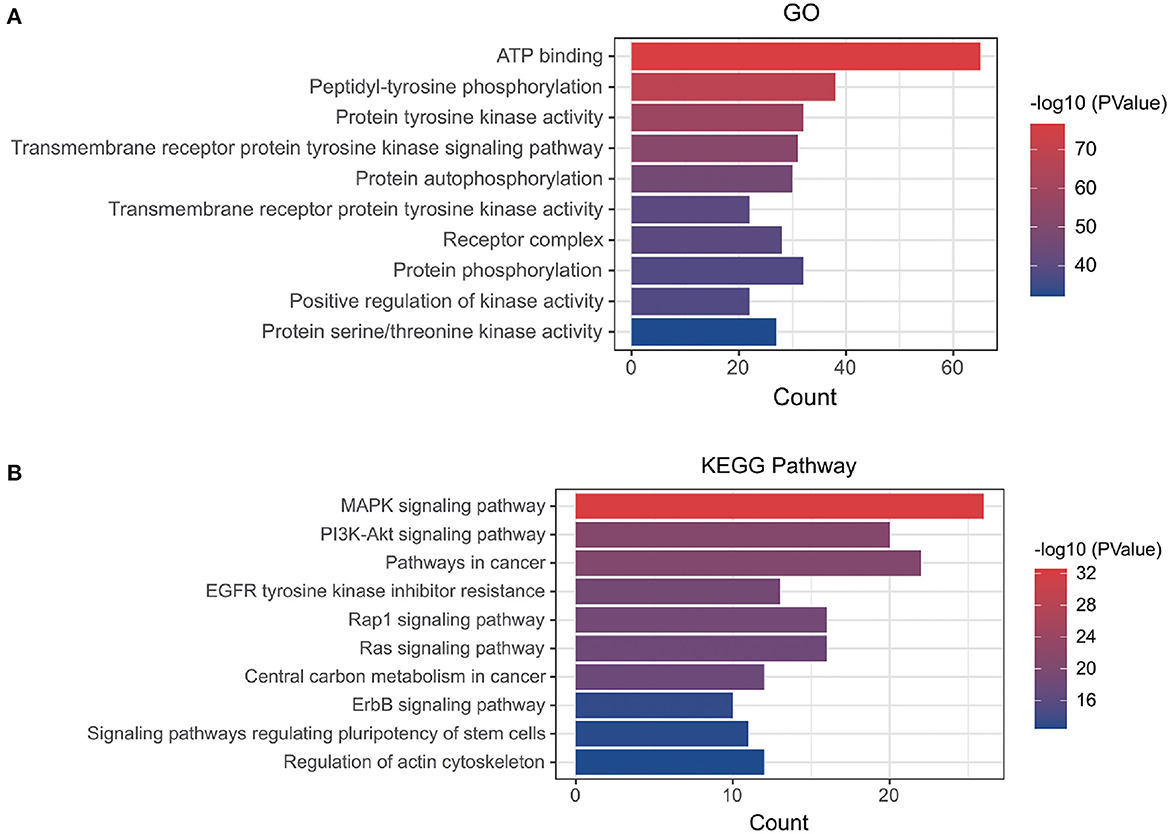
Figure 3. (A) GO and (B) KEGG enrichment analysis of potential targets, Histogram show the enrichment data of the first 10 terms.
To further explore the core targets of Dasatinib activity against radiation skin ulcer, we calculated the top 15 core targets using 6 algorithms in cytohubba (Table 1). By taking the intersection, a total of 6 core targets were obtained, including EGFR, ERBB2, FYN, JAK2, KIT, and SRC (Figure 4A). Table 2 shows their full names and functions. Co-expression network and function analysis of core targets based on GeneMANIA database showed a complex PPI network with the physical interactions of 38.72%, shared protein domains of 26.13%, genetic interactions of 14.21%, pathway of 9.27%, predicted of 8.87%, co-expression of 2.30%, and co-localization of 0.49% (Figure 4B). To further explore the biological functions of core targets, we analyzed and displayed the top 10 GO and KEGG pathway enrichment analysis results (Figure 5). Similar to the previous results, GO analysis results show that these genes were mainly enriched in protein tyrosine kinase activity, peptidyl-tyrosine phosphorylation, protein autophosphorylation, transmembrane receptor protein tyrosine kinase signaling pathway, positive regulation of protein kinase B signaling, MAPK cascade, positive regulation of MAP kinase activity, positive regulation of phosphatidylinositol 3-kinase signaling, ATP binding and ERBB2 signaling pathway. In terms of KEGG Pathway, these genes were mainly enriched in Adherens junction, EGFR tyrosine kinase inhibitor resistance, Focal adhesion, Bladder cancer, PI3K-Akt signaling pathway, Central carbon metabolism in cancer, ErbB signaling pathway, Endocrine resistance, Pathways in cancer and Breast cancer. Finally, a network visualization of dasatinib activity against radiation skin ulcer targets and an interaction diagram for core target-related pathways were generated by Cytoscape software (Figure 6).
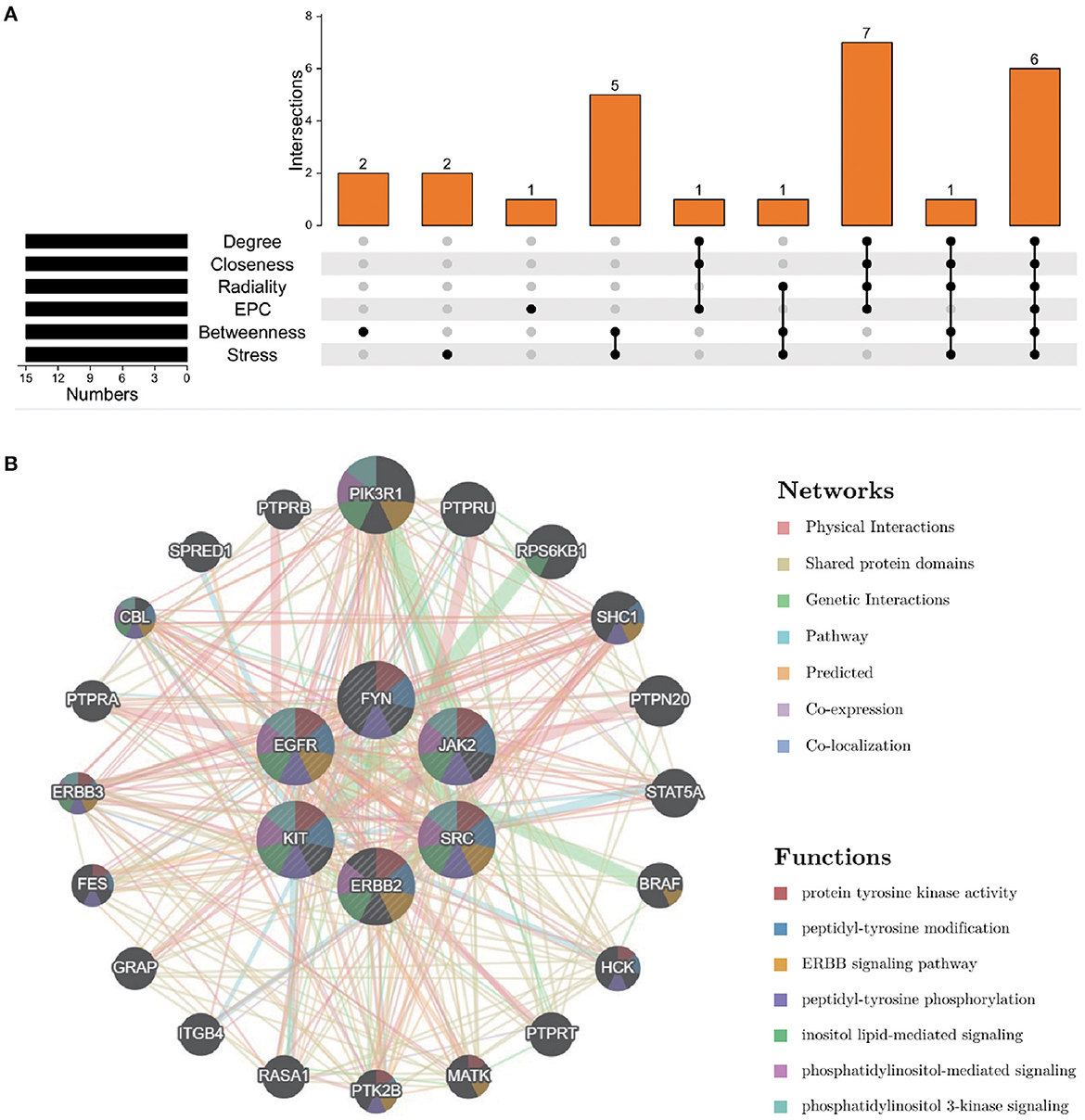
Figure 4. (A) The Venn map drawn show that 8 core targets were screened by 6 algorithms. (B) Core targets and its coexpression genes were analyzed by GeneMANIA.
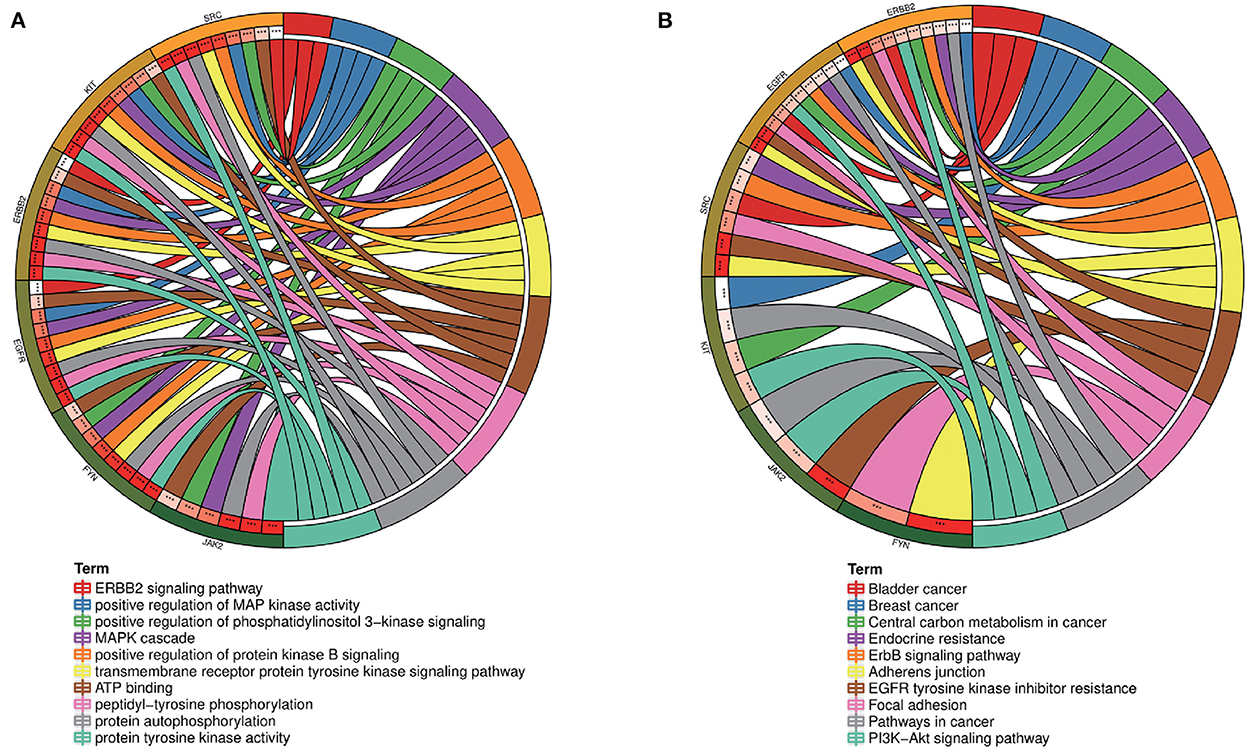
Figure 5. (A) GO and (B) KEGG enrichment analysis of core targets showed the top 10 enrichment data. The outermost circle is term on the right and the inner circle on the left represents the significant P-value of the corresponding pathway of the gene.
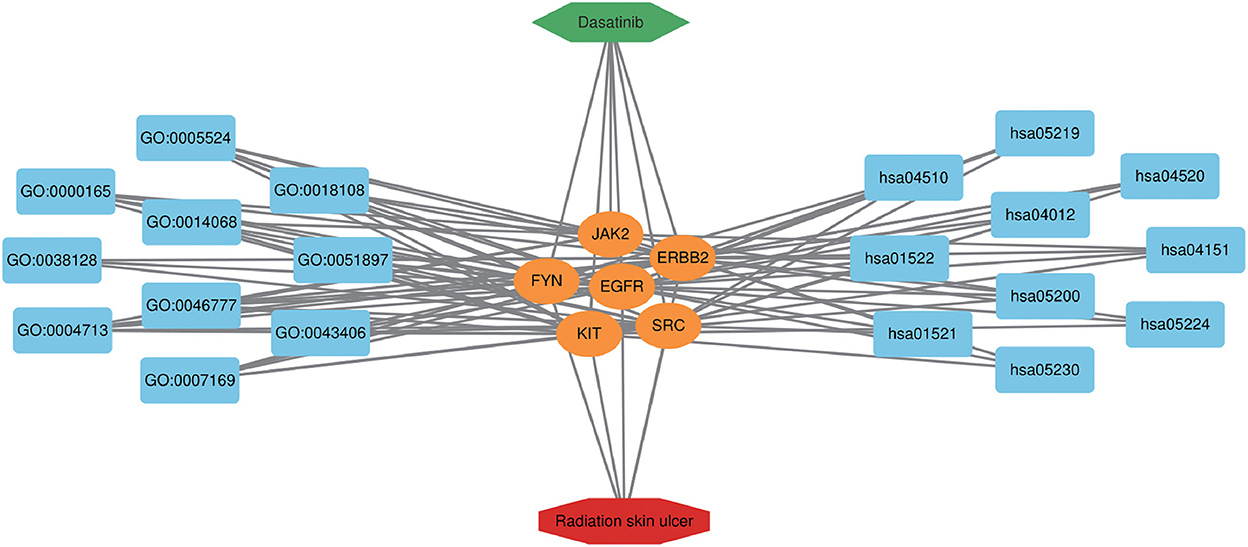
Figure 6. Dasatinib-target-radiation ulcers detailed interaction network diagram. The blue rectangle on the left represents the first 10 BP, and the blue rectangle on the right represents the first 10 KEGG pathways. The orange oval in the middle represents the core targets.
The Dasatinib was molecularly docked with the core genes EGFR, ERBB2, FYN, JAK2, KIT, and SRC, respectively. According to the molecular docking binding energy, each histone-ligand could spontaneously bind (binding energy <0 kcal/mol). The binding energies of EGFR, ERBB2, FYN, JAK2, KIT, and SRC were all ≤ −5 kcal/mol, indicating good protein-ligand binding. See Figure 7 for the visualization of molecular docking. The molecular docking details are shown in Table 3.
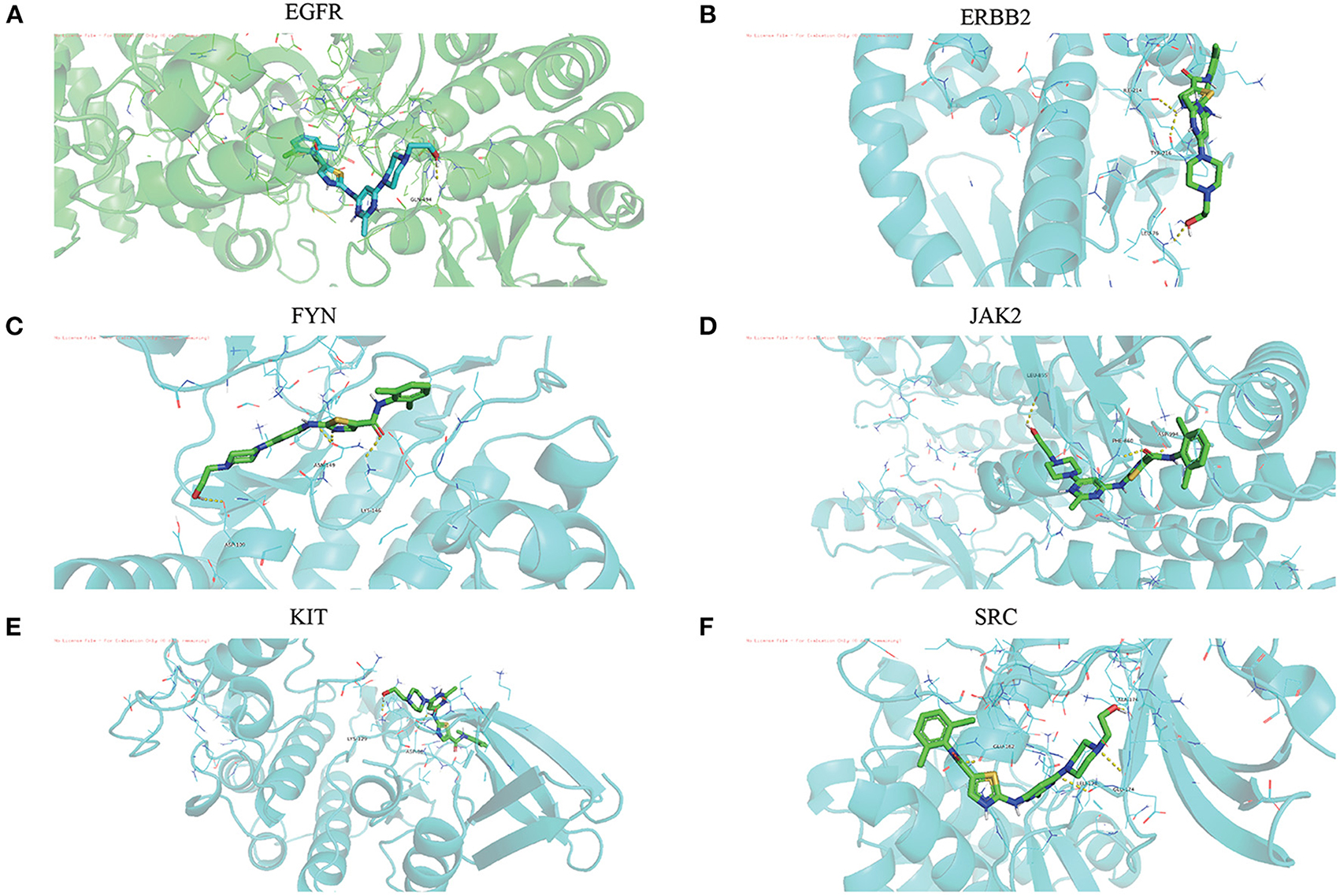
Figure 7. (A-F) The visualization of dasatinib docking with six core targets. The docking results of the core targets are displayed by PyMOL software, and the yellow dotted line represents the interaction line between the target and the compound.
Compared with the original crystal structures, the RSMD values of EGFR, ERBB2, FYN, JAK2, KIT, and SRC were 1.4574, 1.1103, 1.4139, 0.5710, 0.5868, 1.1455A. The ligand conformation in the original crystal structure of the core protein basically overlaps with the docked ligand conformation, and RMSD <2A indicates that the calculation method can accurately predict the binding mode of the original ligand. The molecular docking results of EGFR, ERBB2, FYN, JAK2, KIT, and SRC with Dasatinib are shown in Figure 8. AutoDock vina and Discovery Studio use different algorithms and scoring functions, which may cause differences in screening results. Combining two different algorithms, we believe that EGFR, ERBB2, FYN, JAK2, KIT, and SRC are likely to become core targets.
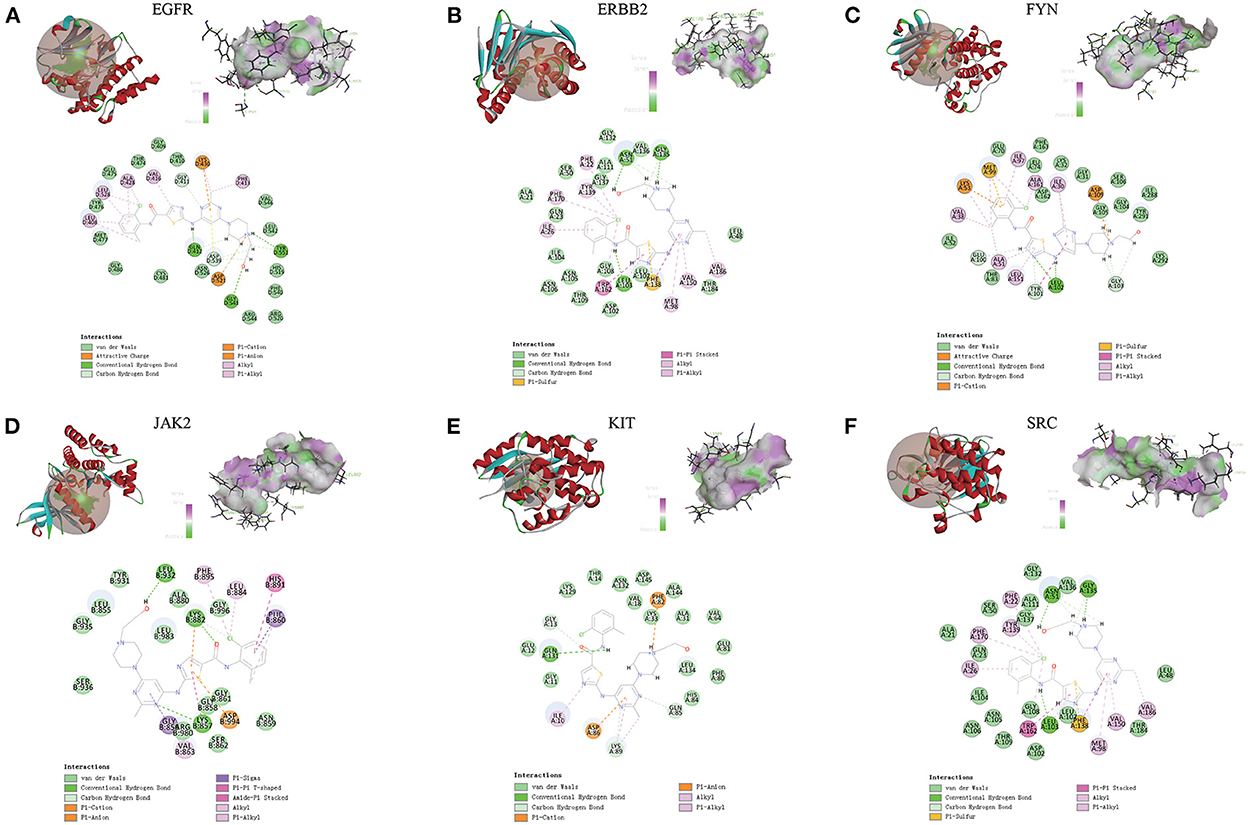
Figure 8. (A-F) Molecular docking results of EGFR, ERBB2, FYN, JAK2, KIT and SRC with dasatinib. (Upper left) Dasatinib docking model with the core targets. (Top right) The interaction diagram of dasatinib and the core targets hydrogen bond residues, the greener the color, the stronger the interaction force, and the more purple the color, the weaker the interaction force. (Bottom) Various interactions of dasatinib with core targets, each color in the circle corresponds to a mode of action.
We extracted the expression values of the six core targets in control skin and post-irradiated skin. T-test was used to verify whether there was a significant difference between the groups. The results showed that the six core targets had significant expression changes at different time points after irradiation (Figure 9; Supplementary Table 6).
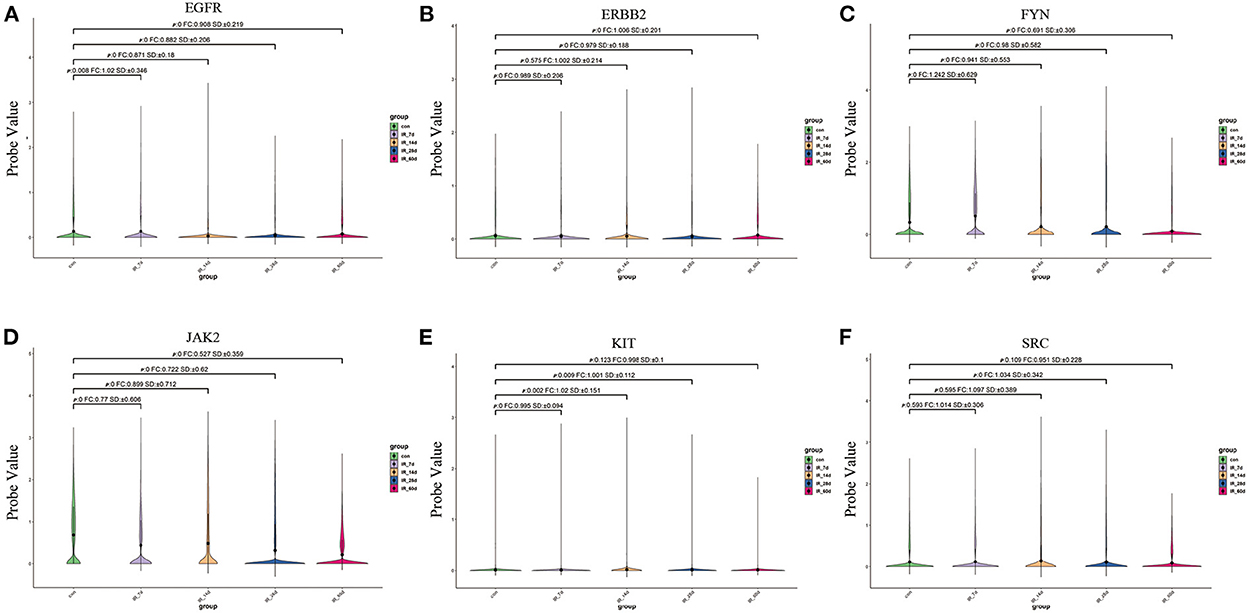
Figure 9. (A–F) Expression of six core targets in rat skin tissue after irradiation was examined by single-cell RNA sequencing. FC > 1 represents up-regulation, and FC <1 represents down-regulation.
Radiation-induced skin damage can be roughly divided into four stages: immediate (0–2 weeks), early (2–8 weeks), mid-term (6–52 weeks), and late (over 40 weeks) according to the time after exposure. In addition, radiation damage response is dose-dependent, and the National Cancer Institute has identified four skin toxicity grades for radiation dermatitis. The most common immediate reaction (dose as low as 2 Gy) is transient erythema (grade 1), which is an inflammatory response with changes in cellular permeability and histamine release (23–25). Early reactions (at doses of 3 to 6 Gy) included temporary alopecia, erythematous reactions, and moderate tissue edema (grade 2). Doses greater than 10 Gy can cause more severe consequences, such as dry or moist peeling (grade 3) (24). Intermediate responses (dose greater than 15 Gy) can include skin atrophy, necrosis, and ulceration (grade 4). Late reactions are characterized by telangiectasia, further thinning of the skin, and deepening of the ulcer (23).
Ionizing radiation causes skin cell death through a variety of pathways, including induction of apoptosis, mitotic disorders, and cellular senescence (26). Reactive oxygen species (ROS) induced by ionizing radiation can induce damage to DNA, endoplasmic reticulum, and mitochondria (27, 28). Recent studies have shown that exposure to radiation also leads to impairment and damage to the immune system, triggering the production of inflammatory cytokines and leading to a systemic inflammatory state (29–31). As previously described, impaired vascular integrity, resulting in transient intra-tissue hemorrhage, promoted cutaneous wound healing in mice with combined platelet GPVI and CLEC2 deficiency (32). Thus, a short-term loss of vascular integrity following dasatinib administration, especially at 5 mg/kg, allowing plasma-derived molecules (including fibrinogen, growth factors, and cytokines) to enter the wound to promote tissue repair (19, 33, 34). High concentrations of dasatinib inhibited platelet aggregation and tyrosine phosphorylation in vitro upon thrombin stimulation (19). More importantly, inhibition of thrombin may affect the repair process, as synthetic thrombin peptides have been shown to promote angiogenesis and promote wound healing in animal skin (35, 36). In addition, high-dose or prolonged exposure to dasatinib may cause vessel wall rupture through its direct effects on endothelial cells (20, 37, 38).
This study focused on the potential mechanism of action of dasatinib in the treatment of radiation ulcers. Network analysis increases the understanding of multiple mechanisms of drug action, while systems pharmacology may provide new targets for drug discovery. By intersecting the potential targets of dasatinib and radiation ulcers in the bioinformatics database, 137 potential therapeutic targets of dasatinib for radiation ulcers were finally obtained. Subsequently, we constructed a PPI network with potential therapeutic targets and further screened six core targets (EGFR, ERBB2, FYN, JAK2, KIT, and SRC).
EGFR is widely expressed in normal skin tissues, such as epidermis, sebaceous glands, glands, eccrine glands, and dendritic cells (39), and plays an important role in the development and physiology of normal epidermis (40). EGFR may be activated through signaling pathways such as MAPK, STATs and PI3K (41). Inhibition of EGFR increases the expression of the cyclin-dependent kinase inhibitor p27-KIP1 (42–44), which leads to cell cycle arrest in the G1 phase of keratinocytes (45). STAT3 is activated by EGFR signaling and is a key molecule in maintaining skin homeostasis. Loss of STAT3 expression in transgenic mice affects wound healing (46), showing impaired hair follicle development and hair growth and a strong inflammatory response (47–49). Children with loss of EGFR expression develop lifelong inflammation in the skin, gut, and lungs, leading to early infant mortality, highlighting the importance of EGFR signaling in establishing and maintaining tissue homeostasis (50). Furthermore, the EGFR signaling pathway is critical for the maintenance of bone precursor cells and the formation of new bone (51). In mice, EGFR-specific inhibitors or inhibition of EGFR in osteoprogenitors and osteoblasts results in a reduction in the number of bone mesenchymal progenitors leading to bone loss (52). It has recently been reported that EGFR signaling inhibits cellular and tissue senescence (53–55). The EGFR signaling pathway in osteoprogenitor cells plays an important role in preventing cellular senescence and regulating cortical bone metabolism. In addition, ERBB2, a member of the EGFR family, is expressed in the epidermis and hair follicle root sheath (56), and overexpression of ERBB2 has also been found in skin cancer patients (57). Furthermore, in a mouse model overexpressing ERBB2, the epidermis and follicles of the skin are hyperproliferative (58–61). All of the above studies suggest that dasatinib may be effective in the treatment of radiation ulcers by targeting EGFR signaling.
Few studies have been performed on other core targets related to radiation ulcers and deserve further consideration in future studies. Given that radiation ulcers are still a major problem facing the global medical system, our study is the first to use network pharmacology to explore the potential core targets and mechanisms of action of dasatinib against radiation ulcers. Their feasibility and possible docking sites are verified by molecular docking technology, which will provide a reference for the study of the molecular mechanism of dasatinib in the treatment of radiation ulcers. However, our study also has certain limitations. First, the core targets and pharmacological mechanisms of dasatinib in the treatment of radiation ulcers need to be verified in vivo and in vitro models, which will be the focus of our future research. Secondly, whether the pharmacological effects of dasatinib are as helpful to human patients as in animal and cell experimental models is also a problem that we need to solve. In follow-up studies, we will further verify the feasibility of treating radiation ulcers with core targets in animal and clinical trials.
Overall, this is the first time that bioinformatics methods such as network pharmacology and molecular docking have been applied to systematically explore the pharmacological and molecular mechanisms of dasatinib in the treatment of radiation skin ulcers. Dasatinib may promote the healing of radiation skin ulcers through EGFR tyrosine kinase inhibitor resistance, PI3K-Akt signaling pathway and ErbB signaling pathway. During treatment, EGFR, ERBB2, FYN, JAK2, KIT, and SRC were its main potential pharmacological targets, and the molecular docking results confirmed our conclusion. The identified potential targets and pathways may be validated in preclinical studies before being used in clinical therapy.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by Ethics Committee of the Second Affiliated Hospital of Chengdu Medical College.
WS and DY: conceptualization. XiC, XuC, and WZ: data curation. WS, DL, and XiC: formal analysis and methodology. DY, DL, and XuC: supervision. WS, XuC, and DL: writing (review and editing). The final manuscript read and approved by all authors. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32071238), the Fundamental Research Funds for the Central Universities and Young Talent Program of China National Nuclear Corporation (CNNC2021136), the Natural Science Project of Chengdu Medical College (CYZYB21-07), and the Medical Research Project of Chengdu 2021 (2021085).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1031038/full#supplementary-material
Supplementary Table 1. The pathological targets of radiation skin ulcer form GeneCards database.
Supplementary Table 2. The dasatinib pharmacological targets form SwissTargetPrediction database.
Supplementary Table 3. The dasatinib pharmacological targets form Drugbank database.
Supplementary Table 4. The dasatinib pharmacological targets form Binding DB database.
Supplementary Table 5. The targets related to dasatinib activity against radiation skin ulcer.
Supplementary Table 6. Expression values of core targets in cells of rats at different time points after irradiation.
DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; PPI, protein-protein interaction; Treg, regulatory T cells; GEO, Gene Expression Omnibus; TBSA, total burn surface area; TOM, topological overlap matrix; GS, gene significance; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MCODE, molecular complex detection technology; ImmuCellAI, The Immune Cell Abundance Identifier; HLA-G, human leukocyte antigen G.
1. Aerts A, Decraene T, van den Oord JJ, Dens J, Janssens S, Guelinckx P, et al. Chronic radiodermatitis following percutaneous coronary interventions: a report of two cases. J Eur Acad Dermatol Venereol. (2003) 17:340–3. doi: 10.1046/j.1468-3083.2003.00687.x
2. Herz-Ruelas ME, Gomez-Flores M, Moxica-Del Angel J, Miranda-Maldonado I, Gutierrez-Villarreal IM, Guerrero-Gonzalez GA, et al. Ulcerated radiodermatitis induced after fluoroscopically guided stent implantation angioplasty. Case Rep Dermatol Med. (2014) 2014:768624. doi: 10.1155/2014/768624
3. Boncher J, Bergfeld WF. Fluoroscopy-induced chronic radiation dermatitis: a report of two additional cases and a brief review of the literature. J Cutan Pathol. (2012) 39:63–7. doi: 10.1111/j.1600-0560.2011.01754.x
4. Spiker A, Zinn Z, Carter WH, Powers R, Kovach R. Fluoroscopy-induced chronic radiation dermatitis. Am J Cardiol. (2012) 110:1861–3. doi: 10.1016/j.amjcard.2012.08.023
5. Nishimoto S, Fukuda K, Kawai K, Fujiwara T, Tsumano T, Fujita K, et al. Supplementation of bone marrow aspirate-derived platelet-rich plasma for treating radiation-induced ulcer after cardiac fluoroscopic procedures: a preliminary report. Indian J Plast Surg. (2012) 45:109–14. doi: 10.4103/0970-0358.96599
6. Otterburn D, Losken A. Iatrogenic fluoroscopy injury to the skin. Ann Plast Surg. (2010) 65:462–5. doi: 10.1097/SAP.0b013e3181d6e2d3
7. Chambers CE, Fetterly KA, Holzer R, Lin PJ, Blankenship JC, Balter S, et al. Radiation safety program for the cardiac catheterization laboratory. Catheter Cardiovasc Interv. (2011) 77:546–56. doi: 10.1002/ccd.22867
8. Miller DL, Balter S, Schueler BA, Wagner LK, Strauss KJ, Vano E. Clinical radiation management for fluoroscopically guided interventional procedures. Radiology. (2010) 257:321–32. doi: 10.1148/radiol.10091269
9. Hirshfeld JW Jr, Balter S, Brinker JA, Kern MJ, Klein LW, Lindsay BD, et al. ACCF/AHA/HRS/SCAI clinical competence statement on physician knowledge to optimize patient safety and image quality in fluoroscopically guided invasive cardiovascular procedures: a report of the American College of Cardiology Foundation/American Heart Association/American College of Physicians Task Force on Clinical Competence and Training. Circulation. (2005) 111:511–32. doi: 10.1161/01.CIR.0000157946.29224.5D
10. Gu Q, Gao Y, Li Y, Yang Z, Zhou J Wang X, et al. Overexpression of p53 and MDM2 proteins in rat radiation-induced skin ulcers. J Environ Pathol Toxicol Oncol. (1999) 18:319–22.
11. Gu Q, Gao Y, Li Y, Yang Z, Wang D, Zhao P. Overexpression of c-erbB-2 and EGF-R proteins in radiation-induced skin ulcers. J Environ Pathol Toxicol Oncol. (1999) 18:313–7.
12. Gu Q, Gao Y, Li Y, Yang Z, Zhou J, Wang X, et al. Overexpression of c-fos and Rb proteins in radiation-induced skin ulcers. J Environ Pathol Toxicol Oncol. (1999) 18:309–12.
13. Wei KC, Yang KC, Mar GY, Chen LW, Wu CS, Lai CC, et al. STROBE–radiation ulcer: an overlooked complication of fluoroscopic intervention: a cross-sectional study. Medicine. (2015) 94:e2178. doi: 10.1097/MD.0000000000002178
14. Winaikosol K, Punyavong P, Jenwitheesuk K, Surakunprapha P, Mahakkanukrauh A. Radiation ulcer treatment with hyperbaric oxygen therapy and haemoglobin spray: case report and literature review. J Wound Care. (2020) 29:452–6. doi: 10.12968/jowc.2020.29.8.452
15. Tejada S, Batle JM, Ferrer MD, Busquets-Cortés C, Monserrat-Mesquida M, Nabavi SM, et al. Therapeutic effects of hyperbaric oxygen in the process of wound healing. Curr Pharm Des. (2019) 25:1682–93. doi: 10.2174/1381612825666190703162648
16. Akita S. Treatment of radiation injury. Adv Wound Care. (2014) 3:1–11. doi: 10.1089/wound.2012.0403
17. Wang Z, Chen Z, Jiang Z, Luo P, Liu L, Huang Y, et al. Cordycepin prevents radiation ulcer by inhibiting cell senescence via NRF2 and AMPK in rodents. Nat Commun. (2019) 10:2538. doi: 10.1038/s41467-019-10386-8
18. Wang Z, Shi C. Cellular senescence is a promising target for chronic wounds: a comprehensive review. Burns Trauma. (2020) 8:tkaa021. doi: 10.1093/burnst/tkaa021
19. Gratacap MP, Martin V, Valera MC, Allart S, Garcia C, Sie P, et al. The new tyrosine-kinase inhibitor and anticancer drug dasatinib reversibly affects platelet activation in vitro and in vivo. Blood. (2009) 114:1884–92. doi: 10.1182/blood-2009-02-205328
20. Kreutzman A, Colom-Fernandez B, Jimenez AM, Ilander M, Cuesta-Mateos C, Perez-Garcia Y, et al. Dasatinib reversibly disrupts endothelial vascular integrity by increasing non-muscle myosin II contractility in a ROCK-dependent manner. Clin Cancer Res. (2017) 23:6697–707. doi: 10.1158/1078-0432.CCR-16-0667
21. Wang H, Wang Z, Huang Y, Zhou Y, Sheng X, Jiang Q, et al. Senolytics (DQ) mitigates radiation ulcers by removing senescent cells. Front Oncol. (2019) 9:1576. doi: 10.3389/fonc.2019.01576
22. Shen W, Song Z, Zhong X, Huang M, Shen D, Gao P, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. (2022) 1:e36. doi: 10.1002/imt2.36
23. Balter S, Hopewell JW, Miller DL, Wagner LK, Zelefsky MJ. Fluoroscopically guided interventional procedures: a review of radiation effects on patients' skin and hair. Radiology. (2010) 254:326–41. doi: 10.1148/radiol.2542082312
24. Fiorilli PN, Kobayashi T, Giri J, Hirshfeld JW Jr. Strategies for radiation exposure-sparing in fluoroscopically guided invasive cardiovascular procedures. Catheter Cardiovasc Interv. (2020) 95:118–27. doi: 10.1002/ccd.28281
25. Kalef-Ezra JA, Karavasilis S, Ziogas D, Dristiliaris D, Michalis LK, Matsagas M. Radiation burden of patients undergoing endovascular abdominal aortic aneurysm repair. J Vasc Surg. (2009) 49:283–7. doi: 10.1016/j.jvs.2008.09.003
26. Wang JS, Wang HJ, Qian HL. Biological effects of radiation on cancer cells. Mil Med Res. (2018) 5:20. doi: 10.1186/s40779-018-0167-4
27. Lomax ME, Folkes LK, O'Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol. (2013) 25:578–85. doi: 10.1016/j.clon.2013.06.007
28. Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. (2019) 25:101084. doi: 10.1016/j.redox.2018.101084
29. Kim JH, Jenrow KA, Brown SL. Mechanisms of radiation-induced normal tissue toxicity and implications for future clinical trials. Radiat Oncol J. (2014) 32:103–15. doi: 10.3857/roj.2014.32.3.103
30. Ratikan JA, Micewicz ED, Xie MW, Schaue D. Radiation takes its toll. Cancer Lett. (2015) 368:238–45. doi: 10.1016/j.canlet.2015.03.031
31. Stoecklein VM, Osuka A, Ishikawa S, Lederer M R, Wanke-Jellinek L, Lederer JA. Radiation exposure induces inflammasome pathway activation in immune cells. J Immunol. (2015) 194:1178–89. doi: 10.4049/jimmunol.1303051
32. Wichaiyo S, Lax S, Montague SJ, Li Z, Grygielska B, Pike JA, et al. Platelet glycoprotein VI and C-type lectin-like receptor 2 deficiency accelerates wound healing by impairing vascular integrity in mice. Haematologica. (2019) 104:1648–60. doi: 10.3324/haematol.2018.208363
33. Lorenz V, Stegner D, Stritt S, Vogtle T, Kiefer F, Witke W, et al. Targeted downregulation of platelet CLEC-2 occurs through Syk-independent internalization. Blood. (2015) 125:4069–77. doi: 10.1182/blood-2014-11-611905
34. Mazharian A, Ghevaert C, Zhang L, Massberg S, Watson S P. Dasatinib enhances megakaryocyte differentiation but inhibits platelet formation. Blood. (2011) 117:5198–206. doi: 10.1182/blood-2010-12-326850
35. Stiernberg J, Norfleet AM, Redin WR, Warner WS, Fritz RR, Carney DH. Acceleration of full-thickness wound healing in normal rats by the synthetic thrombin peptide, TP508. Wound Repair Regen. (2000) 8:204–15. doi: 10.1046/j.1524-475x.2000.00204.x
36. Norfleet AM, Huang Y, Sower LE, Redin WR, Fritz RR, Carney DH. Thrombin peptide TP508 accelerates closure of dermal excisions in animal tissue with surgically induced ischemia. Wound Repair Regen. (2000) 8:517–29. doi: 10.1046/j.1524-475x.2000.00517.x
37. Phan C, Jutant EM, Tu L, Thuillet R, Seferian A, Montani D, et al. Dasatinib increases endothelial permeability leading to pleural effusion. Eur Respir J. (2018) 51:1701096. doi: 10.1183/13993003.01096-2017
38. Liu Y, Dai Y, Xu H, Zhou Q, Li F, Yu B, et al. YQFM alleviates side effects caused by dasatinib through the ROCK/MLC pathway in mice. Evid Based Complement Alternat Med. (2020) 2020:4646029. doi: 10.1155/2020/4646029
39. Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. (2009) 218:460–6. doi: 10.1002/jcp.21635
40. Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. (2002) 3:199–209. doi: 10.1038/nrg758
41. Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. (2005) 97:339–46. doi: 10.1093/jnci/dji055
42. Albanell J, Rojo F, Averbuch S, Feyereislova A, Mascaro JM, Herbst R, et al. Pharmacodynamic studies of the epidermal growth factor receptor inhibitor ZD1839 in skin from cancer patients: histopathologic and molecular consequences of receptor inhibition. J Clin Oncol. (2002) 20:110–24. doi: 10.1200/JCO.2002.20.1.110
43. Baselga J, Rischin D, Ranson M, Calvert H, Raymond E, Kieback DG, et al. Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types. J Clin Oncol. (2002) 20:4292–302. doi: 10.1200/JCO.2002.03.100
44. Malik SN, Siu LL, Rowinsky EK, deGraffenried L, Hammond LA, Rizzo J, et al. Pharmacodynamic evaluation of the epidermal growth factor receptor inhibitor OSI-774 in human epidermis of cancer patients. Clin Cancer Res. (2003) 9:2478–86.
45. Peus D, Hamacher L, Pittelkow MR. EGF-receptor tyrosine kinase inhibition induces keratinocyte growth arrest and terminal differentiation. J Invest Dermatol. (1997) 109:751–6. doi: 10.1111/1523-1747.ep12340759
46. Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. (1999) 18:4657–68. doi: 10.1093/emboj/18.17.4657
47. Lichtenberger BM, Gerber PA, Holcmann M, Buhren BA, Amberg N, Smolle V, et al. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci Transl Med. (2013) 5:199ra111. doi: 10.1126/scitranslmed.3005886
48. Mascia F, Lam G, Keith C, Garber C, Steinberg SM, Kohn E, et al. Genetic ablation of epidermal EGFR reveals the dynamic origin of adverse effects of anti-EGFR therapy. Sci Transl Med. (2013) 5:199ra110. doi: 10.1126/scitranslmed.3005773
49. Sibilia M, Wagner B, Hoebertz A, Elliott C, Marino S, Jochum W, et al. Mice humanised for the EGF receptor display hypomorphic phenotypes in skin, bone and heart. Development. (2003) 130:4515–25. doi: 10.1242/dev.00664
50. Campbell P, Morton PE, Takeichi T, Salam A, Roberts N, Proudfoot LE, et al. Epithelial inflammation resulting from an inherited loss-of-function mutation in EGFR. J Invest Dermatol. (2014) 134:2570–8. doi: 10.1038/jid.2014.164
51. Chandra A, Lan S, Zhu J, Siclari VA, Qin L. Epidermal growth factor receptor (EGFR) signaling promotes proliferation and survival in osteoprogenitors by increasing early growth response 2 (EGR2) expression. J Biol Chem. (2013) 288:20488–98. doi: 10.1074/jbc.M112.447250
52. Zhang X, Tamasi J, Lu X, Zhu J, Chen H, Tian X, et al. Epidermal growth factor receptor plays an anabolic role in bone metabolism in vivo. J Bone Miner Res. (2011) 26:1022–34. doi: 10.1002/jbmr.295
53. Alexander PB, Yuan L, Yang P, Sun T, Chen R, Xiang H, et al. EGF promotes mammalian cell growth by suppressing cellular senescence. Cell Res. (2015) 25:135–8. doi: 10.1038/cr.2014.141
54. Ding C, Zou Q, Wang F, Wu H, Chen R, Lv J, et al. Human amniotic mesenchymal stem cells improve ovarian function in natural aging through secreting hepatocyte growth factor and epidermal growth factor. Stem Cell Res Ther. (2018) 9:55. doi: 10.1186/s13287-018-0781-9
55. Gerber PA, Buhren BA, Schrumpf H, Hevezi P, Bolke E, Sohn D, et al. Mechanisms of skin aging induced by EGFR inhibitors. Support Care Cancer. (2016) 24:4241–8. doi: 10.1007/s00520-016-3254-7
56. De Potter IY, Poumay Y, Squillace KA, Pittelkow MR. Human EGF receptor (HER) family and heregulin members are differentially expressed in epidermal keratinocytes and modulate differentiation. Exp Cell Res. (2001) 271:315–28. doi: 10.1006/excr.2001.5390
57. Liu B, Zhang H, Li S, Chen W, Li R. The expression of c-erbB-1 and c-erbB-2 oncogenes in basal cell carcinoma and squamous cell carcinoma of skin. Chin Med Sci J. (1996) 11:106–9.
58. Kiguchi K, Bol D, Carbajal S, Beltran L, Moats S, Chan K, et al. Constitutive expression of erbB2 in epidermis of transgenic mice results in epidermal hyperproliferation and spontaneous skin tumor development. Oncogene. (2000) 19:4243–54. doi: 10.1038/sj.onc.1203778
59. Xie W, Wu X, Chow LT, Chin E, Paterson AJ, Kudlow JE. Targeted expression of activated erbB-2 to the epidermis of transgenic mice elicits striking developmental abnormalities in the epidermis and hair follicles. Cell Growth Differ. (1998) 9:313–25.
60. Bol D, Kiguchi K, Beltran L, Rupp T, Moats S, Gimenez-Conti I, et al. Severe follicular hyperplasia and spontaneous papilloma formation in transgenic mice expressing the neu oncogene under the control of the bovine keratin 5 promoter. Mol Carcinog. (1998) 21:2–12.
Keywords: dasatinib, radiation ulcer, molecular docking, network pharmacology, core target
Citation: Su W, Chen X, Zhang W, Li D, Chen X and Yu D (2022) Therapeutic targets and signaling mechanisms of dasatinib activity against radiation skin ulcer. Front. Public Health 10:1031038. doi: 10.3389/fpubh.2022.1031038
Received: 29 August 2022; Accepted: 31 October 2022;
Published: 30 November 2022.
Edited by:
Hesham M. H. Zakaly, Al-Azhar University, EgyptReviewed by:
Shams A. M. Issa, Al-Azhar University, EgyptCopyright © 2022 Su, Chen, Zhang, Li, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daojiang Yu, eWRqNTEwODdAMTYzLmNvbQ==; Xiaoming Chen, Y2hlbnhpYW9taW5nODA1NUAxNjMuY29t; Dazhuang Li, bGkxMzYxNjI3OTQ3NkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.