- 1College of Veterinary Medicine, Hawassa University, Hawassa, Ethiopia
- 2Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Addis Ababa, Ethiopia
- 3College of Veterinary Medicine and Agriculture, Addis Ababa University, Addis Ababa, Ethiopia
- 4College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia
- 5College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
Campylobacter is the most common cause of bacterial infectious diarrhea and acute gastroenteritis globally, and is recognized as a significant zoonotic pathogen. Antimicrobial resistance amongst Campylobacter isolates is a significant global concern. A cross-sectional study was conducted to identify and characterize Campylobacter species in humans, animals and water sources in livestock owning households of peri-urban Addis Ababa, Ethiopia; and to characterize antimicrobial resistance. A total of 519 fecal samples from humans (n = 99), livestock (n = 179), poultry (n = 69), and water (n = 172) were collected. Samples were cultured for viable Campylobacter spp. and multiplex PCR utilized for the identification and confirmation. Antimicrobial susceptibility of the isolates was assessed using the Kirby-Bauer disc diffusion method. Campylobacter spp. was detected in 67/519 (13.0%) of the total tested samples, and the household level prevalence of Campylobacter was 42.4%. The prevalence of Campylobacter spp. was: humans (10.1%), cattle (18.5%), poultry (13.0%), sheep (13.3%), goats (7.1%), and water (10.5%). Campylobacter jejuni and C. fetus were the most frequently isolated species, followed by C. coli. The majority of isolates obtained from human samples had co-occurrence with isolates from cattle, poultry or water samples from the same household. The use of stored water, the practice of indoor and outdoor manure collecting, and animal species Campylobacter positivity were significantly associated with greater odds of human Campylobacter spp. positivity. All Campylobacter isolates from humans, poultry, sheep, goats and water, and 96.0% of isolates from cattle were resistant to at least one or more of the tested antimicrobials, with 95.5% of isolates resistant to three or more classes of antimicrobials. A One Health approach is recommended to further investigate Campylobacter species infections, and other zoonotic infectious diseases, in the livestock owning populations in Ethiopia, where there is close interaction between humans, animals and the environment.
Introduction
Diarrheal diseases are the leading cause of childhood illness and death in developing countries (1). Enteric pathogens including rotavirus, norovirus, Campylobacter, Salmonella, E. coli and Shigella species are known causative agents (2, 3). Campylobacter species are the leading zoonotic bacterial causes of human gastroenteritis and foodborne disease globally (1, 2, 4, 5). The genus Campylobacter, comprises of 31 species and 13 subspecies (6); of these, thermophilic Campylobacter, especially C. jejuni and C. coli, are the primary causes of human bacterial gastroenteritis worldwide (2, 7).
Epidemiologically, human campylobacteriosis differs across high and low-income countries. In high-income countries, symptomatic infection occurs in all age groups, whereas in low-income countries, clinical disease mostly affects children under 5 years of age, with adults rarely suffering from the disease, predominately having asymptomatic excretion (8, 9). Most Campylobacter species have been isolated from animals, and direct and indirect contact with animals is a known risk factor for Campylobacter infection, especially in children (10–12). Animals can serve both as reservoirs, and as potential sources for the contamination of food, water, and the environment (4, 13–15). Contamination of different water sources with Campylobacter species presents an opportunity for transmission of environment-adapted genotypes to livestock and humans (7, 13, 14, 16).
Most cases of campylobacteriosis are self-limiting, and present with enteritis, abdominal cramps, fever, nausea, and vomiting (3, 9). However, some cases have been linked to more serious complications including Guillain–Barré syndrome, Reiter's syndrome, bacteremia, and abortion (9). In severe cases of human campylobacteriosis, macrolides, and fluoroquinolones (particularly erythromycin and ciprofloxacin), and intravenous aminoglycosides are the first line antimicrobials of choice (14, 17). There is increasing antimicrobial resistance amongst Campylobacter isolates from various sources (18–20). This situation is more common in developing countries, where there is widespread and largely uncontrolled use of antimicrobials (11, 12, 21).
Studies on the prevalence of Campylobacter in Ethiopia have identified a range of 13.8–50.0% in humans, and 10.6–56.5% in animals (12, 13, 22–27). In addition, studies conducted on the antimicrobial susceptibility profiles of Campylobacter species, in both humans and animals in Ethiopia, have identified increased multidrug resistance of Campylobacter species (4, 25, 28). Most of these studies were conducted in only humans or animals, and utilized only a culture method approach for Campylobacter species identification and characterization. The role of different environments in Ethiopia, including water, as potential risk factors and sources for transmission of Campylobacter infection in humans and animals is far less known. In addition, there is limited literature regarding risk factors associated to human infection with Campylobacter species in Ethiopia. The identification and characterization of Campylobacter species in humans, animals, and water sources, is important in efforts to reduce the risk of human exposure to Campylobacter. The objectives of this study were the identification and characterization of Campylobacter spp. in humans, animals and water sources in livestock owning households of peri-urban Addis Ababa, Ethiopia; and the characterization of antimicrobial resistant isolates.
Materials and Methods
Study Design and Location
A cross-sectional study was conducted between December 2018 and January 2020 in Addis Ababa, Ethiopia. Addis Ababa has 10 sub-cities, of which five are considered as peri-urban areas. Akaki Kality sub-city was selected as it is characterized by a high level of livestock production, and studies have documented a high prevalence of zoonotic enteric infections (29–31). Within Akaki Kality sub-city, two woredas (Woreda 2 and 3) were purposively selected.
Study Population
Study participants were selected from available lists of livestock-owning households in both woredas using simple random selection. Randomization of selected household lists and households was conducted using random numbers generated in a software program (Microsoft Excel 2019, Microsoft Cooperation, USA). Sample sizes were calculated for two independent populations based on an expected Campylobacter species prevalence of 50%, 5% margin of error, and 95% confidence level, using the total number of available livestock-owning households (227 households) in Woreda 2 and 3. The calculated sample size was 143 households and 372 animals. Households were included in the study if they owned at least one free-roaming chicken, or at least one ruminant animal. Humans and animals that were on antimicrobial treatment, or who were treated during the previous 2 weeks at the time of contact, were excluded from the study.
Of the 227 households contacted, 99 consented to participate. Households refused to participate for the following reasons: recently lost, or sold livestock (n = 94), no reason provided (n = 21), and refused to provide human or animal fecal samples (n = 13). Therefore, 99 households participated, providing 99 household pooled human fecal samples, and 248 household pooled livestock fecal samples. The sample size for water samples was conveniently determined based on the available water sources that was in use for both household daily activities, and drinking water for humans and animals in the study location (14).
Questionnaire data on potential risk factors associated with human infection with Campylobacter species were collected. Both closed and open-ended questions were utilized and translated into local languages (Afaan Oromoo and Amharic). The questionnaire was pretested on 20 households from a similar Woreda (8) in Yeka sub-city.
Sample Collection
A total of 347 household pooled fecal samples were collected from households: humans (n = 99), cattle (n = 135), sheep (n = 30), goats (n = 14), and poultry (n = 69). Pooled human stool samples were collected only from individuals who were most closely associated with the management of the livestock. Water samples (n = 172) were collected from the 99 households: surface water (n = 9), municipal pipe water (n = 84), ground water (n = 16), and stored water (n = 63).
A sterile cotton swab moistened with nutrient broth was used to transfer ~7–10 g of the pooled fecal samples into a 15 ml screw-capped falcon tube containing Cary-Blair Transport Medium (Oxoid). Fecal samples from individual animals were collected either directly from rectum or floor of the livestock housing immediately after an animal defecated. Samples were transported to the Microbiology Laboratory of Akililu Lemma Institute of Pathobiology (ALIPB), Addis Ababa University (AAU), and processed within 4–6 h of collection.
Isolation and Identification of Campylobacter Species
A selective enrichment was initially performed on all samples as previously described (32). Water samples were filtered through a 0.45 μm sterile nitrocellulose filter (Pall Corporation, Ann Arbor, MI). Membrane filtered water and homogenized human and animal fecal specimens were inoculated in 30 ml of Bolton enrichment broth containing Campylobacter growth supplement (HiMedia Laboratories; Mumbai, India), with 5% (v/v) defibrinated cattle blood. Tubes were then incubated for 48 h under microaerophilic condition (CO2, 10%; O2, 5%; N2, 85%) using Campy Gene kits (Thermo Scientific, Waltham, MA, USA) as described by Szczepanska et al. (14).
After 48 h of incubation, a loopful of culture from each sample type was spread onto plates containing modified charcoal cefoperazone deoxycholate agar (mCCDA, CM0739-Preston) and Campylobacter selective supplements (SR0174), and incubated under microaerophilic conditions as described earlier for 48–72 h at 42°C. After 48–72 h of incubation, all presumptive Campylobacter colonies were selected from each plate and checked for motility, oxidase, catalase and Gram character as previously described (7). All suspected colonies were subsequently sub-cultured onto Columbia blood agar (Difco, USA) with 5% (v/v) defibrinated cattle blood and incubated under microaerophilic condition at 42°C for 48 h. Isolates with typical colony characteristics of Campylobacter were then subjected to PCR analysis for confirmation and identification of Campylobacter species. Reference strains of Campylobacter jejuni and coli were obtained from the Ethiopian Public Health Institute and used as positive controls.
Identification of Campylobacter by PCR
Genomic DNA was extracted by boiling fresh Campylobacter cultures grown on Columbia blood agar as previously described (33). A loopful of bacterial growth from plates were suspended in 100 μl of sterilized RNase/DNase- free water, boiled at 95°C for 15 min and cooled at 4°C until used. Genus level confirmation and identification of Campylobacter was conducted using multiplex-PCR (mPCR) targeting genus level and species-specific regions of the 16S rRNA, j0414, glyA, cstA, and ask gene sequences, as described previously and in Supplementary Table 1 (34). Campylobacter identification was conducted and isolates that were positive for the genus-specific PCR but negative for the C. lari, C. fetus, C. coli, and C. jejuni- using mPCR assay were designated as unidentified thermophilic Campylobacter species. In this study, a household was considered positive for Campylobacter, if at least one isolate was confirmed by PCR from the pooled samples of any of the animal, human, or water samples obtained.
Antimicrobial Susceptibility of Campylobacter Isolates
All PCR confirmed isolates were subjected to antimicrobial susceptibility testing using the standard Kirby-Bauer disc diffusion assay on Mueller-Hinton agar (Hi Media Laboratories; Mumbai, India) according to Clinical Laboratory Standards Institute (CLSI) guidelines (35). A panel of 11 antimicrobials were used from BD BBL Sensi-Discs: ciprofloxacin (CIP) (5 μg), nalidixic acid (NA) (30 μg), erythromycin (E) (15 μg), azithromycin (AZM) (15 μg), tetracycline (Te) (30 μg), gentamicin (GM) (10 μg), ampicillin (AM) (10 μg), amoxicillin-clavulanic acid (AMC) (10 μg), chloramphenicol (C) (30 μg), and trimethoprim-sulfamethoxazole (SXT) (25 μg), and amikacin (AN) (30 μg). Isolates were defined as multidrug-resistant (MDR) when resistant to three or more classes of antimicrobials (36).
Data Analysis and Ethical Approval
Data were analyzed using SPSS v25 (SPSS Inc, Chicago, Illinois, USA). Descriptive statistics were calculated for Campylobacter species prevalence, antimicrobial susceptibility and household demographic data. Chi-squared and Fisher's tests were used as appropriate to compare differences in the prevalence and antimicrobial resistance profiles between Campylobacter species isolates, and between strains isolated from different sources. Potential risk factors associated with the occurrence of Campylobacter in human samples were analyzed using univariable logistic regression. The outcome measure used was a binary variable reflecting whether a household was PCR positive for Campylobacter. Variables were checked for collinearity, and a backward-stepwise process was used, with covariates remaining in the model if they were statistically significant (p < 0.05). All variables that showed an association with the outcome variable on univariable analysis (p < 0.2) were considered in the final multivariable logistic regression analysis. Ethical approval was obtained (DERC/18/19/10-A) from the Research Ethics Review Committee (RERC) of the Department of Microbiology, Immunology and Parasitology (DMIP), College of Health Sciences, Addis Ababa University. Permission was obtained from the Addis Ababa Bureau of Agriculture and Livestock and Addis Abba Bureau of Health. Written informed consent was obtained prior to enrolment from the head of each household for participation in this study.
Results
Prevalence of Campylobacter Species
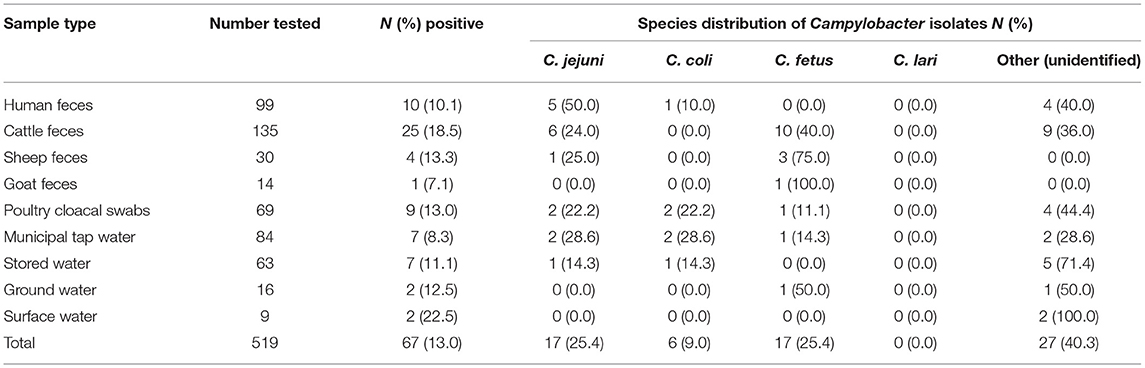
From the 99 households, 42 (42.4%) households were positive for Campylobacter in at least one of the samples tested. Of the 519 samples (347 human and animal fecal samples, and 172 water samples) from these 99 households, 67 (13.0%) were positive for Campylobacter species. The prevalence of Campylobacter species across the different samples was: human, 10 (10.1%), animal, 39 (15.7%), and water, 18 (10.5%) (Table 1). The prevalence of Campylobacter species was highest in surface water samples (22.5%), followed by cattle feces (18.5%), and poultry cloacal swabs (13.0%); with the lowest prevalence in goat feces (7.1%) and municipal tap water (8.3%) (Table 1).
The dominant Campylobacter species isolated in this study were C. jejuni (25.4%), C. fetus (25.4%), and C. coli (9.0%). Twenty-seven isolates were categorized as unidentified Campylobacter species (Table 1). C. jejuni and C. coli were predominately isolated from cattle and water samples, respectively. Of the 10 Campylobacter species isolated from human stool samples, 5 (50%) were C. jejuni, and 1 (10%) was C. coli, with the remaining 4 (40%) unidentified species. Most isolates obtained from human stool samples were C. jejuni. C. fetus and C. jejuni were the dominant species isolated from cattle feces, with C. fetus being the dominant species isolated from sheep feces. From the nine Campylobacter isolates obtained from poultry cloacal swabs, C. jejuni and C. coli were the dominant species (Table 1). Ten Campylobacter species isolated from human feces had co-occurrence in other samples from the same household (Table 2).
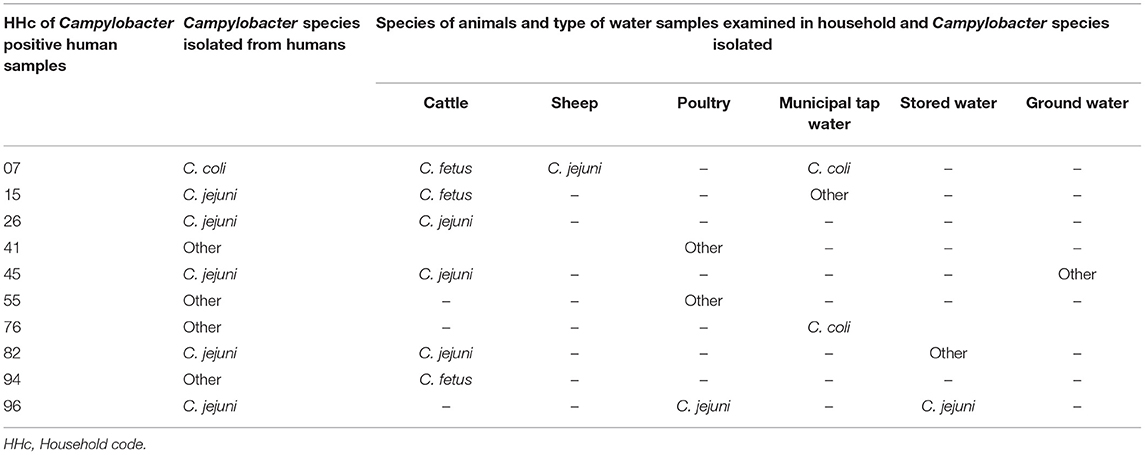
Table 2. Co-occurrence of similar Campylobacter species in human, animals, and water samples within households.
Univariable logistic regression identified 11 risk factors to be associated (p < 0.2) with human Campylobacter species positivity (Table 3). Multivariable logistic regression found no association between human Campylobacter species positivity and self-reported gastrointestinal disease symptoms (Table 4). The collecting of manure indoors and outdoors was significantly associated with human Campylobacter species positivity (p = 0.026; OR: 38.24; 95% CI: 1.54–951.97). The washing of hands with soap before and after cooking, and taking any action to protect oneself while cleaning animal houses were associated with decreased odds of human Campylobacter species positivity (Table 4). In addition, the odds of human Campylobacter species positivity were 28.87 times less in households who did not use stored water for either drinking or food preparation (p = 0.037; OR: 28.87; 95% CI: 1.23–679.70) (Table 4). The multivariable analysis also identified that animal species (cattle and poultry) Campylobacter positivity was associated with human Campylobacter positivity (Table 4).
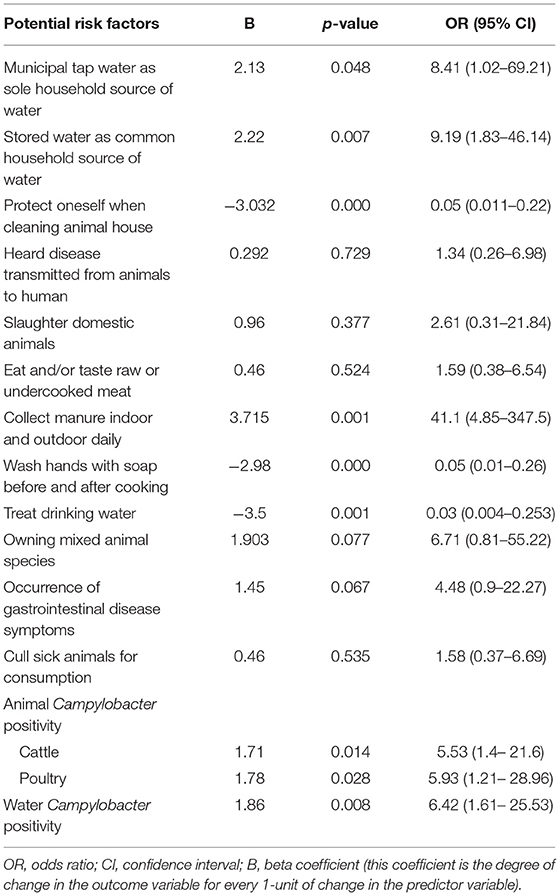
Table 3. Univariable logistic regression analysis of potential risk factors for human Campylobacter species positivity.
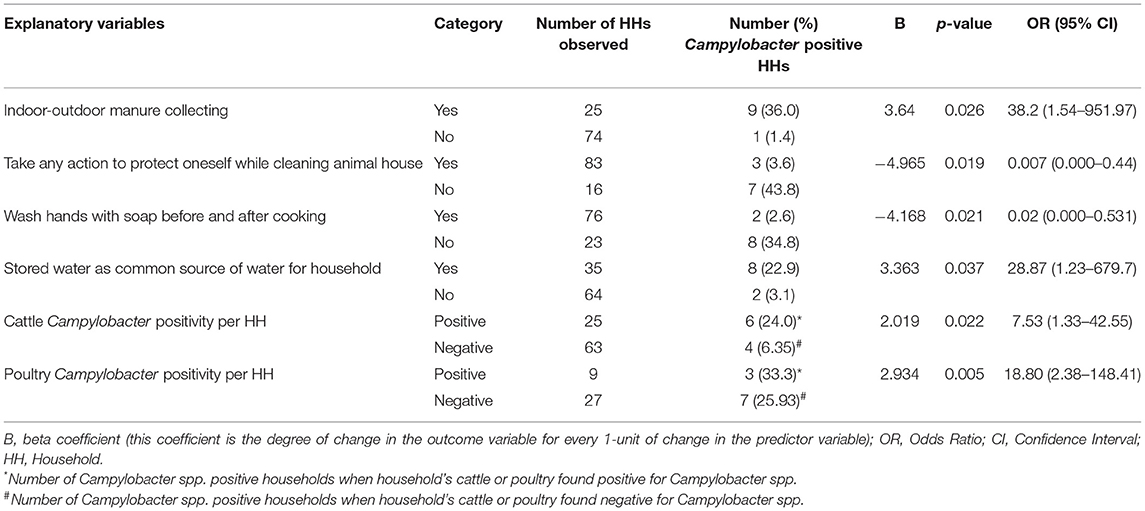
Table 4. Multivariable logistic regression analysis of significantly associated explanatory variables for human Campylobacter species positivity.
Antimicrobial Susceptibility Profiles of Campylobacter Isolates
Over 98% of the isolates were resistant to one or more antimicrobial agents. With the exception of Campylobacter isolates from cattle, where 96% showed resistance to one or more tested antimicrobials, all isolates from humans, sheep, goats, poultry, and water were resistant to at least one of the tested antimicrobials (Table 5). Most isolates were resistant to amikacin (79.1%) followed by amoxicillin-clavulanic acid (70.1%), tetracycline (67.2%), ampicillin (64.2%), and Nalidixic acid (64.2%) (Table 5). A single isolate was found to be resistant to only erythromycin and amikacin, and only one isolate was resistant to only erythromycin, azithromycin and amikacin (Table 5).
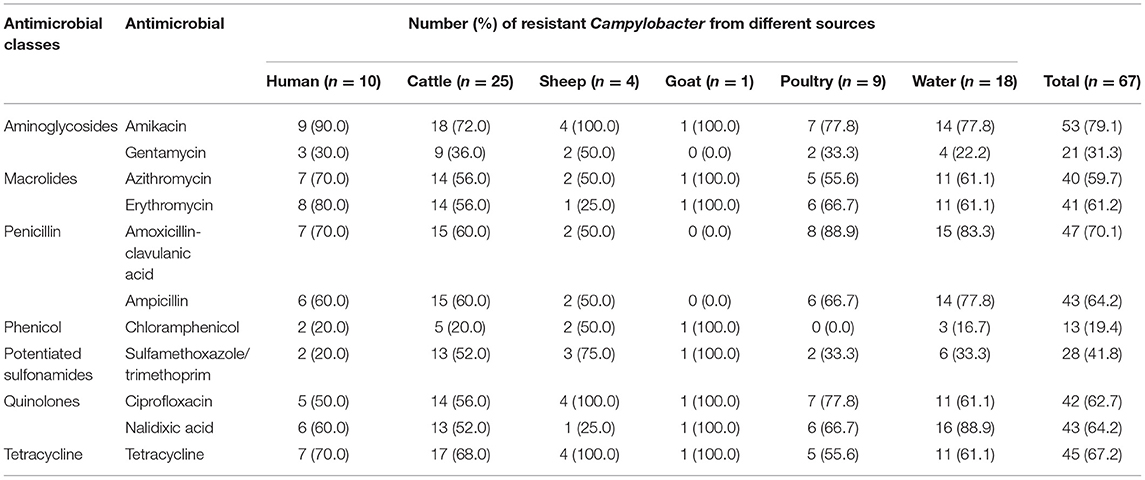
Table 5. Antimicrobial susceptibility profiles of Campylobacter species isolated from various sources.
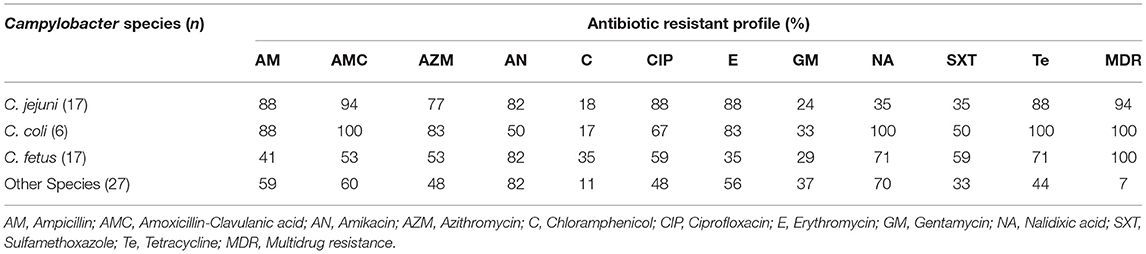
The resistance to individual antimicrobials across Campylobacter species ranging from 11.1 to 100.0% (Table 6). C. coli exhibited wider resistance to most of the tested antimicrobials than C. jejuni and other isolates. There was a significant difference in the resistance of C. jejuni and C. fetus to nalidixic acid, amoxicillin-clavulanic acid, erythromycin, and ampicillin (p < 0.05). However, no statistically significant difference was observed for the other antimicrobials across the different Campylobacter species. All poultry isolates were susceptible to chloramphenicol, with one unidentified Campylobacter species isolate from cattle pan-susceptible, and one C. coli isolate from water resistant to all tested antimicrobials.
Sixty-four (95.5%) of the isolates were found to be resistant to three or more antimicrobial classes, and were thus considered multidrug resistant. They were observed across the following sources: humans, 8 (12.5%); cattle, 24 (37.5%); sheep, 4 (6.3%); goats, 1 (1.6%); poultry, 9 (14.1%); and water, 18 (28.1%) (Table 7). Isolates from humans, cattle and poultry were resistant to 3–6 antimicrobial classes, whereas isolates from water and sheep were resistant to 3–7 and 4–7 antimicrobial classes, respectively. A single isolate from goats was resistant to six different antimicrobial classes.
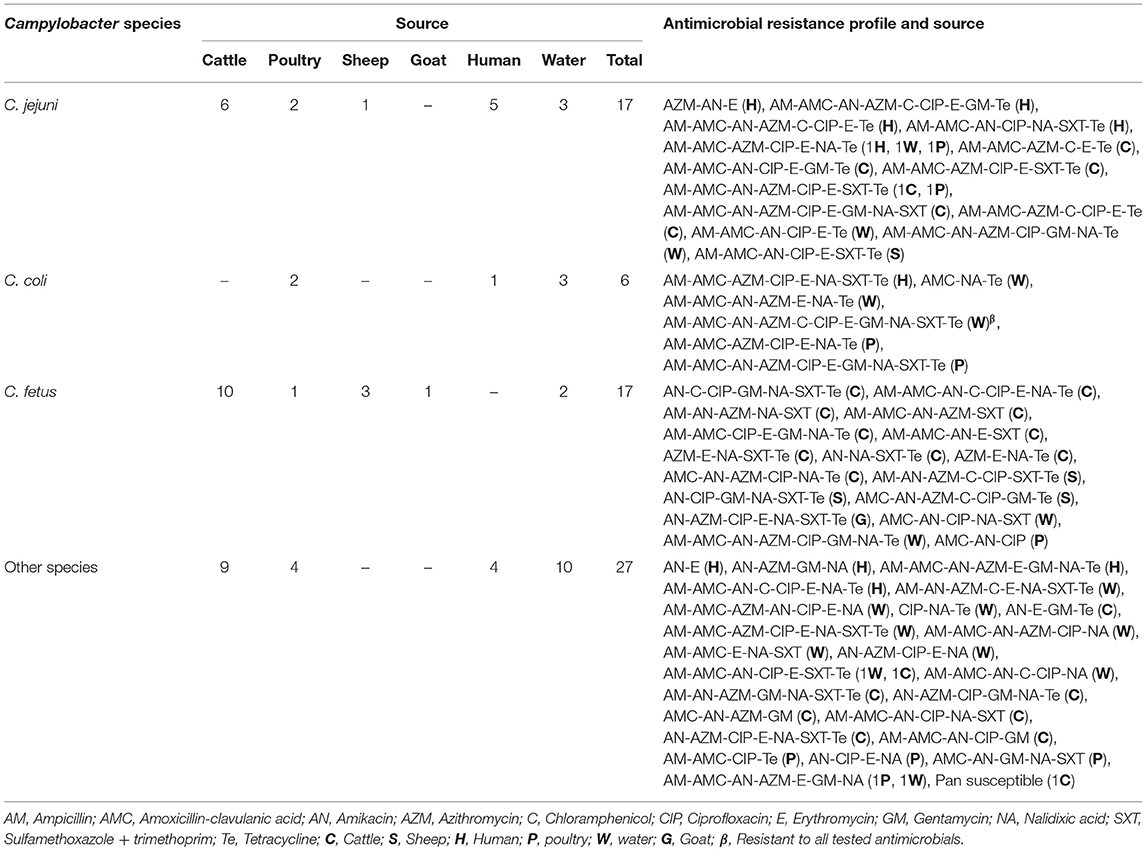
Table 7. Antimicrobial resistance profiles of Campylobacter species isolated from animals, humans, and water in peri-urban Addis Ababa, Ethiopia.
Regardless of the source, all C. coli and C. fetus isolates demonstrated a multidrug resistance profile, whereas a multidrug resistance profile was observed in 94.1% (16/17) of C. jejuni and 92.6% (25/27) of unidentified Campylobacter species (Table 6). Fifty-four different multidrug resistance profiles, ranging from 3 to 7 different antimicrobial classes were observed (Table 7). The most common multidrug resistance profile in isolates was AM-AMC-AZM-CIP-E-NA-Te (7.4%), followed by AM-AMC-AZM-AN-CIP-E-SXT-Te (5.6%), and AM-AMC-AN-CIP-E-SXT-Te (5.6%). Resistance to macrolides (erythromycin and azithromycin), was the most dominant antimicrobial class appearing in 74.1% (37) of the fifty-four observed multidrug resistance patterns (Table 7).
Discussion
Of the 99 households in this study, 42.4% were positive for Campylobacter species in at least one of the household samples tested. The prevalence of Campylobacter species in humans in our study, was similar to a number of previous studies in Ethiopia (20, 38), Poland (14), and Tanzania (39). In contrast, studies in Ethiopia by Terefe et al. (22), Tafa et al. (23), and Lengerh et al. (40) reported higher prevalences than our study. A higher prevalence was also reported by Chuma et al. (41) in Tanzania, and Schiaffino et al. (36) in Peru, whereas Rawat et al. (37) and Meistere et al. (42) reported a lower prevalence in Latvia and India. The variation in prevalence across the studies might be due to differences in study methodologies, periodical and environmental variations, and differences in the study groups (14, 42).
Campylobacter species prevalence observed in cattle in this study is consistent with a previous Ethiopian study (43). However, it is a higher prevalence than reported by Hagos et al. (4) and Kassa et al. (25) in Ethiopia, and a lower prevalence than reported by Abamecha et al. (44) in Ethiopia. Comparable results were also reported from the Republic of Korea (45) and Latvia (42). Prevalence in our study was higher than those reported from Iran (46), and Ghana, but lower than reports from Republic of Korea (47), and the USA (48). The prevalence of Campylobacter species observed in poultry in this study is consistent with the report from Latvia (42), but lower than previous studies in Ethiopia (12, 25), Tanzania (41), Kenya (49), and Cambodia (15). However, the prevalence in our study is higher than the reports by Pires et al. (50) from California, USA, and Rawat et al. (37) from India.
The variation in prevalence of Campylobacter species in animals across these studies might be explained by the differences in study methodology and duration, seasonality, animal management system, sanitary practices, and agro-ecological variations (15, 41). Certain studies, like Hagos et al. (4), reported a prevalence from meat products rather than live animals. Our study was a community level study, utilizing a pooled sampling approach, where all sampling units were asymptomatic at the time of sample collection. In contrast, many previous studies conducted in Ethiopia were based on symptomatic human subjects, and/or farm-level animal studies that potentially resulted in the observed variation in prevalence.
Our study utilized both a culture and molecular approach for the isolation and identification of Campylobacter species. The majority of previous studies in Ethiopia used only the culture and biochemical test approach for the isolation and identification of Campylobacter. This latter approach has the potential to be less sensitive and specific, and could be a reason for the observed differences in prevalence (51–53). Nevertheless, the higher prevalence of zoonotic thermophilic Campylobacter species observed in livestock in our study is of considerable concern, as poultry and other livestock species move freely around the household, contaminating the environment, and are a source of infection for humans, especially children.
Of all the sample types in our study, the prevalence of Campylobacter species was highest in surface water samples (22.5%). The overall prevalence of Campylobacter species observed in all water types in our study, is consistent with a study from Turkey, but lower than studies from South Africa (7) and Poland (14). The variation across studies might be attributed to differences in the sources of water, water collection approach, season, geographical location, and method of isolation. In addition, our study area is impacted by both agricultural operations (cattle, poultry, and vegetable production), and urban wastewater treatment, which may influence the prevalence of Campylobacter and other microorganisms (31, 54). Although the contribution of water to the burden of Campylobacter infection in humans might be unknown (55), the results of our study provide information regarding the potential transmission of Campylobacter species between animals, humans and water sources. More importantly, our study revealed a high prevalence of thermophilic Campylobacter species in municipal tap water, indicating a potential risk for human infection. However, this result should be interpreted with caution, as no information was obtained on whether the source water was treated. The significantly higher isolation of Campylobacter from households using stored water, is consistent with previous studies (7, 56). Studies have shown that untreated stored water is a significant source of Campylobacter infections and outbreaks (16, 55). Risk factors including the source of water and type of storage container have been linked to the poor microbial quality of stored household water (56).
In our study, C. jejuni was the most frequently isolated species from all sample types, except for cattle and poultry isolates. This is consistent with previous studies in Ethiopia (22, 25) and other countries, including South Africa and California, USA (7, 50). C. jejuni is more prevalent, and has a longer viability in the environment compared to other thermophilic Campylobacter species, therefore increasing its chance of recovery (33, 50). This difference could also occur due to variations in the mechanism of pathogenesis and elimination amongst the different thermophilic Campylobacter species within the host cells (14). Differences in the isolation of Campylobacter species might also be related to their actual compositional variations in local environments. C. lari was not recovered in our study, consistent with previous studies in Ethiopia (22, 28, 33, 50). As demonstrated in other studies (7, 12, 39), our study also highlighted that Campylobacter species isolated from human feces also co-occurred in other samples (predominately cattle, poultry, and water) from the same household, suggesting that cattle, water and poultry are potentially the main sources of human Campylobacter infection. It is also possible, that humans could have infected the cattle, poultry and stored water.
The association of indoor and outdoor manure collecting with increased odds of human Campylobacter species positivity is consistent with previous studies (15, 47). Campylobacter can survive at variable rates in stored manure, and even in composted manure [reviewed in (50, 57)]. Consequently, human exposure to Campylobacter species may occur when drinking or ingesting of contaminated products (47). The negative association of taking any specific action to protect oneself while cleaning an animal house, or washing hands with soap before and after cooking, with Campylobacter species positivity is similar to a previous study (15). Poor hygiene and sanitation are associated with increased odds of multiple adverse health outcomes (58). However, our study indicated that there was no significant association between the consumption of raw or under cooked meat, and owning different animal species with human Campylobacter species positivity.
Nearly all isolates in our study were resistant to one or more antimicrobials. Most isolates were resistant to amikacin (79.1%), followed by amoxicillin-clavulanic acid (70.1%), whereas a lower resistance was observed to chloramphenicol (19.4%), followed by gentamicin (31.3%). Similar results have been reported previously in Ethiopia (59) and Iran (60). The level of resistance to nalidixic acid (64.2%) and ciprofloxacin (62.7%) observed in our study are consistent with other studies in Ethiopia (44), Poland (61), and Kenya (49). Resistance to erythromycin (61.2%) and azithromycin (59.7%) observed in this study, is consistent with results found in other previous studies (33, 44, 60), but higher than other studies (14, 23, 40). In our study, resistance observed to tetracycline (67.2%), ampicillin (64.2%), and sulfamethoxazole-trimethoprim (41.8%) is consistent with previous studies (7, 40), but lower resistance has been previously reported (25, 37).
In our study, C. coli was highly resistant to nalidixic acid, amoxicillin-clavulanic acid, azithromycin, and ampicillin, compared to other species. Whereas, C. jejuni showed relatively higher resistance to erythromycin, ciprofloxacin and amikacin than other species. These differences could be due to greater use of these drugs in humans, and that humans are commonly infected with these species of Campylobacter (62). This is consistent with studies from Ethiopia (13), South Africa (7, 63), Tunisia (64), but differs from other studies in Ethiopia (59), Egypt (65), and England (66).
The high number of multidrug resistant Campylobacter isolates observed in our study is consistent with previous studies (7, 44). However, lower antimicrobial resistance has been previously reported in Ethiopia (25), Iran (46), and Tanzania (33). One potential hypothesis for the high resistance exhibited by C. coli, compared to other Campylobacter species, may be that C. coli strains can acquire resistance genes horizontally more effectively than other species, and chromosomally encoded target genes could mutate faster in C. coli (67, 68). However, we need to be mindful about our interpretation, given the low number of C. coli isolates in our study.
The higher antimicrobial resistance observed in our study may potentially be due to the overuse, and inappropriate use, of antimicrobial agents in both human and animals. To our knowledge, oxytetracycline and penicillin (alone or in combination with streptomycin) are used widely in livestock and poultry in Ethiopia. This use contributes to increased selection of resistant Campylobacter species (69, 70). Extensive use of antimicrobials in the animal industry for prophylaxis, and growth promotion, has been associated with increased resistance (14, 65, 71). Due to the close human and animal interaction in our study area, there is the potential for antimicrobial resistant strains, or resistant genetic markers originating from humans, to be transmitted to animals. The differences in the isolation of Campylobacter species across the different sources, may make it challenging to compare the levels of resistance between the sources. Nonetheless, observed resistance discrepancies in our study, compared to previous studies, could be due to differences in exposure rates of the bacteria to the different antimicrobials. The inevitable human-animal-environment interaction in our study, along with inappropriate use of antimicrobials in both animals and humans in Ethiopia, might potentially lead to increased selection pressures for resistant strains.
The challenges presented by infectious diseases and antimicrobial resistance are multifaceted, and it is critically important to address these issues using a One Health approach (72, 73). One Health interventions advocate close intersectoral cooperation, interdisciplinary expertise, and the involvement, and empowerment of multiple stakeholders (74, 75). Our study demonstrated that C. jejuni, C. coli, C. fetus, and unidentified Campylobacter species were prevalent in the study area. The relatively high proportion of Campylobacter species in both livestock and water samples are potential risks for human Campylobacter infection. The high prevalence of Campylobacter species isolated from various water sources, highlights the need for further work to identify for how long Campylobacter species persist in these water sources, and to ascertain the transmission of Campylobacter within the environment. Multi-drug resistant zoonotic Campylobacter species were prevalent in animals, humans and environment in the peri-urban livestock owning households of Addis Ababa, Ethiopia. Recognizing the significant implications of antimicrobial resistance, it is important for Ethiopia to develop and implement a national plan to advance the rational use of antimicrobials utilizing a One Health approach. A One Health approach is recommended to further investigate Campylobacter species infections, and other zoonotic infectious agents, in the livestock owning populations in Ethiopia, where there is close interaction between humans, animals and the environment.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Research Ethics Review Committee (RERC) of the Department of Microbiology, Immunology and Parasitology (DMIP), College of Health Sciences, Addis Ababa University (DERC/18/19/10-A). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin. The animal study was reviewed and approved by the Research Ethics Review Committee (RERC) of the Department of Microbiology, Immunology and Parasitology (DMIP), College of Health Sciences, Addis Ababa University (DERC/18/19/10-A). Permission was obtained from the Addis Ababa Bureau of Agriculture and Livestock and Addis Abba Bureau of Health. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
GC, TE, FA, DA, and AS conceived and designed the study and designed the survey. GC led the sample collection and survey implementation. GC, TE, and DA supervised the laboratory analysis. GC analyzed the data, with supervision from AS and TE. GC and AS wrote the manuscript. All authors contributed to the revision of the manuscript.
Funding
Funding provided by North Carolina State University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge all the livestock owning households who participated in the study, and the Animal Health Assistants of the study woredas for their cooperation and participation. We are also very grateful for the kind cooperation and technical assistance received from the Aklilu Lemma Institute of Pathobiology (ALIPB), Addis Ababa University, especially Mr. Haile Alemayehu and Azeb Teklu for their laboratory technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.750551/full#supplementary-material
References
1. Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. (2015) 12:e1001923. doi: 10.1371/journal.pmed.1001923
2. World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007-2015. (2015). Available online at: https://apps.who.int/iris/handle/10665/199350
3. Whiley H, van den AB, Giglio S, Bentham R. The role of environmental reservoirs in human Campylobacteriosis. Int J Envi Res Public Health. (2013) 10:5886–907. doi: 10.3390/ijerph10115886
4. Hagos Y, Gugsa G, Awol N, Ahmed M, Tsegaye Y, Abebe N, et al. Isolation, identification, and antimicrobial susceptibility pattern of Campylobacter jejuni and Campylobacter coli from cattle, goat, and chicken meats in Mekelle, Ethiopia. PLoS ONE. (2021) 16:e0246755. doi: 10.1371/journal.pone.0246755
5. Bolton DJ. Campylobacter virulence and survival factors. Food Microbiol. (2015) 48:99–108. doi: 10.1016/j.fm.2014.11.017
6. Parte AC, SardàCarbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syste Evol Microbiol. (2020) 70:5607–12. doi: 10.1099/ijsem.0.004332
7. Chukwu MO, Abia ALK, Ubomba-Jaswa E, Obi L, Dewar JB. Characterization and phylogenetic analysis of Campylobacter species isolated from pediatric stool and water samples in the Northwest province, South Africa. Int J Environ Res Public Health. (2019) 16:2205. doi: 10.3390/ijerph16122205
8. FrancËois R, Yori PP, Rouhani S, Siguas SM, Paredes OM, Trigoso DR, et al. The other Campylobacters: not innocent bystanders in endemic diarrhea and dysentery in children in low-income settings. PLoS Negl Trop Dis. (2018) 12:e0006200. doi: 10.1371/journal.pntd.0006200
9. Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. Global epidemiology of campylobacter infection. Clin Microbiol Rev. (2015) 28:687–720. doi: 10.1128/CMR.00006-15
10. Budge S, Barnett M, Hutchings P, Parker A, Tyrrel S, Hassard F, et al. Risk factors and transmission pathways associated with infant Campylobacter spp. prevalence and malnutrition: a formative study in rural Ethiopia. PLoS ONE. (2020) 15:e0232541. doi: 10.1371/journal.pone.0232541
11. Signorini ML, Rossler E, Di'az David DC, Olivero CR, Romero-Scharpen A, Soto LP, et al. Antimicrobial resistance of thermotolerant Campylobacter species isolated from humans, food-producing animals, and products of animal origin: a worldwide meta-analysis. Microbiol Drug Resist. (2018) 24:1174–90. doi: 10.1089/mdr.2017.0310
12. Brena CM, Mekonnen Y, Bettridge MJ, Williams JN, Wigley P, Tessema TS, et al. Changing risk of environmental Campylobacter exposure with emerging poultry production systems in Ethiopia. Epidemiol Infect. (2016) 144:567–75. doi: 10.1017/S0950268815001429
13. Chen D, McKune SL, Singh N, Yousuf HJ, Gebreyes W, Manary MJ, et al. Campylobacter colonization, environmental enteric dysfunction, stunting, and associated risk factors among young children in rural Ethiopia: a cross-sectional study from the campylobacter genomics and environmental enteric dysfunction (CAGED) project. Front Public Health. (2021) 8:615793. doi: 10.3389/fpubh.2020.615793
14. Szczepanska B, Andrzejewska M, Spica D, Klawe JJ. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. (2017) 17:80. doi: 10.1186/s12866-017-0991-9
15. Osbjer K, Tano E, Chhayheng L, Mac-Kwashie AO, Fernstr€om LL, Davun H, et al. Detection of Campylobacter in human and animal field samples in Cambodia. Act Pathol Microbiol ET Immunol Scand. (2016) 124:508–15. doi: 10.1111/apm.12531
16. Nilsson A, Johansson C, Skarp A, Kaden R, Bertilsson S, Rautelin H. Survival of Campylobacter jejuni and Campylobacter coli water isolates in lake and well water. APMIS. (2018) 126:762–70. doi: 10.1111/apm.12879
17. World Health Organization. The Global View of Campylobacteriosis: Report of an Expert Consultation. Utrecht (2013).
18. Abubakar MK, Muigai AWT, Ndung'u P, Kariuki S. Investigating carriage, contamination, antimicrobial resistance and assessment of colonization risk factors of Campylobacter species in broilers from selected farms in Thika, Kenya. J Microbiol Res Int. (2019) 27:1–16. doi: 10.9734/mrji/2019/v27i630119
19. Sproston EL, Wimalarathna HML, Sheppard SK. Trends in fluoroquinolone resistance in Campylobacter. Microb Genom. (2018) 4:e000198. doi: 10.1099/mgen.0.000198
20. Ewnetu D, Mihret A. Prevalence and antimicrobial resistance of Campylobacter isolates from humans and poultry in Bahir Dar, Ethiopia. Foodborne Pathog Dis. (2010) 7:667–70. doi: 10.1089/fpd.2009.0433
21. Rousham EK, Unicomb L, Islam MA. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioral, epidemiological and One Health approaches. Proc R Soc B. (2018) 285:20180332. doi: 10.1098/rspb.2018.0332
22. Terefe Y, Deblais L, Ghanem M, Helmy YA, Mummed B, Chen D, et al. Co-occurrence of Campylobacter species in children from eastern Ethiopia, and their association with environmental enteric dysfunction, diarrhea, and host microbiome. Front Public Health. (2020) 8:99. doi: 10.3389/fpubh.2020.00099
23. Tafa B, Sewunet T, Tassew H, Asrat D. Isolation and antimicrobial susceptibility patterns of Campylobacter species among diarrheic children at Jimma, Ethiopia. Int J Bacterio. (2014) 2014:560617. doi: 10.1155/2014/560617
24. Chanyalew Y, Asrat D, Amavisit P, Loongyai W. Prevalence and antimicrobial susceptibility of thermophilic campylobacter isolated from sheep at Debre Birhan, North-Shoa, Ethiopia. Kasetsart J Nat Sci. (2013) 47:551–60.
25. Kassa T, Gebre-Selassie S, Asrat D. Antimicrobial susceptibility patterns of thermotolerant Campylobacter strains isolated from food animals in Ethiopia. Vet. Microbiol. (2007) 119:82–7. doi: 10.1016/j.vetmic.2006.08.011
26. Gedlu E, Assefa A. Campylobacter enteritis among children in Northwest Ethiopia: a one-year prospective study. Ann Trop Paediat. (1999) 16:207–12. doi: 10.1080/02724936.1996.11747828
27. Dadi L, Asrat D. Prevalence and antimicrobial susceptibility of thermo tolerant Campylobacter strains in retail raw meat products in Ethiopia. Ethiop J Health. Dev. (2008) 22:195–6. doi: 10.4314/ejhd.v22i2.10072
28. Asrat D, Hathaway A, Ekwall E. Studies on enteric Campylobacteriosis in Tikur Anbessa and Ethio-Swedish children's hospital, Addis Ababa, Ethiopia. Ethiop. Med J. (1999) 37:71–84.
29. Agajie LB, Jemal HA, Solomon H. Frequency of diarrheal attack and its predictors among infants and young children from Akaki Kality Sub-city: “A Community Based Study”. EC Paediat. (2020) 9:1–9.
30. Hassen S, Haidar J, Bogale LA. Occurrence of diarrhea and utilization of zinc bundled with ORS among caregivers of children less than five years in Addis Ababa, Ethiopia. J Public Health Epidemiol. (2018) 9:348–55. doi: 10.5897/JPHE2018.1029
31. Gebremichael D, Gebremichael AT, Worku A, Abshare MW, Habte Mariam YM, Balcha G. Building Urban Resilience: Assessing Urban Peri-urban Agriculture in Addis Ababa, Ethiopia. Padgham J, Jabbour J, editor. United Nations Environment Programme (UNEP), Nairobi (2014). Available online at: www.start.org/upa/addis_ababa.pdf (accessed September 5, 2020).
32. Jokinen CC, Koot JM, Carrillo CD, Victor PJ, Gannon VPJ, Jardine CM, et al. An enhanced technique combining pre-enrichment and passive filtration increases the isolation efficiency of Campylobacter jejuni and Campylobacter coli from water and animal fecal samples. J Microbiol Metho. (2012) 91:506–13. doi: 10.1016/j.mimet.2012.09.005
33. Kashoma IP, Kassem II, John J, Kessy BM, Gebreyes W, Kazwala RR, et al. Prevalence and antimicrobial resistance of campylobacter isolated from dressed beef carcasses and raw milk in Tanzania. Microbiol. Drug Resist. (2016) 22:1. doi: 10.1089/mdr.2015.0079
34. Yamazaki-Matsune W, Taguchi M, Seto K, Kawahara R, Kawatsu K, Kumeda Y, et al. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subspecies hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J Med Microbiol. (2007) 56:1467–73. doi: 10.1099/jmm.0.47363-0
35. Clinical Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 3rd ed. CLSI Guideline M45. Wayne, PA (2016). Available online at: https://clsi.org/media/m45ed3_sample.pdf (accessed March 23, 2020).
36. Schiaffino F, Colston JM, Paredes-Olortegui M, François R, Burga PNR, et al. Antibiotic resistance of Campylobacter species in a pediatric cohort study. Antimicrob Agent Chemother. (2019) 63:e01911–8. doi: 10.1128/AAC.01911-18
37. Rawat N, Maansi KD, Upadhyay AK. Virulence typing and antibiotic susceptibility profiling of thermophilic Campylobacters isolated from poultry, animal, and human species. Vet. World. (2018) 11:1698–705. doi: 10.14202/vetworld.2018.1698-1705
38. Beyene G, Haile-Amlak A. Antimicrobial sensitivity pattern of Campylobacter species among children in Jimma University Specialized Hospital, southwest Ethiopia. Ethiop J Health Dev. (2004) 3:185–9. doi: 10.4314/ejhd.v18i3.9958
39. Komba EV, Mdegela RH, Msoffe PL, Nielsen LN, Ingmer H. Prevalence, antimicrobial resistance and risk factors for thermophilic Campylobacter infections in symptomatic and asymptomatic humans in Tanzania. Zoonoses Public Health. (2015) 62:557–68. doi: 10.1111/zph.12185
40. Lengerh A, Moges F, Unakal C, Anagaw B. Prevalence, associated risk factors and antimicrobial susceptibility pattern of Campylobacter species among under five diarrheic children at Gondar University Hospital, Northwest Ethiopia. BMC Paediat. (2013) 13:1–9. doi: 10.1186/1471-2431-13-82
41. Chuma IS, Nonga HE, Mdegela RH, Kazwala RR. Epidemiology and RAPD-PCR typing of thermophilic Campylobacters from children under five years and poultry in Morogoro Municipality, Tanzania. BMC Infect Dis. (2016) 16:692. doi: 10.1186/s12879-016-2031-z
42. Meistere I, Kibilds J, Eglite L, Alksne L, Cibrovska AJA, Makarova S, et al. Campylobacter species prevalence, characterization of antimicrobial resistance (AMR) and analysis of whole-genome sequence (WGS) of isolates from livestock and humans, Latvia, 2008 to 2016. Euro Surveill. (2019) 24:1800357. doi: 10.2807/1560-7917.ES.2019.24.31.1800357
43. Seleshe N, Abebe M, Reta T, Legesse G. Prevalence and drug sensitivity pattern of Campylobacter jejuni isolated from cattle and poultry in and around Gondar Town, Ethiopia. Glob Vet. (2015) 14:43–7. doi: 10.5829/idosi.gv.2015.14.01.9238
44. Abamecha A, Assebe G, Tafa B, Beyene W. Prevalence of thermophilic Campylobacter and their antimicrobial resistance profile in food animals in Lare District, Nuer Zone, Gambella, Ethiopia. J Drug Res Dev. (2015) 1:2. doi: 10.16966/2470-1009.108
45. Kim JS, Lee MY, Kim SJ, Jeon SE, Cha I, Hong S, et al. High-level ciprofloxacin-resistant Campylobacter jejuni Isolates circulating in humans and animals in Incheon, Republic of Korea. Zoonoses Public Health. (2015) 63:545–54. doi: 10.1111/zph.12262
46. Rahimi E, Alipoor-Amroabadi M, Faham K. Investigation of prevalence of thermotolerant Campylobacter species in livestock feces. Can J Anim Sci. (2017) 97:207–13. doi: 10.1139/cjas-2015-0166
47. An J, Ho H, Kim J, Kim W, Kim J, Lee S, et al. Dairy cattle, a potential reservoir of human Campylobacteriosis: epidemiological and molecular characterization of Campylobacter jejuni from cattle farms. Front Microbiol. (2018) 9:3136. doi: 10.3389/fmicb.2018.03136
48. Tang Y, Sahin O, Pavlovic N, LeJeune J, Carlson J, Wu Z, et al. Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci Rep. (2017) 7:494. doi: 10.1038/s41598-017-00584-z
49. Nguyen TNM, Hotzel H, Njeru J, Mwituria J, El-Adawy H, Tomaso H, et al. Antimicrobial resistance of Campylobacter isolates from small scale and backyard poultry in Kenya. Gut Pathog. (2016) 8:39. doi: 10.1186/s13099-016-0121-5
50. Pires AFA, Patterson L, Kukielka EA, Aminabadi P, Navarro-Gonzalez N, Jay-Russell MT. Prevalence and risk factors associated with Campylobacter species and Salmonella enterica in livestock raised on diversified small-scale farms in California. Epidemiol Infect. (2019) 147:e321. doi: 10.1017/S095026881900205X
51. Ahmed R, León-Velarde CG, Odumeru JA. Evaluation of novel agars for the enumeration of Campylobacter spp. in poultry retail samples. J Microbiol Methods. (2012) 88:304–10. doi: 10.1016/j.mimet.2011.12.011
52. Nachamkin I, Nguyen P. Isolation of Campylobacter species from stool samples by use of a filtration method: assessment from a United States-based population. J Clin Microbiol. (2017) 55:2204–7. doi: 10.1128/JCM.00332-17
53. Buss JE, Cresse M, Doyle S, Buchan BW, Craft DW, Young S. Campylobacter culture fails to correctly detect Campylobacter in 30% of positive patient stool specimens compared to non-cultural methods. Eur J Clin Microbiol Infect Dis. (2019) 38:1087–93. doi: 10.1007/s10096-019-03499-x
54. Yohannes H, Elias E. Contamination of rivers and water reservoirs in and around Addis Ababa City and actions to combat it: review. Environ Pollut Climate Change. (2017) 1:116. doi: 10.4172/2753-458X.1000116
55. Pitkänen T. Review of Campylobacter species in drinking and environmental waters. J Microbiol Methods. (2013) 95:39–47. doi: 10.1016/j.mimet.2013.06.008
56. Elfadaly HA, Hassanain NA, Hassanain MA, Barakat AM, Shaapan RM. Evaluation of primitive ground water supplies as a risk factor for the development of major waterborne zoonosis in Egyptian children living in rural areas. J Infect Public Health. (2017) 11:203–8. doi: 10.1016/j.jiph.2017.07.025
57. Hald B, Skov MN, Nielsen EM, Rahbek C, Madsen JJ, Wainø M, et al. Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Vet Scand. (2016) 58:11. doi: 10.1186/s13028-016-0192-9
58. Mbuya MNN, Humphrey JH. Preventing environmental enteric dysfunction through improved water, sanitation and hygiene: an opportunity for stunting reduction in developing countries. Matern Child Nutr. (2016) 12:106–20. doi: 10.1111/mcn.12220
59. Girum F. Identification of Campylobacter species and their antibiotic resistance patterns from raw bovine meat in Addis Ababa, Ethiopia. Int J Microbiol Immunol Res. (2015) 4:1–5.
60. Divsalar G, Kaboosi H, Khoshbakht R, Shirzad-Aski H, Ghadikolaii FP. Antimicrobial resistances, and molecular typing of Campylobacter jejuni isolates, separated from food-producing animals and diarrhea patients in Iran. Comp Immunol Microbiol Infect Dis. (2019) 65:194–200. doi: 10.1016/j.cimid.2019.06.001
61. Wieczorek K, Wołkowicz T, Osek J. flaA-SVR based genetic diversity of multiresistant Campylobacter jejuni Isolated from poultry and humans. Front Microbiol. (2019) 10:1176. doi: 10.3389/fmicb.2019.01176
62. Hlashwayo DF, Sigaúque B, Bila CG. Epidemiology and antimicrobial resistance of Campylobacter species in animals in Sub-Saharan Africa: a systematic review. Heliyon. (2020) 6:e03537. doi: 10.1016/j.heliyon.2020.e03537
63. Igwaran A, Okoh AI. Campylobacteriosis agents in meat carcasses collected from two district municipalities in the Eastern Cape Province, South Africa. Foods. (2020) 9:203. doi: 10.3390/foods9020203
64. Gharbi M., Béjaoui A, Hamda CB, Jouini A, Ghedira K, Zrelli C, et al. Prevalence and antibiotic resistance patterns of Campylobacter species isolated from Broiler Poultry in the North of Tunisia. BioMed Res Inter. (2017) 2018:7943786, 7. doi: 10.1155/2018/7943786
65. Elhadidy M, Ali MM, El-Shibiny A, Miller WG, Elkhatib WF, Botteldoorn N, et al. Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS ONE. (2020) 15:e0227833. doi: 10.1371/journal.pone.0227833
66. Painset A, Day M, Doumith M, Rigby J, Jenkins C, Grant K, et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Campylobacter jejuni and Campylobacter coli isolated from cases of diarrheal disease in England and Wales, 2015-16. J Antimicrob Chemother. (2020) 75:883–9. doi: 10.1093/jac/dkz539
67. Raeisi M, Khoshbakht R, Ghaemi EA, Bayani M, Hashemi M, Seyedghasemi NS, et al. Antimicrobial resistance and virulence-associated genes of Campylobacter species. Isolated from raw milk, fish, poultry, and red meat. Microbial Drug Resist. (2017) 23:925–33. doi: 10.1089/mdr.2016.0183
68. Jamali H, Ghaderpour A, Radmehr B, Wei KSC, Ching CL, Ismail S. Prevalence and antimicrobial resistance of Campylobacter species isolates in ducks and geese. Food Control. (2015) 50:328–30. doi: 10.1016/j.foodcont.2014.09.016
69. Eguale T. Non-typhoidal Salmonella serovars in poultry farms in central Ethiopia: prevalence and antimicrobial resistance. BMC Vet Res. (2018) 14:217. doi: 10.1186/s12917-018-1539-4
70. Beyene T, Endalamaw D, Tolossa Y, Feyisa A. Evaluation of rational use of veterinary drugs especially antimicrobials and anthelminthic in Bishoftu, Central Ethiopia. BMC Res Notes. (2015) 8:482. doi: 10.1186/s13104-015-1466-4
71. Du Y, Wang C, Ye Y, Liu Y, Wang A, Li Y, et al. Molecular identification of multidrug-resistant Campylobacter species from diarrheal patients and poultry meat in Shanghai, China. Front Microbiol. (2018) 9:1642. doi: 10.3389/fmicb.2018.01642
72. Lammie SL, Hughes JM. Antimicrobial resistance, food safety, and one health: the need for convergence. Ann Rev Food Sci Technol. (2016) 7:287–312. doi: 10.1146/annurev-food-041715-033251
73. Bidaisee S, Macpherson CNL. Zoonoses and one health: a review of the literature. J Parasitol Res. (2014) 2014:874345. doi: 10.1155/2014/874345
74. Boqvist S, Söderqvist K, Vågsholm I. Food safety challenges and One Health within Europe. Acta Vet Scand. (2018) 60:1. doi: 10.1186/s13028-017-0355-3
Keywords: Ethiopia, antimicrobial resistance, Campylobacter, livestock, one health, zoonosis
Citation: Chala G, Eguale T, Abunna F, Asrat D and Stringer A (2021) Identification and Characterization of Campylobacter Species in Livestock, Humans, and Water in Livestock Owning Households of Peri-urban Addis Ababa, Ethiopia: A One Health Approach. Front. Public Health 9:750551. doi: 10.3389/fpubh.2021.750551
Received: 30 July 2021; Accepted: 27 September 2021;
Published: 02 December 2021.
Edited by:
Olivier Vandenberg, Laboratoire Hospitalier Universitaire de Bruxelles (LHUB-ULB), BelgiumReviewed by:
Tingting Xu, Jinan University, ChinaAngela Joyce Cornelius, Institute of Environmental Science and Research (ESR), New Zealand
Copyright © 2021 Chala, Eguale, Abunna, Asrat and Stringer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew Stringer, YXBzdHJpbmdlckBuY3N1LmVkdQ==
 Gemechu Chala
Gemechu Chala Tadesse Eguale
Tadesse Eguale Fufa Abunna3
Fufa Abunna3 Andrew Stringer
Andrew Stringer