
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Public Health, 02 August 2021
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 9 - 2021 | https://doi.org/10.3389/fpubh.2021.686789
 Tobias M. Appel1,2*†
Tobias M. Appel1,2*† Natalia Quijano-Martínez3†
Natalia Quijano-Martínez3† Elsa De La Cadena2
Elsa De La Cadena2 María F. Mojica2†
María F. Mojica2† María Virginia Villegas2†
María Virginia Villegas2†The genus Raoultella was established in 2001. Species of Raoultella and Klebsiella share many ecological, biochemical, clinical, and microbiological features. Given the shortcomings of available technology for species identification in the clinical microbiology laboratory, are practically indistinguishable. Since the late 2000s there has been an increase in case reports of human Raoultella infections. Therefore, several authors are postulating that Raoultella spp. are rare and/or emerging pathogens.
Conclusions: Raoultella spp. are very similar to Klebsiella spp. The epidemiology and the clinical relevance of the human Raoultella spp. infections is uncertain and further studies are required. The previous difficulties in the identification of Raoultella spp. and the introduction of more precise identification techniques may explain the recent increase in the number of case reports. Raoultella spp. might be rather underdiagnosed than rare or emerging pathogens.
Since the late 2000s, the number of case reports of human Raoultella-infections available in the literature has been growing remarkably (1). From the 1980s until the 2006, only four case reports of human Raoultella-infections are available on PubMed, in contrast to more than 130 today. Due to this recent increase, several authors have been suggesting that Raoultella spp. are rare and emerging pathogens.
Herein, we review the literature regarding microbiological and clinical aspects of Raoultella spp. and compare key points of this genus with Klebsiella pneumoniae and K. oxytoca, two common human pathogens in the community, as well as in healthcare settings.
We reviewed the literature available as full text in English on PubMed until June 2021, using the search terms “Raoultella,” “Raoultella planticola,” “Raoultella ornithinolytica,” “Raoultella terrigena,” “Klebsiella trevesanii.” All clinical case reports, case series, retro- and prospective studies reporting Raoutella-infections, regardless of the microbiological diagnostic approach were included.
The Enterobacteriaceae, Klebsiella planticola, and K. terrigena were first described in 1981, while K. ornithinolytica was described 8 years later (2–5). K. trevisanii was first described in 1983 but turned out to be identical with K. planticola (3). In the late 1990s, modern taxonomic assays such as DNA sequencing of housekeeping genes catalyzed a variety of modifications in bacterial taxonomy (6). In 2001, based on the molecular analysis of 16S ribosomal RNA (rRNA) and RNA polymerase β subunit encoding genes (rpoB), K. planticola, K. ornithinolytica, and K. terrigena were classified into a new genus. In honor to Didier Raoult it was named Raoultella (3). Thirteen years later, a novel specie was discovered and named R. electrica, which has not been described in human infections (7).
As well as Klebsiella spp., Raoultella spp. are ubiquitous in nature, being found in plants, water and soil, and are known to colonize humans and animals (1). Both genera are facultative anaerobe Gram-negative bacilli, which belong to the family of Enterobacteriaceae and tend to form large mucoid colonies on McConkey agar, due to synthesis of a polysaccharide capsule. Therefore, both genera are morphologically indistinguishable and share several biochemical characteristics, such as the production of catalase, the absence of oxidase, the fermentation of glucose, lactose, sorbose and the reduction of nitrates (8). However, there are a some metabolic features which are present in some species, but not in others, and therefore are useful for their distinction, such as the indole-test, growth at 10°C, the production of histamine, D-melezitose test and the metabolism of ornithine (9) (Table 1), as well as pectate degradation, pigment production on gluconate-ferric citrate agar, and the utilization of gentisate in hydroxybenzoate as a sole carbon source (10, 11). These physiological and biochemical properties can be used for microbiological diagnosis, which are the basis of analytical profile index (API) based systems.
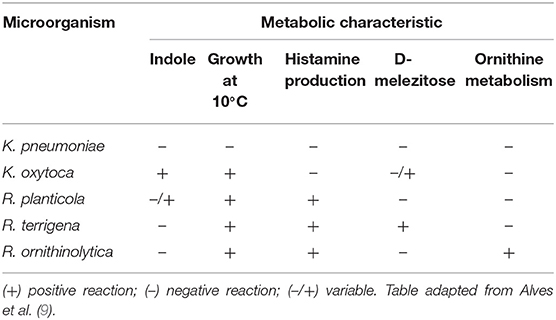
Table 1. Metabolic characteristics of clinically relevant species of Raoultella, K. pneumoniae and K. oxytoca.
Several virulence factors such as hemagglutinins, type 1 and type 3 fimbriae, siderophores, enterobactin and occasionally aerobactin are shared between R. planticola, R. terrigena, and K. pneumoniae (12, 13). Podschun et al. observed that type 1 fimbriae, which are one of the most important proteins for tissue adhesion in Klebsiella spp., are less frequently expressed in R. terrigena. Exopolysaccharide production and biofilm formation was observed in R. planticola and the expression of bacteriocins were described in R. ornithinolytica (14, 15). In R. terrigena, the production of tetrodotoxin, the potent neurotoxin of Takifugu niphobles (commonly known as puffer fish), has been described (16). This is consistent with the hypothesis that tetrodotoxin is not produced by fish. Instead, it might be ingested and accumulated by fish through the food chain, since several tetrodotoxin containing species of fish have been discovered (17).
Species from both genera are intrinsically resistant to penicillins because of the expression of a Ambler class A β-lactamase: LEN-1 or SHV-1 in K. pneumoniae, K1 in K. oxytoca, and PLA-1, ORN-1, and TER-1 in R. planticola, R. ornithinolytica, and R. terrigena, respectively (18, 19). A study by Walckenaer et al. demonstrated that PLA-1 and ORN-1 β-lactamases are chromosomally encoded and are 94% identical in their 291 amino acid sequence. This unusual high homology might be a result of an ancestral horizontal gene transfer. Simultaneously, they share a homology of 66–69% with the amino acid sequence of the chromosomal β-lactamases SHV-1/LEN-1 and 69–71% with the plasmid-mediated TEM-1 (18). TER-1, the β-lactamase described in R. terrigena shares a 78% homology with the amino acid sequence of the β-lactamases of R. planticola and R. ornithinolytica, 69.9% with TEM-1 and 67.5% with SHV-1/LEN-1 (19). PLA-1, ORN-1, and TER-1, as well as LEN-1 can be inhibited in vitro with β-lactam β-lactamase inhibitors, such as clavulanate, sulbactam, and tazobactam (18, 19).
Although the increase in frequency of reported Raoultella spp. isolates is remarkable, the microbiological diagnosis is difficult. The reliable identification is challenging due to the close phylogenetic relationship with Klebsiella spp. (1). For instance, Mori et al. identified among 439 clinical isolates of Klebsiella spp. 18.5% of [R.] planticola using biochemical and physiological methods (10).
Automated identification systems are commonly used in clinical microbiology laboratories. Three studies available in the literature evaluated the diagnostic performance in identifying species of Raoultella of these systems: Park et al. reported that only Vitek 2 system (bioMérieux, Marcyl'Etoile, France) was able to identify correctly all 27 R. ornithinolytica isolates, whereas the MicroScan Neg Combo 32 panel (Beckman Coulter, CA, USA) and API 20E (BioMerieux, Marcy-l'Etoile, France) identified 25 and 24 isolates, respectively, as K. oxytoca (20). Furthermore, Ponce-Alonso et al. found that MicroScan WalkAway and API 20E failed to detect all 11 Raoultella spp. and a study by De Jong reports the misidentification of 3 out of 3 R. ornithinolytica and 2 of 2 R. planticola isolates as K. oxytoca by BD Phoenix (BD, Sparks, USA) (21, 22). In a case report, Demiray et al. described a clinical isolate, which initially was identified as R. planticola by Vitek 2. However, 16S rRNA encoding gene analysis resulted in the identification as R. terrigena (23).
To date, MicroScan (24) and BD Phoenix systems (25) are able to identify R. ornithinolytica, but not R. planticola or R. terrigena. The API 20E system is able to identify R. ornithinolytica and R. terrigena, but requires additional tests for the identification of R. planticola (26). This data strongly suggests that most of the automated identification systems commonly used in clinical microbiology laboratories fail to reliably identify Raoultella spp., thus, the prevalence of this genus in clinical isolates might be underestimated.
The broad introduction of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the identification of microorganisms in hospital scenarios took place in the late 2000s might have further driven the correct identification of R. ornithinolytica and R. planticola case reports as mentioned by Seng et al. (27). Nevertheless, there are reports of the misidentification of Raoultella spp. with MALDI-TOF MS, although only at the species level (22, 28).
Molecular studies are the gold-standard for microbiological identification. Currently, these methods are not routinely used in clinical laboratories for microbiological identification. Sequencing of phylogenetically preserved sequences has been the technique most frequently used. In Raoultella spp. and other Enterobacterales rpoB and the 16S ribosomal RNA encoding gene are analyzed frequently. In addition, the identification is specific variants of gyrA and parC, as well as the presence of blaORN, blaPLA, and blaTER genes has been described to identify Raoultella spp. (19, 29, 30).
Among the 140 case reports of Raoultella-infections, only 65% mention the microbiological diagnostic method used (Supplementary Material 1). Of those reporting on the diagnostic method, 61.9% cases of R. planticola were diagnosed using the Vitek 2 system (6 of 26 the cases were confirmed with MALDI-TOF MS or molecular analysis), 16.7% using MALDI-TOF. MicroScan systems were used in 48.6% of clinical cases for microbiological diagnosis of R. ornithinolytica, followed by Vitek 2 (24.3%) and MALDI-TOF (18.9%). Regarding R. terrigena, API 20E was used in three out of eight cases.
The epidemiology of Raoultella-infections is uncertain since only a few surveillance studies are available in the literature and due to the difficulties with automated diagnostic platforms widely used in clinical microbiology laboratories, the incidence might be underestimated. Chun et al. reported R. planticola and R. ornithinolytica to be isolated in ~0.3 and 0.15% of bacteremia cases, respectively, in a hospital in South Korea. Of note, in about one third of blood samples in which Raoultella spp. were isolated, polymicrobial cultures were obtained (31, 32). In a prospective study from Ethiopia, R. terrigena was isolated in the urine of 8.38% of 429 participants (8.1% in women, 9.1% in men) with urinary tract infections (33).
Since the first description of Raoultella spp., 76 of case reports of R. planticola-, 55 of R. ornithinolytica- and 9 of R. terrigena-infections have been reported (Supplementary Material 1). The recent increase in case reports of Raoultella-infections over the last decade has led some authors to the conclusion that R. planticola and R. ornithinolytica are emerging pathogens (27, 34–36).
In contrast to the case reports, there are 22 studies available on PubMed, which identified 481 isolates of R. planticola, 566 of R. ornithinolytica and 168 of R. terrigena between 1984 and 2015 in several countries in Europe, Asia, Africa and South America (Supplementary Material 1). Most studies evaluated the incidence of isolates of only one Raoultella species, three studies focused specifically on bacteremia, two on urinary tract infections and bacteriuria, one on pneumonia and one exclusively on bile-tract infections.
Regarding colonization, R. planticola was found in 9.9% of the gut microbiome and upper respiratory tract of 131 neonates in a study from Germany (8), while R. terrígena was isolated in feces in 0.9% of 5,377 healthy adults (37). R. ornithinolytica has been identified in human saliva (38, 39), as well as in stool samples of healthy participants in rural Chinese communities (40).
Like Klebsiella spp. and other Enterobacterales, species of Raoultella can cause a broad range of clinical syndromes. There are only few surveillance studies, all are single-center studies, two of them focusing exclusively on bloodstream infections. Boattini et al. reported 30 infections caused by R. planticola and 27 by R. ornithinolytica in a hospital in Portugal between 2010 and 2014 (41). In this study, 29 patients had urinary tract infections, nine pneumonia and five bacteremia. Patient's age ranged from 30 to 99 years (average of 68.9 years) in patients with R. planticola infections and from 28 to 90 years (average of 66.1 years) in the R. ornithinolytica group. Immunodeficiency was reported in 56.3% of patients, such as diabetes (28.1%), malignancy (17.5%), or recipients of a solid organ transplant (14%).
Similarly, a study by Seng et al. reported a series of 112 patients between 1 and 99 years (average of 66 years) with R. ornithinolytica-infections mainly of the urinary tract (32%), respiratory tract (24%) and the bloodstream (5%) (27). Twenty-five percent of the 112 patients were classified as immunodeficient, of these, 25% had a diagnosis of solid cancer and 22% had diabetes.
Among 20 patients with BSI involving R. planticola, the average age was 65 years, while the 16 patients with BSI caused by R. ornithinolytica were 55.7 years on average (31, 32). Chun et al. reported malignancies in 15 of 16 patients and 17 of 20 patients with R. ornithinolytica and R. planticola bacteremia, respectively.
In a case series of 11 patients (ten women, one man) with R. planticola-pneumonia with an age between 51 and 79 years (average of 70 years), five patients had underlying malignancies, two had chronic obstructive pulmonary disease, two cerebral infarctions or hemorrhage, while one patient had end stage renal disease on hemodialysis. Four patients had polymicrobial cultures. In this case series, consolidation (8 out of 11), ground glass opacities (5/11), pleural effusion (5/11), and micronodules were the most common radiological findings (42).
In a study from Ethiopia, R. terrigena was isolated in 8.38% patients (n = 429) with urinary tract infections. The majority of patients were between 30 and 45 years of age (24/36) and female (25/36) (33).
In the studies mentioned above, mortality rates ranged from 8% (9/112) in patients with different types of R. ornithinolytica-infections, to 43.8% (7/16) in patients with R. ornithinolytica bacteremia. Mortality among patients with R. planticola infections was reported between 9.1% (1/11) and 15.6% (5/32) (27, 31, 32, 41, 42).
In the reviewed case reports, age of patients ranges from premature newborns to a 97 year-old patient, with an average of 49.7 years. Cases available in the literature report most frequently bloodstream infections, urinary tract, bile tract, and respiratory tract infections, while skin and soft tissue infections, conjunctivitis, and septic arthritis are reported less frequently (Supplementary Material 1). However, this information should be interpreted with caution, since a publication bias of case reports, favoring the publication of more severe and more interesting cases, might not reflect real-life clinical experience. Most patients in the case reports are in a debilitated condition, such as prematurity, advanced age, comorbidities such as diabetes, cancer, as well as requirement of a critical care unit, long-term antimicrobial therapy and chemotherapy or are exposed to endoscopic procedures and catheters. Of the reviewed case reports, 11.4% of patients died (5.4% of patients with R. ornithinolytica-infections, 11.8% with R. planticola-infections and 44.4% [4/9] with R. terrigena-infections), while 8.6% did not report a clinical outcome.
Unlike Klebsiella spp., species of Raoultella are histamine-producers due to the expression of a histidine-decarboxylase (43, 44). The excess of histamine in ingested fish, especially those of the family scombridae is known as scombroid food poisoning. This condition is associated with exanthema, urticaria, headache, diarrhea, edema, and tachycardia (45). In addition, the production of histamine has been reported to cause red skin flushing in some patients with Raoultella-infections (46, 47). Nevertheless, only a few cases reports mention the presence of flushing, while others explicitly report its absence (48).
As mentioned above, Raoultella spp. are intrinsically resistant to penicillins, due to the expression of a broad-spectrum β-lactamase. Similar to K. pneumoniae and K. oxytoca, wildtype isolates of Raoultella spp. are susceptible to antimicrobials commonly used for the treatment of infections caused by Enterobacterales, such as β-lactams (except penicillins), quinolones, aminoglycosides, tetracyclines, fosfomycin, nitrofurantoin, and polymyxins. Like K. pneumoniae and other Klebsiella species, a variety of acquired genes have been reported to confer resistance to antimicrobials in environmental and clinical isolates of Raoultella spp., including genes encoding for class A, B, and D carbapenemases. Furthermore, the co-expression of different carbapenemases, such as KPC with IMP or NDM, as well as in combination with OXA-48 has been described (35, 49–53). Recently, R. ornithinolytica harboring mcr genes have been described (54, 55).
There are only few studies on clinical isolates of Raoultella spp. available. All studies are from a single center and involved only small numbers of isolates. Seng et al. reported resistance to amoxicillin/clavulanate in 16% of 121 clinical isolates from France, 4% to ceftriaxone, 6% to quinolones, and 10% to trimethoprim (TMP/SMX) (27). The study by Boattini et al. from Portugal reported that all 57 clinical isolates of Raoultella spp. were susceptible to cefotaxime, ciprofloxacin and gentamicin. Three isolates of R. ornithinolytica were resistant to TMP/SMX and cefuroxime, two to nitrofurantoin and one to amoxicillin/clavulanate (41). In the two studies previously mentioned from South Korea, one of the 20 isolates of R. planticola was resistant to cephalothin and two isolates were intermediate for cefoxitin. In addition, two isolates were resistant to TMP/SMX and one to gentamicin (31). Among the 16 strains of R. ornithinolytica, one isolate was resistant to third and fourth-generation cephalosporins, two were resistant to cefazolin and one was resistant to amoxicillin/clavulanate; two isolates were resistant to TMP/SMX (32).
In a study published in 2020, Sękowska et al. evaluated antimicrobial susceptibility and resistance determinants of 79 clinical isolates of R. ornithinolytica and 26 of R. planticola collected from 65 patients from a university hospital in Poland. Overall, susceptibility to all tested antimicrobials was high (≥81.9%), except for amoxicillin/clavulanate (9.5% susceptible; 59.1% intermediate). For β-lactams, the highest susceptibility was observed for imipenem (99%), followed by meropenem (98.1%), and cefepime (88.6%). Regarding non-β-lactams, the highest susceptibility was observed for gentamicin (93.3%) and ciprofloxacin (92.4%). Fourteen isolates (nine R. ornithinolytica and five R. planticola) harbored extended-spectrum β-lactamases, most frequently CTX-M-1 in combination with an unknown variant of TEM. In addition, one strain carried CTX-M-1 plus an unknown variant of VIM. Of note, the high non-susceptibility to amoxicillin/clavulanate (90.5%) in this study is unexpected, since Raoultella β-lactamases, as well as most of the ESBLs belonging to CTX-M, TEM and SHV-families are inhibited by clavulanate. In addition, susceptibility to cephalosporins and piperacillin/tazobactam was high (56). This phenomenon could be either related to automated susceptibility testing or the breakpoints applied in the study.
As presented above, the genera Raoultella and Klebsiella share many ecological, biochemical, clinical, and microbiological features, that given the shortcomings of available technology for species identification in the clinical microbiology laboratory, are indistinguashable in many cases.
The previous difficulties in the identification of Raoultella spp. and the introduction of more precise identification techniques in clinical microbiology laboratories might explain the recent increase in the number of case reports. Hence, like other microorganisms that are identified more frequently in recent years, Raoultella spp. probably should considered as previously underdiagnosed, rather than rare, novel or emerging pathogens.
Reviewing the cases available in the literature, the (although limited) ability of automated identification systems, as well as the introduction of MALDI-TOF MS in microbiological laboratories are probably the main driver of an increasing identification of this genus. Unfortunately, an important number of case reports are not revealing any information on the microbiological diagnosis. Twenty-two studies available in the literature with 1,215 clinical isolates of Raoultella spp. (Supplementary Material 1), suggest that these bacteria probably are not rare pathogens either. Mori et al. described an “unexpectedly high incidence” of R. planticola among clinical isolates of Klebsiella spp. from Japan, using biochemical and metabolic tests for identification, while a study from Ethiopia reported R. terrigena to be isolated in 8.39% of patients with urinary tract infections (10, 33).
Besides the recent increase in case reports of human Raoultella-infections, little is known about the real epidemiology of these infections. Only few surveillance studies are available in the literature, all of which are from a single-center and the majority with retrospective design. In addition, some of these studies focused only on the surveillance of a single species of Raoultella, while others reported only cases of bloodstream-, urinary tract, respiratory tract or bile tract infections.
Regarding the suggested role of Raoultella spp. as opportunistic pathogens, especially in intensive care unit patients, further investigations are required, since in two studies 44 to 75% of patients had no known underlying immunosuppressing condition (27, 41). However, both studies reported an important proportion of polymicrobial infections, enrolled a relatively small numbers of patients, and a homogenous definition of immunosuppression was lacking. On the other hand, there is no standardized and universal definition of “opportunism,” and many species of Klebsiella and even some strains of the omnipresent pathogen K. pneumoniae are considered as opportunistic pathogens.
The clinical implications of histamine productions by species of Raoultella is another relevant feature that requires further investigations. Some case reports mention the presence of a rash which is attributed to histamine production, while other authors explicitly mention its absence (48). High amounts of histamine might have an impact on clinical outcomes and therefore therapeutic implications, especially in blood stream infections and other sever infections in critically ill patients.
Several environmental and clinical isolates of species of Raoultella carrying resistance determinants to last-resort antimicrobials, such as carbapenems and colistin have been reported. Some authors hypothesize that Raoultella spp. could play a role as a reservoir of antimicrobial resistance genes. Nevertheless, only few surveillance studies on antimicrobial susceptibility in Raoultella spp. are available in the literature. Although solid clinical data is lacking, considering microbiological characteristics, the therapeutic approach to patients with Raoultella-infections should be similar to patients with infections caused by Klebsiella spp., considering individual clinical and pharmacological aspects of the patient, as well as the local epidemiology of antimicrobial resistance.
In conclusion, Raoultella spp. are very similar to Klebsiella spp. and are probably not rare, neither emerging. Further studies are required to establish the epidemiology and clinical relevance of this genus.
TA and NQ-M wrote the first draft of the manuscript. MM and ED contributed very insightful suggestions on microbiological aspects. MV contributed with very insightful suggestions on clinical aspects. All authors contributed to the manuscript revision, read, and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.686789/full#supplementary-material
1. Sękowska A. Raoultella spp.-clinical significance, infections and susceptibility to antibiotics. Folia Microbiol. (2017) 62:221–7. doi: 10.1007/s12223-016-0490-7
2. Izard D, Ferragut C, Gavini F, Kersters K, De Ley J, Leclerc H. Klebsiella terrigena, a new species from soil and water. Int J Syst Evol Microbiol. (1981) 31:116–27. doi: 10.1099/00207713-31-2-116
3. Drancourt M, Bollet C, Carta A, Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol. (2001) 51(Pt. 3):925–32. doi: 10.1099/00207713-51-3-925
4. Bagley ST, Seidler RJ, Brenner DJ. Klebsiella planticola sp. nov.: a new species of Enterobacteriaceae found primarily in nonclinical environments. Curr Microbiol. (1981) 6:105–9. doi: 10.1007/BF01569013
5. Sakazaki R, Tamura K, Kosako Y, Yoshizaki E. Klebsiella ornithinolytica sp. nov., formerly known as ornithine-positiveKlebsiella oxytoca. Curr Microbiol. (1989) 18:201–6. doi: 10.1007/BF01570291
6. Janda JM. Clinical decisions: how relevant is modern bacterial taxonomy for clinical microbiologists? Clin Microbiol Newsl. (2018) 40:51–7. doi: 10.1016/j.clinmicnews.2018.03.005
7. Kimura Z, Chung KM, Itoh H, Hiraishi A, Okabe S. Raoultella electrica sp. nov., isolated from anodic biofilms of a glucose-fed microbial fuel cell. Int J Syst Evol Microbiol. (2014) 64(Pt. 4):1384–8. doi: 10.1099/ijs.0.058826-0
8. Podschun R, Acktun H, Okpara J, Linderkamp O, Ullmann U, Borneff-Lipp M. Isolation of Klebsiella planticola from newborns in a neonatal ward. J Clin Microbiol. (1998) 36:2331–2. doi: 10.1128/JCM.36.8.2331-2332.1998
9. Alves MS, Dias RCS, de Castro ACD, Riley LW, Moreira BM. Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. J Clin Microbiol. (2006) 44:3640–6. doi: 10.1128/JCM.00940-06
10. Mori M, Ohta M, Agata N, Kido N, Arakawa Y, Ito H, et al. Identification of species and capsular types of Klebsiella clinical isolates, with special reference to Klebsiella planticola. Microbiol Immunol. (1989) 33:887–95. doi: 10.1111/j.1348-0421.1989.tb00976.x
11. Westbrook GL, O'Hara CM, Roman SB, Miller JM. Incidence and identification of Klebsiella planticola in clinical isolates with emphasis on newborns. J Clin Microbiol. (2000) 38:1495–7. doi: 10.1128/JCM.38.4.1495-1497.2000
12. Podschun R, Fischer A, Ullman U. Expression of putative virulence factors by clinical isolates of Klebsiella planticola. J Med Microbiol. (2000) 49:115–9. doi: 10.1099/0022-1317-49-2-115
13. Podschun R, Fischer A, Ullmann U. Characterization of Klebsiella terrigena strains from humans: haemagglutinins, serum resistance, siderophore synthesis, and serotypes. Epidemiol Infect. (2000) 125:71–8. doi: 10.1017/S0950268899004124
14. Rättö M, Verhoef R, Suihko M-L, Blanco A, Schols HA, Voragen AGJ, et al. Colanic acid is an exopolysaccharide common to many enterobacteria isolated from paper-machine slimes. J Ind Microbiol Biotechnol. (2006) 33:359–67. doi: 10.1007/s10295-005-0064-1
15. Al-Hulu SM, Al-Charrakh AH. Isolation and characterization of Raoultella ornithinolytica from clinical specimens in Hilla city, Iraq. Iraq Med J Babylon. (2009) (7):42–7.
16. Yu VC-H, Yu PH-F, Ho K-C, Lee FW-F. Isolation and Identification of a new tetrodotoxin-producing bacterial species, Raoultella terrigena, from Hong Kong marine puffer fish Takifugu niphobles. Mar Drugs. (2011) 9:2384–96. doi: 10.3390/md9112384
17. Jal S, Khora SS. An overview on the origin and production of tetrodotoxin, a potent neurotoxin. J Appl Microbiol. (2015) 119:907–16. doi: 10.1111/jam.12896
18. Walckenaer E, Poirel L, Leflon-Guibout V, Nordmann P, Nicolas-Chanoine M-H. Genetic and biochemical characterization of the chromosomal class A β-lactamases of Raoultella (formerly Klebsiella) planticola and Raoultella ornithinolytica. Antimicrob Agents Chemother. (2004) 48:305–12. doi: 10.1128/AAC.48.1.305-312.2004
19. Walckenaer E, Delmas J, Leflon-Guibout V, Bonnet R, Nicolas-Chanoine MH. Genetic, biochemical characterization and mutagenesis of the chromosomal class A β-lactamase of Raoultella (formerly Klebsiella) terrigena. Pathol Biol. (2015) 63:158–63. doi: 10.1016/j.patbio.2015.05.002
20. Park JS, Hong KH, Lee HJ, Choi SH, Song SH, Song K-H, et al. Evaluation of three phenotypic identification systems for clinical isolates of Raoultella ornithinolytica. J Med Microbiol. (2011) 60(Pt. 4):492–9. doi: 10.1099/jmm.0.020768-0
21. Ponce-Alonso M, Rodríguez-Rojas L, del Campo R, Cantón R, Morosini M-I. Comparison of different methods for identification of species of the genus Raoultella: report of 11 cases of Raoultella causing bacteraemia and literature review. Clin Microbiol Infect. (2016) 22:252–7. doi: 10.1016/j.cmi.2015.10.035
22. de Jong E, de Jong AS, Smidts-van den Berg N, Rentenaar RJ. Differentiation of Raoultella ornithinolytica/planticola and Klebsiella oxytoca clinical isolates by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Diagn Microbiol Infect Dis. (2013) 75:431–3. doi: 10.1016/j.diagmicrobio.2012.12.009
23. Demiray T, Köroglu M, Özbek A, Hafizoglu T, Altindiş M. The first case of Raoultella terrigena infection in an infant. Turk J Pediatr. (2015) 57:624–8.
24. MicroScan LabPro Software Organism Reference Guide - For Use With LabPro Software v5,.0 (and Later). (2020). Available online at: https://www.beckmancoulter.com/download/file/wsr-316607/C42480AB?type=pdf (accessed June 26, 2021).
25. BBL CrystalTM Identification Systems Enteric/Nonfermenter ID Kit. (2017). Available online at: https://legacy.bd.com/ds/technicalCenter/clsi/clsi-CrysE_nf.pdf (accessed June 26, 2021).
26. API & ID 32 Identification Databases. Available online at: https://www.biomerieux-diagnostics.com/sites/clinic/files/9308960-002-gb-b-apiweb-booklet.pdf
27. Seng P, Boushab BM, Romain F, Gouriet F, Bruder N, Martin C, et al. Emerging role of Raoultella ornithinolytica in human infections: a series of cases and review of the literature. Int J Infect Dis. (2016) 45:65–71. doi: 10.1016/j.ijid.2016.02.014
28. Yan Y, Meng S, Bian D, Quinn C, Li H, Stratton CW, et al. Comparative evaluation of Bruker Biotyper and BD Phoenix systems for identification of bacterial pathogens associated with urinary tract infections. J Clin Microbiol. (2011) 49:3936–9. doi: 10.1128/JCM.05363-11
29. Walckenaer E, Leflon-Guibout V, Nicolas-Chanoine M-H. How to identify Raoultella spp. including R. ornithinolytica isolates negative for ornithine decarboxylase? The reliability of the chromosomal bla gene. J Microbiol Methods. (2008) 75:405–10. doi: 10.1016/j.mimet.2008.07.011
30. Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol. (2001) 51(Pt. 3):915–24. doi: 10.1099/00207713-51-3-915
31. Chun S, Yun JW, Huh HJ, Lee NY. Low virulence? Clinical characteristics of Raoultella planticola bacteremia. Infection. (2014) 42:899–904. doi: 10.1007/s15010-014-0664-7
32. Chun S, Yun JW, Huh HJ, Lee NY. Clinical characteristics of Raoultella ornithinolytica bacteremia. Infection. (2015) 43:59–64. doi: 10.1007/s15010-014-0696-z
33. Seifu WD, Gebissa AD. Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect Dis. (2018) 18:30. doi: 10.1186/s12879-017-2911-x
34. Ershadi A, Weiss E, Verduzco E, Chia D, Sadigh M. Emerging pathogen: a case and review of Raoultella planticola. Infection. (2014) 42:1043–6. doi: 10.1007/s15010-014-0638-9
35. Demiray T, Koroglu M, Ozbek A, Altindis M. A rare cause of infection, Raoultella planticola: emerging threat and new reservoir for carbapenem resistance. Infection. (2016) 44:713–7. doi: 10.1007/s15010-016-0900-4
36. Olivier M, Madruga M, Carlan S. Raoultella planticola, an emerging pathogen. Infect Dis Clin Pract. (2019) 24:3–10. doi: 10.1097/IPC.0000000000000749
37. Podschun R. Isolation of Klebsiella terrigena from human feces: biochemical reactions, capsule types, and antibiotic sensitivity. Zentralblatt Bakteriol Int J Med Microbiol. (1991) 275:73–8. doi: 10.1016/S0934-8840(11)80769-3
38. Heggendorn FL, Gonçalves LS, Dias EP, Silva Junior A, Galvão MM, Lutterbach MTS. Detection of sulphate-reducing bacteria in human saliva. Acta Odontol Scand. (2013) 71:1458–63. doi: 10.3109/00016357.2013.770163
39. Derafshi R, Bazargani A, Ghapanchi J, Izadi Y, Khorshidi H. Isolation and identification of nonoral pathogenic bacteria in the oral cavity of patients with removable dentures. J Int Soc Prev Community Dent. (2017) 7:197–201. doi: 10.4103/jispcd.JISPCD_90_17
40. Wang S, Xu L, Chi X, Li Y, Kou Z, Hou P, et al. Emergence of NDM-1- and CTX-M-3-producing Raoultella ornithinolytica in human gut microbiota. Front Microbiol. (2019) 10:2678. doi: 10.3389/fmicb.2019.02678
41. Boattini M, Almeida A, Cardoso C, Cruz CS, Machado C, Vesza Z, et al. Infections on the rise: Raoultella spp., clinical and microbiological findings from a retrospective study, 2010-2014. Infect Dis. (2016) 48:87–91. doi: 10.3109/23744235.2015.1082619
42. Hong G, Yong HJ, Lee D, Kim DH, Kim YS, Park J-S, et al. Clinical characteristics and treatment outcomes of patients with pneumonia caused by Raoultella planticola. J Thorac Dis. (2020) 12:1305–11. doi: 10.21037/jtd.2020.02.56
43. Kanki M, Yoda T, Tsukamoto T, Shibata T. Klebsiella pneumoniae produces no histamine: Raoultella planticola and Raoultella ornithinolytica strains are histamine producers. Appl Environ Microbiol. (2002) 68:3462–6. doi: 10.1128/AEM.68.7.3462-3466.2002
44. Hwang CC, Kung HF, Lee YC, Wen SY, Chen PY, Tseng DI, et al. Histamine fish poisoning and histamine production by Raoultella ornithinolytica in milkfish surimi. J Food Prot. (2020) 83:874–80. doi: 10.4315/0362-028X.JFP-19-385
45. Taylor SL, Stratton JE, Nordlee JA. Histamine poisoning (scombroid fish poisoning): an allergy-like intoxication. J Toxicol Clin Toxicol. (1989) 27:225–40. doi: 10.3109/15563658908994420
46. Mau N, Ross LA. Raoultella ornithinolytica bacteremia in an infant with visceral heterotaxy. Pediatr Infect Dis J. (2010) 29:477–8. doi: 10.1097/INF.0b013e3181ce9227
47. Solak Y, Gul EE, Atalay H, Genc N, Tonbul HZ. A rare human infection of Raoultella ornithinolytica in a diabetic foot lesion. Ann Saudi Med. (2011) 31:93–4. doi: 10.5144/0256-4947.2011.93a
48. Jellinge ME. Raoultella ornithinolytica diagnosed in a neurointensive patient. A rare case with recovery without antibiotics. J Crit Care Med. (2017) 3:120–2. doi: 10.1515/jccm-2017-0017
49. Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY antimicrobial surveillance program. J Clin Microbiol. (2009) 47:4129. doi: 10.1128/JCM.01502-09
50. Zheng B, Zhang J, Ji J, Fang Y, Shen P, Ying C, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. (2015) 59:7086–9. doi: 10.1128/AAC.01363-15
51. Park SC, Wailan AM, Barry KE, Vegesana K, Carroll J, Mathers AJ, et al. Managing all the genotypic knowledge: approach to a septic patient colonized by different Enterobacteriales with unique carbapenemases. Antimicrob Agents Chemother. (2019) 63:e00029–19. doi: 10.1128/AAC.00029-19
52. Zhao Y, Tang N, Jia R, Hu J, Liu W, Sun Y, et al. Co-existence of the carbapenem resistance genes blaKPC-2 and blaNDM-1 in a Raoultella planticola isolate in China. J Glob Antimicrob Resist. (2020) 23:327–8. doi: 10.1016/j.jgar.2020.09.015
53. Dang B, Zhang H, Li Z, Ma S, Xu Z. Coexistence of the bla NDM-1-carrying plasmid pWLK-NDM and the bla KPC-2-carrying plasmid pWLK-KPC in a Raoultella ornithinolytica isolate. Sci Rep. (2020) 10:2360. doi: 10.1038/s41598-020-59341-4
54. Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. (2019) 45:131–61. doi: 10.1080/1040841X.2018.1492902
55. Wang X, Wang Y, Zhou Y, Wang Z, Wang Y, Zhang S, et al. Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front Microbiol. (2019) 10:228. doi: 10.3389/fmicb.2019.00228
Keywords: Enterobacteriaceae, MALDI-TOF, Raoultella planticola, Raoultella ornithinolytica, Raoultella terrigena, Raoultella electrica
Citation: Appel TM, Quijano-Martínez N, De La Cadena E, Mojica MF and Villegas MV (2021) Microbiological and Clinical Aspects of Raoultella spp. Front. Public Health 9:686789. doi: 10.3389/fpubh.2021.686789
Received: 27 March 2021; Accepted: 09 July 2021;
Published: 02 August 2021.
Edited by:
Leonardo Neves de Andrade, University of São Paulo, BrazilReviewed by:
Dongdong Li, Sichuan University, ChinaCopyright © 2021 Appel, Quijano-Martínez, De La Cadena, Mojica and Villegas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tobias M. Appel, dG9iaWFzbS5hcHBlbEBnbWFpbC5jb20=
†ORCID: Tobias M. Appel orcid.org/0000-0001-6413-5623
Natalia Quijano-Martìnez orcid.org/0000-0002-8592-1727
María F. Mojica orcid.org/0000-0002-1380-9824
María Virginia Villegas orcid.org/0000-0003-1898-9067
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.