- 1National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of Epidemiology and Biostatistics, School of Public Health, Xi'an Jiaotong University Health Science Center, Xi'an, China
- 3Department of Epidemiology & Biostatistics, School of Public Health, Peking University Health Science Center, Beijing, China
Objective: The risk prediction model is an effective tool for risk stratification and is expected to play an important role in the early detection and prevention of esophageal cancer. This study sought to summarize the available evidence of esophageal cancer risk predictions models and provide references for their development, validation, and application.
Methods: We searched PubMed, EMBASE, and Cochrane Library databases for original articles published in English up to October 22, 2021. Studies that developed or validated a risk prediction model of esophageal cancer and its precancerous lesions were included. Two reviewers independently extracted study characteristics including predictors, model performance and methodology, and assessed risk of bias and applicability with PROBAST (Prediction model Risk Of Bias Assessment Tool).
Results: A total of 20 studies including 30 original models were identified. The median area under the receiver operating characteristic curve of risk prediction models was 0.78, ranging from 0.68 to 0.94. Age, smoking, body mass index, sex, upper gastrointestinal symptoms, and family history were the most commonly included predictors. None of the models were assessed as low risk of bias based on PROBST. The major methodological deficiencies were inappropriate date sources, inconsistent definition of predictors and outcomes, and the insufficient number of participants with the outcome.
Conclusions: This study systematically reviewed available evidence on risk prediction models for esophageal cancer in general populations. The findings indicate a high risk of bias due to several methodological pitfalls in model development and validation, which limit their application in practice.
Introduction
Esophageal cancer is one of the most common cancers worldwide, with an estimated 6,04,100 new cases and 5,44,076 cancer deaths in 2020 (1). In China, esophageal cancer is the sixth most common cancer and the fourth most common cause of cancer deaths (2). The two primary histologic subtypes of esophageal cancer are esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), and squamous dysplasia (SD) and Barrett's esophagus (BE) are known precursors of esophageal cancer for ESCC and EAC, respectively (3). Several factors have been found to be associated with an increased risk for esophageal cancer, including older age, male, family history, smoking, and alcohol consumption (4).
The prognosis of esophageal cancer is poor since most cancers were diagnosed at a late stage. Endoscopic screening plays a key role in the prevention of esophageal cancer. It can reduce the incidence and mortality through early detection, early diagnosis, and early treatment of cancer and precancerous lesions (5, 6). However, population-wide endoscopic screening is less cost-effective due to low detection rate and low compliance, while opportunistic screening is limited in clinical practice due to the high requirement for equipment, technic, and personnel (7).
The risk prediction model is an effective tool for risk stratification and prediction and has been widely used in screening for lung cancer (8), colorectal cancer (9), and breast cancer (10). It is also expected to be applied to esophageal cancer screening to optimize the screening strategy and provide patients with individualized screening guidance. By predicting the probability of esophageal cancer and precancerous lesions in the general population based on predictive factors, risk prediction models can identify high-risk individuals eligible for endoscopic screening, which could concentrate the subjects needed to be screened, reduce the cost and increase the efficiency and compliance of screening, as well as avoid over-screening (11, 12).
In recent years, several risk prediction models with various risk factors have been developed for esophageal cancer. However, it is not clear whether these models are developed or validated appropriately or whether they are applicable to screening for the general population. Therefore, it is important to evaluate the methodology underpinning model development and validation, and also risk factors and performance to guide decisions regarding the use and choice of models for risk predictions. The aim of this systematic review was to provide an overview on the models developed to predict the risk of esophageal cancer in general population, including methodological characteristics, predictors, and risk of bias.
Materials and Methods
This systematic review was conducted according to the preferred reporting items for systematic reviews and meta-analyses (PRISMA) (13). A pre-specified protocol was followed when this study was conducted.
Literature Search
We comprehensively searched the PubMed, EMBASE, and Cochrane Library electronic databases to identify relevant publications on December 31, 2020, and updated the search on October 22, 2021. The search strategy consisted of medical subject headings (MeSH) and text words for risk prediction models of esophageal cancer and its precancerous lesions, with the results restricted to human studies. The main search terms were as follows: [(“cancer” OR “carcinoma” OR “neoplasia” OR “tumor” OR “malignancy”) AND (“esophageal” OR “esophagus” OR “gastrointestinal” OR “digestive” OR “alimentary”) OR “esophageal neoplasms” [MeSH]] AND (“predict” OR “predictive” OR “prediction”) AND “model” AND “risk.” Further studies were sought by manually searching for the relevant articles in public databases.
Selection of Studies
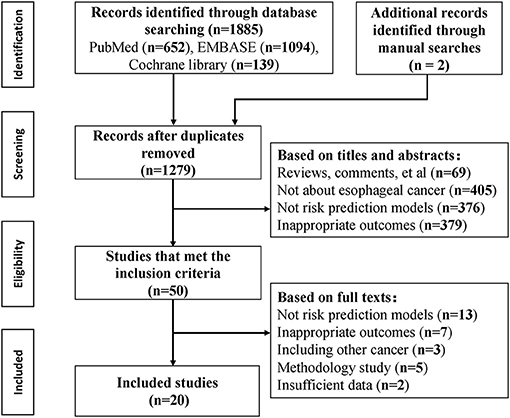
After the removal of duplicates, studies were screened according to the inclusion and exclusion criteria based on titles and abstracts. If a decision could not be made based on abstracts, full articles were retrieved. Studies were selected if they met with the following criteria: (1) published as an original research article in a peer-reviewed journal; (2) developing or validating a risk prediction model of esophageal cancer and/or its precancerous lesions; (3) considering more than one predictors in the prediction model, including traditional and genetic risk factor, laboratory test, or risk scores that combing multiple risk factors; (4) had the outcome as cases with cancer or precancerous lesions vs. non-cases. Two independent authors (RC and JZ) screened the search results to assess conformity with selection criteria, with disagreement resolved with a third author (SW).
Data Extraction and Synthesis
The same authors independently performed data extraction and quality assessment using standardized protocols. The general characteristics of studies were extracted, including first author, publication year, country, outcome, study design, study period, sample size, and model-related information (statistical methods, model performance, validation method, and included predictors). In this process, the methods of studies published for each risk model were classified according to the TRIPOD guidelines (14). For studies which included multiple models, for example separate models for different outcomes or age groups, all were included separately.
Assessment of Risk of Bias
The risk of bias was assessed using PROBAST (Prediction model Risk Of Bias Assessment Tool) (15). The tool contains a total of 20 signaling questions to facilitate structured judgment of risk of bias covering four domains on model development and validation: participants, predictors, outcome, and analysis. Any initial disagreement was resolved through further discussion among the authors.
Results
The initial search identified 1,885 records, and 20 articles were included according to the inclusion criteria (Figure 1). A total of nine cohorts, 11 case-control studies, and two cross-sectional studies were involved since two studies applied two study designs. Three articles (16–18) reported independent models for different outcomes, three articles reported (19–21) independent models with different predictive factors, one article (22) reported sex-specific models, and one article (11) reported four age-stratified models; thus a total of 30 models were identified on risk prediction for esophageal cancer and precancerous lesions.
Overview of Risk Prediction Models
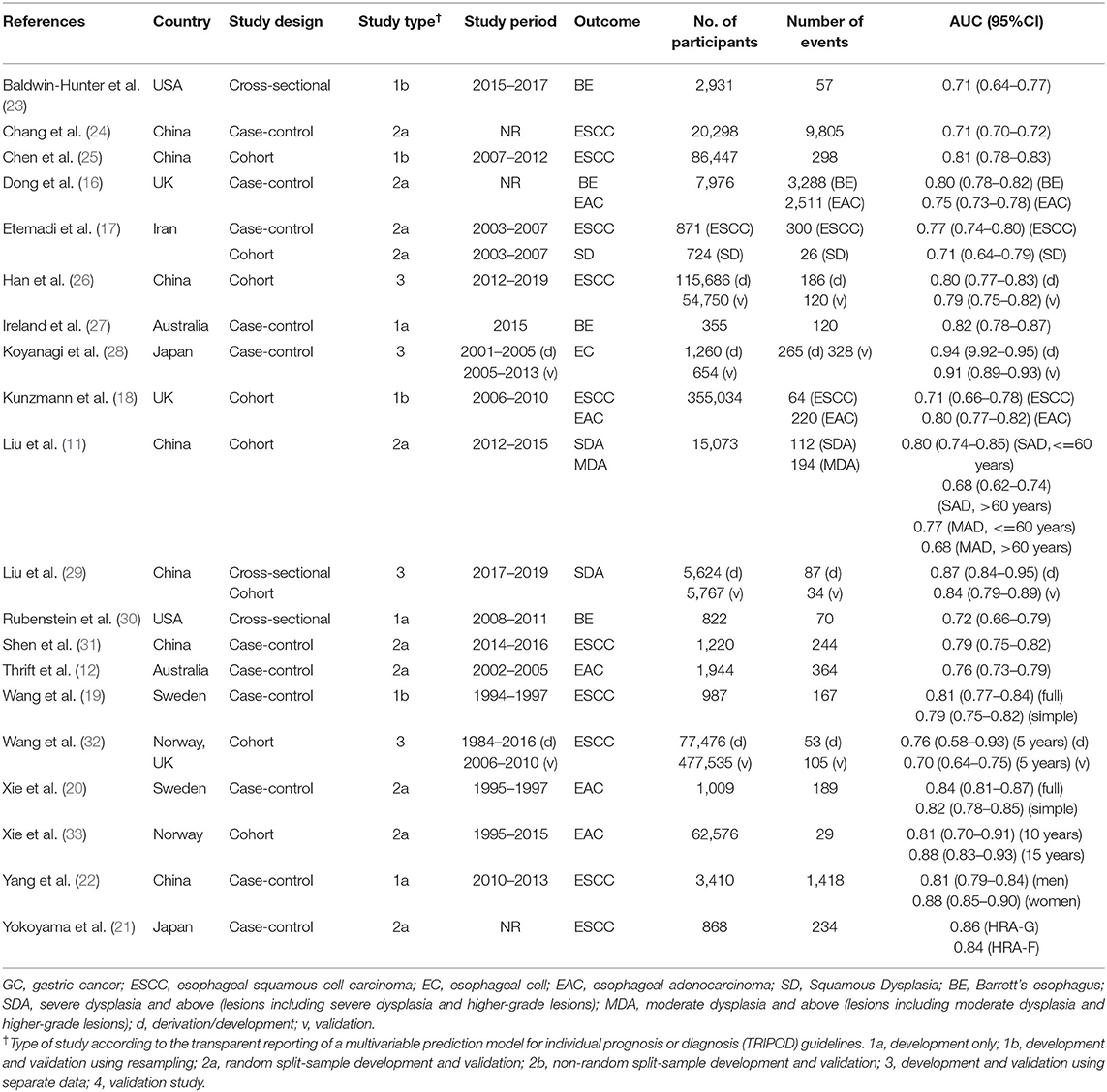
Table 1 presents the primary characteristics of the included studies. The publication year ranged from 2008 to 2021. There are 10 models for EAC and BE, 19 models for ESCC and SD, and one model for unspecified esophageal cancer. The sample size of included studies ranged from 355 to 5,55,011, involving 20,776 cases in total. All models on EAC and BE are for people from Western countries, whereas most models on ESCC and SD are for Asians, especially those in East Asian countries including Japan and China. Among the 30 reported models, model discrimination (C statistic) ranged from 0.68 to 0.94, with a median area under the receiver operating characteristic curve (AUC) of 0.78.
Predictive Factors
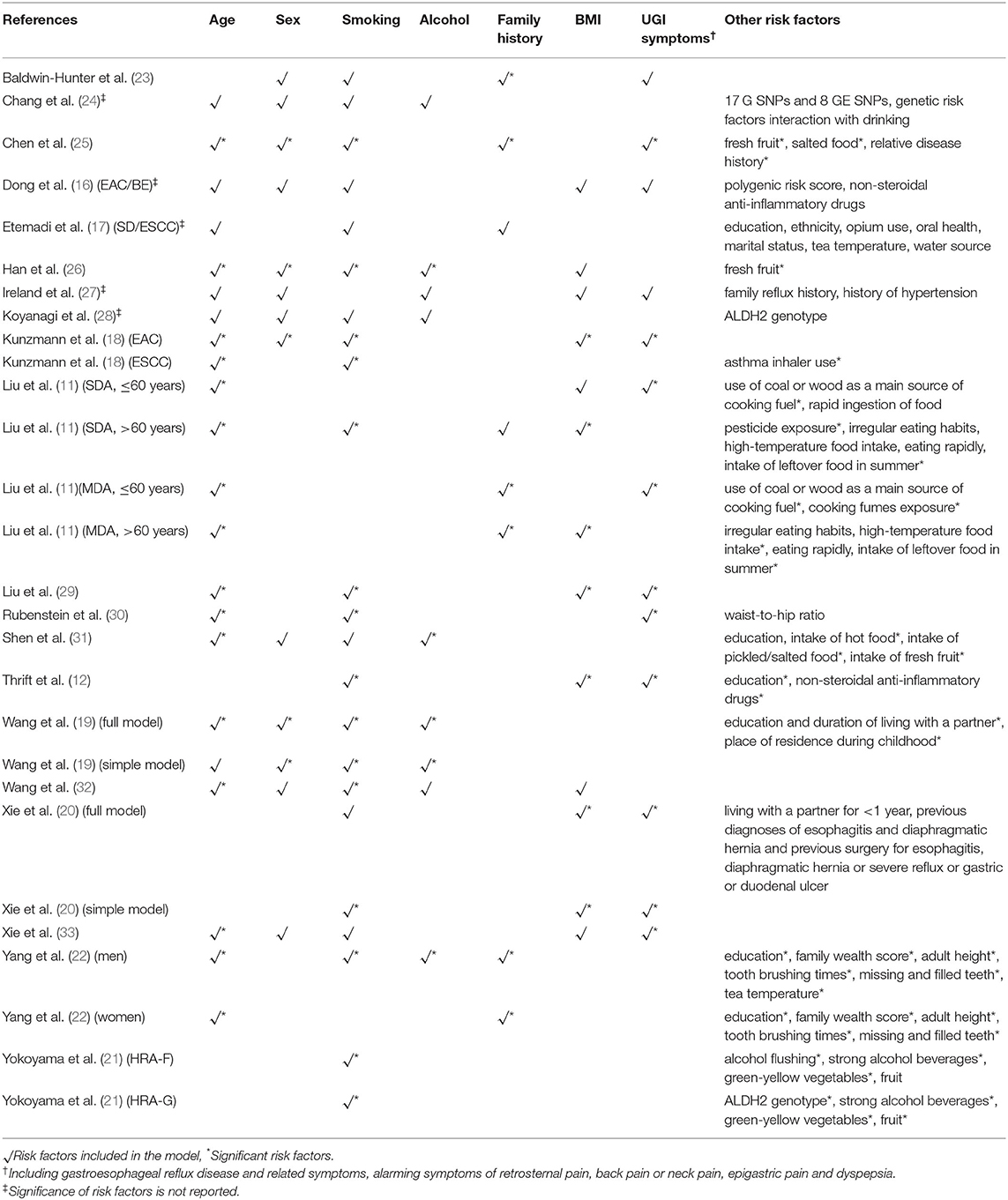
An overview of the predictive factors that were included in the risk prediction models are summarized in Table 2. The number of predictors included in the final models ranged from three to 10, with a median of five predictors. The most common predictors were age, smoking, body mass index (BMI), sex, upper gastrointestinal symptoms, and family history. Besides, genetic variants were included in four studies (16, 21, 24, 28), two of them were ALDH2 genotypes (21, 28), and the other two were single nucleotide polymorphisms (SNPs) or polygenic risk score generated from genome-wide association studies (16, 24).
Methodology of Risk Prediction Models
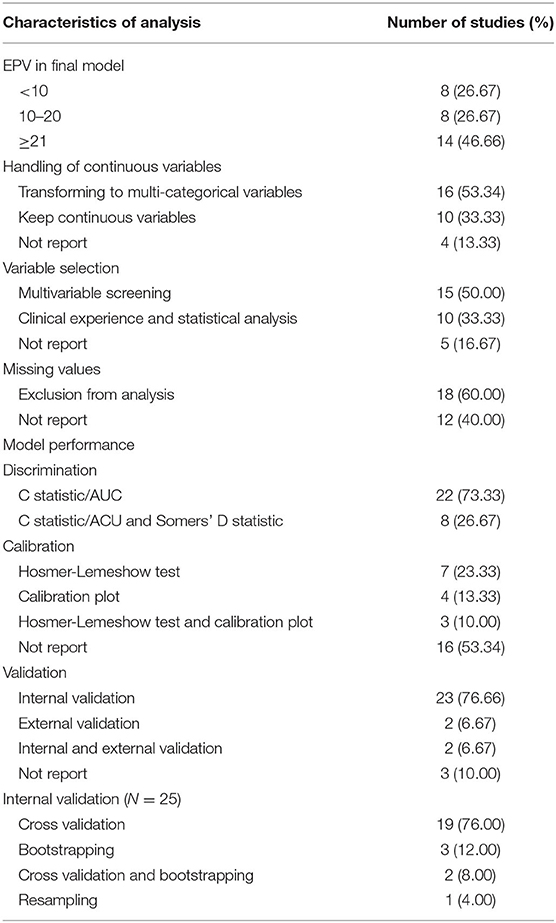
The methodological characteristics of the included models are summarized in Table 3. The number of events per variables (EPVs) in the final multivariate analysis was <10 for eight models, and ranged between 10 and 20 for eight models. One third of the models keep continuous variables whereas about half of the models transformed continuous variables to multicategorical variables. Candidate predictors were selected based on multivariable screening in 15 models and clinical experience combined with statistical analysis in 10 models. Eighteen models reported missing values and excluded them from analysis, whereas other studies did not report the issue of missing data. All models reported discrimination with C statistic, of which eight models reported both C statistic and Somers' D statistic. Model calibration was reported in 10 models with Hosmer-Lemeshow test and seven models with calibration plot. Twenty-three models were internally validated using cross validation, bootstrapping, or resampling. Only four models performed independent external validation.
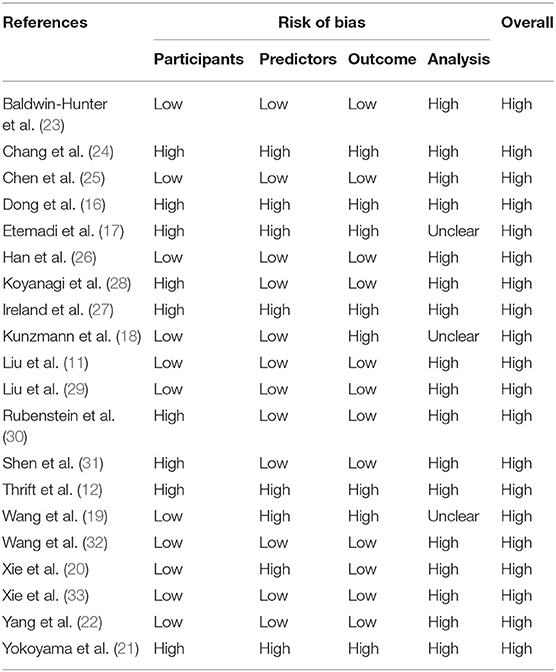
Risk of Bias
A summary of the risk of bias analysis is shown in Table 4. None of the models were identified as at low risk of bias. The high risk of bias in participants is mainly due to inappropriate date source, such as cases and non-cases from different populations in case-control studies, which also causes inconsistent definition of predictors and outcomes and makes the domain of predictors and outcome high risk. Major deficiencies in the analysis domain relate to number of participants with the outcome and evaluation of model performance. The relatively recommended prediction model for subjects with high risk of ESCC were Chen's model (25) and Han's model (26), and the model by Kunzmann was recommended for identifying individuals at risk for EAC (18).
Discussion
This systematic review summarizes the available evidence on risk prediction for esophageal cancer in general populations and evaluated the methodology of the included models comprehensively. The findings indicate a high risk of bias in the existing models due in part to limitations in study design and poor reporting and are expected to guide efforts to improve the development, validation, and transparent reporting of risk prediction models for esophageal cancer as well as other related diseases.
Unlike breast cancer (34), colorectal cancer (35, 36), and lung cancer (37), which already have a large number of models with externally validations, the development and validation of risk prediction models of esophageal cancer is still insufficient. Although the included models showed a relatively acceptable performance on discrimination, less than half models evaluated the calibration, and few models have independent external validation. The results of risk of bias assessment indicated that the predictive performance may be overestimated in the derivation dataset and is probably lower than that reported when used in practice. In addition, all EAC/BE models were developed in Western countries, and half ESCC/SD models were developed in China. Due to difference in risk factors among populations, the existing models may not be applicable for people in other high risk areas of esophageal cancer such as Eastern Asia, Southern and Eastern Africa. To ensure the generalizability of models in populations with different characteristics, more models with external validation are needed in further studies (38).
The most common predictors included in the models are well-known risk factors of esophageal cancer, such as age, sex, smoking, and BMI, although there was a variety of predictive factors between different pathological types. For example, ESCC is more related to eating habits including eating rapidly, irregular eating, intake of hot food, intake of fresh fruit, and so on, whereas the more common predictor of EAC is gastroesophageal reflux disease (GERD) (39). However, none of these risk factors are included as predictors by all models. Approximately 80% of the models included age and smoking as predictors, and about half of the models involved sex and BMI. One possible explanation is that these known risk factors did not reach statistical significance in the derivation dataset. A better method to handle these well-established predictors and those with clinical credibility in multivariate modeling is to include them in a prediction model regardless of any statistical significance (40). Some models also incorporate less common predictive factors related to data set features and population characteristics. The predictors only reported by one study include asthma inhaler use (18), waist-to-hip ratio (30), use of coal or wood as the main source of cooking fuel (11), pesticide exposure (24), opium use (17), oral health (12), and alcohol flushing (21). More evidence is needed to verify that their selection is not based on accidental association with esophageal cancer. In addition, there are models that combined genetic risk factors such as genetic polymorphisms or polygenic risk score as predictors to produce better performance. When applied to the general population in both population-based screening and opportunistic screening, the predictors included in the models must be readily obtained in primary care settings. The inclusion of genetic risk predictors may reduce the applicability of the models because they require additional measurement and costs. In summary, the selection of a suitable risk prediction model should comprehensively take the performance, the quality, and the feasibility into consideration.
The results of the risk of bias assessment show that none of the models meet all the requirements of PROBAST (40). The risk of bias in a section of participants, predictors, and outcomes is mainly caused by pitfalls in the study design. In a non-nested case–control study, case patients from a hospital with advanced conditions may be overrepresented, leading to overestimated model performance and reduced model applicability. The methodological limitations in the analysis section are as follows: (1) About 60% of the studies have an EPV lower than 10 and are likely to be overfitting in general (41, 42). For predictors with low-prevalence presented in a model, such as non-steroidal anti-inflammatory drugs (NSAIDs) use (16) and pesticide exposure (11), higher EPV is needed to eliminate bias in regression coefficients and improve predictive accuracy (41). (2) Continuous variables are transformed to multicategorical variables in more than half of the prediction models, the most common of which is categorizing age into groups. However, categorization of continuous predictors might lead to loss of information and weaken the performance of risk prediction models (43). (3) Most of the models either did not report any missing information or exclude missing value in the analysis, which may reduce the available sample size and cause selection bias (44, 45). It should be noted that risk assessment relies on the transparent reporting of studies, and poor reporting could cause the items to be evaluated as high risk or unknown risk. However, to be clear, the clinicians and policy makers, who decide which clinical prediction models should be promoted in evidence-based guidelines or implemented in practice, should refer to reviews like current study at the very beginning. Meanwhile, the high prevalence of models with a high risk of bias emphasizes the need to improve methodological quality of prediction research.
To our knowledge, this is the first review that summarizes the risk prediction models of esophageal cancer in the average-risk population. We used the recently published tool PROBAST to assess the risk of bias and applicability of the included prediction model studies and then evaluated their methodological characteristics and performance. Risk prediction models by Chen (25), Han (26), and Kunzmann (18) were recommended based on the relatively standardized measurement of predictors and outcome, large sample size, complete evaluation, and good result of model performance. The results of this study will provide a reference for the further development, validation, and application of risk prediction models in the target subjects for endoscopic screening.
A limitation of the present study is the methodological shortcomings and inadequate reporting in the included articles. It is not possible to objectively evaluate and directly compare the models based on the poor reporting. Due to the lack of validation studies, we cannot perform meta-analysis of calibration measures for the prediction models. In addition, the variation of predictors and ethnic differences contribute to the poor implementation of the existing models.
Conclusions
There have been a few risk prediction models developed for esophageal cancer with acceptable predictive ability, although certain methodological limitations, especially the low rate of external validation, should be noted. Further researches are needed to develop a reliable risk prediction model for esophageal cancer based on general population and validate the existing models in an independent dataset.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
RC, RZ, SW, and WW designed and supervised the study. RC, JZ, and SW carried out the literature research, extracted data, and wrote the original draft. All authors critically reviewed the manuscript and approved the final draft.
Funding
This study was supported by the Beijing Natural Science Foundation (7204294), National Natural Science Foundation of China (81903403 and 81974493), and the National Key R&D Program of China (2016YFC0901404 and 2018YFC1311704).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Zhang S, Sun K, Zheng R, Zeng H, Wang S, Chen R, et al. Cancer incidence and mortality in China, 2015. J Natl. Cancer Center. (2020) 1:2–11. doi: 10.1016/j.jncc.2020.12.001
3. Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. (2017) 390:2383–96. doi: 10.1016/S0140-6736(17)31462-9
4. Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. (2018) 154:360–73. doi: 10.1053/j.gastro.2017.08.023
5. Chen R, Liu Y, Song G, Li B, Zhao D, Hua Z, et al. Effectiveness of one-time endoscopic screening programme in prevention of upper gastrointestinal cancer in China: a multicentre population-based cohort study. Gut. (2021) 70:251–60. doi: 10.1136/gutjnl-2019-320200
6. Guan CT, Song GH, Li BY, Gong YW, Hao CQ, Xue LY, et al. Endoscopy screening effect on stage distributions of esophageal cancer: a cluster randomized cohort study in China. Cancer Sci. (2018) 109:1995–2002. doi: 10.1111/cas.13606
7. Kim JA, Shah PM. Screening and prevention strategies and endoscopic management of early esophageal cancer. Chin Clin Oncol. (2017) 6:50. doi: 10.21037/cco.2017.09.05
8. Ten Haaf K, Jeon J, Tammemagi MC, Han SS, Kong CY, Plevritis SK, et al. Risk prediction models for selection of lung cancer screening candidates: a retrospective validation study. PLoS Med. (2017) 14:e1002277. doi: 10.1371/journal.pmed.1002277
9. Cooper JA, Ryan R, Parsons N, Stinton C, Marshall T, Taylor-Phillips S. The use of electronic healthcare records for colorectal cancer screening referral decisions and risk prediction model development. BMC Gastroenterol. (2020) 20:78. doi: 10.1186/s12876-020-01206-1
10. Harkness EF, Astley SM, Evans DG. Risk-based breast cancer screening strategies in women. Best Pract Res Clin Obstet Gynaecol. (2020) 65:3–17. doi: 10.1016/j.bpobgyn.2019.11.005
11. Liu M, Liu Z, Cai H, Guo C, Li X, Zhang C, et al. A model to identify individuals at high risk for esophageal squamous cell carcinoma and precancerous lesions in regions of high prevalence in China. Clin Gastroenterol Hepatol. (2017) 15:1538–46.e7. doi: 10.1016/j.cgh.2017.03.019
12. Thrift AP, Kendall BJ, Pandeya N, Whiteman DC. A model to determine absolute risk for esophageal adenocarcinoma. Clin Gastroenterol Hepatol. (2013) 11:138–44.e2. doi: 10.1016/j.cgh.2012.10.026
13. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
14. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD). Ann Intern Med. (2015) 162:735–6. doi: 10.7326/L15-5093-2
15. Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. (2019) 170:51–8. doi: 10.7326/M18-1376
16. Dong J, Buas MF, Gharahkhani P, Kendall BJ, Onstad L, Zhao S, et al. Determining Risk of Barrett's esophagus and esophageal adenocarcinoma based on epidemiologic factors and genetic variants. Gastroenterology. (2018) 154:1273–81.e3. doi: 10.1053/j.gastro.2017.12.003
17. Etemadi A, Abnet CC, Golozar A, Malekzadeh R, Dawsey SM. Modeling the risk of esophageal squamous cell carcinoma and squamous dysplasia in a high risk area in Iran. Arch Iran Med. (2012) 15:18–21. Available online at: https://pubmed.ncbi.nlm.nih.gov/22208438/
18. Kunzmann AT, Thrift AP, Cardwell CR, Lagergren J, Xie S, Johnston BT, et al. Model for identifying individuals at risk for esophageal adenocarcinoma. Clin Gastroenterol Hepatol. (2018) 16:1229–36.e4. doi: 10.1016/j.cgh.2018.03.014
19. Wang QL, Lagergren J, Xie SH. Prediction of individuals at high absolute risk of esophageal squamous cell carcinoma. Gastrointest Endosc. (2019) 89:726–32.e2. doi: 10.1016/j.gie.2018.10.025
20. Xie SH, Lagergren J. A model for predicting individuals' absolute risk of esophageal adenocarcinoma: moving toward tailored screening and prevention. Int J Cancer. (2016) 138:2813–9. doi: 10.1002/ijc.29988
21. Yokoyama T, Yokoyama A, Kumagai Y, Omori T, Kato H, Igaki H, et al. Health risk appraisal models for mass screening of esophageal cancer in Japanese men. Cancer Epidemiol Biomarkers Prev. (2008) 17:2846–54. doi: 10.1158/1055-9965.EPI-08-0397
22. Yang X, Suo C, Zhang T, Yin X, Man J, Yuan Z, et al. A nomogram for screening esophageal squamous cell carcinoma based on environmental risk factors in a high-incidence area of China: a population-based case-control study. BMC Cancer. (2021) 21:343. doi: 10.1186/s12885-021-08053-7
23. Baldwin-Hunter BL, Knotts RM, Leeds SD, Rubenstein JH, Lightdale CJ, Abrams JA. Use of the electronic health record to target patients for non-endoscopic barrett's esophagus screening. Digest Dis Sci. (2019) 64:3463–70. doi: 10.1007/s10620-019-05707-2
24. Chang J, Huang Y, Wei L, Ma B, Miao X, Li Y, et al. Risk prediction of esophageal squamous-cell carcinoma with common genetic variants and lifestyle factors in Chinese population. Carcinogenesis. (2013) 34:1782–6. doi: 10.1093/carcin/bgt106
25. Chen W, Li H, Ren J, Zheng R, Shi J, Li J, et al. Selection of high-risk individuals for esophageal cancer screening: a prediction model of esophageal squamous cell carcinoma based on a multicenter screening cohort in rural China. Int J Cancer. (2021) 148:329–39. doi: 10.1002/ijc.33208
26. Han J, Wang L, Zhang H, Ma S, Li Y, Wang Z, et al. Development and validation of an esophageal squamous cell carcinoma risk prediction model for rural chinese: multicenter cohort study. Front Oncol. (2021) 11:729471. doi: 10.3389/fonc.2021.729471
27. Ireland CJ, Fielder AL, Thompson SK, Laws TA, Watson DI, Esterman A. Development of a risk prediction model for Barrett's esophagus in an Australian population. Dis Esophagus. (2017) 30:1–8. doi: 10.1093/dote/dox033
28. Koyanagi YN, Ito H, Oze I, Hosono S, Tanaka H, Abe T, et al. Development of a prediction model and estimation of cumulative risk for upper aerodigestive tract cancer on the basis of the aldehyde dehydrogenase 2 genotype and alcohol consumption in a Japanese population. Eur J Cancer Prev. (2017) 26:38–47. doi: 10.1097/CEJ.0000000000000222
29. Liu Z, Guo C, He Y, Chen Y, Ji P, Fang Z, et al. A clinical model predicting the risk of esophageal high-grade lesions in opportunistic screening: a multicenter real-world study in China. Gastrointest Endosc. (2020) 91:1253–60.e3. doi: 10.1016/j.gie.2019.12.038
30. Rubenstein JH, Morgenstern H, Appelman H, Scheiman J, Schoenfeld P, McMahon LF Jr, et al. Prediction of Barrett's esophagus among men. Am J Gastroenterol. (2013) 108:353–62. doi: 10.1038/ajg.2012.446
31. Shen Y, Xie S, Zhao L, Song G, Shao Y, Hao C, et al. Estimating individualized absolute risk for esophageal squamous cell carcinoma: a population-based study in high-risk Areas of China. Front Oncol. (2020) 10:598603. doi: 10.3389/fonc.2020.598603
32. Wang QL, Ness-Jensen E, Santoni G, Xie SH, Lagergren J. Development and validation of a risk prediction model for esophageal squamous cell carcinoma using cohort studies. Am J Gastroenterol. (2020) 116:683–91. doi: 10.14309/ajg.0000000000001094
33. Xie SH, Ness-Jensen E, Medefelt N, Lagergren J. Assessing the feasibility of targeted screening for esophageal adenocarcinoma based on individual risk assessment in a population-based cohort study in Norway (The HUNT Study). Am J Gastroenterol. (2018) 113:829–35. doi: 10.1038/s41395-018-0069-9
34. Anothaisintawee T, Teerawattananon Y, Wiratkapun C, Kasamesup V, Thakkinstian A. Risk prediction models of breast cancer: a systematic review of model performances. Breast Cancer Res Treat. (2012) 133:1–10. doi: 10.1007/s10549-011-1853-z
35. Peng L, Weigl K, Boakye D, Brenner H. Risk scores for predicting advanced colorectal neoplasia in the average-risk population: a systematic review and meta-analysis. Am J Gastroenterol. (2018) 113:1788–800. doi: 10.1038/s41395-018-0209-2
36. Usher-Smith JA, Walter FM, Emery JD, Win AK, Griffin SJ. Risk prediction models for colorectal cancer: a systematic review. Cancer Prev Res. (2016) 9:13–26. doi: 10.1158/1940-6207.CAPR-15-0274
37. Gray EP, Teare MD, Stevens J, Archer R. Risk prediction models for lung cancer: a systematic review. Clin Lung Cancer. (2016) 17:95–106. doi: 10.1016/j.cllc.2015.11.007
38. Moons KG, Kengne AP, Grobbee DE, Royston P, Vergouwe Y, Altman DG, et al. Risk prediction models: II. external validation, model updating, and impact assessment. Heart. (2012) 98:691–8. doi: 10.1136/heartjnl-2011-301247
39. Shiota S, Singh S, Anshasi A, El-Serag HB. Prevalence of Barrett's esophagus in asian countries: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. (2015) 13:1907–18. doi: 10.1016/j.cgh.2015.07.050
40. Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. (2019) 170:W1–33. doi: 10.7326/M18-1377
41. Ogundimu EO, Altman DG, Collins GS. Adequate sample size for developing prediction models is not simply related to events per variable. J Clin Epidemiol. (2016) 76:175–82. doi: 10.1016/j.jclinepi.2016.02.031
42. van Smeden M, de Groot JA, Moons KG, Collins GS, Altman DG, Eijkemans MJ, et al. No rationale for 1 variable per 10 events criterion for binary logistic regression analysis. BMC Med Res Methodol. (2016) 16:163. doi: 10.1186/s12874-016-0267-3
43. Royston P, Altman DG, Sauerbrei W. Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med. (2006) 25:127–41. doi: 10.1002/sim.2331
44. Vergouwe Y, Royston P, Moons KG, Altman DG. Development and validation of a prediction model with missing predictor data: a practical approach. J Clin Epidemiol. (2010) 63:205–14. doi: 10.1016/j.jclinepi.2009.03.017
Keywords: methodology, risk of bias, systematic review, esophageal cancer, risk prediction model
Citation: Chen R, Zheng R, Zhou J, Li M, Shao D, Li X, Wang S and Wei W (2021) Risk Prediction Model for Esophageal Cancer Among General Population: A Systematic Review. Front. Public Health 9:680967. doi: 10.3389/fpubh.2021.680967
Received: 15 March 2021; Accepted: 29 October 2021;
Published: 01 December 2021.
Edited by:
Kevin Lu, University of South Carolina, United StatesReviewed by:
Damilola Olajide, University of Nottingham, United KingdomShaowen Tang, Nanjing Medical University, China
Copyright © 2021 Chen, Zheng, Zhou, Li, Shao, Li, Wang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengfeng Wang, c2hlbmdmZW5nMTk4NEAxMjYuY29t; Wenqiang Wei, d2Vpd3FAY2ljYW1zLmFjLmNu
†These authors share first authorship
 Ru Chen
Ru Chen Rongshou Zheng
Rongshou Zheng Jiachen Zhou
Jiachen Zhou Minjuan Li
Minjuan Li Dantong Shao
Dantong Shao Xinqing Li
Xinqing Li Shengfeng Wang
Shengfeng Wang Wenqiang Wei
Wenqiang Wei