
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Public Health , 23 November 2020
Sec. Environmental Health and Exposome
Volume 8 - 2020 | https://doi.org/10.3389/fpubh.2020.569209
 Andreina Baj1*
Andreina Baj1* Lorenzo Azzi2
Lorenzo Azzi2 Daniela Dalla Gasperina3
Daniela Dalla Gasperina3 Angelo Genoni1
Angelo Genoni1 Antonio Tamborini1
Antonio Tamborini1 Cinzia Gambarini4
Cinzia Gambarini4 Giulio Carcano5
Giulio Carcano5 Paolo Grossi3
Paolo Grossi3 Fausto Sessa6
Fausto Sessa6Only 4 months after the beginning of SARS-CoV-2 epidemic, the world is facing a global pandemic due to a complex and insidious virus that today constantly poses new challenges. In this study, we highlight a persistent shedding of SARS-CoV-2 RNA into the urine, even in patients with a negative nasopharyngeal swab and in patients considered recovered. What does it mean? Besides the fact that the kidney is a probable site of viral replication, the prolonged viral excretion is a matter of great concern for our drainage system contamination.
After having looked at China as the place of onset of an epidemic in December 2019, on February 18, 2020 we recorded the first case of COronaVIrus Disease 2019 (COVID-19) in Italy. Since then, in only 2 months, our reports have accounted for 233,000 cases and more than 33,000 deaths (1).
The viral agent, SARS-CoV-2, belongs to Coronaviridae family and, together with SARS-CoV and MERS-CoV, is the third zoonotic coronavirus that, in recent history, has emerged in the human population causing severe and unexpected disease outbreaks (2, 3).
In this storm that has caught everyone unprepared, we have tried to cope with many issues related to the diagnosis and management of this disease, and now many others are coming to light. The gold standard to detect the virus in the patients affected by COVID-19 is an rRT-PCR on their nasopharyngeal swabs, both for diagnosis and follow-up. After two negative swabs, patients are discharged but the SARS-CoV-2 RNA could be still present in the saliva (4, 5), and probably in other body districts, possibly acting as a viral reservoir (6).
In this study, we demonstrate that SARS-CoV-2 RNA is detected in urine of COVID-19 patients even when the patient is no longer symptomatic and in some cases for a long time.
From this perspective, a prolonged viral shedding into the urine could be a serious environmental problem because the virus enters the drainage system of urban and non-urban centers, making its presence everlasting (7). Taking into account that, according to the United Nations, more than 80% of the world's wastewater flows back into the environment without treatment (8), it is not difficult to imagine sewage as a viral reservoir that continuously contaminates the environment.
Of interest, even if fecal shedding of SARS-CoV-2 RNA is frequently observed in COVID-19 patients and it supports a productive infection of enterocytes, it is rapidly inactivated after its release into the intestinal lumen (9). This observation raises the need for further studies to evaluate the viability of SARS-CoV-2 in different environments to assess its infectivity.
Twenty-two SARS-CoV-2–infected patients with severe or very severe disease were recruited into the study following the consecutive sampling technique, in agreement with the Helsinki declaration. Patients were admitted to our hospital (ASST Sette Laghi, Lombardia, Italy) after being diagnosed with COVID-19 through an rRT-PCR on their nasopharyngeal swabs (GeneFinder COVID-19 Plus Real Amp Kit; Elitech, Puteaux, France). Their clinical situation was classified according to the Diagnosis and Treatment Plan of COVID-19 issued by the Chinese National Health Commission (10).
A single urine sample was obtained from each patient, in conjunction with the second nasopharyngeal swab.
One milliliter of urine specimens was centrifuged at 1,500 rpm for 5 min; the obtained pellets, resuspended in 140 μl of phosphate buffered saline, were subjected to RNA extraction by QIAmp Viral RNA mini kit (Qiagen) according to the manufacturer's instructions and eluted in 60 μl. One-step rRT-PCR was performed using Luna Universal qPCR Master Mix (New England Biolabs) from 5 μl of extracted RNA and a SYBR GREEN detection method. Forward (5′-ACCTTCCCAGGTAACAAACCA-3′) and reverse (5′-TTACCTTTCGGTCACACCCG-3′) primers targeting the 5′UTR region of SARS-CoV-2, designed using CLC genomics workbench (QIAGEN), were used (4, 5).
All samples were run in four replicates, together with urine from healthy people as a negative control, and a quantified positive control (Twist Bioscience SARS-CoV-2 RNA Control). Provided at a concentration of 106 copies μl, serial dilutions of RNA control from 105 to 10 copies/μl were used to construct a standard curve (Figure 1).
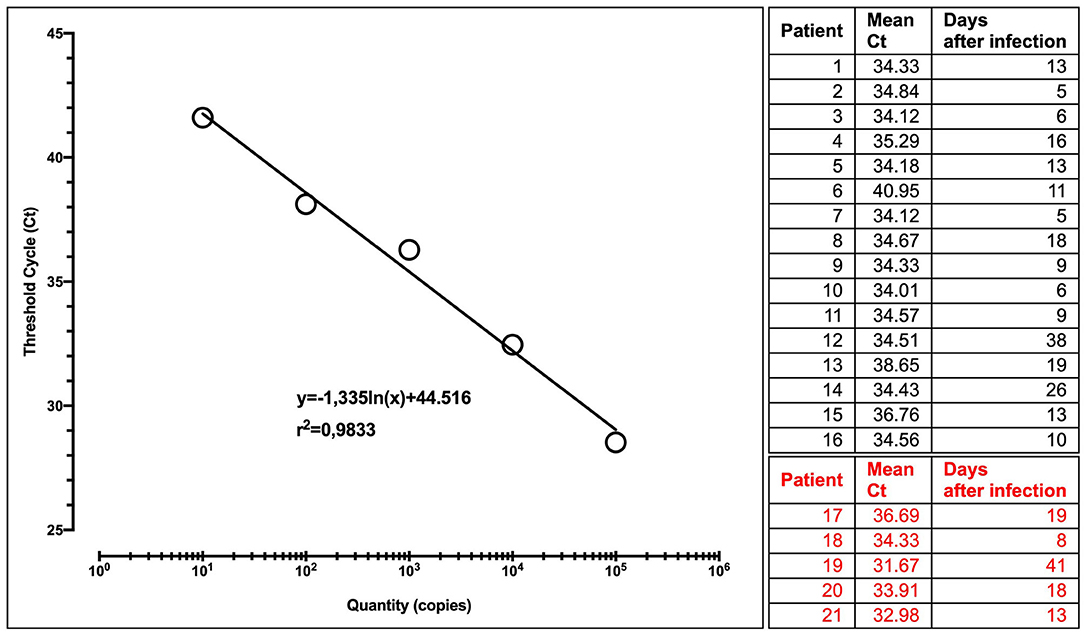
Figure 1. Calibration curve with the regression equation (characterized by the slope and intercept) and regression coefficient. Patients tested at different times after infection are shown. In red, patients with a precedent or contextually negative nasopharyngeal swab.
RT-PCR was performed using Abi Prism 7000 sequence detection system (Applied Biosystems), with an annealing temperature of 60°C.
In case of uncertain result, an endpoint RT-PCR and subsequent sequencing of 5′UTR region was performed on the same RNA. To this end, the 251-bp amplicon was detected with LabChipGx Touch24 (Perkin Elmer) and sequenced by the Sanger method (SeqStudio Genetic Analyzer; Thermo Fisher Scientific).
A total of 22 subjects, 14 males and 6 females, were analyzed in this study. Age values ranged from 37 to 82 years, with a mean age of 61 ± 12.5 years.
All patients were affected by severe or very severe COVID-19 and hospitalized in the Intensive Care Unit, in the Unit of Infectious and Tropical Diseases, and in Pneumology.
In all observed patients, microhematuria was present, associated with proteinuria in 70% and hypoalbuminemia in 89% of the subjects. Serum creatinine and urea level were higher than normal in 45 and 62% of the patients, respectively (data not shown).
Interestingly, in all patients but one (95%), we found RNA of SARS-CoV-2 in urine sediment, in a period lasting until 41 days after the onset of COVID-19 symptoms (Figure 1).
Of note, in five patients (23%), viral RNA was present in urine but no longer detectable in nasopharyngeal swab at the time of enrollment in the study. Among them, a patient with a constantly negative nasopharyngeal swab for 30 days but with a high urine viral load after 41 day after the onset of symptoms is of particular interest.
With time, many aspects of SARS-CoV-2 infection are coming to light. In these last few months, scientific evidence has shown us that it is a respiratory virus, but the resulting infection is not localized, and clinical pictures are suggestive of extrapulmonary viral dissemination. Furthermore, virus shedding could occur from different sites and not only through respiratory droplets (6, 7).
In our study, we demonstrated a significant presence of viral genome in the urine of COVID-19 patients, even when the virus RNA itself was no longer detectable at nasopharyngeal level and also when symptoms had disappeared.
This paper is part of an ongoing study and the number of patients analyzed at this moment does not allow to establish a correlation between renal damage and viral presence in urine. Nevertheless, the presence of virus receptor ACE2 on podocytes and tubular cells was described; thus, we could presume a possible direct infection of the kidney (11) and additional studies reported the kidney as specific target for SARS-CoV-2 infection (12). In a recent study, acute kidney injury developed in 36.6% of the patients admitted with COVID-19, with high rates of proteinuria and hematuria (13).
With this preliminary work, we would like to draw close attention to the growing concern for the environment, especially in some cities and areas with a high population density. A prolonged shedding of the virus into the urine, even when the patient is no longer symptomatic and with negative nasopharyngeal swabs, could mean a virus release into the sewage system and, through it, its spread in the environment.
Many aspects of SARS-CoV-2 remain to be considered: its viability in different body fluids, its environmental stability, and its transmission via different routes, but this finding raises a concern about the management of recovering patients and, above all, the treatment of wastewater. Strict monitoring of this process is essential to prevent viral contamination. Further studies are needed to investigate these hypotheses.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by This study was carried out in agreement with the Helsinki declaration, authorized by the Hospital Direction, due to the situation of emergency and approved by the Ethical Committee of the University of Insubria (study n° 68/2000). The patients/participants provided their written informed consent to participate in this study.
AB, LA, DD, and FS contributed to study design, data collection, data interpretation, the literature search, and writing of the report. CG, GC, and PG contributed to patients' recruitment, data collection, and clinical management. AT managed the laboratory diagnostic procedures and collected data. AG managed the laboratory experimental procedures. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Professor Marina Tettamanti supervised the revision process in the English language.
1. COVID-19 Situation Report. Available online at: http://www.salute.gov.it/portale/nuovocoronavirus/homeNuovoCoronavirus.jsp?lingua=english
2. Saif ur Rehman, Laiba Shafique, Awais Ihsan QL. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. (2020) 9:240. doi: 10.3390/pathogens9030240
3. Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Singh Malik Y, et al. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev. (2020) 33:4:e00028–20. doi: 10.1128/CMR.00028-20
4. Azzi L, Carcano G, Gianfagna F, Grossi P, Dalla Gasperina D, Genoni A, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. (2020) 81:E45–50. doi: 10.1016/j.jinf.2020.04.005
5. Azzi L, Carcano G, Dalla Gasperina D, Sessa F, Maurino V, Baj A. Two cases of COVID-19 with positive salivary and negative pharyngeal or respiratory swabs at hospital discharge: a rising concern. Oral Dis. (2020) 1–3. doi: 10.1111/odi.13368
6. SanJuan-Reyes S, Gómez-Oliván LM, Islas-Flores H. COVID-19 in the environment. Chemosphere. (2020) 263:127973. doi: 10.1016/j.chemosphere.2020.127973
7. United Nations. UN World Water Development Report, Wastewater: The Untapped Resource, Paris: UNESCO (2017).
8. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. (2020) 5:eabc3582. doi: 10.1126/sciimmunol.abc3582
9. Kataki S, Chatterjee S, Vairale MG, Sharma S, Dwivedi SK. Concerns and strategies for wastewater treatment during COVID-19 pandemic to stop plausible transmission. Resour Conserv Recycl. (2021) 164:105156. doi: 10.1016/j.resconrec.2020.105156
10. Peng F, Tu L, Yang Y, Sun J, Xu G, Chang C, et al. Management and treatment of COVID-19: the Chinese experience. Can J Cardiol. (2020) 36:6:915–30. doi: 10.1016/j.cjca.2020.04.010
11. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. (2020) 14:85–192.12. doi: 10.1007/s11684-020-0754-0
12. Sharma P, Uppal NN, Wanchoo R, Shah HH, Yang Y, Parikh R, et al. COVID-19-associated kidney injury: a case series of kidney biopsy findings. J Am Soc Nephrol. (2020) 9:1948–58. doi: 10.1681/ASN.2020050699
Keywords: shedding, wastewater, urine, COVID-19, SARS CoV-2
Citation: Baj A, Azzi L, Dalla Gasperina D, Genoni A, Tamborini A, Gambarini C, Carcano G, Grossi P and Sessa F (2020) Pilot Study: Long-Term Shedding of SARS-CoV-2 in Urine: A Threat for Dispersal in Wastewater. Front. Public Health 8:569209. doi: 10.3389/fpubh.2020.569209
Received: 11 June 2020; Accepted: 22 October 2020;
Published: 23 November 2020.
Edited by:
Alesia Coralie Ferguson, North Carolina Agricultural and Technical State University, United StatesReviewed by:
Dongyang Deng, North Carolina Agricultural and Technical State University, United StatesCopyright © 2020 Baj, Azzi, Dalla Gasperina, Genoni, Tamborini, Gambarini, Carcano, Grossi and Sessa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreina Baj, YW5kcmVpbmEuYmFqQHVuaW5zdWJyaWEuaXQ=; YW5kcmVpbmEuYmFqQGFzc3Qtc2V0dGVsYWdoaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.