- 1Infectious Diseases Division, International Center for Diarrhoeal Disease Research, Dhaka, Bangladesh
- 2Department of Biotechnology and Genetic Engineering, Noakhali Science and Technology University, Noakhali, Bangladesh
- 3Department of International Health, Johns Hopkins Bloomberg School of Public Health Baltimore, Baltimore, MD, United States
- 4Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, Bangladesh
Recurrent cholera causes significant morbidity and mortality in cholera endemic estuarine areas of Bangladesh. There have been limited studies to investigate the transmission patterns of V. cholerae associated with cholera in Bangladesh. In this study, we characterized V. cholerae serogroup O1 isolated from 30 cholera patients, 76 household contacts, 119 stored drinking water samples, and 119 water source samples in Bakerganj and Mathbaria, two cholera endemic coastal regions in Bangladesh. Results of phenotypic and molecular characterization of V. cholerae isolates (n = 56) confirmed them to be toxigenic belonging to serogroup O1 biotype El Tor (ET), and possessing cholera toxin of the classical biotype (altered ET). Molecular fingerprinting of the V. cholerae O1 of clinical and water origins determined by PFGE of Not-I- digested genomic DNA showed them to be closely related, as the PFGE banding patterns were highly homogenous. Phylogenetic analysis using dendrogram of cholera patients, household contacts, and household groundwater sources showed isolates within households to be clonally linked, suggesting water as an important vehicle of transmission of cholera in the coastal villages of Bangladesh. Transmission of toxigenic V. cholerae O1 through drinking water in cholera endemic rural settings underscores the urgent need for evidence based water, sanitation, and hygiene interventions promoting safe drinking water to prevent morbidity and mortality related to cholera and other enteric infections in Bangladesh.
Introduction
Cholera is an acute dehydrating, potentially life-threatening diarrheal disease, transmitted through contaminated drinking water and poor WASH infrastructure and practices in low resource settings (1). Worldwide cholera remains a major public health problem can cause up to an estimated 4.0 million cases and 95,000 deaths annually (2).
Cholera is a severe form of acute diarrhea, caused by the gamma-proteobacterium Vibrio cholerae. Of the more than 200 serogroups of V. cholerae, only serogroups O1 and O139 which possess potent cholera toxin (CT) encoded by a filamentous prophage lysogenizing into the genome of the bacterium are responsible for the epidemic and pandemic cholera worldwide (3). Serogroup O1 has two biotypes: classical (CL) and El Tor (ET) which differ in major phenotypic and genetic traits (4).
V. cholerae O1classical biotype caused the sixth and presumably earlier pandemics out of seven cholera pandemics before being replaced with the ET biotype which has been responsible for the ongoing seventh pandemic since 1961 (4, 5). The El Tor biotype strains have undergone genetic changes such as a new hybrid El Tor carrying the classical biotype CT (6). These El Tor variant strains are referred to as “hyper-virulent” owing to their ability to produce more cholera toxin, greater spreading ability during epidemics, and increased competitive fitness for colonization than many ET isolates (7–9).
The actual mode of transmission of V. cholerae among cholera cases remains debated. Some study suggests that V. cholerae transmits fecal-orally through contaminated water (7, 10–12) following a “slow” human-to-aquatic environment-to-human pathway (13). However, a key element in transmission may be a recently recognized hyper infectious phase, which persists for hours after passage in diarrheal feces and can transmit rapidly among the household members of cholera patients following a faster “human-to-human” transmission route through fecal-oral contamination (13, 14). Cholera patients and the environmental reservoir can be considered as a potential source of outbreak but their respective relations to cholera transmission have been heavily disputed. Household members of cholera patients are at more than 100 times higher risk of a cholera infection than the general population (15). Furthermore, in spite of the substantial work done in this field, the exact source and mode of transmission for amplification of the disease to reach an epidemic status is still not completely elucidated.
Previous studies have shown the estuarine aquatic environment of the Bay of Bengal to be natural habitat for toxigenic V. cholerae (16–19). In these regions, the low salinity of rivers or shallow wells (averaging between 2.8 and 8.2 ppt) and temperatures between 26° and 35°C during the dry season (20) favors the growth and multiplication of V. cholerae. People living in this region are at high risk of cholera as salt intrusion (20) into the surface water bodies supports the growth and survival of toxigenic V. cholerae. The high population densities present an ideal environment for people to be infected with V. cholerae which causes deadly disease cholera (13, 18, 21). In addition, previous studies have found contaminated household water as an important risk factor for cholera among the household members and are likely attributed to sharing contaminated environmental sources or secondary transmission through poor hygiene practices (22–24).
In our recent cohort study investigating risk factors for cholera infections among 76 household contacts of cholera patients in the rural Bakerganj and Mathbaria, Bangladesh, we found that 37% of cholera patient households had a household member with a cholera infection by bacterial culture. Furthermore, 27% of stored household drinking water, and 13% of water sources in patient households had V. cholerae. In addition, household contacts with V. cholerae in their source water had a significantly higher odds of a symptomatic cholera infection 25. Rafique et al. (26), by analyzing V. cholerae O1 isolated from clinical cholera cases, their household contacts and drinking and water source samples as part of a randomized controlled trial of hospital based hand washing with soap and water treatment intervention [Cholera-Hospital-Based Intervention-for-7-Days (CHoBI7) Trial] (27) by pulsed-field gel electrophoresis (PFGE), showed water as the source of transmission of cholera among the study households in Dhaka city, Bangladesh. However, there are no studies, to our knowledge, that have investigated cholera transmission patterns in patient households in a rural setting, especially in coastal villages of Bangladesh which is important for identifying interventions to reduce cholera transmission. In the present study, we investigate person to person and environmental transmission routes for cholera infection among household contacts of cholera cases using pulsed-field gel electrophoresis (PFGE) of selected V. cholerae O1 isolated from coastal villages of Bangladesh namely Mathbaria and Bakerganj to identify the transmission routes that should be targeted in future interventions.
Materials and Methods
Description of Study Sites
Our study was conducted in Bakerganj and Mathbaria, Bangladesh. Bakerganj is in the southern district of Barisal, about 70 km north of the coast of the Bay of Bengal and approximately 300 km southwest of Dhaka, the capital city of Bangladesh (Figure 1). Additional details on this study site are published in Alam et al. (16). Previous studies have shown significant correlations between the ecology and epidemiology of V. cholerae and selected environmental parameters in Bakerganj (21, 28).
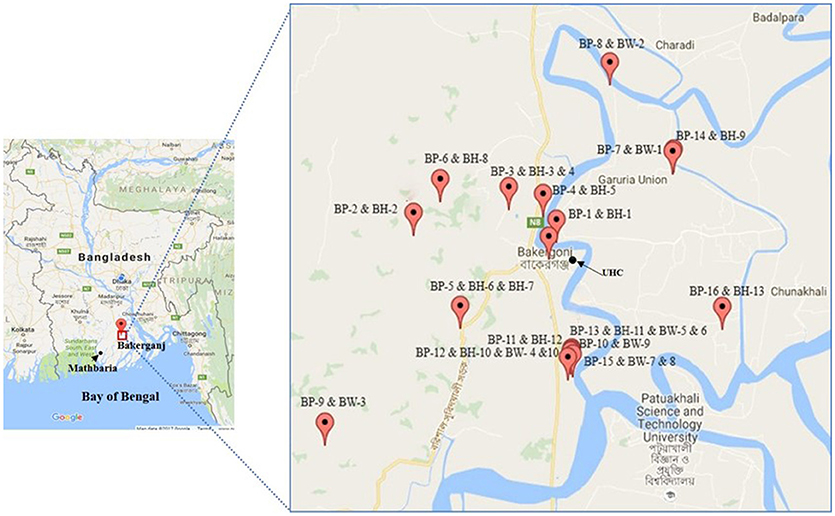
Figure 1. Map of Bakerganj with exact GPS (global positioning system) point for the location of households of cholera patients (BP, Bakerganj patient; BH, Bakerganj household contacts; BW, Bakerganj water; UHC, Upazila Health Complex). This map shows the distribution of representative (n = 16) households of cholera patients.
The second study site is Mathbaria, which is located adjacent to the Bay of Bengal, in close proximity to the Sunderban mangrove forest and approximately 400 km southwest of Dhaka (Figure 2). The major river, Baleshwar, flows along the western boundary of Mathbaria, next to a tropical mangrove forest of the Sundarbans, the temporary island system of that part of the Bay of Bengal. A previous study found the presence of toxigenic V. cholerae in the aquatic environment of Mathbaria which has been responsible for the endemicity of cholera in this area (17).
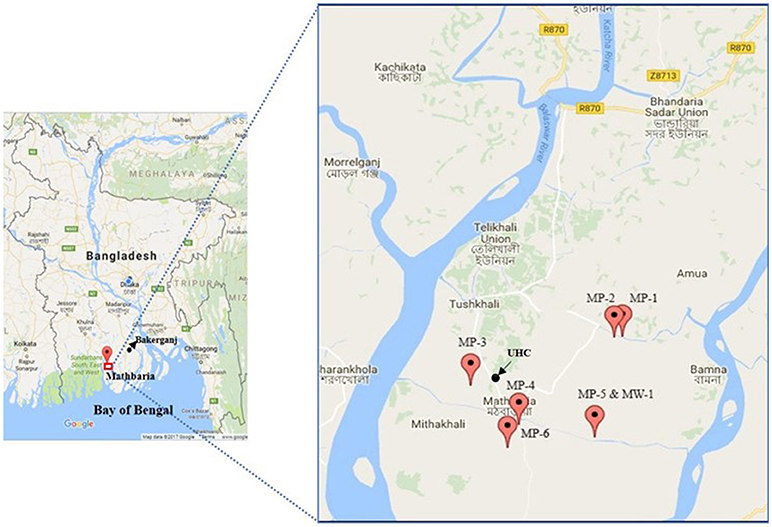
Figure 2. Map of Mathbaria with exact GPS (global positioning system) point for the location of households of cholera patients (MP, Mathbaria patient; MW, Mathbaria household; UHC, Upazila Health Complex). This map shows the distribution of representative (n = 6) households of cholera patients.
Research Procedures
Enrollment
Suspected cholera patients of any age and both sexes were recruited from hospitals in Bakerganj and Mathbaria. Suspected cholera patients (irrespective of age) were defined as anyone reporting diarrhea (3 or more loose stools within a 24-h period) and moderate to severe clinical dehydration (using WHO classification). All those individuals meeting these criteria who were admitted to Mathbaria and Bakerganj upazila government health complexes were screened for cholera using the Crystal VC Rapid Dipstick test. All individuals with a dipstick result positive for either V. cholerae O1 or O139, had their stool samples tested by bacterial culture for V. cholerae. A cholera patient was defined as an individual with diarrhea and a stool culture result positive for V. cholerae.
Once the index cholera patient was enrolled, their household contacts were invited to participate in the study. Household Contacts were defined as individuals sharing the same cooking pot with the cholera patients for the past 3 days. Household contacts of cholera patients residing were recruited and prospectively followed for 7 days after the presentation of the cholera patient at the local Government Upazila Health Complex (UHC). Eligible household contacts present in the health facility at the time of case enrollment were invited to participate. In addition, a household visit was made to recruit additional household contacts within 24–36 h of enrollment of the case. Household contacts were visited at 1, 3, 5, and 7 days for clinical and environmental surveillance.
Written informed consent was obtained from all study participants (household contacts and index cholera cases), this included adult participant (≥18 years of age) signing an informed consent and/or parental consent form and children between the ages of 12 and 17 years old signing an assent form. If a study participant could not read, the consent form was read to the participant and was asked to document their consent with an x in the presence of a witness. All study procedures were approved by the research Ethical Review Committee of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) and IRB of the Johns Hopkins Bloomberg School of Public Health.
Clinical Surveillance
For clinical surveillance, household contacts were asked if they had diarrhea or vomiting during the surveillance period, and fecal specimens were collected at each household visit to capture the majority of cholera infections that develop during the 7-day study period.
Environmental Surveillance
Environmental surveillance was conducted during all household visits. Water samples were collected from the household's primary drinking water source and from drinking water stored in the home to measure V. cholerae by bacterial culture.
Sample Collection and Processing
Fecal specimens (stool) from index patients and their household contacts were collected aseptically and placed immediately in Cary-Blair media and transported to laboratory maintaining cold-chain. The water samples were collected using sterile 500 mL dark Nalgene bottles (Nalgene Nunc International, St. Louis, Mo.) and transported to the icddr,b laboratory putting in an insulated cool box (with ice packs). Stool samples from the cholera patients (index patient) were collected from Mathbaria and Bakerganj upazila government health complexes. Water samples were filtered through 0.22 μm filter papers and the membrane filters were then enriched in alkaline peptone water (APW) (pH 8.4) at 37°C for 4–6 h, and then cultured on selective media, as described previously (29). Stool samples were also subjected to APW enrichment and subsequent culture following same procedure. Clinical and water isolates were preserved for microbiological analysis.
Detection and Characterization of V. Cholerae
After culturing on selective media, isolation and identification of typical V. cholerae colonies were performed according to standard methods (30). The serogroups of the V. cholerae strains were determined serologically by a slide agglutination test using specific polyvalent antisera for V. cholerae O1 and O139. The serotypes of these strains were confirmed using serotype-specific monovalent Inaba and Ogawa antisera (29).
All phenotypically identified V. cholerae isolates were further confirmed by PCR targeting the V. cholerae species-specific gene ompW (31). Multiplex PCR assays were performed to identify genes encoding O1 (rfbO1) and O139 (rfbO139)-specific O biosynthetic genes, as well as the major virulence gene ctxA (32). To complement the biotype characterization by phenotypic traits, PCR assays were performed to detect the tcpA allele (CL and ET) (33), the type of the rstR gene, the presence of rstC gene encoding the phage transcriptional regulator (34), and the rtxC gene of RTX (repeat in toxin) (35) according to previously published methods. Double mismatch amplification mutation assay (DMAMA)-PCR was used to distinguish between the CL (ctxB genotype 1), ET (ctxB genotype 3) and Haitian types (ctxB genotype 7) of ctxB alleles by focusing on nucleotide positions 58 and 203 of the ctxB gene. DMAMA-PCR was performed in this study to detect the ctxB genotype using the primers and conditions described elsewhere (36).
Pulsed-Field Gel Electrophoresis (PFGE)
The whole agarose-embedded genomic DNA for V. cholerae was prepared. PFGE was carried out with a contour-clamped homogeneous electrical field (CHEF-DR II) apparatus (Bio-Rad), according to procedures described elsewhere (37). Genomic DNAs of the test strains were digested by the NotI restriction enzyme (Gibco-BRL), and Salmonella enterica serovar Braenderup was digested by XbaI, with the fragments being used as molecular size markers. The restriction fragments were separated in 1% pulsed-field-certified agarose in 0.5X TBE (Tris/borate-EDTA) buffer. In the post electrophoresis gel treatment step, the gel was stained and de-stained. The DNA was visualized using a UV trans illuminator, and images were digitized by a 1D gel documentation system (Bio-Rad). The fingerprint pattern in the gel was analyzed using the Bionumeric software (Version 3.1). A dendrogram was constructed on the basis of banding similarity and dissimilarity using the Dice similarity coefficient and unweighted-pair group method (UPGMA) as recommended by the manufacturer.
Results
Occurrence and Distribution of V. Cholerae Among Study Households
We performed genetic characterization of 56 V. cholerae O1 isolated from 30 cholera patients; 76 household contacts and 238 water samples (119 stored drinking water samples, and 119 water source samples) collected from 30 households. We have included the location of 22 representative households (16 in Bakerganj and 6 in Mathbaria) in Figures 1, 2. The map shows that majority of households with multiple V. cholerae infections in Bakerganj were located at the western and southern side of the Bighai river which is used for drinking, bathing and other household activities (e.g., washing kitchen utensils, vegetables) by nearby residents. In Mathbaria, all the households of cholera patients were located at the eastern side of Baleshwar river connected to the Bay of Bengal. From both maps, it was also found that households with multiple cholera infections were clustered around the Upazila Health Complexes (UHC) of Bakerganj and Mathbaria.
Molecular Characteristics of V. Cholerae
All isolates were serologically confirmed to be V. cholerae O1, and all possessed “O” serogoup-specific gene rfbO1 thereby complementing the serological results (Table 1). All V. cholerae strains belonged to serotype Ogawa; and all possessed the cholera toxin gene, ctxA. In addition, an ET biotype-specific toxin co-regulated pilus (tcpAET), a phage transcription regulation gene (rstRET), a phage transcription anti-repressor gene (rstC), and a repeat in toxin (rtxC) were found among in all V. cholerae O1 isolates, confirming the El Tor biotype traits. All V. cholerae strains possessed ctxB genotype 1, which is the classical type CT, confirming that the bacterium was El Tor but possessed classical biotype CT, characteristics of altered El Tor strains.
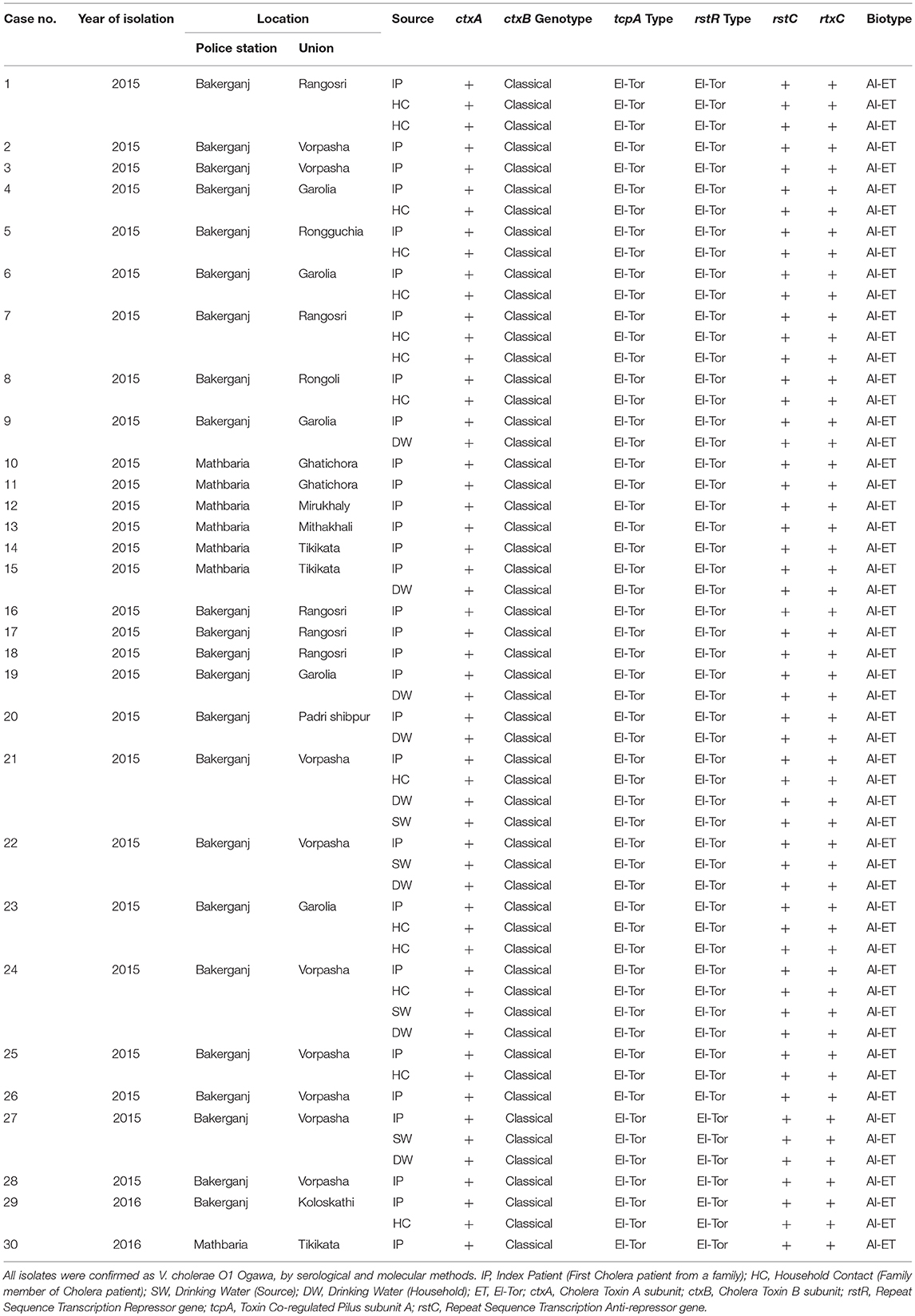
Table 1. Molecular characteristics of V. cholerae O1 isolated from cholera patients, their household contacts, stored drinking water and water sources.
PFGE of noti-Digested Genomic DNA
PFGE analysis of Not-I- digested genomic DNA of 32 representative strains from hospitalized cholera patients, household contacts, stored drinking water and source water showed them to be closely related, as the PFGE banding patterns were highly homogenous.
The majority of the strains (n = 29) belonged to two major clusters A and B while rest of the strains (n = 3) showed discrete patterns and thus fell into a different minor cluster C (Figure 3). Cluster A, B, and C share 95–100% similarity co-efficient among them. Cluster A includes 16 isolates from index patients, household contacts, and drinking water sources from Bakerganj and Mathbaria of which 93.75% (15/16) isolates were found to be identical in their banding pattern. Cluster B comprised of 13 isolates of the same type and share 97% similarity. The minor cluster C comprised isolates from index patients and household contacts only. The N16961 (V. cholerae prototype El Tor) and O395 (V. cholerae prototype classical) reference strain showed significant differences with the altered El Tor strains analyzed in the present study.
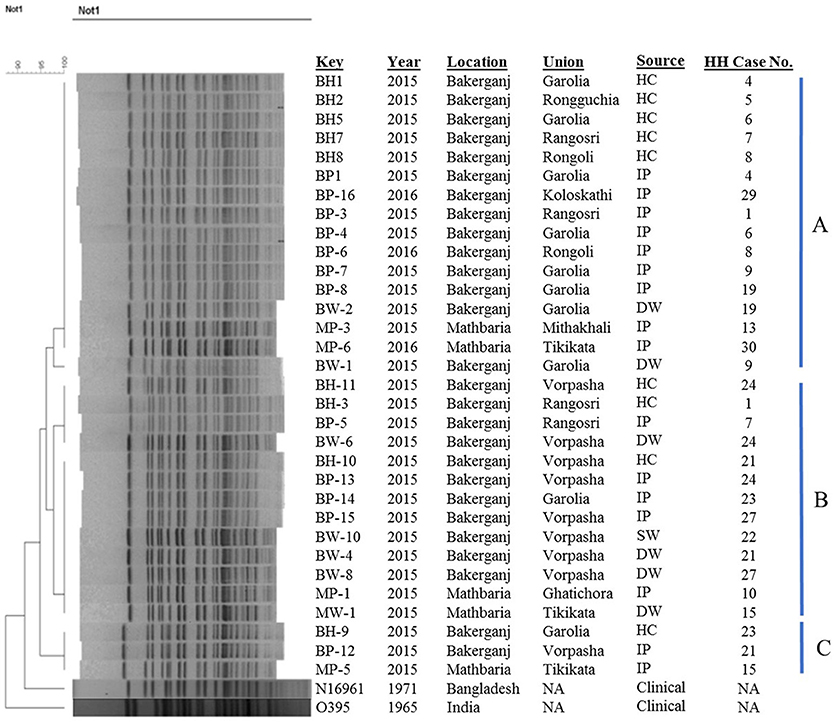
Figure 3. Genomic fingerprinting of V. cholerae O1 strains collected from environmental (water) and clinical (stool) sources of the household of index cholera patients. Thirty-Two V. cholerae O1 strains were isolated in Bakerganj and Mathbaria from hospitalized cholera patients (n = 16), their household contacts (n = 9), drinking water (n = 6), and source water (n = 1). The dendrogram was constructed by Dice similarity coefficient and mainstream hierarchical clustering (UPGMA) using PFGE images of Not1-digested genomic DNA. The scale bar at the top left indicates the similarity coefficient (range: 90–100%). IP, Index Patient; HC, Household contact; SW, Source Water; DW, Drinking Water; NA, Not Applicable.
Discussion
Our phenotypic and genetic characterization data suggests that drinking water is an important transmission route for toxigenic Vibrio cholerae in cholera patient households in coastal villages in Mathbaria and Bakerganj, Bangladesh. This is consistent with our recent findings in Dhaka, Bangladesh showing a single V. cholerae ancestral clone and cholera transmission through drinking water in households of cholera patients (26). Most previous studies on household transmission of cholera in Bangladesh took place either in urban settings (15, 26, 38, 39) or in rural areas far away from coastal regions (22). This study is the first to report household transmission of toxigenic V. cholerae through drinking water in the cholera endemic coastal villages of Bakerganj and Mathbaria, Bangladesh.
Phenotypic and genetic characterizations of V. cholerae showed all belonged to serogroup O1, biotype El Tor (ET), and serotype Ogawa. All strains also had cholera toxin (CT) specific for classical biotype (40), which was replaced in the 1980s by ET biotype strains responsible for the ongoing 7th pandemic of cholera worldwide (41). Our results also confirmed all strains to be altered ET which has been associated with endemic and epidemic cholera in this region (16, 17, 40).
Historically, cholera has been endemic in coastal villages in the Bay of Bengal in both Bangladesh and India for centuries (2, 42). In the Ganges Delta of the Bay of Bengal recurrent cholera often turns into epidemics causing significant morbidity and mortality each year (16, 41). Previous studies have shown the aquatic environments of estuarine villages of Mathbaria to be an important reservoir for V. cholerae serogroups O1 and O139 which are responsible for epidemic cholera in this region (16). In coastal villages of Bangladesh, accessing drinking water remains a longstanding issue as the underground water is saline rich (16). As a result, coastal villages sometimes rely on surface water for drinking (16).
Mapping cholera patient households allowed us to investigate the spatial distribution of cholera patients in Bakerganj and Mathbaria. Through this analysis we found cholera patients to be highly clustered, with most residing near the Upazila Health Complexes (UHC) of Bakerganj and Mathbaria. Many cholera patient households also resided near ponds (water reservoirs) with small channels (canals) carrying water from the major rivers Bighai in Bakerganj and Baleshwar in Mathbaria. Both these rivers are in the tidal zone and are connected directly to seawater of the Bay of Bengal. Although the two study sites are geographically apart, both represent estuarine habitats favoring survival and multiplication of toxigenic V. cholerae (16, 17). Since only one household reported using pond water we suspect bathing in ponds and rivers and using pond water for household activities (e.g., washing kitchen utensils, vegetables) may also be a potential exposure route to V. cholerae that should be investigated in future studies.
Cholera transmission in coastal communities is of urgent public health importance because the warming climate globally could increase the number of coastal areas exposed to saline-rich water carrying V. cholerae and other pathogenic bacteria that flourish in elevated temperatures (43). Currently there is no standard of care for highly susceptible household contacts of cholera patients, instead the focus is on the use of oral rehydration solution (ORS) for the discharged cholera patient (42). George et al. (27) in a randomized control trial of the CHoBI7 intervention (Cholera Hospital-Based Intervention for 7 days), a hospital based handwashing with soap and water treatment intervention for cholera patient households, showed a 47% reduction in the incidence of cholera among household contacts of cholera patients with delivery of this intervention in Dhaka, Bangladesh (27). Therefore, the CHoBI7 intervention could present a promising approach to reduce cholera in rural coastal regions of Bangladesh and should be tested in future studies.
Conclusion
Cholera remains a major public health problem worldwide especially in cholera endemic estuarine areas of Bangladesh where the estuarine habitat favors the survival and multiplication of toxigenic V. cholerae. This study provides evidence to support that toxigenic V. cholerae O1 is spread through contaminated drinking water among household contacts of cholera patients in the Bay of Bengal villages of Bangladesh. Transmission of toxigenic V. cholerae O1 through drinking water in cholera endemic rural settings underscores the urgent need for evidence based water, sanitation, and hygiene interventions promoting safe drinking water to prevent morbidity and mortality related to cholera and other enteric infections in Bangladesh.
Author Contributions
MA and CG are the Principal Investigators of the project and contributed to the design of the study, manuscript revision and final approval of the version to be published. SM is the functional Principal Investigator of the project and contributed to the design of the study, manuscript review and critical revision. ZR and MR designed and implemented the study. ZR performed the study in the laboratory and wrote the first draft of the manuscript. MAR, MM, and F-TJ performed the study in the laboratory and reviewed the manuscript. NI was involved in data analysis and reviewed the manuscript. KS-U-R oversaw the collection of data in the hospital/field and reviewed manuscript. FZ and TP were involved in data collection and contributed to manuscript writing. SB and KH were involved in database construction, data analysis and reviewed the manuscript. All authors read and approved the final manuscript. The authors have agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Funding
This research was supported by the Center for Global Health at Johns Hopkins University and the National Institute of Allergy and Infectious Diseases (NIAID 1K01AI110526-01A1 & Grant 1 RO1 A13912901).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the study participants and the following research assistants who conducted the fieldwork for this study: Ismat Minhaz Uddin, Rafiqul Islam, Al-Mamun, Maynul Hasan, Kalpona Akhter, Khandokar Fazilatunnessa, Sadia Afrin Ananya, Akhi Sultana, Sohag Sarker, Jahed Masud, Abul Sikder, Shirin Akter, and Laki Das.
We also want to thank Mr. Abdus Sadique and Md. Golam Mostafa for their continuous support in the laboratory. This research was supported by the Center for Global Health at Johns Hopkins University and the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The icddr,b thanks the governments of Bangladesh, Canada, Sweden and United Kingdom for providing unrestricted core support.
References
1. WHO. Diarrhoeal Disease [Online] (2017). Available online at: http://www.who.int/mediacentre/factsheets/fs330/en/ (Accessed May 01, 2017).
2. Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis. (2015) 9:e0003832. doi: 10.1371/journal.pntd.0003832
3. Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. Cholera. Lancet (2017) 390:1539–49. doi: 10.1016/S0140-6736(17)30559-7
4. Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. (2010) 18:46–54. doi: 10.1016/j.tim.2009.10.003
5. Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, et al. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci USA. (2016) 113:E7730–39. doi: 10.1073/pnas.1608732113
6. Nair GB, Qadri F, Holmgren J, Svennerholm A-M, Safa A, Bhuiyan NA, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. (2006) 44:4211–3. doi: 10.1128/JCM.01304-06
7. Alam M, Islam A, Bhuiyan NA, Rahim N, Hossain A, Khan GY, et al. Clonal transmission, dual peak, and off-season cholera in Bangladesh. Infect Ecol Epidemiol. (2011) 1. doi: 10.3402/iee.v1i0.7273
8. Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J Clin Microbiol. (2011) 49:3739–49. doi: 10.1128/JCM.01286-11
9. Satchell KJ, Jones CJ, Wong J, Queen J, Agarwal S, Yildiz FH. Phenotypic analysis reveals that the 2010 haiti cholera epidemic is linked to a hypervirulent strain. Infect Immun. (2016) 84:2473–81. doi: 10.1128/IAI.00189-16
11. Tamplin M, Carrillo PC. Environmental spread of Vibrio cholerae in Peru. Lancet (1991) 338:1216. doi: 10.1016/0140-6736(91)92089-K
12. Colwell RR. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int Microbiol. (2004) 7:285–9.
13. Morris J. Cholera—Modern Pandemic disease of ancient lineage. Emerg Infect Dis. J. CDC (2011) 17:2099–104. doi: 10.3201/eid1711.111109
14. Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, et al. Host-induced epidemic spread of the cholera bacterium. Nature (2002) 417:642–5. doi: 10.1038/nature00778
15. Weil AA, Khan AI, Chowdhury F, LaRocque RC, Faruque A, Ryan ET, et al. Clinical outcomes in household contacts of patients with cholera in Bangladesh. Clin Infect Dis. (2009) 49:1473–9. doi: 10.1086/644779
16. Alam M, Hasan NA, Sadique A, Bhuiyan N, Ahmed KU, Nusrin S, et al. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ Microbiol. (2006a) 72:4096–104. doi: 10.1128/AEM.00066-06
17. Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique A, et al. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ Microbiol. (2006b) 72:2849–55. doi: 10.1128/AEM.72.4.2849-2855.2006
18. Stine OC, Alam M, Tang L, Nair GB, Siddique AK, Faruque SM, et al. Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerging Infect Dis. (2008) 14:831. doi: 10.3201/eid1405.071116
19. Boucher Y, Orata FD, Alam M. The out-of-the-delta hypothesis: dense human populations in low-lying river deltas served as agents for the evolution of a deadly pathogen. Front Microbiol. (2015) 6:1120. doi: 10.3389/fmicb.2015.01120
20. Khan AE, Ireson A, Kovats S, Mojumder SK, Khusru A, Rahman A, et al. Drinking water salinity and maternal health in coastal Bangladesh: implications of climate change. Environ Health Perspect. (2011) 119:1328–32. doi: 10.1289/ehp.1002804
21. Sack RB, Siddique AK, Longini IM, Nizam A, Yunus M, Islam MS, et al. A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J Infect Dis. (2003) 187:96–101. doi: 10.1086/345865
22. Spira WM, Khan MU, Saeed YA, Sattar MA. Microbiological surveillance of intra-neighbourhood E1 Tor cholera transmission in rural Bangladesh. Bull World Health Organ. (1980) 58:731–40.
23. Hughes JM, Boyce JM, Levine RJ, Khan M, Aziz K, Huq M, et al. Epidemiology of eltor cholera in rural Bangladesh: importance of surface water in transmission. Bull World Health Organ. (1982) 60:395.
24. Burrowes V, Perin J, Monira S, Sack DA, Rashid MU, Mahamud T, et al. Risk Factors for Household Transmission of Vibrio cholerae in Dhaka, Bangladesh (CHoBI7 Trial). (2017). 96:1382–7. doi: 10.4269/ajtmh.16-0871
25. George CM, Hasan K, Monira S, Rahman Z, Saif-Ur-Rahman KM, Rashid M-ur, et al. A prospective cohort study comparing household contact and water Vibrio cholerae isolates in households of cholera patients in rural Bangladesh. PLoS Negl Trop Dis. (2018) 12:e0006641. doi: 10.1371/journal.pntd.0006641
26. Rafique R, Rashid MU, Monira S, Rahman Z, Mahmud MT, Mustafiz M, et al. Transmission of infectious Vibrio cholerae through drinking water among the household contacts of cholera patients (CHoBI7 Trial). Front Microbiol. (2016) 7:1635. doi: 10.3389/fmicb.2016.01635
27. George CM, Monira S, Sack DA, Rashid MU, Saif-Ur-Rahman K, Mahmud T, et al. Randomized controlled trial of hospital-based hygiene and water treatment intervention (CHoBI7) to reduce cholera. Emerging Infect. Dis. (2016) 22:233. doi: 10.3201/eid2202.151175
28. Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, et al. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol. (2005) 71:4645–54. doi: 10.1128/AEM.71.8.4645-4654.2005
29. Alam M, Sultana M, Nair GB, Siddique A, Hasan NA, Sack RB, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci USA. (2007) 104:17801–6. doi: 10.1073/pnas.0705599104
30. Huq A, Haley BJ, Taviani E, Chen A, Hasan NA, Colwell RR. Detection, isolation, and identification of Vibrio cholerae from the environment. Curr Protocols Microbiol. (2006) 6:6A. 5. doi: 10.1002/9780471729259.mc06a05s02
31. Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification ofVibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. (2000) 38:4145–51.
32. Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. (1998) 20:201–7. doi: 10.1111/j.1574-695X.1998.tb01128.x
33. Rivera IN, Chun J, Huq A, Sack RB, Colwell RR. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl Environ Microbiol. (2001) 67:2421–9. doi: 10.1128/AEM.67.6.2421-2429.2001
34. Kimsey HH, Waldor MK. CTXϕ immunity: application in the development of cholera vaccines. Proc Natl Acad Sci USA. (1998) 95:7035–9. doi: 10.1073/pnas.95.12.7035
35. Chow K, Ng T, Yuen K, Yam W. Detection of RTX toxin gene in Vibrio cholerae by PCR. J Clin Microbiol. (2001) 39:2594–7. doi: 10.1128/JCM.39.7.2594-2597.2001
36. Naha A, Pazhani G, Ganguly M, Ghosh S, Ramamurthy T, Nandy RK, et al. Development and evaluation of a PCR assay for tracking the emergence and dissemination of Haitian variant ctxB in Vibrio cholerae O1 strains isolated from Kolkata, India. J Clin Microbiol. (2012) 50:1733–6. doi: 10.1128/JCM.00387-12
37. Cooper K, Luey C, Bird M, Terajima J, Nair G, Kam K, et al. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodbourne Pathogens Dis. (2006) 3:51–8. doi: 10.1089/fpd.2006.3.51
38. Kendall EA, Chowdhury F, Begum Y, Khan AI, Li S, Thierer JH, et al. Relatedness of Vibrio cholerae O1/O139 isolates from patients and their household contacts, determined by multilocus variable-number tandem-repeat analysis. J Bacteriol. (2010) 192:4367–76. doi: 10.1128/JB.00698-10
39. George CM, Rashid M, Almeida M, Saif-Ur-Rahman K, Monira S, Bhuyian MSI, et al. Genetic relatedness of Vibrio cholerae isolates within and between households during outbreaks in Dhaka, Bangladesh. BMC Genom. (2017) 18:903. doi: 10.1186/s12864-017-4254-9
40. Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. (2002) 40:3296–9. doi: 10.1128/JCM.40.9.3296-3299.2002
41. Siddique A, Nair G, Alam M, Sack D, Huq A, Nizam A, et al. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect. (2010) 138:347–52. doi: 10.1017/S0950268809990550
42. Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet (2004) 363:223–33. doi: 10.1016/S0140-6736(03)15328-7
Keywords: cholera, Vibrio cholerae, household contacts, water, clonal transmission, PFGE, Bangladesh
Citation: Rahman Z, Rahman MA, Rashid M, Monira S, Johura F-T, Mustafiz M, Bhuyian SI, Zohura F, Parvin T, Hasan K, Saif-Ur-Rahman KM, Islam NN, Sack DA, George CM and Alam M (2018) Vibrio cholerae Transmits Through Water Among the Household Contacts of Cholera Patients in Cholera Endemic Coastal Villages of Bangladesh, 2015–2016 (CHoBI7 Trial). Front. Public Health 6:238. doi: 10.3389/fpubh.2018.00238
Received: 28 January 2018; Accepted: 08 August 2018;
Published: 30 August 2018.
Edited by:
Vitali Sintchenko, University of Sydney, AustraliaReviewed by:
Khwaja M. Sultanul Aziz, Bangladesh Academy of Sciences (BAS), BangladeshRajat Dhakal, The University of Queensland, Australia
Copyright © 2018 Rahman, Rahman, Rashid, Monira, Johura, Mustafiz, Bhuyian, Zohura, Parvin, Hasan, Saif-Ur-Rahman, Islam, Sack, George and Alam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Munirul Alam, bXVuaXJ1bEBpY2RkcmIub3Jn
 Zillur Rahman
Zillur Rahman Md. Anisur Rahman
Md. Anisur Rahman Mahamud-ur Rashid
Mahamud-ur Rashid Shirajum Monira
Shirajum Monira Fatema-Tuz Johura
Fatema-Tuz Johura Munshi Mustafiz
Munshi Mustafiz Sazzadul I. Bhuyian1
Sazzadul I. Bhuyian1 Khaled Hasan
Khaled Hasan K. M. Saif-Ur-Rahman
K. M. Saif-Ur-Rahman David A. Sack
David A. Sack Christine M. George
Christine M. George Munirul Alam
Munirul Alam