
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Public Health, 09 April 2018
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 6 - 2018 | https://doi.org/10.3389/fpubh.2018.00104
During the last two decades, scientists have grown increasingly aware that viruses are emerging from the human–animal interface. In particular, respiratory infections are problematic; in early 2003, World Health Organization issued a worldwide alert for a previously unrecognized illness that was subsequently found to be caused by a novel coronavirus [severe acute respiratory syndrome (SARS) virus]. In addition to SARS, other respiratory pathogens have also emerged recently, contributing to the high burden of respiratory tract infection-related morbidity and mortality. Among the recently emerged respiratory pathogens are influenza viruses, coronaviruses, enteroviruses, and adenoviruses. As the genesis of these emerging viruses is not well understood and their detection normally occurs after they have crossed over and adapted to man, ideally, strategies for such novel virus detection should include intensive surveillance at the human–animal interface, particularly if one believes the paradigm that many novel emerging zoonotic viruses first circulate in animal populations and occasionally infect man before they fully adapt to man; early detection at the human–animal interface will provide earlier warning. Here, we review recent emerging virus treats for these four groups of viruses.
During the last two decades, scientists have grown increasingly aware that viruses are emerging from the human–animal interface. In order to combat this increasingly complex problem, the One Health approach or initiative has been proposed as a way of working across disciplines to incorporate human, animal, and environmental health. Of particular concern are emerging respiratory virus infections; in a recent seminar given by the National Institute of Health on emerging and re-emerging pathogens, nearly 18% were respiratory viruses (1). Among the recently emerged respiratory pathogens contributing to the high burden of respiratory tract infection-related morbidity and mortality, displayed graphically in Figure 1, are influenza viruses, coronaviruses, enteroviruses (EVs), and adenoviruses (Ads). In this report, we summarize the emerging threat characteristics of these four groups of viruses.
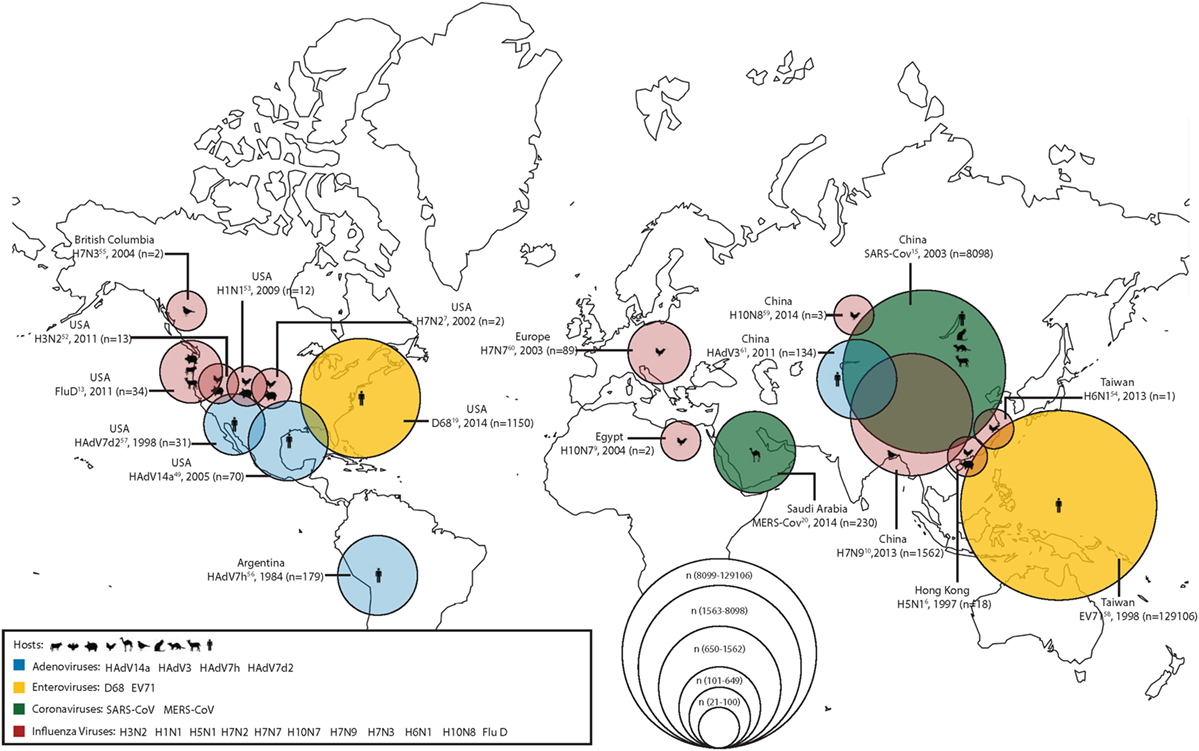
Figure 1. The geographical location of first detections (with known reservoirs) for recently emerged adenoviruses (Ads), enteroviruses (EVs), coronaviruses, and influenza viruses. Zoonotic (coronaviruses and influenza viruses) and non-zoonotic viruses (Ads and EVs) are shown. For zoonotic viruses, the hosts range from cattle, bats, chickens, camels, wild birds, cats, ferrets, goats, and humans (from left to right). The different sizes of the circles represent the number of human cases during the first outbreaks of the emerging respiratory viruses. Human cases of adenoviral infections are shown in blue; human cases of enteroviral infections are shown in yellow; human cases of coronaviral infections are shown in green; and human cases of influenza viral infections are shown in red.
Influenza viruses are RNA viruses that are members of the orthomyxovirus family and classified into four types: A, B, C, and D (2). As shown in Table 1, these four types of viruses are characterized by their immunologically distinct nucleoprotein and matrix protein antigens. Influenza A and B viruses consist of hemagglutinin (HA), which binds a sialic acid receptor, allowing the virus to enter the host cell, and neuraminidase (NA), which cleaves the sialic acid to release the virus. Similarly, influenza C and D viruses contain HA-esterase fusion glycoproteins that also allow for the attachment of viral and cellular membranes. Antigenic shifts (influenza A only) in HA, NA, and the HA-esterase proteins contribute to the generation of novel viral strains. The host range of influenza viruses includes humans, birds, pigs, bats, and other livestock animals such as cattle and goats. The network of the influenza viral transmission is complex with both inter- and intraspecies transmission. As the viruses continue to change in their genetic sequences, ongoing research is imperative in investigating the ecology of these viruses at the human–animal interface to control further spread of infections and prevent the risk of future pandemics.
H3N2
Influenza A virus H3N2 subtypes are frequently reported in swine, avian, and canine hosts that are responsible for highly infectious respiratory diseases in pigs and have been examined as a potential cause of influenza in humans. One study, examining the role of IAV in pigs at USA agricultural fairs, reported an average influenza A prevalence of 77.5% among 161 swine across all seven fairs (3). The genomic sequences of the viruses isolated from the swine were ≥99.89% similar to the H3N2 viruses isolated in humans. At these fairs, IAVs were detected at least 1 day before symptoms of the virus were observed in humans, indicating that H3N2 was transmitted from pigs to humans in this case.
H1N1
Since 2009, H1N1 virus has posed a significant threat to livestock workers and the greater community and has now become a seasonal influenza virus which circulates in humans. To explore the role of swine production facilities in the development of new swine-like influenza viruses, the spatiotemporal association between weekly influenza-like illnesses (ILIs) in humans and the location of pig farms was investigated in North Carolina over four influenza seasons (4). Analyses showed that the years of H1N1 pandemic, 2009–2010 and 2010–2011, were closely related with earlier peaking of ILI cases. These findings suggest that increased exposure to pigs was associated with earlier observations of the greatest number of human H1N1 cases.
In China, the transmission of influenza A between humans and pigs in six farms is being examined using a One Health approach, taking into consideration the interconnectedness of humans, animals, and the environment (5). Findings suggest that both A(H1N1)pdm09-like and swine-lineage H1N1 and swine-lineage H3N2 viruses are circulating in swine workers and that these viruses likely reassort and cross species within the pig farms; as such, additional research is needed to understand the relationship between cross species transmission of viruses in humans and pigs.
Avian influenza viruses are the largest group of influenza A viruses reservoired in aquatic birds or poultry. Although infrequently transmitted to humans, many cases have now been reported. Human infection with avian influenza can lead to serious health conditions, including death. The first outbreak of an IAV strain, H5N1, in humans occurred in Hong Kong SAR, China, in 1997, infecting 18 humans (6). The first identified cases of human infection with H7N2, another avian influenza, occurred in North America with two human cases reported in 2002 (7). Another variation of the virus, H7N7, was the first avian influenza strain reported in Europe; it infected 89 humans in 2003. In 2004, the first human cases of H10N7 infections were observed in Africa (8). It is important to note that H5N1 virus outbreak occurred again in 2004, 7 years after its first outbreak in humans, infecting more than 650 humans and causing more than 386 deaths worldwide (9). The avian influenza viruses have continuously evolved, causing serious infections among humans across the world.
H7N9
H7N9 virus, a sporadic subtype of an avian influenza A virus, was first reported in humans in China in 2013. Since the first outbreak, China has been experiencing epidemics annually, with a cumula-tive number of 1,562 reported cases, 40% of which have led to deaths as of September 2017 (10). The incidence of the H7N9 infections has been increasing in both humans and poultry and in 2017 alone 764 infections have been reported (11). Although H7N9 was first recognized as a low pathogenic avian influenza, two divergent lineages were detected in 2016—including a highly pathogenic avian influenza variant (12). According to the Center for Disease Control and Prevention (CDC), H7N9 is now recognized as the virus with the greatest potential to cause a pandemic due to its rapid genetic changes over the last 5 years. This further supports the need to improve disease control strategies and increase efforts to develop an effective vaccination strategy in the future as the spread of the H7N9 infection poses a threat to the poultry business.
Influenza D virus (IDV) is a novel influenza virus that is structurally different from the other influenza viruses. IDV was first isolated in 2001 from pigs in USA and since the first report, viral infection has been reported in various locations in USA, Europe, and Asia. In a serological study, cattle workers and non cattle-exposed adults in Florida were screened for IDV antibodies (13). Of the cattle workers, 97% of IDV seroprevalence was observed, while less than 20% was observed in non-cattle-exposed adults, suggesting a greater risk of IDV infection for cattle workers. During a swine respiratory disease outbreak in Northern Italy in 2015, the IDV genome was detected and isolated in both pigs and cattle herds (14). The viral genome isolated from the pigs was closely related to the viral genome isolated in USA in 2011. Additionally, the archived serum samples from 2009 had lower IDV antibody titers compared to the serum samples collected in 2015. These findings suggest that the incidence of IDV infections in pigs may have increased over time, and therefore, IDV may pose a public health threat to the community.
Coronaviruses are single-stranded, RNA viruses with a large genome in which mutations are very common. There are six human types of coronavirus: 229E, OC43, NL63, HKU1, which are often associated with mild upper respiratory tract infections, as well as the virus causing severe acute respiratory syndrome (SARS-CoV) and Middle East respiratory syndrome (MERS-CoV), both of which are zoonotic and have previously caused human disease. Interspecies transmission and the resulting emergent coronaviruses have been important factors in emerging respiratory disease as coronaviruses are known to infect feline, swine, canine, and bat species. Both MERS-CoV and SARS-CoV emerged from animal reservoirs and are now increasingly important respiratory virus threats.
SARS-CoV first emerged as a global problem in 2003, when China informed the World Health Organization (WHO) of 305 cases of atypical pneumonia in Guangdong Province. The WHO estimates that a total of 8,098 people worldwide became sick with SARS-CoV during the 2003 outbreak, and of them 774 died (15). In USA, only eight people had laboratory evidence of SARS-CoV infection and, in all cases, infection was travel related. The secondary spread of SARS-CoV was characterized by transmission between patients and nurses, requiring intervention from hospital infection control (16). Horseshoe bats are the host reservoir for SARS-CoV, but it is also postulated that other intermediate hosts such as civet cats, domestic cats, rodents, and swine may play an important role in transmission (17, 18). Recently, it has also been suggested that bats may play a role in the direct human transmission as bat SARS-like coronaviruses have been identified in some species (19).
In the past decade, teams from the Sabin Vaccine Institute and Baylor College of Medicine have been working toward the development of a vaccine for SARS-CoV. Although initial reports indicated that a vaccine may be ready for human clinical trials in 2017, progress has been slow and few human SARS vaccine trials have been conducted to date.
Middle East respiratory syndrome was first recognized in Saudi Arabia in 2012. Many cases were linked to travel to or residence in countries in and near the Arabian Peninsula. Symptoms include severe acute respiratory illness with fever, cough, and shortness of breath. There is limited human-to-human transmission of MERS-CoV, but exposure to camels is a risk factor for infection, with seroprevalences 15–23 times higher in camel exposed individuals (20). Despite this, major health care-associated transmission of MERS-CoV was reported in the Middle East and Korea, with outbreaks characterized by interhospital spread related to overcrowding and a lack of personal protective equipment (21). The total number of worldwide cases reported to the WHO as of January 9, 2017 was 2,067 MERS-CoV cases (22).
The cocirculation of CoVs in its animal reservoirs (camels and bats) raises important questions about the evolution of MERS-CoV. In a study conducted between 2014 and 2015 in Saudi Arabia, researchers found that dromedary camels share three CoV species with humans, including betacoronavirus 1, MERS-COV, and a CoV 229E-related virus (23). With the aim of reducing MERS-CoV transmission to humans, Haagmans et al. developed a vaccine for camels using a poxvirus vehicle (24). This vaccine has significantly reduced virus excretion among camels and conferred cross-immunity to camelpox infections (25).
Enteroviruses are small, positive-sense, single-stranded RNA viruses in the Picornaviridae family. There are 12 species of EVs found globally, including EV A-J (EV-A, B, C, D, E, F, G, H, and J) and rhinovirus A–C (RV-A, B, and C). With low replication fidelity and frequent recombination, EVs have viral genetic diversity and a potential for cross-species infection. In February 2013, the International Committee on Taxonomy of Viruses (ICTV) approved changes to EV and rhinovirus species names after many of the human EV species were identified and isolated in non-human hosts. Based on an analysis of Picornaviridae hosts listed in the ICTV database and subsequent studies of EV infection in non-human primates, there is growing evidence to indicate a potential for future zoonotic transmission between animals and humans (26, 27). Among the most important emerging respiratory viruses are EV68, EV71, coxsackieviruses, echoviruses, rhinoviruses, and polioviruses.
Enterovirus transmission occurs year-round, with seasonal peaks occurring in the summer and fall (June–October). Infants less than 1 year of age are most susceptible to infection, and males are at an increased risk for infection until the age of 20 years (28). The predominant mode of transmission is through a direct or indirect fecal–oral route; however, certain serotypes are transmitted via the respiratory route, in tears, and via fomites (29). Immunity to EVs is serotype specific with most causing mild respiratory infections.
Rhinoviruses are small, single-stranded RNA viruses in the picornavirus family that are responsible for more than half of all upper respiratory tract infections. In addition to exacerbating asthma and chronic obstructive pulmonary disease, rhinoviruses have also been associated with acute respiratory hospitalizations among children (30). In a large prospective study of US pneumonias, rhinoviruses have been identified as the second most prevalent etiology of pneumonia in children after respiratory syncytial virus and the first most common etiology among adults (31). There are more than 150 unique types of rhinoviruses. Among the three genotypes (A, B, and C) types A and C are most often associated with increased morbidity and bacterial secondary infection. In animals, rhinovirus type C has been associated with morbidity in chimpanzees (32). With an array of unique serotypes no vaccines or approved antiviral therapies have been commercially produced; however, experiments have suggested that vaccines and antiviral therapy may be possible (33, 34).
Enterovirus D68 has caused sporadic respiratory disease outbreaks across Asia, Europe, and USA since 1960s; however, in 2014, a nationwide outbreak of D68 was associated with severe respiratory illness in USA, resulting in 14 deaths out of a known 1,150 cases (35). The CDC found 36% of all EVs tested during this outbreak were D68 and that patients with a history of asthma were found to be at a disproportionately increased risk of infection (36). One study of the 2014 outbreak found 59% of patients seen with EV-D68 in hospitals across Missouri, Illinois, and Colorado were admitted to intensive care units and 28% received ventilator support (35). In a study evaluating EVs in non-human primates, EV-D68 was detected as a recombinant zoonotic strain (37).
While there are several strains of coxsackievirus and EVs that can cause hand-foot-and-mouth disease (HFMD), EV71 is most commonly associated with severe disease outcomes. HFMD predominantly affects young children and is found worldwide but especially in the Asia-Pacific region. Although EV71 is not typically detected in animals, recent research has indicated that it infects non-human primates (38). Various antiviral therapies are currently under study, including small molecules, monoclonals, and antivirals. Vaccine candidates are also in development, with two vaccines currently available in China, which involve recombinant proteins, attenuated strains, inactivated whole-virus and virus-like particles, and DNA vaccines (39).
First discovered in 1953 by Rowe et al., Ads are non-enveloped, double-stranded DNA viruses with 57 unique serotypes, some of which are specific for attacking the respiratory track, conjunctiva, or gastrointestinal track (40). Key features of Ad infections include various symptoms of disease, including rhinorrhea, nasal congestion, cough, sneezing, pharyngitis, keratoconjunctivitis, pneumonia, meningitis, gastroenteritis, cystitis, and encephalitis. Illnesses may be asymptomatic, mild, or severe; however, immunocompromised patients and infants are at increased risk of severe morbidity and death.
Outbreaks of respiratory Ad infection are common in both military recruits and other large training groups, such as police trainees. Large persistent epidemics of Ad type 4-associated respiratory disease have been documented in various military trainees (41–43).
In response to the increased disease burden from Ad4 and Ad7 in military recruits, Teva has made a vaccine available to military recruits in USA (42). Despite a 12-year hiatus from use, in late 2011 oral Ad4 and Ad7 vaccines were reintroduced as an infection control measure for military recruits (42). After reintroduction, military recruits experienced a 100-fold decline in Ad disease burden, which accounted for the prevention of approximately 1 death, 1,100–2,700 hospitalizations, and 13,000 febrile Ad cases per year among trainees (44).
Outbreaks of Ad in the general population have been characterized by infection due to novel viruses such as Ad7h, Ad7d2, Ad14a, and Ad3 variants. These novel viruses are sometimes associated with high attack rates and a high prevalence of pneumonia. Severe mortality is also prevalent among patients with chronic disease and in the elderly.
One of the most important novel serotypes, Ad14, previously rarely reported, is now considered as an emerging Ad type causing severe and sometimes fatal respiratory illness in patients of all ages (45). Beginning in 2005, Ad14 cases were suddenly identified in four locations across USA (46); the strain associated with this outbreak was different than the original Ad14 strain isolated in 1950s. The novel strain, Ad14a, has now spread to numerous US states and is associated with a higher rate of severe illness when compared to other Ad strains.
Novel Ad species have also been recently detected in cross-species infections from non-human primates to man in USA and between psittacine birds and man in China (47). These cross-species infections indicate that Ads should be monitored for their potential to cause cross-species outbreaks. In a recent review of the risks of potential outbreaks associated with zoonotic Ad (48), it was noted that intense human–animal interaction is likely to increase the probability of emergent cross-species Ad infection. Additionally, the recombination of AdVs with latent “host-specific” AdVs is the most likely scenario for adaptation to a new host, either human or animal.
Currently, there are no FDA approved antivirals for Ad infection; however, the best antiviral success has been seen with ribavirin, cidofovir, and most recently brincidofovir an analog of cidofovir (49).
As it is clear that many emerging respiratory viruses have zoono-tic reservoirs, the design and implementation of effective control strategies are increasingly important. It has been suggested that avoiding direct contact with animals known to be zoonotic reservoirs for these viruses is one potential strategy (50); however, in populations where contact at the human–animal interface is common this may not be an acceptable solution.
Complex disease problems cannot be solved by one institution or one discipline; as such, this presents opportunities to incorporate the One Health approach of working across disciplines to incorporate human, animal, and environmental health to solve complex problems. Although some of the respiratory viruses described here are found almost exclusively in humans (Ad strains), many of the most important emerging respiratory viruses are found at the human/animal interface. This suggests that strategies for novel virus detection should incorporate global surveillance at the human–animal interface to detect potentially emerging zoonotic viruses. This surveillance will require collaboration and cooperation among many stakeholders in order to address emerging and novel viral diseases.
EB, JF, and JC conducted the literature review and wrote the manuscript; GG conceived the idea of the review and helped revise the manuscript to add important scientific content and refine the interpretation of the results. All the authors reviewed the final version of the manuscript and agreed to its submission.
The authors declare that this research was conducted in the absence of any potential financial or commercial conflicts of interest.
This work was supported in part by NIH/NIAID grant R01AI108993-01A1 (Gregory Gray PI).
1. National Institute of Health. Current Infectious Disease Challenges (2017) [cited 2018 Jan 02]. Available from: http://afludiary.blogspot.com/2017/05/nih-videocast-current-infectious.html
2. Centers for Disease Control and Prevention. Types of Influenza Viruses (2017) [cited 2017 Sep 23]. Available from: https://www.cdc.gov/flu/about/viruses/types.htm
3. Bowman AS, Walia RR, Nolting JM, Vincent AL, Killian ML, Zentkovich MM, et al. Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg Infect Dis (2017) 23(9):5. doi:10.3201/eid2309.170847
4. Lantos PM, Hoffman K, Hohle M, Anderson B, Gray GC. Are people living near modern swine production facilities at increased risk of influenza virus infection? Clin Infect Dis (2016) 63(12):6. doi:10.1093/cid/ciw646
5. Ma MJ, Wang GL, Anderson BD, Bi ZQ, Lu B, Wang XJ, et al. Evidence for cross-species influenza A virus transmission within swine farms, China. Clin Infect Dis (2018) 66(4):533–40. doi:10.1093/cid/cix823
6. WHO. Avian and Other Zoonotic Influenza (2016). [cited 2017 Sep 23]. Available from: http://www.who.int/mediacentre/factsheets/avian_influenza/en/
7. Centers for Disease Control and Prevention. H7N2 Questions & Answers (2016) [updated 2017 May 26; cited 2017 Dec 23]. Available from: https://www.cdc.gov/flu/fluincats/h7n2-cat-questions-answers.htm
8. WHO. Avian influenza virus A (H10N7) circulating among humans in Egypt. Emerg Infect Dis (2004) 2(18):1.
9. Li X, Zhang Z, Yu A, Ho SY, Carr MJ, Zheng W, et al. Global and local persistence of influenza A(H5N1) virus. Emerg Infect Dis (2014) 20(8):1287–95. doi:10.3201/eid2008.130910
10. Centers for Disease Control Prevention. Asian Lineage Avian Influenza A (H7N9) Virus (2016). [updated 2017 Sep 26; cited 2017 Sep 20]. Available from: https://www.cdc.gov/flu/avianflu/h7n9-virus.htm
11. Food and Agriculture Organization of the United Nations. H7N9 Situation Update (2017). [cited 2017 Sep 20]. Available from: http://www.fao.org/ag/againfo/programmes/en/empres/h7n9/situation_update.html
12. Centers for Disease Control and Prevention. Summary of Influenza Risk Assessment Tool (IRAT) Results (2017). [updated 2017 Oct 23; cited 2017 Oct 23]. Available from: https://www.cdc.gov/flu/pandemic-resources/monitoring/irat-virus-summaries.htm#H7N9_hongkong
13. White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J Clin Virol (2016) 81:31–3. doi:10.1016/j.jcv.2016.05.017
14. Foni E, Chiapponi C, Baioni L, Zanni I, Merenda M, Rosignoli C, et al. Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci Rep (2017) 7(1):11660. doi:10.1038/s41598-017-12012-3
15. Centers for Disease Control and Prevention. Update: outbreak of severe acute respiratory syndrome – worldwide, 2003. MMWR Morb Mortal Wkly Rep (2003) 52(13):269–72.
16. Reynolds MG, Anh BH, Thu VH, Montgomery JM, Bausch DG, Shah JJ, et al. Factors associated with nosocomial SARS-CoV transmission among healthcare workers in Hanoi, Vietnam, 2003. BMC Public Health (2006) 6:207. doi:10.1186/1471-2458-6-207
17. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science (2003) 302(5643):276–8. doi:10.1126/science.1087139
18. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A (2005) 102(39):14040–5. doi:10.1073/pnas.0506735102
19. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature (2013) 503(7477):535–8. doi:10.1038/nature12711
20. Kayali G, Peiris M. A more detailed picture of the epidemiology of Middle East respiratory syndrome coronavirus. Lancet Infect Dis (2015) 15(5):495–7. doi:10.1016/S1473-3099(15)70128-3
21. Balkhy HH, Perl TM, Arabi YM. Preventing healthcare-associated transmission of the Middle East Respiratory Syndrome (MERS): our Achilles heel. J Infect Public Health (2016) 9(3):208–12. doi:10.1016/j.jiph.2016.04.006
23. Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science (2016) 351(6268):81–4. doi:10.1126/science.aac8608
24. Haagmans BL, van den Brand JM, Raj VS, Volz A, Wohlsein P, Smits SL, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science (2016) 351(6268):77–81. doi:10.1126/science.aad1283
25. Alharbi NK. Vaccines against Middle East respiratory syndrome coronavirus for humans and camels. Rev Med Virol (2017) 27(2):e1917. doi:10.1002/rmv.1917
26. Daszak KJO, Parviez RH, Carlos Z-T, Noam R, Tiffany LB, Peter D. Host and viral traits predict zoonotic spillover from mammals. Nature (2017) 546(7660):646. doi:10.1038/nature22975
27. Oberste MS, Feeroz MM, Maher K, Nix WA, Engel GA, Hasan KM, et al. Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J Virol (2013) 87(1):558–71. doi:10.1128/jvi.00837-12
28. Khetsuriani N, Lamonte-Fowlkes A, Fau–Oberst S, Oberst S, Pallansch MA; CDC. Enterovirus surveillance – United States, 1970-2005. MMWR Surveill Summ (2006) 55(8):1–20.
29. Couch RB, Douglas RG Jr, Lindgren KM, Gerone PJ, Knight V. Airborne transmission of respiratory infection with coxsackievirus A type 21. Am J Epidemiol (1970) 91(1):78–86. doi:10.1093/oxfordjournals.aje.a121115
30. Miller EK, Lu X, Erdman DD, Poehling KA, Zhu Y, Griffin MR, et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis (2007) 195(6):773–81. doi:10.1086/511821
31. Jain A, Humar A, Lien D, Weinkauf J, Kumar D. Strategies for safe living among lung transplant recipients: a single-center survey. Transpl Infect Dis (2015) 17(2):185–91. doi:10.1111/tid.12354
32. Scully EJ, Basnet S, Wrangham RW, Muller MN, Otali E, Hyeroba D, et al. Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerg Infect Dis (2018) 24(2):267–74. doi:10.3201/eid2402.170778
33. Patel MC, Pletneva LM, Boukhvalova MS, Vogel SN, Kajon AE, Blanco JCG. Immunization with live human rhinovirus (HRV) 16 induces protection in cotton rats against HRV14 infection. Front Microbiol (2017) 8:1646. doi:10.3389/fmicb.2017.01646
34. Williams GR, Kubajewska I, Glanville N, Johnston SL, McLean GR. The potential for a protective vaccine for rhinovirus infections. Expert Rev Vaccines (2016) 15(5):569–71. doi:10.1586/14760584.2016.1142375
35. Midgley CM, Watson JT, Nix WA, Curns AT, Rogers SL, Brown BA, et al. Severe respiratory illness associated with a nationwide outbreak of entero-virus D68 in the USA (2014): a descriptive epidemiological investigation. Lancet Respir Med (2015) 3(11):879–87. doi:10.1016/S2213-2600(15)00335-5
36. Centers for Disease Control and Prevention. Non-Polio Enterovirus | About EV-D68 | Enterovirus D68. CDC (2017).
37. Mombo IM, Lukashev AN, Bleicker T, Brunink S, Berthet N, Maganga GD, et al. African non-human primates host diverse enteroviruses. PLoS One (2017) 12(1):e0169067. doi:10.1371/journal.pone.0169067
38. Nagata N, Shimizu H, Ami Y, Tano Y, Harashima A, Suzaki Y, et al. Pyramidal and extrapyramidal involvement in experimental infection of cynomolgus monkeys with enterovirus 71. J Med Virol (2002) 67(2):207–16. doi:10.1002/jmv.2209
39. Yi E-J, Shin Y-J, Kim J-H, Kim T-G, Chang S-Y. Enterovirus 71 infection and vaccines. Clin Exp Vaccine Res (2017) 6(1):4–14. doi:10.7774/cevr.2017.6.1.4
40. Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med (1953) 84(3):570–3. doi:10.3181/00379727-84-20714
41. Hwang SM, Park DE, Yang YI, Park SJ, Lee HK, Kim MJ, et al. Outbreak of febrile respiratory illness caused by adenovirus at a south Korean military training facility: clinical and radiological characteristics of adenovirus pneumonia. Jpn J Infect Dis (2013) 66(5):359–65. doi:10.7883/yoken.66.359
42. Gray GC, Erdman DD. Adenovirus vaccines. 7th ed. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. Philadelphia: Saunders (2017). p. 113–26.
43. Yusof MA, Rashid TRTA, Thayan R, Othman KA, Abu Hasan N, Adnan N, et al. Human adenovirus type 7 outbreak in police training center, Malaysia, 2011. Emerg Infect Dis (2012) 18(5):852–4. doi:10.3201/eid1805.110865
44. Radin JM, Hawksworth AW, Blair PJ, Faix DJ, Raman R, Russell KL, et al. Dramatic decline of respiratory illness among US military recruits after the renewed use of adenovirus vaccines. Clin Infect Dis (2014) 59(7):962–8. doi:10.1093/cid/ciu507
45. Centers for Disease Control and Prevention. Acute respiratory disease associated with adenovirus serotype 14 – four states, 2006-2007. MMWR Morb Mortal Wkly Rep (2007) 56(45):1181–4.
46. Louie JK, Kajon AE, Holodiny M, Guardia-LaBar L, Lee B, Petru AM, et al. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin Infect Dis (2008) 46(3):421–5. doi:10.1086/525261
47. To KKW, Tse H, Chan WM, Choi GKY, Zhang AJX, Sridhar S, et al. A novel psittacine adenovirus identified during an outbreak of avian chlamydiosis and human psittacosis: zoonosis associated with virus-bacterium coinfection in birds. Plos Neglect Trop Dis (2014) 8(12):e3318. doi:10.1371/journal.pntd.0003318
48. Benko M, Harrach B, Kremer EJ. Do nonhuman primate or bat adenoviruses pose a risk for human health? Future Microbiol (2014) 9(3):269–72. doi:10.2217/fmb.13.170
49. Hiwarkar P, Amrolia P, Sivaprakasam P, Lum SH, Doss H, O’Rafferty C, et al. Brincidofovir is highly efficacious in controlling adenoviremia in pediatric recipients of hematopoietic cell transplant. Blood (2017) 129(14):2033–7. doi:10.1182/blood-2016-11-749721
50. Gautret P, Gray GC, Charrel RN, Odezulu NG, Al-Tawfiq JA, Zumla A, et al. Emerging viral respiratory tract infections – environmental risk factors and transmission. Lancet Infect Dis (2014) 14(11):1113–22. doi:10.1016/S1473-3099(14)70831-X
51. Danilov SM, Balyasnikova IV, Albrecht RF II, Kost OA. Simultaneous determination of ACE activity with 2 substrates provides information on the status of somatic ACE and allows detection of inhibitors in human blood. J Cardiovasc Pharmacol (2008) 52(1):90–103. doi:10.1097/FJC.0b013e31817fd3bc
52. Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011-2012. Clin Infect Dis (2013) 57(Suppl 1):S4–11. doi:10.1093/cid/cit272
53. Presanis AM, Lipsitch M, Daniela De A; Swine Flu Investigation Team, New York City Department of Health and Mental Hygiene, Mental H, Hagy A, et al. The severity of pandemic H1N1 influenza in the United States, April–July 2009. PLoS Curr (2009) 1:RRN1042. doi:10.1371/currents.RRN1042
54. Yan Z, Li H, Chen F. Novel influenza A (H6N1) virus that infected a person in Taiwan. Iran J Public Health (2014) 43(11):1582–4.
55. Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis (2004) 10(12):2196–9. doi:10.3201/eid1012.040961
56. Kajon A, Wadell G. Genome analysis of South American adenovirus strains of serotype 7 collected over a 7-year period. J Clin Microbiol (1994) 32(9):2321–3.
57. Gerber SI, Erdman DD, Pur SL, Diaz PS, Segreti J, Kajon AE, et al. Outbreak of adenovirus genome type 7d2 infection in a pediatric chronic-care facility and tertiary-care hospital. Clin Infect Dis (2001) 32(5):694–700. doi:10.1086/319210
58. Lin TY, Chang LY, Hsia SH, Huang YC, Chiu CH, Hsueh C, et al. The 1998 enterovirus 71 outbreak in Taiwan: pathogenesis and management. Clin Infect Dis (2002) 34(Suppl 2):S52–7. doi:10.1086/338819
59. Zhang T, Bi Y, Tian H, Li X, Liu D, Wu Y, et al. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis (2014) 20(12):2076–9. doi:10.3201/eid2012.140911
60. Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A (2004) 101(5):1356–61. doi:10.1073/pnas.0308352100
Keywords: emerging viruses, respiratory viruses, influenza viruses, adenoviruses, coronaviruses, enteroviruses, one health
Citation: Bailey ES, Fieldhouse JK, Choi JY and Gray GC (2018) A Mini Review of the Zoonotic Threat Potential of Influenza Viruses, Coronaviruses, Adenoviruses, and Enteroviruses. Front. Public Health 6:104. doi: 10.3389/fpubh.2018.00104
Received: 17 January 2018; Accepted: 27 March 2018;
Published: 09 April 2018
Edited by:
Margaret Ip, The Chinese University of Hong Kong, ChinaReviewed by:
Peng Yang, Beijing Center for Disease Prevention and Control, ChinaCopyright: © 2018 Bailey, Fieldhouse, Choi and Gray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily S. Bailey, ZW1pbHkuYmFpbGV5MkBkdWtlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.