- 1Scientific Advice Coordination Section, Office of the Chief Scientist, European Centre for Disease Prevention and Control, Stockholm, Sweden
- 2Microbiology Coordination Section, Office of the Chief Scientist, European Centre for Disease Prevention and Control, Stockholm, Sweden
In an era of global health threats caused by epidemics of infectious diseases and rising multidrug resistance, microbiology laboratories provide essential scientific evidence for risk assessment, prevention, and control. Microbiology has been at the core of European infectious disease surveillance networks for decades. Since 2010, these networks have been coordinated by the European Centre for Disease Prevention and Control (ECDC). Activities delivered in these networks include harmonization of laboratory diagnostic, antimicrobial susceptibility and molecular typing methods, multicentre method validation, technical capacity mapping, training of laboratory staff, and continuing quality assessment of laboratory testing. Cooperation among the European laboratory networks in the past 7 years has proved successful in strengthening epidemic preparedness by enabling adaptive capabilities for rapid detection of emerging pathogens across Europe. In partnership with food safety authorities, international public health agencies and learned societies, ECDC-supported laboratory networks have also progressed harmonization of routinely used antimicrobial susceptibility and molecular typing methods, thereby significantly advancing the quality, comparability and precision of microbiological information gathered by ECDC for surveillance for zoonotic diseases and multidrug-resistant pathogens in Europe. ECDC continues to act as a catalyst for sustaining continuous practice improvements and strengthening wider access to laboratory capacity across the European Union. Key priorities include optimization and broader use of rapid diagnostics, further integration of whole-genome sequencing in surveillance and electronic linkage of laboratory and public health systems. This article highlights some of the network contributions to public health in Europe and the role that ECDC plays managing these networks.
Introduction
Facing global epidemics of infectious diseases and rising multidrug resistance, microbiology laboratories provide pivotal information trough surveillance, from local to global levels, as specified in the International Health Regulations (1). At international level, the World Health Organization (WHO) operates laboratory networks that are part of epidemic preparedness and response programs as well as monitor communicable disease elimination and eradication programs (2, 3). In the European Union (EU), the European Centre for Disease Prevention and Control (ECDC), a public health agency financed by the EU, is tasked with detection, surveillance, and risk assessment of threats to human health from communicable diseases (4, 5). ECDC has a multidisciplinary workforce providing scientific advice, epidemic intelligence, disease surveillance, outbreak response support, preparedness support, microbiology support health communication, and training activities in collaboration with public health experts and national agencies in EU countries. It does not operate its own microbiology laboratories but relies instead on laboratory information provided at national level. EU countries report notifiable diseases to ECDC using EU case definitions (6). ECDC is mandated to “foster the development of sufficient capacity within the Community for the diagnosis, detection, identification, and characterization of infectious agents which may threaten public health, by encouraging cooperation between expert and reference laboratories” (4). This mandate builds upon decades of professional collaboration in Europe between infectious disease experts, microbiologists, and epidemiologists.
This article highlights ECDC key activities supporting the coordination of laboratory networks targeting the diseases which ECDC monitors at EU level. It discusses the effectiveness of laboratory response across Europe to recent public health events and indicates future directions for enhancing public health microbiology.
ECDC’s Support to EU Member States Laboratory Capacities
Since 2007–2010, ECDC has gradually supported the coordination of 12 EU-wide networks of microbiology laboratories embedded in disease specific networks. These primarily contribute to integrated epidemiological and microbiological surveillance for EU notifiable communicable diseases as well as to detection of emerging diseases (Table 1). Within these networks, ECDC supports microbiology activities ranging from EU-wide laboratory network coordination, capability and external quality assessments (EQA), laboratory staff training, reference microbial strain collections establishment, supranational reference services, outbreak investigations and risk assessments support, technology assessment, method harmonization and development of standard procedures, and integration of molecular typing into surveillance programs. Since 2014, ECDC also operated the EU laboratory capability (EULabCap) system for monitoring the capacities and capabilities of microbiology laboratories in EU countries (7).

Table 1. European Union (EU) public health microbiology laboratory networks supported by ECDC by disease group or health issue, 2017.
To strengthen the quality of surveillance and threat detection, ECDC has commissioned in the last 7 years 121 EQA exercises for it’s laboratory network members (Table 2). These EQAs covered a range of methods from pathogen detection and/or identification, molecular typing, to antimicrobial susceptibility testing (AST). EQAs was rated as one of the highest valued ECDC capacity building activities by external stakeholders (8), with its certificates being used for laboratory accreditation at the national level. EQA results over the years indicate better performance across countries, even though gaps remain (9–12). Having the networks centralized at ECDC has facilitated harmonization of EQA practice and cost efficiency across networks.
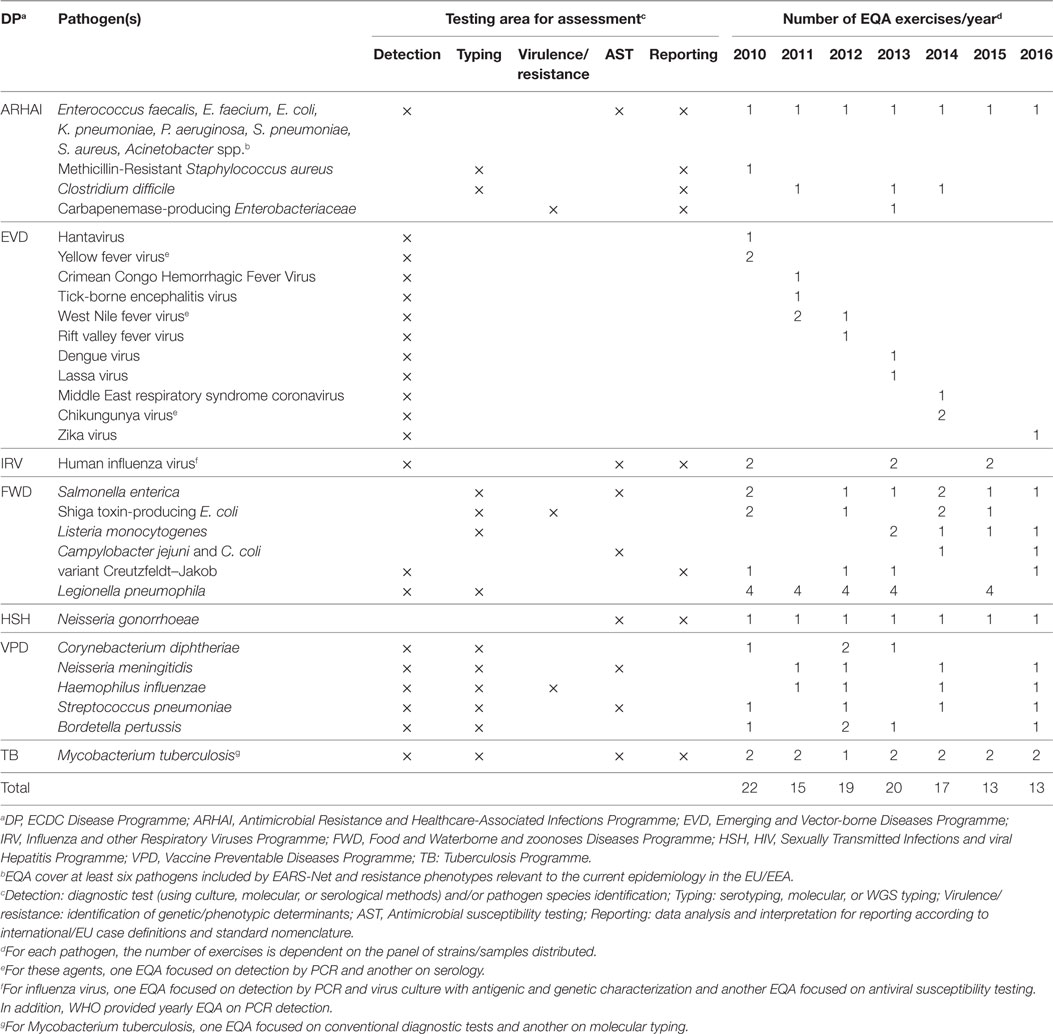
Table 2. Overview of ECDC- supported laboratory external quality assessments schemes by Disease Programme (DP), target pathogen, testing area and year, 2010–2016.
Rapid Detection of Emerging Infectious Threats, 2012–2017
At the request of the European Commission or the Member States, EU laboratory networks together with ECDC participate in investigations and risk assessment of potential cross-border health threats caused by emerging diseases or outbreaks. Recent events illustrate how laboratory detection capacity developed across networks contributed to a coordinated public health management of threats in Europe and beyond.
In September 2012, a novel coronavirus was isolated from two cases of acute severe respiratory illness who had traveled to or resided in Saudi Arabia (13, 14). Within a few weeks, more cases were identified in patients with links to the Middle East. This virus was named Middle East respiratory syndrome coronavirus (MERS-CoV) (15). In November 2012, real-time reverse-transcriptase (RT)-polymerase chain reaction (PCR) detection and identification tests were developed to ensure rapid detection capability (14). Technical protocols and positive RNA control material were made available by the European Commission funded European Virus Archive project and distributed within the former ECDC funded “European Network for Diagnostics of Imported Viral Diseases” (ENIVD). ECDC, in collaboration with WHO Regional Office for Europe (WHO/Europe), surveyed detection capability for MERS-CoV by virology reference laboratories (16). Ten months after the virus discovery, laboratories had diagnostic capabilities in 24 of the 30 EU and European Economic Area (EEA) countries (17). In 2013, an ECDC EQA exercise showed correct performance in laboratories across the ENIVD (18). In 2015, 28 EU/EEA countries had capability to screen and confirm MERS-CoV cases for appropriate management (7). By October 2015, 14 cases of MERS had been diagnosed across seven EU countries among patients with connection with the Middle-East. Thanks to rapid diagnosis, patients were promptly isolated and secondary transmission to household members or hospital patients occurred only rarely (19).
In March 2013, fatal cases of human infection with novel reassortant avian influenza virus strain A(H7N9) following contact with infected poultry were reported in China (20). This was the first time that human infection and deaths due to a low-pathogenicity avian influenza virus had been identified. Within 1 month of this event, ECDC, the European Reference Laboratory Network for Human Influenza (ERLI-Net) coordinated by ECDC, and the WHO/Europe released a joint technical note on diagnostic preparedness for detection of these viruses (21). In May 2013, the capability of ERLI-Net laboratories to detect and subtype the novel avian influenza A(H7N9) viruses was jointly assessed by ERLI-Net, ECDC, and WHO/Europe (22). The survey showed that the generic influenza A virus detection and H7 and N9 subtyping assays used in 24 laboratories in 19 EU/EEA countries were adequate. Later in 2013, the results of an ECDC EQA confirmed that 33 of the 36 ERLI-Net laboratories correctly detected, typed, and subtyped the novel A(H7N9) influenza viruses (23). In 2015, EULabCap survey results documented that diagnostic capability for avian influenza A(H7N9) virus infection existed in 28 of 29 EU/EEA countries, suggesting that the ERLI-Net response support had facilitated European-wide laboratory compliance with ECDC/WHO influenza surveillance guidance (7). No human influenza case caused by this strain has yet been diagnosed in Europe but annual epidemics of human infections in China indicate a persisting risk.
In December 2013, the largest ever epidemic of Ebola virus disease (EVD), started in Guinea and quickly spread to neighboring West African countries where it caused over 10,000 deaths until controlled March 2016. In March 2014, as part of global assistance efforts, the EU allocated funding and deployed medical and laboratory staff and supplies in West Africa. The European Mobile field Laboratory (EMLab), an initiative by the International Cooperation and Development Office of the European Commission, established field diagnostic facilities in the affected countries to support patient screening (24). Between March 2014 and October 2015, more than 19,000 samples were tested in these laboratories (24). Deployment of an EMLab unit close to an Ebola Treatment Unit decreased the average turnaround time from reception of a sample to diagnostic result to 4 h instead of several days. Several EU-funded laboratory networks worked to ensure capacity for testing suspected cases of EVD both in Africa and in travelers returning from the epidemic affected areas. These included the ENIVD network and the Joint Action “Quality Assurance Exercises and Networking on the Detection of Highly Infectious Pathogens” (QUANDHIP) which was co-funded by the European Commission and Member States. Both networks contributed expert staff to the EMLab activities in Africa. The ENIVD members participated in an ECDC EQA exercise for rapid molecular diagnosis of EVD (25). Until September 2015, QUANDHIP expert laboratories provided tests for 692 patients with suspected EVD in Europe (26). During the epidemic, five cases of EVD were confirmed in repatriated patients from West Africa and three new cases were diagnosed in the EU, including two travel-associated cases and one nosocomial case in a healthcare provider (27). These data confirmed that EVD patient isolation measures based on rapid diagnostics interrupt virus transmission and mitigate the risk of spread from patients evacuated to EU countries.
In 2015, a novel healthcare-associated disease was discovered in Switzerland by Sax et al., who reported a hospital outbreak of cardiovascular infection caused by Mycobacterium chimaera in surgery patients, linked to contaminated heater-cooler units used in surgery (28). This environmental mycobacterium is difficult to detect (i.e., fastidious) and identify (i.e., need for DNA sequence analysis). In April 2015, following the report of similar cases of infection in the Netherlands, Germany and the UK, ECDC evaluated the risk across Europe (29). It advised performing diagnostic investigations to ascertain possible cases of post-surgical infections by this organism (29). In collaboration with experts from the affected countries, ECDC followed-up national investigations and published an EU case-definition and technical protocol for case detection, laboratory diagnosis, environmental testing, and molecular typing of M. chimaera infection (30). This protocol provided a basis for harmonized data collection across Europe to facilitate the sharing of information on the extent and the molecular epidemiology of the outbreak. In 2015 and 2016, M. chimaera strains were included in the EQA panel of the ECDC coordinated European Reference Laboratory Network for Tuberculosis (ERLTB-Net). An updated risk assessment by ECDC indicated that, by November 2016, 52 cases of invasive cardiovascular infection by M. chimaera had been documented across seven European countries (31). Investigations using whole-genome sequencing (WGS) of clinical and environmental isolates were conducted in several countries by national public health agencies in collaboration with device manufacturers. Their concordant findings consistently pointed to airborne dispersal of M. chimaera from a particular model of heater–cooler device as the most likely infection source in most cases. The device was found to be contaminated at the manufacturing plant with M. chimaera of the same genotype as in outbreak-related patients (32–34). Following replacement or repositioning of the device in the operating theater, no further case has been detected in the affected surgical care facilities so far.
Advancing Laboratory-Based Surveillance for Communicable Diseases and Antimicrobial Resistance
To overcome inconsistency in categorizing antimicrobial resistance across countries, the European Committee for Antimicrobial Susceptibility Testing (EUCAST) operating under the joint auspices of ECDC and the European Society of Clinical Microbiology and Infectious Diseases has developed standard methods and nomenclature and advocated their use for in vitro AST of bacteria and fungi of medical interest. They have published evidence-based criteria for categorization of clinical isolates as wild-type/non-wild phenotype or clinically susceptible/resistant to antimicrobial agents (35). In 2012, EU case definitions enforced the EUCAST clinical breakpoints for surveillance of antimicrobial resistance in humans (6). Between 2011 and 2015, use of EUCAST breakpoints has rapidly progressed in clinical microbiology laboratories across Europe (36). The percentage of EU clinical laboratories reporting to the ECDC coordinated European Antimicrobial Resistance Surveillance Network (EARS-Net) which are using EUCAST susceptibility breakpoints increased from 29 to 84% over this 5-year period (37, 38). Likewise, the percentage of national reference laboratories participating in the ECDC coordinated European Gonococcal Antimicrobial Surveillance Programme (Euro-GASP) that use EUCAST breakpoints increased from 62 to 85% from 2014 to 2016 (12, 39). This harmonization across laboratories, encouraged by ECDC, improves the quality of surveillance of antimicrobial resistance in Europe.
Because antimicrobial resistance in zoonotic pathogens is transferable from food animals to humans, One-Health monitoring, covering microbiota from human, animal, and environmental sectors in a holistic approach, is essential for containment. Unfortunately, comparisons between antimicrobial resistance data from humans, food, and animals have long been hampered by the use of different test methods and interpretative criteria in each health sector. Whereas AST results on human isolates is interpreted with clinical breakpoints, those from healthy animal and food isolates monitoring are interpreted based on epidemiological cut-off values (ECOFFs). These cut-off values distinguish normally susceptible isolates (wild-type phenotype) from those who have acquired resistance to the antimicrobial (non-wild-type phenotype). In 2013, ECDC together with national representatives from its Food- and Waterborne Diseases and Zoonoses (FWD) network developed an EU protocol for harmonized monitoring of antimicrobial resistance in Salmonella enterica and Campylobacter jejuni and C. coli from human isolates, aiming at increasing the quality and comparability of data collected in the EU Member States by ECDC with those collected by the European Food Safety Authority (EFSA) from the veterinary sector. This protocol defines the panel of antimicrobials, the test methods, and reporting of quantitative susceptibility data to allow direct comparison with animal isolates’ data interpreted with ECOFF values (40). Between 2013 and 2015, the proportion of reporting countries compliant with the EU protocol doubled from 30 to 60%, improving comparability of antimicrobial resistance surveillance data among sectors (41).
Limiting the dissemination of multidrug-resistant bacterial pathogens requires a better understanding of the emergence and mode of spread of resistance determinants. To this end, ECDC is undertaking pan-European molecular epidemiology surveys. In 2012, addressing the most threatening carbapenem resistance problem, the ECDC commissioned the European Survey of Carbapenemase-producing Enterobacteriaceae (EuSCAPE) among a consortium of hospitals to assess the prevalence and geographical distribution of carbapenemase-producing Enterobacteriaceae (CPE) characterized at the genetic level across 38 countries (42, 43). The project demonstrated the feasibility of conducting integrated epidemiological and microbiological sentinel multi-center surveys and of collecting quality-assured data for EU level analyses (43–45). It also built national capacities across Europe for standardized laboratory detection, identification, and surveillance of CPE (42). In 2015, all EU/EEA countries had nominated a national reference laboratory and operated national surveillance for CPE (44). The EuSCAPE surveys revealed a rapid dissemination of carbapenemase-producing Klebsiella pneumoniae around Europe, with wide variations by country in the type of carbapenemase, and the number of countries reaching an inter-regional or endemic level doubling from 6 to 13 countries over the period 2013–2015 (44, 45).
Rising multidrug resistance also imperils the control of gonococcal infections worldwide. Since 2009, Euro-GASP monitors emerging resistance in Neisseria gonorrhoeae to therapeutic antimicrobials. The program is using a standardized methodology across the EU/EEA which integrates epidemiological and microbiological data to understand the risk factors associated with resistance and thereby inform intervention strategies (39). For AST, the Euro-GASP uses a hybrid approach of EU centralized and decentralized national testing (39). EQA and training activities helped standardize surveillance across Europe: between 2009 and 2014, the number of EU/EEA countries participating in Euro-GASP increased from 17 to 23 countries while the number of countries providing quality-assured (decentralized) testing increased from 3 to 17. In addition, Euro-GASP also piloted molecular typing to unravel associations between clonal type and antimicrobial resistance profiles that could aid understanding of the dissemination of resistance within at risk populations (46). Euro-GASP results have revealed rapidly changing patterns of N. gonorrhoeae resistance to drugs of choice across Europe, leading to the revision of European and national gonorrhea treatment guidelines and development by ECDC of the “Response plan to control and manage the threat of multidrug-resistant gonorrhoea in Europe” (47).
Integration of Molecular and Genomic Typing in European Surveillance and Epidemic Investigations
Whole-genome sequencing is empowering high-precision infectious disease surveillance and control (48). ECDC developed multi-annual roadmaps for the integration of molecular typing data on high-priority pathogens into its EU surveillance and epidemic preparedness systems, fact-checking the feasibility, and capacity in the EU countries, in synergy with third-party activities along a global One-Health partnership (49–51). The added-value of molecular surveillance and WGS-based outbreak investigation was demonstrated following the introduction among the ECDC FWD public health laboratory surveillance network of a novel Multi-Locus Variable Number Tandem Repeat Analysis (MLVA) typing scheme for Salmonella Enteritidis (52). Increases in salmonellosis cases with an uncommon MLVA profile were posted first by Scotland and then by the Netherlands in January and August 2016 in the Epidemic Intelligence Information System (53, 54). Several other countries reported detection of this MLVA profile. WGS analysis performed either by national reference laboratories or in a central facility through ECDC support confirmed a multi-country outbreak involving two distinct Salmonella Enteritidis genomic types. An outbreak investigation team involving affected countries, ECDC, and EFSA agreed on the outbreak case definition based on WGS and MLVA types, assessed the magnitude of the outbreak, and identified response options. At least 18 EU/EEA countries were affected by the outbreak. Environmental and food investigations by the food safety authorities showed that food establishments or retail shops in eight EU countries had received eggs contaminated with Salmonella Enteritidis of the epidemic MLVA or WGS types from farms located in one EU country (53).
Outlook
Whether for health protection or for food safety, we rely on microbiology laboratories to detect, identify, and characterize human pathogens of public health significance. Although health services are a national responsibility in the EU, a cost–benefit analysis study by the European Commission has concluded that the benefits of maintaining EU public health reference laboratory networks were likely to outweigh the costs, both from a Member State and from an EU perspective (55). As outlined in the examples above, EU laboratory networks coordinated by ECDC as well as the European Commission over the last years have brought public health added value through fit-for-purpose harmonization and validation of laboratory methods, quality assurance and training activities as well as contribution to preparedness and response for biological threats. The EULabCap surveys concluded that the EU/EEA as a whole has a strong and improving public health microbiology system (7). Its key assets include harmonized methods for AST, extensive reference laboratory services, and laboratory inputs within national and EU surveillance networks. The use of molecular typing for surveillance and laboratory participation in outbreak response showed significant progress. In an era of rapid progress in electronic recording and transmission of health data, it is noticeable that half of the EU/EEA countries have implemented automated electronic reporting of microbiology laboratory data to their national communicable disease or antimicrobial resistance surveillance systems (7). To track the transmission of resistance genes needed for targeted control measures, ECDC has developed a protocol for genomic-based surveillance of carbapenem-resistant and/or colistin-resistant Enterobacteriaceae at the EU level (56). To deliver their full potential, these encouraging developments require sustained innovation in molecular diagnostic and typing methods and the collaborative engineering of wider connectivity between laboratory and public health information systems from the local to the global level. Collaboration between clinical and public health practitioners, government agencies, and academic institutions is key to develop integrated clinical, epidemiological, and molecular microbiological data collection, recording and exchange protocols as well as build the integrated information technology infrastructure enabling real time, molecular epidemiologic surveillance, and alert systems for infectious diseases. ECDC as convener, funding source and coordinator in close partnership with multidisciplinary experts from EU disease networks continues to act as catalyst for translating these novel approaches into practice and further strengthening the collective laboratory capability for disease control in the EU.
Author Contributions
BA: data collation, analysis, and writing the initial draft. JR and KL: data collation and reviewing the initial draft. MS: conceptualization and rewriting the final draft.
Conflict of Interest Statement
The authors are employees of the European Centre for Disease Prevention and Control. They declare no commercial or financial conflict of interest.
Acknowledgments
The dedicated work of the project managers, coordinators, and members of the ECDC-supported disease networks is gratefully acknowledged. We thank Laura Espinosa, Amanda Ozin, Polya Rosin, and Daniel Palm for collating data for the annual ECDC Microbiology Activity Reports 2010–2014. We are grateful to ECDC experts and heads of the ECDC Diseases Programmes for their input and critical review of the manuscript.
References
2. Mulders MN, Rota PA, Icenogle JP, Brown KE, Takeda M, Rey GJ, et al. Global measles and rubella laboratory network support for elimination goals, 2010-2015. MMWR Morb Mortal Wkly Rep (2016) 65(17):438–42. doi:10.15585/mmwr.mm6517a3
3. Laszlo A, Rahman M, Espinal M, Raviglione M, WHO/IUATLD Network of Supranational Reference Laboratories. Quality assurance programme for drug susceptibility testing of Mycobacterium tuberculosis in the WHO/IUATLD Supranational Reference Laboratory Network: five rounds of proficiency testing, 1994-1998. Int J Tuberc Lung Dis (2002) 6(9):748–56.
4. European Commission. Regulation (EC) No 851/2004 of the European Parliament and of the Council of 21 April 2004 Establishing a European Centre for Disease Prevention and Control. Strasbourg: OJEU (2004). Available from: https://publications.europa.eu/en/publication-detail/-/publication/77d98344-249e-486a-a9a9-88931b01aa7c/language-en
5. European Commission. Decision No 1082/2013/EU of the European Parliament and of the Council of 22 October 2013 on Serious Cross-Border Threats to Health and Repealing Decision No 2119/98/EC Text with EEA Relevance (2013). Available from: http://ec.europa.eu/health/sites/health/files/preparedness_response/docs/decision_serious_crossborder_threats_22102013_en.pdf
6. European Commission. Decision No 2012/506/EU. Commission Implementing Decision of 8 August 2012 Amending Decision 2002/253/EC Laying Down Case Definitions for Reporting Communicable Diseases to the Community Network under Decision No 2119/98/EC of the European Parliament and of the Council (2012). Available from: http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32012D0506&qid=1428573336660&from=EN#page=30
7. European Centre for Disease Prevention and Control. EU Laboratory Capacity Monitoring System (EULabCap) – Report on 2015 Survey of EU/EEA Capabilities and Capacities. Stockholm: ECDC (2017). Available from: https://ecdc.europa.eu/sites/portal/files/documents/EULabCap_report-for-2015.pdf
8. Economisti Associati Srl. The Second Independent Evaluation of the ECDC in Accordance with its Founding Regulation (European Parliament and Council Regulation (EC) no 851/2004) Bologna (2014). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/aboutus/Key%20Documents/ECDC-external-evaluation-2014.pdf
9. Nikolayevskyy V, Hillemann D, Richter E, Ahmed N, van der Werf MJ, Kodmon C, et al. External quality assessment for tuberculosis diagnosis and drug resistance in the European Union: a five year multicentre implementation study. PLoS One (2016) 11(4):e0152926. doi:10.1371/journal.pone.0152926
10. de Beer JL, Kodmon C, van Ingen J, Supply P, van Soolingen D. Global network for molecular surveillance of tuberculosis. Second worldwide proficiency study on variable number of tandem repeats typing of Mycobacterium tuberculosis complex. Int J Tuberc Lung Dis (2014) 18(5):594–600. doi:10.5588/ijtld.13.0531
11. Pas SD, Koopmans MP, Niedrig M. Clinical implications of and lessons learnt from external assessment of MERS-CoV diagnostics. Expert Rev Mol Diagn (2016) 16(1):7–9. doi:10.1586/14737159.2016.1116943
12. European Centre for Disease Prevention and Control. Euro-GASP External Quality Assessment (EQA) Scheme for Neisseria gonorrhoeae Antimicrobial Susceptibility Testing. Stockholm: ECDC (2017). Available from: https://ecdc.europa.eu/sites/portal/files/documents/EQA%20Report%202016%20final.pdf
13. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med (2012) 367(19):1814–20. doi:10.1056/NEJMoa1211721
14. Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill (2012) 17(39):20285. doi:10.2807/ese.17.39.20285-en
15. van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio (2012) 3(6):e473–412. doi:10.1128/mBio.00473-12
16. Palm D, Pereyaslov D, Vaz J, Broberg E, Zeller H, Gross D, et al. Laboratory capability for molecular detection and confirmation of novel coronavirus in Europe, November 2012. Euro Surveill (2012) 17(49):20335. doi:10.2807/ese.17.49.20335-en
17. Pereyaslov D, Rosin P, Palm D, Zeller H, Gross D, Brown CS, et al. Laboratory capability and surveillance testing for Middle East respiratory syndrome coronavirus infection in the WHO European Region, June 2013. Euro Surveill (2014) 19(40):20923. doi:10.2807/1560-7917.ES2014.19.40.20923
18. Pas SD, Patel P, Reusken C, Domingo C, Corman VM, Drosten C, et al. First international external quality assessment of molecular diagnostics for Mers-CoV. J Clin Virol (2015) 69:81–5. doi:10.1016/j.jcv.2015.05.022
19. European Centre for Disease Prevention and Control. Severe Respiratory Disease Associated with Middle East Respiratory Syndrome Coronavirus. Stockholm: ECDC (2015). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/MERS-rapid-risk-assessment-update-october-2015.pdf.
20. Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med (2013) 368(20):1888–97. doi:10.1056/NEJMoa1304459
21. European Centre for Disease Prevention and Control. Diagnostic Preparedness in Europe for Detection of Avian Influenza A(H7N9) Viruses. Technical Briefing Note 23 April 2013. Stockholm (2013). Available from: http://ecdc.europa.eu/en/publications/Publications/avian-influenza-H7N9-microbiology-diagnostic-preparedness-for-detection.pdf
22. Broberg E, Pereyaslov D, Struelens M, Palm D, Meijer A, Ellis J, et al. Laboratory preparedness in EU/EEA countries for detection of novel avian influenza A(H7N9) virus, May 2013. Euro Surveill (2014) 19(4):20682. doi:10.2807/1560-7917.ES2014.19.4.20682
23. European Centre for Disease Prevention and Control. External Quality Assessment Scheme for Influenza Virus Detection, Isolation and Culture for the European Reference Laboratory Network for Human Influenza: 2013. Stockholm: ECDC (2014). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/influenza-virus-detection-isolation-culture-EQA-2013.pdf
24. Wolfel R, Stoecker K, Fleischmann E, Gramsamer B, Wagner M, Molkenthin P, et al. Mobile diagnostics in outbreak response, not only for Ebola: a blueprint for a modular and robust field laboratory. Euro Surveill (2015) 20(44). doi:10.2807/1560-7917.ES.2015.20.44.30055
25. Ellerbrok H, Jacobsen S, Patel P, Rieger T, Eickmann M, Becker S, et al. External quality assessment study for ebolavirus PCR-diagnostic promotes international preparedness during the 2014 – 2016 Ebola outbreak in West Africa. PLoS Negl Trop Dis (2017) 11(5):e0005570. doi:10.1371/journal.pntd.0005570
26. Nisii C, Vincenti D, Fusco FM, Schmidt-Chanasit J, Carbonnelle C, Raoul H, et al. The contribution of the European high containment laboratories during the 2014-2015 Ebola virus disease emergency. Clin Microbiol Infect (2017) 23(2):58–60. doi:10.1016/j.cmi.2016.07.003
27. European Centre for Disease Prevention and Control. Annual Epidemiological Report 2016 – Ebola and Marburg Fevers. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/documents/Ebola%20and%20Marburg%20fevers%20AER_0.pdf
28. Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, et al. Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis (2015) 61(1):67–75. doi:10.1093/cid/civ198
29. European Centre for Disease Prevention and Control. Invasive Cardiovascular Infection by Mycobacterium chimaera – 30 April 2015. Stockholm: ECDC (2015). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/mycobacterium-chimaera-infection-associated-with-heater-cooler-units-rapid-risk-assessment-30-April-2015.pdf
30. European Centre for Disease Prevention and Control. EU Protocol for Case Detection, Laboratory Diagnosis and Environmental Testing of Mycobacterium chimaera Infections Potentially Associated with Heater-Cooler Units: Case Definition and Environmental Testing Methodology. Stockholm: ECDC (2015). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/EU-protocol-for-M-chimaera.pdf
31. European Centre for Disease Prevention and Control. Invasive Cardiovascular Infection by Mycobacterium chimaera Associated with 3T Heater-Cooler System Used during Open-Heart Surgery – 18 November 2016. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/RRA-mycobacterium-chimaera-November-2016.pdf
32. Haller S, Holler C, Jacobshagen A, Hamouda O, Abu Sin M, Monnet DL, et al. Contamination during production of heater-cooler units by Mycobacterium chimaera potential cause for invasive cardiovascular infections: results of an outbreak investigation in Germany, April 2015 to February 2016. Euro Surveill (2016) 21(17):30215. doi:10.2807/1560-7917.ES.2016.21.17.30215
33. Chand M, Lamagni T, Kranzer K, Hedge J, Moore G, Parks S, et al. Insidious risk of severe Mycobacterium chimaera infection in cardiac surgery patients. Clin Infect Dis (2017) 64(3):335–42. doi:10.1093/cid/ciw754
34. van Ingen J, Kohl TA, Kranzer K, Hasse B, Keller PM, Katarzyna Szafranska A, et al. Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect Dis (2017) 17(10):1033–41. doi:10.1016/S1473-3099(17)30324-9
35. Kahlmeter G. Defining antibiotic resistance-towards international harmonization. Ups J Med Sci (2014) 119(2):78–86. doi:10.3109/03009734.2014.901446
36. Brown D, Canton R, Dubreuil L, Gatermann S, Giske C, MacGowan A, et al. Widespread implementation of EUCAST breakpoints for antibacterial susceptibility testing in Europe. Euro Surveill (2015) 20(2):21008. doi:10.2807/1560-7917.ES2015.20.2.21008
37. European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe: Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net): 2010. Stockholm: ECDC (2011). Available from: http://ecdc.europa.eu/en/publications/Publications/1111_SUR_AMR_data.pdf.pdf
38. European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe: Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net): 2015. Stockholm: ECDC (2017). Available from: http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-europe-2015.pdf
39. European centre for Disease Prevention and Control. Gonococcal Antimicrobial Susceptibility Surveillance in Europe, 2014. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/gonococcal-antimicrobial-susceptibility-surveillance-Europe-2014.pdf
40. European Center for Disease Prevention and Control. EU Protocol for Harmonised Monitoring of Antimicrobial Resistance in Human Salmonella and Campylobacter Isolates – June 2016. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/antimicrobial-resistance-Salmonella-Campylobacter-harmonised-monitoring.pdf
41. European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J (2017) 15(2):4694. doi:10.2903/j.efsa.2017.4694
42. European Centre for Disease Prevention and Control. Carbapenemase-Producing Bacteria in Europe: Interim Results from the European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Project. Stockholm: ECDC (2013). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/antimicrobial-resistance-carbapenemase-producing-bacteria-europe.pdf
43. Glasner C, Albiger B, Buist G, Tambic Andrasevic A, Canton R, Carmeli Y, et al. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill (2013) 18(28):20525. doi:10.2807/1560-7917.ES2013.18.28.20525
44. Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae Working Group. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill (2015) 20:45. doi:10.2807/1560-7917.ES.2015.20.45.30062
45. Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasevic AT, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis (2017) 17(2):153–63. doi:10.1016/S1473-3099(16)30257-2
46. Jacobsson S, Golparian D, Cole M, Spiteri G, Martin I, Bergheim T, et al. WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC >2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J Antimicrob Chemother (2016) 71(11):3109–16. doi:10.1093/jac/dkw279
47. European Centre for Disease Prevention and Control. Response Plan to Control and Manage the Threat of Multidrug-Resistant Gonorrhoea in Europe. Stockholm: ECDC (2012). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/1206-ECDC-MDR-gonorrhoea-response-plan.pdf
48. European Centre for Disease Prevention and Control. Surveillance of Communicable Diseases in Europe – A Concept to Integrate Molecular Typing Data into EU-level surveillance. Version 2.4, 7 September 2011. Stockholm: ECDC (2013). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/surveillance-concept-molecular%20typing-sept2011.pdf
49. European Centre for Disease Prevention and Control. Expert Opinion on Whole Genome Sequencing for Public Health Surveillance. Stockholm: ECDC (2016). Available from: http://ecdc.europa.eu/en/publications/Publications/whole-genome-sequencing-for-public-health-surveillance.pdf
50. European Centre for Disease Prevention and Control. ECDC Roadmap for Integration of Molecular and Genomic Typing into European-Level Surveillance and Epidemic Preparedness – Version 2.1, 2016-19. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/molecular-typing-EU-surveillance-epidemic-preparedness-2016-19-roadmap.pdf
51. European Centre for Disease Prevention and Control. ECDC Roadmap for Integration of Molecular Typing into European-Level Surveillance and Epidemic Preparedness – Version 1.2, 2013. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/molecular-typing-EU-surveillance-epidemic-preparedness-2013.pdf
52. European Centre for Disease Prevention and Control. Laboratory Standard Operating Procedure for Multiple-Locus Variable-Number Tandem Repeat Analysis of Salmonella enterica Serotype Enteritidis. Stockholm: ECDC (2016). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/Salmonella-Enteritidis-Laboratory-standard-operating-procedure.pdf.
53. European Centre for Disease Prevention and Control, European Food Safety Authority. Multi-Country Outbreak of Salmonella Enteritidis Phage Type 8, MLVA profile 2-9-7-3-2 and 2-9-6-3-2 Infections, 7 March 2017. Stockholm and Parma: ECDC and EFSA (2017). Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/rapid-outbreak-assessment-Salmonella-Enteritidis-7-mar-2017.pdf
54. Nadon CA, Trees E, Ng LK, Moller Nielsen E, Reimer A, Maxwell N, et al. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro Surveill (2013) 18(35):20565. doi:10.2807/1560-7917.ES2013.18.35.20565
55. European Commission. Study on Cost-Benefit Analysis of Reference Laboratories for Human Pathogens. Luxembourg (2016). Available from: https://ec.europa.eu/health/sites/health/files/preparedness_response/docs/2016_laboratorieshumanpathogens_frep_en.pdf.
56. European Centre for Disease Prevention and Control. ECDC Study Protocol for Genomic-Based Surveillance of Carbapenem-Resistant and/or Colistin-Resistant Enterobacteriaceae at the EU Level. Version 1.1. Stockholm: ECDC (2017). Available from: http://ecdc.europa.eu/en/publications/Publications/Protocol-genomic-surveillance-resistant-Enterobacteriaceae.pdf
Keywords: public health laboratory networks, public health microbiology, external quality assessment, molecular diagnostics, antimicrobial susceptibility testing, molecular typing, communicable disease surveillance, emerging diseases
Citation: Albiger B, Revez J, Leitmeyer KC and Struelens MJ (2018) Networking of Public Health Microbiology Laboratories Bolsters Europe’s Defenses against Infectious Diseases. Front. Public Health 6:46. doi: 10.3389/fpubh.2018.00046
Received: 22 September 2017; Accepted: 07 February 2018;
Published: 26 February 2018
Edited by:
Margaret Ip, The Chinese University of Hong Kong, Hong KongReviewed by:
Sergey Eremin, World Health Organization, SwitzerlandAleksandra Barac, University of Belgrade, Serbia
Copyright: © 2018 Albiger, Revez, Leitmeyer and Struelens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc J. Struelens, bWFyYy5zdHJ1ZWxlbnMmI3gwMDA0MDtlY2RjLmV1cm9wYS5ldQ==
 Barbara Albiger
Barbara Albiger Joana Revez
Joana Revez Katrin Claire Leitmeyer
Katrin Claire Leitmeyer Marc J. Struelens
Marc J. Struelens