
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
DATA REPORT article
Front. Public Health, 12 December 2017
Sec. Epidemiology
Volume 5 - 2017 | https://doi.org/10.3389/fpubh.2017.00336
 Nazli Ayhan1,2
Nazli Ayhan1,2 Bulent Alten3
Bulent Alten3 Vladimir Ivovic4
Vladimir Ivovic4 Franjo Martinkovic5
Franjo Martinkovic5 Ozge E. Kasap3
Ozge E. Kasap3 Yusuf Ozbel6
Yusuf Ozbel6 Xavier de Lamballerie1,2
Xavier de Lamballerie1,2 Remi N. Charrel1,2*
Remi N. Charrel1,2*
Toscana virus (TOSV) is a sandfly borne virus (genus Phlebovirus, family Phenuiviridae) which shows a wide distribution in the Mediterranean basin (1). TOSV was first discovered from Phlebotomus perniciosus and P. perfiliewi sand flies in 1971 in Italy (2). Since then TOSV has been isolated or detected in France, Spain, Portugal, Morocco, Algeria, Tunisia, Croatia, Greece, Turkey, Cyprus, and Corsica either from sand flies or human samples (3). During the warm season, TOSV is recognized as one of the main causes of aseptic meningitis within endemic countries due to increased vector activity during the warm season. TOSV is the most pathogenic among the phleboviruses transmitted by sand flies; it can affect the central nervous system and cause meningitis and meningoencephalitis (1).
Three distinct lineages of TOSV have been identified so far: A, B, and C, at the outset of this study, only lineage C had been identified in Croatia in the cerebral spinal fluid (CSF) of a patient presenting with meningitis (4). Serological studies demonstrated that TOSV is circulating at a high rate in the islands and along the Adriatic coast of Croatia (5). Serology does not discriminate between the three genetic lineages, the presence of TOSV strains belonging to lineages A and/or B had never been reported. In this study, sand flies collected in Croatia were tested for the presence of TOSV RNA and for subsequent identification of TOSV genetic lineages.
A total of 1,453 sand flies were collected from 5 locations in Croatia in July 2015 using modified CO2-CDC traps (Table 1). A total of 78 pools, each containing up to 30 insects, were analyzed according to location, sex, and date of trapping (Table 1). They were tested by real-time RT-qPCR for TOSV RNA (6). Two pools, C63 and C64, were positive with respective Ct values at 22.5 and 35.3. Since sequence analysis of the corresponding PCR product did not allow to identify the lineage, two other PCR assays were used for partial sequencing of the nucleoprotein gene (7, 8). Colinearization of the two sequences obtained from pool C63 resulted in a 576-nt long sequence (GenBank acc no KY867756) (9). From pool C64, only the Sandfly Fever Naples Virus (SFNV) nested PCR was positive and resulted in a 320-nt long sequence (GenBank acc no KY867757) (10).
Virus isolation was attempted by inoculating 50 µL of the homogenate supernatant onto Vero cells as previously described (11). After six blind passages, TOSV was not isolated.
C63 and C64 sequences were aligned using CLUSTAL X (MEGA 6.06) with homologous sequences of other TOSV strains and selected phleboviruses belonging to the Sandfly fever Naples species obtained from GenBank (see Supplementary Material) (12). Amino acid and nucleotide identities were calculated with the p-distance algorithm. Phylogenetic studies were performed using the neighbor-joining method in MEGA6 (Figure 1). The robustness of the nodes was tested by 1,000 bootstrap replications.
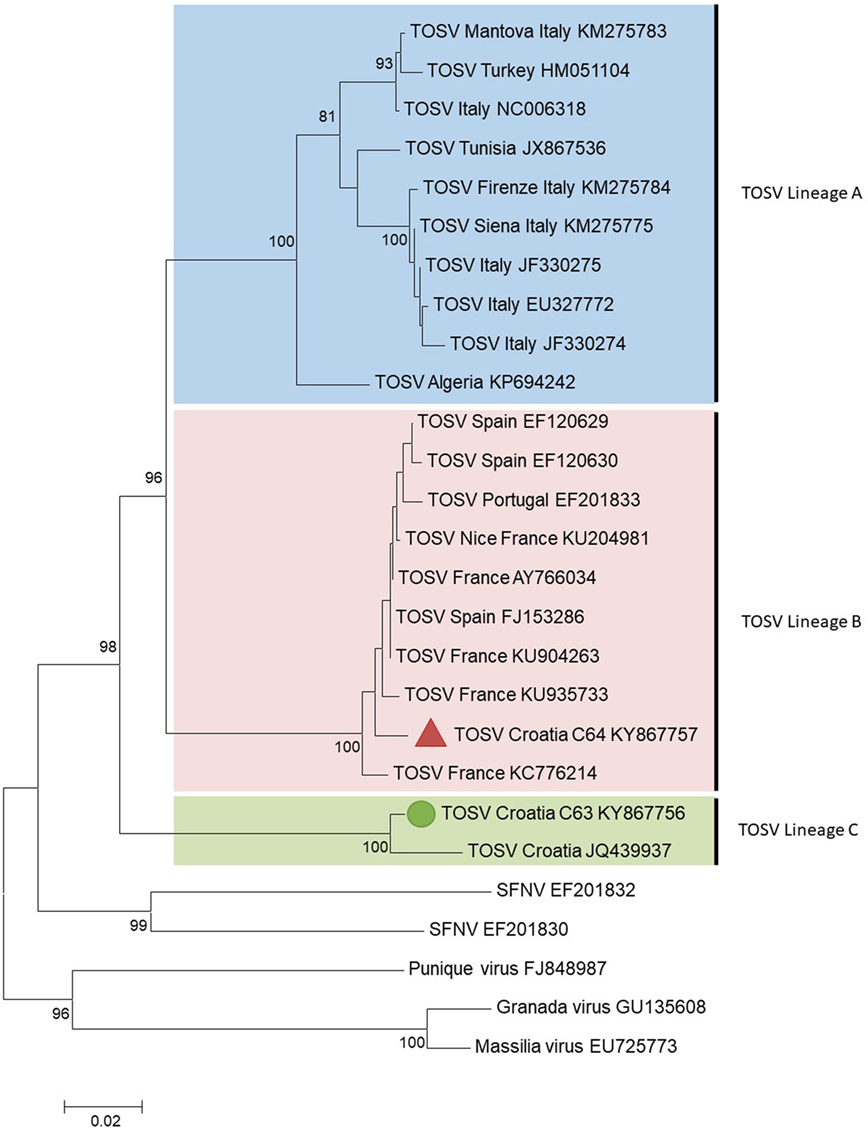
Figure 1. Phylogenetic analysis of Toscana virus (TOSV) based on a 576-nt long sequences between positions 88 and 663 (numbered after strain IssPhL3, Acc No X53794) located in the nucleoprotein gene. Distances and groupings between the N protein sequences were determined by the p-distance algorithm and the neighbor-joining method with the MEGA 6.06 software program (12). Bootstrap values are indicated and correspond to 1,000 pseudoreplications.
The two TOSV sequences were clearly different from each other with 3.8 and 16.5% genetic divergence at amino acid and nucleotide level, respectively. C63 sequence was grouped with the unique Croatian TOSV sequence (corresponding to the CSF sample of a patient presenting with TOSV meningitis in 2008) (4); hence, C63 sequence was identified as belonging to the lineage C. Surprisingly, C64 sequence was most closely related to the sequence of TOSV strain 113/Nice (GenBank acc no KU204981) isolated from sand flies trapped in southeastern France, belonging to the lineage B. To the best of our knowledge, this is the first evidence that TOSV strains belonging to the lineage B are present in Croatia and probably more largely in the Balkan Peninsula.
Sand fly species from the TOSV positive pools were identified based on cytochrome b and cytochrome c oxidase subunit I as previously described (13). C63 and C64 pools consisted exclusively of Phlebotomus neglectus (Table 2). These results are congruent with the results of a larger study which aimed at building a species inventory of sand flies in the Balkans (VectorNet project supported by ECDC/EFSA) (14). Three sand fly species, P. neglectus, P. tobbi, and Sergentomyia minuta, were identified from exactly the same location with high dominancy of P. neglectus (Vidonje, Tables 1 and 2) in 2015 (Alten et al., unpublished data). P. perniciosus and P. perfiliewi are the most recognized vectors of TOSV. P. sergenti and P. longicuspis are also suspected TOSV vectors (15). Here, we provide the first evidence of P. neglectus as a probable vector of TOSV. This is a very important finding since this species is present at high density in regions where other TOSV vectors are not present, particularly in the Balkan Peninsula, Eastern Europe, and Turkey. Confirmation of P. neglectus as a competent vector of TOSV would considerably increase the size of the exposed human populations.
Our results demonstrated that the two TOSV lineages can cohabitate sympatrically within the study area showing no exclusion/interference of one virus by one another (Massilia/Toscana, Punique/Utique, Fermo/Toscana) (16, 17). The same vector species can transmit different types of viruses in the same area. P. neglectus is the vector of Corfou virus (18), Balkan virus (19), and TOSV in the same region (this study). Whether a single insect can be coinfected by two viruses is not proven, but is likely to happen and this could drive to the production of a recombinant or reassortant virus (20).
With the assumption that only one insect in each pool is infected, the infection rate of TOSV in Croatia was 0.1% which is higher than rates observed in Tunisia (0.03%), Spain (0.05%), and Algeria (0.004%) (11, 21).
Despite the efforts, virus isolation was not obtained probably because of virion degradation during field collection. Due to the low viral load in pool C64 (Ct value 35.3) only the SFNV nested PCR was positive and resulted in a 320-nt long sequence. However, the length of the sequence did not affect neither the quality of the alignment nor the topology of the phylogenetic tree.
In Croatia, 37.5% of 755 healthy residents of the coastal regions and islands had TOSV IgG (5). Accordingly, TOSV should be included in the repertoire of pathogens to be explored in patients presenting with neuroinvasive infections during the warm season.
In conclusion, this study showed that (i) strains of TOSV lineage B are present in coastal Croatia where they cocirculate with lineage C strains; (ii) P. neglectus is the most probable vector of both lineages of TOSV in the region, (iii) Croatia is, after France and Turkey, the third country where two lineages of TOSV are sympatric; (iv) physicians should consider testing patients who present with neuroinvasive infections in Croatia during the season of sand fly activity for TOSV.
Sequences generated in this study are available in the GenBank database under the accession numbers KY867756 and KY867757.
NA performed the sample collection, administered experiments, and wrote the manuscript. BA, VI, FM, OK, and YO organized the sample collection and contributed to the manuscript. XL designed the experiments and contributed to the manuscript. RC designed the experiments and wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Karine Almani for excellent technical assistance. The work of RNC was done under the frame of EurNegVec (TD1303) COST Action. NA is a PhD student supported by a grant from Fondation Mediterranee Infection.
This work was supported by funds received from (i) VectorNet, a European network for sharing data on the geographic distribution of arthropod vectors, transmitting human and animal disease agents (Contract OC/EFSA/AHAW/2013/02-FWC1) funded by the European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC) (http://ecdc.europa.eu/en/healthtopics/vectors/VectorNet/Pages/VectorNet.aspx), (ii) the European Virus Archive goes Global (EVAg) project in the European Union’s Horizon 2020 research and innovation program under grant agreement No 653316 (http://global.european-virus-archive.com/). NA is a PhD student supported by a grant from Fondation Mediterranee Infection. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fpubh.2017.00336/full#supplementary-material.
1. Charrel RN, Gallian P, Navarro-Mari JM, Nicoletti L, Papa A, Sánchez-Seco MP, et al. Emergence of Toscana virus in Europe. Emerg Infect Dis (2005) 11(11):1657–63. doi:10.3201/eid1111.050869
2. Verani P, Ciufolini MG, Nicoletti L, Balducci M, Sabatinelli G, Coluzzi M, et al. Ecological and epidemiological studies of Toscana virus, an arbovirus isolated from Phlebotomus. Ann Ist Super Sanita (1982) 18(3):397–9.
3. Alkan C, Bichaud L, de Lamballerie X, Alten B, Gould E, Charrel RN. Sandfly-borne phleboviruses of Eurasia and Africa: epidemiology, genetic diversity, geographic range, control measures. Antiviral Res (2013) 100(1):54–74.
4. Punda-Polić V, Mohar B, Duh D, Bradarić N, Korva M, Fajs L, et al. Evidence of an autochthonous Toscana virus strain in Croatia. J Clin Virol (2012) 55(1):4–7. doi:10.1016/j.jcv.2012.06.006
5. Punda-Polić V, Jerončić A, Mohar B, Šiško Kraljević K. Prevalence of Toscana virus antibodies in residents of Croatia. Clin Microbiol Infect (2012) 18(6):E200–3. doi:10.1111/j.1469-0691.2012.03840.x
6. Pérez-Ruiz M, Collao X, Navarro-Marí JM, Tenorio A. Reverse transcription, real-time PCR assay for detection of Toscana virus. J Clin Virol (2007) 39(4):276–81. doi:10.1016/j.jcv.2007.05.003
7. Lambert AJ, Lanciotti RS. Consensus amplification and novel multiplex sequencing method for S segment species identification of 47 viruses of the orthobunyavirus, phlebovirus, and nairovirus genera of the family Bunyaviridae. J Clin Microbiol (2009) 47(8):2398–404. doi:10.1128/JCM.00182-09
8. Charrel RN, Izri A, Temmam S, Delaunay P, Toga I, Dumon H, et al. Cocirculation of 2 genotypes of Toscana virus, southeastern France. Emerg Infect Dis (2007) 13(3):465–8. doi:10.3201/eid1303.061086
9. Ayhan N, Alten B, Ivovic V, Martinkovic F, Kasap OE, Ozbel Y, et al. Nucleotide. Accession No. KY867756, Toscana Virus Strain C63 Nucleocapsid Protein Gene, Partial CDS. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information (1988). Available from: https://www.ncbi.nlm.nih.gov/nuccore/KY867756
10. Ayhan N, Alten B, Ivovic V, Martinkovic F, Kasap OE, Ozbel Y, et al. Nucleotide. Accession No. KY867757, Toscana Virus Strain C64 Nucleocapsid Protein Gene, Partial CDS. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information (1988). Available from: https://www.ncbi.nlm.nih.gov/nuccore/KY867757
11. Alkan C, Allal-Ikhlef AB, Alwassouf S, Baklouti A, Piorkowski G, de Lamballerie X, et al. Virus isolation, genetic characterization and seroprevalence of Toscana virus in Algeria. Clin Microbiol Infect (2015) 21(11):1040.e1–9. doi:10.1016/j.cmi.2015.07.012
12. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol (2013) 30(12):2725–9. doi:10.1093/molbev/mst197
13. Ayhan N, Velo E, de Lamballerie X, Kota M, Kadriaj P, Ozbel Y, et al. Detection of Leishmania infantum and a novel phlebovirus (Balkan virus) from sand flies in Albania. Vector Borne Zoonotic Dis (2016) 16(12):802–6. doi:10.1089/vbz.2016.2002
14. European Centre for Disease Prevention and Control. VectorNet. (2014). Available from: http://ecdc.europa.eu/en/healthtopics/vectors/VectorNet/
15. Es-Sette N, Nourlil J, Hamdi S, Mellouki F, Lemrani M. First detection of Toscana virus RNA from sand flies in the genus Phlebotomus (Diptera: phlebotomidae) naturally infected in Morocco. J Med Entomol (2012) 49(6):1507–9. doi:10.1603/ME12042
16. Charrel RN, Moureau G, Temmam S, Izri A, Marty P, Parola P, et al. Massilia virus, a novel phlebovirus (Bunyaviridae) isolated from sand flies in the Mediterranean. Vector Borne Zoonotic Dis (2009) 9(5):519–30. doi:10.1089/vbz.2008.0131
17. Remoli ME, Fortuna C, Marchi A, Bucci P, Argentini C, Bongiorno G, et al. Viral isolates of a novel putative phlebovirus in the Marche region of Italy. Am J Trop Med Hyg (2014) 90(4):760–3. doi:10.4269/ajtmh.13-0457
18. Rodhain F, Madulo-Leblond G, Hannoun C, Tesh RB. Le virus corfou: un nouveau Phlebovirusisolé de phlébotomes en Grèce. Ann Inst Pasteur Virol (1985) 136(2):161–6. doi:10.1016/S0769-2617(85)80042-3
19. Ayhan N, Alten B, Ivovic V, Dvořák V, Martinkovic F, Omeragic J, et al. Direct evidence for an expanded circulation area of the recently identified Balkan virus (Sandfly Fever Naples Virus species) in several countries of the Balkan archipelago. Parasit Vectors (2017) 10(1):402. doi:10.1186/s13071-017-2334-y
20. Collao X, Palacios G, de Ory F, Sanbonmatsu S, Pérez-Ruiz M, Navarro JM, et al. Granada virus: a natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans. Am J Trop Med Hyg (2010) 83(4):760–5. doi:10.4269/ajtmh.2010.09-0697
Keywords: phlebovirus, Toscana virus, sandfly fever, Sandfly Fever Naples Virus, arbovirus
Citation: Ayhan N, Alten B, Ivovic V, Martinkovic F, Kasap OE, Ozbel Y, de Lamballerie X and Charrel RN (2017) Cocirculation of Two Lineages of Toscana Virus in Croatia. Front. Public Health 5:336. doi: 10.3389/fpubh.2017.00336
Received: 04 May 2017; Accepted: 27 November 2017;
Published: 12 December 2017
Edited by:
Joanna Lankester, Stanford University, United StatesReviewed by:
Fang Wang, St. Jude Children’s Research Hospital, United StatesCopyright: © 2017 Ayhan, Alten, Ivovic, Martinkovic, Kasap, Ozbel, de Lamballerie and Charrel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Remi N. Charrel, cmVtaS5jaGFycmVsQHVuaXYtYW11LmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.