- 1Department of Psychiatry, First Hospital/First Clinical Medical College of Shanxi Medical University, Taiyuan, China
- 2Basic Medical College, Shanxi Medical University, Taiyuan, China
- 3Shanxi Key Laboratory of Artificial Intelligence Assisted Diagnosis and Treatment for Mental Disorder, First Hospital of Shanxi Medical University, Taiyuan, China
- 4Shanxi Province Mental Health Center, Taiyuan Psychiatric Hospital, Taiyuan, China
- 5The Eighth Affiliated Hospital, Sun Yat-Sen University, Shenzhen, China
Background: The severe functional impairment and poor prognosis of early-onset schizophrenia (EOS) create a great need to identify effective biomarkers for early diagnosis in young psychiatric patients. Current research indicates a potential link between loss of autophagy function and emotional and behavioral abnormalities in individuals with psychiatric disorders.
Materials and Methods: This study aimed to explore diagnostic autophagy-related endogenous competitive RNA (ceRNA) networks for EOS patients. The messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) expression profiles were obtained from peripheral blood mononuclear cells of 18 EOS patients and 12 healthy controls (HC). A co-expression analysis was performed between 365 core lncRNAs and 55 differentially expressed autophagy-related genes (ARGs) to identify differentially expressed autophagy-related lncRNAs. Subsequently, five diagnostic autophagy-related lncRNAs were identified as candidate genes to construct a ceRNA regulatory network using least absolute shrinkage and selection operator (LASSO) Cox regression, and receiver operating characteristic (ROC) curve analysis was performed to evaluate their predictive accuracy. Then, putative interactions among lncRNA-microRNAs (miRNAs)-mRNA were determined based on the lncRNASNP2 and TarBase databases.
Results: Three lncRNAs, twenty miRNAs, and ten mRNAs were selected to construct an autophagy-associated ceRNA network associated with EOS occurrence. Through protein-protein interaction network analysis, five hub mRNAs were identified, which exhibited good predictive ability in distinguishing EOS patients from healthy individuals. ROC curve analysis demonstrated that integrating three diagnostic lncRNAs (RP1-135L22.1, RP5-884C9.2, RP11-390F4.3) along with five hub mRNAs (EIF4G1, AKT1, BAX, WIPI2, MAPT) appeared to yield better diagnostic accuracy compared to using either lncRNAs or mRNAs alone. Furthermore, all three diagnostic lncRNAs and five hub mRNAs were positively correlated with at least two types of immune infiltration.
Conclusion: Through transcriptome analysis, we searched for diagnostic autophagy-related ceRNA networks, which provided valuable candidates for the early diagnosis of EOS.
1 Introduction
Schizophrenia (SCZ) is a devastating mental illness that significantly impacts a substantial portion of the global population (1). SCZ not only affects patients’ mental health but also elevates the risk of physical disorders, leading to a higher loss of life. Compared to the general population, individuals diagnosed with SCZ have a shortened life expectancy of 15-20 years (2). While SCZ typically emerges during late adolescence or early adulthood, its occurrence in childhood and adolescence represents an important subset of psychoses. Early-onset schizophrenia (EOS), characterized by the first appearance of psychotic symptoms before age 18, is considered to have more severe neurodevelopmental abnormalities, a worse clinical course and outcome, a heightened genetic loading, and a greater familial burden of SCZ and associated spectrum disorders than adult-onset SCZ (3). The severe functional impairment and poor prognosis of EOS create a great need to identify effective biomarkers for early diagnosis in young psychiatric patients.
Although many hypotheses related to the etiology of SCZ have been proposed, its neuropathological mechanism has not been established. Cellular autophagy, a unique intracellular degradation process in lysosomes, is essential for neuronal survival and function by maintaining a balance between cell survival and death (4). Current research indicates that SCZ is broadly associated with prominent dysregulation of autophagy function. Studies on autopsy brain tissue of patients with SCZ have shown abnormal expression of key genes involved in regulating neuronal autophagy processes (5). Loss of autophagy function may be related to emotional and behavioral abnormalities in individuals with psychiatric disorders (6). Therefore, identifying biomarkers related to disease progression, prognosis, and treatment responsiveness by exploring the relationship between autophagy and EOS is necessary for early diagnosis and treatment.
Long non-coding RNAs (lncRNAs), a class of non-coding RNAs comprised of more than 200 nucleotides, participate in numerous physiological and pathological processes such as autophagy, apoptosis, and development by regulating transcription, epigenetic modifications, protein translation, etc (7–9). For SCZ, lncRNAs are considered important clues in understanding the molecular mechanisms because they are involved in many steps in the occurrence and development of the disease (10, 11). Currently, endogenous competitive RNAs (ceRNAs) have drawn increasing attention in the field of SCZ. LncRNAs and microRNAs (miRNAs) are important components of the ceRNA networks. LncRNAs act as “molecular sponges”, binding with miRNAs to prevent their inhibitory effect on targeted messenger RNAs (mRNAs). The balance between lncRNAs, miRNAs, and mRNAs can regulate various biological processes, and when disrupted, pathological processes occur (12). The lncRNA-miRNA-mRNA axis is an important regulatory network for SCZ, so they may serve as diagnostic biomarkers for SCZ patients. Current studies have explored the role of autophagy-related ceRNA networks on the diagnosis of SCZ patients, but not all of them included the same lncRNAs/miRNAs/mRNAs, and the accuracy of each model was different (13). In addition, the impact of autophagy-related ceRNAs in EOS has not been clearly defined. Therefore, it is worth exploring diagnostic autophagy-related ceRNAs for EOS patients.
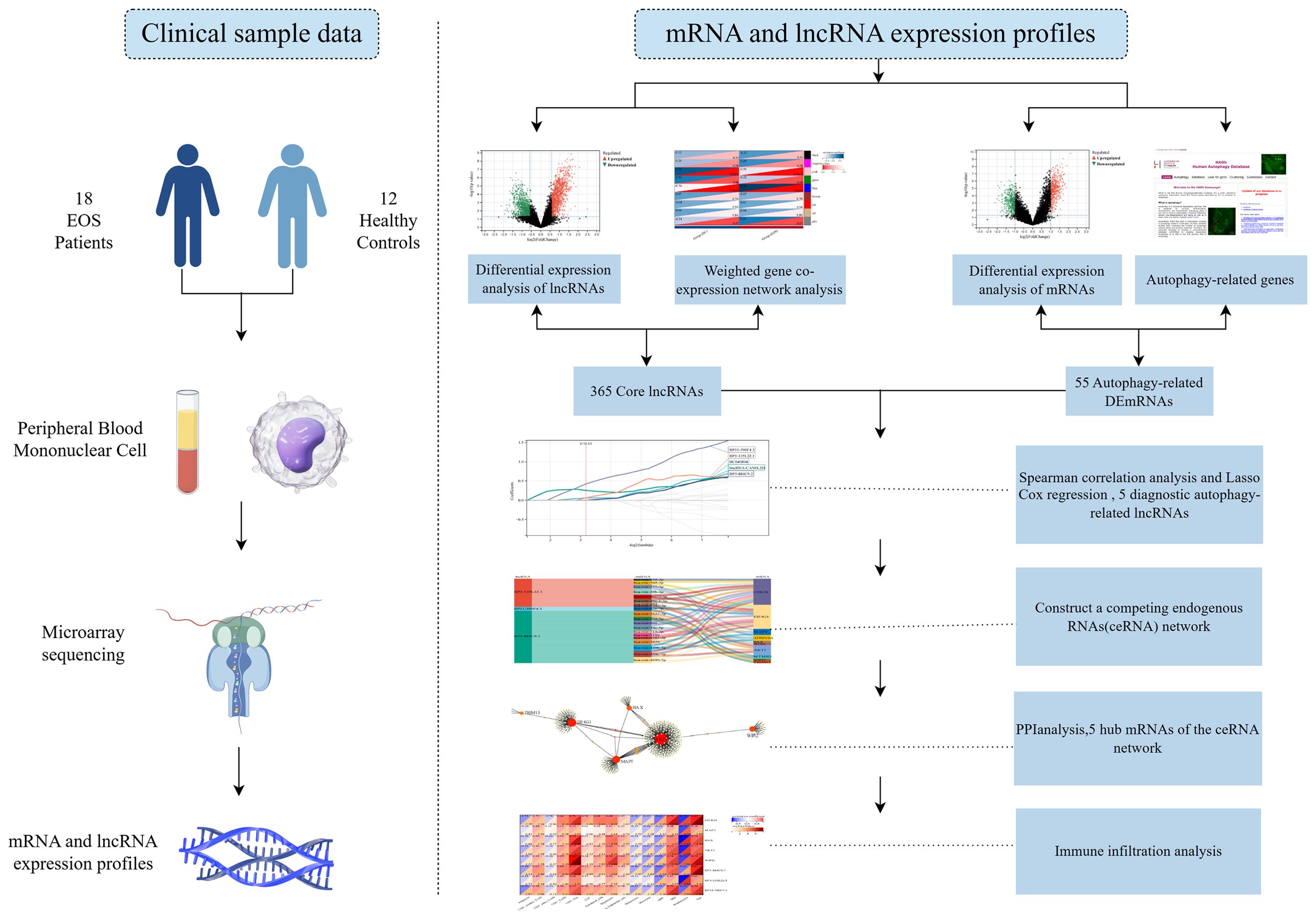
This study aimed to explore diagnostic autophagy-related ceRNA networks for EOS patients. In a first step, we screened 365 core lncRNAs and 55 differentially expressed autophagy-related genes (DEARGs), and identified 365 differentially expressed autophagy-related lncRNAs by a co-expression analysis. Subsequently, five diagnostic autophagy-related lncRNAs were identified as candidate genes to construct a ceRNA regulatory network using least absolute shrinkage and selection operator (LASSO) Cox regression, and receiver operating characteristic (ROC) curve analysis was performed to evaluate their predictive accuracy. Finally, putative interactions among lncRNA-miRNA-mRNA were determined based on the lncRNASNP2 and TarBase databases. Figure 1 is the technical roadmap of this study (By Figdraw).
2 Materials and methods
2.1 Data source
The dataset comes from the GSE54913 dataset, which was previously made public by our research group in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds). This study recruited 18 EOS patients (10 females and 8 males, aged 14.78 ± 1.70 years) and 12 healthy controls (6 females and 6 males, aged 14.75 ± 2.14 years) (14). We collected peripheral blood mononuclear cell (PBMC) samples from EOS patients and healthy controls. The mRNA and lncRNA expression profiles were obtained using microarrays. All subjects are from the First Hospital of Shanxi Medical University. EOS patients are diagnosed by at least two psychiatrists independently based on the criteria for SCZ in the Diagnosis and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV). The study was approved by the Medical Research Ethics Committee of Shanxi Medical University, and both the subjects and their parents signed informed consent forms.
2.2 Identification of differentially expressed mRNAs and lncRNAs
The differential expression analysis was performed via the R package Limma. LncRNAs with a p-value < 0.05 and |logFC|> 2, as well as mRNAs with a p-value < 0.05 and |logFC|> 1.5, were considered to be statistically significant. The final results were visualized as volcano maps and heat maps using Sangerbox tools (http://sangerbox.com/home.html).
2.3 Identification of key trait-module lncRNAs of EOS
The WGCNA (Weighted Correlation Network Analysis) package was used to screen for co-expression modules and key trait-module lncRNAs related to EOS. We chose a soft threshold power β = 14 to construct the scale-free topology module. Next, we performed hierarchical clustering to identify co-expression modules, and each module contained at least 100 lncRNAs. Then, we selected co-expression modules that were highly correlated with traits. Pearson’s correlation was used to analyze the module–trait relationships, with a p-value < 0.05 defined as a significant correlation. Using gene significance (GS) and module membership (MM), the key trait-module lncRNAs were identified for further analysis. The core lncRNAs of EOS depended on the overlap of differentially expressed lncRNAs (DELs) and key trait-module lncRNAs.
2.4 Identification of differentially expressed autophagy-related mRNAs
By intersecting differentially expressed mRNAs (DEGs) with ARGs from the Human Autophagy Database and the GO_AUTOPHAGY gene set in the Gene Set Enrichment Analysis website, we obtained a group of DEARGs. Subsequently, Gene Ontology (GO) function analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were used to analyze the biological function of DEARGs.
2.5 Identification of differentially expressed autophagy-related lncRNAs
Since the expression data of core lncRNAs and DEARGs did not conform to a normal distribution, we used Spearman correlation to analyze the correlation between them. The lncRNAs with |r| > 0.7 and a p-value <0.05 were defined as differentially expressed autophagy-related lncRNAs. The interaction relationship network was visualized using Cytoscape software (Version 3.9.1).
2.6 Identification of diagnostic-related lncRNAs
We used LASSO Cox regression to screen for autophagy-related lncRNAs with diagnostic value. Then, the predictive accuracy of core lncRNAs was further evaluated using ROC curve analysis.
2.7 Construction of the lncRNA-miRNA-mRNA ceRNA network
The lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP) was utilized to predict the lncRNA-miRNA interaction, while the TarBase v8.0 database (https://microrna.gr/tarbase) was used to predict the targeting miRNAs of DEARGs. The lncRNAs, miRNAs, and mRNAs were visualized as a network using the Sangerbox tools.
2.8 Analysis of protein-protein interactions and identification of hub genes
A PPI (Protein-Protein Interaction Networks) network of DEARGs included in the ceRNA network was built using the Search Tool for the Retrieval of Interacting Genes (STRING) database via the Network Analyst v3.0 (https://www.networkanalyst.ca/) web tool, with a confidence score cut-off set at 900. We used degree ≥8 as the screening parameter to discover hub genes. Subsequently, principal component analysis (PCA) was performed to differentiate EOS patients from HC based on the level of hub genes.
2.9 Immune infiltration analysis
The xCell algorithm was used to estimate the infiltration abundance of 64 immune and stroma cells in EOS patients and HC. In addition, Pearson correlation analysis was performed to analyze the correlation between hub genes and significantly infiltrated immune cells.
3 Results
3.1 Identification of DELs and DEGs between EOS and HC
We initially employed limma differential analysis to screen for DELs and DEGs. A total of 876 DELs and 1929 DEGs were identified in patients with EOS compared to HC. Among these, 461 lncRNAs were upregulated, and 415 were downregulated, while 1033 mRNAs were upregulated, and 896 were downregulated. The volcano plots and heat maps for the DELs and DEGs are presented in Figures 2a-d.
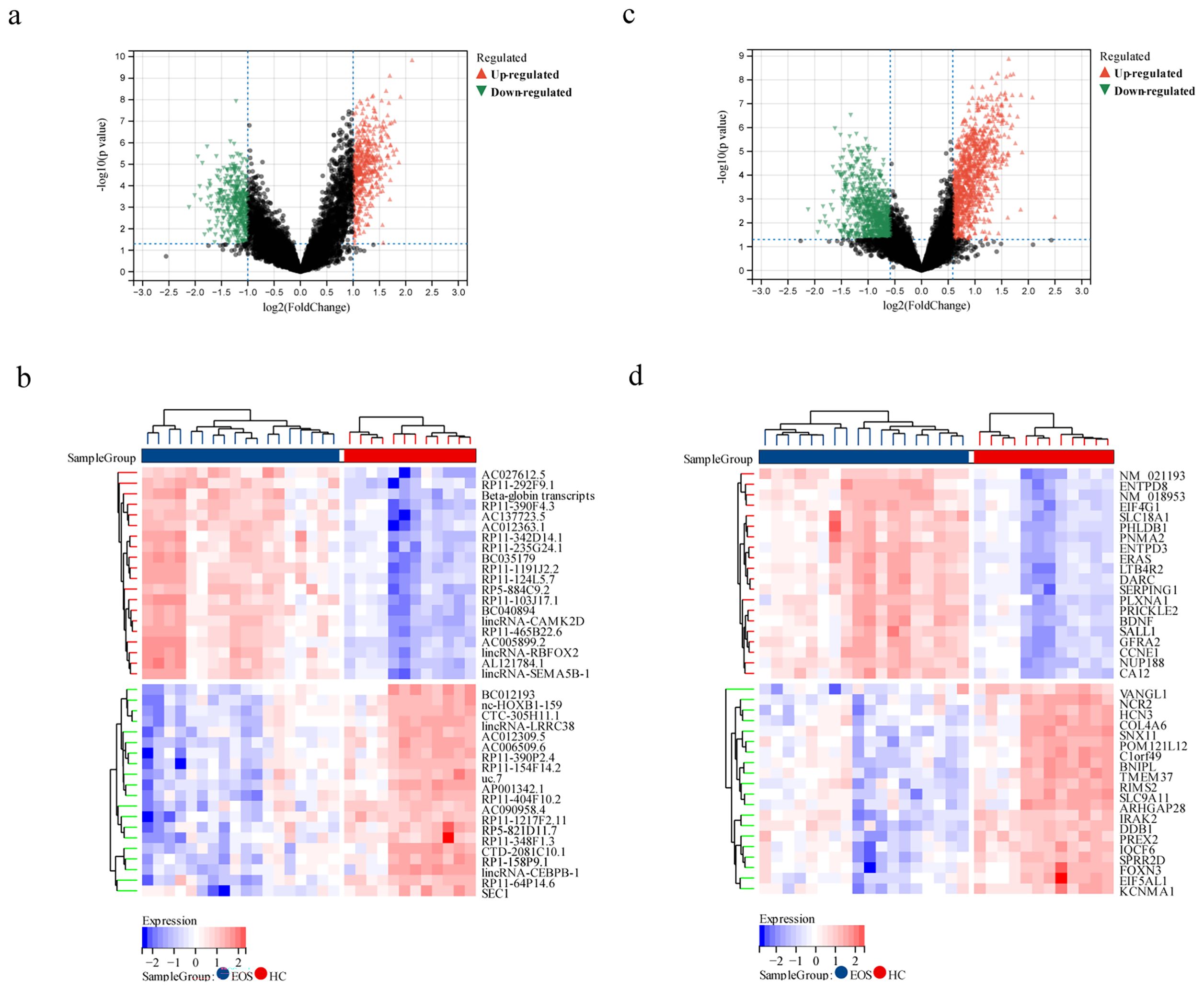
Figure 2. Identification of DELs and DEGs between EOS patients and HC. (a, b) Volcano map and heat map depicting DELs between EOS and HC. (c, d) Volcano map and heat map illustrating DEGs between EOS and HC.
3.2 Identification of key trait-module lncRNAs
Then, we employed WGCNA to identify characteristic genes within co-expression modules in the lncRNA expression profile. We set the soft threshold power β at 14 to construct the scale-free topology module (Figure 3a). A total of 9 modules were identified, and the number of lncRNAs in each module was more than 100. The cluster dendrogram of module eigengenes is shown in Figure 3b. The module–trait relationship represented a correlation between modules and clinical traits (Figure 3c). The gray module included lncRNAs that did not belong to any module, and the blue module, which has the strongest positive correlation with EOS, was selected as the clinically significant module for further analysis. A scatter plot of MM and GS is shown in Figure 3d. Using MM>0.8 and GS>0.1 as thresholds, 598 lncRNAs in the blue module were screened as key trait-module lncRNAs. Finally, 365 core lncRNAs of EOS were selected from the union set between DELs and key trait-module lncRNAs, which was visualized in the Venn diagram (Figure 3e).
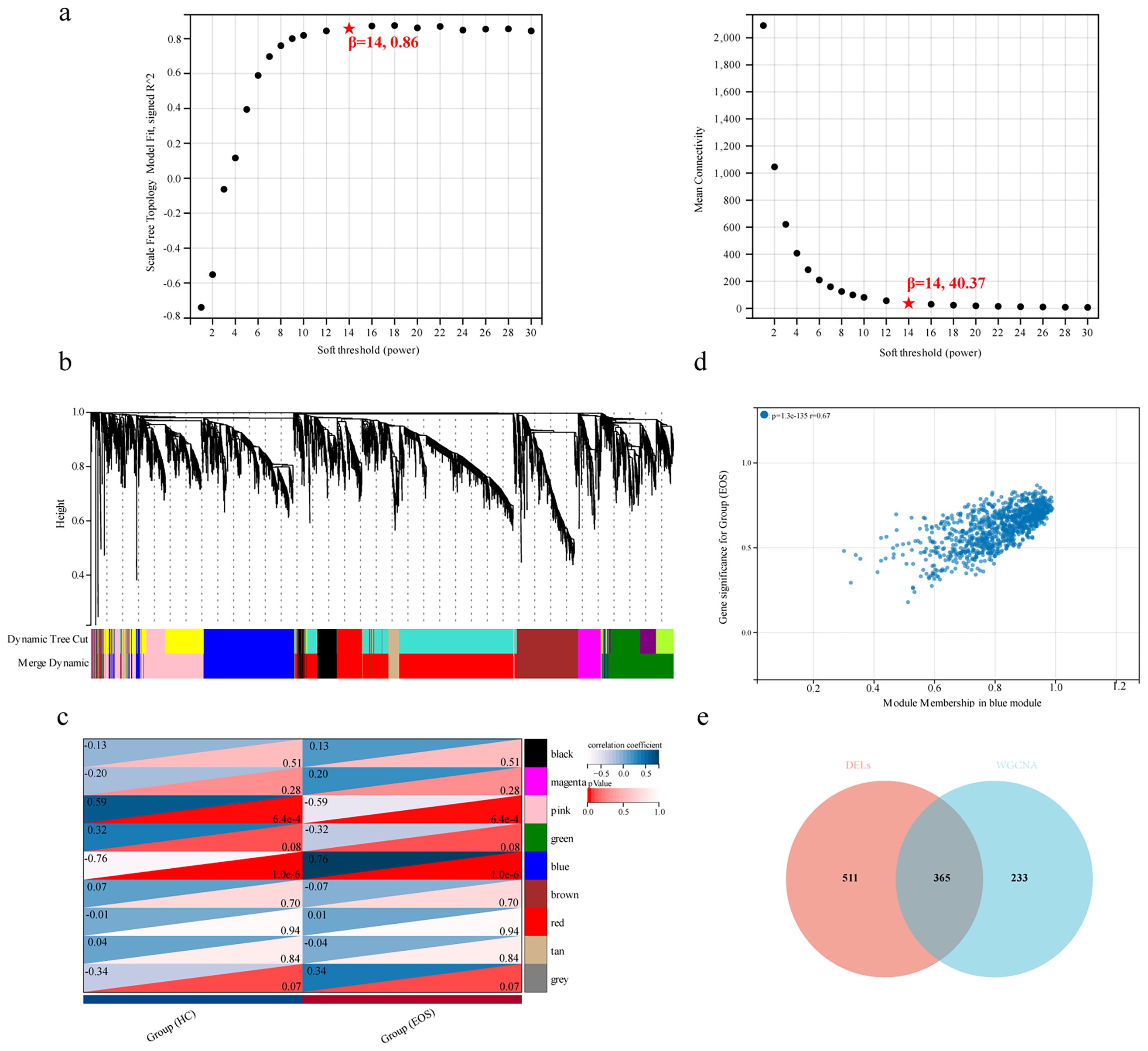
Figure 3. Identification of key trait-module lncRNAs. (a) Scale-free index (left panel) and mean connectivity (right panel) of different soft thresholding powers. (b) Cluster dendrogram of module eigengenes based on the dissimilarity of topological overlap measure. (c) Heatmap showing the correlation between modules and clinical traits. (d) Scatter plot of MM and GS for the group (EOS) in the blue module. (e) Venn diagram of DELs and key trait-module lncRNAs.
3.3 Identification of differentially expressed autophagy-related mRNAs
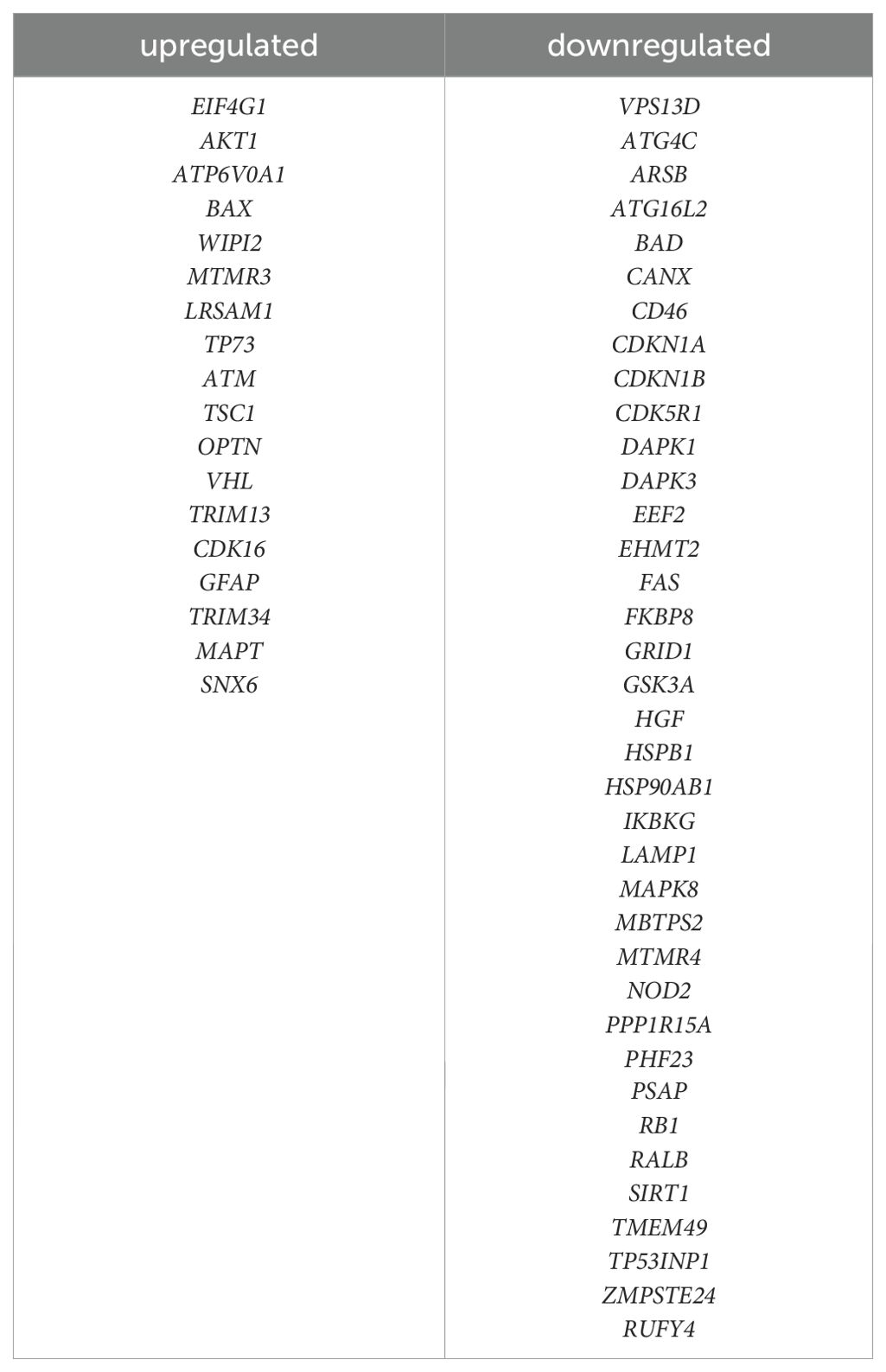
To identify DEGs involved in autophagy, we obtained a list of 518 ARGs from the Human Autophagy Database (http://autophagy.lu/clustering/index.html) and the GO_AUTOPHAGY gene set in the Gene Set Enrichment Analysis website (http://software.broadinstitute.org/gsea/index.jsp). The Venn diagram between ARGs and DEGs was analyzed, and 55 overlapping genes were defined as DEARGs in EOS, including 18 upregulated mRNAs and 37 downregulated mRNAs (Figure 4a, Table 1). Next, we performed GO enrichment analysis and KEGG pathway analysis of DEARGs. DEARGs were primarily enriched in biological processes such as autophagy and macro-autophagy, as well as cell components such as the autophagosome and inclusion body (Figure 4b). They were also enriched in molecular functions such as protein phosphatase binding and heat shock protein binding. Furthermore, these DEARGs were mainly enriched in biological pathways such as autophagy-animal (Figure 4c).
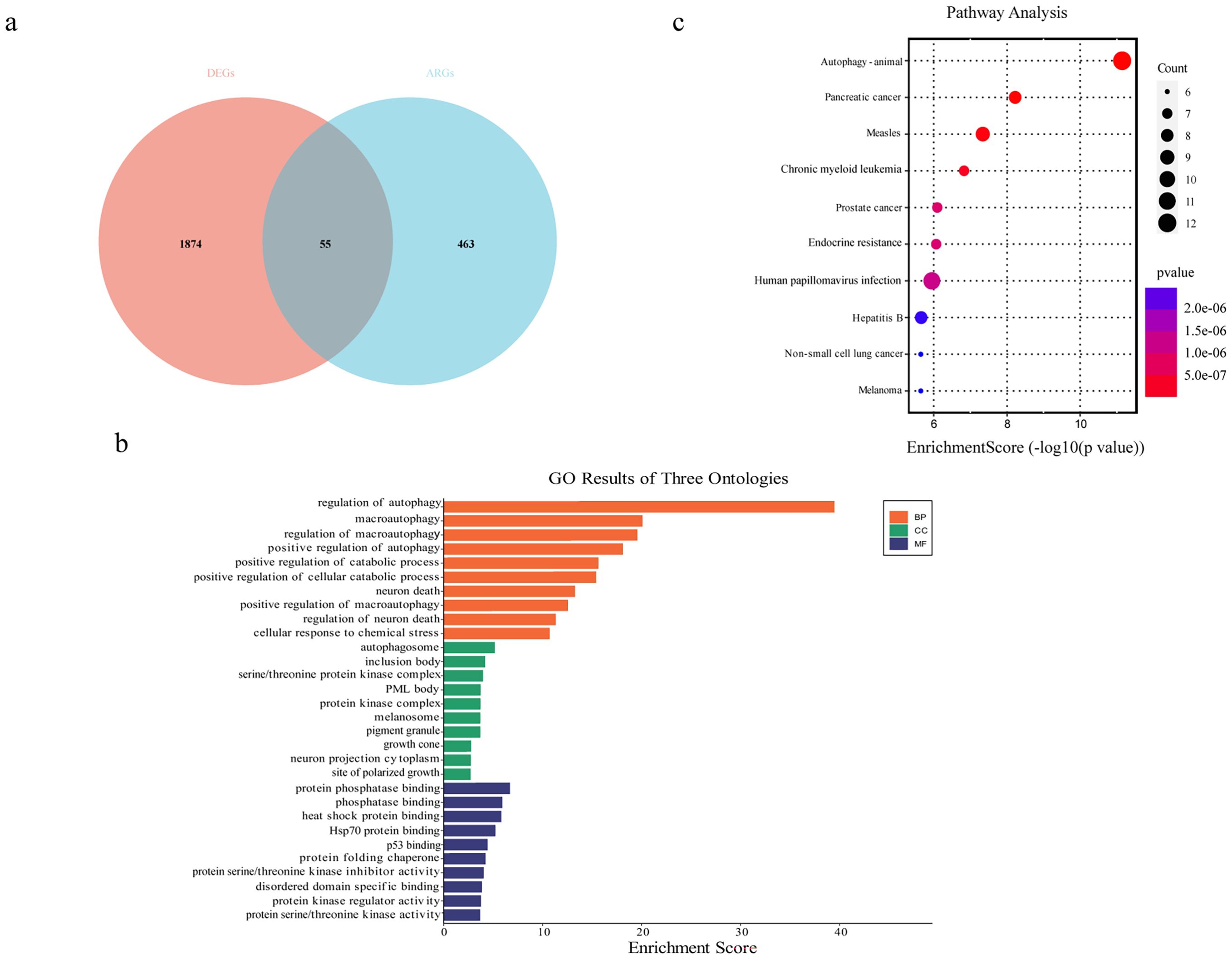
Figure 4. Identification of differentially expressed autophagy-related mRNAs. (a) Venn diagram of DEGs and autophagy-related genes. (b) GO enrichment analysis of DEARGs. (c) KEGG enrichment analysis of DEARGs.
3.4 The co-expression network indicated the relationship between diagnostic autophagy-related lncRNAs and DEARGs
To identify lncRNAs associated with autophagy, a Spearman correlation analysis (with |r| > 0.7 and a p-value < 0.05) between core lncRNAs and DEARGs was performed, resulting in 365 differentially expressed autophagy-related lncRNAs. These 365 lncRNAs were then utilized for identifying candidate diagnostic autophagy-related lncRNAs by using LASSO Cox regression. We set the Lambda value to 0.110293666617702 and five lncRNAs (BC040894, lincRNA-CAMK2D, RP5-884C9.2, RP1-135L22.1 and RP11-390F4.3) were identified as potential diagnostic factors (Figures 5a, b). The area under the ROC curve was separately for BC040894 = 0.995, lincRNA-CAMK2D = 1.000, RP5-884C9.2 = 0.981, RP1-135L22.1 = 1.000, RP11-390F4.3 = 0.981 (Figure 5c). A co-expression network between 5 diagnostic autophagy-related lncRNAs and 49 DEARGs was built using Cytoscape software (Figure 5d).
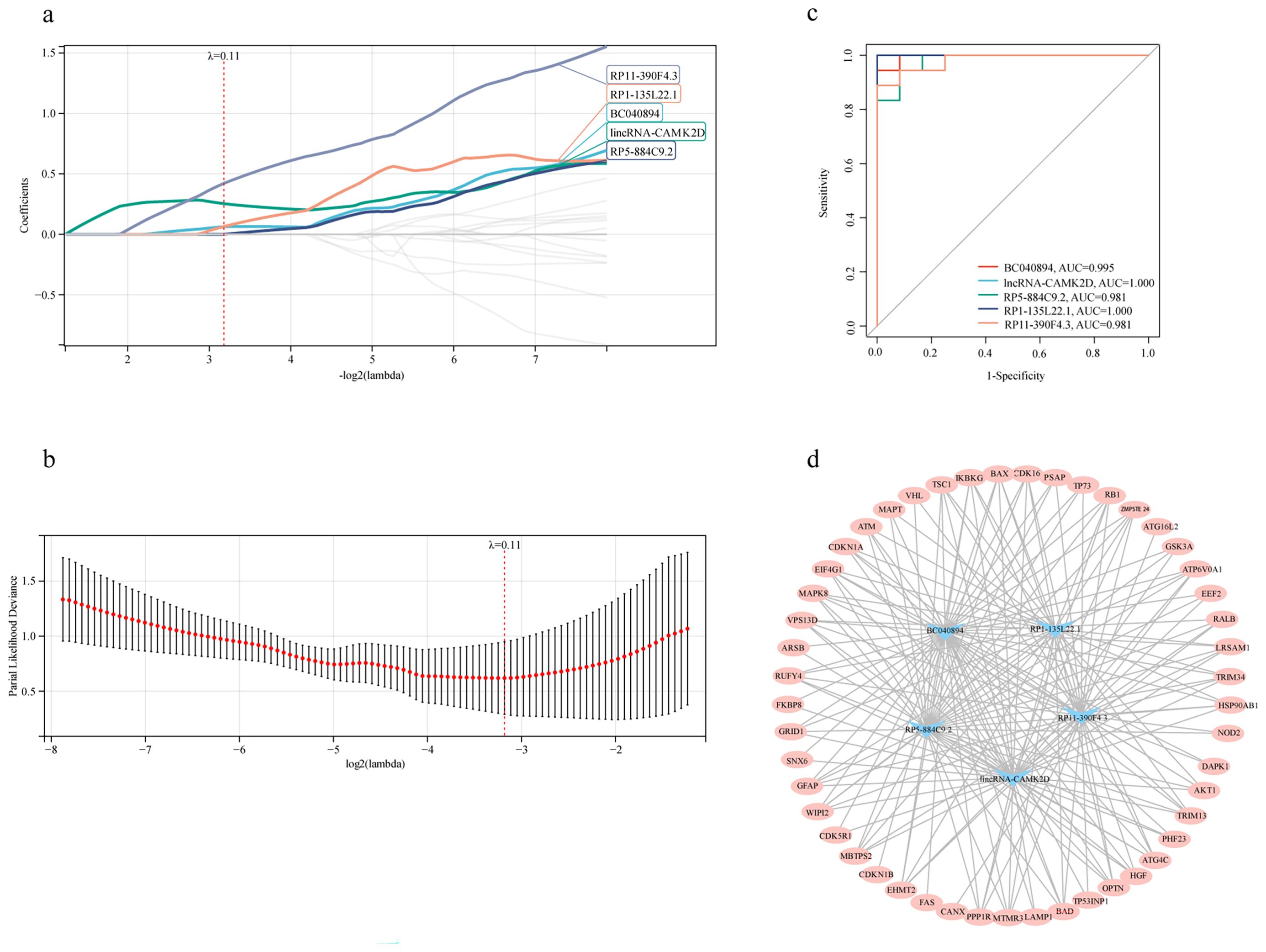
Figure 5. The co-expression network indicated the relationship between diagnostic autophagy-related lncRNAs and DEARGs. (a, b) LASSO Cox regression analysis for the diagnostic autophagy-related lncRNAs. (c) ROC curves of five diagnostic autophagy-related lncRNAs. (d) The co-expression network between diagnostic autophagy-related lncRNAs and DEARGs.
3.5 Construction of a diagnostic lncRNA-mediated ceRNA network for EOS
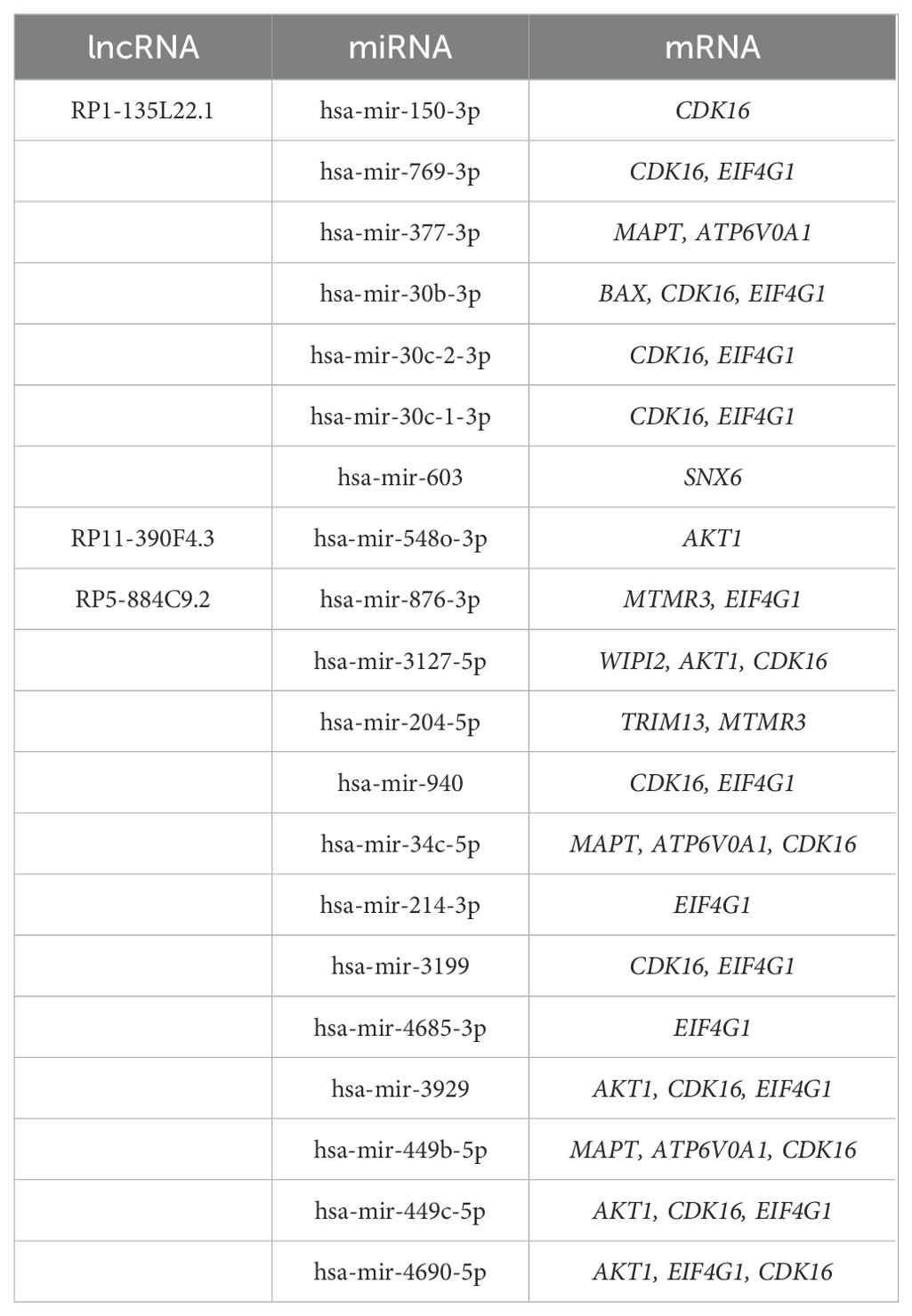
We constructed an lncRNA-mediated ceRNA network to explore the regulatory mechanism between lncRNAs and mRNAs. Interestingly, the expression of all five diagnostic autophagy-related lncRNAs was significantly upregulated in EOS (Figure 6a). Based on the assumption of ceRNA, the expression of mRNAs should be positively correlated with lncRNAs. We selected 18 DEARGs upregulated in EOS (EIF4G1, AKT1, ATP6V0A1, BAX, WIPI2, MTMR3, LRSAM1, TP73, ATM, TSC1, OPTN, VHL, TRIM13, CDK16, GFAP, TRIM34, MAPT, and SNX6) (Table 2) for the construction of a ceRNA network. As described in the Methods section, the lncRNASNP2 database was used to screen the interaction between 4 diagnostic autophagy-related lncRNAs and 152 predicted miRNAs, and the TarBase database was used to identify 436 predicted miRNAs of 17 DEARGs. Finally, the miRNA-mRNA pairs and the lncRNA-miRNA pairs were integrated to construct a diagnostic lncRNA-miRNA-mRNA ceRNA network for EOS which was composed of three diagnostic autophagy-related lncRNAs (RP5-884C9.2, RP1-135L22.1, and RP11-390F4.3), twenty predicted miRNAs, and ten DEARGs (AKT1, EIF4G1, MAPT, WIPI2, BAX, CDK16, ATP6V0A1, SNX6, MTMR3, and TRIM13) (Figure 6b, Table 3).
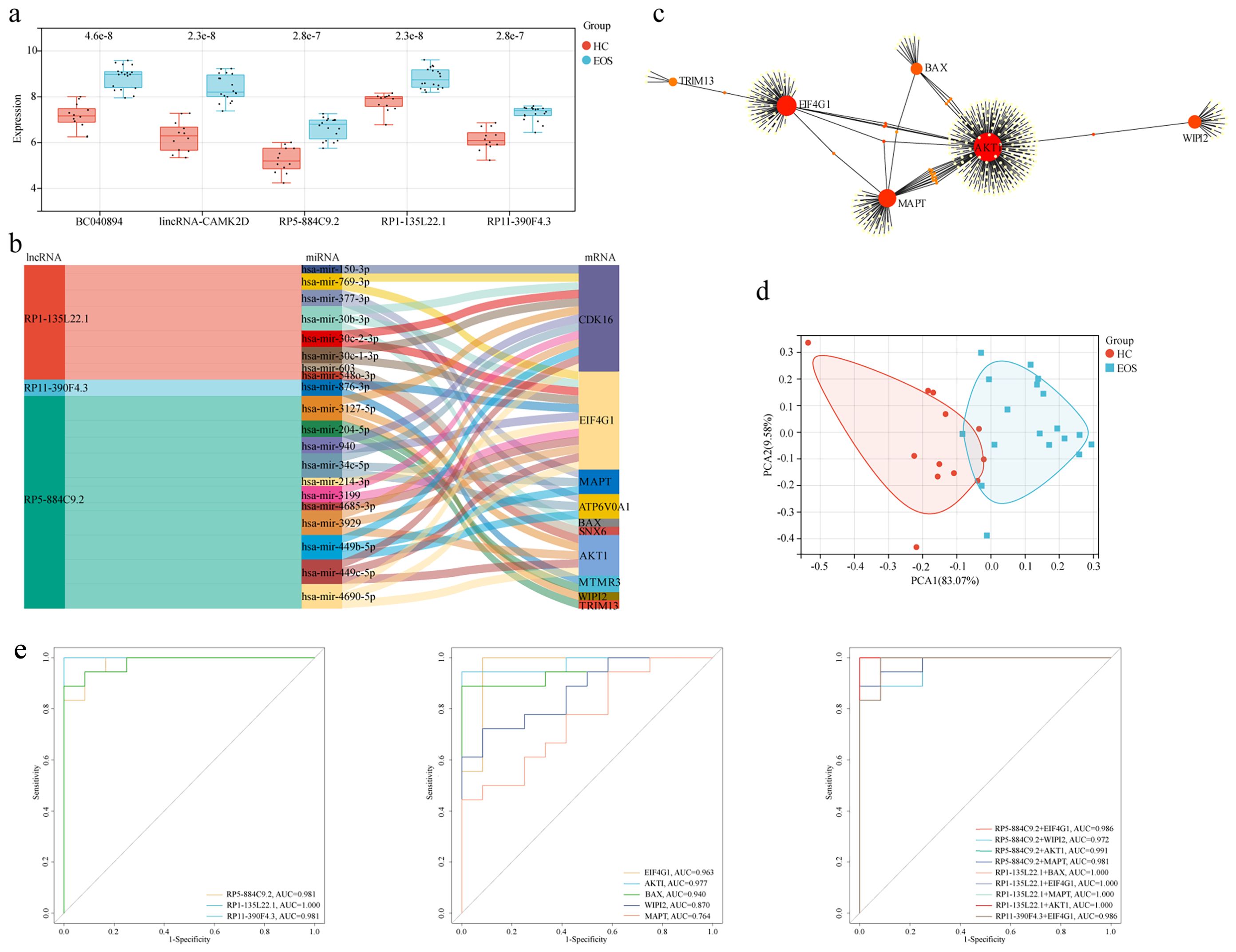
Figure 6. Construction of a diagnostic lncRNA-mediated ceRNA network and identification of hub mRNAs. (a) The box diagram showed the expression level differences of diagnostic autophagy-related lncRNAs between EOS and HC. (b) The Sankey diagram of the ceRNA network included three diagnostic autophagy-related lncRNAs, twenty predicted miRNAs, and ten DEARGs. (c) PPI network of DEARGs in the ceRNA network. (d) PCA of EOS and HC samples. (e) ROC curves of integrated lncRNAs-mRNAs, lncRNAs alone, and mRNAs alone.
3.6 Identification of hub mRNAs of the ceRNA network
A PPI network of DEARGs included in the ceRNA network was constructed using the STRING database (Figure 6c). We used degree ≥8 as the screening parameter and screened five hub proteins (including AKT1, EIF4G1, MAPT, WIPI2, and BAX). The PCA analysis indicated that these hub mRNAs can distinguish EOS patients from HC significantly (Figure 6d). The area under the ROC curve was separately for EIF4G1 = 0.963, AKT1 = 0.977, BAX = 0.940, WIPI2 = 0.870, MAPT = 0.764 (Figure 6e). Furthermore, ROC curve analysis demonstrated that integrating three diagnostic lncRNAs along with five hub mRNAs appeared to yield better diagnostic accuracy compared to using either lncRNAs or mRNAs alone.
3.7 Immune cell infiltration in EOS
We evaluated the differences in immune cell infiltration between EOS and HC by the xCell algorithm. As shown in Figure 7a, significant differences were demonstrated in the infiltration of 15 immune cell types such as CD4(+) memory T cells, CD4(+) naive T cells, and regulatory T cells (p < 0.05). It indicated that the infiltration of these immune cells may be closely related to the disease state. Through performing a correlation analysis between immune cell infiltration and lncRNAs/mRNAs expression levels, we found that all three diagnostic lncRNAs and five hub mRNAs were positively correlated with at least two types of immune infiltration (Figure 7b). This indicated that these autophagy-related ceRNAs may provide valuable information for guiding immunotherapy.
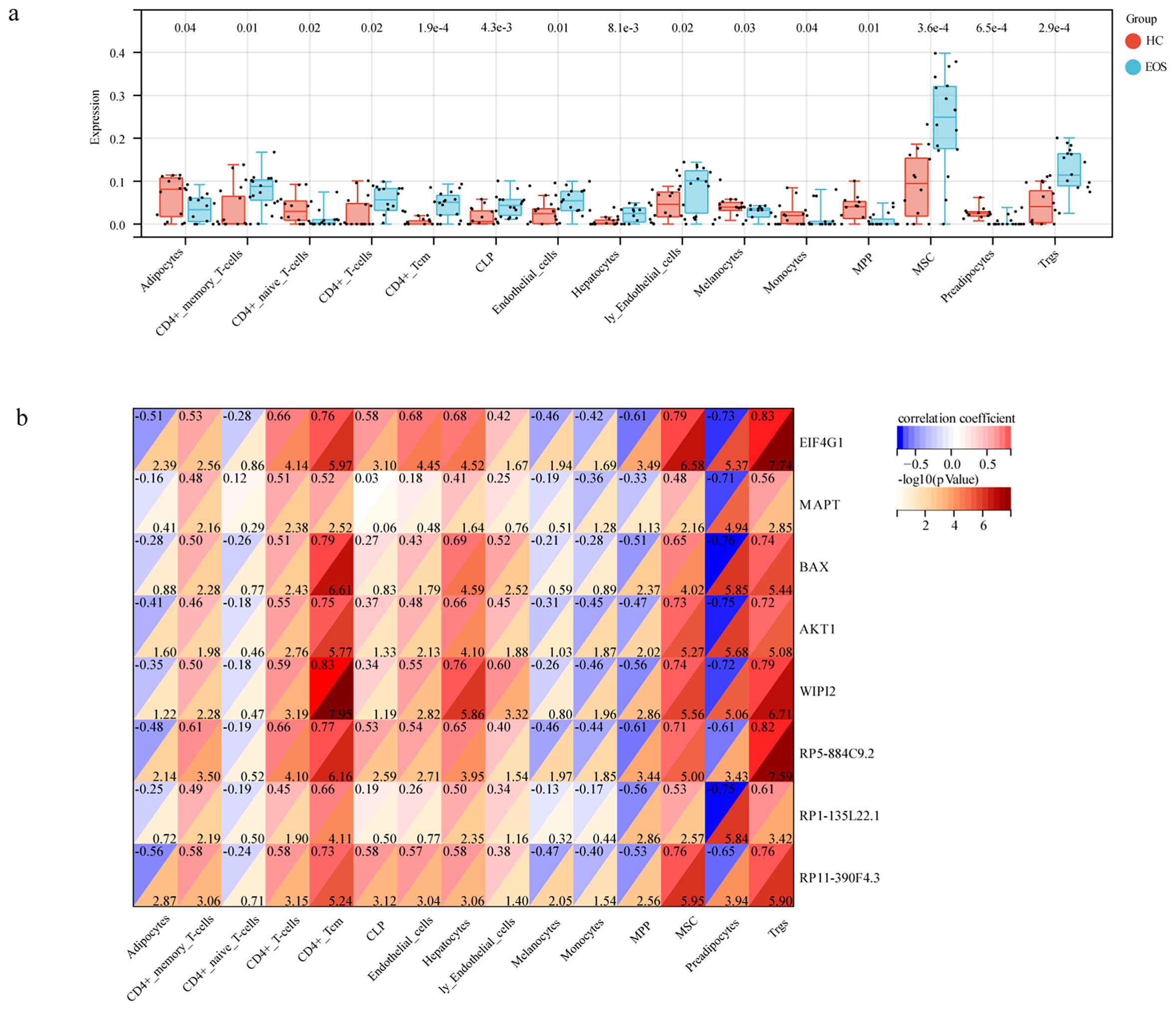
Figure 7. Immune cell infiltration in EOS. (a) The box diagram showed the differences in immune cell infiltrates between EOS and HC. (b) Correlation analysis between hub genes and immune cells.
3.8 Conclusion
In conclusion, through transcriptome analysis, we systematically explored the autophagy-related ceRNA network associated with diagnosis and constructed a network related to the occurrence of EOS, which includes 3 lncRNAs, 20 miRNAs, and 10 mRNAs. Through PPI network analysis, we ultimately identified 5 key mRNAs that demonstrated good predictive ability in distinguishing EOS patients from HC. The combined use of 3 diagnostic lncRNAs and 5 key mRNAs appears to enhance diagnostic efficacy. Furthermore, all 3 diagnostic lncRNAs and 5 key mRNAs showed a positive correlation with at least two types of immune infiltration. We identified an autophagy-related ceRNA network for the early diagnosis of EOS, where the lncRNA-miRNA-mRNA network plays a role in the regulation of cellular autophagy, which may contribute to a deeper understanding of the pathogenesis of EOS.
4 Discussion
SCZ has a poor clinical course and outcome, partly due to a lack of effective early diagnosis. Unlike most diagnosable diseases, the diagnosis of SCZ is still based on clinical symptoms, which are subjective and variable, and may lead to delayed diagnosis or misdiagnosis (15). However, the prognosis of EOS which begins in childhood or adolescence is even worse. Therefore, searching for sensitive and specific biomarkers is meaningful for the early diagnosis and treatment of EOS. Increasing evidence has indicated that impaired autophagy is involved in the pathophysiology of psychiatric disorders such as SCZ (16, 17). We speculated that lncRNAs/mRNAs involved in the autophagy process may be potential diagnostic biomarkers or therapeutic targets for EOS.
In the study, a total of 365 core lncRNAs and 55 DEARGs were identified in EOS patients compared with HC. By implementing co-expression analysis and Lasso Cox regression, we identified five diagnostic autophagy-related lncRNAs (BC040894, lincRNA-CAMK2D, RP5-884C9.2, RP1-135L22.1, and RP11-390F4.3) and used ROC curve analysis to evaluate the predictive accuracy. By predicting the targeted miRNAs of autophagy-related lncRNAs and upregulated DEARGs, we constructed an autophagy-related lncRNA-miRNA-mRNA ceRNA network for the diagnosis of EOS which was composed of three diagnostic autophagy-related lncRNAs (RP1-135L22.1, RP5-884C9.2, and RP11-390F4.3), twenty predicted miRNAs, and ten DEARGs.
The functioning of lncRNAs as competitive endogenous RNAs mainly occurs in the cytoplasm (18). We used the lncRNA subcellular localization predictor (lncLocator) to understand the subcellular localization of these three lncRNAs. All of the lncRNAs were predicted to reside in the cytoplasm, including RP1-135L22.1 (53.82%), RP5-884C9.2 (35.68%), and RP11-390F4.3 (42.67%), suggesting that they were involved in the ceRNA regulatory network in the cytoplasm. Among them, two lncRNAs RP5-884C9.2 and RP1-135L22.1 have never been reported before. It was reported that RP11-390F4.3 is induced by hypoxia/HIF-1α and is involved in epithelial-mesenchymal transition and tumor metastasis (19). These three autophagy-related lncRNAs may be new biomarkers for EOS that have to be tested by further experiments.
Recent research findings support the important role of ARGs and pathways in SCZ. The research conducted by Lappas et al. found abnormal expression of ARGs in the hippocampus of patients with chronic SCZ, indicating that the autophagy pathway plays a crucial role in the pathological processes associated with SCZ (20). Bjornson et al. (21) emphasize the important role of ARGs’ activity in patients with SCZ (21). We further discovered five hub mRNAs of the ceRNA network (EIF4G1, AKT1, MAPT, WIPI2, and BAX) with good predictive accuracy of EOS, and the PCA analysis showed that these hub mRNAs can distinguish EOS patients from HC significantly. Eukaryotic Translation Initiation Factor 4 Gamma 1 (EIF4G1) is involved in the control process of translation which affects cellular survival and cellular death. The high levels of EIF4G1 may increase cell proliferation and prevent autophagy (22). Some studies implied that EIF4G1 may be related to EOS. Proteomic analysis revealed that EIF4G1 was a potential target of Cullin 3 (CUL3) deficiency, a risk factor for autism spectrum disorder (ASD) and SCZ that caused cellular and behavioral defects. The level of EIF4G1 was increased in CUL3-deficient brains (23). A study exploring peripheral blood miRNA biomarkers in patients with first-episode schizophrenia (FES) found that miR-4467 may be a non-invasive biomarker and EIF4G1 was a hub target gene of miR-4467 (24). The research conducted by Tan et al. (25) underscores the critical role of ARGs in the onset and advancement of SCZ (25). Our results align with the conclusions drawn by Tan et al., particularly concerning the role of EIF4G1, which we similarly identified as a central mRNA within the ceRNA network. Tan et al. illustrated the substantial significance of EIF4G1 in the context of SCZ, thereby corroborating our findings that underscore its potential as a diagnostic biomarker. Serine/Threonine Kinase 1 (AKT1) plays an indispensable role in autophagy. The physiological AKT-MTORC1 signaling pathway is considered a key node in the regulation of macroautophagy/autophagy. Under nutrient-rich conditions, AKT can negatively regulate autophagy by activating the mechanistic target of rapamycin complex 1 (mTORC1), inhibiting the activity of the transcription factor forkhead box class O (FoxO), and inhibiting the expression of autophagy genes (26, 27). In current studies, the changing trend of AKT1 expression in SCZ patients was inconsistent. A study found that the expression levels of AKT1 significantly increased in acute schizophrenia patients (28), which is consistent with our result. Another case-control study showed significantly decreased expression of AKT1 in male recent-onset SCZ patients (29). The results of this study were contrary to ours, possibly because the subjects were ethnically different, coming from the Netherlands and China respectively. BCL2 Associated X (BAX) is a pro-apoptotic gene that plays an indispensable role in regulating the mitochondrial apoptosis pathway. Recent studies have revealed cross-talk between the apoptotic and autophagic pathways, and many genes can play a role in both responses. BAX was identified as an important gene with the ability to induce mitochondrial autophagy in the process of apoptosis (30, 31). Evidence has indicated that the expression level of the BAX gene is upregulated in SCZ rats, which was consistent with our result (32). The upregulated BAX may be involved in apoptosis and autophagy processes in EOS. WD Repeat Domain, Phosphoinositide Interacting 2 (WIPI2), the mammalian homolog of the yeast ATG18 gene, plays a critical role in autophagosome biogenesis (33). It was reported that WIPI2 promoted mature autophagosome biogenesis, and when depleted, cells fail to form autophagosomes and engulfment of pathogenic bacteria (34). WIPI2 has never been reported in SCZ before. The study found that the mutation of the WIPI2 gene was associated with other neurodevelopmental diseases, such as global developmental abnormalities. Microtubule-associated protein tau (MAPT) encodes tau protein, the main component of neurofibrillary tangles (NFTs). MAPT protein aggregation inhibits the fusion of autophagosome and lysosome, resulting in autophagy disorders (35). MAPT has been reported to be associated with the onset of neuropsychiatric disorders, such as SCZ, Alzheimer’s disease, and Parkinson’s disease (36). Patients with SCZ have genetic variants, differential expression, and abnormal methylation in the MAPT gene that are linked with the disease risk (37, 38). A study has suggested the existence of tau-related neurodevelopmental disorders in adolescent psychosis (39). Therefore, these five hub genes may be effective biomarkers for the diagnosis of EOS.
Autophagy plays a critical role in the development, maintenance, and survival of various immune cell types, such as macrophages, neutrophils, T lymphocytes, and B lymphocytes (40). Dysregulation of autophagy can lead to impaired immune responses and contribute to the development of various diseases, including psychiatric disorders such as SCZ (41). In our study, immune cell infiltration analysis revealed significant differences in the infiltration of 15 immune cell types, such as CD4(+) memory T cells, CD4(+) naive T cells, and regulatory T cells between EOS patients and HC. Furthermore, all three diagnostic lncRNAs and five hub mRNAs were positively correlated with at least two types of immune infiltration. EIF4G1 plays a critical role in the regulation of translation, which in turn influences cellular survival and the process of autophagy. Elevated levels of EIF4G1 have the potential to enhance cellular proliferation while inhibiting autophagy, thereby possibly leading to immune dysregulation (42). AKT1 is integral to the regulation of autophagy. The AKT-mTORC1 signaling pathway exerts a negative regulatory effect on autophagy in nutrient-abundant environments, thereby impacting the functionality and viability of immune cells (43). BAX is a pro-apoptotic gene that plays a critical role in the regulation of the mitochondrial apoptosis pathway. BAX is also implicated in mitochondrial autophagy, and its increased expression in patients with EOS may influence both apoptotic and autophagic processes, thereby affecting the functionality of immune cells (44). WIPI2 plays a pivotal role in the biogenesis of autophagosomes, facilitating their maturation, which is essential for cellular responses to immunological challenges (34, 45). MAPT plays a role in the development of neurofibrillary tangles. The aggregation of MAPT proteins disrupts the fusion of autophagosomes and lysosomes, resulting in autophagy dysfunction and possibly affecting immune responses (46). These results highlight the crucial involvement of autophagy in the pathophysiological mechanisms underlying EOS and its possible interactions with the immune system.
However, this study has some limitations, which need further discussion in future work. First, the sample size in this study was relatively small, so the results obtained need further validation in a large sample. Second, the results obtained were only based on bioinformatics analytical methods, so we need to verify the findings through animal experiments and cellular experiments in future research. Finally, in this study, we utilized PBMC to analyze mRNA and lncRNA expression profiles. Although PBMC serve as a convenient and minimally invasive source for biomarker identification, they may not comprehensively represent the molecular alterations occurring within the brain. Therefore, future research should contemplate the incorporation of brain tissue samples or the application of advanced imaging methodologies to corroborate the findings observed in peripheral analyses. By addressing these limitations in subsequent research, we can enhance our comprehension of the role of autophagy in EOS and facilitate the advancement of precise diagnostic instruments and targeted therapeutic strategies. Notwithstanding these constraints, our study establishes a significant basis for investigating autophagy-related ceRNA networks as prospective biomarkers for the early diagnosis and treatment of EOS.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE54913.
Ethics statement
The studies involving humans were approved by Medical Research Ethics Committee of Shanxi Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
WH: Writing – original draft. XD: Writing – original draft. XW: Writing – review & editing. KZ: Writing – review & editing. JL: Methodology, Writing – review & editing. YG: Methodology, Writing – review & editing. TA: Methodology, Writing – review & editing. HZ: Methodology, Writing – review & editing. YZ: Methodology, Writing – review & editing. ZR: Conceptualization, Writing – review & editing. YX: Conceptualization, Writing – review & editing. SL: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (82271546, 82371511 and 81701326); Special fund for Science and Technology Innovation Teams of Shanxi Province (202304051001049); Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (20240041); Research project supported by Shanxi Scholarship Council of China (2022-190); Shanxi Province Higher Education “Billion Project” Science and Technology Guidance Project (BYJL062); Shanxi Provincial Nature Foundation (202403021211162); Taiyuan Science and Technology Fund (Y2024008); National Natural Science Foundation of China (82371511); National Key Research and Development Program of China (2023YFC2506201); Shenzhen Natural Science Foundation Project (JCYJ20240813150616022); Shenzhen Futian District Health System Research Project (FTWS042); Clinical Research and Cultivation Project of the Eighth Affiliated Hospital of Sun Yat-sen University (PY-2024-001).
Acknowledgments
The authors would like to express their gratitude to all participants for their contributions to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Campeau A, Mills RH, Stevens T, Rossitto LA, Meehan M, Dorrestein P, et al. Multi-omics of human plasma reveals molecular features of dysregulated inflammation and accelerated aging in schizophrenia. Mol Psychiatry. (2022) 27:1217–25. doi: 10.1038/s41380-021-01339-z
2. Pillinger T, D’Ambrosio E, McCutcheon R, Howes OD. Is psychosis a multisystem disorder? A meta-review of central nervous system, immune, cardiometabolic, and endocrine alterations in first-episode psychosis and perspective on potential models. Mol Psychiatry. (2019) 24:776–94. doi: 10.1038/s41380-018-0058-9
3. Kumra S, Charles Schulz S. Editorial: research progress in early-onset schizophrenia. Schizophr Bull. (2008) 34:15–7. doi: 10.1093/schbul/sbm123
4. Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E, et al. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry. (2015) 20:126–32. doi: 10.1038/mp.2013.174
5. Horesh Y, Katsel P, Haroutunian V, Domany E. Gene expression signature is shared by patients with alzheimer’s disease and schizophrenia at the superior temporal gyrus. Eur J Neurol. (2011) 18:410–24. doi: 10.1111/j.1468-1331.2010.03166.x
6. Polajnar M, Zerovnik E. Impaired autophagy: A link between neurodegenerative and neuropsychiatric diseases. J Cell Mol Med. (2014) 18:1705–11. doi: 10.1111/jcmm.12349
7. Bridges MC, Daulagala AC, Kourtidis A. Lnccation: lncrna localization and function. J Cell Biol. (2021) 220(2):e202009045. doi: 10.1083/jcb.202009045
8. Mercer TR, Dinger ME, Mattick JS. Long non-coding rnas: insights into functions. Nat Rev Genet. (2009) 10:155–9. doi: 10.1038/nrg2521
9. Yang L, Froberg JE, Lee JT. Long noncoding rnas: fresh perspectives into the rna world. Trends Biochem Sci. (2014) 39:35–43. doi: 10.1016/j.tibs.2013.10.002
10. Mukhopadhyay A, Deshpande SN, Bhatia T, Thelma BK. Significance of an altered lncrna landscape in schizophrenia and cognition: clues from a case-control association study. Eur Arch Psychiatry Clin Neurosci. (2023) 273(8):1677–91. doi: 10.1007/s00406-023-01596-9
11. Jovčevska I, Videtič Paska A. Neuroepigenetics of psychiatric disorders: focus on lncrna. Neurochemistry Int. (2021) 149:105140. doi: 10.1016/j.neuint.2021.105140
12. Karreth FA, Pandolfi PP. Cerna cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discovery. (2013) 3:1113–21. doi: 10.1158/2159-8290.Cd-13-0202
13. Li R, Wang Q, Qiu Y, Meng Y, Wei L, Wang H, et al. A potential autophagy-related competing endogenous rna network and corresponding diagnostic efficacy in schizophrenia. Front Psychiatry. (2021) 12:628361. doi: 10.3389/fpsyt.2021.628361
14. Sun L, Cheng Z, Zhang F, Xu Y. Gene expression profiling in peripheral blood mononuclear cells of early-onset schizophrenia. Genomics Data. (2015) 5:169–70. doi: 10.1016/j.gdata.2015.04.022
15. Modai S, Shomron N. Molecular risk factors for schizophrenia. Trends Mol Med. (2016) 22:242–53. doi: 10.1016/j.molmed.2016.01.006
16. Bar-Yosef T, Damri O, Agam G. Dual role of autophagy in diseases of the central nervous system. Front Cell Neurosci. (2019) 13:196. doi: 10.3389/fncel.2019.00196
17. Merenlender-Wagner A, Shemer Z, Touloumi O, Lagoudaki R, Giladi E, Andrieux A, et al. New horizons in schizophrenia treatment: autophagy protection is coupled with behavioral improvements in a mouse model of schizophrenia. Autophagy. (2014) 10:2324–32. doi: 10.4161/15548627.2014.984274
18. Bai Y, Long J, Liu Z, Lin J, Huang H, Wang D, et al. Comprehensive analysis of a cerna network reveals potential prognostic cytoplasmic lncrnas involved in hcc progression. J Cell Physiol. (2019) 234:18837–48. doi: 10.1002/jcp.28522
19. Peng PH, Chieh-Yu-Lai J, Hsu KW, Wu KJ. Hypoxia-induced lncrna rp11-390f4.3 promotes epithelial-mesenchymal transition (Emt) and metastasis through upregulating emt regulators. Cancer Lett. (2020) 483:35–45. doi: 10.1016/j.canlet.2020.04.014
20. Lappas AS, Ioannou M, Christodoulou NG. Histopathological evidence of cellular alterations in the dentate gyrus is associated with aberrant rb1cc1-atg16l1 expression in the hippocampus among older adults with chronic schizophrenia: A pilot post-mortem study. Schizophr Res. (2025) 275:14–24. doi: 10.1016/j.schres.2024.11.009
21. Bjornson KJ, Vanderplow AM, Bhasker AI, Cahill ME. Increased regional activity of a pro-autophagy pathway in schizophrenia as a contributor to sex differences in the disease pathology. Cell Rep Med. (2024) 5:101652. doi: 10.1016/j.xcrm.2024.101652
22. Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. Eif4gi links nutrient sensing by mtor to cell proliferation and inhibition of autophagy. J Cell Biol. (2008) 181:293–307. doi: 10.1083/jcb.200710215
23. Dong Z, Chen W, Chen C, Wang H, Cui W, Tan Z, et al. Cul3 deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron. (2020) 105:475–90.e6. doi: 10.1016/j.neuron.2019.10.035
24. Jin M, Zhu X, Sun Y, Li Z, Li X, Ai L, et al. Identification of peripheral blood mirna biomarkers in first-episode drug-free schizophrenia patients using bioinformatics strategy. Mol Neurobiol. (2022) 59:4730–46. doi: 10.1007/s12035-022-02878-4
25. Tan Y, Zhu J, Hashimoto K. Autophagy-related gene model as a novel risk factor for schizophrenia. Transl Psychiatry. (2024) 14:94. doi: 10.1038/s41398-024-02767-5
26. Wang H, Liu Y, Wang D, Xu Y, Dong R, Yang Y, et al. The upstream pathway of mtor-mediated autophagy in liver diseases. Cells. (2019) 8. doi: 10.3390/cells8121597
27. Cheng Z. The foxo-autophagy axis in health and disease. Trends Endocrinol metabolism: TEM. (2019) 30:658–71. doi: 10.1016/j.tem.2019.07.009
28. Liu L, Luo Y, Zhang G, Jin C, Zhou Z, Cheng Z, et al. The mrna expression of drd2, pi3kcb, and akt1 in the blood of acute schizophrenia patients. Psychiatry Res. (2016) 243:397–402. doi: 10.1016/j.psychres.2016.07.010
29. van Beveren NJ, Buitendijk GH, Swagemakers S, Krab LC, Röder C, de Haan L, et al. Marked reduction of akt1 expression and deregulation of akt1-associated pathways in peripheral blood mononuclear cells of schizophrenia patients. PloS One. (2012) 7:e32618. doi: 10.1371/journal.pone.0032618
30. Lindqvist LM, Frank D, McArthur K, Dite TA, Lazarou M, Oakhill JS, et al. Autophagy induced during apoptosis degrades mitochondria and inhibits type I interferon secretion. Cell Death differentiation. (2018) 25:784–96. doi: 10.1038/s41418-017-0017-z
31. Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. Puma- and bax-induced autophagy contributes to apoptosis. Cell Death differentiation. (2009) 16:1135–45. doi: 10.1038/cdd.2009.28
32. Talukdar PM, Abdul F, Maes M, Binu VS, Venkatasubramanian G, Kutty BM, et al. Maternal Immune Activation Causes Schizophrenia-Like behaviors in the Offspring through Activation of Immune-Inflammatory, Oxidative and Apoptotic Pathways, and Lowered Antioxidant Defenses and Neuroprotection. Mol Neurobiol. (2020) 57:4345–61. doi: 10.1007/s12035-020-02028-8
33. Lu G, Tan HWS, Schmauck-Medina T, Wang L, Chen J, Cho YL, et al. Wipi2 positively regulates mitophagy by promoting mitochondrial recruitment of vcp. Autophagy. (2022) 18:2865–79. doi: 10.1080/15548627.2022.2052461
34. Jelani M, Dooley HC, Gubas A, Mohamoud HSA, Khan MTM, Ali Z, et al. A mutation in the major autophagy gene, wipi2, associated with global developmental abnormalities. Brain. (2019) 142:1242–54. doi: 10.1093/brain/awz075
35. Feng Q, Luo Y, Zhang XN, Yang XF, Hong XY, Sun DS, et al. Mapt/tau accumulation represses autophagy flux by disrupting ist1-regulated escrt-iii complex formation: A vicious cycle in alzheimer neurodegeneration. Autophagy. (2020) 16:641–58. doi: 10.1080/15548627.2019.1633862
36. Wiberg A, Ng M, Al Omran Y, Alfaro-Almagro F, McCarthy P, Marchini J, et al. Handedness, language areas and neuropsychiatric diseases: insights from brain imaging and genetics. Brain: J Neurol. (2019) 142:2938–47. doi: 10.1093/brain/awz257
37. Li C, Pang D, Lin J, Yang T, Shang H. Shared genetic links between frontotemporal dementia and psychiatric disorders. BMC Med. (2022) 20:131. doi: 10.1186/s12916-022-02335-y
38. Jahn K, Heese A, Kebir O, Groh A, Bleich S, Krebs MO, et al. Differential methylation pattern of schizophrenia candidate genes in tetrahydrocannabinol-consuming treatment-resistant schizophrenic patients compared to non-consumer patients and healthy controls. Neuropsychobiology. (2021) 80:36–44. doi: 10.1159/000507670
39. Andreou D, Jørgensen KN, Nerland S, Smelror RE, Wedervang-Resell K, Johannessen CH, et al. Lower plasma total tau in adolescent psychosis: involvement of the orbitofrontal cortex. J Psychiatr Res. (2021) 144:255–61. doi: 10.1016/j.jpsychires.2021.10.031
40. Zuo H, Chen C, Sa Y. Therapeutic potential of autophagy in immunity and inflammation: current and future perspectives. Pharmacol Rep. (2023) 75:499–510. doi: 10.1007/s43440-023-00486-0
41. Toh PP, Luo S, Menzies FM, Raskó T, Wanker EE, Rubinsztein DC. Myc inhibition impairs autophagosome formation. Hum Mol Genet. (2013) 22:5237–48. doi: 10.1093/hmg/ddt381
42. Li F, He Z, Zhang X, Gao D, Xu R, Zhang Z, et al. Usp10 promotes cell proliferation, migration, and invasion in nsclc through deubiquitination and stabilization of eif4g1. Sci Rep. (2024) 14:23685. doi: 10.1038/s41598-024-74490-6
43. Liu D, Wang H, Li J, Sheng S, Wang S, Tian Y. Non-lethal sonodynamic therapy mitigates hypertensive renal fibrosis through the pi3k/akt/mtorc1-autophagy pathway. Sci Rep. (2025) 15:4534. doi: 10.1038/s41598-025-86973-1
44. Mao R, Zong N, Hu Y, Chen Y, Xu Y. Neuronal death mechanisms and therapeutic strategy in ischemic stroke. Neurosci Bull. (2022) 38:1229–47. doi: 10.1007/s12264-022-00859-0
45. Qiao Q, Wang Y, Zhang R, Pang Q. Autophagy related DNA methylation signature predict clinical prognosis and immune microenvironment in low-grade glioma. Transl Cancer Res. (2022) 11:2157–74. doi: 10.21037/tcr-22-310
Keywords: early-onset schizophrenia, autophagy, long non-coding RNAs, endogenous competitive RNAs, diagnosis
Citation: Hu W, Du X, Wang X, Zhang K, Li J, Gao Y, An T, Zhang H, Zhang Y, Ren Z, Xu Y and Liu S (2025) Explore autophagy-related lncRNA-miRNA-mRNA ceRNA networks for diagnosis of early-onset schizophrenia through transcriptome analysis. Front. Psychiatry 16:1567148. doi: 10.3389/fpsyt.2025.1567148
Received: 26 January 2025; Accepted: 10 February 2025;
Published: 26 February 2025.
Edited by:
Tianhong Zhang, Shanghai Jiao Tong University, ChinaReviewed by:
Cunyou Zhao, Southern Medical University, ChinaHao Yu, Jining Medical University, China
Jun Li, Peking University, China
Copyright © 2025 Hu, Du, Wang, Zhang, Li, Gao, An, Zhang, Zhang, Ren, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sha Liu, bGl1c2hhQHN4bXUuZWR1LmNu; Yong Xu, eHV5b25nc211QHZpcC4xNjMuY29t; Zhiyong Ren, cmVuenkyMDExQDE2My5jb20=
†These authors have contributed equally to this work
 Wei Hu1,2†
Wei Hu1,2† Xinzhe Du
Xinzhe Du Yao Gao
Yao Gao Yu Zhang
Yu Zhang Yong Xu
Yong Xu Sha Liu
Sha Liu