
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Psychiatry , 17 May 2024
Sec. Molecular Psychiatry
Volume 15 - 2024 | https://doi.org/10.3389/fpsyt.2024.1390366
Background: Evidence has suggested that microRNAs (miRNAs) may play an important role in the pathogenesis of psychiatric disorders (PDs), but the results remain inconclusive. We aimed to identify specific differentially expressed miRNAs and their overlapping miRNA expression profiles in schizophrenia (SZ), major depression disorder (MDD), and bipolar disorder (BD), the three major PDs.
Methods: The literatures up to September 30, 2023 related to peripheral blood miRNAs and PDs were searched and screened from multiple databases. The differences in miRNA levels between groups were illustrated by the standardized mean difference (SMD) and 95% confidence interval (95% CI).
Results: In total, 30 peripheral blood miRNAs were included in the meta-analysis, including 16 for SZ, 12 for MDD, and 2 for BD, each was reported in more than 3 independent studies. Compared with the control group, miR-181b-5p, miR-34a-5p, miR-195-5p, miR-30e-5p, miR-7-5p, miR-132-3p, miR-212-3p, miR-206, miR-92a-3p and miR-137-3p were upregulated in SZ, while miR-134-5p, miR-107 and miR-99b-5p were downregulated. In MDD, miR-124-3p, miR-132-3p, miR-139-5p, miR-182-5p, miR-221-3p, miR-34a-5p and miR-93-5p were upregulated, while miR-144-5p and miR-135a-5p were downregulated. However, we failed to identify statistically differentially expressed miRNAs in BD. Interestingly, miR-132-3p and miR-34a-5p were upregulated in both SZ and MDD.
Conclusions: Our study identified 13 differentially expressed miRNAs in SZ and 9 in MDD, among which miR-132-3p and miR-34a-5p were upregulated in both SZ and MDD by systematically analyzing qualified studies. These miRNAs may be used as potential biomarkers for the diagnosis of SZ and MDD in the future.
Systematic Review Registration: http://www.crd.york.ac.uk/PROSPERO, identifier CRD42023486982.
Psychiatric disorders (PDs) are debilitating disease with unknown etiology and pathogenesis, characterized by the dysfunction of complex emotional and cognitive processes (1). Many patients with PDs require long-term treatment to maintain social function and prevent symptom relapse, causing heavy public health and economic burden (2). Schizophrenia (SZ), major depression disorder (MDD) and bipolar disorder (BD) are the three major PDs with high disability and lethality (3). SZ is the most severe PDs characterized by hallucinations, delusions, disturbed emotions, and social withdrawal, with a lifetime prevalence of approximately 1% worldwide (4). MDD is characterized by depressed mood and anhedonia, with a lifetime prevalence of 2-21% worldwide (5). BD is characterized by recurrent episodes of mania and depression, as well as impairments in cognitive performance, which occurs with a lifetime prevalence of 1-2% (6). Currently, the diagnosis of PDs mainly relies on patient’s statements and doctor’s subjective judgment of clinical symptoms, rather than on pathological and physiological indicators, and many PDs have overlapping symptoms, resulting in high rates of misdiagnosis and missed diagnosis. Thus, there is an urgent need to seek objective, effective, convenient and feasible early molecular diagnostic biomarkers for PDs.
Both genetic and environmental factors are thought to contribute to PDs (7). Epigenetic mechanisms, which combine genetic and environmental factors by translating the environmental information into a genetic code, have been reported to regulate pathways affecting PDs (8). Epigenetic mechanisms, which include DNA methylation, histone modification, and noncoding RNA (ncRNA), can regulate the gene expression without perturbation of DNA sequences (9). Among them, microRNAs (miRNAs), as a class of small ncRNA molecules, have been given great attention for their potential role in the etiology and pathophysiology of many diseases (10, 11). miRNAs negatively regulate gene expression at the post-transcriptional level by inhibiting translation and/or activating messenger RNAs (mRNAs) degradation through binding to the 3’-untranslated region (3’-UTR) of target mRNAs (12). miRNAs have strong cell and tissue specificity, and these specific expressions are not only the basis for its functional study, but also good disease markers. Evidence indicates that miRNAs regulate several aspects of neurodevelopment, including neurogenesis, neuronal differentiation, and synaptic plasticity through complex genetic networks (13).
Recent studies have revealed that disturbances in miRNAs may contribute to the etiology of SZ, MDD and BD, but there were conflicting results between these studies (10, 14), which may be due to differences in study design, small sample size, different specimen types. Therefore, the purpose of the present study was to comprehensively analyze the expression profiles of peripheral blood miRNAs associated with the pathogenesis or development of SZ, MDD and BD, and identify their specific differentially expressed miRNAs and their overlapping miRNAs expression profiles, so as to explore whether one or more miRNAs are promising biomarkers for their early diagnosis.
The study protocol and registration information are available at http://www.crd.york.ac.uk/PROSPERO/ (registration number: CRD42023486982).
This study was followed by recommendations from the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guideline. Literature search was conducted using Cochrane Library, PubMed, Embase, Medline, Wanfang, CNKI, and Weipu for studies published from February 2007 to September 2023, investigating differentially expressed miRNAs in SZ, MDD, or BD patients versus controls. The Search was performed using the following key terms: (“microRNA” OR “miRNA” OR “miR”) AND (“psychiatric disorders” and “schizophrenia” OR “SZ” OR “major depressive disorder” OR MDD OR “bipolar disorder” OR BD). A manual search of reference lists from relevant articles was conducted to uncover more potential studies.
Studies were included if they met the criteria below: 1) case-control studies; 2) studies on differential expression of peripheral blood miRNAs in SZ, MDD or BD; 3) the relative miRNA expression was detected by real-time quantitative polymerase chain reaction (RT-qPCR) or miRNA PCR panel or microarray or sequencing; 4) the mean and standard deviation (SD) of miRNA expression in the case group and control group could be obtained, or the relevant data could be used to calculate the above indicators. The exclusion criteria were as follows: 1) studies were not conducted in human subjects; 2) incomplete data; 3) duplicate data; 4) reviews, meta-analyses, letters or conference.
Two authors independently manually screened and extracted the data from included studies. Any inconsistencies were discussed with a third author until consensus was reached. The following items for each included study were extracted: 1) first author; 2) year of publication; 3) country; 4) specimen type; 5) sample size; 6) age; 7) miRNA detection methods; 8) mean and SD of the identified miRNAs in each group. If the mean and SD couldn’t be extracted from studies, we tried to contact their authors. The studies we didn’t receive a response were listed in Supplementary Table 1. If different specimen types were involved in the same study, data extraction and corresponding analysis were performed separately.
The quality of included studies was assessed by using the Newcastle–Ottawa Scale (NOS) (15), which consists of three dimensions: selection, comparability and exposure. The studies with a score ≥ 5 are regarded as high quality.
TargetScan and miRanda were used to predict the target genes for common differentially expressed miRNAs in SZ, MDD or BD. TargetScan algorithms eliminated genes with context scores < 50%. miRanda algorithms eliminated genes with maximum energy > -10. Genes co-identified by both databases were potential target genes for a given miRNA. Functional enrichment analysis of the predicted target genes was implemented with kyoto encyclopedia of genes and genomes (KEGG). We performed enrichment analysis with the cut-off criterion of P < 0.05.
All analyses were conducted by Stata 12.0. The standard mean difference (SMD) and its 95% confidence interval (CI) were used to combine the miRNA expression results. The between-study heterogeneity was evaluated by a Cochran’s Q-statistic and quantified by I2 metric value. If I2<50% and P>0.10, the fixed-effects model was conducted, otherwise, the random-effects model was applied. Subgroup analyses were performed based on specimen types. The potential for publication bias was examined by Begg’s test and Egger’s test. Leave-one-out sensitivity analysis was performed to detect the stability of the results. P<0.05 was considered statistically significant.
According to the search strategy, 5572 studies were identified in the database. After an initial screen, 2532 duplicate studies were removed. Next, 2877 studies were excluded based on titles and/or abstracts. The remaining 163 studies were evaluated in detail of which 50 studies were excluded due to incomplete data (n=27), no healthy controls (n=8), reviews (n=13) and meta-analysis (n=2). Of the remaining 113 studies, 35 were not included in the meta-analysis after data extraction because the mean and SD of the miRNAs reported in these studies could not be extracted from more than 3 independent studies (Supplementary Table 2). Finally, 78 studies were suitable for quantitative meta-analysis, of which 29 were identified for SZ (16–44), 45 for MDD (45–89), and 6 for BD (21, 24, 90–93). 2 studies (21, 24) were for both SZ and BD. The flowchart of the study screening process was shown in Figure 1. The NOS results showed that all the included studies were of high quality, with scores ranging from 5 to 9. The characteristics of the eligible studies were summarized in Table 1.
In our meta-analysis, we analyzed the expression of 16 miRNAs from 29 studies for SZ. The results showed that SZ patients had higher miRNA levels than control group in miR-181b-5p, miR-34a-5p, miR-195-5p, miR-30e-5p, miR-7-5p, miR-132-3p, miR-212-3p, miR-206, miR-92a-3p and miR-137-3p, while lower miRNA levels than control group in miR-134-5p, miR-107, and miR-99b-5p. Besides, miR-432-5p, miR-346 and miR-22-3p were not dysregulated (Table 2, Figure 2). 9 of 16 miRNAs were included in subgroup analysis stratified by specimen type in SZ patients. The results revealed that miR-34a-5p, miR-30e-5p, miR-7-5p and miR-212-3p were both upregulated in plasma and PBMC. miR-195-5p was upregulated in plasma, PBMC and whole blood. miR-181b-5p was upregulated in plasma, but not in PBMC and whole blood. miR-132-3p was upregulated in plasma, but not in PBMC. miR-346 was downregulated in PBMC, but not in plasma. miR-432-5p was not dysregulated both in plasma and PBMC (Figure 2).
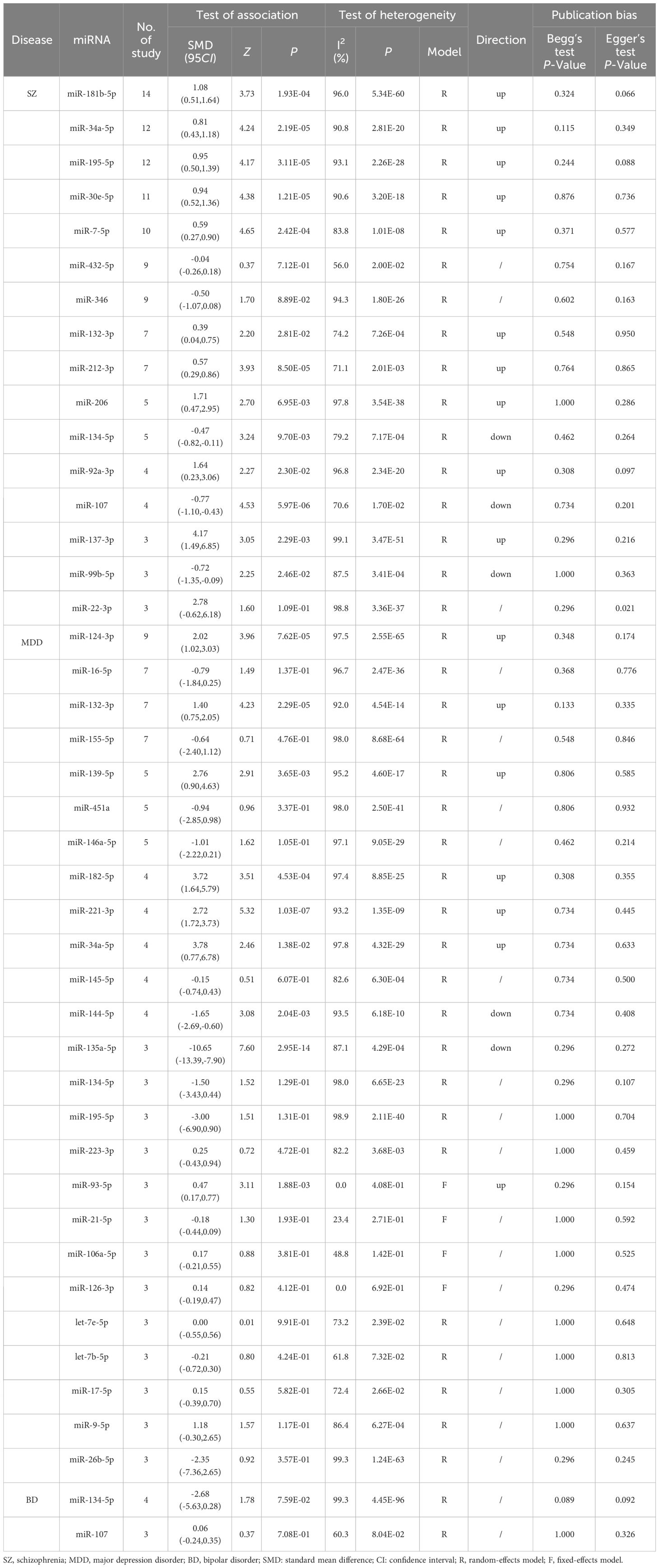
Table 2 Meta-analysis results of differentially expressed miRNAs in PDs reported in three or more studies.

Figure 2 Forest plot of the meta-analysis of peripheral blood microRNAs of SZ patients versus controls.
We analyzed the expression of 25 miRNAs from 45 studies for MDD. The results suggested that MDD patients had higher miRNA levels than control group in miR-124-3p, miR-132-3p, miR-139-5p, miR-182-5p, miR-221-3p, miR-34a-5p and miR-93-5p, while lower miRNA levels than control group in miR-144-5p and miR-135a-5p. Besides, miR-16-5p, miR-155-5p, miR-451a, miR-146a-5p, miR-145-5p, miR-134-5p, miR-195-5p, miR-223-3p, miR-21-5p, miR-106a-5p, miR-126-3p, let-7e-5p, let-7b-5p, miR-17-5p, miR-9-5p and miR-26b-5p were not dysregulated (Table 2, Figure 3). 3 of 25 miRNAs were included in subgroup analysis stratified by specimen type in MDD patients. The results showed that miR-124-3p was upregulated in serum, but not in plasma and PBMC. miR-16-5p was not dysregulated in plasma, whole blood and PBMC. miR-132-3p was upregulated in serum, plasma and whole blood, but not in PBMC. (Figure 3).
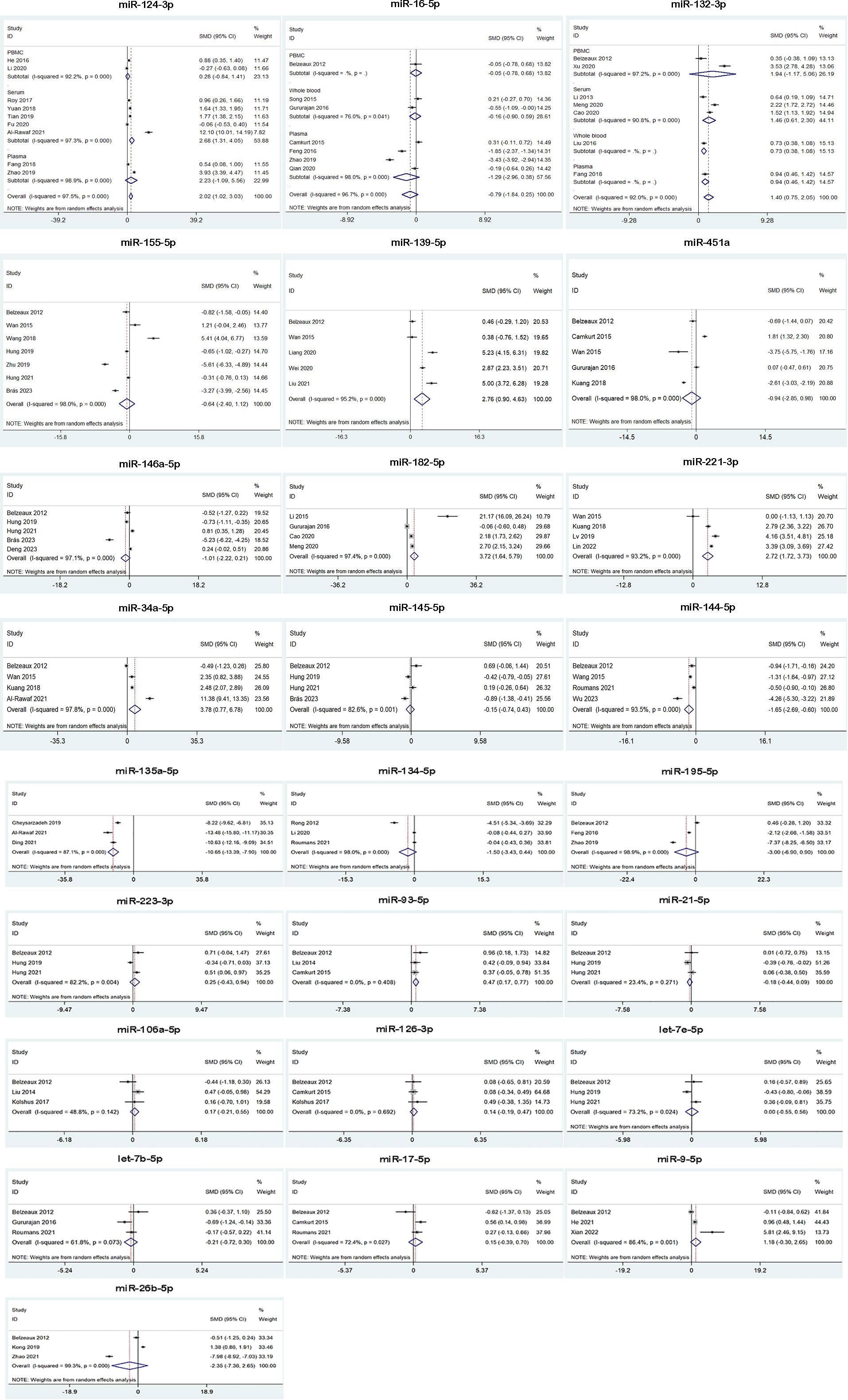
Figure 3 Forest plot of the meta-analysis of peripheral blood microRNAs of MDD patients versus control.
We analyzed the expression of miR-134-5p and miR-107 from 6 studies for BD, but neither of them were dysregulated in BD patients (Table 2, Figure 4).
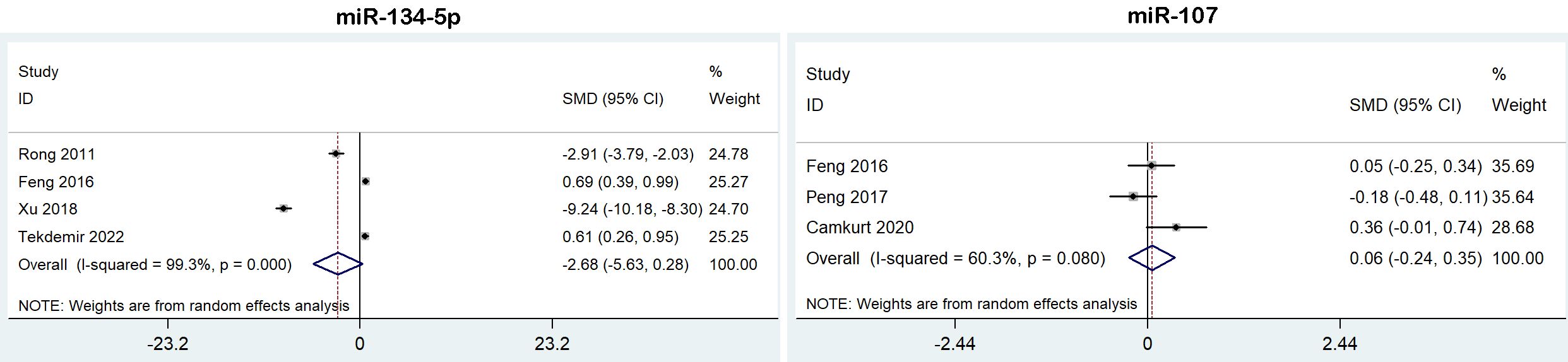
Figure 4 Forest plot of the meta-analysis of peripheral blood microRNAs of BD patients versus controls.
Our results found that miR-132-3p and miR-34a-5p were upregulated in both SZ and MDD patients, suggesting that they may likely share some common molecular mechanisms.
To get insight into the possible roles of the miR-132-3p and miR-34a-5p, we performed target gene prediction and KEGG pathway analysis. A total number of 4138 target genes from miR-132-3p and miR-34a-5p were identified by using TargetScan and miRanda. Figure 5 showed the top 20 significant enriched terms identified for KEGG pathway analysis, including axon guidance, neurotrophin signaling pathway, ErbB signaling pathway, FoxO signaling pathway, etc.
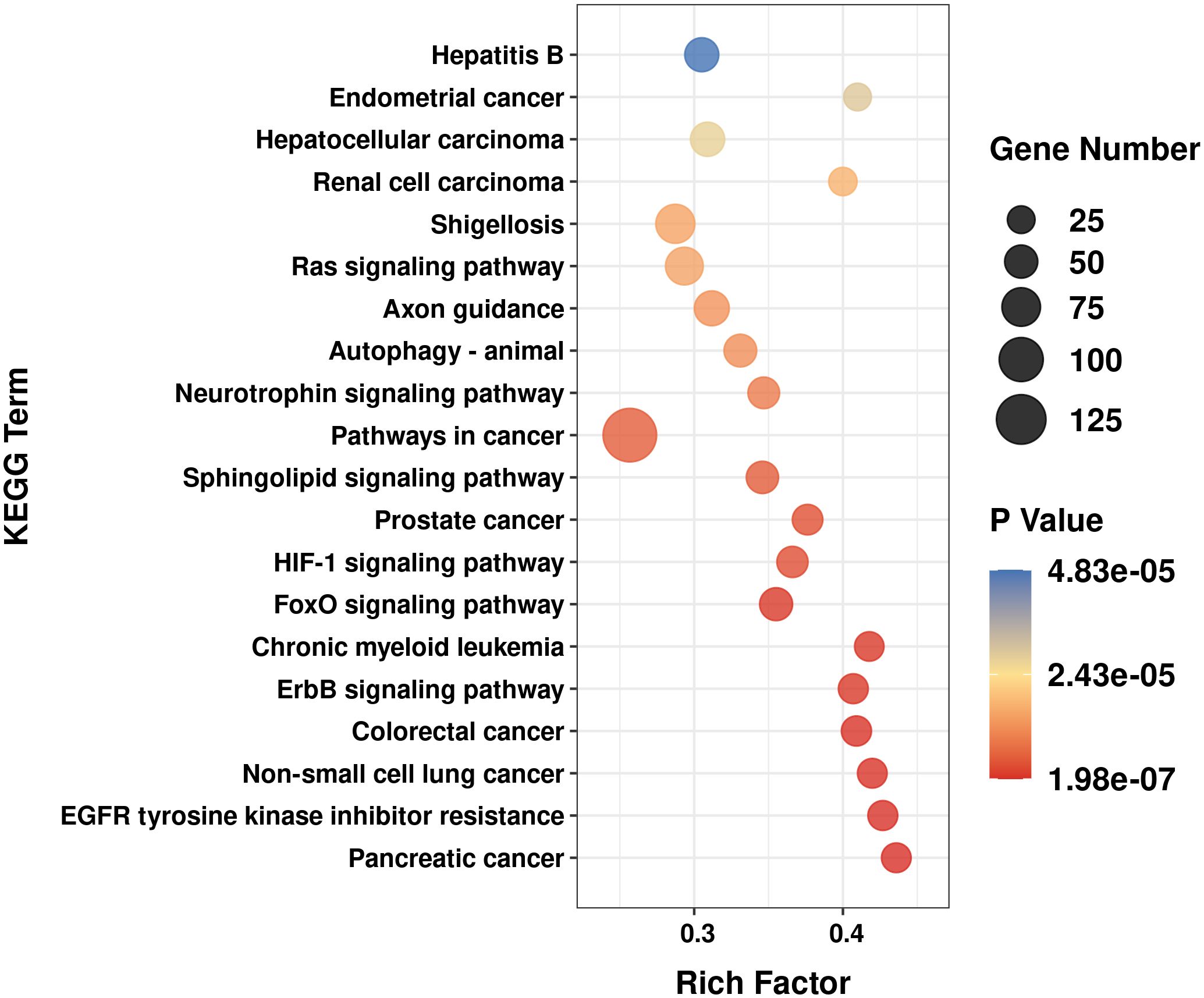
Figure 5 Top 20 significant enriched kyoto encyclopedia of genes and genomes (KEGG) signal pathway for the predicted target genes of miR-132-3p and miR-34a-5p.
Begg’s test and Egger’s test results indicated that there were no publication bias in this meta-analysis except miR-22-3p in SZ (Table 2). Sensitivity analysis showed that none of individual study could obviously influenced the pooled ORs except miR-22-3p in SZ, miR-223-3p and miR-17-5p in MDD (Supplementary Figures 1-3). For miR-22-3p, miR-223-3p and miR-17-5p, when Du et al.’s study, Huang et al.’s study, Belzeaux et al.’s were removed, respectively, the levels of these 3 miRNAs were all upregulated, but only 2 studies remained for each miRNA, so we wouldn’t further discuss them.
A major goal of psychiatric research is to identify biomarkers for early and reliable diagnosis of PDs and guide their effective clinical treatment. In recent years, miRNAs, a key regulator of neurogenesis, neuronal differentiation, and synaptic plasticity, have received widespread attention as potential biomarkers of PDs (94, 95). However, as the literature reviews on miRNAs in PDs were merely narrative, or only meta-analysis for single disease (such as SZ or MDD), and no relatively comprehensive data was available. In this study we conducted a comprehensive and systematic meta-analysis for the first time to simultaneously identify dysregulated miRNAs expression profiles in SZ, MDD and BD.
In our meta-analysis, we focused on differentially expressed miRNAs derived from peripheral blood, excluding studies from brain tissue, as the method for extracting miRNAs from brain tissue has limited sample sources and are difficult to apply in clinical practice. Studies have found that brain disease-specific miRNAs can also be detected in peripheral blood, where their levels were highly correlated with those in the brain (96, 97). Interestingly, in SZ patients, miR-181b-5p and miR-132-3p were significantly increased in the pooled results, but only in certain blood elements in subgroup analysis based on specimen type. However, miR-346 was significantly decreased in PBMC, but not in plasma or the pooled results. In MDD patients, miR-124-3p was significantly increased in serum, but not in plasma and PBMC. These results indicated that the expression patterns of miRNAs could be affected by different specimen types. Previous evidence showed that the miRNA expression profiles in different blood elements may vary due to element-specific miRNAs released by specific tissues (98), unique miRNA features from unique lineage (99), different biological specimen processing conditions, and variation in reference miRNA levels (100).
With respect to SZ, Liu et al. (23) revealed that miR-181b-5p, miR-21-5p, miR-195-5p, miR-137, miR-346 and miR-34a-5p in PBMCs had high diagnostic sensitivity and specificity in SZ based on their meta-analysis of six diagnostic studies. Han et al. (101) found 27 significant differentially expressed miRNAs in SZ, of which 5 were downregulated, whereas 22 were upregulated. In our meta-analysis, we found the levels of miR-181b-5p, miR-34a-5p, miR-195-5p, miR-30e-5p, miR-7-5p, miR-132-3p, miR-212-3p, miR-206, miR-92a-3p and miR-137-3p were increased in SZ patients, while the levels of miR-134-5p, miR-107 and miR-99b-5p were decreased. Our research findings were not entirely consistent with the two previous meta-analysis, such as miR-195-5p was increased in SZ patients in our study but not in Han et al.’s study. The reasons for the inconsistent conclusion may be due to differences in inclusion and exclusion criteria, outcome measures, and the number of studies included. Of the thirteen differentially miRNAs in our study, miR-181b-5p was the most commonly reported one. Increased levels of miR-181b-5p have been detected in plasma (17, 19, 22), serum (43), as well as in brain of SZ patients (102). Of note, miR-181b-5p targeted a-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate acid (AMPA) glutamate ionotropic receptor type subunit 2 (GRIA2) and the calcium sensor protein gene visinin like 1 (VSNL1) in SZ patients (102); both of these targets were themselves suspected to have a role in the pathology of SZ (103, 104). Guo et al. (105) constructed a miRNA-transcription factors regulatory network for SZ and found that miR-195-5p was one of the core regulators in this regulatory network. Many of the predicted target genes of miR-195-5p, such as regulator of G-protein signaling 4 (RGS4), N-methyl-D-aspartate (NMDA) glutamate ionotropic receptor type subunit 3A (GRIN3A), and reelin (RELN), have been reported to correlate with SZ (106, 107). Brain Derived Neurotrophic Factor (BDNF) was involved in neuronal plasticity, and multiple studies supported its close association with SZ (108). Mellios et al. showed that miR-195-5p regulated BDNF, thereby affecting the expression of downstream gamma-aminobutyric acid (GABA)ergic transcripts, such as parvalbumin (PV), somatostatin (SST), and neuropeptide Y (NPY) in SZ (109, 110). Xu et al. (111) indicated that a potentially functional variant that affected pre-miR-30-5p played a role in SZ susceptibility. Overexpression of miR-30e-5p in the rat brain could lead to cognitive impairment, resulting in anxiety, depression, and SZ like symptoms (112). Abnormal expression of miR-7-5p could inhibit the protein kinase AKT1 gene, which has been confirmed to be a susceptibility gene for SZ (113). In addition, Zhang et al. (114) found that miR-7-5p was overexpressed in plasma of SZ and the overexpression of miR-7-5p significantly inhibited the expression levels of SH3 and multiple ankyrin repeat domains protein 3 (SHANK3), which in turn may play an essential role in the pathological process of SZ. It was found that miR-212-3p was co-transcribed with miR-132-3p, the miR-132-3p/miR-212-3p family influenced genes associated with circadian clock entrainment (115), which was consistent with the defective circadian synchronization observed in SZ. A recent study suggested that miR-206 may contribute to SZ risk through allele-dependent regulation of the genome-wide significant gene NT5C2 (116). Du et al. (32) showed significantly increased miR-206 levels and decreased BDNF levels in SZ, and antipsychotics restored the dysregulations of miR-206 and BDNF in SZ, suggesting that upregulation of miR-206 may contribute to the dysfunction of BDNF in SZ. miR-92a-3p was related to synaptic transmission (117). Studies have confirmed that miR-137-3p was closely related to the development and maturation of the nervous system, and can regulate multiple neural development signaling pathways and target gene expression through cascade effects (118). Wright et al. (119) identified the possible regulatory signaling pathways involved in SZ by miR-137-3p through functional enrichment analysis, including axonal guidance, Ephrin receptor signaling, long-term regulation, Sertoli cell junction, and protein kinase A signaling. Kwon et al. (120) confirmed that susceptibility genes of SZ, such as transcription factor 4 gene (TCF4), calcium voltage-gated channel subunit alpha1 C gene (CACNA1C), CUB, and Sushi multiple domains 1 gene (CSMD1), WW domain binding protein 1 like gene (C10orf26) were target genes for miR-137-3p. miR-134-5p was a brain-specific miRNA that presented in the synaptic dendrite chamber of hippocampal neurons, which repressed dendritic spine size by inhibiting the translation of Lim kinase 1 (Limk1) mRNA, thereby affecting the strength of excitatory synapses (121). More recently, it has been shown that the expression of silent information regulator 1 (SIRT1), which modulates synaptic plasticity and memory formation, is regulated by cAMP-response element-binding protein (CREB), which itself is translationally repressed by miR-134-5p (121, 122). Beveridge et al. (107) suggested that miR-107 were highly enriched in pathways involved in neural connectivity and synaptic plasticity, such as axon guidance, long-term potentiation. Scarr et al. (123) demonstrated that miR-107 could regulate the expression of cortical muscarinic M1 receptors (CHRM1), which was involved in the pathophysiology of SZ (124). Kaurani et al. (125) reported that miR-99b-5p regulated Z-DNA binding protein 1 (Zbp1) to control inflammatory response in microglia, which may contributed to the pathogenesis of SZ.
Regarding MDD, Li et al. (126) showed that 17 miRNAs had high sensitivity and specificity in diagnosing MDD based on 7 studies. We found the levels of miR-124-3p, miR-132-3p, miR-139-5p, miR-182-5p, miR-221-3p, miR-34a-5p and miR-93-5p were increased, while the level of miR-144-5p and miR-135a-5p were decreased. Our research findings were not entirely consistent with Li et al.’s meta-analysis, such as miR-16-5p was not dysregulated in MDD patients in our study but was upregulated in their study. The most possible reason for the inconsistent conclusion may be their meta-analysis based on diagnostic studies and all mentioned miRNAs only reported in single study, but our present study included miRNAs from at least 3 independent studies. Moreover, we conducted subgroup analysis stratified by specimen type. Of the seven differentially expressed miRNAs in our study, miR-124-3p was the most frequently reported one. Increased levels of miR-124-3p have been consistently detected in serum (58, 79) and plasma (60, 69). miR-124-3p was a rich brain-specific miRNA that inhibited serotonin induced synaptic facilitation by regulating CREB, thereby negatively regulating synaptic plasticity (127). Studies indicated that miR-124-3p could inhibit the expression of BDNF in the hippocampus of depression model rats (128). BDNF was a validated miR-124-3p target (47) and low expression levels of BDNF played a predominant role in the pathophysiology of MDD (129). miR-139-5p might act as a negative regulator for neural stem cell proliferation and neuronal differentiation, and modulated cortical neuronal migration by targeting lissencephaly-1 (Lis1) (77, 130). Wei et al. (77) found stress-induced elevation of miR-139-5p caused impairment of hippocampal neurogenesis and depressive-like behaviors in adult mice. miR-182-5p has been proven to be an important regulatory factor in the nervous system, involved in various biological processes such as neuronal survival (131), axonogenesis (132), and protein signal transduction (133). Studies have provided evidence for miR-182-5p as a modulator of the endogenous circadian clock (134). Disruption of circadian rhythms has long been implicated in the pathophysiology of MDD (135). Li et al. (47) found the serum levels of miR-182-5p were increased and BDNF levels were reduced in MDD patients, which supported that miR-182-5p could negatively regulate BDNF expression and might be related to the development of MDD. Although miR-221-3p was commonly considered as a tumor regulator, in recent years, some researchers have been repeatedly reported abnormally high levels of miR-221-3p in the cerebrospinal fluid (CSF) and serum of MDD patients (49, 136), suggesting that miR-221-3p may also be involved in the pathogenesis of MDD. Studies revealed that miR-221-3p was closely related to neuronal development and axon growth (137, 138). In addition, Lian et al. (139) demonstrated that miR-221-3p could promote the development of MDD by modulating Wnt2/CREB/BDNF axis. Wu et al. (89) demonstrated that miR-144-5p influenced synaptic plasticity by targeting phosphatase and tensin homolog (PTEN), and miR-144-5p exerted anti-inflammatory effects in patients with MDD. miR-135a-5p regulated axon growth/regeneration and mediated long-term depression (140, 141). Ding et al. (78) demonstrated that miR−135a-5p regulated apoptosis and inflammatory response in mouse hippocampal neurons by regulating the expression of Toll like receptor 4 (TLR 4), thereby alleviating the depressive behavior of mice and playing a protective role in depression. Valiuliene et al. (142) revealed that miR-93-5p may regulate the expression of the pro-inflammatory cytokine IL-18, involving in the pathophysiology of MDD.
Our results found that miR-132-3p and miR-34a-5p were increased in both SZ and MDD patients, suggesting that they may likely share some common molecular mechanisms. miR-132-3p was a miRNA enriched in the brain and participated in axonal growth, proliferation and synaptic plasticity (115). Neuronal plasticity and its related pathways have shown to be disturbed in SZ and MDD (143, 144). miR-132-3p targeted important genes that regulate neuronal plasticity, including BDNF, methyl-CpG-binding protein 2 (MeCP2), GTPase activating protein (p250GAP) (145–147). Su et al. (148) demonstrated that miR−132-3p was significantly increased in the peripheral blood of MDD patients, while BDNF and MeCP2 were decreased, and the level of miR-132-3p was negatively correlated with the protein expression levels of MeCP2 and BDNF. Low BDNF level was also detected in CSF and plasma of SZ patients (149). Besides, MeCP2 has been repeatedly reported as a risk gene for SZ (150, 151). p250GAP was a brain-enriched NDMA receptor-interacting RhoGAP. Studies have shown that the p250GAP gene was associated with risk for SZ and MDD (152, 153). miR-34a-5p suppressed SIRT1, leading to increased acetylated p53, a regulator of the cell cycle progression and cellular senescence (154). It has also been shown that miR-34a-5p was a transcriptional target of p53, thus establishing a positive feedback loop between miR-34a-5p, p53, and SIRT1 (154, 155). Oxidative stress induced the upregulation of p53 activity, consequently increasing the expression levels of miR-34a-5p (155, 156). Both SZ and MDD were associated with high oxidative stress levels (157, 158), which could elucidate the upregulated miR-34a-5p found in these patients. In addition, SZ and MDD also were genetically associated with the SIRT1 gene (159–161). Xu et al. (162) indicated that miR-34a-5p targeted the NMDA receptors (including Grin1, Grin2a, and Grin2b), providing evidence of a post-transcriptional mechanism of SZ and MDD associated glutamatergic and synaptic dysfunction (163–165). Moreover, KEGG pathway analysis in the present study indicated that the identified signaling pathways enriched by the predicted target genes of miR-132-3p and miR-34a-5p, such as axon guidance, neurotrophin signaling pathway, ErbB signaling pathway, FoxO signaling pathway, were closely related to the pathologic mechanisms of SZ and MDD (166–171). Interestingly, enriched KEGG pathways also contained cancer pathways, which may be involved in shared pathogenesis of SZ and MDD. For example, PI3K/Akt pathway, which modulated by miR-132-3p, was a prototypic cancer pathway (172). Many genes on PI3K/Akt pathway were considered to be potentially susceptible genes for the development of SZ (173). The levels of Akt1 were decreased in the brain, as well as in the peripheral lymphocytes of individuals with SZ (174). P13K/Akt signaling cascade also was strongly linked with the neurobiology of MDD (175). Reduced Akt1 activity was found in the brain of MDD patients (176). Evidence showed that p53, which could regulate the transcription of miR-34a-5p, was one of the most important tumor suppressor genes (177). Catts et al. (178) proposed that p53 might be a candidate susceptibility gene for SZ by regulating apoptosis. Mahmood et al. (179) suggested the protective effect of minor allele 72C of p53 gene towards MDD.
The following limitations of the study should be considered. Firstly, between-study heterogeneity remained substantial although we performed subgroup analyses to explore their sources. The possible causes of heterogeneity may be related to the duration, severity, and treatment of patient’s disease. Due to the limited information provided by the included studies, further analysis was not possible. Secondly, the majority of the population included in the study came from China, which may limit the broad applicability of the findings. Thirdly, potential publication bias may affect the present results due to the relative small number of studies included for some miRNAs. Finally, most of miRNAs included in the present meta-analysis were detected by qPCR, which may also result in bias.
In summary, our study identified 13 differentially expressed miRNAs in SZ, 9 differentially expressed miRNAs in MDD, among which miR-132-3p and miR-34a-5p were upregulated in both SZ and MDD by systematically analyzing qualified studies. These miRNAs may be used as potential biomarkers for the diagnosis of SZ and MDD in the future. Further validation in large patient cohorts is required to confirm the findings.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
XL: Conceptualization, Data curation, Formal Analysis, Validation, Writing – original draft. LD: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. ZJ: Investigation, Resources, Validation, Writing – original draft. MS: Software, Validation, Visualization, Writing – review & editing. PY: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Zhejiang Provincial Natural Science Foundation (Grant Nos. LGF22H090014), and Science and Technology Project of Medicine and Health of Zhejiang Province (Grant Nos. 2020KY222).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2024.1390366/full#supplementary-material
1. Mandal PK, Gaur S, Roy RG, Samkaria A, Ingole R, Goel A. Schizophrenia, bipolar and major depressive disorders: overview of clinical features, neurotransmitter alterations, pharmacological interventions, and impact of oxidative stress in the disease process. ACS Chem Neurosci. (2022) 13:2784–802. doi: 10.1021/acschemneuro.2c00420
2. Collaborators GDaIIaP. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. (2018) 392:1789–858. doi: 10.1016/S0140-6736(18)32279-7
3. Collaborators GMD. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. (2022) 9:137–50. doi: 10.1016/S2215-0366(21)00395-3
4. Stepnicki P, Kondej M, Kaczor AA. Current concepts and treatments of schizophrenia. Molecules. (2018) 23:2087. doi: 10.3390/molecules23082087
5. Gutierrez-Rojas L, Porras-Segovia A, Dunne H, Andrade-Gonzalez N, Cervilla JA. Prevalence and correlates of major depressive disorder: a systematic review. Braz J Psychiatry. (2020) 42:657–72. doi: 10.1590/1516-4446-2020-0650
6. Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. (2016) 387:1561–72. doi: 10.1016/S0140-6736(15)00241-X
7. Toth M. Mechanisms of non-genetic inheritance and psychiatric disorders. Neuropsychopharmacology. (2014) 40:129–40. doi: 10.1038/npp.2014.127
8. Varela RB, Cararo JH, Tye SJ, Carvalho AF, Valvassori SS, Fries GR, et al. Contributions of epigenetic inheritance to the predisposition of major psychiatric disorders: Theoretical framework, evidence, and implications. Neurosci Biobehav Rev. (2022) 135:104579. doi: 10.1016/j.neubiorev.2022.104579
9. Panariello F, Fanelli G, Fabbri C, Atti AR, De Ronchi D, Serretti A. Epigenetic basis of psychiatric disorders: A narrative review. CNS Neurol Disord Drug Targets. (2022) 21:302–15. doi: 10.2174/1871527320666210825101915
10. van den Berg MMJ, Krauskopf J, Ramaekers JG, Kleinjans JCS, Prickaerts J, Briedé JJ. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog Neurobiol. (2020) 185:101732. doi: 10.1016/j.pneurobio.2019.101732
11. Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagnosis Ther. (2016) 20:509–18. doi: 10.1007/s40291-016-0221-4
12. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci. (2019) 20:6249. doi: 10.3390/ijms20246249
13. Olde Loohuis NFM, Kos A, Martens GJM, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. (2011) 69:89–102. doi: 10.1007/s00018-011-0788-1
14. Micale V, Di Bartolomeo M, Di Martino S, Stark T, Dell'Osso B, Drago F, et al. Are the epigenetic changes predictive of therapeutic efficacy for psychiatric disorders? A translational approach towards novel drug targets. Pharmacol Ther. (2023) 241:108279. doi: 10.1016/j.pharmthera.2022.108279
15. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
16. Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. (2012) 17:827–40. doi: 10.1038/mp.2011.78
17. Wang TY, Xie SP, Xiao ZD, Du JL, Yang K. MicroRNA-181b plasma levels before and after antipsychotic treatment for male patients with schizophrenia. J Clin Psychiatry. (2012) 22:105–7.
18. Zhang L, Guo W, Sun XY, Song HT, Zhao L, Zhong AF, et al. Partial microRNA profiling differences between peripheral plasma and mononuclear leucocytes in schizophrenia patients. Chin J Behav Med Brain Sci. (2014) 23:694–7. doi: 10.3760/cma.j.issn.1674-6554.2014.08.007
19. Song HT, Sun XY, Zhang L, Zhao L, Guo ZM, Fan HM, et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J Psychiatr Res. (2014) 54:134–40. doi: 10.1016/j.jpsychires.2014.03.008
20. Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, et al. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci. (2015) 22:570–4. doi: 10.1016/j.jocn.2014.08.018
21. Feng SS, Zhong L, Yang M, Peng Y, Tang M, Li BL, et al. Expression of circulating miRNA in WBC of the patients with schizophrenia and its clinical significance. Chin J Clin Lab. (2016) 34:247–50. doi: 10.13602/j.cnki.jcls.2016.04.03
22. Su XD, Fan CL, Yu L, Li DF, Xie JJ, Zhang SL, et al. Association of expression with the level of microRNA in plasma and mononuclear cells in patients with schizophrenia. J Mod Lab Med. (2017) 32:45–48+52. doi: 10.3969/j.issn.1671-7414.2017.02.012
23. Liu S, Zhang F, Wang X, Shugart YY, Zhao Y, Li X, et al. Diagnostic value of blood-derived microRNAs for schizophrenia: results of a meta-analysis and validation. Sci Rep. (2017) 7:15328. doi: 10.1038/s41598-017-15751-5
24. Peng Y, Feng SS, Tian G, Yang M, Deng ZH, Wang KZ. Diagnostic value of plasma microRNA-107 and microRNA-99b-5p in schizophrenia. Shandong Med J. (2017) 57:98–100. doi: 10.3969/j.issn.1002-266X.2017.04.034
25. Qu HF, Zhu YC, Wang F, Zhou J, Zhang W, Chang XL, et al. Expression and significance of peripheral blood leucocyte miR-198 and miR-206 in schizophrenics. J Jiangsu University( Med Edition). (2017) 27:420–422+427. doi: 10.13312/j.issn.1671-7783.y160210
26. Ma J, Shang S, Wang J, Zhang T, Nie F, Song X, et al. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. (2018) 265:70–6. doi: 10.1016/j.psychres.2018.03.080
27. Bao CX, Lv QY, Zhao J, Geng RJ, Cheng XY, Wang YY, et al. An analysis of miR-195 in PBMC expression differences before and after antipsychotic treatment in patients with schizophrenia. J Clin Psychiatry. (2018) 28:81–3. doi: 10.3969/j.issn.1005-3220.2018.02.003
28. Feng SS, Luo B, Yin MG, Zhong L, Wang KZ, Yang M, et al. The ROC curve combined Logistic regression analysis evaluate the diagnostic value of plasma miR-31-5p and miR-134-5p for schizophrenia. Chin J Clin Lab Sci. (2018) 36:116–9. doi: 10.13602/j.cnki.jcls.2018.02.10
29. Fu GH, Xu JT, Li Y, Fang F, Xie SF. Relationship between plasma microRNA-30e expression and symptoms in patients with first-episode schizophrenia. Chin J Nerv Ment Dis. (2018) 44:489–92. doi: 10.3969/j.issn.1002-0152.2018.08.010
30. Wang YH, Wang J, Zhou WX, Guo TH, Luo BR, Cheng F, et al. Research on microRNAs in plasma and peripheral blood mononuclear cell in the diagnosis of schizophrenia. J Clin Psychiatry. (2019) 29:175–9.
31. Shi YJ, Zhang XM. Value analysis of plasma miR-7, miR-30e and miR-195 in the diagnosis of schizophrenia. J Psychiatry. (2019) 32:273–6. doi: 10.3969/j.issn.2095-9346.2019.04.009
32. Du Y, Yu Y, Hu Y, Li XW, Wei ZX, Pan RY, et al. Genome-wide, integrative analysis implicates exosome-derived microRNA dysregulation in schizophrenia. Schizophr Bull. (2019) 45:1257–66. doi: 10.1093/schbul/sby191
33. Guan J, Liu J, Qin W. Diagnostic value of miRNA expression in peripheral blood for schizophrenia. J North Sichuan Med Coll. (2021) 36:237–41. doi: 10.3969/j.issn.1005-3697.2021.02.025
34. Lu YN, Wang GQ, Zhang FQ, Yin JJ. Effect of antipsychotic treatment on the expression of miR-92a-3p in peripheral blood mononuclear cells. J Clin Psychiatry. (2021) 31:306–8. doi: 10.3969/j.issn.1005-3220.2021.04.017
35. Jiang YL, Wu JQ, Chen ZJ, Zhu JY. Expression and Clinical Significance of miR-92a-3p and miR-193a-3p in Peripheral Blood of First-episode Schizophrenia. Chin And Foreign Med Res. (2021) 19:72–4. doi: 10.14033/j.cnki.cfmr.2021.12.026
36. Zhang D, Li H, Ding K, Zhang Z, Luo S, Li G. Polymorphisms in microRNA genes associated with schizophrenia susceptibility but not with effectiveness of MECT. Comput Math Methods Med. (2021) 2021:1959172. doi: 10.1155/2021/1959172
37. Chen BY, Lin JJ, Lu MK, Tan HP, Jang FL, Lin SH. Neurodevelopment regulators miR-137 and miR-34 family as biomarkers for early and adult onset schizophrenia. NPJ Schizophr. (2021) 7:35. doi: 10.1038/s41537-021-00164-1
38. Gou M, Pan S, Tong J, Zhou Y, Han J, Xie T, et al. Effects of microRNA-181b-5p on cognitive deficits in first-episode patients with schizophrenia: Mediated by BCL-2. J Psychiatr Res. (2021) 136:358–65. doi: 10.1016/j.jpsychires.2021.02.003
39. Pan S, Feng W, Li Y, Huang J, Chen S, Cui Y, et al. The microRNA-195 - BDNF pathway and cognitive deficits in schizophrenia patients with minimal antipsychotic medication exposure. Transl Psychiatry. (2021) 11:117. doi: 10.1038/s41398-021-01240-x
40. Fu X, Liu Y, Baranova A, Zhang F. Deregulatory miRNA-BDNF network inferred from dynamic expression changes in schizophrenia. Brain Sci. (2022) 12:167. doi: 10.3390/brainsci12020167
41. Lu QZ, Gao N. The effect of Qingxin Dingzhi Tang on serum miR-132, miR-137, and miR-206 levels in first-episode schizophrenia patients. Contemp Med. (2022) 28:175–7. doi: 10.3969/j
42. Huang Y, Li YJ, Gong HF, Jin Y, Shi DH, Zhu N, et al. Correlation analysis of serum miR-15a,miR-206 levels and cognitive function in female first-episode schizophrenia. Int J Lab Med. (2022) 43:67–72. doi: 10.3969/j.issn.1673-4130.2022.01.014
43. Wang L, Cao F, Chen H. Expression and significance of microRNA-21 and microRNA-181b in peripheral blood of patients with schizophrenia. Chin J Prim Med Pharm. (2023) 30:596–600. doi: 10.3969/j.issn.1009-4393.2022.04.066
44. Jin M, Liu Y, Hu G, Li X, Jia N, Cui X, et al. Establishment of a schizophrenia classifier based on peripheral blood signatures and investigation of pathogenic miRNA-mRNA regulation. J Psychiatr Res. (2023) 159:172–84. doi: 10.1016/j.jpsychires.2023.01.035
45. Rong H, Liu TB, Yang HC, Feng F, Wei ZG. Expression levels of pre- and post-treatment plasma microRNA-134 in depression patients. J Clin Psychosom Dis. (2012) 18:300–2. doi: 10.3969/j.issn.1672-187X.2012.04.005-0300-03
46. Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Treziny C, Verrier L, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry. (2012) 2:e185. doi: 10.1038/tp.2012.112
47. Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang YX, et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PloS One. (2013) 8:e63648. doi: 10.1371/journal.pone.0063648
48. Liu X, Zhang L, Cheng K, Wang X, Ren G, Xie P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J Affect Disord. (2014) 163:133–9. doi: 10.1016/j.jad.2013.12.035
49. Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J, et al. Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PloS One. (2015) 10:e0121975. doi: 10.1371/journal.pone.0121975
50. Camkurt MA, Acar Ş, Coşkun S, Güneş M, Güneş S, Yılmaz MF, et al. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res. (2015) 69:67–71. doi: 10.1016/j.jpsychires.2015.07.023
51. Li J, Meng H, Cao W, Qiu T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci Lett. (2015) 606:167–72. doi: 10.1016/j.neulet.2015.08.038
52. Song MF, Dong JZ, Wang YW, He J, Ju X, Zhang L, et al. CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J Affect Disord. (2015) 178:25–31. doi: 10.1016/j.jad.2015.02.022
53. Wang X, Sundquist K, Hedelius A, Palmér K, Memon AA, Sundquist J. Circulating microRNA-144-5p is associated with depressive disorders. Clin Epigenet. (2015) 7:69. doi: 10.1186/s13148-015-0099-8
54. Gururajan A, Naughton ME, Scott KA, O'Connor RM, Moloney G, Clarke G, et al. MicroRNAs as biomarkers for major depression: a role for let-7b and let-7c. Transl Psychiatry. (2016) 6:e862. doi: 10.1038/tp.2016.131
55. He S, Liu X, Jiang K, Peng D, Hong W, Fang Y, et al. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J Psychiatr Res. (2016) 78:65–71. doi: 10.1016/j.jpsychires.2016.03.015
56. Liu Y, Yang X, Zhao L, Zhang J, Li T, Ma X. Increased miR-132 level is associated with visual memory dysfunction in patients with depression. Neuropsychiatr Dis Treat. (2016) 12:2905–11. doi: 10.2147/ndt.s116287
57. Feng Q, Tang Z, Zhou H, Sun QY, Zhu H, Qiu CH, et al. Study on the expression levels of plasma microRNA-16 and microRNA-195 in patients with depression before treatment. Chin Ment Health J. (2016) 30:758–60. doi: 10.3969/j.issn.1000-6729.2016.10.007
58. Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of microRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology. (2017) 42:864–75. doi: 10.1038/npp.2016.175
59. Kolshus E, Ryan KM, Blackshields G, Smyth P, Sheils O, McLoughlin DM. Peripheral blood microRNA and VEGFA mRNA changes following electroconvulsive therapy: implications for psychotic depression. Acta Psychiatr Scand. (2017) 136:594–606. doi: 10.1111/acps.12821
60. Fang Y, Qiu Q, Zhang S, Sun L, Li G, Xiao S, et al. Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J Affect Disord. (2018) 227:745–51. doi: 10.1016/j.jad.2017.11.090
61. Kuang WH, Dong ZQ, Tian LT, Li J. MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as predictors of response to antidepressant treatment. Braz J Med Biol Res. (2018) 51:e7212. doi: 10.1590/1414-431x20187212
62. Wang X, Wang B, Zhao J, Liu C, Qu X, Li Y. MiR-155 is involved in major depression disorder and antidepressant treatment via targeting SIRT1. Biosci Rep. (2018) 38:BSR20181139. doi: 10.1042/bsr20181139
63. Gheysarzadeh A, Sadeghifard N, Afraidooni L, Pooyan F, Mofid MR, Valadbeigi H, et al. Serum-based microRNA biomarkers for major depression: MiR-16, miR-135a, and miR-1202. J Res Med Sci. (2018) 23:69. doi: 10.4103/jrms.JRMS_879_17
64. Yuan MJ, Bi XF. Changes and clinical significance of microRNA-124 in peripheral blood of patients with first-episode depression. J Psychiatry. (2018) 31:27–9. doi: 10.3969/j.issn.2095-9346.2018.01.008
65. Hung YY, Wu MK, Tsai MC, Huang YL, Kang HY. Aberrant Expression of Intracellular let-7e, miR-146a, and miR-155 Correlates with Severity of Depression in Patients with Major Depressive Disorder and Is Ameliorated after Antidepressant Treatment. Cells. (2019) 8:647. doi: 10.3390/cells8070647
66. Zhu SY, Zhang LN, Liu CY, Wu Z. Correlation analysis of serum miR-155 and sex hormone levels in adolescent female patients with depression. Maternal Child Health Care China. (2019) 34:5397–9. doi: 10.7620/zgfybj.j.issn.1001-4411.2019.23.24
67. Lv R, Cao YC, Wang XQ. Expression analysis of serum miR-221-3p in patients of severe depression. Clin Focus. (2019) 34:999–1003. doi: 10.3969/j.issn.1004-583X.2019.11.008
68. Tian T, Duan FR, Dai LL, Du H, Yang MH. Relationship between expression of BDNF, IL-6 and miR-124 and disease severity in patients with depression. J Psychiatry. (2019) 32:454–7. doi: 10.3969/j.issn.2095-9346.2019.06.013
69. Zhao Y, Tan ZL, Jiang XY. Study on the expression of endoplasmic reticulum stress-related genes and microRNA-16, microRNA-124 and microRNA-195 in peripheral blood of depressive patients. Chin J Gen Pract. (2019) 17:619–21. doi: 10.16766/j.cnki.issn.1674-4152.000750
70. Kong LM, Yao GF, He MJ, Zhu XL, Zhang LY. Effects of antidepressants on suicide in depressive patients and its association with expression level of miRNA in peripheral blood mononuclear cells. J Prev Med Chin PLA. (2019) 37:9–15. doi: 10.13704/j.cnki.jyyx.2019.07.005
71. Meng JH, Zhao Y, Liang P, Yang XN, Cao Y, Yang Y, et al. Effect of duloxetine on serum microRNA and cytokine levels in depressive patients. J China Prescription Drug. (2020) 18:110–2.
72. Fu KD, Li SH, Hu YY, Chen ME. Effects of high-frequency repetitive transcranial magnetic stimulation on serum levels of mir-124, mir-214 and inflammatory factors in patients with severe depression. China Modern Doctor. (2020) 58:103–6.
73. Cao YC, Lv R, Wang XQ. Expression of miR-132, miR-182 and their relationship with neurotrophic factor in elderly patients with depression. Pract Geriatr. (2020) 34:1146–9. doi: 10.3969/j.issn.1003-9198.2020.11.011
74. Liang JQ, Liao HR, Xu CX, Li XL, Wei ZX, Xie GJ, et al. Serum exosome-derived miR-139-5p as a potential biomarker for major depressive disorder. Neuropsychiatr Dis Treat. (2020) 16:2689–93. doi: 10.2147/ndt.s277392
75. Xu XH, Yu ZH, Li J, Li Y, Wang SQ, Yan P, et al. Expression levels of miRNA132 in depression patients and chronic unpredictable mild stress rats. Acta Academiae Medicinae Sinicae. (2020) 42:573–7. doi: 10.3881/j.issn.1000-503X.12958
76. Qian SX, Fang Y, Sun L, Chou Q, Lin ZG, Xiao SF, et al. Effect of citalopram on miRNA-16/serotonin transporter pathway in peripheral blood of patients with depression. J Shanghai Jiaotong Univ. (2020) 40:815–819+814. doi: 10.3969/j.issn.1674-8115.2020.06.017
77. Wei ZX, Xie GJ, Mao X, Zou XP, Liao YJ, Liu QS, et al. Exosomes from patients with major depression cause depressive-like behaviors in mice with involvement of miR-139-5p-regulated neurogenesis. Neuropsychopharmacology. (2020) 45:1050–8. doi: 10.1038/s41386-020-0622-2
78. Ding Y, Zhong M, Qiu B, Liu C, Wang J, Liang J. Abnormal expression of miR-135a in patients with depression and its possible involvement in the pathogenesis of the condition. Exp Ther Med. (2021) 22:726. doi: 10.3892/etm.2021.10158
79. Al-Rawaf HA, Alghadir AH, Gabr SA. Circulating microRNAs and molecular oxidative stress in older adults with neuroprogression disorders. Dis Markers. (2021) 2021:4409212. doi: 10.1155/2021/4409212
80. Hung YY, Chou CK, Yang YC, Fu HC, Loh EW, Kang HY. Exosomal let-7e, miR-21-5p, miR-145, miR-146a and miR-155 in Predicting Antidepressants Response in Patients with Major Depressive Disorder. Biomedicines. (2021) 9:1428. doi: 10.3390/biomedicines9101428
81. Liu Z, Yang J, Fang Q, Shao H, Yang D, Sun J, et al. MiRNA-199a-5p targets WNT2 to regulate depression through the CREB/BDNF signaling in hippocampal neuron. Brain Behav. (2021) 11:e02107. doi: 10.1002/brb3.2107
82. He C, Bai Y, Wang Z, Fan D, Wang Q, Liu X, et al. Identification of microRNA-9 linking the effects of childhood maltreatment on depression using amygdala connectivity. Neuroimage. (2021) 224:117428. doi: 10.1016/j.neuroimage.2020.117428
83. Roumans S, Sundquist K, Memon AA, Hedelius A, Sundquist J, Wang X. Association of circulating let-7b-5p with major depressive disorder: a nested case-control study. BMC Psychiatry. (2021) 21:616. doi: 10.1186/s12888-021-03621-4
84. Zhao Y, Mi QR, Wang BH, Ren ZY, Kong LP, Zhao HW, et al. Clinical significance of detecting miRNA-26a/b expression in peripheral blood of patients with depression. Contemp Med Symposium. (2021) 19:44–5. doi: 10.3969/j.issn.2095-7629.2021.16.024
85. Xian X, Cai LL, Li Y, Wang RC, Xu YH, Chen YJ, et al. Neuron secrete exosomes containing miR-9-5p to promote polarization of M1 microglia in depression. J Nanobiotechnology. (2022) 20:122. doi: 10.1186/s12951-022-01332-w
86. Lin H, Xie YX, Li QR, Lin WC, Zhong SM. Correlation Analysis of Serum miR-135a and miR-221 Expression Levels with Cognitive Function, Event-Related Potential P300 and Inflammatory Cytokines in Patients With Depression. Prog Modern Biomedicine. (2022) 22:173–6. doi: 10.13241/j.cnki.pmb.2022.01.033
87. Brás JP, Pinto S, von Doellinger O, Prata J, Coelho R, Barbosa MA, et al. Combining inflammatory miRNA molecules as diagnostic biomarkers for depression: a clinical study. Front Psychiatry. (2023) 14:1227618. doi: 10.3389/fpsyt.2023.1227618
88. Deng Y, Gong P, Han S, Zhang J, Zhang S, Zhang B, et al. Reduced cerebral cortex thickness is related to overexpression of exosomal miR-146a-5p in medication-free patients with major depressive disorder. Psychol Med. (2023) 53:6253–60. doi: 10.1017/s0033291722003567
89. Wu X, Zhang Y, Wang P, Li X, Song Z, Wei C, et al. Clinical and preclinical evaluation of miR-144-5p as a key target for major depressive disorder. CNS Neurosci Ther. (2023) 29:3598–611. doi: 10.1111/cns.14291
90. Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. (2011) 45:92–5. doi: 10.1016/j.jpsychires.2010.04.028
91. Xu D, Rong H, Liu TB, Zhao J, Lai WT. Expressions research of microRNA-134 and LIMK-1 before and after treatment in the plasma of bipolar mania. China Prac Med. (2018) 13:34–6. doi: 10.14163/j.cnki.11-5547/r.2018.20.015
92. Camkurt MA, Karababa İF, Erdal ME, Kandemir SB, Fries GR, Bayazıt H, et al. MicroRNA dysregulation in manic and euthymic patients with bipolar disorder. J Affect Disord. (2020) 261:84–90. doi: 10.1016/j.jad.2019.09.060
93. Tekdemir R, Selvi Y, Altınbaş K, Koçak N. Decreased miR-15b-5p/miR-155-5p levels and increased miR-134-5p/miR-652-3p levels among BD patients under lithium treatment. J Affect Disord. (2022) 317:6–14. doi: 10.1016/j.jad.2022.08.034
94. Roy B, Yoshino Y, Allen L, Prall K, Schell G, Dwivedi Y. Exploiting circulating microRNAs as biomarkers in psychiatric disorders. Mol Diagn Ther. (2020) 24:279–98. doi: 10.1007/s40291-020-00464-9
95. Tsermpini EE, Kalogirou CI, Kyriakopoulos GC, Patrinos GP, Stathopoulos C. miRNAs as potential diagnostic biomarkers and pharmacogenomic indicators in psychiatric disorders. Pharmacogenomics J. (2022) 22:211–22. doi: 10.1038/s41397-022-00283-7
96. Zhang HC, Du Y, Chen L, Yuan ZQ, Cheng Y. MicroRNA schizophrenia: Etiology, biomarkers and therapeutic targets. Neurosci Biobehav Rev. (2023) 146:105064. doi: 10.1016/j.neubiorev.2023.105064
97. Jin M, Zhu X, Sun Y, Li Z, Li X, Ai L, et al. Identification of peripheral blood miRNA biomarkers in first-episode drug-free schizophrenia patients using bioinformatics strategy. Mol Neurobiol. (2022) 59:4730–46. doi: 10.1007/s12035-022-02878-4
98. Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. (2013) 6:39. doi: 10.3389/fnmol.2013.00039
99. Yu H-R, Hsu T-Y, Huang H-C, Kuo H-C, Li S-C, Yang KD, et al. Comparison of the functional microRNA expression in immune cell subsets of neonates and adults. Front Immunol. (2016) 7:615. doi: 10.3389/fimmu.2016.00615
100. Pfeffer S, Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, et al. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PloS One. (2011) 6:e24145. doi: 10.1371/journal.pone.0024145
101. Han S, Li Y, Gao J. Peripheral blood MicroRNAs as biomarkers of schizophrenia: expectations from a meta-analysis that combines deep learning methods. World J Biol Psychiatry. (2023) 25:65–81. doi: 10.1080/15622975.2023.2258975
102. Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. (2008) 17:1156–68. doi: 10.1093/hmg/ddn005
103. Eastwood SL, McDonald B, Burnet PWJ, Beckwith JP, Kerwin RW, Harrison PF. Decreased expression of mRNAs encoding non-NMDA glutamate receptors GluR1 and GluR2 in medial temporal lobe neurons in schizophrenia. Brain Res Mol Brain Res. (1995) 29:211–23. doi: 10.1016/0169-328X(94)00247-C
104. Bernstein HG, Braunewell KH, Spilker C, Danos P, Baumann B, Funke S, et al. Hippocampal expression of the calcium sensor protein visinin-like protein-1 in schizophrenia. Neuroreport. (2002) 13:393. doi: 10.1097/00001756-200203250-00006
105. Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol. (2010) 4:10. doi: 10.1186/1752-0509-4-10
106. Yu Y, Lin Y, Takasaki Y, Wang C, Kimura H, Xing J, et al. Rare loss of function mutations in N-methyl-d-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Trans Psychiatry. (2018) 8:12. doi: 10.1038/s41398-017-0061-y
107. Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. (2010) 15:1176–89. doi: 10.1038/mp.2009.84
108. Nieto R, Kukuljan M, Silva H. BDNF and schizophrenia: from neurodevelopment to neuronal plasticity, learning, and memory. Front Psychiatry. (2013) 4:45. doi: 10.3389/fpsyt.2013.00045
109. Mellios N, Huang H-S, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. (2008) 17:3030–42. doi: 10.1093/hmg/ddn201
110. Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. (2009) 65:1006–14. doi: 10.1016/j.biopsych.2008.11.019
111. Xu Y, Li F, Zhang B, Zhang K, Zhang F, Huang X, et al. MicroRNAs and target site screening reveals a pre-microRNA-30e variant associated with schizophrenia. Schizophr Res. (2010) 119:219–27. doi: 10.1016/j.schres.2010.02.1070
112. Xu Y, Liu Z, Song XI, Zhang K, Li X, Li J, et al. Cerebralcare Granule® attenuates cognitive impairment in rats continuously overexpressing microRNA-30e. Mol Med Rep. (2015) 12:8032–40. doi: 10.3892/mmr.2015.4469
113. Chow TJ, Tee SF, Loh SY, Yong HS, Abu Bakar AK, Tang PY. Analysis of variants of AKT1 in schizophrenia multiplex families. Asian J Psychiatr. (2020) 49:101957. doi: 10.1016/j.ajp.2020.101957
114. Zhang J, Sun XY, Zhang LY. MicroRNA-7/Shank3 axis involved in schizophrenia pathogenesis. J Clin Neurosci. (2015) 22:1254–7. doi: 10.1016/j.jocn.2015.01.031
115. Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. (2012) 40:4742–53. doi: 10.1093/nar/gks151
116. Hauberg ME, Holm-Nielsen MH, Mattheisen M, Askou AL, Grove J, Børglum AD, et al. Schizophrenia risk variants affecting microRNA function and site-specific regulation of NT5C2 by miR-206. Eur Neuropsychopharmacol. (2016) 26:1522–6. doi: 10.1016/j.euroneuro.2016.06.014
117. Siedlecki-Wullich D, Català-Solsona J, Fábregas C, Hernández I, Clarimon J, Lleó A, et al. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimers Res Ther. (2019) 11:46. doi: 10.1186/s13195-019-0501-4
118. Mahmoudi E, Cairns MJ. MiR-137: an important player in neural development and neoplastic transformation. Mol Psychiatry. (2017) 22:44–55. doi: 10.1038/mp.2016.150
119. Wright C, Calhoun VD, Ehrlich S, Wang L, Turner JA, Bizzozero NIP. Meta gene set enrichment analyses link miR-137-regulated pathways with schizophrenia risk. Front Genet. (2015) 6:147. doi: 10.3389/fgene.2015.00147
120. Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. (2013) 18:11–2. doi: 10.1038/mp.2011.170
121. Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. (2006) 439:283–9. doi: 10.1038/nature04367
122. Gao J, Wang W-Y, Mao Y-W, Gräff J, Guan J-S, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. (2010) 466:1105–9. doi: 10.1038/nature09271
123. Scarr E, Craig JM, Cairns MJ, Seo MS, Galati JC, Beveridge NJ, et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl Psychiatry. (2013) 3:e230. doi: 10.1038/tp.2013.3
124. Dean B, Bakker G, Ueda HR, Tobin AB, Brown A, Kanaan RAA. A growing understanding of the role of muscarinic receptors in the molecular pathology and treatment of schizophrenia. Front Cell Neurosci. (2023) 17:1124333. doi: 10.3389/fncel.2023.1124333
125. Kaurani L, Islam MR, Heilbronner U, Krüger DM, Zhou J, Methi A, et al. Regulation of Zbp1 by miR-99b-5p in microglia controls the development of schizophrenia-like symptoms in mice. EMBO J. (2024) 43:1420–44. doi: 10.1038/s44318-024-00067-8
126. Li W, Li X, Li Y, Chen Y, Zhu L, Guo R. Diagnostic value of MicroRNAs for depression: A systematic review and meta-analysis. J Psychiatr Res. (2023) 157:132–40. doi: 10.1016/j.jpsychires.2022.11.028
127. Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, et al. Characterization of Small RNAs in Aplysia Reveals a Role for miR-124 in Constraining Synaptic Plasticity through CREB. Neuron. (2009) 63:803–17. doi: 10.1016/j.neuron.2009.05.029
128. Bahi A, Chandrasekar V, Dreyer JL. Selective lentiviral-mediated suppression of microRNA124a in the hippocampus evokes antidepressants-like effects in rats. Psychoneuroendocrinology. (2014) 46:78–87. doi: 10.1016/j.psyneuen.2014.04.009
129. Carniel BP, da Rocha NS. Brain-derived neurotrophic factor (BDNF) and inflammatory markers: Perspectives for the management of depression. Prog Neuropsychopharmacol Biol Psychiatry. (2021) 108:110151. doi: 10.1016/j.pnpbp.2020.110151
130. Huang Y, Jiang J, Zheng GUO, Chen J, Lu H, Guo HU, et al. miR-139-5p modulates cortical neuronal migration by targeting Lis1 in a rat model of focal cortical dysplasia. Int J Mol Med. (2014) 33:1407–14. doi: 10.3892/ijmm.2014.1703
131. Roser A-E, Caldi Gomes L, Halder R, Jain G, Maass F, Tönges L, et al. miR-182-5p and miR-183-5p act as GDNF mimics in dopaminergic midbrain neurons. Mol Ther Nucleic Acids. (2018) 11:9–22. doi: 10.1016/j.omtn.2018.01.005
132. Gao L, S-b Y, Yang J, Kong J-l, Shi K, Ma F-c, et al. MiR-182-5p and its target HOXA9 in non-small cell lung cancer: a clinical and in-silico exploration with the combination of RT-qPCR, miRNA-seq and miRNA-chip. BMC Med Genomics. (2020) 13:3. doi: 10.1186/s12920-019-0648-7
133. Du X, Wei J, Tian D, Wu M, Yan C, Hu P, et al. miR-182-5p contributes to intestinal injury in a murine model of Staphylococcus aureus pneumonia-induced sepsis via targeting surfactant protein D. J Cell Physiol. (2019) 235:563–72. doi: 10.1002/jcp.28995
134. Saus E, Soria V, Escaramís G, Vivarelli F, Crespo JM, Kagerbauer B, et al. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. (2010) 19:4017–25. doi: 10.1093/hmg/ddq316
135. Mendlewicz J. Disruption of the circadian timing systems. CNS Drugs. (2009) 23:15–26. doi: 10.2165/11318630-000000000-00000
136. Cui Y, Ma G, Kong F, Song L. Diagnostic values of miR-221-3p in serum and cerebrospinal fluid for post-stroke depression and analysis of risk factors. Iran J Public Health. (2021) 50:1241–9. doi: 10.18502/ijph.v50i6.6423
137. Lang Y, Li Y, Yu H, Lin L, Chen X, Wang S, et al. HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson's disease by elevating NPTX2 via miR-221-3p binding. Aging (Albany NY). (2020) 12:7660–78. doi: 10.18632/aging.103028
138. Sun Q, Zeng QC, Chen YQ, Zhang M, Wei LL, Chen P. Long intergenic noncoding RNA p21 suppresses the apoptosis of hippocampus neurons in streptozotocin-diabetic mice by sponging microRNA-221 through upregulation of FOS. J Cell Physiol. (2019) 234:21113–25. doi: 10.1002/jcp.28714
139. Lian N, Niu Q, Lei Y, Li X, Li Y, Song X. MiR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell Cycle. (2018) 17:2745–55. doi: 10.1080/15384101.2018.1556060
140. Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, et al. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun. (2014) 5:3263. doi: 10.1038/ncomms4263
141. van Battum EY, Verhagen MG, Vangoor VR, Fujita Y, Derijck AAHA, O'Duibhir E, et al. An Image-Based miRNA Screen Identifies miRNA-135s As Regulators of CNS Axon Growth and Regeneration by Targeting Krüppel-like Factor 4. J Neurosci. (2018) 38:613–30. doi: 10.1523/jneurosci.0662-17.2017
142. Valiuliene G, Valiulis V, Zentelyte A, Dapsys K, Germanavicius A, Navakauskiene R. Anti-neuroinflammatory microRNA-146a-5p as a potential biomarker for neuronavigation-guided rTMS therapy success in medication resistant depression disorder. BioMed Pharmacother. (2023) 166:115313. doi: 10.1016/j.biopha.2023.115313
143. Liu W, Ge T, Leng Y, Pan Z, Fan J, Yang W, et al. The role of neural plasticity in depression: from hippocampus to prefrontal cortex. Neural Plasticity. (2017) 2017:6871089. doi: 10.1155/2017/6871089
144. Voineskos D, Rogasch NC, Rajji TK, Fitzgerald PB, Daskalakis ZJ. A review of evidence linking disrupted neural plasticity to schizophrenia. Can J Psychiatry. (2013) 58:86–92. doi: 10.1177/070674371305800205
145. Mouillet-Richard S, Baudry A, Launay JM, Kellermann O. MicroRNAs and depression. Neurobiol Dis. (2012) 46:272–8. doi: 10.1016/j.nbd.2011.12.035
146. Zhang R, Huang M, Cao Z, Qi J, Qiu Z, Chiang L-Y. MeCP2 Plays an Analgesic Role in Pain Transmission through Regulating CREB / miR-132 Pathway. Mol Pain. (2015) 11:19. doi: 10.1186/s12990-015-0015-4
147. Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U.S.A. (2005) 102:16426–31. doi: 10.1073/pnas.0508448102
148. Su M, Hong J, Zhao Y, Liu S, Xue X. MeCP2 controls hippocampal brain-derived neurotrophic factor expression via homeostatic interactions with microRNA−132 in rats with depression. Mol Med Rep. (2015) 12:5399–406. doi: 10.3892/mmr.2015.4104
149. Pillai A, Kale A, Joshi S, Naphade N, Raju MSVK, Nasrallah H, et al. Decreased BDNF levels in CSF of drug-naive first-episode psychotic subjects: correlation with plasma BDNF and psychopathology. Int J Neuropsychopharmacol. (2009) 13:535. doi: 10.1017/s1461145709991015
150. Chen C-H, Cheng M-C, Huang A, Hu T-M, Ping L-Y, Chang Y-S. Detection of rare methyl-cpG binding protein 2 gene missense mutations in patients with schizophrenia. Front Genet. (2020) 11:476. doi: 10.3389/fgene.2020.00476
151. Wong EHM, So H-C, Li M, Wang Q, Butler AW, Paul B, et al. Common variants on xq28 conferring risk of schizophrenia in han Chinese. Schizophr Bull. (2014) 40:777–86. doi: 10.1093/schbul/sbt104
152. Hashimoto K, Ohi K, Hashimoto R, Nakazawa T, Okada T, Yasuda Y, et al. The p250GAP gene is associated with risk for schizophrenia and schizotypal personality traits. PloS One. (2012) 7:e35696. doi: 10.1371/journal.pone.0035696
153. Qi S, Yang X, Zhao L, Calhoun VD, Perrone-Bizzozero N, Liu S, et al. MicroRNA132 associated multimodal neuroimaging patterns in unmedicated major depressive disorder. Brain. (2018) 141:916–26. doi: 10.1093/brain/awx366
154. Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U.S.A. (2008) 105:13421–6. doi: 10.1073/pnas.0801613105
155. Li X, Khanna A, Li N, Wang E. Circulatory miR-34a as an RNA-based, noninvasive biomarker for brain aging. Aging. (2011) 3:985–1002. doi: 10.18632/aging.v3i10
156. Bhatnagar S, Chertkow H, Schipper HM, Yuan Z, Shetty V, Jenkins S, et al. Increased microRNA-34c abundance in Alzheimer's disease circulating blood plasma. Front Mol Neurosci. (2014) 7:2. doi: 10.3389/fnmol.2014.00002
157. Palta P, Samuel LJ, Miller ER, Szanton SL. Depression and oxidative stress. Psychosomatic Med. (2014) 76:12–9. doi: 10.1097/psy.0000000000000009
158. Ermakov EA, Dmitrieva EM, Parshukova DA, Kazantseva DV, Vasilieva AR, Smirnova LP, et al. Oxidative stress-related mechanisms in schizophrenia pathogenesis and new treatment perspectives. Oxid Med Cell Longev. (2021) 2021:1–37. doi: 10.1155/2021/8881770
159. Herskovits AZ, Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. (2014) 81:471–83. doi: 10.1016/j.neuron.2014.01.028
160. Kishi T, Fukuo Y, Kitajima T, Okochi T, Yamanouchi Y, Kinoshita Y, et al. SIRT1 gene, schizophrenia and bipolar disorder in the Japanese population: an association study. Genes Brain Behav. (2011) 10:257–63. doi: 10.1111/gbb.2011.10.issue-3
161. Kishi T, Yoshimura R, Kitajima T, Okochi T, Okumura T, Tsunoka T, et al. SIRT1 gene is associated with major depressive disorder in the Japanese population. J Affect Disord. (2010) 126:167–73. doi: 10.1016/j.jad.2010.04.003
162. Xu Y, Chen P, Wang X, Yao J, Zhuang S. miR-34a deficiency in APP/PS1 mice promotes cognitive function by increasing synaptic plasticity via AMPA and NMDA receptors. Neurosci Lett. (2018) 670:94–104. doi: 10.1016/j.neulet.2018.01.045
163. Khoodoruth MAS, Estudillo-Guerra MA, Pacheco-Barrios K, Nyundo A, Chapa-Koloffon G, Ouanes S. Glutamatergic system in depression and its role in neuromodulatory techniques optimization. Front Psychiatry. (2022) 13:886918. doi: 10.3389/fpsyt.2022.886918
164. Snyder MA, Gao WJ. NMDA receptor hypofunction for schizophrenia revisited: Perspectives from epigenetic mechanisms. Schizophr Res. (2020) 217:60–70. doi: 10.1016/j.schres.2019.03.010
165. Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. (2007) 8:413–26. doi: 10.1038/nrn2153
166. Buonanno A. The neuregulin signaling pathway and schizophrenia: from genes to synapses and neural circuits. Brain Res Bull. (2010) 83:122–31. doi: 10.1016/j.brainresbull.2010.07.012
167. Gu S, Cui F, Yin J, Fang C, Liu L. Altered mRNA expression levels of autophagy- and apoptosis-related genes in the FOXO pathway in schizophrenia patients treated with olanzapine. Neurosci Lett. (2021) 746:135669. doi: 10.1016/j.neulet.2021.135669
168. Li X, Zhao J, Li Z, Zhang L, Huo Z. Applications of acupuncture therapy in modulating the plasticity of neurodegenerative disease and depression: do microRNA and neurotrophin BDNF shed light on the underlying mechanism? Neural Plast. (2020) 2020:8850653. doi: 10.1155/2020/8850653
169. Fiori LM, Kos A, Lin R, Théroux JF, Lopez JP, Kühne C, et al. miR-323a regulates ERBB4 and is involved in depression. Mol Psychiatry. (2021) 26:4191–204. doi: 10.1038/s41380-020-00953-7
170. Rana T, Behl T, Sehgal A, Mehta V, Singh S, Sharma N, et al. Elucidating the possible role of foxO in depression. Neurochem Res. (2021) 46:2761–75. doi: 10.1007/s11064-021-03364-4
171. Wang Z, Li P, Wu T, Zhu S, Deng L, Cui G. Axon guidance pathway genes are associated with schizophrenia risk. Exp Ther Med. (2018) 16:4519–26. doi: 10.3892/etm.2018.6781
172. Moghbeli M, Zangouei AS, Nasrpour Navaii Z, Taghehchian N. Molecular mechanisms of the microRNA-132 during tumor progressions. Cancer Cell Int. (2021) 21:439. doi: 10.1186/s12935-021-02149-7
173. Enriquez-Barreto L, Morales M. The PI3K signaling pathway as a pharmacological target in Autism related disorders and Schizophrenia. Mol Cell Ther. (2016) 4:2. doi: 10.1186/s40591-016-0047-9
174. Emamian ES. AKT/GSK3 signaling pathway and schizophrenia. Front Mol Neurosci. (2012) 5:33. doi: 10.3389/fnmol.2012.00033
175. Matsuda S, Ikeda Y, Murakami M, Nakagawa Y, Tsuji A, Kitagishi Y. Roles of PI3K/AKT/GSK3 pathway involved in psychiatric illnesses. Diseases. (2019) 7:22. doi: 10.3390/diseases7010022
176. Karege F, Perroud N, Burkhardt S, Schwald M, Ballmann E, La Harpe R, et al. Alteration in kinase activity but not in protein levels of protein kinase B and glycogen synthase kinase-3β in ventral prefrontal cortex of depressed suicide victims. Biol Psychiatry. (2007) 61:240–5. doi: 10.1016/j.biopsych.2006.04.036
177. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. (2012) 12:613–26. doi: 10.1038/nrc3318
178. Catts VS, Catts SV. Apoptosis and schizophrenia: is the tumour suppressor gene, p53, a candidate susceptibility gene? Schizophr Res. (2000) 41:405–15. doi: 10.1016/s0920-9964(99)00077-8
Keywords: psychiatric disorders, microRNAs, biomarkers, systematic review, meta-analysis
Citation: Liu X, Dong L, Jiang Z, Song M and Yan P (2024) Identifying the differentially expressed peripheral blood microRNAs in psychiatric disorders: a systematic review and meta-analysis. Front. Psychiatry 15:1390366. doi: 10.3389/fpsyt.2024.1390366
Received: 28 February 2024; Accepted: 25 April 2024;
Published: 17 May 2024.
Edited by:
Stefania Schiavone, University of Foggia, ItalyReviewed by:
Luisella Bocchio Chiavetto, eCampus University, ItalyCopyright © 2024 Liu, Dong, Jiang, Song and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pan Yan, Ynp5eHl5YW5wYW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.