
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry , 13 May 2022
Sec. Behavioral and Psychiatric Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fpsyt.2022.857574
This article is part of the Research Topic The Genetics and Epigenetics of Mental Health View all 8 articles
Obsessive-compulsive disorder (OCD) is a deliberating disorder with complex genetic and environmental etiologies. Hypotheses about OCD mainly include dysregulated neurotransmitters, especially serotonin, and disturbed neurodevelopment. Single nucleotide polymorphism (SNP) association studies regarding OCD are often met with inconsistent results. However, stratification by age of onset may sometimes help to limit the heterogenicity of OCD patients. Therefore, we conducted a stratified SNP association study enrolling 636 patients and 612 healthy controls. Patients were stratified by age of onset as early-onset (EO-OCD) and late-onset (LO-OCD). Blood extracted from the patients was used to genotype 18 loci, including serotonin system genes, Slitrk1, Slitrk5, and DMRT2 and related miRNA genes. Logistic regression was used to compare allele and genotype frequencies of variants. A general linear model was used to evaluate the association between variants and trait anxiety. In our study, rs3824419 in DMRT2 was associated with EO-OCD, G allele was the risk allele. Rs2222722 in miR-30a-5p was associated with EO-OCD, with the C allele being the risk allele. Rs1000952 in HTR3D was found associated with trait anxiety in OCD patients. The significance disappeared after FDR correction. Our results supported neurodevelopment-related genes, DMRT2 and miR-30a-5p, to be related to EO-OCD. However, we cannot prove serotonin genes to be directly associated with EO-OCD. While an association between HTR3D and trait anxiety was discovered, comparisons based on biological or clinical traits may be helpful in future studies. As our detective powers were limited, more large-scale studies will be needed to confirm our conclusion.
Obsessive-compulsive disorder (OCD) is one of the most common mental disorders, with a prevalence of 2–3% worldwide (1), and 2.4% in China (2). Genetic and environmental factors are both involved in the etiology and pathogenesis with the heritability as high as 26–45% (3). It is characterized by recurrent intrusive thoughts (obsessions) and corresponding repetitive behavior (compulsions), which may be explained by the chaos of neurotransmitters (1).
Due to the early finding that OCD patients respond to selective serotonin reuptake inhibitors (SSRIs), most of the etiological studies in OCD focused on serotonin-related systems. Numerous studies were conducted on serotonin-related candidate genes, including serotonin receptors (HTR), serotonin transporter (SERT, or SLC6A4, or 5-HTT), tryptophan hydroxylase 2 (TPH2), and so on. Meta-analyses showed that serotonin 2a receptor (HTR2A) and serotonin transporter-linked polymorphic region (5-HTTLPR) got the most evidence in single nucleotide polymorphisms (SNPs) studies in OCD (4, 5). However, the results of genetic association studies exploring other serotonin-related genes were inconsistent.
After stratified analysis by age of onset, some clearer clues appeared. Early-onset OCD (EO-OCD), as a putative subtype of OCD, is increasingly regarded to have unique biological characteristics (6). Given its higher heritability and distinct comorbidity patterns, EO-OCD was more suggested to be a neurodevelopmental disorder caused by the perturbation in neurodevelopment processes than late-onset OCD (LO-OCD) (7). Neurodevelopment processes include axon guidance and dendrite development prenatally, as well as synaptic plasticity postnatally.
Brain-derived neurotrophic factor (BDNF) is one of the most studied biomarkers based on the neurodevelopmental hypothesis. It is a protein involved in the formation of the nervous system, playing a key role in cortisol development and synaptic plasticity (8). It may interact with the neurotransmitter systems (9), underlying the pathogenesis of a wide range of mental disorders. Altered levels of BDNF were observed in OCD. But results were different between studies in adults and children, while BDNF levels decreased in adult OCD patients and increased in children with OCD (10–13). These clues suggested that there are likely some complex links between BDNF and OCD. SNP rs6265 (or Val66Met), which was the most studied SNP in BDNF, is the only known functional SNP in the BDNF gene. Its relationship with OCD was uncertain. Meta-analyses of previous studies detected no or ethnicity-specific weak association between OCD and rs6265 (14, 15). Homogenous samples are needed for further certification.
Slitrks (SLIT and NTRK like family members), comprising a series of neurogenesis-related proteins including Slitrk1-6, are also promising candidate genes in OCD. All of them were highly expressed in the human brain, regulating neuronal outgrowth, neuronal survival, and synapse formation (16). Slitrk1 was discovered to be related to Tourette syndrome (16). Tourette syndrome is a neurodevelopmental disorder that may show some overlap with OCD in pathogenesis. Slitrk5 was also thought to be related to OCD. A study, which showed that slitrk5 knock-out mice manifested OCD-like phenotypes, suggested strongly that Slitrk5 may be involved in the onset of OCD (17).
DMRTs (doublesex and mab-3-related transcription factors), including DMRT1-8, were also potential candidate genes. They play a conserved role in sexual development and other developmental processes like neural development (18). According to current literature, DMRTs were essential genes for the central nervous system during early embryonic development, playing critical roles in brain patterning, corticogenesis, and other neurogenesis processes (19, 20). Previous studies suggested a putative gene in distal 9p associated with OCD (21, 22). DMRT2 is right located in the DMRT1-DMRT3-DMRT2 gene cluster in 9p24.3, the most susceptive location. In mice, it was detected to mostly express in adult testis and brain (23) but its expression condition and its polymorphisms in the OCD population have not yet been studied.
MiRNAs are non-coding RNAs regulating gene expression post-transcriptionally via mRNA degradation or transcription inhibition. MiRNAs are increasingly getting attention in mental disorders because they regulate up to 60% of protein-coding genes (24) and are highly expressed in the developing central nervous system (25). Since its function of orchestrating genetic spatiotemporal expression in the transitional processes during neurodevelopment, it provides a different perspective to elucidate the complex role of neurotransmitter chaos in mental disorders. Also, it was found to be related to several neurodevelopmental diseases such as Autism Spectrum Disorder (ASD) and schizophrenia for its continuous effect on the process of neurogenesis and synaptic plasticity (26, 27). A few studies revealed the value of circulating miRNAs supposed to be biomarkers of OCD (28, 29). Evidence has shown that miRNA-directed regulation in behavioral disorders can be affected by SNPs (30). Nevertheless, miRNA gene polymorphisms in OCD had not yet been reported.
In this study, we chose candidate genes, including serotonin-related genes (HTR1B, HTR3A/B/D/E, SERT, TPH2), BDNF, DMRT2, Slitrk1, and Slitrk5, in order to verify and explore the SNP association study in OCD. To elucidate the potential influence of MiRNA, we selected MiRNA which regulates these candidate genes. The selected MiRNA were hsa-miR-96 (HTR1B), hsa-mir-497 (HTR2A), hsa-mir-544a (HTR3D), hsa-mir-544b (HTR3D), hsa-mir-195 (BDNF), and hsa-mir-30a-5p (BDNF)1. To better demonstrate the research landscape of our chosen SNPs, literature about these SNPs in human sapiens is presented in Supplementary Table 1.
Samples from 636 OCD patients and 612 healthy controls were collected from the biobank of Shanghai Mental Health Center (31). All the patients were diagnosed by a psychiatrist according to DSM-IV in the Chinese Han population. The Mini-International Neuropsychiatric Interview (M.I.N.I.) was used to check the diagnosis (32). Those OCD participants were included if they met the following criteria: (1) met the DSM-IV diagnostic threshold OCD; (2) had a Yale-Brown Obsessive-compulsive Scale (Y-BOCS) total score ≥16 (33); (3) were between 18 and 64 years old; (4) had at least an elementary school education; (5) were in sufficient health to complete the research. Exclusion criteria include: (1) being pregnant; (2) being previously or contemporarily diagnosed as other mental disorders (e.g., affective disorder, schizophrenia); (3) having a history of intellectual disability or other neurological disorder. (4) having a severe physical health condition. (5) tic disorder.
All the patients underwent clinical assessment before blood extraction employing (1) the Yale-Brown Obsessive Compulsive Scale (Y-BOCS) to evaluate the severity of obsessive-compulsive symptoms (33); (2) the State–Trait Anxiety Inventory (STAI) including subscale State Anxiety Inventory (SAI) and Trait Anxiety Inventory (TAI) to evaluate state anxiety and trait anxiety (34); (3) the 24-item Hamilton Depression Rating Scale (HAMD24) to rate the severity of depression (35); (4) the Hamilton Anxiety Rating Scale (HAMA) to rate the severity of anxiety (36).
The study was approved by the ethics committee of Shanghai Mental Health Center. Written informed consents were obtained from all participants before enrollment.
Nine SNPs, namely rs1000952 in HTR3D, rs7627615 in HTR3E, rs13212041, and rs6296 in HTR1B, rs6265 in BDNF, rs1176744 in HTR3B, rs1062613 in HTR3A, rs4570625 in TPH2, and rs1042173 in SERT(SLC6A4) were selected as they were SNPs reported to be related to OCD with mixed results or related to other psychiatric disorders, putative as risk SNP in OCD.
Other SNPs were selected according to the following steps. We selected SNPs in Slitrk1, Slitrk5, DMRT2 that presented: (1) MAF ≥0.10; (2) located within 5 kb to the 5'UTR/3'UTR of the target gene; (3) r2 >0.8 in the HapMap database (https://www.ncbi.nlm.nih.gov/snp/) with Chinese Han origin in the Beijing population. As none of the SNPs were reported before, one or two SNP in each gene was selected and rs3824419, rs17641078, rs9531519, rs9582391, in total four SNPs were selected from the preliminary results.
Candidate SNPs of miRNAs, including has-mir-544b(HTR3D), hsa-mir-30a-5p(BDNF), has-miR-96(HTR1B), has-mir-544a(HTR3D), hsa-mir-497 (HTR2A)/hsa-mir-195 (BDNF), were filtered in National Center for Biotechnology Information (NCBI) variation database (http://www.ncbi.nlm.nih.gov/variation/) according to: (1) located within 5 kb to the 5'UTR of target gene; (2) 1,000 Genomes MAF ≥ 0.05. As none of SNPs were reported before, one SNP in each gene was selected and finally rs10934682 in hsa-mir-544b, rs2222722 in hsa-mir-30a-5p, rs4421293 in hsa-miR-96, rs10144193 in hsa-mir-544a, rs78312845 in hsa-mir-497/hsa-mir-195, totally five SNPs were selected from preliminary results.
Genomic DNA was extracted from leukocytes in venous blood using the QiaAmp® Isolation system (Qiagen, Basel, Switzerland) with the guidance of standard protocol. SNP genotyping was performed on the MassARRAY® SNP IPLEX platform (Agena Bioscience™, San Diego, CA, USA). Detailed information about primer design and polymerase chain reaction process is available upon request. Quality control was performed by excluding individual SNPs or samples with genotype call rates lower than 95%, as well as SNP assays with poor quality spectra or cluster plots. Ten percent of samples were randomly tested on the same platform and no inconsistency was found.
We used a two-tailed t-test, Mann–Whitney U-test, or Chi-square test to reveal the differences in demographic and clinical data. These results were presented as raw p-values. Hardy-Weinberg equilibrium (HWE) analyses were conducted using Haploview v4.2 (http://www.broad.mit.edu/mpg/haploview). Those which deviated from HWE were ruled out in the following analysis. In the following analysis, the OCD group was stratified as EO-OCD and LO-OCD by age of onset. According to previous studies and our clinical practice (37, 38), the cutoff between the EO and LO groups was set at 18 years old. General linear model analysis was used to evaluate SNP effects on TAI scores (adjust for sex and age of onset). All the single-locus analyses were analyzed by logistic regression (sex as a covariate) analysis employing an additive model for genotype analysis. The above process was conducted using SPSS Statistics v26.0 (IBM Corp., Chicago, IL, USA). We applied FDR correction for multiple testing. α was set as 0.05. Linkage disequilibrium and haplotype analysis were conducted using the SHEsis calculator (39, 40). Generalized multifactor dimensionality reduction (GMDR) analysis following 5,000 permutation tests was conducted to explore the gene*gene or gene*gene*sex interaction. The flowchart of our work was shown in Figure 1.
The demographic and clinical characteristics of participants were presented in Table 1. Sex (p < 0.001) and age (p < 0.001) were different between OCD and control. Sex (p < 0.001), age (p < 0.001), age of onset (p < 0.001), course of disease (p < 0.001), HAMD24 (p = 0.001), HAMA (p = 0.003), and TAI (p = 0.006) was different between EO-OCD and LO-OCD. The characteristics of SNPs are summarized in Table 2. Four SNPs including rs6296, rs4570625, rs10144193, and rs1042173 deviated from HWE (p < 0.05) and were excluded from the following analyses.
The genotype frequencies and outcomes of logistic regression are presented in Supplementary Table 2. There was no statistical difference observed in genotypic or allelic frequencies between OCD and the control group. No significant gene*gene or gene*gene*sex model was detected in GMDR analysis after the permutation test (data not shown).
Data from EO-OCD was compared with the healthy control. The results were demonstrated in Supplementary Table 2.
A significant difference was found both in the allelic and genotypic frequency of rs3824419 (genotype association p = 0.011; allele association p = 0.007). However, all the differences disappeared after FDR correction.
With the significance of both allele and genotype analyses of rs3824419, we conducted further analysis on it, and the results are presented in Table 3. We observed a higher risk of G allele (OR = 1.375, 95%CI 1.089–1.736) and the increased risk of G carriers (CG, OR 1.341, 95%CI 0.925–1.943; GG, OR 1.935, 95% CI 1.256–2.982). Only the comparison in men manifested significance. These results are presented in Figure 2.
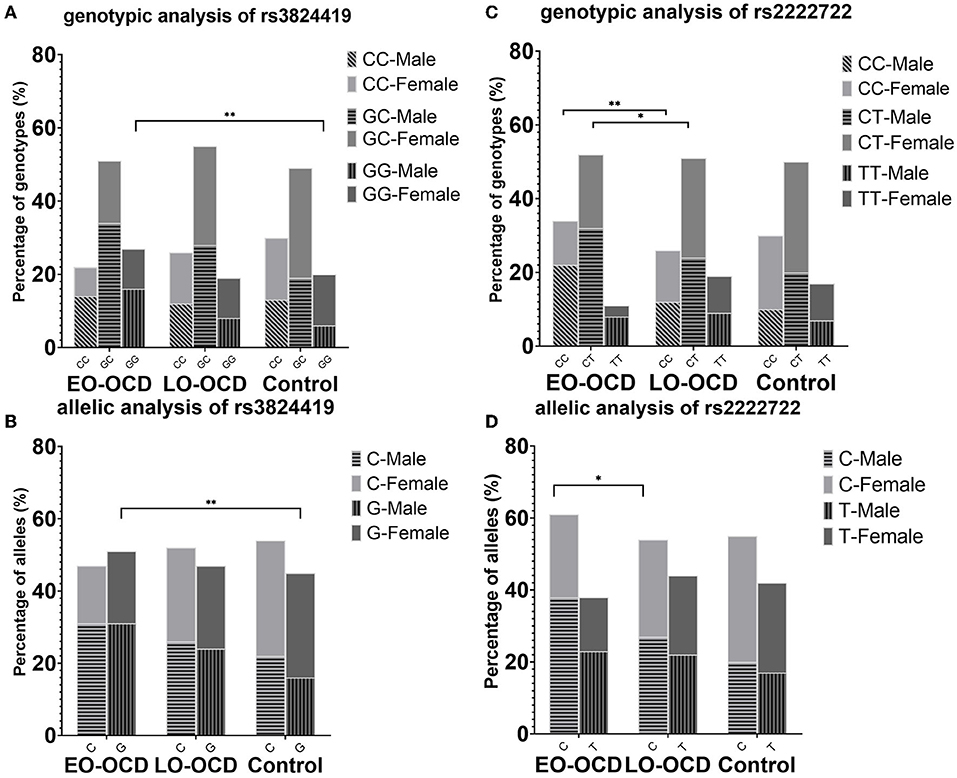
Figure 2. Genotype and allele association results of rs3824419 and rs2222722, adjusted and separately presented by sex. (A) Genotype association results of rs3824419. Between EO-OCD and control, GG homozygotes showed significantly higher risk. (B) Allele association results of rs3824419. Between EO-OCD and control, allelic distribution was significant difference and G allele showed higher risk. (C) Genotype association results of rs2222722. Between EO-OCD and LO-OCD, both CC homozygotes and CC heterozygotes showed significantly higher risk. (D) Allele association results of rs2222722. Between EO-OCD and LO-OCD, allelic distribution was significant difference and C allele showed higher risk.
In the following linkage disequilibrium analysis, rs3824419–rs17641078 showed different distribution between EO and control. The frequency of G-G is 0.523 in the EO group and 0.457 in the control (OR = 1.302, 95% CI 1.059–1.600) and that of C-G is 0.343 in the EO group and 0.412 in the control (OR = 0.744, 95%CI 0.601–0.922).
No significant gene*gene or gene*gene*sex model was detected (data not show).
Data from LO-OCD were compared with healthy control. The results were demonstrated in Supplementary Table 2.
No association of significance was observed in any analysis between LO and the control group (Supplementary Table 2). No significant gene*gene or gene*gene*sex model was detected (data not show).
Single locus analysis revealed rs2222722 in miR-30a-5p to distribute differently between EO-OCD and LO-OCD (genotype association p = 0.013, allele association p = 0.038), while rs9582391 in Slitrk1 have only marginal allele and genotype association. All the significance disappeared after FDR correction (Supplementary Table 2).
We conducted further analysis on rs2222722 for the co-exist of genotype and allele association. We detected a higher risk of C allele in allele association (OR = 1.296, 95%CI 1.014–1.657) and an increased risk of C carriers (CT, OR 1.749, 95%CI 1.081–2.830; CC, OR 2.161, 95% CI 1.292–3.614). Both comparisons in men and women were of no significance. However, both comparisons showed a rather large OR value (Table 3). These results are presented in Figure 2.
No significant gene*gene or gene*gene*sex model was detected (data not shown).
Comparison of TAI scores among genotype groups after adjusting for sex and age of onset were presented in Supplementary Table 2. Only rs1000952 in HTR3D demonstrated significance (p = 0.024), whereas the significance disappeared after FDR correction. The results were shown in Figure 3. In post-hoc analysis, patients carrying CC genotype presented higher TAI sore (60.68 ± 5.033) than CT (47.177 ± 0.695, Bonferroni's P = 0.03) or TT (47.740 ± 0.332, Bonferroni's P =0.018) while CT and TT were not different from each other (Bonferroni's P > 1). As OCD patients with CC genotype were extremely rare (n = 3), we did not make stratified analyses based on sex.
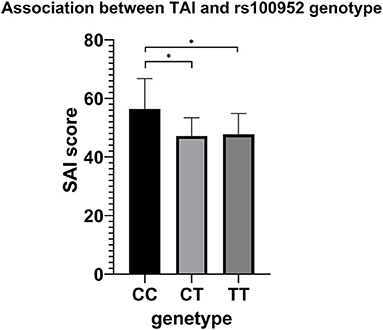
Figure 3. Association results between TAI and rs100952 genotype, adjusted by sex and age of onset. The general linear model revealed a significant difference (F = 3.744, p = 0.024). *p < 0.05 before FDR correction.
Our results revealed rs2222722 in miR-30a-5p to be related to EO-OCD for the first time. Though the significance disappeared after FDR correction. In contrast, none of the positive results were observed between the overall OCD group or LO-OCD group and the control group. Our results corroborated the genetic heterogenicity of OCD and emphasized the importance of stratification by age of onset. Results from women and men showed similar trends when stratified by sex and the association was more significant in men. The impact of our relatively insufficient sample size in women should be considered. The reason why there was relatively fewer women than men in the EO-OCD group may be that EO-OCD presented a higher prevalence in men for unknown reasons (7). In the future, a larger sample size will be needed in female EO-OCD patients.
Our study found the DMRT2 haplotype (rs3824419–rs17641078) in strong linkage disequilibrium and its distribution was significantly different between EO-OCD and the control group. Both rs3824419 and rs17641078 are missense variants of the DMRT2 gene. The association analysis showed rs3824419 can well represent rs3824419-rs17641078 haplotype and it may be an important variant in DMRT2. To date, DMRTs are most famous for sex determination, and brain-related studies targeting DMRTs mainly focused on animal experiments. Among DMRTs, DMRT3 and DMRT5 were found to be highly expressed in the central nervous system, encoding nuclear proteins and playing an important role in neurogenesis (41). Evidence of DMRT2 was limited in the early linkage studies and 9p deletion-related studies. As was observed, catastrophic deletion of DMRTs may lead to severe functional disorders like intellectual disability in 9p deletion patients, or lead to abnormality in the macro-structure of the brain in knock-out mice (19, 20). SNPs with minor effects may also affect the brain at a smaller scale prenatally. There were several studies reporting that ASD was found in 9p deletion patients (42, 43). In fact, ASD was a well-known neurodevelopmental disorder sharing a genetic basis with OCD (44). Concerning the previous studies reporting a putative gene in distal 9p associated with OCD (21, 22), we deduced that DMRTs may play a role in the OCD pathogenesis. However, the exact effect of genetic factors and how they interact with prenatal environment factors remains to be explored. It is becoming clearer that DMRTs contribute to multiple aspects of CNS development, the mechanisms and pathologic relationship with human disease are still poorly understood. What exactly is the role of DMRT2 and other DMRT genes, especially DMRT1/3/5, in the onset of OCD, remained unknown. Our results suggested this gene family to be promising etiological genes of OCD, awaiting confirmation and mechanism research in the future.
We did not observe the significance of rs6265, the only known functional SNP in BDNF. This result was in accordance with our previous study (45). Though rs6265 may influence BDNF level with certain mechanisms, previous meta-analyses detected no or only ethnicity-specific weak association (14, 15). The locus rs2222722 came out instead. It is located in the miR-30a-5p gene in 6q13, downgrading the expression level of BDNF. MiR-30a-5p was previously found to present distinct developmental and lamina-specific expression in the human prefrontal cortex (46). Researchers found that miR-30a-5p in the prefrontal cortex was related to alcohol consumption (47) and miR-30a-5p in the striatum was related to Huntington's disease (48). EO-OCD occurs in a period when the brain is in intense development including neurogenesis, increased synaptic formation, and increased dendritic pruning. The dynamical fluctuation of miRNAs in different regions throughout their lifetime makes them switches or fine tuners in brain remodeling and neural plasticity before the age of 18 (49). Concerning neuroimaging studies that suggest distinct patterns of subcortical abnormalities in heterogeneous OCD patients (50), there may be different expressions of BDNF regulated by miRNA in different regions. Exploration of miRNAs has made great progress in other neurodevelopmental disorders like schizophrenia, Tourette Syndrome, and ASD (51, 52), but they were poorly studied in OCD. In our study, though we detected the risk allele, the pathway from SNP to miR-30a-5p, to region-specific expression of BDNF, and finally to the onset of OCD, remains to be clarified. In the context of the negative results of functional variant rs6265, our results called for attention to the potential role of miRNA and post-transcriptionally regulation of BDNF.
In our study, rs9582391 in Slitrk5 manifested marginal significance before FDR correction, but rs9531519 in Slitrk1 did not. While evidence in animal models was implicating the promising outlook of Slitrks (16, 53, 54), studies in human were disproportionately inadequate. A newly-published study distinguished Slitrk5 as the most significant single-gene result via exome sequencing of 1313 OCD participants (55), shedding light on the promising outlook of Slitrks. As we tested only two loci in this family, important SNPs may be missed. We were yet unable to determine the potential role of Slitrks in the pathogenesis of OCD in our study. This is the first time these two loci were reported in OCD. More explorations are needed, perhaps toward other SNPs, copy number variants, or rare variants.
None of the serotonin-related SNPs was directly associated with OCD in our study, which may be due to the genetic complexity and phenotypic heterogeneity. On the one hand, though our analysis covered a wide range of common genes in the serotonin system, detected SNPs were limited in each gene. Important SNPs may be missed. On the other hand, a single gene may not play a vital role in OCD. For example, polymorphism of HTR1B was reported to be associated with a concentration of different neuro-metabolites in the anterior cingulate cortex in pediatric OCD patients (56), which indicates its relevance to the underlying pathology of OCD, but meta-analysis reported only a trend (4). While our analysis did not find any direct association between HTR1B and OCD, we conducted an analysis between related miRNA and OCD. Rs13212041 was a functional variant affecting miRNA-mediated HTR1B regulation in aggression-related behavior (30) and was associated with some neuropsychiatry characteristics (57) but its role in OCD has not yet been reported. Our result did not support its direct effect, either. There may be other important SNPs in the HTR1B system acting in other ways to affect the onset of OCD.
HTR3 genes were studied in very limited OCD studies. Rs1176144 in HTR3B was reported to be relevant to EO-OCD (58), rs7627615 in HTR3E, and rs1000952 in HTR3D were both reported to be associated with the washing phenotype of OCD (59), but we did not observe any significant direct association between them and OCD. Besides, our result on rs1062613 in HTR3A, a functional variant widely concerned to be associated with alcohol addiction, was in accordance with previous studies reporting a negative result on OCD (58, 60).
Trait anxiety is a personality trait presenting the predisposition of an individual to anxiety-related feelings or behaviors. It was found to correlate with reduced thickness in the medial orbitofrontal cortex in healthy volunteers (61). In OCD patients, trait anxiety may play a mediated role between OCD-related stress and functional disability (62). Though we did not find serotonin-related genes to be directly associated with OCD, we found rs1000952 in HTR3D related to trait anxiety and this SNP may exert influence on OCD through trait anxiety.
According to existing studies, comparisons based on specific bio-medical traits may be beneficial to limit the heterogeneity. A large-sample genetic association study made in a population-based, pediatric sample found 5-HTTLPR and HTR2A to be associated with rumination, one of the obsessive-compulsive trait symptoms (63), which also highlighted the importance of addressing symptom dimensions in OCD study. There were 14 subtypes of serotonin receptors identified, but it remained unclear whether and how each receptor affected OCD. Our analysis suggested an optional path to study the underlying pathology of serotonin receptors of OCD.
All the loci in our study were scarcely studied or never reported before in mental disorders. Though none of the results reached the FDR threshold, their different distribution in serotonin-related genes and neurodevelopment-related genes manifested an interesting trend. Besides the methodological reason, the underlying distinct neurobiochemical profiles are worthy of concern. Though serotonin disturbance is the most studied etiological theory in OCD, serotonin-related genes may cooperate with other neurotransmitter-related genes. Clinically, EO-OCD patients were thought to be less responsive to serotonin reuptake inhibitor therapy, requiring an augmentation treatment with antipsychotics (6). Neuroimaging data about serotonin transporter availability also suggested less serotonergic pathology in EO-OCD than LO-OCD (64), indicating more exploration from other perspectives. We were unable to decide the role of our chosen serotonin-related genes in OCD according to available data. However, neurogenesis-related genes, including BDNF-related genes, DMRTs, and Slitrks, may act in a more systematic way affecting considerable neurons and synapsis in neurotransmitter systems. Therefore, they may play a more vital role in the onset of EO-OCD with effects easier to detect.
Firstly, it was hard to avoid recall bias with regards to the age of onset under the retrospective background of our study. Secondly, as an exploratory analysis, this study covered a series of candidate genes but SNPs in each gene were limited, which may lead to the miss of important SNPs or haplotypes. Thirdly, due to the unavailable data, we did not make further analysis employing serum levels of biomarkers to draw the links between SNPs and neurochemicals. Fourthly, our power was limited in that the significance disappeared after FDR correction, though the study was conducted with a rather large sample size of 1,248 participants. As a result, confirmation with larger sample sizes was needed.
To sum up, we found rs3824419 in DMRT2 and rs2222722 in miR-30a-5p to be related to EO-OCD. Our results suggested the merits of neurodevelopment-related genes in EO-OCD, supporting the neurodevelopmental hypothesis of EO-OCD and suggesting a distinct treatment strategy for it. In contrast, we did not find evidence of serotonin-related SNPs directly associated with OCD. Correlation between HTR3D and trait anxiety was observed, suggesting comparisons based on specific bio-medical traits in the OCD study. Finally, as our detection power was limited, more large-scale studies are needed to confirm our conclusion. To sum up, we found DMRT2, miR-30a-5p to be related to EO-OCD. Our results suggested the merits of neurodevelopment-related genes in EO-OCD, supporting the neurodevelopmental hypothesis of EO-OCD and suggesting a distinct treatment strategy for it.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Ethics Committee of Shanghai Mental Health Center. The patients/participants provided their written informed consent to participate in this study.
MD is responsible for original draft, visualization, and data curation. YW is responsible for the review, editing, funding acquisition, and data curation. SY is responsible for methodology. JQ and QF are responsible for resources collecting. ZW is responsible for project administration and funding acquisition. ZX is responsible for conceptualization, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81971261), Hospital Project of Shanghai Mental Health Center (2019-YJ15), and the National Natural Science Foundation of China (82071518) in the sample collection and genotyping.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thanked for the substantial support from all the patients and healthy volunteers. Also, we thank every member in our group for exploring the mechanisms of OCD to better strategies for OCD treatment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2022.857574/full#supplementary-material
1. ^The gene name in the following bracket is the corresponding gene thought to be regulated by this miRNA.
1. Stein DJ, Costa DLC, Lochner C, Miguel EC, Reddy YCJ, Shavitt RG, et al. Obsessive-compulsive disorder. Nat Rev Dis Primers. (2019) 5:52. doi: 10.1038/s41572-019-0102-3
2. Huang Y, Wang Y, Wang H, Liu Z, Yu X, Yan J, et al. Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry. (2019) 6:211–24. doi: 10.1016/S2215-0366(18)30511-X
3. Smoller JW, Andreassen OA, Edenberg HJ, Faraone SV, Glatt SJ, Kendler KS. Psychiatric genetics and the structure of psychopathology. Mol Psychiatry. (2019) 24:409–20. doi: 10.1038/s41380-017-0010-4
4. Taylor S. Molecular genetics of obsessive–compulsive disorder: a comprehensive meta-analysis of genetic association studies. Mol Psychiatry. (2012) 18:799–805. doi: 10.1038/mp.2012.76
5. Mattina GF, Samaan Z, Hall GB, Steiner M. The association of HTR2A polymorphisms with obsessive-compulsive disorder and its subtypes: A meta-analysis. J Affect Disord. (2020) 275:278–89. doi: 10.1016/j.jad.2020.06.016
6. Grassi G, Cecchelli C, Mazzocato G, Vignozzi L. Early onset obsessive-compulsive disorder: the biological and clinical phenotype. CNS Spectr. (2021) 1:1–7. doi: 10.1017/S1092852921000122 [Epub ahead of print].
7. Burchi E, Pallanti S. Diagnostic issues in early-onset obsessive-compulsive disorder and their treatment implications. Curr Neuropharmacol. (2019) 17:672–80. doi: 10.2174/1570159X16666180426151746
8. Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. (2001) 24:677–736. doi: 10.1146/annurev.neuro.24.1.677
9. Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. (2006) 311:864–8. doi: 10.1126/science.1120972
10. Bilgic A, Sertdemir M, Kilinc I, Akca OF. Increased serum brain-derived neurotrophic factor and adrenocorticotropic hormone levels are associated with obsessive compulsive disorder in medicationfree children. Eur Child Adolesc Psychiatry. (2021) 31:325–35. doi: 10.1007/s00787-020-01690-6
11. Maina G, Rosso G, Zanardini R, Bogetto F, Gennarelli M, Bocchio-Chiavetto L. Serum levels of brain-derived neurotrophic factor in drug-naive obsessive-compulsive patients: a case-control study. J Affect Disord. (2010) 122:174–8. doi: 10.1016/j.jad.2009.07.009
12. Simşek S, Gençoglan S, Yüksel T, Kaplan I, Alaca R. Cortisol and brain-derived neurotrophic factor levels prior to treatment in children with obsessive-compulsive disorder. J Clin Psychiatr. (2016) 77:e855–9. doi: 10.4088/JCP.15m10146
13. Wang Y, Mathews CA, Li Y, Lin Z, Xiao Z. Brain-derived neurotrophic factor (BDNF) plasma levels in drug-naive OCD patients are lower than those in healthy people, but are not lower than those in drug-treated OCD patients. J Affect Disord. (2011) 133:305–10. doi: 10.1016/j.jad.2011.04.002
14. Wang J, Zhang F, Zhu W, Liu Y, Zhou Z. Meta-analysis of the association of brain-derived neurotrophic factor Val66Met polymorphism with obsessive-compulsive disorder. Acta Neuropsychiatr. (2015) 27:327–35. doi: 10.1017/neu.2015.38
15. Wang S, Xu X, Yan P, Song M, Li J, Wang S. Is brain-derived neurotrophic factor (bdnf) val66met polymorphism associated with obsessive-compulsive disorder? A meta-analysis. Psychiatr Danub. (2019) 31:141–7. doi: 10.24869/psyd.2019.141
16. Proenca CC, Gao KP, Shmelkov SV, Rafii S, Lee FS. Slitrks as emerging candidate genes involved in neuropsychiatric disorders. Trends Neurosci. (2011) 34:143–53. doi: 10.1016/j.tins.2011.01.001
17. Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. (2010) 16:598–602. doi: 10.1038/nm.2125
18. Bellefroid EJ, Leclere L, Saulnier A, Keruzore M, Sirakov M, Vervoort M, et al. Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell Mol Life Sci. (2013) 70:3829–45. doi: 10.1007/s00018-013-1288-2
19. Kikkawa T, Osumi N. Multiple functions of the dmrt genes in the development of the central nervous system. Front Neurosci. (2021) 15:789583. doi: 10.3389/fnins.2021.789583
20. Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev Biol. (2007) 310:1–9. doi: 10.1016/j.ydbio.2007.07.035
21. Willour VL, Yao Shugart Y, Samuels J, Grados M, Cullen B, Bienvenu OJ, 3rd, et al. Replication study supports evidence for linkage to 9p24 in obsessive-compulsive disorder. Am J Hum Genet. (2004) 75:508–13. doi: 10.1086/423899
22. Hanna GL, Veenstra-VanderWeele J, Cox NJ, Boehnke M, Himle JA, Curtis GC, et al. Genome-wide linkage analysis of families with obsessive-compulsive disorder ascertained through pediatric probands. Am J Med Genet. (2002) 114:541–52. doi: 10.1002/ajmg.10519
23. Kim S, Kettlewell JR, Anderson RC, Bardwell VJ, Zarkower D. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Express. Patt. (2003) 3:77–82. doi: 10.1016/s1567-133x(02)00071-6
24. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
25. Juvale IIA, Chehas AT. The potential role of miRNAs as predictive biomarkers in neurodevelopmental disorders. J Mol Neurosci. (2021) 71:1356. doi: 10.1007/s12031-021-01854-2
26. Sun E, Shi Y. MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Exp Neurol. (2015) 268:46–53. doi: 10.1016/j.expneurol.2014.08.005
27. Hicks SD, Middleton FA. A comparative review of microRNA expression patterns in autism spectrum disorder. Front Psychiatry. (2016) 7:176. doi: 10.3389/fpsyt.2016.00176
28. Yue J, Zhang B, Wang H, Hou X, Chen X, Cheng M, et al. Dysregulated plasma levels of miRNA-132 and miRNA-134 in patients with obsessive-compulsive disorder. Br J Med Psychol. (2020) 8:996. doi: 10.21037/atm-20-5217
29. Kandemir H, Erdal ME, Selek S, Izci Ay O, Karababa IF, Ay ME, et al. Microribonucleic acid dysregulations in children and adolescents with obsessive-compulsive disorder. Neuropsychiatr Dis Treat. (2015) 11:1695–701. doi: 10.2147/NDT.S81884
30. Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. (2009) 14:381–9. doi: 10.1038/mp.2008.15
31. The biobank unit of shanghai mental health center. Biopreserv Biobank. (2011) 9:136–7. doi: 10.1089/bio.2011.9225
32. Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatr. (1998) 59:22–33.
33. Goodman WK, Price LH, Rasmussen SA, Mazure C, Fleischmann RL, Hill CL, et al. The yale-brown obsessive compulsive scale: i. development, use, and reliability. Arch Gen Psychiatry. (1989) 46:1006–11.
34. Spielberger CD, Gorsuch RL. Manual for the State-Trait Anxiety Inventory (STAI : Form Y). Palo Alto, CA: Consulting Psychologists Press (1983).
35. Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol. (1967) 6:278–96.
37. Huang X, Liu J, Cong J, Zhang X. Association between the slc1a1 glutamate transporter gene and obsessive-compulsive disorder in the chinese han population. Neuropsychiatr Dis Treat. (2021) 17:347–54. doi: 10.2147/NDT.S281623
38. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. (2005) 15:97–8. doi: 10.1038/sj.cr.7290272
39. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
40. Li ZT, Tan SZ, Lyu ZH, Zou LQ. Olfactory identification impairment in early-and late-onset obsessive-compulsive disorder. Early Interv Psychiatry. (2022) 16:133–38. doi: 10.1111/eip.13136
41. Konno D, Kishida C, Maehara K, Ohkawa Y, Kiyonari H, Okada S, et al. Dmrt factors determine the positional information of cerebral cortical progenitors via differential suppression of homeobox genes. Development. (2019) 146:dev174243. doi: 10.1242/dev.174243
42. Ounap K, Uibo O, Zordania R, Kiho L, Ilus T, Oiglane-Shlik E, et al. Three patients with 9p deletions including DMRT1 and DMRT2: a girl with XY complement, bilateral ovotestes, and extreme growth retardation, and two XX females with normal pubertal development. Am J Med Genet A. (2004) 130A:415–23. doi: 10.1002/ajmg.a.30269
43. Vinci G, Chantot-Bastaraud S, El Houate B, Lortat-Jacob S, Brauner R, McElreavey K. Association of deletion 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Mol Human Reprod. (2007) 13:685–9. doi: 10.1093/molehr/gam045
44. O'Connell KS, McGregor NW, Lochner C, Emsley R, Warnich L. The genetic architecture of schizophrenia, bipolar disorder, obsessive-compulsive disorder and autism spectrum disorder. Mol Cell Neurosci. (2018) 88:300–7. doi: 10.1016/j.mcn.2018.02.010
45. Wang Y, Zhang H, Li Y, Wang Z, Fan Q, Yu S, et al. BDNF Val66Met polymorphism and plasma levels in Chinese Han population with obsessive-compulsive disorder and generalized anxiety disorder. J Affect Disord. (2015) 186:7–12. doi: 10.1016/j.jad.2015.07.023
46. Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. (2008) 17:3030–42. doi: 10.1093/hmg/ddn201
47. Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. (2015) 20:1219–31. doi: 10.1038/mp.2014.120
48. Müller S. In silico analysis of regulatory networks underlines the role of miR-10b-5p and its target BDNF in huntington's disease. Transl Neurodegener. (2014) 3:17. doi: 10.1186/2047-9158-3-17
49. Rao YS, Pak TR. microRNAs and the adolescent brain: Filling the knowledge gap. Neurosci Biobehav Rev. (2016) 70:313–22. doi: 10.1016/j.neubiorev.2016.06.008
50. Boedhoe PS, Schmaal L, Abe Y, Ameis SH, Arnold PD, Batistuzzo MC, et al. Distinct subcortical volume alterations in pediatric and adult OCD: A worldwide meta- and mega-analysis. Am J Psychiatry. (2017) 174:60–9. doi: 10.1176/appi.ajp.2016.16020201
51. Paul S, Reyes PR, Garza BS, Sharma A. MicroRNAs and child neuropsychiatric disorders: A brief review. Neurochem Res. (2020) 45:232–40. doi: 10.1007/s11064-019-02917-y
52. Smigielski L, Jagannath V, Rossler W, Walitza S, Grunblatt E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol Psychiatry. (2020) 25:1718–48. doi: 10.1038/s41380-019-0601-3
53. Song M, Giza J, Proenca CC, Jing D, Elliott M, Dincheva I, et al. Slitrk5 mediates BDNF-dependent trkb receptor trafficking and signaling. Dev Cell. (2015) 33:690–702. doi: 10.1016/j.devcel.2015.04.009
54. Kang H, Han KA, Won SY, Kim HM, Lee YH, Ko J, et al. Slitrk missense mutations associated with neuropsychiatric disorders distinctively impair slitrk trafficking and synapse formation. Front Mol Neurosci. (2016) 9:104. doi: 10.3389/fnmol.2016.00104
55. Halvorsen M, Samuels J, Wang Y, Greenberg BD, Fyer AJ, McCracken JT, et al. Exome sequencing in obsessive-compulsive disorder reveals a burden of rare damaging coding variants. Nat Neurosci. (2021) 24:1071–6. doi: 10.1038/s41593-021-00876-8
56. Ortiz AE, Gasso P, Mas S, Falcon C, Bargallo N, Lafuente A, et al. Association between genetic variants of serotonergic and glutamatergic pathways and the concentration of neurometabolites of the anterior cingulate cortex in paediatric patients with obsessive-compulsive disorder. World J Biol Psychiatry. (2016) 17:394–404. doi: 10.3109/15622975.2015.1111524
57. Varga G, Szekely A, Antal P, Sarkozy P, Nemoda Z, Demetrovics Z, et al. Additive effects of serotonergic and dopaminergic polymorphisms on trait impulsivity. Am J Med Genet B Neuropsychiatr Genet. (2012) 159B:281–8. doi: 10.1002/ajmg.b.32025
58. Kim HW, Kang JI, Lee SH, An SK, Sohn SY, Hwang EH, et al. Common variants of HTR3 genes are associated with obsessive-compulsive disorder and its phenotypic expression. Sci Rep. (2016) 6:32564. doi: 10.1038/srep32564
59. Lennertz L, Wagner M, Grabe HJ, Franke PE, Guttenthaler V, Rampacher F, et al. 5-HT3 receptor influences the washing phenotype and visual organization in obsessive-compulsive disorder supporting 5-HT3 receptor antagonists as novel treatment option. Eur Neuropsychopharmacol. (2014) 24:86–94. doi: 10.1016/j.euroneuro.2013.07.003
60. Mössner R, Döring N, Scherag A, Schäfer H, Herpertz-Dahlmann B, Remschmidt H, et al. Transmission disequilibrium analysis of the functional 5-HT3A receptor variant C178T in early-onset obsessive—compulsive disorder. J Psychopharm. (2007) 21:833–6. doi: 10.1177/0269881106073560
61. Kuhn S, Schubert F, Gallinat J. Structural correlates of trait anxiety: reduced thickness in medial orbitofrontal cortex accompanied by volume increase in nucleus accumbens. J Affect Disord. (2011) 134:315–9. doi: 10.1016/j.jad.2011.06.003
62. Storch EA, Abramowitz JS, Keeley M. Correlates and mediators of functional disability in obsessive-compulsive disorder. Depress Anxiety. (2009) 26:806–13. doi: 10.1002/da.20481
63. Sinopoli VM, Erdman L, Burton CL, Park LS, Dupuis A, Shan J, et al. Serotonin system genes and obsessive-compulsive trait dimensions in a population-based, pediatric sample: a genetic association study. J Child Psychol Psychiatry. (2019) 60:1289–99. doi: 10.1111/jcpp.13079
Keywords: single locus polymorphism, obsessive-compulsive disorder, BDNF, DMRT, serotonin
Citation: Deng M, Wang Y, Yu S, Fan Q, Qiu J, Wang Z and Xiao Z (2022) Exploring Association Between Serotonin and Neurogenesis Related Genes in Obsessive-Compulsive Disorder in Chinese Han People: Promising Association Between DMRT2, miR-30a-5p, and Early-Onset Patients. Front. Psychiatry 13:857574. doi: 10.3389/fpsyt.2022.857574
Received: 18 January 2022; Accepted: 04 April 2022;
Published: 13 May 2022.
Edited by:
Qiang Wang, Sichuan University, ChinaCopyright © 2022 Deng, Wang, Yu, Fan, Qiu, Wang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Wang, d2FuZ3poZW5Ac21oYy5vcmcuY24=; Zeping Xiao, eGlhb3plcGluZzg4QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.