
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry , 05 October 2021
Sec. Molecular Psychiatry
Volume 12 - 2021 | https://doi.org/10.3389/fpsyt.2021.715481
 LiHua Cui1†
LiHua Cui1† WenRan Du2†
WenRan Du2† Ning Xu1
Ning Xu1 JingYi Dong2
JingYi Dong2 BingJie Xia3
BingJie Xia3 JingYi Ma1
JingYi Ma1 RuoTong Yan1
RuoTong Yan1 LanYing Wang2
LanYing Wang2 FuMin Feng1*
FuMin Feng1*Background: This study aimed to explore the main effects of environmental risk factors as well as their interaction effects with miRNA on the risk of autism spectrum disorder (ASD).
Methods: One hundred fifty-nine ASD children (ASD group) and 159 healthy children (control group), aged 2–6 years, were included in this study. ASD diagnoses were based on DSM-5 criteria. The extensive medical and demographic characterization of the two groups were recorded. MicroRNAs (miRNAs) in serum were detected by qRT-PCR.
Results: Compared with the control group, the ASD group had significantly higher rates of maternal stress during pregnancy (p < 0.001), maternal drinking during pregnancy (p = 0.006), threatened abortion (p = 0.011), pregnancy-induced hypertension (p = 0.032), gestational diabetes (p = 0.039), maternal anemia during pregnancy (p < 0.001), umbilical cord knot (p < 0.001), neonatal jaundice (p < 0.001), family psychiatric history (p = 0.001), and much lower birth weight (p = 0.012). Furthermore, the ASD group had much lower expression levels of hsa-miR-181b-5p (p < 0.001) and hsa-miR-320a (p < 0.001) and significantly higher levels of hsa-miR-19b-3p (p < 0.001). The interactions of hsa-miR-320a and maternal stress during pregnancy (OR = 39.42, p < 0.001), hsa-miR-19b-3p and neonatal jaundice (OR = 2.44, p < 0.001), and hsa-miR-181b-5p and family psychiatric history (OR = 8.65, p = 0.001) could increase ASD risk.
Conclusions: The dysregulation of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p could interact with environmental factors, such as maternal stress during pregnancy, neonatal jaundice, and family psychiatric history, to impact the risk of ASD.
As a heterogeneous brain-based neurodevelopmental disorder, autism spectrum disorder (ASD) is characterized by a continuum of deficits in communication, social interaction, behavior, and restricted interests (1, 2). It is reported that the annual incidence of ASD is between 1 and 3% (3). The prevalence of ASD is sex imbalance with a distribution of three males to one female (4, 5). ASD often impairs social skills and autonomy, causing ASD children with difficulty in social, speech, and behavioral skill development.
The etiological factors of ASD remain largely unknown. However, it has been reported that risk factors, such as genetics, environmental factors, prenatal and perinatal factors, are involved in the development of ASD (6). Among these, genetics occupies the main factor. ASD has a complex genetic background (7). Approximately 10% of ASD patients are reported to have an identifiable genetic cause (8). The characteristics of highly genetic heterogeneity made the pathophysiology of ASD really elusive (4). MicroRNAs (miRNAs) are short noncoding RNAs with 18–25 nucleotides, playing an important role in regulating gene expression of ASD patients (9). Previously, miRNAs were known to be essential for normal brain development and function, making them attractive biomarker candidates for central nervous system disorders (2). It is reported that miRNAs are also closely associated with the pathogenesis of ASD (2). MicroRNAs, such as miR-146a, miR-19b, miR-181b, hsa-miR-320a, and miR-107 in brain tissues, serum, and/or saliva, could be used as diagnostic biomarkers of ASD (10).
Environmental factors are also important for the etiology of ASD. According to several previous studies, environmental factors, including drugs, toxic exposures, parental age, nutrition, and fetal environment, make up 40–50% of variance in ASD liability (11). For example, it is revealed by a meta-analysis of 27 studies that parental age is associated with the risk of ASD in children (12). Two meta-analyses focus on the associations between ASD risk and obstetric factors; they find that factors such as umbilical cord complications, maternal hemorrhage, low birth weight, and genital malformation were associated with the risk of ASD (13, 14).
A recent study shows that the interaction of genetic and environmental risk factors could exacerbate ASD symptoms (15), suggesting that gene–environment interaction may be a potential mechanism to reveal the etiology of ASD. At present, few studies focus on the interaction of miRNAs and environmental risk factors in children with ASD. Thus, we conducted this study to explore the main effects of miRNAs as well as their interaction effects with well-replicated ASD environmental risk factors on the risk of ASD.
This multicenter, cross-sectional study included ASD and healthy children, aged 2–6 years. The recruitment occurred between June 2018 and June 2020 at Tangshan Maternal and Child Health Care Hospital and two Special Training Centers. Children were diagnosed as ASD according to the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) criteria (16) assessed by two developmental pediatricians with at least 10 years of experience. The inclusion criteria for ASD children (ASD group) were clinical diagnosis of ASD and the absence of other medical, neurological, genetic, or metabolic condition. Healthy children for the same period in Tangshan Maternal and Child Health Care Hospital child health care department without any history of ASD were enrolled as the control group.
This study was approved by the ethical committee of North China University of Science and Technology, and written informed consent was obtained from the parents.
For all subjects in the two groups, extensive medical and demographic characteristics were collected from existing medical records, including sex, age, paternal/maternal age, maternal stress during pregnancy (the psychosocial stress of pregnant woman), maternal smoking during pregnancy, maternal drinking during pregnancy, toxic exposure during pregnancy, threatened abortion (threatened abortion was diagnosed when vaginal bleeding with or without abdominal pain occurred before 28 weeks of gestation), premature birth (premature birth was defined as the birth occurring after 28 weeks and before 37 weeks of gestation), pregnancy-induced hypertension, gestational diabetes, maternal anemia during pregnancy, multivitamin intake during pregnancy, and family psychiatric history. Autistic traits have been screened with the Childhood Autism Rating Scale (CARS) (17): a (raw) score of ≥30 indicates the probability of an ASD.
Blood samples were collected from all children in a nonfasting state; 4 mL of peripheral venous blood was collected and centrifuged at 3,000 rpm for 10 min at 4°C to separate the pellet and serum. The serum was stored at −80°C until analysis.
The TRIZOL reagent (Invitrogen, USA) was used to isolate serum total RNA. The miRcute miRNA cDNA First-Strand Synthesis Kit (Tiangen, China) was used to synthesize the cDNA. The expressions of hsa-miR-181b-5p, hsa-miR-19b-3p, and hsa-miR-320a were detected by SYBR® Premix Ex Taq TM II (Takara, Japan). The PCR upstream primers were shown as follows: hsa-miR-181b-5p, 5′-AACAUUCAUUGCUGUCGGUGGGU-3′; hsa-miR-19b-3p, 5′-UGUGCAAAUCCAUGCAAAACUGA-3′; hsa-miR-320a, 5′-AAAAGCUGGGUUGAGAGGGCGA-3′; U6, 5′-GCAAGGATGACACGCCAAT-3′. The downstream primers were mRQ 3′ universal primers provided by the reverse transcription kit. The primers were synthesized by Shanghai Shenggong Bioengineering Co., LTD. The qRT-PCR was run on ABI StepOne Plus (Applied Biosystems, CA, USA) using a two-step PCR protocol: an initial denaturation step at 95°C for 10 min, followed by 40 cycles with a denaturation step at 95°C for 2 min and an annealing/extension step at 60°C for 60 s. The miRNA expression was calculated using the 2−ΔΔCt method.
SPSS 22.0 (IBM Corp., NY, USA) was used to analyze the data. Demographic characteristics were compared between the ASD and control groups using t-test (two-tailed) for normal distribution continuous variables, Kruskal–Wallis rank test for nonnormal distribution continuous variables, or χ2 test for categorical variables when appropriate. The comparison of miRNA expression between the two groups also used two-tailed t-tests. Multivariable logistic regression was used to assess possible risk factors for ASD. If the variables exhibited statistically significant contributions shown by the likelihood ratio test, they could be included in the model. P < 0.05 were considered statistically significant.
A total of 318 children aged 2–6 years were recruited to this study. Of them, 159 children with ASD were assigned to the ASD group, and the remaining 159 healthy children were enrolled as controls. The demographic characteristics of the ASD and control groups are shown in Table 1. Compared with the control group, the ASD group had much higher scores of CARS (p < 0.001), significantly higher rates of maternal stress during pregnancy (p < 0.001), maternal drinking during pregnancy (p = 0.006), threatened abortion (p = 0.011), pregnancy-induced hypertension (p = 0.032), gestational diabetes (p = 0.039), maternal anemia during pregnancy (p < 0.001), umbilical cord knot (p < 0.001), neonatal jaundice (p < 0.001), family psychiatric history (p = 0.001), and much lower birth weight (p = 0.012).
The relative expressions of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p in the ASD and control groups were detected by qRT-PCR. As shown in Figure 1, the ASD group had much lower expression levels of hsa-miR-181b-5p (0.774 ± 0.131 vs. 0.991 ± 0.037, p < 0.001) and hsa-miR-320a (0.805 ± 0.121 vs. 1.010 ± 0.043, p < 0.001) and a significantly higher level of hsa-miR-19b-3p (1.502 ± 0.413 vs. 1.004 ± 0.044, p < 0.001) compared with the control group.
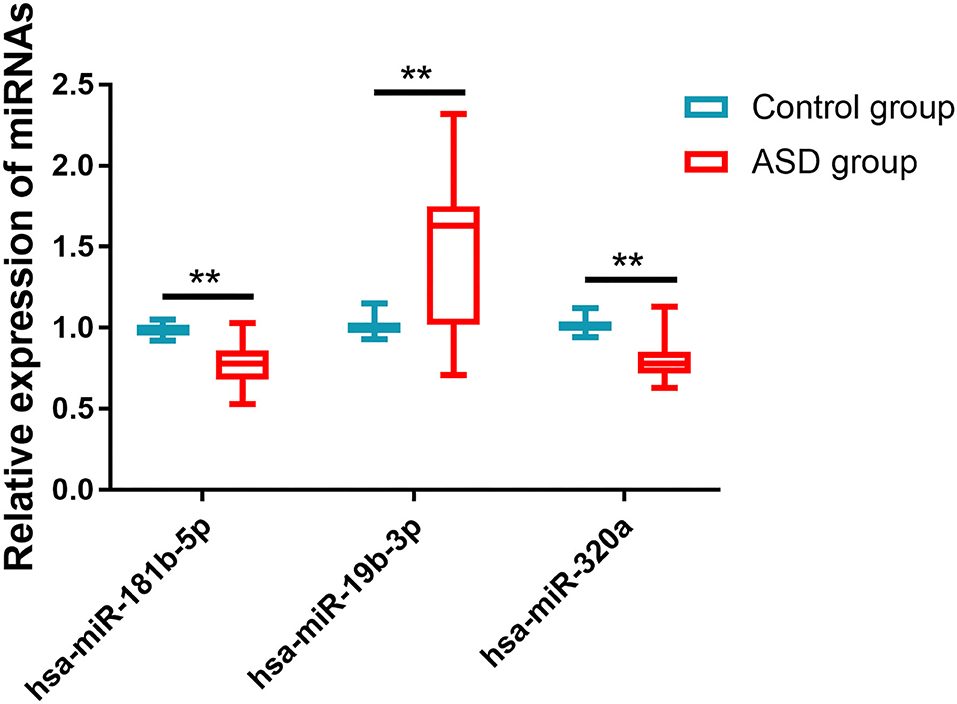
Figure 1. The expressions of miRNAs in two groups. Compared with the control group, the ASD group had much lower expression levels of hsa-miR-181b-5p and hsa-miR-320a and significantly higher levels of hsa-miR-19b-3p. **p < 0.001 vs. control group.
Based on the results of univariate analysis, risk factors, including maternal stress during pregnancy, maternal drinking during pregnancy, threatened abortion, pregnancy-induced hypertension, gestational diabetes, maternal anemia during pregnancy, umbilical cord knot, neonatal jaundice, much lower birth weight and family psychiatric history and their interactions with hsa-miR-181b-5p, hsa-miR-320a, or hsa-miR-19b-3p were included in the multivariable logistic regression model. As shown in Table 2, the expressions of has-miR-181b-5p (OR = 0.002, 95% CI: 0–0.014; p < 0.001) and hsa-miR-320a (OR = 0.001, 95% CI: 0–0.005; p < 0.001) decreased the ASD risk. Some environmental factors could interact with miRNAs and alter the effect of miRNAs on ASD. Having maternal stress during pregnancy was associated with hsa-miR-320a expression with 39.42-fold increased odds of ASD risk (OR = 39.42, 95% CI: 6.07–255.84; p < 0.001), having neonatal jaundice was associated with hsa-miR-19b-3p expression with a 2.44-fold increase (OR = 2.44, 95% CI: 1.55–3.83; p < 0.001), and having a family psychiatric history was associated with hsa-miR-181b-5p expression with a 8.65-fold increase (OR = 8.65, 95% CI: 2.23–33.58; p = 0.001).
In this study, we enrolled 318 ASD and healthy children to explore the main effects of miRNAs as well as their interaction effects with well-replicated ASD environmental risk factors on the risk of ASD. We find that the dysregulation of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p could interact with environmental factors to impact the risk of ASD.
Previous studies demonstrate that ASD is a heritable disorder involving multiple gene networks (5). MicroRNAs could influence gene expression, playing important roles in neurodevelopment. It is reported that miRNAs could influence neurogenesis and synaptogenesis and participate in ASD pathogenesis, serving as the biomarkers of ASD (5). MicroRNAs in salivary or serum show high accuracies to differentiate control and ASD subjects (18, 19). Several miRNAs, including miR-181b-5p, miR-320a, miR-19b-3p, miR-106b, miR-140, and miR-199b are regarded as candidates to identify ASD (5). In the study conducted by Mundalil et al. (19), miR-181b-5p and miR-320a were downregulated, and miR-19b-3p was upregulated in ASD patients compared with controls. Due to the results of Mundalil's study, miR-181b-5p, miR-320a, and miR-19b-3p were chosen in this study to detect the interaction effects of miRNAs and well-replicated ASD environmental risk factors on the risk of ASD. Our results are in line with Mundalil's findings. We find that the serum levels of hsa-miR-181b-5p and hsa-miR-320a in ASD children were much lower than those in healthy controls, and the serum levels of hsa-miR-19b-3p in ASD children were much higher. The possible molecular mechanisms to underly miRNA upregulation or downregulation in ASD are still being investigated. A review published previously reveals that the location of specific miRNAs at copy number variant (CNV) loci in ASD may lead to their dysregulation (5). Another potential mechanism is that individual miRNA sequences are altered in ASD children (20).
Previously investigated environmental risk factors for ASD include paternal and maternal age, fetal environment, perinatal and obstetric events (e.g., hypoxia), smoking and alcohol use, nutrition, and toxic exposures (21). In this study, we find that the ASD group had significantly higher rates of maternal stress during pregnancy, maternal drinking during pregnancy, threatened abortion, pregnancy-induced hypertension, gestational diabetes, maternal anemia during pregnancy, umbilical cord knot, neonatal jaundice, family psychiatric history, and much lower birth weight compared with the control group. Parental or maternal age is a well-established risk factor for ASD (21). Unfortunately, in the present study, parental, and maternal age were not significantly different between the two groups. Lacking a large enough sample size may contribute to this result.
Gene–environment interaction is an emerging hypothesis to expound the increased incidence of ASD (22). MicroRNAs may be one of the factors to explain the gene–environment interaction. Hicks et al. (2) find that salivary miRNAs were associated with environmental factors to affect the risk of ASD. Nakata et al. (23) identified that the downregulation of miR-6126 was correlated with the severity of social deficits in ASD patients. In our study, we find that the dysregulation of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p could interact with environmental factors, such as maternal stress during pregnancy, neonatal jaundice, and family psychiatric history to impact the risk of ASD.
Maternal stress during pregnancy susceptibility appears to affect offspring neurodevelopment (24). The extent of this risk for ASD is investigated in a number of studies (25). Previously, maternal stress exposure was found to be associated with dysregulation of miRNAs in offspring brain in rats (26). This result indicates that miRNAs might be associated with maternal stress exposure, contributing to the ASD risk. As expected, we find that hsa-miR-320a could interact with maternal stress exposure to affect ASD risk in this study. The OR of this interaction is as high as 39.42. It is reported that total bilirubin levels can result in infants' neuronal injury (27). A recently published meta-analysis shows that neonatal jaundice is a potential risk factor for ASD (28). The results of this study confirm that the interaction of neonatal jaundice and hsa-miR-19b-3p might increase the risk of ASD. Children with a family psychiatric history were more likely to be diagnosed with ASD. Interestingly, in this study, we find that family psychiatric history could interact with hsa-miR-181b-5p, playing a role in increasing ASD risk.
Previous studies (29) and our study show that miRNAs in both serum and saliva are dysregulated in patients with ASD, indicating that miRNAs are promising diagnostic and prognostic biomarkers for ASD. Whether miRNAs could be used to diagnose ASD has not reach a consensus (29). Thus, further studies are needed to identify if miRNAs could be used to diagnose ASD. Whether miRNAs could be used as therapeutic targets for ASD should also be investigated.
The present study has several limitations. First, this study had a relatively small sample size. Second, most of the patients and healthy controls were from a single hospital. Third, we only used qRT-PCR to analyze the differentially expressed miRNAs. Other methods are also needed to confirm our results. Fourth, the environmental and biochemical risk factors for ASD mentioned in this study were not comprehensive. Risk factors, such as prenatal viral infection, zinc deficiency, and oxidative stress (30, 31) should also be discussed in further studies. Furthermore, in this study, the target genes of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p were predicted based on previous studies (5). The role of other miRNAs should be also investigated. Finally, how miRNA affects ASD were not detected in this study either. Further studies are required to figure out this issue.
Our study finds that the serum levels of hsa-miR-181b-5p and hsa-miR-320a in ASD children are much lower than those in healthy controls, and the serum levels of hsa-miR-19b-3p in ASD children were much higher. The dysregulation of hsa-miR-181b-5p, hsa-miR-320a, and hsa-miR-19b-3p could interact with environmental factors, such as maternal stress during pregnancy, neonatal jaundice, and family psychiatric history to impact the risk of ASD.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
This study was conducted with approval from the Ethics Committee of North China University of Science and Technology. The patients/participants provided their written informed consent to participate in this study.
LC and WD have made substantial contributions to conception and design. NX, JD, BX, and JM acquisition of data, analysis, and interpretation of data. LC, RY, and LW have been involved in drafting the manuscript and revising it critically for important intellectual content. FF have given final approval of the version to be published. All authors contributed to the article and approved the submitted version.
This work was supported by Major Project of Medical Science of Hebei Provincial Commission of Health and Family Planning (No. 20180734).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Vaccaro TDS, Sorrentino JM, Salvador S, Veit T, Souza DO, Almeida RF. Alterations in the microRNA of the blood of autism spectrum disorder patients: effects on epigenetic regulation and potential biomarkers. Behav Sci (Basel). (2018) 8:75. doi: 10.3390/bs8080075
2. Hicks SD, Carpenter RL, Wagner KE, Pauley R, Barros M, Tierney-Aves C, et al. Saliva microRNA differentiates children with autism from peers with typical and atypical development. J Am Acad Child Adolesc Psychiatry. (2020) 59:296–308. doi: 10.1016/j.jaac.2019.03.017
3. Li HH, Wang CX, Feng JY, Wang B, Li CL, Jia FY. A Developmental profile of children with autism spectrum disorder in china using the Griffiths Mental Development Scales. Front Psychol. (2020) 11:570923. doi: 10.3389/fpsyg.2020.570923
4. Tong Z, Zhou Y, Wang J. Identification and functional analysis of long non-coding RNAs in autism spectrum disorders. Front Genet. (2020) 11:849. doi: 10.3389/fgene.2020.00849
5. Hicks SD, Middleton FA. A comparative review of microRNA expression patterns in autism spectrum disorder. Front Psychiatry. (2016) 7:176. doi: 10.3389/fpsyt.2016.00176
6. Kheirouri S, Alizadeh M. Maternal excessive gestational weight gain as a risk factor for autism spectrum disorder in offspring: a systematic review. BMC Pregnancy Childbirth. (2020) 20:645. doi: 10.1186/s12884-020-03324-w
7. Tonacci A, Bagnato G, Pandolfo G, Billeci L, Sansone F, Conte R, et al. MicroRNA cross-involvement in autism spectrum disorders and atopic dermatitis: a literature review. J Clin Med. (2019) 8:88. doi: 10.3390/jcm8010088
8. RK CY, Merico D, Bookman M, HoweJ L, Thiruvahindrapuram B, Pate R V, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. (2017) 20:602–11. doi: 10.1038/nn.4524
9. Yoon SH, Choi J, Lee WJ, Do JT. Genetic and epigenetic etiology underlying autism spectrum disorder. J Clin Med. (2020) 9:966. doi: 10.3390/jcm9040966
10. Wu X, Li W, Zheng Y. Recent progress on relevant microRNAs in autism spectrum disorders. Int J Mol Sci. (2020) 21:5904. doi: 10.3390/ijms21165904
11. Masini E, Loi E, Vega-Benedetti AF, Carta M, Doneddu G, Fadda R, et al. An overview of the main genetic, epigenetic and environmental factors involved in autism spectrum disorder focusing on synaptic activity. Int J Mol Sci. (2020) 21:8290. doi: 10.3390/ijms21218290
12. Wu S, Wu F, Ding Y, Hou J, Bi J, Zhang Z. Advanced parental age and autism risk in children: a systematic review and meta-analysis. Acta Psychiatr Scand. (2017) 135:29–41. doi: 10.1111/acps.12666
13. Gardener H, Spiegelman D, Buka SL. Prenatal risk factors for autism: comprehensive meta-analysis. Br J Psychiatry. (2009) 195:7–14. doi: 10.1192/bjp.bp.108.051672
14. Gardener H, Spiegelman D, Buka SL. Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis. Pediatrics. (2011) 128:344–55. doi: 10.1542/peds.2010-1036
15. Kim JW, Park K, Kang RJ, Gonzales EL, Oh HA, Seung H, et al. Gene-environment interaction counterbalances social impairment in mouse models of autism. Sci Rep. (2019) 9:11490. doi: 10.1038/s41598-019-47680-w
16. Lee PF, Thomas RE, Lee PA. Approach to autism spectrum disorder: using the new DSM-V diagnostic criteria and the CanMEDS-FM framework. Can Fam Physician. (2015) 61:421–4.
17. Schopler E, Van Bourgondien ME, Wellman GJ, Love SR. Childhood Autism Rating Scale, 2nd ed. (CARS-2). Los Angeles, CA: Western Psychological Services (2010). doi: 10.1080/15374410701817808
18. Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. (2016) 16:52. doi: 10.1186/s12887-016-0586-x
19. Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. Serum microRNA profiles in children with autism. Mol Autism. (2014) 5:40. doi: 10.1186/2040-2392-5-40
20. Toma C, Torrico B, Hervás A, Salgado M, Rueda I, Valdés-Mas R, et al. Common and rare variants of microRNA genes in autism spectrum disorders. World J Biol Psychiatry. (2015) 16:376–86. doi: 10.3109/15622975.2015.1029518
21. Bölte S, Girdler S, Marschik PB. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell Mol Life Sci. (2019) 76:1275–97. doi: 10.1007/s00018-018-2988-4
22. Vasu MM, Sumitha PS, Rahna P, Thanseem I, Anitha A. microRNAs in autism spectrum disorders. Curr Pharm Des. (2019) 25:4368–78. doi: 10.2174/1381612825666191105120901
23. Nakata M, Kimura R, Funabiki Y, Awaya T, Murai T, Hagiwara M. MicroRNA profiling in adults with high-functioning autism spectrum disorder. Mol Brain. (2019) 12:82. doi: 10.1186/s13041-019-0508-6
24. Beversdorf DQ, Stevens HE, Margolis KG, Van de Water J. Prenatal stress and maternal immune dysregulation in autism spectrum disorders: potential points for intervention. Curr Pharm Des. (2019) 25:4331–43. doi: 10.2174/1381612825666191119093335
25. Beversdorf DQ, Stevens HE, Jones KL. Prenatal Stress, Maternal immune dysregulation, and their association with autism spectrum disorders. Curr Psychiatry Rep. (2018) 20:76. doi: 10.1007/s11920-018-0945-4
26. Zucchi FC, Yao Y, Ward ID, Ilnytskyy Y, Olson D M, Benzies K, et al. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS One. (2013) 8:e56967. doi: 10.1371/journal.pone.0056967
27. Johnson L, Bhutani VK. The clinical syndrome of bilirubin-induced neurologic dysfunction. Semin Perinatol. (2011) 35:101–13. doi: 10.1053/j.semperi.2011.02.003
28. Jenabi E, Bashirian S, Khazaei S. Association between neonatal jaundice and autism spectrum disorders among children: a meta-analysis. Clin Exp Pediatr. (2020) 63:8–13. doi: 10.3345/kjp.2019.00815
29. Salloum-Asfar S, Satheesh NJ, Abdulla SA. Circulating miRNAs, small but promising biomarkers for autism spectrum disorder. Front Mol Neurosci. (2019) 12:253. doi: 10.3389/fnmol.2019.00253
30. Grabrucker AM. Environmental factors in autism. Front Psychiatry. (2013) 3:118. doi: 10.3389/fpsyt.2012.00118
Keywords: autism spectrum disorder, microRNA, environment, risk, etiology
Citation: Cui L, Du W, Xu N, Dong J, Xia B, Ma J, Yan R, Wang L and Feng F (2021) Impact of MicroRNAs in Interaction With Environmental Factors on Autism Spectrum Disorder: An Exploratory Pilot Study. Front. Psychiatry 12:715481. doi: 10.3389/fpsyt.2021.715481
Received: 31 May 2021; Accepted: 03 August 2021;
Published: 05 October 2021.
Edited by:
Stefania Schiavone, University of Foggia, ItalyReviewed by:
Moein Dehbashi, University of Isfahan, IranCopyright © 2021 Cui, Du, Xu, Dong, Xia, Ma, Yan, Wang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: FuMin Feng, ZnVtaW5mZW5nZHJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.