- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Psychiatric Clinics, Center for Affective, Stress and Sleep Disorders, University of Basel, Basel, Switzerland
- 4Exercise Neuroscience Research Laboratory, The University of Alabama at Birmingham, Birmingham, AL, United States
- 5Sleep Disorders Research Center, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 6Substance Abuse Prevention Research Center, Health Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 7Division of Sport Science and Psychosocial Health, Department of Sport, Exercise and Health, University of Basel, Basel, Switzerland
- 8Department of Psychiatry, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
Schizophrenia is a destructive neuropsychiatric disease with a median prevalence of 4.0 per 1,000 during the whole life. Genome-wide association studies have shown the role of copy number variants (generally deletions) and certain alleles of common single nucleotide polymorphisms in the pathogenesis of schizophrenia. This disorder predominantly follows the polygenic inheritance model. Schizophrenia has also been linked with various alterations in the transcript and protein content of the brain tissue. Recent studies indicate that alterations in non-coding RNAs (ncRNAs) signature underlie a proportion of this dysregulation. High throughput microarray investigations have demonstrated momentous alterations in the expression of long non-coding RNAs (lncRNA) and microRNAs (miRNAs) in the circulation or post-mortem brain tissues of patients with schizophrenia compared with control samples. While Gomafu, PINT, GAS5, TCONS_l2_00021339, IFNG-AS1, FAS-AS1, PVT1, and TUG1 are among down-regulated lncRNAs in schizophrenia, MEG3, THRIL, HOXA-AS2, Linc-ROR, SPRY4-IT1, UCA1, and MALAT1 have been up-regulated in these patients. Moreover, several miRNAs, such as miR-30e, miR-130b, hsa-miR-130b, miR-193a-3p, hsa-miR-193a-3p, hsa-miR-181b, hsa-miR-34a, hsa-miR-346, and hsa-miR-7 have been shown to be dysregulated in blood or brain samples of patients with schizophrenia. Dysregulation of these transcripts in schizophrenia not only provides insight into the pathogenic processes of this disorder, it also suggests these transcripts could serve as diagnostic markers for schizophrenia. In the present paper, we explore the changes in the expression of miRNAs and lncRNAs in patients with schizophrenia.
Introduction
Schizophrenia is a destructive neuropsychiatric disease with a median prevalence of 4.0 per 1,000 during the whole life and a lifetime morbid risk of 7.2 per 1,000 (1). The onset of disorder is usually in youth or early adulthood (2) and rarely after the forties or in the childhood (3, 4). In spite of comparable prevalence in both sexes (1), the disorder is usually begins earlier with a more severe course in male subjects (2, 5). The disorder is associated with an increased mortality rate (1) due to suicide and cardiovascular comorbidities. Schizophrenia is diagnosed by the detection of symptoms in three major domains: positive symptoms, such as hallucination, delusion, and disorganized thought and movement, negative symptoms, i.e., lack of interest and enthusiasm, poverty of content of speech, lack of motivation or aptitude to do tasks and affective annihilation, and cognitive symptoms including deficiencies in executive function and attention (6). Genome-wide association studies have shown the role of copy number variants (generally deletions) and certain alleles of common single nucleotide polymorphisms (SNPs) in the pathogenesis of schizophrenia. This disorder predominantly follows the polygenic inheritance model with a significant level of overlap in genetic factors with other psychiatric disorders namely autism and bipolar disorder (7).
Schizophrenia has also been associated with dysregulation of several transcripts and proteins in the brain tissue. These alterations are caused by an intricate dysregulation of gene expression and protein synthesis. Both spatial and temporal elements contribute in determination of this dysregulation. Several studies indicate that alterations in non-coding RNAs (ncRNAs) signature underlie the mechanisms of this dysregulation (8). Changes in expression profile of ncRNAs in the brain tissues of patients with schizophrenia and the observed association between this disorder and certain SNPs in genomic regions coding these transcripts further highlight the role of ncRNAs in the pathogenesis of schizophrenia (8). These transcripts have been shown to govern the complex pattern of gene expression, thus being considered as one of the principal epigenetic mechanisms of gene expression (8). It is worth mentioning epigenetic alterations in some brain areas and neural tracks characterize an important route through which environmental parameters interplay with personal genetic composition to influence susceptibility to psychiatric disorders during the lifespan (9). Moreover, two classes of ncRNAs namely microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are extremely abundant in the human brain, implying their crucial role in the appropriate function of this tissue (10, 11). While miRNAs mainly repress gene expression at post-transcriptional stage (12), lncRNAs can either enhance or suppress expression of gene through acting at chromatin, transcriptional an post-transcriptional levels (13). The important roles of these two classes of ncRNAs in the regulation of expression of genes and their developmental and tissue-specific signature suggest that they might underlie the observed aberrations in the transcriptome and proteome of brain tissue in patients with schizophrenia. Besides, alterations in the expression of lncRNAs might explain the plasticity of the organization of evolving neurons and their role in development of brain (14). In the current review, we explore the changes in the expression of miRNAs and lncRNAs in patients with schizophrenia. For the purpose of preparing this narrative review, we searched PubMed and google scholar with the key words “microRNA” or “miRNA” or “long non-coding RNA” or “lncRNA” AND “schizophrenia.” Subsequently, we assessed the relevance of the obtained material through reading the full texts of the articles. Finally, we tabulated the retrieved data in distinct tables.
LncRNAs and Schizophrenia
LncRNAs are a huge and dissimilar group of ncRNA which have more than 200 nucleotides. LncRNAs signify the bulk of the non-coding transcriptome and utilize numerous mechanisms to exert their regulatory functions on gene expression among them being suppression or recruitment of transcription factors, modulation of chromatin structure and regulation of the stability of transcripts (15, 16). High throughput microarray investigations have demonstrated significant changes in lncRNA signature in the peripheral blood or post-mortem brain tissues of patients with schizophrenia compared with control samples (17–20).
LncRNA Profile in Central Tissues
Expression of the lncRNA Gomafu has been regulated by neuronal activation. This lncRNA has been shown to directly interact with two splicing factors namely QKI and SRSF1. Therefore, aberrant expression of Gomafu changes the splicing patterns of DISC1 and ERBB4 genes to a pattern which is similar to what is reported in schizophrenia. Expression of Gomafu is substantially decreased in post-mortem cortical gray matter of patients with schizophrenia (20). Level of Gomafu activity has been correlated with neuronal structural plasticity which is strong in the course of development and subsequently is decreased in the adulthood (14). Therefore, abnormal activity of Gomafu in the brain tissues of subjects with schizophrenia might reflect abnormal neurodevelopment. Moreover, abnormal levels of this lncRNA might also affect human behavior, since it has been associated with susceptibility to substance abuse (21).
Hu et al. have assessed RNA profile of post-mortem brain samples in patients with schizophrenia and those with bipolar disorder and control subjects. They reported differential expression of 20 long intergenic non-coding RNAs (lincRNAs) in orbitofrontal cortex of bipolar patients and aberrant expression of 34 and 1 lincRNAs in anterior cingulate cortex and dorsolateral prefrontal cortex of patients with schizophrenia, respectively. Thus, they reported brain area-specific profiles for lincRNAs. Differentially expressed lincRNAs were enriched in pathways namely immune system development and oligodendrocyte differentiation. Moreover, they reported altered DNA methylation as a possible mechanism for dysregulation of lincRNAs (18).
LncRNA Profile in Peripheral Blood
Chen et al. have examined expression profile of lncRNAs in the peripheral blood mononuclear cells (PBMCs) of patients with schizophrenia compared with healthy subjects. They reported differential expression of 125 lncRNAs between these two subgroups. Notably, expression levels of ENST00000394742, TCONS_l2_00025502, ENST00000563823, ENST00000521622, and TCONS_l2_00021339 were suggestively down-regulated in patients (17). LncRNA profiling has also revealed up-regulation of three lncRNAs in schizophrenia, down-regulation of six lncRNAs in major depressive disorder, and up-regulation of three lncRNAs in generalized anxiety disorder (GAD). Notably, lncRNAs observed to be up-regulated in schizophrenia were significantly decreased in patients with GAD. Furthermore, down-regulated lncRNAs in major depressive disorder were up-regulated in patients with schizophrenia. Finally, there were significant differences in the expression levels of lncRNAs between patients with schizophrenia and GAD. Therefore, a number of lncRNAs are putative biomarkers for differentiation of schizophrenia from major depressive disorder and generalized anxiety disorder (19). Sudhalkar et al. have examined expression levels of MEG3, PINT, and GAS5 in the PBMCs of patients with psychosis compared with healthy controls. They reported diagnostic differences with MEG3, PINT, and GAS5, and symptom acuity effect with MEG3 and GAS5. Moreover, there was significant difference in the expression of MEG3 between drug naïve patients and patients received risperidone (22). IFNG-AS1 expression has been shown to be down-regulated in patients with schizophrenia compared with healthy subjects in correlation with IFNG expression indicating a putative role for inflammation in this disorder (23). We have recently demonstrated down-regulation of FAS-AS1, PVT1, and TUG1 in patients with schizophrenia compared with controls. Yet, expressions of GAS5, NEAT1, and OIP5-AS1 were similar between patients and controls (24).
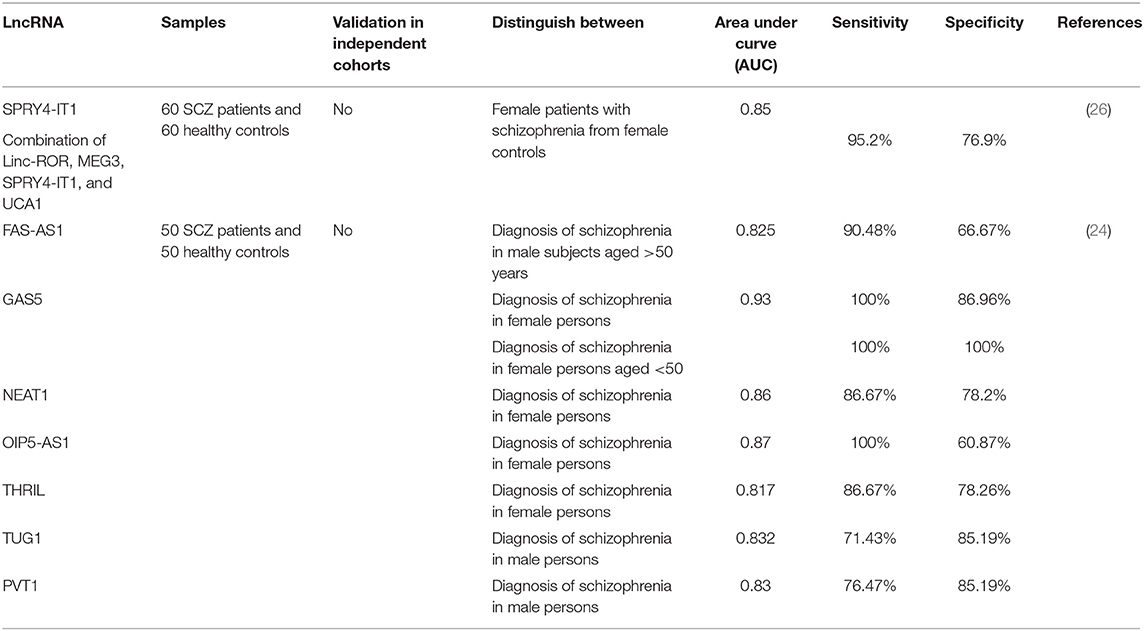
Table 1 exhibits the results of researches which demonstrated down-regulation of lncRNAs in schizophrenia.
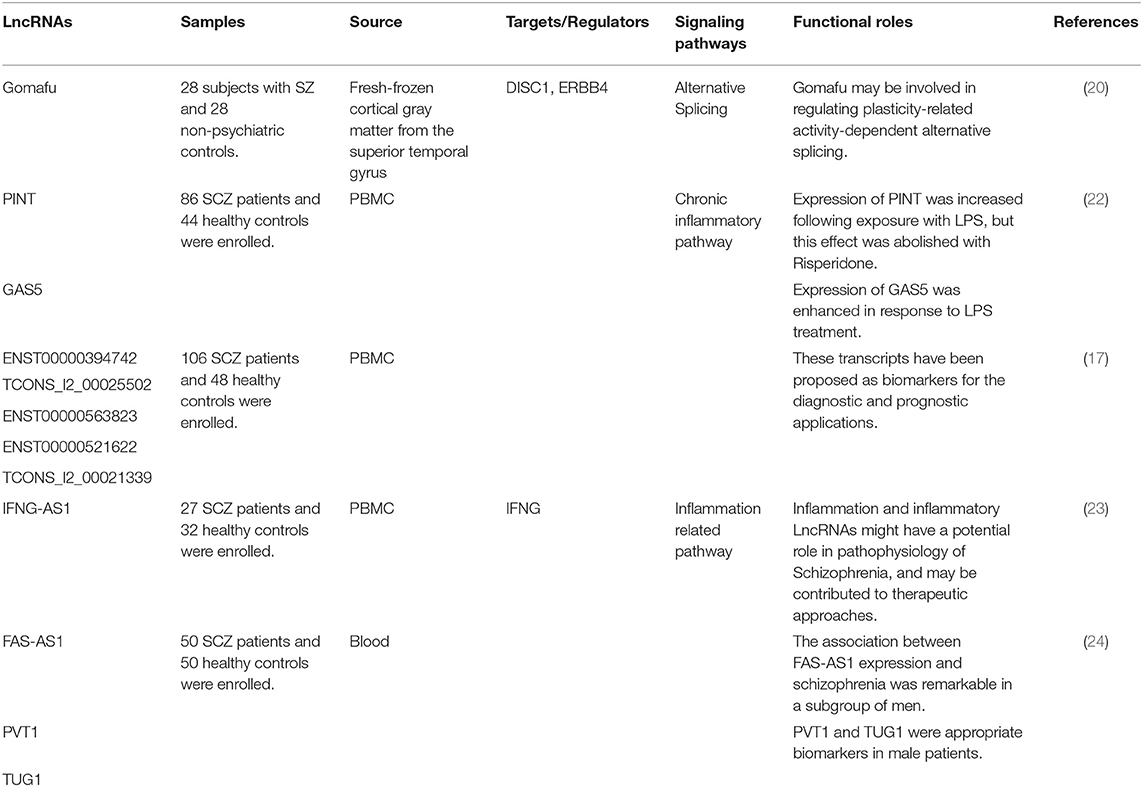
Table 1. Down-regulated lncRNAs in schizophrenia (Empty cells show that this information has not been provided in the main articles).
A high throughput microarray-based strategy has demonstrated 125 dysregulated lncRNAs in patients with schizophrenia compared with including 62 over-expressed and 63 under-expressed lncRNAs in patients. Antipsychotic treatment has resulted in reduction in NONHSAT089447 and NONHSAT041499 levels, parallel with decrease in the post-treatment Positive And Negative Syndrome Scale (PANSS) scores. Moreover, reduction in NONHSAT041499 levels have been associated with improvement of several clinical manifestations and better response to therapies (17).
Chen et al. have reported the effects of olanzapine treatment in suppression of expression of the NONHSAT089447 lncRNA. Small interfering RNA-mediated NONHSAT089447 silencing has reduced expression of dopamine receptors DRD3 and DRD5. In addition, Western blot studies verified the role of this lncRNA in regulation of DRD signaling (25). We have demonstrated up-regulation of HOXA-AS2, Linc-ROR, MEG3, SPRY4-IT1, and UCA1 in patients with schizophrenia compared with healthy subjects. Yet, when assessing their expressions in sex-based subclasses, the differences in their expressions were significant just among females. Moreover, we reported correlations between expressions of Linc-ROR and SPRY4-IT1 and age of patients (26). Ni et al. have profiled peripheral blood transcriptome of monozygotic twins discordant for schizophrenia. Using this approach, authors have demonstrated up-regulation of AC006129.1 lncRNA in patients. This lncRNA regulates inflammatory reactions through promoting expression of SOCS3 and CASP1. Further experiments showed that AC006129.1 interacts with the promoter region of the transcriptional repressor Capicua (CIC) to enhance the interactions of DNA methyltransferases with its promoter and decrease CIC expression, thus reversing CIC-associated SOCS3 and CASP1 suppression. Activation of SOCS3 increases the anti-inflammatory reactions by obstructing JAK/STAT pathway (27). Table 2 exhibits the list of up-regulated lncRNAs in schizophrenia.
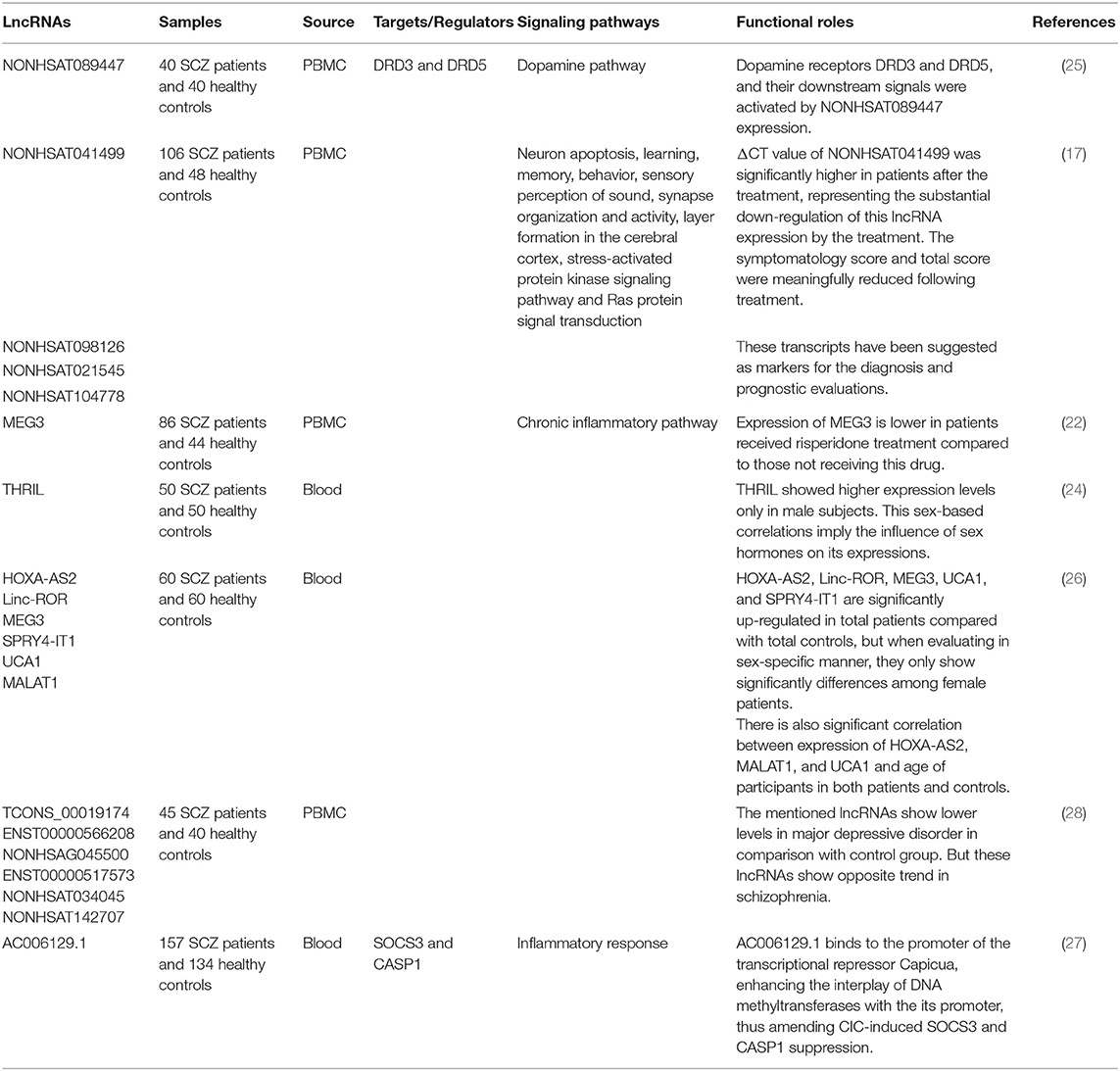
Table 2. Up-regulated lncRNAs in schizophrenia (Empty cells show that this information has not been provided in the main articles).
Association Between LncRNA(s) SNP(s) and Risk of Schizophrenia
Few studies have appraised association between lncRNAs SNPs and risk of schizophrenia. Rao et al. have performed a two-phase association study on 8 tag SNPs that encompass the entire MIAT region in two independent cohorts from Han Chinese population. They demonstrated significant association between paranoid schizophrenia and the rs1894720. Moreover, there was a weak association between rs4274 and this condition. No specific haplotype was detected that modulate risk of paranoid schizophrenia in the assessed population (29).
Diagnostic Value of LncRNAs in Schizophrenia
Expression levels of lncRNAs could be used as diagnostic markers for schizophrenia. We have assessed this possibility in a cohort of Iranian patients with schizophrenia (24, 26). Based on the obtained results in a limited number of patients, GAS5 and OIP5-AS1 have been proposed as appropriate biomarkers in female subjects (24). Table 3 summarizes the results of these studies.
miRNAs and Schizophrenia
These small transcripts have intricate temporospatial signature in the brain tissue and can modulate expressions of myriad of genes by working as the specificity elements for gene-silencing apparatus in the cells. Based on the observed association between miRNA dysregulation and substantial alterations in the network organization in the course of neurodevelopment, miRNAs are considered as important regulators of several neurological processes (30). Numerous studies have indicated aberrant expression of miRNAs in the brain and peripheral blood of patients with schizophrenia.
miRNA Levels in Peripheral Blood
Lai et al. have used the same strategy to assess miRNA profile of PBMCs in patients with schizophrenia. They reported association between expression profile of 7 miRNAs and negative symptoms as well as neurocognitive performance scores (31). Lai et al. have demonstrated aberrant expression of a panel of miRNAs in patients with schizophrenia and correlation between expression pattern of some miRNAs and the presence of negative symptoms, level of neurocognitive function, and event-related potentials (31).
Gardiner et al. have assessed the miRNA signature in peripheral blood mononuclear cells (PBMCs) obtained from patients with schizophrenia and control individuals. Using microarray technique, they detected down-regulation of 33 miRNAs, dysregulation of seven of them being verified by real-time PCR technique as well. Notably, 17 down-regulated miRNAs have been shown to be transcribed from a particular imprinted locus at the maternally expressed DLK1-DIO3 area. This distinctive miRNA signature in PBMCs might represent a fundamental genetic or epigenetic mechanism for the pathogenesis of schizophrenia (32). Shi et al. have reported up-regulation of miR-181b, miR-219-2-3p, miR-1308, and let-7g whereas down-regulation of miR-195 in serum samples obtained from patients with schizophrenia compared with controls (33). Sun et al. have demonstrated over-expression of miR-132, miR-195, miR-30e, and miR-7 in plasma samples of patients with schizophrenia, and up-regulation of miR-212, miR-34a, and miR-30e in their PBMCs {#164}. They suggested that miRNA signature is more distinctive in plasma samples compared with PBMCs (34). A high throughput miRNA profiling revealed up-regulation of eight miRNAs in plasma samples obtained from schizophrenia patients, among them were miR-130b and miR-193a-3p which were up-regulated in schizophrenia but not in non-schizophrenia psychotic disorders. These results indicated these miRNAs as state-independent biomarkers for schizophrenia (35).
Lai et al. have subsequently assessed the impact of hospitalization on the expression levels of these miRNAs. Notably, expression of none of these miRNAs did not change after 2 months hospitalization of patients even when clinical symptoms were remarkably ameliorated. Thus, these miRNAs have been suggested as trait biomarkers instead of state-dependent biomarkers. Assessment of expression profile of hsa-miR-34a and hsa-miR-548d in post-mortem brain samples showed no difference between patients and controls (36).
miRNA Profile in Central Tissues/Cell Lines
Expression profile of miRNAs has also been assessed in the olfactory epithelium as one of the limited available neural tissues that have neurons and neural stem cells. Mor et al. have detected over-expression of miR-382-5p in cultured olfactory cells obtained from schizophrenia patients compared with controls. Up-regulation of this miRNA was also verified in microdissected olfactory epithelium neuronal tissues of these patients. However, miR-382 was not expressed in lymphoblastoid cell lines originated from either patients with schizophrenia or control subjects. This miRNA has been shown to target FGFR1 and SPRY4, two genes whose expressions were decreased in the olfactory cells obtained from patients with schizophrenia (37).
Other experiments in HEK293 and SH-SY5Y cell lines have validated CALN1 as a target of miR-137 (38). A number of other miRNAs can also regulate development of dendrites, thus being implicated in the functionality of synapses and neuronal interactions. For instance, miR-214 has been shown to increase dendrite dimension, complexity and morphogenesis. Its involvement in the pathogenesis of schizophrenia has been suggested through the observed interaction between this miRNA and quaking (Qki), a candidate gene in schizophrenia. The miR-214-Qki axis is a fundamental axis in the modulation of dendritic development in the neurons (39). Experiments in a mouse neuroblastoma cell line (Neuro2A) have shown the regulatory role of miR-137 on the expression of several important proteins in the PI3K-Akt-mTOR axis which have functions downstream of neuregulin/ErbB and BDNF. Therefore, this miRNA controls neuronal reactions to these factors and dendritic development, thus contributing in the risk of schizophrenia (40). Figure 1 shows the mechanism of involvement of miR-214 in the pathogenesis of schizophrenia.
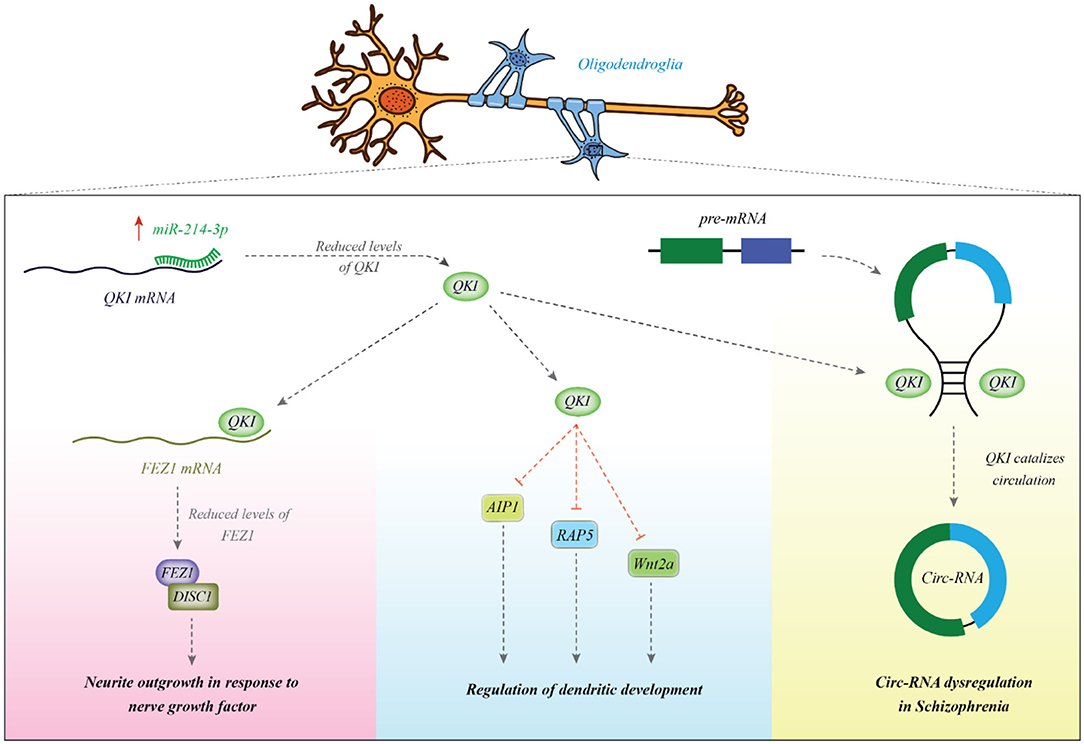
Figure 1. miR-214 is overexpressed in patients with schizophrenia. This miRNA binds with the 3′ UTR of Qki transcript to reduce its expression. Qki is an RNA-binding protein which modulates stability of several mRNAs among them are AIP1, Wnt2a, and Rab5. These transcripts are involved in the dendritic development (39). Qki can also bind with FEZ1 mRNA to enhance its stability (41). FEZ1 binds with DISC1 and increases neurite outgrowth in response to NGF (42). Qki can bind with intronic regions and participate in the biogenesis of circRNAs (43). Abnormal expression of Qki might be associated with the observed dysregulation of cricRNAs in schizophrenia (44).
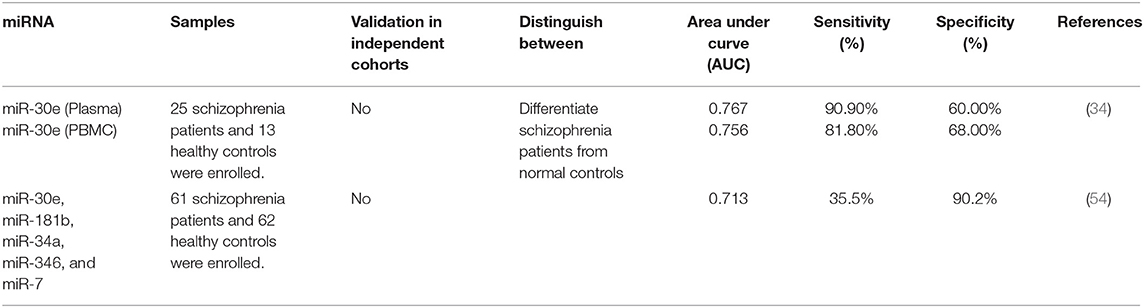
Tables 4, 5 show the list of down-/up-regulated miRNAs in the schizophrenia patients, respectively.
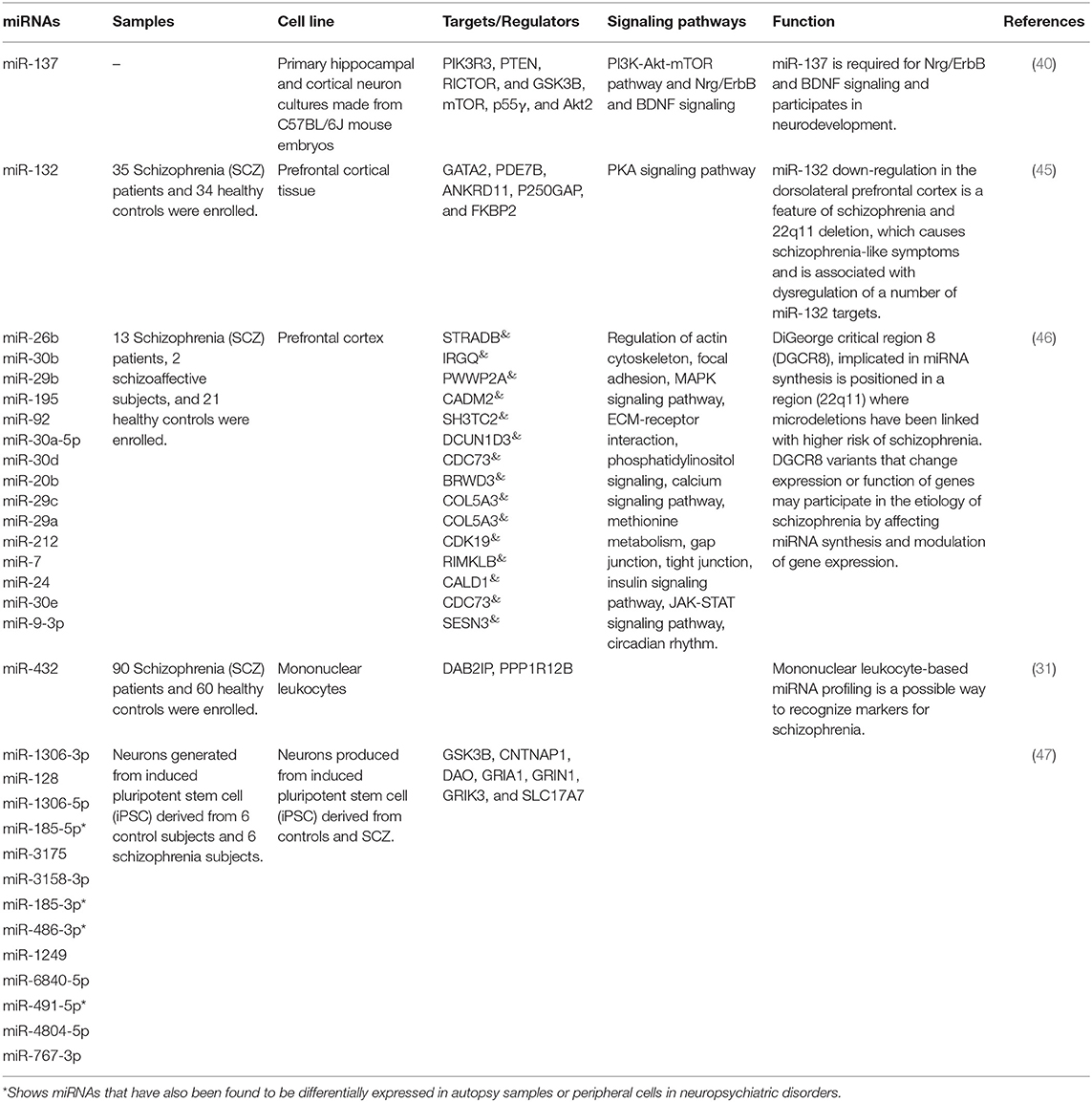
Table 4. Down-regulated miRNAs in schizophrenia [&Top targets based on the predictions of miRDB (http://www.mirdb.org/), empty cells show that this information has not been provided in the main articles].
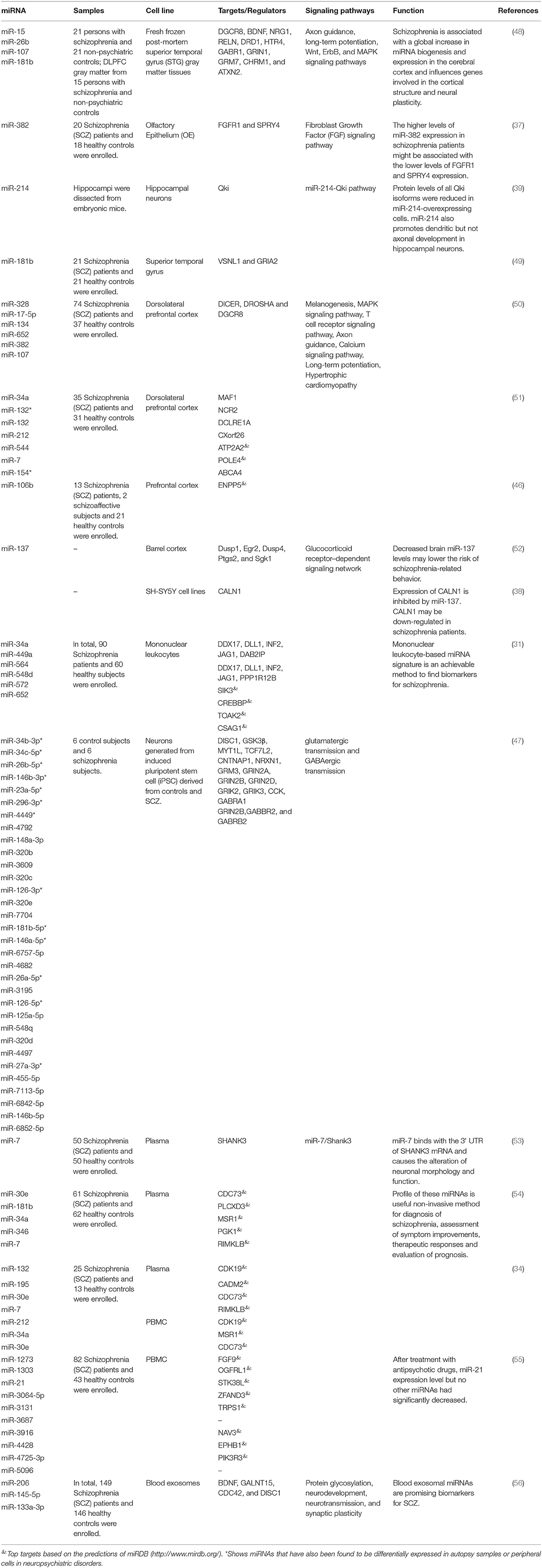
Table 5. Up-regulated microRNAs in schizophrenia (Empty cells show that this information has not been provided in the main articles).
miRNAs Variations in Schizophrenia
Some studies have verified associations between SNPs within miRNA-coding genes and risk of schizophrenia. For instance, in a genome-wide association study, Ripke et al. have shown significant association between the rs1625579 of the miR-137 and risk of schizophrenia (57). Notably, others have confirmed associations between miR-137 target genes CUB, CSMD1, C10orf26, CACNA1C, TCF4, and ZNF804A, and risk of schizophrenia (58–61). Notably, the mentioned SNP in miR-137 has been shown to influence cognitive activity. The T/T genotype of this SNP is correlated with working memory defects in patients with schizophrenia as reflected by their reduced scores in the brief assessment of cognition in schizophrenia instrument (62). The risk genotype has also been associated with dorsolateral prefrontal cortex hyperactivation (DLPFC) in patients with schizophrenia (63). In addition, the risk allele of this SNP has been associated with down-regulation of miR-137 in schizophrenia and is potentially involved in the modulation of expression of the schizophrenia risk locus TCF4, more emphasizing on the participation of miR-137 and its downstream molecules in this disorder (64). These studies further support the role of this miRNA in the pathogenesis of schizophrenia.
Diagnostic Value of miRNAs in Schizophrenia
Diagnostic value of miRNAs has been assessed in different biological sources of patients with schizophrenia using statistical methods, such as plotting the receiver operating characteristic curve and calculation of the area under the curve (AUC) values. Such method has shown the diagnostic values of 0.767 and 0.756 for plasma and PBMC expression patterns of miR-30e, respectively. Plasma levels of this miRNA had sensitivity and specificity of 90.90 and 60.00%, respectively. These values for its PBMC levels were 81.80 and 68.00%, respectively. Moreover, logistic regression analysis showed higher sensitivity of plasma levels of this miRNA in differentiating schizophrenia patients from healthy subjects compared with its PBMC levels (34). The diagnostic value of a panel of over-expressed miRNAs including miR-30e, miR-181b, miR-34a, miR-346, and miR-7 has been reported to be 0.713. Notably, suitable pharmacotherapy resulted in down-regulation of miR-132, miR-181b, miR-432, and miR-30e expressions. Besides, there was a remarkable correlation between amelioration of clinical symptoms and alterations in the expression levels of miR-132, miR-181b, miR-212, and miR-30e (54). A certain molecular axis including the transcription factor the early growth response protein 1 (EGR1), miR-30a-5p, and its target gene NEUROD1 has been shown to differentiate schizophrenia patients from healthy subjects with diagnostic accuracy of 0.962 which was far higher than the diagnostic power of miR-30a-5p alone. This axis has also been proved useful for monitoring of patients in acute psychotic phase (65). Table 6 shows the results of studies which assessed diagnostic role of miRNAs in the schizophrenia.
Discussion
Schizophrenia is complex disorder caused by interaction between several genomic loci. The speculation that this disorder is caused by one or a few common principal gene effects has been experimentally examined in genome-wide linkage studies yet results generally showed no genome-wide significance (66). In addition to genomic variants that contribute in the pathogenesis of this disorder, some other mechanisms might modify the risk of development of schizophrenia. Expressions of ncRNAs are modulated by neuronal activation (20), suggesting a role for these transcripts in the pathophysiology of neuropsychiatric disorders. In the current study, we reviewed the literature about the role of ncRNAs in the pathophysiology of schizophrenia. Based on the above-mentioned evidence, several lncRNAs and miRNAs have been dysregulated in blood or brain samples of patients with schizophrenia. Not discounting the role of ncRNAs as biomarkers for schizophrenia, peripheral expression profile of these transcripts does not necessarily reflect their expression in the brain tissues (36). Aberrant expression of a number of these transcripts has been associated with PANSS score (17). Moreover, clinical diagnoses of psychosis and symptom severity have been shown to alter expression of a number of lncRNAs (22) indicating a substantial role for these transcripts in the pathogenesis of schizophrenia.
Although there is a considerable level of overlap between psychiatric disorders in the terms of contributing genetic factors, expression levels of some ncRNAs could be used to differentiate a number of these conditions (19). Moreover, expression profile of certain ncRNAs can distinguish patients with schizophrenia from healthy subjects. At the present, schizophrenia is principally diagnosed based on the clinical symptoms and signs instead of on the pathophysiological biomarkers (67). Identification of such biomarkers would facilitate detection of malingering, thus has practical significance in the forensic medicine in the assessment of cases pretending psychological disorders for a particular gain. Moreover, molecular biomarkers have implications in the establishment of targeted therapies. Notably, expression profile of ncRNAs in the circulation has the potential to improve the diagnostic and prognostic assessment of patients with schizophrenia (67). A former study has demonstrated up-regulation of NONHSAT089447, NONHSAT021545, and NONHSAT041499 lncRNAs in patients with schizophrenia, while down-regulation of these transcripts in patients with GAD. Notably, authors have reported six lncRNAs with opposite expression patterns in schizophrenia and major depressive disorder. Moreover, they have identified three GAD-related lncRNAs whose expressions were significantly differences between patients with schizophrenia and GAD patients (28). However, most of these studies do not test if expression profile of ncRNAs can distinguish people with schizophrenia from people with other disorders, such as major depression, bipolar disorder, or autism. Based on the similarities in many of clinical symptoms between subjects with schizophrenia and other disorders, including bipolar disorder, autism, and major depression, identification of specific markers for each disease has practical significance. Therefore, this field should be explored in future investigations.
Particularly, miRNA signature has been correlated with clinical course, patients' response to pharmacologic interventions and prognosis of patients with schizophrenia (54). Perhaps, the most intensively assessed miRNA in this regard is miR-30 family. Consistent with this observation, expression of EGR1 which regulates expressions of this miRNA family has been decreased in PBMCs of patients with schizophrenia. On the other hand, expression of NEUROD1 as a target gene of miR-30a-5p has been increased in these patients (65). Based on the results of this study, identification of transcription factor/ miRNA/ target gene axes would be a practical method for recognition of molecular pathways in the pathogenesis of schizophrenia and development of diagnostic/ prognostic panels for this disorder.
Several schizophrenia-associated ncRNAs have been shown to modulate immune responses and inflammatory pathways. Recent studies have highlighted the presence of intricate interplay between the immune system, systemic inflammatory responses, and the central nervous system, which can result alterations in mood, cognitive functions, and behavior. All of these aspects contribute in the pathogenesis of schizophrenia (68). Immune responses can influence activity of neurotransmitters as well as neurodegenerative and neurodevelopmental processes all of which are related with this disorder (68). Assessment of lncRNA signature in Amygdala samples from schizophrenia patients has further endorsed dysregulation of immune-associated lncRNAs in these samples (69). Thus, the functional axes between ncRNAs and immune-related genes provide an explanation for involvement of these transcripts in the pathophysiology of schizophrenia.
Besides, high throughput studies in schizophrenia subjects have shown a tendency toward global up-regulation of miRNAs normally abundant in infants, whereas down-regulation of those normally abundant in prepuberty. Therefore, dysregulation of miRNAs dynamic changes might be involved in the pathogenesis of schizophrenia (70). A number of ncRNAs might explain the observed dissimilarity in the brain activation modes between patients with schizophrenia and controls. For instance, Gomafu has been shown to be intensely regulated in reaction to neuronal activation (71). This lncRNA is also implicated in schizophrenia-related alternative splicing (71). Therefore, aberrant expression of this lncRNA might reflect the abnormal brain activity in these patients.
Finally, it is worth mentioning that miRNAs have distinctive roles as trait-dependent markers or state-dependent markers. This speculation is supported by amelioration of dysregulated expression of a number of miRNAs after pharmacotherapy (54), while no change in the expression of other miRNAs following suitable treatments (36). Therefore, it is necessary to define the role of each miRNA as trait- or state-dependent marker to design distinctive diagnostic/ prognostic miRNA panels for schizophrenia.
Taken together, lncRNAs and miRNAs are potential transcripts that can explain the difference in expression profile of protein coding genes in brain and blood tissues of patients with schizophrenia and healthy subjects. Moreover, as a number of above-mentioned ncRNAs are functionally related with dopamine neurotransmission, these ncRNAs might alter response of patients to some types of antipsychotic drugs. In addition, several ncRNAs can be used as disease markers in schizophrenia.
Several questions should be addressed about the role of ncRNAs in the development of schizophrenia. Studies reviewed in this article have mostly assessed expression of ncRNAs in adult patients. Although differentially expressed ncRNAs among patients and controls have been enriched in neurodevelopment, neurotransmission, and synaptic plasticity, the functional impact of these ncRNAs in the development of neurons should be assessed through knock-out/-in studies in animal models and cell lines.
Finally, most of studies reviewed here have not appraised the effects of antipsychotic drugs on expression of ncRNAs. The data regarding the therapeutic regimens and types of antipsychotic drugs have not been presented in the main articles. This is possibly because patients have been under treatment with different antipsychotic drugs. We mention this point as a limitation of these studies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. (2008) 30:67–76. doi: 10.1093/epirev/mxn001
2. Messias EL, Chen CY, Eaton WW. Epidemiology of schizophrenia: review of findings and myths. Psychiatr Clin North Am. (2007) 30:323–38. doi: 10.1016/j.psc.2007.04.007
3. Girard C, Simard M. Clinical characterization of late-and very late-onset first psychotic episode in psychiatric inpatients. Am J Geriatr Psychiatry. (2008) 16:478–87. doi: 10.1097/JGP.0b013e31816c7b3c
4. Remschmidt H, Theisen FM. Schizophrenia and related disorders in children and adolescents. In: Fleischhacker WW, Brooks DJ, editors. Neurodevelopmental Disorders. Vienna: Springer (2005). doi: 10.1007/3-211-31222-6_7
5. Leung A, Chue P. Sex differences in schizophrenia, a review of the literature. Acta Psychiatr Scand Suppl. (2000) 401:3–38. doi: 10.1111/j.0065-1591.2000.0ap25.x
6. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). Washington, DC: American Psychiatric Pub (2013). doi: 10.1176/appi.books.9780890425596
7. Gejman PV, Sanders AR, Duan J. The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am. (2010) 33:35–66. doi: 10.1016/j.psc.2009.12.003
8. Gibbons A, Udawela M, Dean B. Non-coding RNA as novel players in the pathophysiology of schizophrenia. Noncoding RNA. (2018) 4:11. doi: 10.3390/ncrna4020011
9. Ibi D, González-Maeso J. Epigenetic signaling in schizophrenia. Cell Signal. (2015) 27:2131–6. doi: 10.1016/j.cellsig.2015.06.003
10. Tang Y, Liu D, Zhang L, Ingvarsson S, Chen H. Quantitative analysis of miRNA expression in seven human foetal and adult organs. PLoS ONE. (2011) 6:e28730. doi: 10.1371/journal.pone.0028730
11. Nie JH, Li TX, Zhang XQ, Liu J. Roles of non-coding RNAs in normal human brain development, brain tumor, and neuropsychiatric disorders. Noncoding RNA. (2019) 5:36. doi: 10.3390/ncrna5020036
12. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. (2014) 2014:970607. doi: 10.1155/2014/970607
13. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. (2019) 20:5573. doi: 10.3390/ijms20225573
14. Merelo V, Durand D, Lescallette AR, Vrana KE, Hong LE, Faghihi MA, et al. Associating schizophrenia, long non-coding RNAs and neurostructural dynamics. Front Mol Neurosci. (2015) 8:57. doi: 10.3389/fnmol.2015.00057
15. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. (2017) 3:eaao2110. doi: 10.1126/sciadv.aao2110
16. Fernandes JCR, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA. (2019) 5:17. doi: 10.3390/ncrna5010017
17. Chen S, Sun X, Niu W, Kong L, He M, Li W, et al. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med Sci Monit. (2016) 22:3340–51. doi: 10.12659/MSM.896927
18. Hu J, Xu J, Pang L, Zhao H, Li F, Deng Y, et al. Systematically characterizing dysfunctional long intergenic non-coding RNAs in multiple brain regions of major psychosis. Oncotarget. (2016) 7:71087. doi: 10.18632/oncotarget.12122
19. Cui X, Niu W, Kong L, He M, Jiang K, Chen S, et al. Can lncRNAs be indicators for the diagnosis of early onset or acute schizophrenia and distinguish major depressive disorder and generalized anxiety disorder?—A cross validation analysis. Am J Med Genet B Neuropsychiatr Genet. (2017) 174:335–41. doi: 10.1002/ajmg.b.32521
20. Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. (2014) 19:486–94. doi: 10.1038/mp.2013.45
21. Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. (2006) 31:2304–12. doi: 10.1038/sj.npp.1301089
22. Sudhalkar N, Rosen C, Melbourne JK, Park MR, Chase KA, Sharma RP. Long non-coding RNAs associated with heterochromatin function in immune cells in psychosis. Noncoding RNA. (2018) 4:43. doi: 10.3390/ncrna4040043
23. Ghafelehbashi H, Pahlevan Kakhki M, Kular L, Moghbelinejad S, Ghafelehbashi SH. Decreased expression of IFNG-AS1, IFNG and IL-1B inflammatory genes in medicated schizophrenia and bipolar patients. Scand J Immunol. (2017) 86:479–85. doi: 10.1111/sji.12620
24. Safari MR, Komaki A, Arsang-Jang S, Taheri M, Ghafouri-Fard S. Expression pattern of long non-coding RNAs in schizophrenic patients. Cell Mol Neurobiol. (2019) 39:211–21. doi: 10.1007/s10571-018-0640-3
25. Chen S, Zhu X, Niu W, Yao G, Kong L, He M, et al. Regulatory role of lncRNA NONHSAT089447 in the dopamine signaling pathway in schizophrenic patients. Med Sci Monit. (2019) 25:4322–32. doi: 10.12659/MSM.915684
26. Fallah H, Azari I, Neishabouri SM, Oskooei VK, Taheri M, Ghafouri-Fard S. Sex-specific up-regulation of lncRNAs in peripheral blood of patients with schizophrenia. Sci Rep. (2019) 9:12737. doi: 10.1038/s41598-019-49265-z
27. Ni C, Jiang W, Wang Z, Wang Z, Zhang J, Zheng X, et al. LncRNA-AC006129.1 reactivates a SOCS3-mediated anti-inflammatory response through DNA methylation-mediated CIC downregulation in schizophrenia. Mol Psychiatry. (2020). doi: 10.1038/s41380-020-0662-3. [Epub ahead of print].
28. Cui X, Niu W, Kong L, He M, Jiang K, Chen S, et al. Long noncoding RNA as an indicator differentiating schizophrenia from major depressive disorder and generalized anxiety disorder in nonpsychiatric hospital. Biomarkers Med. (2017) 11:221–8. doi: 10.2217/bmm-2016-0317
29. Rao SQ, Hu HL, Ye N, Shen Y, Xu Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res. (2015) 166:125–30. doi: 10.1016/j.schres.2015.04.032
30. Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. (2012) 46:263–71. doi: 10.1016/j.nbd.2011.12.029
31. Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE. (2011) 6:e21635. doi: 10.1371/journal.pone.0021635
32. Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. (2012) 17:827–40. doi: 10.1038/mp.2011.78
33. Shi W, Du J, Qi Y, Liang G, Wang T, Li S, et al. Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res. (2012) 46:198–204. doi: 10.1016/j.jpsychires.2011.09.010
34. Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, et al. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci. (2015) 22:570–4. doi: 10.1016/j.jocn.2014.08.018
35. Wei H, Yuan Y, Liu S, Wang C, Yang F, Lu Z, et al. Detection of circulating miRNA levels in schizophrenia. Am J Psychiatry. (2015) 172:1141–7. doi: 10.1176/appi.ajp.2015.14030273
36. Lai CY, Lee SY, Scarr E, Yu YH, Lin YT, Liu CM, et al. Aberrant expression of microRNAs as biomarker for schizophrenia: from acute state to partial remission, and from peripheral blood to cortical tissue. Transl Psychiatry. (2016) 6:e717. doi: 10.1038/tp.2015.213
37. Mor E, Kano S, Colantuoni C, Sawa A, Navon R, Shomron N. MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol Dis. (2013) 55:1–10. doi: 10.1016/j.nbd.2013.03.011
38. Xia S, Zhou X, Wang T, Zhang Q, Li Q, Liu Y, et al. Experimental validation of candidate schizophrenia gene CALN1 as a target for microRNA-137. Neurosci Lett. (2015) 602:110–4. doi: 10.1016/j.neulet.2015.07.001
39. Irie K, Tsujimura K, Nakashima H, Nakashima K. MicroRNA-214 promotes dendritic development by targeting the schizophrenia-associated gene quaking (Qki). J Biol Chem. (2016) 291:13891–904. doi: 10.1074/jbc.M115.705749
40. Thomas KT, Anderson BR, Shah N, Zimmer SE, Hawkins D, Valdez AN, et al. Inhibition of the schizophrenia-associated MicroRNA miR-137 disrupts Nrg1alpha neurodevelopmental signal transduction. Cell Rep. (2017) 20:1–12. doi: 10.1016/j.celrep.2017.06.038
41. Chen X, Ku L, Mei R, Liu G, Xu C, Wen Z, et al. Novel schizophrenia risk factor pathways regulate FEZ1 to advance oligodendroglia development. Transl Psychiatry. (2017) 7:1–10. doi: 10.1038/s41398-017-0028-z
42. Matsuzaki S, Tohyama M. Molecular mechanism of schizophrenia with reference to disrupted-in-schizophrenia 1 (DISC1). Neurochem Int. (2007) 51:165–72. doi: 10.1016/j.neuint.2007.06.018
43. Gasparini S, Licursi V, Presutti C, Mannironi C. The secret garden of neuronal circRNAs. Cells. (2020) 9:1815. doi: 10.3390/cells9081815
44. Li Z, Liu S, Li X, Zhao W, Li J, Xu Y. Circular RNA in schizophrenia and depression. Front Psychiatry. (2020) 11:392. doi: 10.3389/fpsyt.2020.00392
45. Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci USA. (2012) 109:3125–30. doi: 10.1073/pnas.1113793109
46. Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. (2007) 8:R27. doi: 10.1186/gb-2007-8-2-r27
47. Zhao D, Lin M, Chen J, Pedrosa E, Hrabovsky A, Fourcade HM, et al. MicroRNA profiling of neurons generated using induced pluripotent stem cells derived from patients with schizophrenia and schizoaffective disorder, and 22q11.2 Del. PLoS ONE. (2015) 10:e0132387. doi: 10.1371/journal.pone.0132387
48. Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. (2010) 15:1176–89. doi: 10.1038/mp.2009.84
49. Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. (2008) 17:1156–68. doi: 10.1093/hmg/ddn005
50. Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. (2011) 69:180–7. doi: 10.1016/j.biopsych.2010.09.030
51. Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. (2010) 124:183–91. doi: 10.1016/j.schres.2010.07.002
52. Valles A, Martens GJ, De Weerd P, Poelmans G, Aschrafi A. MicroRNA-137 regulates a glucocorticoid receptor-dependent signalling network: implications for the etiology of schizophrenia. J Psychiatry Neurosci. (2014) 39:312–20. doi: 10.1503/jpn.130269
53. Zhang J, Sun XY, Zhang LY. MicroRNA-7/Shank3 axis involved in schizophrenia pathogenesis. J Clin Neurosci. (2015) 22:1254–7. doi: 10.1016/j.jocn.2015.01.031
54. Sun XY, Zhang J, Niu W, Guo W, Song HT, Li HY, et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am J Med Genet B. (2015) 168:170–8. doi: 10.1002/ajmg.b.32292
55. Chen SD, Sun XY, Niu W, Kong LM, He MJ, Fan HM, et al. A preliminary analysis of microRNA-21 expression alteration after antipsychotic treatment in patients with schizophrenia. Psychiatry Res. (2016) 244:324–32. doi: 10.1016/j.psychres.2016.04.087
56. Du Y, Yu Y, Hu Y, Li XW, Wei ZX, Pan RY, et al. Genome-wide, integrative analysis implicates exosome-derived MicroRNA dysregulation in schizophrenia. Schizophr Bull. (2019) 45:1257–66. doi: 10.1093/schbul/sby191
57. Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. (2011) 43:969. doi: 10.1038/ng.940
58. Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. (2013) 18:708–12. doi: 10.1038/mp.2012.67
59. Navarrete K, Pedroso I, De Jong S, Stefansson H, Steinberg S, Stefansson K, et al. TCF4 (e2-2; ITF2): A schizophrenia-associated gene with pleiotropic effects on human disease. Am J Med Genet B. (2013) 162:1–16. doi: 10.1002/ajmg.b.32109
60. Kwon E, Wang W, Tsai L. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. (2013) 18:11–2. doi: 10.1038/mp.2011.170
61. Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. (2012) 141:60–4. doi: 10.1016/j.schres.2012.06.038
62. Kuswanto CN, Sum MY, Qiu A, Sitoh YY, Liu J, Sim K. The impact of genome wide supported microRNA-137 (MIR137) risk variants on frontal and striatal white matter integrity, neurocognitive functioning, and negative symptoms in schizophrenia. Am J Med Genet B. (2015) 168:317–26. doi: 10.1002/ajmg.b.32314
63. van Erp TG, Guella I, Vawter MP, Turner J, Brown GG, McCarthy G, et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol Psychiatry. (2014) 75:398–405. doi: 10.1016/j.biopsych.2013.06.016
64. Guella I, Sequeira A, Rollins B, Morgan L, Torri F, van Erp TG, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. (2013) 47:1215–21. doi: 10.1016/j.jpsychires.2013.05.021
65. Liu S, Zhang F, Shugart YY, Yang L, Li X, Liu Z, et al. The early growth response protein 1-miR-30a-5p-neurogenic differentiation factor 1 axis as a novel biomarker for schizophrenia diagnosis and treatment monitoring. Transl Psychiatry. (2017) 7:e998. doi: 10.1038/tp.2016.268
66. Ng MY, Levinson DF, Faraone SV, Suárez BK, DeLisi LE, Arinami T, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. (2009) 14:774–85. doi: 10.1038/mp.2008.135
67. He K, Guo C, He L, Shi Y. MiRNAs of peripheral blood as the biomarker of schizophrenia. Hereditas. (2018) 155:9. doi: 10.1186/s41065-017-0044-2
68. Khandaker GM, Cousins L, Deakin J, Lennox BR, Yolken R, Jones PB. Inflammation and immunity in schizophrenia: implications for pathophysiology and treatment. Lancet Psychiatry. (2015) 2:258–70. doi: 10.1016/S2215-0366(14)00122-9
69. Tian T, Wei Z, Chang X, Liu Y, Gur RE, Sleiman PMA, et al. The long noncoding RNA landscape in amygdala tissues from schizophrenia patients. EBioMedicine. (2018) 34:171–81. doi: 10.1016/j.ebiom.2018.07.022
70. Hu Z, Gao S, Lindberg D, Panja D, Wakabayashi Y, Li K, et al. Temporal dynamics of miRNAs in human DLPFC and its association with miRNA dysregulation in schizophrenia. Transl Psychiatry. (2019) 9:1–17. doi: 10.1038/s41398-019-0538-y
Keywords: lncRNA, miRNA, review, schizophrenia spectrum disorder, development of schizophrenia
Citation: Ghafouri-Fard S, Eghtedarian R, Taheri M, Beatrix Brühl A, Sadeghi-Bahmani D and Brand S (2021) A Review on the Expression Pattern of Non-coding RNAs in Patients With Schizophrenia: With a Special Focus on Peripheral Blood as a Source of Expression Analysis. Front. Psychiatry 12:640463. doi: 10.3389/fpsyt.2021.640463
Received: 11 December 2020; Accepted: 06 May 2021;
Published: 18 June 2021.
Edited by:
Sabina Berretta, Harvard Medical School, United StatesReviewed by:
Harry Pantazopoulos, University of Mississippi Medical Center, United StatesMurray John Cairns, The University of Newcastle, Australia
Shahram Arsang-Jang, Zanjan University of Medical Sciences, Iran
Seyed Javad Mowla, Tarbiat Modares University, Iran
Copyright © 2021 Ghafouri-Fard, Eghtedarian, Taheri, Beatrix Brühl, Sadeghi-Bahmani and Brand. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Serge Brand, c2VyZ2UuYnJhbmRAdW5pYmFzLmNo
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Reyhane Eghtedarian
Reyhane Eghtedarian Mohammad Taheri
Mohammad Taheri Annette Beatrix Brühl
Annette Beatrix Brühl Dena Sadeghi-Bahmani
Dena Sadeghi-Bahmani