- 1Psychosis Studies Department, Institute of Psychiatry, Psychology and Neuroscience, King's College London, London, United Kingdom
- 2Department of Psychiatry, University of Oxford, Oxford, United Kingdom
Background: Clozapine is the recommended antipsychotic for treatment-resistant schizophrenia (TRS) but there is significant variability between patients in the degree to which clozapine will improve symptoms. The biological basis of this variability is unknown. Although clozapine has efficacy in TRS, it can elicit adverse effects and initiation is often delayed. Identification of predictive biomarkers of clozapine response may aid initiation of clozapine treatment, as well as understanding of its mechanism of action. In this article we systematically review prospective or genetic studies of biological predictors of response to clozapine.
Methods: We searched the PubMed database until 20th January 2018 for studies investigating “clozapine” AND (“response” OR “outcome”) AND “schizophrenia.” Inclusion required that studies examined a biological variable in relation to symptomatic response to clozapine. For all studies except genetic-studies, inclusion required that biological variables were measured before clozapine initiation.
Results: Ninety-eight studies met the eligibility criteria and were included in the review, including neuroimaging, blood-based, cerebrospinal fluid (CSF)-based, and genetic predictors. The majority (70) are genetic studies, collectively investigating 379 different gene variants, however only three genetic variants (DRD3 Ser9Gly, HTR2A His452Tyr, and C825T GNB3) have independently replicated significant findings. Of the non-genetic variables, the most consistent predictors of a good response to clozapine are higher prefrontal cortical structural integrity and activity, and a lower ratio of the dopamine and serotonin metabolites, homovanillic acid (HVA): 5-hydroxyindoleacetic acid (5-HIAA) in CSF.
Conclusions: Recommendations include that future studies should ensure adequate clozapine trial length and clozapine plasma concentrations, and may include multivariate models to increase predictive accuracy.
Introduction
Approximately one third of patients with schizophrenia do not respond to standard antipsychotic treatment and are classified as having treatment resistant schizophrenia (TRS) (1). Clozapine has efficacy in reducing symptoms in patients who have not responded to other antipsychotics (2–4), but carries risk of serious side effects and requires regular blood monitoring. Unfortunately, clozapine will still fail to improve symptoms in 40 to 70% of TRS patients (2, 5), and currently this can only be determined through a trial of clozapine treatment. For these reasons patients and clinicians are often reluctant to initiate clozapine treatment. For example, a recent study found that there was a delay of around 4 years between patients meeting TRS criteria and the initiation of clozapine, and that during this period patients were often treated with alternative drug regimens that are not evidence-based and are associated with adverse effects, such as antipsychotics at doses higher than the maximum recommended, and antipsychotic polypharmacy (6). If tests could be developed to help clinicians predict in advance whether or not a given patient is likely to respond to clozapine, this could substantially reduce the delay before clozapine initiation, and clozapine could be selectively employed in the subset of patients in whom it is likely to be effective.
Of course, clozapine response first requires adequate dosing; patients who have clozapine plasma concentrations of 350 ng/mL or above are more likely to show improvements in symptoms, with reported sensitivity and specificity of 64–86 and 55–78% (7–10). Nonetheless a significant proportion of patients do not improve despite having adequate clozapine plasma concentrations (9), which may be termed “clozapine resistant schizophrenia” (11). An emerging number of cross-sectional studies that have compared treatment-resistant to treatment responsive schizophrenia report biological differences at group level, which may suggest that TRS is a categorically distinct illness subtype (12), and it is possible that clozapine-resistant schizophrenia may reflect a further biological subtype. Overall, this suggests that individual biological variability may play an important role in determining the degree of clozapine response in the context of adequate dosing. This raises the possibility that biological markers may be able to predict the likelihood that symptoms will improve with clozapine treatment in advance of clozapine initiation.
Numerous studies have investigated biological predictors of response to non-clozapine antipsychotics, including symptomatic response to initial antipsychotic administration in patients with first-episode psychosis [for recent review see (13)]. The degree of antipsychotic response may be related to brain structure (14), neurochemistry (15), or activity (16–19) before starting antipsychotic treatment, or associated with genetic variability (20). However, it is unknown whether similar factors may be predictive of response to clozapine, and this is a particularly important question for clinical practice as it may encourage earlier clozapine initiation in those patients most likely to benefit, or avoidance of clozapine exposure in those unlikely to respond. Recent studies indicate that there are two distinct patterns of treatment-resistance onset, with some patients developing resistance later in their illness but the majority demonstrating resistance from illness onset (21, 22), further supporting the need to promptly identify these patients and establish their likelihood of responding to clozapine.
The purpose of this article is to provide a systematic review of studies that have investigated biological predictors of response to clozapine, in order to provide an update on the research in the area and identify the most promising areas for further investigation. We limit our scope to biological variables as predictors of response. Demographic and clinical factors may also be important in understanding some aspects of clozapine response, and these have been comprehensively reviewed elsewhere (23, 24).
Methods
Search Strategy
The search was performed in the PubMed database on 20th January 2018 using the keywords “clozapine” AND (“response” OR “outcome”) AND “schizophrenia.” The search was limited to the titles and abstracts of the papers, with additional filters set to human studies and English language.
Abstracts were reviewed against study inclusion and exclusion criteria (below), and independently reviewed; there was an inter-rater reliability kappa of 0.914. The full text of the remaining potentially eligible studies were reviewed independently by authors RS and AG; there was 100% agreement on inclusion of the final studies. Reference lists were hand-searched to identify additional studies.
Study Selection
Inclusion required that studies were published in English in peer-reviewed academic journals. Inclusion also required that studies examined a biological variable in relation to clozapine response. Only studies that measured clozapine response as a change in positive, negative or overall symptom severity or global functioning were included. For biological variables such as brain activity or metabolite concentrations in blood, which may be affected by clozapine treatment, inclusion required that these measures were acquired prospectively, before clozapine initiation. For genetic variables, cross-sectional studies of clozapine response were also included. Studies were included if they investigated either clozapine monotherapy or clozapine in combination with other pharmacological or non-pharmacological interventions, as is reflective of clinical practice.
Data reported only in editorials, review articles, conference abstracts, conference reports, news articles, meta-analyses, or other non-primary data formats were excluded. Where more than one article reported data in overlapping patient samples, only the study with the largest sample was included. Studies were also excluded if the samples included a combination of patients taking only non-clozapine antipsychotics and clozapine-treated patients, without reporting results for clozapine-treated patients separately.
Data Extraction
Data were extracted into an Excel database. The following data were extracted: the biological predictor variable(s), sample size, availability of plasma clozapine concentrations (yes/no), mean plasma clozapine concentrations, mean clozapine dose, duration of clozapine treatment (months), the clozapine response criteria used, and whether results were statistically significant.
For review, articles were categorized into neuroimaging, blood-based, cerebrospinal fluid-based, cardiac, and genetic markers.
Results
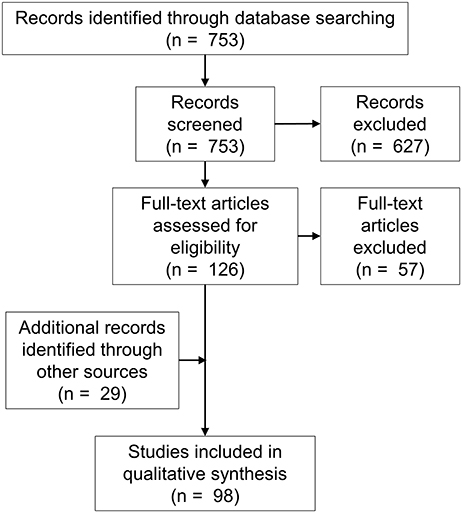
The search returned 753 articles. Abstract review identified 126 potentially eligible studies, and subsequent full-text screening identified 69 eligible studies. The excluded studies are listed in Table 1. Twenty-nine additional eligible articles were identified via other means including hand-searches of reference lists (Figure 1).
Study Characteristics
Ninety-eight studies met the inclusion criteria, for which the methodological details are provided in Tables 2, 4, 6, and 8. Of these, 70 studies investigated genetic variables, 16 studies investigated blood or CSF-based variables, 11 studies investigated neuroimaging markers, and 1 investigated a cardiac variable. Sample sizes ranged from 7 (42) to 591 participants (43). Studies included participants from across Europe (Britain, Turkey, Italy, Spain, Germany), America, Canada, and Asia (China, Israel, India, Taiwan, Pakistan).
As detailed in Tables 2, 4, 6, and 8, clozapine trial length varied from 4 weeks (44) to 16 months (45). Only nine studies (9%) reported clozapine plasma levels; of these, six gave a group mean (46–51) and three reported the mean dose for a responder and non-responder group separately (52–54). Sixty-three studies (64%) reported data on clozapine doses. Of these, 21 reported the dose range across the sample (e.g., 150–600 mg) while 36 reported the group mean and 6 reported the mean dose for a responder and non-responder group separately.
The primary outcome variables for determining clozapine response varied considerably (Tables 2, 4, 6, and 8). Thirteen studies used a combination of outcome measures to define clozapine response, and one used different outcome measures for different participants (55).
Neuroimaging Predictors of Clozapine Response
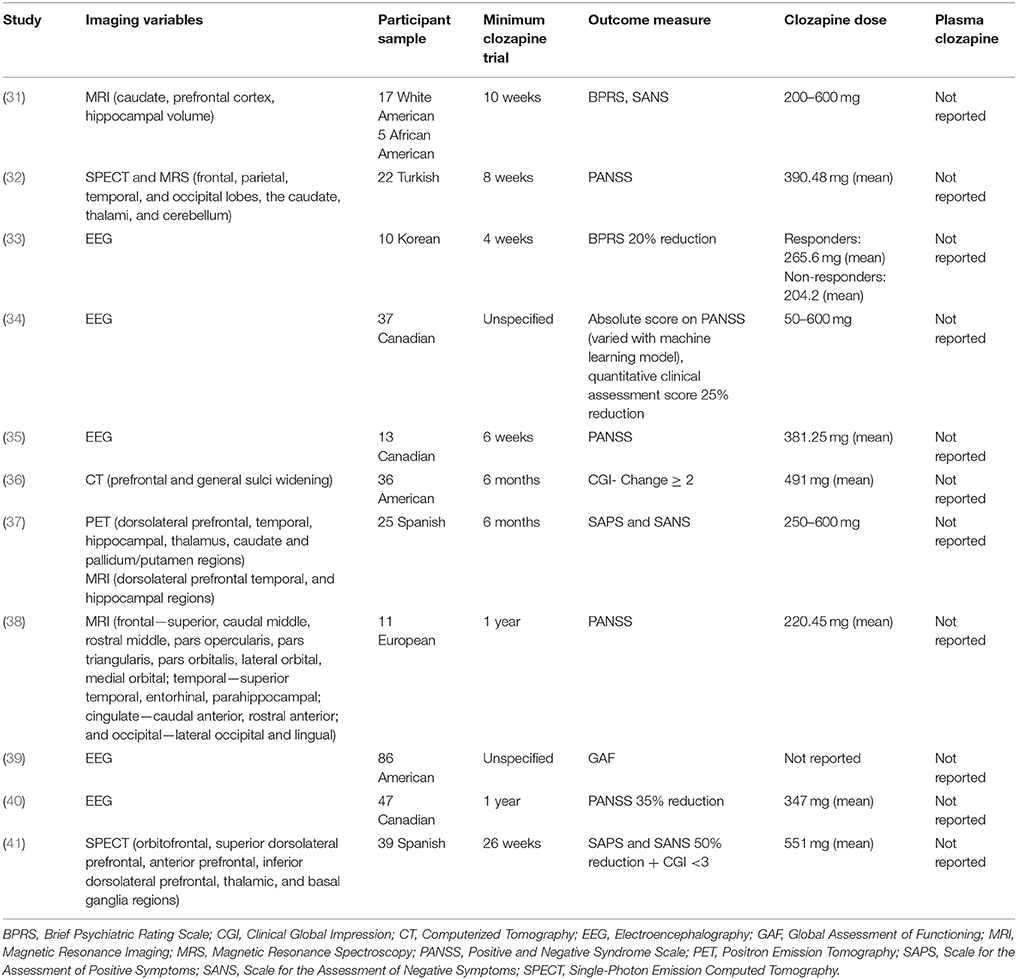
Eleven neuroimaging studies met the inclusion criteria (Tables 2, 3). These included four structural imaging studies, three single photon emission computerized tomography or positron emission tomography (SPECT/PET) studies of brain perfusion or metabolism, one proton magnetic resonance spectroscopy (1H-MRS) study of brain metabolite concentrations, and five electro-encephalography (EEG) studies. The length of clozapine treatment in the neuroimaging studies ranged from 4 weeks (33) to 1 year (37, 38), but none reported plasma clozapine levels.
Brain Structure
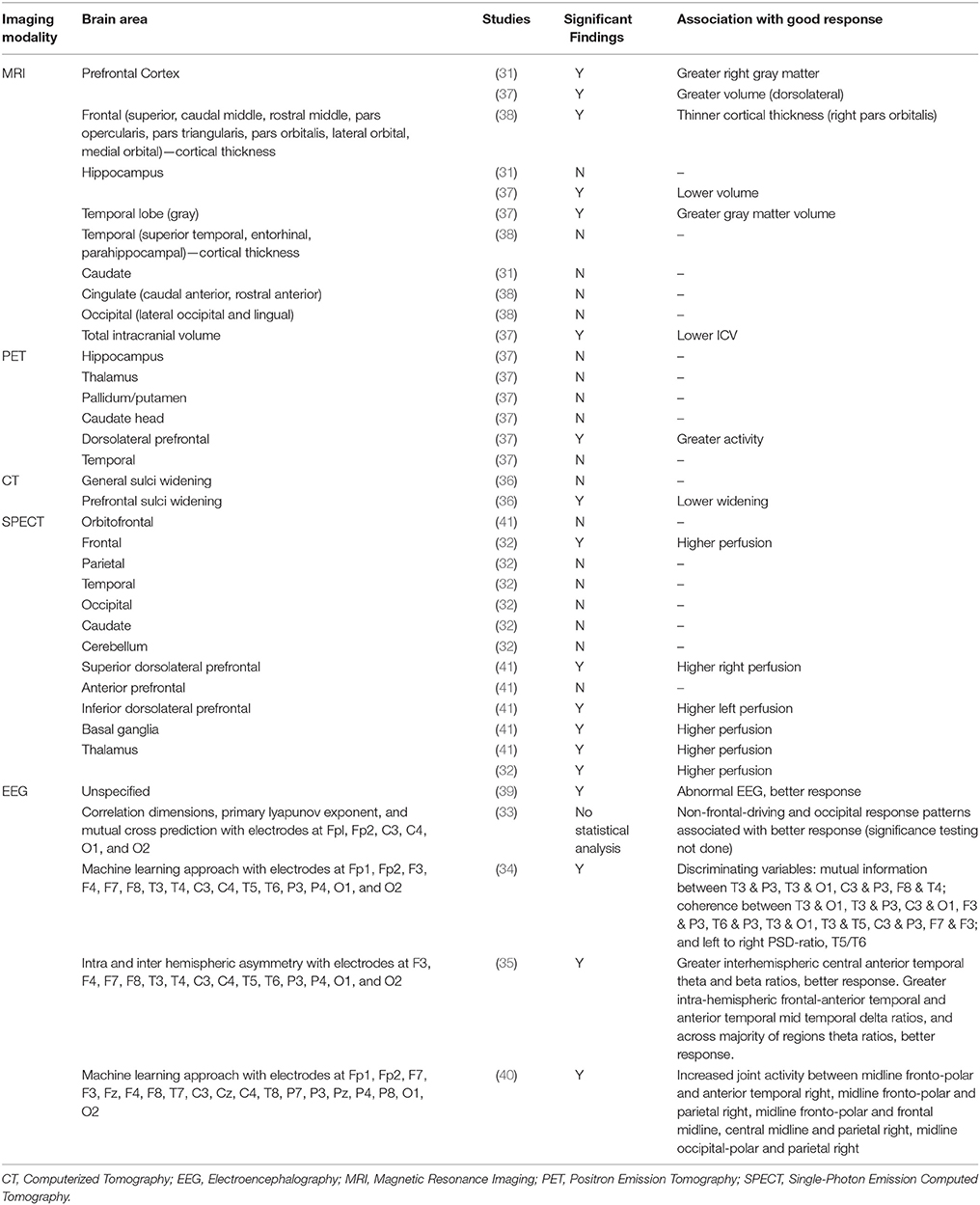
The first published study used computerized tomography (CT) to examine sulcal widening as a predictor of clozapine response (36). A good clozapine response was associated with significantly lower widening scores in the prefrontal sulci compared to a poor response, suggesting that poor clozapine response may be associated with a higher degree of frontal atrophy. Three more recent studies used structural magnetic resonance imaging (MRI) to predict clozapine response. In a clinical trial of clozapine vs. haloperidol, Arango et al. (31) found that larger right prefrontal cortical gray matter volumes were associated with greater reduction in SANS total scores after treatment in the clozapine group. No associations were found with positive symptoms, or for relationships between symptoms and caudate, hippocampal or total intracranial volumes. Molina et al. (37) investigated associations between regional brain volume and clozapine response. Temporal cortex volume was directly associated with improvement in positive symptoms, whereas dorsolateral prefrontal cortical (DLPFC) cerebrospinal fluid (CSF) content was inversely associated with improvement in positive symptoms. DLPFC volume was directly associated with improvement in negative symptom severity, and the intracranial volume was negatively related to improvement in disorganization syndrome.
These studies therefore provide a generally consistent picture that greater volumes, particularly in frontal cortical regions, are associated with a better response to clozapine treatment. However, Molina et al. (38) found that thinner baseline right pars orbitalis cortex predicted greater improvement in PANSS scores following at least 1 year clozapine use in antipsychotic-naïve first-episode patients. This difference might be explained by the different patient populations, with the two former studies including treatment-resistant patients with previous antipsychotic exposure and the latter including antipsychotic-naïve patients who may have responded to conventional antipsychotics.
Brain Perfusion and Metabolism
Regional brain perfusion and metabolism were also investigated as predictors of clozapine response. Rodriguez et al. (41), in an extension of an earlier report (56), used 99mTc-HMPAO single photon emission computed tomography (SPECT) to measure regional brain perfusion as a predictor of response to clozapine. Compared to the non-responder group, responders had higher baseline perfusion in right lower DLPFC, left upper DLPFC, thalamus, and left and right basal ganglia. Discriminant analysis showed that perfusion in the thalamus and right DLPFC distinguished between responders and non-responders with 78.9% accuracy. Similarly, Ertugrul et al. (32), also employing Tc-99m HMPAO SPECT imaging, reported that increased levels of perfusion in the right frontal cortex and thalamus were associated with greater improvement in PANSS score with clozapine treatment. Molina et al. (37), using 18F-deoxyglucose (18F-DG) positron emission tomography (PET), found that baseline metabolic rate in the DLPFC was directly related to improvement in negative symptoms, however no associations were found between metabolism in other brain regions, or with improvement in positive or disorganization symptoms. This finding of a direct association between DLPFC metabolic rate and clozapine response is consistent with findings of a direct association between DLPFC perfusion and clozapine response (32, 41).
Magnetic Resonance Spectroscopy
One 1H-MRS study investigated whether metabolite concentrations in the DLPFC may predict response to clozapine (32). In this sample of 22 patients, neither the concentration of n-acetyl aspartate (NAA) nor choline was predictive of the subsequent degree of change in symptoms on the PANSS. Relationships with other metabolites in the 1H-MRS spectrum, including glutamate, were not reported.
Electroencephalography
Five EEG studies, investigating a range of variables related to brain electrical activity, including EEG abnormalities and hemispheric asymmetry, were included (33–35, 39, 40). The first EEG study (39) investigated whether clozapine response was predicted by the presence of minor EEG abnormalities, defined as focal or generalized slowing or sharp waves, focal dysrhythmias, spikes, and spike-wave patterns. There were no overall differences in clozapine response between patients with normal compared to abnormal EEG, however secondary analysis found that in female participants, improvements in GAF score were greater in those with a normal EEG before clozapine treatment. Knott et al. (35) reported that improvements in PANSS positive, negative symptoms and global psychopathology were related to greater intrahemispheric frequency asymmetries. Kang et al. (33) ran mutual cross-prediction analysis to identify if activity in one channel was driving the dynamics of another channel. The sample was too small to conduct significant testing, but they observed that the group of participants without a frontal-driving system and occipital response system had a higher proportion of responders to clozapine. A fourth EEG study of clozapine response (34) applied a machine-learning algorithm to distinguish clozapine responders and non-responders based on their pre-treatment EEG measures, using first the leave-one-out cross-validation procedure and then two independent datasets to train and test the classifiers. This algorithm successfully distinguished these groups with more than 85% accuracy. The authors reported a list of 20 EEG measures that were found to have the greatest predictive value, which mainly included measures of the left temporal areas. Similarly, Ravan et al. (40) applied a machine-learning algorithm to patients' EEG data from before and after a year of clozapine treatment. The most-responsive patients had five “discriminating features” at baseline; these were predominantly in the beta-band, with the most dominant features joint activity between the pre-frontal and right parietal or right anterior temporal regions.
CSF-Based Predictors of Clozapine Response
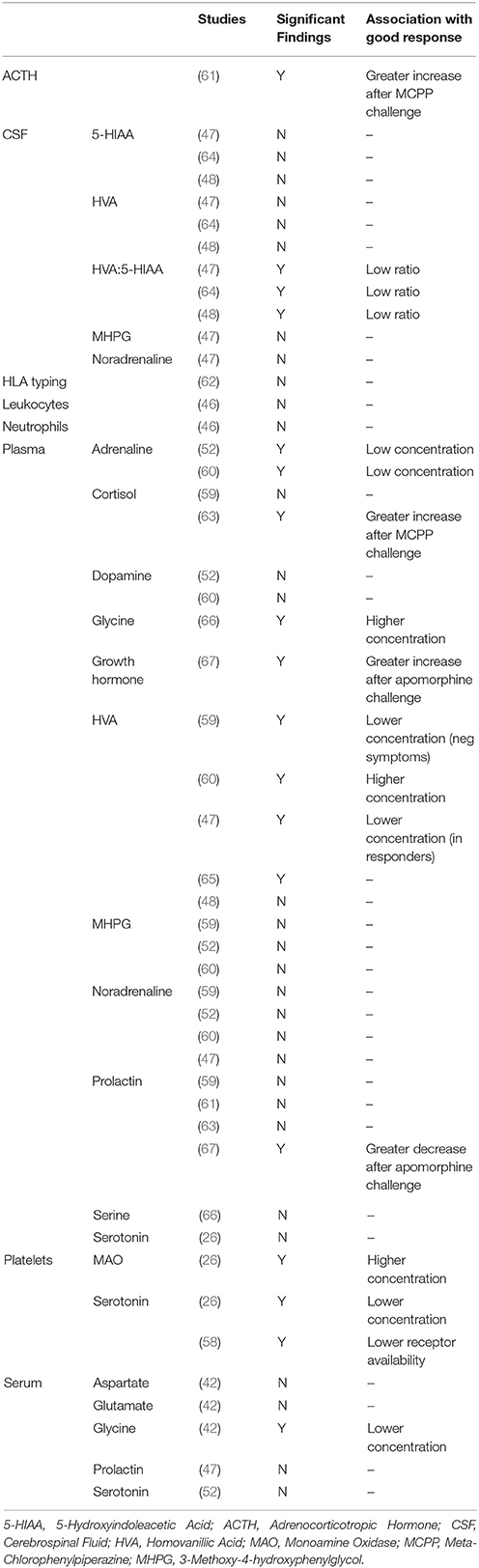
A priori selection of CSF- and peripheral predictive biomarkers of clozapine response has been driven by clozapine's “atypical” pharmacological profile of high affinity at serotonin 5-HT2A receptors in combination with lower affinity at dopamine D2 receptors (57). Our search returned three studies of CSF biochemicals in predicting clozapine response (Tables 4, 5). Two of these studies provided data on plasma clozapine concentrations (47, 48). Sample sizes in these studies ranged from 10 (64) to 21 participants (47), and all used the BPRS to measure symptomatic improvement.
CSF Monoamines
All three studies investigated the dopamine metabolite homovanillic acid (HVA), and the serotonin metabolite 5-hydroxyindoleacetic acid (5-HIAA) (47, 48, 64). None of these studies found that HVA nor 5-HIAA concentrations alone were predictive of clozapine response. The ratio between HVA and 5-HIAA was also investigated. In all studies, lower HVA/5-HIAA concentration ratios before clozapine were associated with a greater degree of subsequent symptomatic improvement, both in the short- and longer-term (47, 48, 64). This suggests that the balance between dopamine and serotonin metabolism before clozapine administration may be predictive of clozapine response, with lower levels of dopamine metabolism relative to higher levels of serotonin metabolism being associated with better outcomes.
One study also investigated concentrations of the noradrenaline metabolite 3-methoxy-4-hydroxyphenylglycol (MHPG) in relation to clozapine response, and found no association (47).
CSF Hormones
A single study investigated CSF prolactin concentrations as a predictor of clozapine response and found no association (47).
Blood-Based Predictors of Clozapine Response
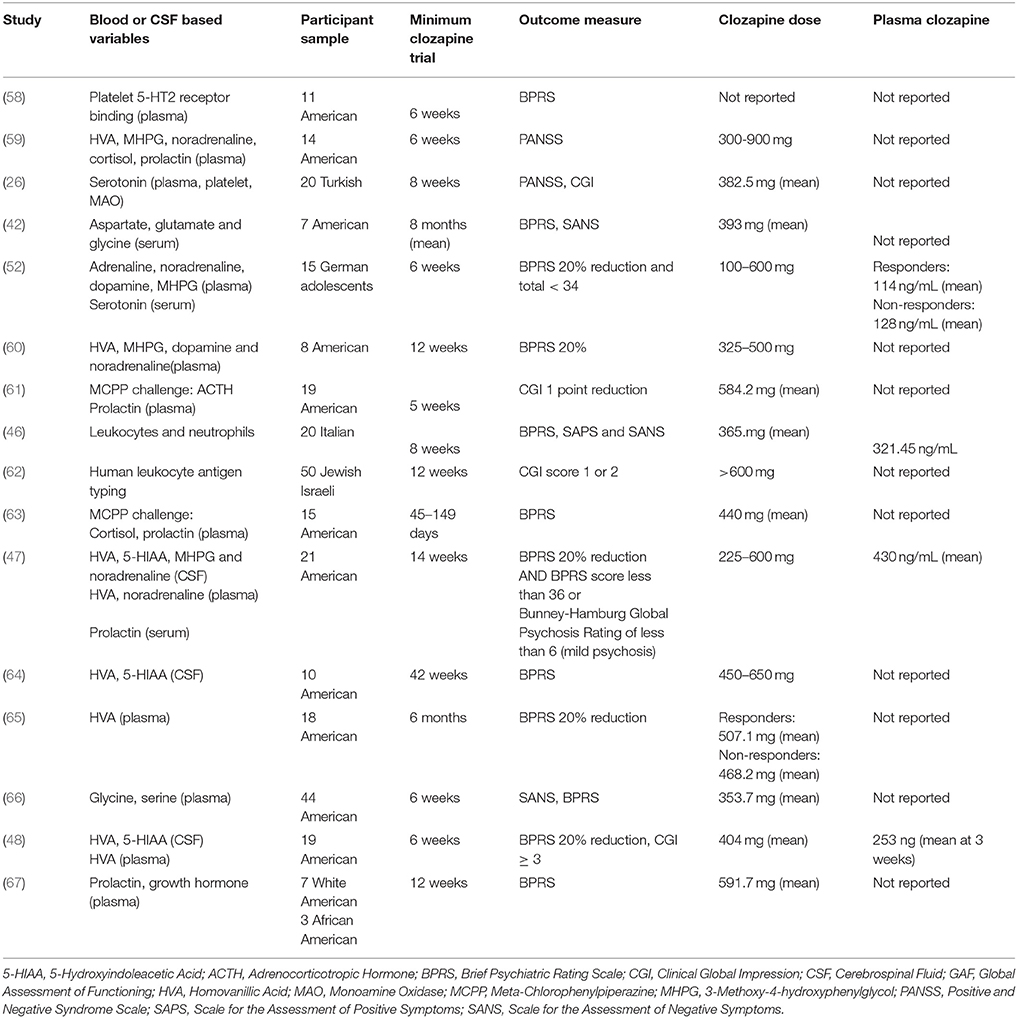
Our search returned 11 studies which investigated biochemicals in plasma, serum or platelets as predictors of clozapine response (Tables 4, 5). As for CSF approaches, these peripheral studies have also focussed on dopaminergic and serotonergic measures. Sample sizes in these studies ranged from 7 (42) to 50 participants (62), and all except Kahn et al. (61) and Ertugrul et al. (26) used the BPRS to measure symptomatic improvement. Data on plasma clozapine concentrations were unavailable in all but three of the studies (47, 48, 52).
Blood Monoamines
Several studies have investigated peripheral dopaminergic variables as predictors of clozapine response, with overall negative or inconclusive findings. Two studies investigated plasma dopamine concentrations, both with negative findings (52, 60). The five studies which investigated concentrations of the dopamine metabolite HVA in plasma have reported mixed findings. Pickar et al. (47) initially reported that lower baseline plasma HVA concentrations were associated with greater reductions in symptoms, but three later studies reported that higher baseline plasma HVA concentrations were associated with greater symptom reduction (59, 60, 65), although one study found this only for negative symptoms (59) and one study found this only as a correlation with positive symptoms within the clozapine responder group (65). A further study found no association between plasma HVA and clozapine response (48). One study investigated concentrations of platelet monoamine oxidase B (MAO-B) which metabolizes dopamine (68), and found a positive association with symptom improvements following clozapine (26). Finally, as a dopaminergic pharmacological challenge, apomorphine-induced prolactin suppression and growth hormone secretion predicted better clozapine response in a preliminary study (67).
In terms of peripheral serotonergic studies, Ertugrul et al. (26) found no association with plasma serotonin concentrations and clozapine response as did an earlier study of serum serotonin concentrations in children and adolescents (52). However, Ertugrul et al. (26) also reported a negative correlation between platelet serotonin concentrations, (reflecting uptake of plasma serotonin through platelet serotonin transporters) and improvement in positive symptoms following clozapine. Arora and Meltzer (58) measured platelet 5HT2 receptor binding in platelet-rich plasma and reported that a lower number of 5HT2 binding sites before clozapine initiation was associated with poorer treatment outcomes.
Pharmacological serotonin challenge using the non-selective 5-HT receptor agonist m-chlorophenylpiperazine (mCPP) has also been employed to investigate clozapine response (61, 63). mCPP-induced adrenocorticotropic hormone (ACTH) release (61) and plasma cortisol (63) were directly associated with improvement in symptoms. In contrast, there was no association between MCPP-induced prolactin increase and clozapine response in either study (61, 63). The finding of increased MCPP-responses would suggest that elevated 5-HT system function is associated with better clinical responses to clozapine.
Finally, four studies investigated adrenaline, noradrenaline or MHPG concentrations. Two studies reported that low plasma adrenaline concentrations associate with better clozapine response (52, 60). In contrast, studies have found no association between plasma noradrenaline concentrations (47, 52, 59, 60), or plasma MHPG and clozapine response (52, 59, 60).
Blood Glutamatergic Amino Acids
The glutamatergic amino acids glycine and serine act as endogenous co-agonists at the N-methyl-D-aspartate (NMDA) glutamate receptor complex, which is thought to be hypofunctional in schizophrenia and therefore increasing glycine or serine levels may have therapeutic potential (69). Two studies (42, 66) investigated glycine and serine concentrations in relation to clozapine response, from serum and plasma respectively, and have produced conflicting evidence. In a sample of 7 patients, Evins et al. (42) found that lower serum glycine concentrations predicted a better response to clozapine, whereas in the larger and longer-term study of Sumioyshi et al. (66) higher plasma glycine concentrations and higher plasma glycine/serine ratios predicted greater negative symptom improvements, whereas no associations were found between serine concentrations and clozapine response. Evins et al. (42) also measured glutamate and aspartate concentrations and report no significant associations.
Blood Hormones
One study investigated serum prolactin levels (47) and another investigated plasma prolactin and cortisol levels (59). Neither of these studies reported significant associations with clozapine response.
Blood Immunological Variables
Two studies have looked at immological variables as predictors of clozapine response. Mauri et al. (46) measured neutrophil and leukocyte numbers before 8 weeks of clozapine treatment in 20 patients. They do not report significance testing but provide summary statistics; independent t-tests using this data indicates no association with response to clozapine. Meged et al. (62) investigated human leukocyte antigen (HLA) type in 50 Israeli patients but found no association between HLA type and response to clozapine after 12 weeks.
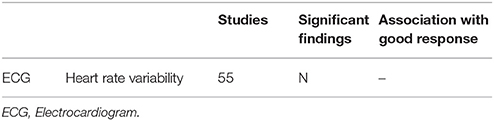
Cardiac Predictors of Clozapine Response
One study investigated heart rate variability in 40 participants with treatment-resistant schizophrenia using ECG (70) but did not find any pre-clozapine differences in heart rate variability associated with changes in BPRS after 8 weeks of clozapine treatment (Tables 6, 7).
Genetic Predictors of Clozapine Response
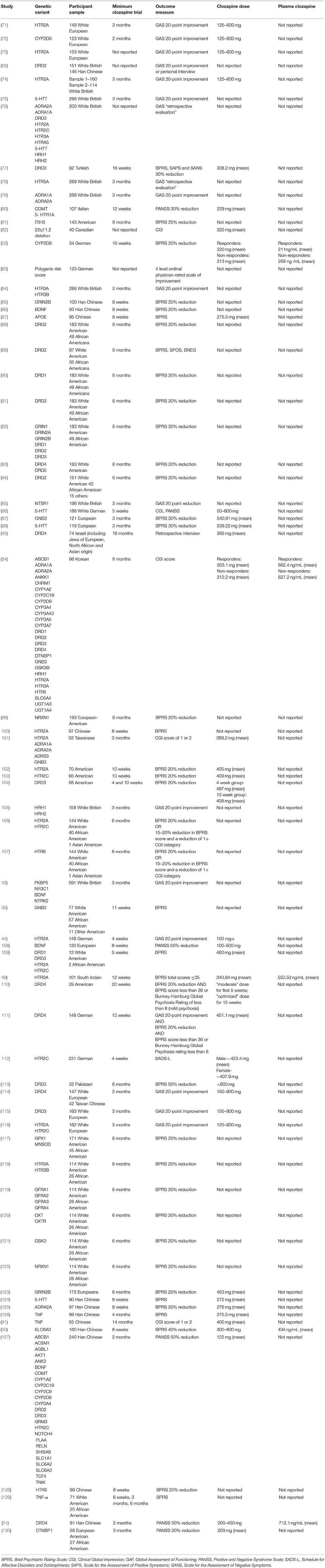
We identified a total of 70 studies investigating associations between genetic variants and clozapine response (Tables 8–11). In the first study of its kind, Frank et al. (83) recently reported that higher genetic risk of schizophrenia, calculated as the schizophrenia polygenic risk score (131), was associated with a poorer degree of response to clozapine1. Butcher et al. (82) recently reported that individuals with a large chromosomal deletion (22q11.2) respond as well to clozapine as patients with schizophrenia who do not have this deletion.
Of the other genetic studies, seven reported significant associations between genetic haplotypes of DRD1, DRD2, DRD3, FKBP5, GFRA2, HTR3A, and NTRK2 (43, 88–91, 119) (see Table 10) and clozapine response, but none of these have been replicated. Two unreplicated studies also reported significant associations between gene-gene interactions of DRD1 and DRD2, DRD2 and DRD3, DRD1 and GRIN2A, and GFRA1, GFRA2, and GFRA3 (92, 119) and clozapine response (see Table 11).
Two studies reported the predictive validity of multivariate genetic models. One study investigated a logistic regression analysis with a combination of six polymorphisms (T102C and His452Tyr of HTR2A gene, G-330T/ C-244T repeat and Cys23Ser of HTR2C gene, HTTLPR of SLC6A4 gene, G-1018A of HRH1) which was able to predict clozapine response with the retrospective positive predictive value of 76.7%, negative predictive value of 82%, a sensitivity of 95% and specificity of 38% (76). A more recent study used an artificial neural network analysis to combine five genetic polymorphisms (T102C of the HTR2A gene, Arg347Cys of the ADRA1A gene, −1291 C>G of the ADRA2A gene, Trp64Arg of the ADRB3 gene, and 825 C>T of the GNB3 gene), which were insignificant individually, with clinical predictor variables (gender, age, height, baseline body weight, baseline body mass index) (101). This approach was able to retrospectively identify all clozapine responders and 76.5% clozapine non-responders.
However, our search mainly returned studies that have employed candidate gene approaches to investigation of clozapine response. Overall, these studies have investigated associations with clozapine response for a total of 379 different gene variants, 362 of which relate to single nucleotide polymorphisms (SNPs). For these studies, we limit comment to significant findings with at least one replication. Of the 379 different gene variants investigated, significant findings have been reported for 40 variants, 8 of which have been replicated. 28 variants have replicated null results with no significant findings, including the rs6275 and rs6277 polymorphisms of DRD2 (54, 88, 127) and the val66met polymorphism in BDNF (43, 86, 108, 127). The details for all genetic studies, including those with non-significant or non-replicated findings, are provided in Table 9.
Dopaminergic Genes
The DRD3 gene, encoding the D3 dopamine receptor, has been investigated in nine studies, all of which have investigated the Ser9Gly polymorphism of rs6280. While two initial studies independently reported that the Gly allele was associated with a good response to clozapine (113, 115), all seven subsequent studies found non-significant results (54, 76, 77, 91, 104, 109, 127), including the two studies with the largest sample size (76, 91).
Serotonergic Genes
The HTR2A gene, encoding the 5-HT2A receptor at which clozapine has high affinity, has been investigated in 12 studies. The His allele of His452Tyr has been associated with good response to clozapine in four studies conducted by two research groups (73, 74, 76, 106), although two studies did not detect this association (44, 102). Within the same gene, the T allele of the T102C polymorphism has been associated with good response to clozapine in three studies by the same research group (71, 76, 116), although seven studies by other groups have failed to replicate these findings (44, 54, 100–102, 106, 109). The G-1438A SNP also significantly predicted clozapine response in two studies by the same group (74, 76) but these results were not replicated in a second sample analyzed by the same research group (74) or in separate samples from two independent research groups (54, 106).
The HTR3A gene has been investigated in five studies (49, 54, 76, 84, 118); the only SNP which has been reported more than once, across all five studies, is rs1062613, with one study finding that good response to clozapine was associated with the T allele (49), another finding that good clozapine response was associated with the C allele (118) and the other three studies reporting no association.
The 5HTT (or SLC6A4) gene, encoding the serotonin transporter, has been investigated in six studies by five independent groups (54, 75, 76, 96, 98, 124), with the only independently replicated finding for an association of the HTTLPR polymorphism at rs25531 with clozapine response; Kohlrausch et al. (98) found an association between good response and the long allele, but Arranz et al. (76) do not report the direction of effect.
Other Gene Variants
An association between the C allele of the C825T polymorphism in the gene encoding G-protein subunit-beta 3 (GNB3) and a good response to clozapine has been reported in two studies performed by independent research groups (30, 97), though two separate studies by two other research groups have found no association (54, 101).
Discussion
Since 1992, ninety-eight published studies have tested biological predictors of symptomatic response to clozapine. While this highlights the potential clinical importance of identifying good clozapine responders in advance of starting treatment, these 25 years of research have failed to produce biomarkers with sufficient accuracy for clinical decision making. The most consistent findings are that a good response to clozapine is associated with greater structural integrity and activity in prefrontal cortical areas, possibly reflecting less severe brain pathophysiology than in poor responders, and a lower ratio of the dopamine metabolite HVA to the serotonin metabolite 5-HIAA in CSF before clozapine initiation, reflecting higher serotonergic compared to dopaminergic turnover. However, there have been relatively few studies investigating these biomarkers prospectively and further replication is required.
Regarding prefrontal cortical areas, prospective studies have found consistent evidence that higher prefrontal cortical volumes before clozapine initiation are directly associated with a greater degree of symptomatic response to clozapine (31, 36, 37), with some suggestion of specificity to improvements in negative symptom severity (31, 37). Studies examining perfusion or metabolism have similarly associated higher levels of prefrontal activity with a higher degree of symptomatic response (32, 37, 41). These results are consistent with the majority, but not all (132) of cross-sectional studies finding that clozapine responders have higher prefrontal cortical volumes than non-responders (25, 27, 28). In addition, some evidence indicates that integrity/activity of the thalamus may also be important in predicting clozapine response (32, 37, 41). Importantly, the jack-knifed classification of Rodriguez et al. (41) using DLPFC and thalamic activity correctly identified 78.9% cases according to clozapine response, and the effect size of the difference in prefrontal sulcal widening score between clozapine responders and non-responders reported by Konicki et al. (36) can be calculated as a large effect size of d = 3.8.
It is unclear whether prefrontal structural integrity or activity may be predictive of clozapine response specifically, or whether prefrontal integrity is non-specifically prognostic of outcome. Findings relating prefrontal volume to symptom outcomes in non-clozapine treated patients are mixed (37, 133–135), with the largest study finding no relationships between gray matter volume at illness onset and outcome 2 years later (135). Some studies indicate that clozapine has greater ability to modulate prefrontal activity than other antipsychotic compounds (29, 109, 136–138), but we are not aware of any studies that have specifically compared the ability of prefrontal cortical variables to predict response to clozapine vs. other antipsychotics. Determination of treatment specificity would be important for clinical decision-making around clozapine initiation.
The other most replicated finding is that the ratio of the dopamine to serotonin metabolites HVA:5-HIAA in CSF at baseline predicted clozapine response (47, 48, 64). Where available, the effect sizes calculated for these studies are large [d = 0.8 (47) and 1.2 (48)]. CSF HVA and 5-HIAA respectively reflect brain dopaminergic and serotonergic turnover, with some evidence that lumbar CSF HVA is primarily from the striatum (139) and 5-HIAA from the frontal cortex (47). These findings in the absence of predictive value of CSF HVA or 5-HIAA alone suggest that the dopamine-serotonin balance is predictive of clozapine response. One report that CSF HVA/5-HIAA ratio was not predictive of response to olanzapine (140) may be suggestive of clozapine specificity, although further confirmation is needed.
In terms of genetic predictors of clozapine response, our results highlight the overall inability of candidate gene approaches to reproducibly predict clozapine response. Of the 379 polymorphisms investigated in relation to clozapine response, replication by two or more independent research groups is only available for the DRD3 Ser9Gly (113, 115), HTR2A His452Tyr (73, 74, 76, 106), 5HTT rs25531 (76, 98), and C825T GNB3 (30, 97) polymorphisms. Furthermore, findings of no association with clozapine response were also reported for DRD3 (76, 77, 91, 104, 109), HTR2A His452Tyr (44, 102), C825T GNB3 (54) and no findings were replicated by more than two independent groups. However, as is the case for schizophrenia, clozapine response is unlikely to be dictated by a single gene variant, and more likely reflects additive or interacting effects at multiple genetic loci. One study investigating a combination of six polymorphisms predicted clozapine response with the retrospective positive predictive value of 76.7% and a sensitivity of 95% (76) on which basis a pharmacogenetic test was developed, although it is no longer available. Similarly, using an artificial neural network to combine five polymorphisms with clinical data retrospectively identified all clozapine responders and 76.5% of non-responders (101).
Since many of these studies were done, technology has advanced to genome-wide association studies (GWAS), which take a hypothesis-free approach but require very large samples. GWAS is being applied to identify polymorphisms contributing to response to non-clozapine antipsychotics (141) and may be applied to clozapine in the future. This approach is encouraged by reports that polygenic risk scores for schizophrenia may associate with the degree of clozapine response (83). However, genome-wide approaches specifically comparing good vs. poor responders to clozapine are required because many of the candidate gene studies identified by our review investigated polymorphisms previously associated with non-clozapine antipsychotic response with minimal success or without replication [e.g., NRXN1: (122); ABCB1: (54, 127)], indicating that clozapine research would benefit from approaches able to identify novel genetic associations. Another avenue to explore is epigenetic variation, in the form of chemical modifications associated with differing gene expression such as DNA or histone methylation, which may play a role in clozapine response above and beyond genetic variation; evidence indicates both that variation in these modifications is associated with schizophrenia (142) and that clozapine induces changes in these modifications (143).
Our review also highlights several methodological considerations for future studies examining predictive biomarkers of clozapine response. First, there are overall relatively few studies that have prospectively examined non-genetic biological predictors of clozapine response despite their potential clinical importance. This likely reflects several practical factors. In our own experience, patients who are about to start clozapine can be difficult to recruit to research involving neuroimaging or invasive procedures, because they are often very unwell and may lack capacity to consent. Additionally, research participation needs to be approached and timed carefully around clinical conversations regarding clozapine initiation. This may partly explain the relatively few studies overall, and small sample sizes in some studies.
Secondly, although a response to clozapine will require adequate dosing, only nine of the ninety-eight studies included in our review reported clozapine plasma concentrations. Without this information it is not possible to determine the extent to which poor response may reflect sub-therapeutic plasma clozapine concentrations rather than clozapine inefficacy. There was also significant variability in criteria used to determine clozapine response/non-response as well as variability in clozapine treatment duration. Clinical trials indicate that the majority of patients who will respond to clozapine will do so in the first 6 weeks of treatment, which is associated with ~30% response (57, 144). By 12 weeks of clozapine treatment, a response is seen in 40–50% of patients (145, 146). Therefore, studies of less than 12 weeks duration may have been too short to establish clozapine response or non-response. To address some of this inconsistency, the Treatment Response and Resistance in Psychosis (TRRIP) Working Group have recently provided consensus guidelines for determining and reporting adequate treatment and treatment response (11); this includes a recommendation that clozapine therapy be maintained for a minimum of 3 months after therapeutic plasma levels are reached before determining response.
As with other biomarker research, technical constraints, and cost may impede the translation of some markers to clinical practice. Broadly, blood-based biomarkers may be more readily implemented than biomarkers requiring advanced neuroimaging techniques, lumbar puncture or specialized analysis. However, this should be balanced against the high economic costs of treatment resistant schizophrenia. Models based on clinical or demographic factors may be easier to implement. However, as for biological markers, previous reviews of clinical predictors of clozapine response have failed to identify any with “adequate reproducibility, sensitivity and specificity for clozapine,” instead suggesting that a combination of factors may be most fruitful (24). Another broader challenge is the lack of established biological underpinnings for schizophrenia and the subsequent heterogeneity in patients, which may obscure identification of biological predictors. Research indicates potential categorical differences between patients with treatment-responsive and treatment-resistant schizophrenia (12), as well as potential sub-groups within treatment-resistant patients (21), with further sub-groups likely. Such differences may contribute to the lack of reproducible research findings, and future research could explore whether predictors of outcome are specific to sub-groups within the schizophrenia diagnosis.
In conclusion, this review supports the notion that biological measures might be useful in predicting response to clozapine, and that higher prefrontal structural integrity and activity and lower ratios of HVA/5-HIAA in CSF may be associated with a better response. Future research should confirm these findings, investigate treatment-specificity, and apply genome-wide approaches. If these approaches are to aid clinical decision making, future studies will also need to address the accuracy of prediction at the individual patient level, which may be facilitated by statistical models combining neuroimaging, CSF-based, blood-based, genetic, clinical, or demographic measures.
Author Contributions
RS and AE designed the study and protocol. RS and AG conducted the systematic review. RS, AG, and AE jointly wrote the first draft of the manuscript. GM, K-VS, and JM provided additional intellectual contributions, and all authors contributed to and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by a UK Medical Research Council (MRC) grant MR/ L003988/1 (AE). This study presents independent research funded in part by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley National Health Service (NHS) Foundation Trust and King's College London. AG received a Biomedical Research Studentship from the NIHR Biomedical Research Centre. The views expressed are those of the authors and do not necessarily represent those of the NHS, NIHR, or Department of Health.
Abbreviations
ACTH, adrenocorticotropic hormone; BPRS, brief psychiatric rating Scale; CGI, clinical global impression; CSF, cerebrospinal fluid; CT, computerized tomography; DLPFC, dorsolateral prefrontal cortex; ECG, electrocardiogram; EEG, electroencephalogram; GWAS, genome-wide association studies; HLA, human leukocyte antigen; 5-HIAA, ty5-hydroxyindoleacetic acid; HVA, homovanillic acid; MAO-B, monoamine oxidase B; MCPP, m-chlorophenylpiperazine; MHPG, 3-methoxy-4-hydroxyphenylglycol; MRI, magnetic resonance imaging; MRS, magnetic resonance spectroscopy; NAA, n-acetyl aspartate; NMDA, N-methyl-D-aspartate; PANSS, positive and negative syndrome scale; PET, positron emission tomography; SANS, scale for the assessment of negative symptoms; SAPS, scale for the assessment of positive Symptoms; SNPs, single nucleotide polymorphisms; SPECT, single photon emission computerized tomography; TRS, treatment resistant schizophrenia.
Footnotes
1. ^This was clarified directly with the corresponding author for this paper due to discrepancies between the text and figure in the paper.
reference
1. Meltzer HY. Treatment-resistant schizophrenia–the role of clozapine. Curr Med Res Opin. (1997) 14:1–20. doi: 10.1185/03007999709113338
2. Kane J, Honigfeld G, Singer J, Meltzer H. Clozapine for the treatment-resistant schizophrenic: a double-blind comparison with chlorpromazine. Arch Gen Psychiatry (1988) 45:789–96. doi: 10.1001/archpsyc.1988.01800330013001
3. Lewis SW, Barnes TRE, Davies L, Murray RM, Dunn G, Hayhurst KP, et al. Randomized controlled trial of effect of prescription of clozapine versus other second-generation antipsychotic drugs in resistant schizophrenia. Schizophr Bull. (2006) 32:715–23. doi: 10.1093/schbul/sbj067
4. Rosenheck R, Cramer J, Xu W, Thomas J, Henderson W, Frisman L, et al. A comparison of clozapine and haloperidol in hospitalized patients with refractory schizophrenia. N Engl J Med. (1997) 337:809–15. doi: 10.1056/NEJM199709183371202
5. Meltzer HY, Bobo WV, Roy A, Jayathilake K, Chen Y, Ertugrul A, et al. A randomized, double-blind comparison of clozapine and high-dose olanzapine in treatment-resistant patients with schizophrenia. J Clin Psychiatry (2008) 69:274–85. doi: 10.4088/JCP.v69n0214
6. Howes OD, Vergunst F, Gee S, McGuire P, Kapur S, Taylor D. Adherence to treatment guidelines in clinical practice: study of antipsychotic treatment prior to clozapine initiation. Br J Psychiatry (2012) 201:481–5. doi: 10.1192/bjp.bp.111.105833
7. Kronig MH, Munne RA, Szymanski S, Safferman AZ, Pollack S, Cooper T, et al. Plasma clozapine levels and clinical response for treatment-refractory schizophrenic patients. Am J Psychiatry (1995) 152:179–82. doi: 10.1176/ajp.152.2.179
8. Miller DD, Fleming F, Holman TL, Perry PJ. Plasma clozapine concentrations as a predictor of clinical response: a follow-up study. J Clin Psychiatry (1994) 55(Suppl. B):117–21.
9. Perry PJ, Miller DD, Arndt SV, Cadoret RJ. Clozapine and norclozapine plasma concentrations and clinical response of treatment-refractory schizophrenic patients. Am J Psychiatry (1991) 148:231–5.
10. Spina E, Avenoso A, Facciolà G, Scordo MG, Ancione M, Madia AG, et al. Relationship between plasma concentrations of clozapine and norclozapine and therapeutic response in patients with schizophrenia resistant to conventional neuroleptics. Psychopharmacology (2000) 148:83–9. doi: 10.1007/s002130050028
11. Howes OD, McCutcheon R, Agid O, de Bartolomeis A, van Beveren NJ, Birnbaum ML, et al. Treatment-resistant schizophrenia: Treatment Response and Resistance in Psychosis (TRRIP) working group consensus guidelines on diagnosis and terminology. Am J Psychiatry (2016) 174:216–29. doi: 10.1176/appi.ajp.2016.16050503
12. Gillespie AL, Samanaite R, Mill J, Egerton A, MacCabe JH. Is treatment-resistant schizophrenia categorically distinct from treatment-responsive schizophrenia? a systematic review BMC Psychiatry (2017) 17:12. doi: 10.1186/s12888-016-1177-y
13. Malhotra AK. Dissecting the Heterogeneity of Treatment Response in First-Episode Schizophrenia. Schizophr Bull. (2015) 41:1224–6. doi: 10.1093/schbul/sbv117
14. Dazzan P. Neuroimaging biomarkers to predict treatment response in schizophrenia: the end of 30 years of solitude? Dialogues Clin Neurosci. (2014) 16:491–503. doi: 10.1016/S0924-977X(16)30935-X
15. Szulc A, Galinska B, Tarasów E, Kubas B, Dzienis W, Konarzewska B, et al. N-acetylaspartate (NAA) levels in selected areas of the brain in patients with chronic schizophrenia treated with typical and atypical neuroleptics: a proton magnetic resonance spectroscopy (1H MRS) study. Med Sci Monit. (2007) 13:17–22. Available online at: https://www.medscimonit.com/download/index/idArt/482274
16. Ehlis AC, Pauli P, Herrmann MJ, Plichta MM, Zielasek J, Pfuhlmann B, et al. Hypofrontality in schizophrenic patients and its relevance for the choice of antipsychotic medication: an event-related potential study. World J Biol Psychiatry (2012) 13:188–99. doi: 10.3109/15622975.2011.566354
17. Hadley JA, Nenert R, Kraguljac NV, Bolding MS, White DM, Skidmore FM, et al. Ventral tegmental area/midbrain functional connectivity and response to antipsychotic medication in schizophrenia. Neuropsychopharmacology (2014) 39:1020–30. doi: 10.1038/npp.2013.305
18. Kraguljac NV, White DM, Hadley N, Hadley JA, Hoef L, Ver Davis E, et al. Aberrant hippocampal connectivity in unmedicated patients with schizophrenia and effects of antipsychotic medication: a longitudinal resting state functional mri study. Schizophr Bull. (2016) 42:1046–55. doi: 10.1093/schbul/sbv228
19. Sarpal DK, Robinson DG, Lencz T, Argyelan M, Ikuta T, Karlsgodt K, et al. Antipsychotic treatment and functional connectivity of the striatum in first-episode schizophrenia. JAMA Psychiatry (2015) 72:5–13. doi: 10.1001/jamapsychiatry.2014.1734
20. Reynolds GP. The pharmacogenetics of symptom response to Antipsychotic drugs. Psychiatry Investig. (2012) 9:1–7. doi: 10.4306/pi.2012.9.1.1
21. Demjaha A, Lappin JM, Stahl D, Patel MX, MacCabe JH, Howes OD, et al. Antipsychotic treatment resistance in first-episode psychosis: prevalence, subtypes and predictors. Psychol Med. (2017) 47:1981–9. doi: 10.1017/S0033291717000435
22. Lally J, Ajnakina O, Di Forti M, Trotta A, Demjaha A, Kolliakou A, et al. Two distinct patterns of treatment resistance: clinical predictors of treatment resistance in first-episode schizophrenia spectrum psychoses. Psychol Med. (2016) 46:3231–40. doi: 10.1017/S0033291716002014
23. Chung C, Remington G. Predictors and markers of clozapine response. Psychopharmacology (2005) 179:317–35. doi: 10.1007/s00213-005-2174-x
24. Suzuki T, Uchida H, Watanabe K, Kashima H. Factors associated with response to clozapine in schizophrenia: a review. Psychopharmacol Bull. (2011) 44:32–60.
25. Ahmed M, Cannon DM, Scanlon C, Holleran L, Schmidt H, McFarland J, et al. Progressive brain atrophy and cortical thinning in schizophrenia after commencing clozapine treatment. Neuropsychopharmacology (2015) 40:1–30. doi: 10.1038/npp.2015.90
26. Ertugrul A, Ucar G, Basar K, Demir B, Yabanoglu S, Ulug B. Influence of clozapine on platelet serotonin, monoamine oxidase and plasma serotonin levels. Psychiatry Res. (2007) 149:49–57. doi: 10.1016/j.psychres.2005.12.009
27. Friedman L, Knutson L, Shurell M, Meltzer HY. Prefrontal sulcal prominence is inversely related to response to clozapine in schizophrenia. Biol Psychiatry (1991) 29:865–77. doi: 10.1016/0006-3223(91)90053-O
28. Honer WG, Smith GN, Lapointe JS, Macewan GW, Kopala L, Altman S. Regional cortical anatomy and clozapine response in refractory schizophrenia. Neuropsychopharmacology (1995) 13:85–7. doi: 10.1016/0893-133X(95)00017-8
29. Lahti AC, Holcomb HH, Weiler MA, Medoff DR, Frey KN, Hardin M, et al. Clozapine but not haloperidol Re-establishes normal task-activated rCBF patterns in schizophrenia within the anterior cingulate cortex. Neuropsychopharmacology (2004) 29:171–8. doi: 10.1038/sj.npp.1300312
30. Müller DJ, De Luca V, Sicard T, King N, Hwang R, Volavka J, et al. Suggestive association between the C825T polymorphism of the G-protein 3 subunit gene (GNB3) and clinical improvement with antipsychotics in schizophrenia. Eur Neuropsychopharmacol. (2005) 15:525–31. doi: 10.1016/j.euroneuro.2005.02.001
31. Arango C, Breier A, McMahon R, Carpenter WT, Buchanan RW. The relationship of clozapine and haloperidol treatment response to prefrontal, hippocampal, and caudate brain volumes. Am J Psychiatry (2003) 160:1421–7. doi: 10.1176/appi.ajp.160.8.1421
32. Ertugrul A, Volkan-Salanci B, Basar K, Oguz KK, Demir B, Ergun EL, et al. The effect of clozapine on regional cerebral blood flow and brain metabolite ratios in schizophrenia: Relationship with treatment response. Psychiatry Res. (2009) 174:121–9. doi: 10.1016/j.pscychresns.2009.04.007
33. Kang UG, Park KT, Ahn YM, Koo YJ, Yoon SC, Yi SH, et al. Non-linear dynamic analysis of clozapine-induced electroencephalographic changes in schizophrenic patients-a preliminary study. Prog Neuropsychopharmacol Biol Psychiatry (2001) 25:1229–39. doi: 10.1016/S0278-5846(01)00183-X
34. Khodayari-Rostamabad A, Hasey GM, MacCrimmon DJ, Reilly JP, de Bruin H. A pilot study to determine whether machine learning methodologies using pre-treatment electroencephalography can predict the symptomatic response to clozapine therapy. Clin Neurophysiol. (2010) 121:1998–2006. doi: 10.1016/j.clinph.2010.05.009
35. Knott V, Labelle A, Jones B, Mahoney C. EEG hemispheric asymmetry as a predictor and correlate of short-term response to clozapine treatment in schizophrenia. Clin Electroencephalogr. (2000) 31:145–52. doi: 10.1177/155005940003100308
36. Konicki PE, Kwon KY, Steele V, White J, Fuller M, Jurjus GJ, et al. Prefrontal cortical sulcal widening associated with poor treatment response to clozapine. Schizophr Res. (2001) 48:173–6. doi: 10.1016/S0920-9964(00)00130-4
37. Molina V, Reig S, Sarramea F, Sanz J, Artaloytia JF, Luque R, et al. Anatomical and functional brain variables associated with clozapine response in treatment-resistant schizophrenia. Psychiatry Res. (2003) 124:153–61. doi: 10.1016/S0925-4927(03)00108-2
38. Molina V, Taboada D, Aragüés M, Hernández JA, Sanz-Fuentenebro J. Greater clinical and cognitive improvement with clozapine and risperidone associated with a thinner cortex at baseline in first-episode schizophrenia. Schizophr Res. (2014) 158:223–9. doi: 10.1016/j.schres.2014.06.042
39. Pillay SS, Stoll AL, Weiss MK, Tohen M, Zarate CA, Banov MD, et al. EEG abnormalities before clozapine therapy predict a good clinical response to clozapine. Ann Clin Psychiatry (1996) 8:1–5. doi: 10.3109/10401239609149083
40. Ravan M, Hasey G, Reilly JP, MacCrimmon D, Khodayari-Rostamabad A. A machine learning approach using auditory odd-ball responses to investigate the effect of Clozapine therapy. Clin Neurophysiol Int Feder Clin Neurophysiol. (2015) 126:721–30. doi: 10.1016/j.clinph.2014.07.017
41. Rodriguez VM, Andree RM, Perez Castejon MJ, Luisa Catalina Zamora M, Alvaro PC, Luis Carreras Delgado J, et al. Fronto-striato-thalamic perfusion and clozapine response in treatment- refractory schizophrenic patients. A 99mTc-HMPAO study. Psychiatry Res. (1997) 76:51–61. doi: 10.1016/S0925-4927(97)00057-7
42. Evins A, Amico E, Shih V, Goff DC. Clozapine treatment increases serum glutamate and aspartate compared to conventional neuroleptics. J Neural Transm. (1997) 104:761–6. doi: 10.1007/BF01291892
43. Mitjans M, Catalán R, Vázquez M, González-Rodríguez A, Penadés R, Pons A, et al. Hypothalamic-pituitary-adrenal system, neurotrophic factors and clozapine response: association with FKBP5 and NTRK2 genes. Pharmacogenet Genomics (2015) 25:274–7. doi: 10.1097/FPC.0000000000000132
44. Nöthen MM, Rietschel M, Erdmann J, Oberländer H, Möller HJ, Nober D, et al. Genetic variation of the 5-HT2A receptor and response to clozapine. Lancet (1995) 346:908–9. doi: 10.1016/S0140-6736(95)92756-5
45. Kohn Y, Ebstein RP, Heresco-Levy U, Shapira B, Nemanov L, Gritsenko I, et al. Dopamine D4 receptor gene polymorphisms: relation to ethnicity, no association with schizophrenia and response to clozapine in Israeli subjects. Eur Neuropsychopharmacol. (1997) 7:39–43. doi: 10.1016/S0924-977X(96)00380-X
46. Mauri MC, Volonteri LS, Dell'Osso B, Regispani F, Papa P, Baldi M, et al. Predictors of clinical outcome in schizophrenic patients responding to clozapine. J Clin Psychopharmacol. (2003) 23:660–4. doi: 10.1097/01.jcp.0000095351.32154.3a
47. Pickar D, Owen RR, Litman RE, Konicki E, Gutierrez R, Rapaport MH. Clinical and biologic response to clozapine in patients with schizophrenia. Crossover comparison with fluphenazine. Arch Gen Psychiatry (1992) 49:345–53. doi: 10.1001/archpsyc.1992.01820050009001
48. Szymanski S, Lieberman J, Pollack S, Munne R, Safferman A, Kane J, et al. The dopamine-serotonin relationship in clozapine response. Psychopharmacology (1993) 112(1 Suppl.):S85–9. doi: 10.1007/BF02245011
49. Rajkumar AP, Poonkuzhali B, Kuruvilla A, Srivastava A, Jacob M, Jacob KS. Outcome definitions and clinical predictors influence pharmacogenetic associations between HTR3A gene polymorphisms and response to clozapine in patients with schizophrenia. Psychopharmacology (2012) 224:441–9. doi: 10.1007/s00213-012-2773-2
50. Xu M, Xing Q, Li S, Zheng Y, Wu S, Gao R, et al. Pharacogenetic effects of dopamine transporter gene polymorphisms on response to chlorpromazine and clozapine and on extrapyramidal syndrome in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry (2010) 34:1026–32. doi: 10.1016/j.pnpbp.2010.05.017
51. Zhao AL, Zhao JP, Zhang YH, Xue ZM, Chen JD, Chen XG. Dopamine D4 receptor gene exon III polymorphism and interindividual variation in response to clozapine. Int J Neurosci. (2005) 115:1539–47. doi: 10.1080/00207450590957863
52. Fleischhaker C, Schulz E, Remschmidt H. Biogenic amines as predictors of response to clozapine treatment in early-onset schizophrenia. J Psychiatr Res. (1998) 32:325–33. doi: 10.1016/S0022-3956(98)00012-0
53. Dettling M, Sachse C, Brockmoller J, Schley J, Muller-Oerlinghausen B, Pickersgill I, et al. Long-term therapeutic drug monitoring of clozapine and metabolites psychiatric in- and outpatients. Psychopharmacology (2000) 152:80–6. doi: 10.1007/s002130000503
54. Lee ST, Ryu S, Kim SR, Kim MJ, Kim S, Kim JW, et al. Association study of 27 annotated genes for clozapine pharmacogenetics: validation of preexisting studies and identification of a new candidate gene, ABCB1, for treatment response. J Clin Psychopharmacol. (2012) 32:441–8. doi: 10.1097/JCP.0b013e31825ac35c
55. Arranz MJ, Li T, Munro J, Liu X, Murray R, Collier DA, et al. Lack of association between a polymorphism in the promoter region of the dopamine-2 receptor gene and clozapine response. Pharmacogenetics (1998) 8:481–4. doi: 10.1097/00008571-199812000-00004
56. Rodriguez VM, Andree RM, Castejón M, Garcia EC. SPECT study of regional cerebral perfusion in neuroleptic-resistant schizophrenic patients who responded or did not respond to clozapine. Am J Psychiatry (1996) 153:1343–6. doi: 10.1176/ajp.153.10.1343
57. Meltzer HY, Bastani B, Kwon KY, Ramirez LF, Burnett S, Sharpe J. A prospective study of clozapine in treatment-resistant schizophrenic patients. I. Preliminary report. Psychopharmacology (1989) 99(Suppl.):S68–72. doi: 10.1007/BF00442563
58. Arora RC, Meltzer HY. Effect of clozapine treatment on serotonin-2—receptor binding in the blood platelets of schizophrenic patients. Neuropsychopharmacology (1994) 10:109–14. doi: 10.1038/npp.1994.12
59. Brown AS, Gewirtz G, Harkavy-Friedman J, Cooper T, Brébion G, Amador XF, et al. Effects of clozapine on plasma catecholamines and relation to treatment response in schizophrenia: a within-subject comparison with haloperidol. Neuropsychopharmacology (1997) 17:317–25. doi: 10.1016/S0893-133X(97)00073-0
60. Green AI, Alam MY, Sobieraj JT, Pappalardo KM, Waternaux C, Salzman C, et al. Clozapine response and plasma catecholamines and their metabolites. Psychiatry Res. (1993) 46:139–49. doi: 10.1016/0165-1781(93)90016-A
61. Kahn RS, Davidson M, Siever L, Gabriel S, Apter S, Davis KL. Serotonin function and treatment response to clozapine in schizophrenic-patients. Am J Psychiatry (1993) 150:1337–42. doi: 10.1176/ajp.150.9.1337
62. Meged S, Stein D, Sitrota P, Melamed Y, Elizur A, Shmuelian I, et al. Human leukocyte antigen typing, response to neuroleptics, and clozapine-induced agranulocytosis in Jewish Israeli schizophrenic patients. Int Clin Psychopharmacol. (1999) 14:305–12. doi: 10.1097/00004850-199909000-00005
63. Owen RR, Gutierrez-Esteinou R, Hsiao J, Hadd K, Lawlor BA, Murphy DL, et al. Effects of clozapine and fluphenazine treatment on responses to m-chlorophenylpiperazine infusions in schizophrenia. Arch Gen Psychiatry (1993) 50:636–44. doi: 10.1001/archpsyc.1993.01820200046005
64. Risch SC, Lewine RRJ. Low cerebrospinal fluid homovanillic acid-5-hydroxyindoleacetic acid ratio predicts clozapine efficacy: a replication. Arch Gen Psychiatry (1993) 50:670. doi: 10.1001/archpsyc.1993.01820200088011
65. Sumiyoshi T, Hasegawa M, Jayathilake K, Meltzer HY. Prediction of short-term changes in symptom severity by baseline plasma homovanillic acid levels in schizophrenic patients receiving clozapine. Psychiatry Res. (1997) 69:113–21. doi: 10.1016/S0165-1781(96)02993-9
66. Sumiyoshi T, Jin D, Jayathilake K, Lee M, Meltzer HY. Prediction of the ability of clozapine to treat negative symptoms from plasma glycine and serine levels in schizophrenia. Int J Neuropsychopharmacol. (2005) 8:451–5. doi: 10.1017/S1461145705005237
67. Szymanski S, Lieberman J, Pollack S, Safferman A, Munne R, Umbricht D, et al. Clozapine effects on neuroendocrine response to apomorphine challenge testing in chronic neuroleptic nonresponsive schizophrenia: preliminary findings. Biol. Psychiatry (1995) 37:52–5. doi: 10.1016/0006-3223(94)00191-5
68. Tipton KF, Boyce S, O'Sullivan J, Davey GP, Healy J. Monoamine oxidases: certainties and uncertainties. Curr Med Chem. (2004) 11:1965–82. doi: 10.2174/0929867043364810
69. Javitt DC. Glutamate as a therapeutic target in psychiatric disorders. Mol Psychiatry (2004) 9:984–97, 979. doi: 10.1038/sj.mp.4001551
70. Kim JH, Yi SH, Lee J, Kim YS. Effects of clozapine on heart rate dynamics and their relationship with therapeutic response in treatment-resistant schizophrenia. J Clin Psychopharmacol. (2013) 33:69–73. doi: 10.1097/JCP.0b013e31827d14e3
71. Arranz MJ, Collier DA, Sodhi M, Ball D, Roberts GW, Sham P, et al. Association between clozapine response and allelic variation in 5-HT2A receptor gene. Lancet (1995) 346:281–2. doi: 10.1016/S0140-6736(95)92168-0
72. Arranz M, Dawson E, Shaikh S, Sham P, Sharma T, Aitchison K, et al. Cytochrome P4502D6 genotype does not determine response to clozapine. Br J Clin Pharmacol. (1995) 39:417–20. doi: 10.1111/j.1365-2125.1995.tb04471.x
73. Arranz M, Collier D, Munro J, Sham P. Analysis of a structural polymorphism in the 5-HT 2A receptor and clinical response to clozapine. Neuroscience (1996) 217:177–8.
74. Arranz M, Munro J, Owen M. Evidence for association between polymorphisms in the promoter and coding regions of the 5-HT2A receptor gene and response to clozapine. Mol Psychiatry (1998) 3:61–6. doi: 10.1038/sj.mp.4000348
75. Arranz MJ, Bolonna AA, Munro J, Curtis CJ, Collier DA, Kerwin RW. The serotonin transporter and clozapine response. Mol Psychiatry (2000) 5:124–5. doi: 10.1038/sj.mp.4000652
76. Arranz MJ, Munro J, Birkett J, Bolonna A, Mancama D, Sodhi M, et al. Pharmacogenetic prediction of clozapine response. Lancet (2000) 355:1615–6. doi: 10.1016/S0140-6736(00)02221-2
77. Barlas IO, Cetin M, Erdal ME, Semiz UB, Basoglu C, Ay ME, et al. Lack of association between DRD3 gene polymorphism and response to clozapine in Turkish schizoprenia patients. Am J Med Genet Part B Neuropsychiatr Genet. (2009) 150:56–60. doi: 10.1002/ajmg.b.30770
78. Birkett JT, Arranz MJ, Munro J, Osbourn S, Kerwin RW, Collier DA. Association analysis of the 5-HT5A gene in depression, psychosis and antipsychotic response. Neuroreport (2000) 11:2017–20. doi: 10.1097/00001756-200006260-00042
79. Bolonna AA, Arranz MJ, Munro J, Osborne S, Petouni M, Martinez M, et al. No influence of adrenergic receptor polymorphisms on schizophrenia and antipsychotic response. Neurosci Lett. (2000) 280:65–8. doi: 10.1016/S0304-3940(99)01000-9
80. Bosia M, Pigoni A, Pirovano A, Lorenzi C, Spangaro M, Buonocore M, et al. COMT and STH polymorphisms interaction on cognition in schizophrenia. Neurol Sci. (2015) 36:215–20. doi: 10.1007/s10072-014-1936-9
81. Brandl EJ, Lett TA, Chowdhury NI, Tiwari AK, Bakanidze G, Meltzer HY, et al. The role of the ITIH3 rs2535629 variant in antipsychotic response. Schizophr Res. (2016) 176:131–5. doi: 10.1016/j.schres.2016.06.032
82. Butcher NJ, Fung WLA, Fitzpatrick L, Guna A, Andrade DM, Lang AE, et al. Response to clozapine in a clinically identifiable subtype of schizophrenia. Br J Psychiatry (2015) 206:484–91. doi: 10.1192/bjp.bp.114.151837
83. Frank J, Lang M, Witt SH, Strohmaier J, Rujescu D, Cichon S, et al. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol Psychiatry (2015) 20:150. doi: 10.1038/mp.2014.56
84. Gutiérrez B, Arranz MJ, Huezo-Diaz P, Dempster D, Matthiasson P, Travis M, et al. Novel mutations in 5-HT3A and 5-HT3B receptor genes not associated with clozapine response. Schizophr Res. (2002) 58:93–7. doi: 10.1016/S0920-9964(02)00205-0
85. Hong CJ, Yu YW, Lin CH, Cheng CY, Tsai SJ. Association analysis for NMDA receptor subunit 2B (GRIN2B) genetic variants and psychopathology and clozapine response in schizophrenia. Psychiatr Genet. (2001) 11:219–22. doi: 10.1097/00041444-200112000-00007
86. Hong CJ, Yu YWY, Lin CH, Tsai SJ. An association study of a brain-derived neurotrophic factor Val66Met polymorphism and clozapine response of schizophrenic patients. Neurosci Lett. (2003) 349:206–8. doi: 10.1016/S0304-3940(03)00828-0
87. Hong CJ, Yu YW, Lin CH, Song HL, Lai HC, Yang KH, et al. Association Study of Apolipoprotein Eε4with Clinical Phenotype and Clozapine Response in Schizophrenia. Neuropsychobiology (2000) 42:172–4. doi: 10.1159/000026689
88. Hwang R, Shinkai T, De Luca V, Müller DJ, Ni X, Macciardi F, et al. Association study of 12 polymorphisms spanning the dopamine D2 receptor gene and clozapine treatment response in two treatment refractory/intolerant populations. Psychopharmacology (2005) 181:179–87. doi: 10.1007/s00213-005-2223-5
89. Hwang R, Shinkai T, Deluca V, Macciardi F, Potkin S, Meltzer HY, et al. Dopamine D2 receptor gene variants and quantitative measures of positive and negative symptom response following clozapine treatment. Eur Neuropsychopharmacol. (2006) 16:248–59. doi: 10.1016/j.euroneuro.2005.09.004
90. Hwang R, Shinkai T, De Luca V, Ni X, Potkin SG, Lieberman JA, et al. Association study of four dopamine D1 receptor gene polymorphisms and clozapine treatment response. J Psychopharmacol. (2007) 21:718–27. doi: 10.1177/0269881106072341
91. Hwang R, Zai C, Tiwari A, Müller DJ, Arranz MJ, Morris AG, et al. Effect of dopamine D3 receptor gene polymorphisms and clozapine treatment response: exploratory analysis of nine polymorphisms and meta-analysis of the Ser9Gly variant. Pharmacogenomics (2010) 10:200–18. doi: 10.1038/tpj.2009.65
92. Hwang R, Souza RP, Tiwari AK, Zai CC, Müller DJ, Potkin SG, et al. Gene-gene interaction analyses between NMDA receptor subunit and dopamine receptor gene variants and clozapine response. Pharmacogenomics (2011) 12:277–91. doi: 10.2217/pgs.10.182
93. Hwang R, Tiwari AK, Zai CC, Felsky D, Remington E, Wallace T, et al. Dopamine D4 and D5 receptor gene variant effects on clozapine response in schizophrenia: replication and exploration. Prog Neuropsychopharmacol Biol Psychiatry (2012) 37:62–75. doi: 10.1016/j.pnpbp.2011.11.018
94. Huang E, Maciukiewicz M, Zai CC, Tiwari AK, Li J, Potkin SG, et al. Preliminary evidence for association of genome-wide significant DRD2 schizophrenia risk variant with clozapine response. Pharmacogenomics (2016) 17:103–9. doi: 10.2217/pgs.15.155
95. Huezo-Diaz P, Arranz MJ, Munro J, Osborne S, Makoff A, Kerwin RW, et al. An association study of the neurotensin receptor gene with schizophrenia and clozapine response. Schizophr Res. (2004) 66:193–5. doi: 10.1016/S0920-9964(03)00128-2
96. Kaiser R, Tremblay PB, Schmider J, Henneken M, Dettling M, Müller-Oerlinghausen B, et al. Serotonin transporter polymorphisms: no association with response to antipsychotic treatment, but associations with the schizoparanoid and residual subtypes of schizophrenia. Mol Psychiatry (2001) 6:179–85. doi: 10.1038/sj.mp.4000821
97. Kohlrausch FB, Salatino-Oliveira A, Gama CS, Lobato MI, Belmonte-de-Abreu P, Hutz MH. G-protein gene 825C>T polymorphism is associated with response to clozapine in Brazilian schizophrenics. Pharmacogenomics (2008) 9:1429–36. doi: 10.2217/14622416.9.10.1429
98. Kohlrausch FB, Salatino-Oliveira A, Gama CS, Lobato MI, Belmonte-de-Abreu P, Hutz MH. Influence of serotonin transporter gene polymorphisms on clozapine response in Brazilian schizophrenics. J Psychiatr Res. (2010) 44:1158–62. doi: 10.1016/j.jpsychires.2010.04.003
99. Lett TAP, Tiwari AK, Meltzer HY, Lieberman JA, Potkin SG, Voineskos AN, et al. The putative functional rs1045881 marker of neurexin-1 in schizophrenia and clozapine response. Schizophr Res. (2011) 132:121–4. doi: 10.1016/j.schres.2011.08.007
100. Lin CH, Tsai SJ, Yu YW, Song HL, Tu PC, Sim CB, et al. No evidence for association of serotonin-2A receptor variant (102T/C) with schizophrenia or clozapine response in a Chinese population. Neuroreport (1999) 10:57–60. doi: 10.1097/00001756-199901180-00011
101. Lin CC, Wang YC, Chen JY, Liou YJ, Bai YM, Lai IC, et al. Artificial neural network prediction of clozapine response with combined pharmacogenetic and clinical data. Comput Methods Programs Biomed. (2008) 91:91–9. doi: 10.1016/j.cmpb.2008.02.004
102. Malhotra AK, Goldman D, Ozaki N, Breier A, Buchanan R, Pickar D. Lack of association between polymorphisms in the 5-HT(2A) receptor gene and the antipsychotic response to clozapine. Am J Psychiatry (1996) 153:1092–4. doi: 10.1176/ajp.153.8.1092
103. Malhotra AK, Goldman D, Ozaki N, Rooney W, Clifton A, Buchanan RW, et al. Clozapine response and the 5HT2C Cys23Ser polymorphism. Neuroreport (1996) 7:2100–2. doi: 10.1097/00001756-199609020-00007
104. Malhotra A, Goldman D, Buchanan R, Rooney W, Clifton A, Kosmidis MH, et al. The dopamine D3 receptor (DRD3) Ser9Gly polymorphism and schizophrenia: a haplotype relative risk study and association with clozapine response. Mol Psychiatry (1998) 3:72–5. doi: 10.1038/sj.mp.4000288
105. Mancama D, Arranz MJ, Munro J, Osborne S, Makoff A, Collier D, et al. Investigation of promoter variants of the histamine 1 and 2 receptors in schizophrenia and clozapine response. Neurosci Lett. (2002) 333:207–11. doi: 10.1016/S0304-3940(02)00178-7
106. Masellis M, Basile V, Meltzer HY, Lieberman JA, Sevy S, Macciardi FM, et al. Serotonin subtype 2 receptor genes and clinical response to clozapine in schizophrenia patients. Neuropsychopharmacology (1998) 19:123–32. doi: 10.1016/S0893-133X(98)00007-4
107. Masellis M, Basile VS, Meltzer HY, Lieberman JA, Sevy S, Goldman DA, et al. Lack of association between the T/C 267 serotonin 5-HT6 receptor gene (HTR6) polymorphism and prediction of response to clozapine in schizophrenia. Schizophr Res. (2001) 47:49–58. doi: 10.1016/S0920-9964(00)00016-5
108. Perkovic M, Nedic Erjavec G, Zivkovic M, Sagud M, Uzun S, Mihaljevic-Peles A, et al. Association between the brain-derived neurotrophic factor Val66Met polymorphism and therapeutic response to olanzapine in schizophrenia patients. Psychopharmacology (2014) 231:3757–64. doi: 10.1007/s00213-014-3515-4
109. Potkin SG, Basile VS, Jin Y, Masellis M, Badri F, Keator D, et al. D1 receptor alleles predict PET metabolic correlates of clinical response to clozapine. Mol Psychiatry (2003) 8:109–13. doi: 10.1038/sj.mp.4001191
110. Rao PA, Pickar D, Gejman PV, Ram A, Gershon ES, Gelernter J. Allelic variation in the D4 dopamine receptor (DRD4) gene does not predict response to clozapine. Arch Gen Psychiatry (1994) 51:912–7. doi: 10.1001/archpsyc.1994.03950110072009
111. Rietschel M, Naber D, Oberländer H, Holzbach R, Fimmers R, Eggermann K, et al. Efficacy and side-effects of clozapine: testing for association with allelic variation in the dopamine D4 receptor gene. Neuropsychopharmacology (1996) 15:491–6. doi: 10.1016/S0893-133X(96)00090-5
112. Rietschel M, Naber D, Fimmers R, Möller HJ, Propping P, Nöthen MM. Efficacy and side-effects of clozapine not associated with variation in the 5-HT2C receptor. Neuroreport (1997) 8:1999–2003. doi: 10.1097/00001756-199705260-00040
113. Scharfetter J, Chaudhry HR, Hornik K, Fuchs K, Sieghart W, Kasper S, et al. Dopamine D3 receptor gene polymorphism and response to clozapine in schizophrenic Pakistani patients. Eur Neuropsychopharmacol. (1999) 10:17–20. doi: 10.1016/S0924-977X(99)00044-9
114. Shaikh S, Collier DA, Sham P, Pilowsky L, Sharma T, Lin LK, et al. Analysis of clozapine response and polymorphisms of the dopamine D4 receptor gene (DRD4) in schizophrenic patients. Am J Med Genet Neuropsychiatr Genet. (1995) 60:541–5. doi: 10.1002/ajmg.1320600611
115. Shaikh S, Collier DA, Sham PC, Ball D, Aitchison K, Vallada H, et al. Allelic association between a Ser-9-Gly polymorphism in the dopamine D3 receptor gene and schizophrenia. Hum Genet. (1996) 97:714–9. doi: 10.1007/BF02346178
116. Sodhi MS, Arranz MJ, Curtis D, Ball DM, Sham P, Roberts GW, et al. Association between clozapine response and allelic variation in the 5-HT2C receptor gene. Neuroreport (1995) 7:169–72. doi: 10.1097/00001756-199512000-00041
117. Souza RP, Tampakeras M, Basile V, Shinkai T, Rosa DVF, Potkin S, et al. Lack of association of GPX1 and MnSOD genes with symptom severity and response to clozapine treatment in schizophrenia subjects. Hum Psychopharmacol. (2009) 24:676–9. doi: 10.1002/hup.1076
118. Souza RP, de Luca V, Meltzer HY, Lieberman JA, Kennedy JL. Influence of serotonin 3A and 3B receptor genes on clozapine treatment response in schizophrenia. Pharmacogenet Genomics (2010) 20:274–6. doi: 10.1097/FPC.0b013e328337ce3e
119. Souza RP, Romano-Silva MA, Lieberman JA, Meltzer HY, MacNeil LT, Culotti JG, et al. Genetic association of the GDNF alpha-receptor genes with schizophrenia and clozapine response. J Psychiatr Res. (2010) 44:700–6. doi: 10.1016/j.jpsychires.2010.01.002
120. Souza RP, de Luca V, Meltzer HY, Lieberman JA, Kennedy JL. Schizophrenia severity and clozapine treatment outcome association with oxytocinergic genes. Int J Neuropsychopharmacol. (2010) 13:793–8. doi: 10.1017/S1461145710000167
121. Souza RP, Romano-Silva MA, Lieberman JA, Meltzer HY, Wong AH, Kennedy JL. Association study of GSK3 gene polymorphisms with schizophrenia and clozapine response. Psychopharmacology (2008) 200:177. doi: 10.1007/s00213-008-1193-9
122. Souza RP, Meltzer HY, Lieberman JA, Le Foll B, Kennedy JL. Influence of neurexin 1 (NRXN1) polymorphisms in clozapine response. Hum Psychopharmacol Clin Exp. (2010) 25:582–5. doi: 10.1002/hup.1146
123. Taylor DL, Tiwari AK, Lieberman JA, Potkin SG, Meltzer HY, Knight J, et al. Genetic association analysis of N-methyl-d-aspartate receptor subunit gene GRIN2B and clinical response to clozapine. Hum Psychopharmacol. (2016) 31:121–34. doi: 10.1002/hup.2519
124. Tsai SJ, Hong CJ, Yu YWY, Lin CH, Song HL, Lai HC, et al. Association study of a functional serotonin transporter gene polymorphism with schizophrenia, psychopathology and clozapine response. Schizophr Res. (2000) 44:177–81. doi: 10.1016/S0920-9964(99)00170-X
125. Tsai SJ, Wang YC, Yu Younger WY, Lin CH, Yang KH, Hong CJ. Association analysis of polymorphism in the promoter region of the alpha2a-adrenoceptor gene with schizophrenia and clozapine response. Schizophr Res. (2001) 49:53–8. doi: 10.1016/S0920-9964(00)00127-4
126. Tsai S-J, Hong C-J, Yu YW-Y, Lin C-H, Liu L-L. No association of tumor necrosis factor alpha gene polymorphisms with schizophrenia or response to clozapine. Schizophr Res. (2003) 65:27–32. doi: 10.1016/S0920-9964(02)00531-5
127. Xu Q, Wu X, Li M, Huang H, Minica C, Yi Z, et al. Association studies of genomic variants with treatment response to risperidone, clozapine, quetiapine and chlorpromazine in the Chinese Han population. Pharmacogenomics (2016) 16:357–65. doi: 10.1038/tpj.2015.61
128. Yu YW, Tsai SJ, Lin CH, Hsu CP, Yang KH, Hong CJ. Serotonin-6 receptor variant (C267T) and clinical response to clozapine. Neuroreport (1999) 10:1231–3. doi: 10.1097/00001756-199904260-00014
129. Zai G, Müller DJ, Volavka J, Czobor P, Lieberman JA, Meltzer HY, et al. Family and case–control association study of the tumor necrosis factor-alpha (TNF-α) gene with schizophrenia and response to antipsychotic medication. Psychopharmacology (2006) 188:171–82. doi: 10.1007/s00213-006-0482-4
130. Zuo L, Luo X, Krystal JH, Cramer J, Charney DS, Gelernter J. The efficacies of clozapine and haloperidol in refractory schizophrenia are related to DTNBP1 variation. Pharmacogenet Genomics (2009) 19:437–46. doi: 10.1097/FPC.0b013e32832b9cfc
131. Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. (2011) 43:969–76. doi: 10.1038/ng.940
132. Anderson VM, Goldstein ME, Kydd RR, Russell BR. Extensive gray matter volume reduction in treatment-resistant schizophrenia. Int J Neuropsychopharmacol. (2015) 18:pyv016. doi: 10.1093/ijnp/pyv016
133. Kasparek T, Prikryl R, Schwarz D, Tronerova S, Ceskova E, Mikl M, et al. Movement sequencing abilities and basal ganglia morphology in first-episode schizophrenia. World J Biol Psychiatry (2009) 10:752–62. doi: 10.1080/15622970701882433
134. Jaaskelainen E, Juola P, Kurtti J, Haapea M, Kyllonen M, Miettunen J, et al. Associations between brain morphology and outcome in schizophrenia in a general population sample. Eur Psychiatry (2014) 29:456–62. doi: 10.1016/j.eurpsy.2013.10.006
135. Van Haren NEM, Cahn W, Hulshoff Pol HE, Schnack HG, Caspers E, Lemstra A, et al. Brain volumes as predictor of outcome in recent-onset schizophrenia: a multi-center MRI study. Schizophr Res. (2003) 64:41–52. doi: 10.1016/S0920-9964(03)00018-5
136. Cohen RM, Nordahl TE, William E, Andreason P, Litman RE, Pickar D. Clozapine- fluphenazine-treated schizophrenia. Arch Gen Psychiatry (1997) 54:481–6. doi: 10.1001/archpsyc.1997.01830170107014
137. Cohen RM, Nordahl TE, Semple WE, Pickar D. The brain metabolic patterns of clozapine- and fluphenazine-treated female patients with schizophrenia: evidence of a sex effect. Neuropsychopharmacology (1999) 21:632–40. doi: 10.1016/S0893-133X(99)00065-2
138. Lahti AC, Holcomb HH, Weiler MA, Medoff DR, Tamminga CA. Functional effects of antipsychotic drugs: comparing clozapine with haloperidol. Biol. Psychiatry (2003) 53:601–8. doi: 10.1016/S0006-3223(02)01602-5
139. Amin F, Davidson M, Davis KL. Homovanillic acid measurement in clinical research: a review of methodology. Schizophr Bull. (1992) 18:123–48. doi: 10.1093/schbul/18.1.123
140. Scheepers FE, Gispen-de Wied CC, Westenberg HG, Kahn RS. The effect of olanzapine treatment on monoamine metabolite concentrations in the cerebrospinal fluid of schizophrenic patients. Neuropsychopharmacology (2001) 25:468–75. doi: 10.1016/S0893-133X(01)00250-0
141. McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry (2011) 16:76–85. doi: 10.1038/mp.2009.89
142. Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. (2016) 17:176. doi: 10.1186/s13059-016-1041-x
143. Boks MP, de Jong NM, Kas MJ, Vinkers CH, Fernandes C, Kahn RS, et al. Current status and future prospects for epigenetic psychopharmacology. Epigenetics (2012) 7:20–8. doi: 10.4161/epi.7.1.18688
144. Suzuki T, Remington G, Arenovich T, Uchida H, Agid O, Graff-Guerrero A, et al. Time course of improvement with antipsychotic medicationin treatment-resistant schizophrenia. Br J Psychiatry (2011) 199:275–80. doi: 10.1192/bjp.bp.110.083907
145. Rosenheck R, Evans D, Herz L, Cramer J, Xu W, Thomas J, et al. How long to wait for a response to clozapine: a comparison of time course of response to clozapine and conventional antipsychotic medication in refractory schizophrenia. Schizophr Bull. (1999) 25:709–19. doi: 10.1093/oxfordjournals.schbul.a033412
Keywords: clozapine, treatment response, schizophrenia, treatment-resistance, response biomarker
Citation: Samanaite R, Gillespie A, Sendt K-V, McQueen G, MacCabe JH and Egerton A (2018) Biological Predictors of Clozapine Response: A Systematic Review. Front. Psychiatry 9:327. doi: 10.3389/fpsyt.2018.00327
Received: 22 March 2018; Accepted: 29 June 2018;
Published: 26 July 2018.
Edited by:
Thomas W. Weickert, University of New South Wales, AustraliaReviewed by:
Jose Antonio Apud, National Institute of Mental Health (NIMH), United StatesRoberto Cavallaro, Università Vita-Salute San Raffaele, Italy
Cherrie Ann Galletly, University of Adelaide, Australia
Copyright © 2018 Samanaite, Gillespie, Sendt, McQueen, MacCabe and Egerton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alice Egerton, YWxpY2UuZWdlcnRvbkBrY2wuYWMudWs=
†Joint first authors.
‡Joint last authors.
 Ruta Samanaite
Ruta Samanaite Amy Gillespie
Amy Gillespie Kyra-Verena Sendt
Kyra-Verena Sendt Grant McQueen
Grant McQueen James H. MacCabe
James H. MacCabe Alice Egerton
Alice Egerton