- State Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University, Beijing, China
Alzheimer’s disease (AD) is the most common form of dementia. As an incurable, progressive, and neurodegenerative disease, it causes cognitive and memory deficits. However, the biological mechanisms underlying the disease are not thoroughly understood. In recent years, non-invasive neuroimaging and neurophysiological techniques [e.g., structural magnetic resonance imaging (MRI), diffusion MRI, functional MRI, and EEG/MEG] and graph theory based network analysis have provided a new perspective on structural and functional connectivity patterns of the human brain (i.e., the human connectome) in health and disease. Using these powerful approaches, several recent studies of patients with AD exhibited abnormal topological organization in both global and regional properties of neuronal networks, indicating that AD not only affects specific brain regions, but also alters the structural and functional associations between distinct brain regions. Specifically, disruptive organization in the whole-brain networks in AD is involved in the loss of small-world characters and the re-organization of hub distributions. These aberrant neuronal connectivity patterns were associated with cognitive deficits in patients with AD, even with genetic factors in healthy aging. These studies provide empirical evidence to support the existence of an aberrant connectome of AD. In this review we will summarize recent advances discovered in large-scale brain network studies of AD, mainly focusing on graph theoretical analysis of brain connectivity abnormalities. These studies provide novel insights into the pathophysiological mechanisms of AD and could be helpful in developing imaging biomarkers for disease diagnosis and monitoring.
Introduction
Alzheimer’s disease (AD) is the most common form of dementia, comprising 50–70% of all dementia cases (Kukull and Bowen, 2002). Currently, 35.6 million people suffer from AD globally and the number is predicted to rise to 115.4 million by 20501. As an incurable, progressive, and neurodegenerative disease, it causes memory loss and other cognitive deficits.
In recent years, modern magnetic resonance imaging [MRI; e.g., structural MRI (sMRI), functional MRI (fMRI), and diffusion MRI] and neurophysiological (e.g., electroencephalograph and magnetoencephalograph, usually referred as EEG/MEG) techniques have provided an efficient, feasible, and non-invasive way to investigate the biological mechanisms of AD in vivo. A large quantity of studies have found focal structural and functional abnormalities of the brains of patients with AD, including disturbed functional activation and reduced gray matter volume or thickness in regions of the brain including the posterior cingulate, the medial temporal lobe, the hippocampus, and the parahippocampal gyrus (Rombouts et al., 2000; Frisoni et al., 2002; Busatto et al., 2003; Sperling et al., 2003). Recent studies have suggested that AD is not only associated with regional disturbance of brain structure and function but also with abnormalities in the connections between different regions. De Lacoste and White (1993) suggested that neurofibrillary tangles and neuritic plaques (the two principle neuropathological biomarkers of AD) are usually distributed in the regions where corticocortical connections begin or end. Disruptive alterations in white matter tracts have been observed in AD and involve the cingulum, the uncinate fasciculus, the splenium, and the genu of the corpus callosum (Rose et al., 2000; Bozzali et al., 2002; Naggara et al., 2006; Xie et al., 2006; Fellgiebel et al., 2008; Ukmar et al., 2008; Kiuchi et al., 2009). Abnormal functional connectivities have also been found, including abnormal interhemispheric and intrahemispheric (frontoparietal, frontotemporal, and temporoparietal) connections (Wada et al., 1998a,b; Berendse et al., 2000; Grady et al., 2001; Greicius et al., 2004; Pijnenburg et al., 2004; Koenig et al., 2005; Celone et al., 2006; Stam et al., 2006; Wang et al., 2007). All of these studies proposed that AD is a syndrome of disconnection in neuronal networks (for reviews, see Delbeuck et al., 2003; He et al., 2009a; Filippi and Agosta, 2011).
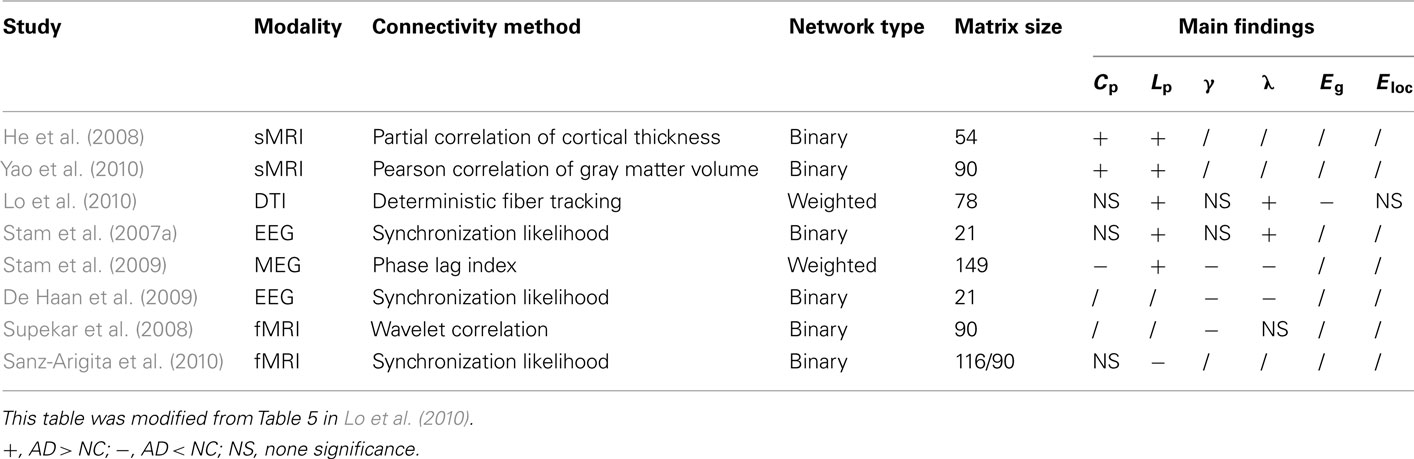
Despite the number of studies of AD-related alterations in structural and functional connections between brain regions, there is increasing evidence that AD is also characterized by large-scale brain system disruptions. Sporns et al. (2005) proposed the notion of the “connectome” to describe the detailed structural and functional connectivity pattern of the human brain. Since then, many studies have utilized multi-modal neuroimaging and neurophysiological techniques as well as advanced graph theoretical approaches to investigate the human brain connectome in health and disease. These studies have discovered many important topological characteristics of the brain system such as efficient small-worldness and distributed network hubs in the medial frontal and parietal regions (for reviews, see Bullmore and Sporns, 2009; He and Evans, 2010; Stam, 2010; Sporns, 2011). Such topology-based approaches have also been used to study the neuronal systems of patients with AD and have revealed a disruption of the typical organizational pattern of brain networks, including shifts in small-world topology and redistribution of hub regions (Stam et al., 2007a, 2009; He et al., 2008; Supekar et al., 2008; De Haan et al., 2009; Lo et al., 2010; Sanz-Arigita et al., 2010; Yao et al., 2010). Moreover, these methods have also been used to study topological organization of brain networks in the apolipoprotein E epsilon 4 allele (APOE-4) carriers (APOE-4 is a major genetic determinant for AD; Brown et al., 2011). These findings have provided new insights into the understanding of the biological mechanism of AD and could lead to the use of a network based imaging biomarker for disease diagnosis and monitoring.
In this review, we will summarize recent advances on graph theory based network analysis of the brain connectome in AD. First, we will briefly introduce several basic concepts of graph-based network analysis and human connectomics. Then we will review recent studies of graph theoretical analysis of AD brain networks derived from different imaging modalities including sMRI, diffusion MRI, EEG/MEG, and fMRI. Next we will have a short discussion regarding the effects of genetics on brain connectome in AD. Finally, we will propose further considerations for future studies of AD connectomics.
Graph Theory and Human Connectomics
Graph Theory
Generally speaking, a graph G (or a network) consists of N nodes linked by K edges. Depending on whether the edges have a direction or not, the graphs can be classified into directed or undirected. Furthermore, the graph is classified as weighted or unweighted based on whether the edges are weighted. Graphs (networks) can be described by an adjacent matrix A(n, n) in which n is the number of nodes and the value of Aij refers to the edge linking node i and node j.
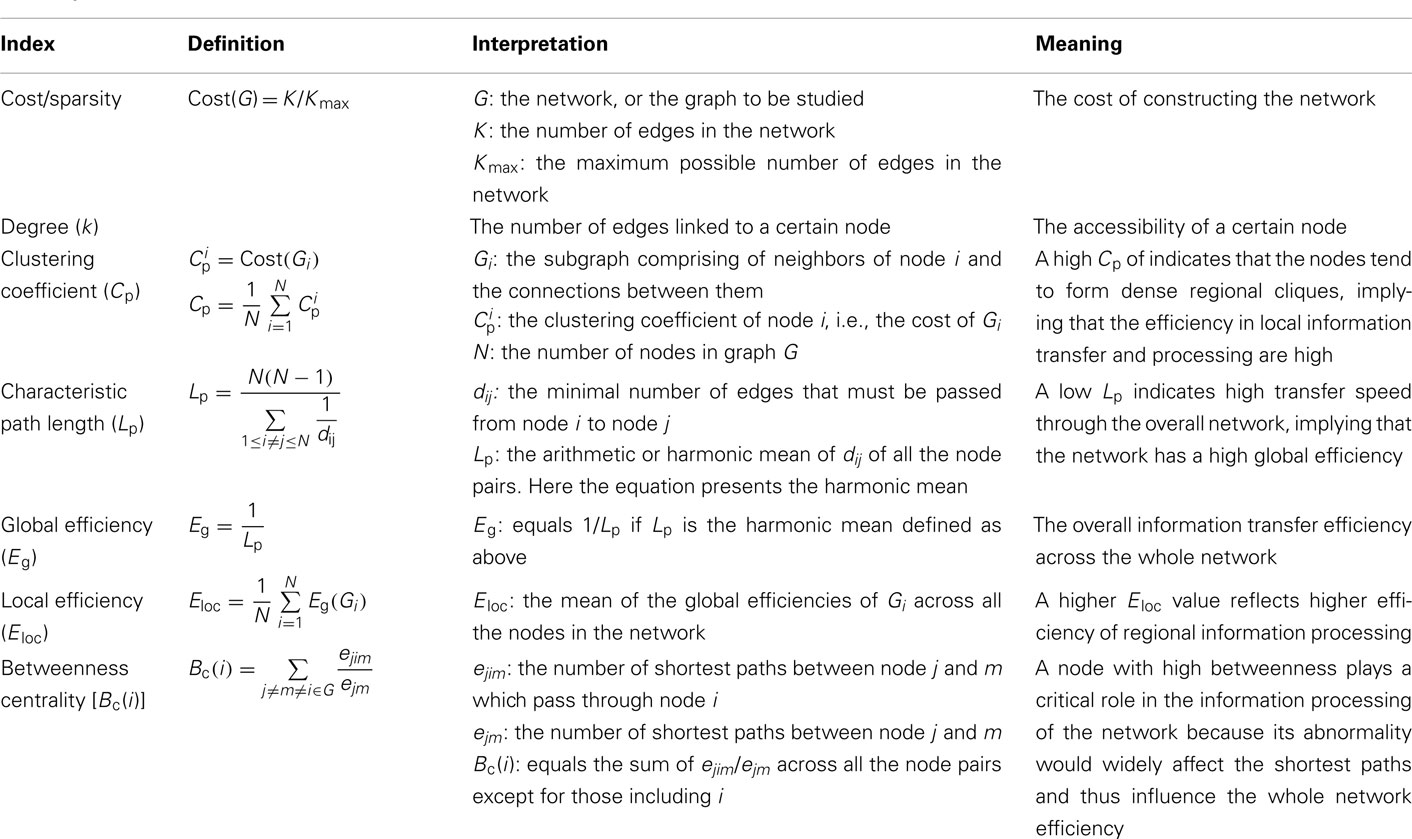
There are many graph metrics that can be used to describe the topological properties of a network, including cost/sparsity, clustering coefficient (Cp), characteristic path length (Lp), normalized clustering coefficient (γ), normalized characteristic path length (λ), small-worldness (σ), global efficiency (Eg), local efficiency (Eloc), degree (k), nodal efficiency (Enodal), and betweenness centrality (Bc; Table 1). In this review we will only focus on undirected and unweighted networks. For a detailed description of network metrics in directed or weighted networks, please see Boccaletti et al. (2006) and Rubinov and Sporns (2010).
The cost/sparsity of a network is the ratio of K to the possible maximum number of edges in the network Kmax, which equals N(N − 1)/2. The Cp of node i is the cost/sparsity of the subgraph Gi consisting of the nodes directly linked with i (the neighbors of node i). The Cp of a network is the mean Cp across all the nodes. The distance between node i and j (noted as dij), also known as the shortest path length, refers to the minimum number of edges that must be passed from i to j, and Lp is the arithmetic mean or the harmonic mean of the shortest path lengths between all pairs of nodes in G. The Cp and Lp of a network reveals the local and global efficiency of information transfer and processing, respectively. According to Cp and Lp, networks can be assigned to three different categories: regular networks with high Cp and Lp, random networks with low Cp and Lp, and small-world networks with high Cp (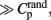 the mean Cp of a number of matched random networks) and low Lp (
the mean Cp of a number of matched random networks) and low Lp (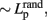 the mean Lp of a number of matched random networks; Watts and Strogatz, 1998). Small-world is a common organizational structure of networks in lots of fields such as airline networks, social networks, physiological networks, and neuronal networks and has been proved to support highly efficient segregated and integrated information processing with low wiring costs (Watts and Strogatz, 1998). Three secondary parameters, γ
the mean Lp of a number of matched random networks; Watts and Strogatz, 1998). Small-world is a common organizational structure of networks in lots of fields such as airline networks, social networks, physiological networks, and neuronal networks and has been proved to support highly efficient segregated and integrated information processing with low wiring costs (Watts and Strogatz, 1998). Three secondary parameters, γ 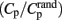 , λ
, λ 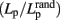 , and σ (γ/λ) can reveal the network’s small-worldness (Watts and Strogatz, 1998; Humphries et al., 2006). The efficiency of information processing in a graph can be measured with Eg and Eloc (Latora and Marchiori, 2001). Eg equals the inverse of Lp if Lp is the harmonic mean of distances over all pairs of nodes and Eloc is the average of Eg(Gi) in which i ranges from 1 to N.
, and σ (γ/λ) can reveal the network’s small-worldness (Watts and Strogatz, 1998; Humphries et al., 2006). The efficiency of information processing in a graph can be measured with Eg and Eloc (Latora and Marchiori, 2001). Eg equals the inverse of Lp if Lp is the harmonic mean of distances over all pairs of nodes and Eloc is the average of Eg(Gi) in which i ranges from 1 to N.
While the metrics mentioned above contain information about the organizational properties of the comprehensive network, several nodal metrics such as k, Enodal, and Bc can be further used to indicate the different roles of the nodes. The degree, k, refers to the number of edges linking to a particular node and reveals the accessibility of the node. Enodal of node i is the inverse of the harmonic mean distance between i and all other nodes (Achard and Bullmore, 2007). The definition of Bc is much more complex. To get the Bc of a certain node i [i.e., Bc(i) in Table 1], we should first select a pair of nodes, noted as m and n, calculate the number of shortest paths between them passing through i, divide that number by the total number of shortest paths between m and n, and then sum the ratios across all pairs of nodes in the network (Freeman, 1977). Bc(i) measures the extent to which the node i is a necessity of the shortest paths between any pair of nodes excluding i in the network. Nodes with high k, Bc, or with short average path length to other nodes (and thus with high Enodal) are considered of high importance to the information processing efficiency of the network and are called hubs. Because the hubs tend to have lots of connections to other nodes or on the way of lots of shortest paths, removal of hubs can cause significant changes in the organization of the network.
Human Connectomics
Human connectomics is an emerging scientific concept that is used to represent the comprehensive descriptions of structural and functional connectivity patterns of the human brain (Sporns et al., 2005). The human connectome can be constructed on different scales: the microscale, the mesoscale, and the macroscale. The main difference between the three scales is the definition of the network node. A single neuron represents the node when using the microscale. For the mesoscale the nodes are a group of neurons and for the macroscale the nodes are anatomically separate brain regions (Sporns et al., 2005). The edges are then determined by analyzing multi-modal imaging data, for example by measuring the properties of white matter tracts derived from diffusion MRI images, the correlations of time courses from EEG/MEG/fMRI data and the association of brain morphometry obtained from sMRI. Currently, it is hard to obtain microscale and mesoscale network data on the human brain in vivo. To date, existing studies mainly focused on undirected and unweighted macroscale matrices. All the networks mentioned in this review are undirected and unweighted brain networks if not noted specifically. Once the brain networks are constructed using neuroimaging data, a threshold is usually used to transform the initial connectivity matrix into a binary adjacent matrix. Either the correlation coefficient or the cost/sparsity can be used to set the threshold. The flowchart of brain network construction is shown in Figure 1.
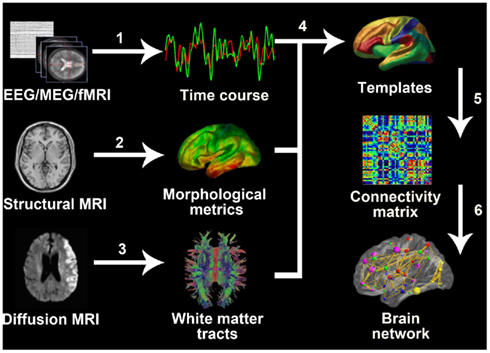
Figure 1. General process of whole-brain network construction. 1, Extract time course from EEG/MEG records or fMRI images. 2, Calculate morphological metrics such as cortical thickness (the picture showed in Figure 1) and gray matter volume. 3, Define white matter fiber bundles using tractography. 4, Extract regional information from the original voxel- or vertex-based MRI data according to templates. 5, For EEG/MEG, fMRI, and sMRI, the connectivity matrix usually refers to the correlation matrix; for diffusion MRI, it can be a matrix consisting of numbers of fibers regions or the connectivity strength. 6, Generate the whole-brain network using further modification of the connectivity matrix, for example by using thresholds.
On the basis of the connectome analysis, many studies have demonstrated that healthy human brain networks derived from different modalities are small-world networks with high Cp and short Lp (for reviews, see Reijneveld et al., 2007; Stam and Reijneveld, 2007; Bullmore and Sporns, 2009; He and Evans, 2010; Sporns, 2011). Considering the traits of a small-world network, it can be inferred that the human brain has evolved into the optimal architecture that maximizes the local and global information processing efficiency in the human brain while lowering the wiring cost. Existing studies also have demonstrated coincident areas as hubs in human brain networks such as the precuneus, the posterior cingulate cortex, the dorsal superior frontal gyrus, the precentral gyrus, and the middle and superior occipital gyri (Achard et al., 2006; He et al., 2007; Hagmann et al., 2008; Buckner et al., 2009; Gong et al., 2009; Tomasi and Volkow, 2010). In addition, significant genetic effects on the brain connectome of healthy people have been demonstrated by two recent studies on twins. Using resting-state fMRI, Fornito et al. (2011) illustrated that in functional brain network, 60% of the variation of the cost–efficiency, which is an index measuring the difference between the network cost and efficiency, was attributed to additive genetic effects. Using sMRI, Schmitt et al. (2008) demonstrated that genetically mediated neuroanatomic network derived from cortical thickness correlations follows a small-world architecture, suggesting that genetic factors are involved in the correlative patterning of the human cortex in this manner.
Brain Connectomics in AD
Structural Connectomics in AD
Using sMRI and diffusion tensor imaging (DTI), several studies have demonstrated abnormal topological properties in the structural brain networks of patients with AD. In this section, we will review the existing studies of AD structural connectomics.
Gray matter networks
Gray matter morphometric information (gray matter density, gray matter volume, and cortical thickness) revealed by sMRI provides a promising way to explore human brain anatomy. Coordinate variations of brain morphometry measurements between functionally- or anatomically-connected areas have been found in recent sMRI studies, in the visual areas (Andrews et al., 1997) and in the frontotemporal (Bullmore et al., 1998; Lerch et al., 2006), frontoparietal (Wright et al., 1999), and symmetrical interhemispheric regions (Mechelli et al., 2005; He et al., 2007; Zielinski et al., 2010). Human brain structural networks can be established from sMRI images based on gray matter volume or cortical thickness correlations between different areas. He et al. (2007) used graph theoretical network analysis (GRETNA) to examine the macroscale cortical thickness correlation network of 124 normal adults and described it as small-world. Networks based on gray matter volume correlations also revealed a similar topology (Bassett et al., 2008). Gray matter-based network analysis technique has gained more and more attention in the AD research field.
He et al. (2008) was the first group to use sMRI and graph theory tools to investigate structural brain networks in AD patients. Their study included 97 healthy older adults and 92 AD patients. The cortical thickness coordination networks at large-scale were constructed for both groups. The networks consisted of 54 nodes each, referring to 54 regions from the automated non-linear image matching and anatomical labeling (ANIMAL) template. GRETNA, as used in their previous study (He et al., 2007), was then applied to the two structural networks. They found that the AD group had decreased interregional correlations of cortical thickness between the bilateral postcentral gyri and between the bilateral superior parietal lobes. Increased correlations were also discovered within regions such as the medial prefrontal cortex, the cingulate regions, the supramarginal gyrus, the superior temporal gyrus, and the inferior temporal gyrus. These regions were mostly located in the so-called default mode network (DMN), which is a neuronal network closely related to episodic memory, comprising of the posterior cingulate cortex/precuneus, the lateral temporal and parietal cortex, the hippocampus, and the medial frontal cortex regions (Raichle et al., 2001). While the networks derived from both groups demonstrated small-world characteristics, significant differences in network parameters were observed over binary networks using a wide range of sparsity thresholds (Figures 2A,B). The brain networks in AD showed increased Cp and Lp compared with those of healthy adults, indicating a less optimal topological structure. They also found decreased betweenness centrality in the right superior temporal gyrus and the bilateral angular gyri. Increases were also found in the left lingual gyrus, the left lateral occipitotemporal gyrus, and the right cingulate gyrus in the network of patients with AD (Figure 2C). All of these regions were identified as hubs in either the health network or in the AD network by this study. In addition, they discovered that the AD network was more vulnerable to targeted attack, that is, the absence of hub regions had a greater influence on the AD network.
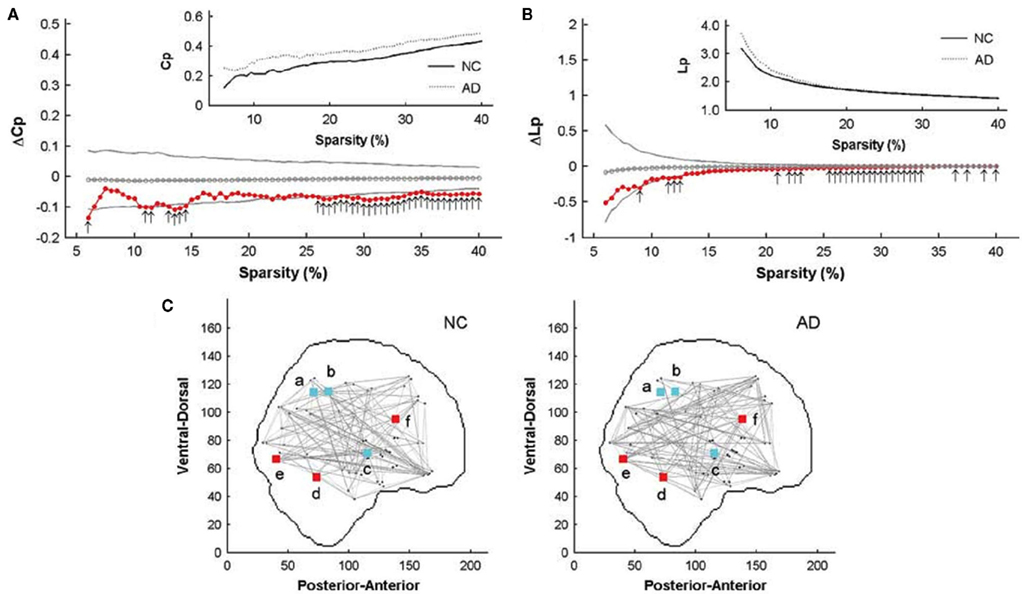
Figure 2. Comparison of structural connectome between patients with AD and healthy controls (He et al., 2008). Significant differences (p < 0.05) in Cp and Lp between patients with AD and healthy controls with different sparsity thresholds are shown in (A,B) with arrows. The gray lines represent mean values and 95% confidence intervals of between-group differences obtained by permutation tests, while red dots show the real value of the differences. (C) Shows hub regions with significantly different betweenness in the AD group compared with the healthy control group. Red regions have significantly increased betweenness in the AD group and cyan regions have decreased values of betweenness. The small letters a through f represented the right angular gyrus, the left angular gyrus, the right superior temporal gyrus, the left lateral occipitotemporal gyrus, the right lingual gyrus, and the left cingulate gyrus, respectively.
Another recent study explored changes in the topological properties of the structural brain network in patients with AD and mild cognitive impairment (MCI; Yao et al., 2010). MCI is considered an intermediate stage between normal aging and AD, and people with MCI are at high risk developing AD. The dataset for this study was acquired from the Alzheimer’s Disease Neuroimaging Initiative2,3 and included 98 normal controls, 113 subjects with MCI, and 91 AD patients. In this work, Yao and colleagues constructed a 90 by 90 gray matter volume correlation network for each of the three groups using an automated anatomical labeling (AAL) template with multiple sparsity thresholds ranging from 15 to 30%. Permutation testing revealed a significant increase in Cp over a wide range of thresholds and a larger Lp on higher thresholds in the AD networks compared with the healthy networks, implying a weakening of small-worldness. This result was consistent with previous study based on cortical thickness correlation networks (He et al., 2008). The Cp and Lp values of MCI network were intermediate between AD group and normal control group but no significant changes were found. They further identified the middle temporal gyrus, temporal pole, lingual gyrus, orbital frontal gyrus, and superior parietal gyrus as hub regions in the network of the normal control group, and the orbital frontal gyrus, inferior frontal gyrus, cingulate, and medial orbital frontal gyrus in the AD group. The hubs of MCI network largely overlapped with AD network. The alteration in hub regions revealed the disturbed large-scale brain connectome integration in AD. Regions including the parahippocampal gyrus, temporal pole, fusiform, cingulate, superior parietal region, and orbital frontal gyrus showed significant changes in the interregional correlations between the normal control and AD groups.
In summary, these sMRI-based studies have consistently demonstrated that patients with AD had aberrant morphological organization in gray matter structural networks. Specifically, the patients were found to have higher Cp and Lp in the brain structural networks, suggesting a tendency from the optimal small-world organization toward a regular-like connectivity pattern in the AD brain connectome. However, it needs to note that the biological mechanisms underlying topological alterations of morphological networks in AD remain largely unclear, although several previous studies have suggested that these morphological correlations among regions might be associated with the mutually tropic effects, environment-related plasticity, and genetic effects (Mechelli et al., 2005; He et al., 2007).
White matter networks
Different from sMRI, diffusion MRI captures the movement of water molecule in brain tissues, revealing the orientation of white matter fiber bundles by deterministic (Mori et al., 1999) or probabilistic (Behrens et al., 2003) tractography. Studies using the DTI technique have found faithful white matter fiber bundles known as real anatomical connections (Catani et al., 2002; Wakana et al., 2004). Relating to the AD research, DTI-based studies have reported widespread disruptions of white matter integrity in the corpus callosum, the superior longitudinal fasciculus, and cingulum (Rose et al., 2000; Bozzali et al., 2002; Naggara et al., 2006; Xie et al., 2006; Fellgiebel et al., 2008; Ukmar et al., 2008; Kiuchi et al., 2009).
Studies on the brain’s white matter are extremely important for the human connectome because white matter tracts connect functionally related regions and therefore might underlie functional states of the brain. Several recent studies have utilized DTI to construct human whole-brain white matter networks and demonstrated small-world topological properties (Hagmann et al., 2007; Iturria-Medina et al., 2008; Gong et al., 2009). Several hub regions have also been identified in the white matter structural networks in healthy adults, including the precuneus, the medial frontal cortex, the middle occipital gyrus, and the cingulate gyrus (Hagmann et al., 2008; Gong et al., 2009).
Lo et al. (2010) published the first research on the AD network based on the DTI technique. They used a dataset of 25 AD patients and 30 age- and gender-matched normal controls. They performed fiber tracking via the fiber assignment by continuous tracking algorithm (Mori et al., 1999). The fiber number between two cortical regions multiplied by the mean fractional anisotropy of the fiber bundles was calculated as the weight of edge. After constructing an undirected weighted network for each participant according to the AAL template, they calculated the Cp, Lp, γ, λ, σ, Eg, Eloc, and Enodal to investigate the topological differences between the normal control group and the AD group. It turned out that both normal and AD networks showed prominent small-worldness. No significant differences were found for the values of Cp, γ, and σ between the two groups. However the AD group did have larger Lp and λ values. The increased Lp was in accordance with previous structural connectomics studies of AD (He et al., 2008; Yao et al., 2010). As to the efficiency measurements, Eg was significantly reduced in AD network, while Eloc was not significantly different. These differences revealed a less optimal organization of the brain network in patients with AD. The researchers further identified nodes with high Enodal values as hubs and compared the Enodal of the hubs in AD with those of normal controls. They found that AD-related Enodal reduction was limited to several prefrontal areas including the medial superior frontal gyrus, the middle frontal gyrus, the orbital part of the inferior frontal gyrus, and the temporal pole of the middle temporal gyrus in the temporal lobe (Figure 3). The researchers correlated the network properties with the cognitive performance of the patients with AD and found significant correlations between some of the network metrics and memory test scores.
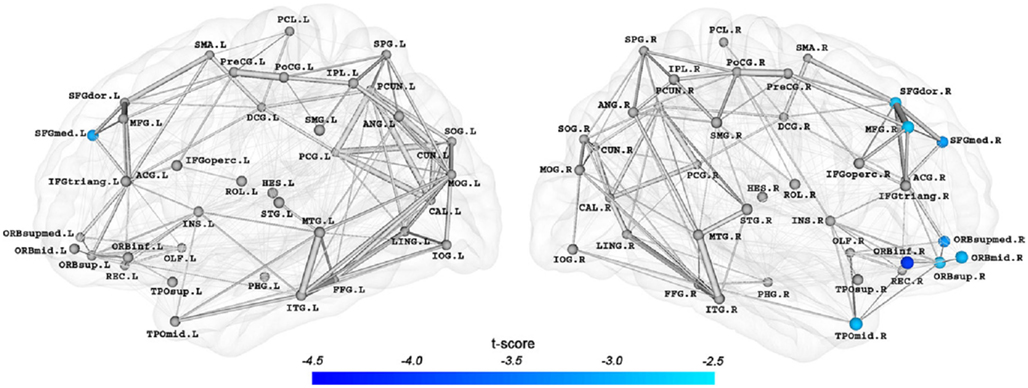
Figure 3. Brain regions with significant differences (p < 0.05, FDR corrected) in Enodal in the AD networks (Lo et al., 2010). The regions included the medial part of the superior frontal gyrus (SFGmed.F and SFGmed.R), the right dorsolateral part of the superior frontal gyrus (SFGdor.R), the right middle frontal gyrus (MFG.R), the right orbital part of the inferior frontal gyrus (ORBinf.R), the orbital and the medial orbital part of the superior frontal gyrus (ORBsup.R and ORBsupmed.R), the orbital part of the middle frontal gyrus (ORBmid.R), and the right temporal pole of the middle temporal gyrus (TPOmid.R). The connection strengths between nodes were represented by the edge widths, removing the effects of age and gender.
Functional Connectomics in AD
Modern functional neuroimaging (e.g., fMRI) and neurophysiological techniques (e.g., EEG/MEG) can non-invasively measure human brain activities and provides valuable information about human brain networks. In this section we will summarize recent advances in AD functional connectomics.
EEG/MEG networks
EEG/MEG records the electric and magnetic field changes caused by the neuronal activities during a task or during the resting-state. These neurophysiological techniques also provide powerful approaches with high temporal resolution to investigate human brain function in health and disease. Functional brain connectome analysis based on EEG/MEG data have uncovered small-world topology in healthy people (Stam, 2004; Bassett et al., 2006; Micheloyannis et al., 2006; Ferri et al., 2007; Smit et al., 2008). The techniques have also been applied studies of AD and have demonstrated abnormal functional connectivity, both in interhemispheric and intrahemispheric connections (Berendse et al., 2000; Knott et al., 2000; Adler et al., 2003; Pijnenburg et al., 2004; Koenig et al., 2005; Stam et al., 2006).
Stam et al. (2007a) used EEG to conduct brain network analysis on 15 patients with AD and 13 control subjects with only subjective memory complains. They computed synchronization likelihood in the beta band (13–30 Hz) between any pairs of 21 nodes and constructed a binary brain network for each participant. Their study showed that the AD group had significant increases in Lp both under synchronization likelihood thresholds and sparsity thresholds, implying impaired large-scale brain functional integration. However, they barely found significant changes in Cp below either type of the thresholds. This might imply that the local connectivity of the brain network in AD was relatively spared. Further analysis revealed significant negative Pearson’s correlations between Lp and the mini mental state examination (MMSE) score. The results demonstrated the altered brain functional connectivity pattern associated with AD.
In a later work, Stam et al. (2009) used resting-state MEG data to investigate the human brain connectomics in AD. The study included 18 healthy people and 18 patients with AD. They produced a 149-node weighted brain network based on the phase lag index (PLI, see Stam et al., 2007b). The AD group showed significant mean PLI reduction in the beta band and the lower alpha band (8–10 Hz). Significant decreases were also observed in the left frontoparietal, the frontotemporal, the parietooccipital, and the temporooccipital PLIs in the lower alpha band, and in the interhemispheric frontal and right frontoparietal PLIs in the beta band. The findings supported AD as a disconnection syndrome. Statistical analysis on small-world indices revealed significantly higher Lp and lower Cp, γ, and λ in the brain networks of the lower alpha band of patients with AD, leaving no discovery in the beta band. The alteration of small-world indices showed that the AD brain network exhibited a random-like pattern. Putting all the participants together, the MMSE score was positively correlated with mean PLI in the beta band and γ in the lower alpha band.
De Haan et al. (2009) conducted another EEG study of AD and frontotemporal dementia. They acquired resting-state EEG records from 20 patients with AD and 23 healthy people with only subjective cognitive complaints. Binary synchronization likelihood brain networks were constructed with synchronization likelihood thresholds and sparsity thresholds. In the beta band, σ was significantly decreased in AD networks while small-worldness was demonstrated both in healthy and AD networks across all band frequencies. Cp and γ decreased in the AD group in the lower alpha and beta bands. The λ value of AD networks also decreased in the lower alpha and gamma (30–45 Hz) bands. These results implied a disturbance in the balance of localized and integrated information processing and a random-oriented shift of the AD brain networks. The degree correlation, which refers to the mean Pearson correlation coefficient of the degree between each pair of directly linked nodes, was decreased in AD in the lower and upper (10–13 Hz) alpha bands. Taken together, all of these findings supported the conclusion that AD is a disconnection syndrome. The researchers also found that λ was positively correlated with MMSE score in AD patients in the lower alpha band.
More recently, Ahmadlou et al. (2010) studied EEG networks in AD using a visibility graph method (Lacasa et al., 2008). The basic idea of visibility graph is to transform time series into a network whose structure is related to the self-similarity and complexity of the time series. The complexity in the visibility graph of AD patients was significantly decreased in the alpha and delta bands compared with the normal elderly group. They further derived classifiers based on the discriminative complexity measurements and yielded an average accuracy of 97.75% at best. This study demonstrated the possibility of using graph metrics as biomarkers for the diagnosis of AD.
In summary, the EEG/MEG network analysis demonstrated abnormal brain connectome from a functional perspective. All the networks presented a random-like reconstruction in patients with AD, characterized by lower Cp/γ or shorter Lp. The alpha and beta bands showed the highest consistency in detecting AD-related changes in network metrics. A previous study combining EEG and fMRI (Laufs et al., 2003) indicated that the power of the alpha band (8–12 Hz) was correlated with spontaneous neuronal activities of attention-related brain regions, and the power of part of the beta band (17–23 Hz) was correlated with activities in DMN regions. Thus, we speculate that the alterations of network indices in the alpha and beta bands might reflect the underlying mechanism of functional deficits observed in patients with AD.
Functional MRI networks
Functional MRI captures blood–oxygen level dependent signal and indirectly describe the brain activity. fMRI has a relatively low temporal resolution (∼2 s) but a high special resolution (∼2 mm). Using resting-state fMRI (R-fMRI; Biswal et al., 1995), Salvador et al. (2005) first performed the graph theoretical analysis of the functional networks of the human brain. They constructed a 90-node undirected binary network for each participant. Graph theoretical analysis showed that the healthy human brain connectome is a small-world network with hierarchical organization. Later studies found similar topological structure in the human brain, studied the efficiency of the connectome (Achard and Bullmore, 2007) and identified several hub regions such as the precuneus, the middle temporal gyrus, the middle frontal gyrus, and the medial superior frontal gyrus (Achard et al., 2006; Buckner et al., 2009; He et al., 2009b; Zuo et al., 2011; for a review, see Wang et al., 2010). These findings made the understanding of brain network topology more clear and detailed.
Studies of AD based on R-fMRI data have found altered brain functional connectivity in patients with AD (Wang et al., 2006; Allen et al., 2007). Some task-based fMRI studies also found aberrant brain activity in the DMN of patients with AD during simple motor tasks (Greicius et al., 2004) and tasks of associative memory (Celone et al., 2006). Buckner et al. (2009) found a correlation between the locations of hub regions of fMRI brain networks in healthy adults and the sites of Aβ deposition in the brains of patients with AD. These regions included the inferior/superior parietal lobule, the medial superior frontal cortex, the medial prefrontal cortex, and the posterior cingulate/precuneus (Figure 4), implying that the hubs are preferentially affected in the progress of AD.
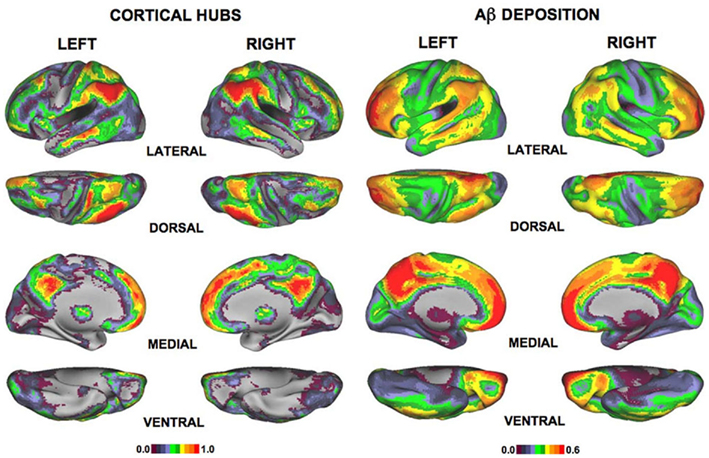
Figure 4. The spatial distribution of functional brain hubs in normal controls (the two columns on the left) and Aβ deposition in AD (the two columns on the right; Buckner et al., 2009). The left color bar shows the Z score of degree. The right color bar reveals the extent of Aβ deposition.
Supekar et al. (2008) published the first R-fMRI study of the functional brain connectome in AD using the topological network analysis method. The researchers recruited 21 patients with AD and 18 healthy volunteers matched for age, gender, and education. R-fMRI brain networks were established using wavelet correlation. The researchers computed small-world metrics of the 90-node networks based on the AAL template and found that both γ and σ of the functional networks were significantly lower in the AD group, indicating that the functional network in AD lost small-worldness. Further investigation showed that using γ as a biomarker to diagnose AD would yield 72% sensitivity and 78% specificity at best, suggesting that the topological network indices could serve as biomarkers of AD. Nodal Cp values were significantly decreased in the hippocampus bilaterally, demonstrating that intrinsic brain functional organization was disrupted. The researchers also found decreased intratemporal connections and weakened connectivity strength (i.e., correlation coefficients) between the thalamus and the frontal, temporal, and occipital lobes. Conversely, the connections within the frontal lobe were enhanced. The analysis was repeated on a second fMRI dataset acquired from the same subjects and produced similar results, suggesting that this analysis technique is reproducible.
In a more recent study, Sanz-Arigita et al. (2010) compared 18 patients with mild AD to 21 healthy controls to explore the loss of small-worldness in AD brain networks. According to the AAL template, a region-based synchronization likelihood matrix was established for each subject and then binarized by a series of thresholds ranged from 0.01 to 0.05 with increments of 0.01. Cp and Lp, along with γ and λ were calculated as the indices of small-worldness. The Cp of patients with AD did not show significant differences from the healthy control group, but the Lp was significantly decreased in the AD group across a wide range of thresholds, implying a trend toward random networks. They found significant synchronization differences between the AD and control groups. Similar to the findings of Supekar et al. (2008), these changes included increases in the functional connectivity within the frontal cortices, between the frontal cortices and the corpus striatum and between the frontal cortices and the thalamus, as revealed by synchronization likelihood and decreases between the temporal lobe, the parietal cortex and the occipital cortex. The long-distance connectivity loss supported the conclusion that AD is a disconnection syndrome, while the strengthened connections suggested that a compensatory mechanism might be responsible for reserving cognitive functions.
Other than the whole-brain network analysis studies, several graph theoretical DMN studies based on fMRI have been conducted. Ciftci (2011) utilized the minimum spanning tree (the subgraph of a network with the minimum cost while connecting all the nodes) to investigate the alteration of DMN connectivity during AD. Their study included 14 young subjects, 14 healthy elderly subjects, and 13 subjects with AD. Significantly lower connection density was observed in the AD and elderly groups compared with the younger group, although the minimum spanning tree of the three groups all presented a similar chain-like structure. Cluster analysis on the three spanning trees revealed much more fragmented functioning organization in AD, which was most notable in the hippocampus/parahippocampus and the precuneus/posterior cingulate complex. They also found a decreased correlation coefficient between the hippocampus/parahippocampus and the inferior temporal gyrus and between the precuneus/posterior cingulate gyrus and the angular gyrus. Another study by Miao et al. (2011) used independent component analysis to identify DMN in 12 normal young adults, 16 older adult controls, and 15 patients with AD. The researchers further constructed directed brain networks using Granger causality modeling and examined the proportion of edges connected with hubs compared to all edges. They found the proportion to be significantly decreased in patients with AD, implying impaired directed DMN connectivity in AD. Utilizing this ratio as a diagnostic tool for AD yielded a specificity of 81.25% and a sensitivity of 80.00%.
In summary, the AD brain connectome studies based on fMRI data demonstrated disrupted network connectivity pattern in patients with AD. The lower Cp, Lp, γ, or λ revealed a random-toward transition of brain connectome in the disease, which were consistent with EEG/MEG studies (Stam et al., 2007a, 2009; De Haan et al., 2009). These less optimized reconfigurations of functional brain network supported the theory that AD is a disconnection syndrome and might imply the functional basis of cognitive deficits.
The studies of structural and functional brain connectomics in AD have illustrated that the brain network configuration in patients with AD was significantly altered compared with normal controls. However, it needs to note that the alterations of topological metrics in the brain networks such as Cp and Lp showed distinct patterns in different modalities (see Table 2 for detail information). These discrepancies could be attributed to different imaging modality, network size, and population size applied in these studies (Table 2). In spite of these differences, we noticed that all of the studies pointed to a less optimized connectivity pattern in AD brain networks. Correlation analysis also revealed that cognitive performances of patients with AD were correlated to topological network indices. As to the nodal properties, the existing studies found aberrant changes in Bc and connectivity strength involving DMN regions. These regions were closely associated with episodic memory and showed significant gray matter atrophy and abnormal functional activities in AD (Rombouts et al., 2000; Frisoni et al., 2002; Busatto et al., 2003; Sperling et al., 2003; Buckner et al., 2005). Although the biological mechanism underlying disrupted topological properties in AD brain networks still remains unclear, we speculate that the disruption could be attributed to the neuron loss, amyloid deposition, or metabolic abnormalities.
AD Connectome and Genetics
Researchers have demonstrated that numerous genes have been associated with late-onset AD, including amyloid precursor protein, presenilin 1, presenilin 2, and APOE (for reviews, see Bookheimer and Burggren, 2009; Bekris et al., 2010). Of these genes, APOE is one of the major genetic risk factors for developing AD. Studies on normal people have revealed APOE-4 effects on the brain structure and function but controversy exists. For example, some studies reported smaller gray matter volume or thinner cortex in APOE-4 carriers in the hippocampus (Tohgi et al., 1997; Den Heijer et al., 2002; Honea et al., 2009) and the entorhinal cortex (Shaw et al., 2007; Burggren et al., 2008), yet others found no such differences (Reiman et al., 1998; Jak et al., 2007; Cherbuin et al., 2008). As to the studies of brain function, decreased activities were reported in the APOE-4 carriers in regions such as the medial prefrontal cortex, the hippocampus, and the posterior cingulate (Reiman et al., 1996; Small et al., 2000; Persson et al., 2008; Pihlajamaki and Sperling, 2009; Adamson et al., 2011), but enhanced activities were also found in these regions (Bookheimer et al., 2000; Wishart et al., 2006; Han et al., 2007; Filippini et al., 2009). Notably, APOE was also found to modulate disease phenotype. For example, several studies have demonstrated greater gray matter atrophy in the hippocampus and entorhinal cortex in APOE-4 carriers with AD as compared to APOE-4 non-carriers with AD (Lehtovirta et al., 1996; Geroldi et al., 1999; Bigler et al., 2000; Hashimoto et al., 2001; Wolk and Dickerson, 2010), while evidences for non-significant volume differences in hippocampus were also reported (Jack et al., 1998; Drzezga et al., 2009). The discrepancies of the results could be attributable to the sample size and the demographic differences of subjects.
There are also evidences indicating that APOE-4 alters the brain connectivity in normal participants. For example, several studies magnified abnormal functional connectivity associated with APOE-4 in DMN (Filippini et al., 2009; Fleisher et al., 2009; Sheline et al., 2010; Machulda et al., 2011). DTI studies found aberrant white matter tracts with descended fractional anisotropy in APOE-4 carriers, including the posterior corpus callosum and the medial temporal lobe (Persson et al., 2006) and the parahippocampal white matter (Nierenberg et al., 2005; Honea et al., 2009). So far, there’s only one study using graph theoretical analysis to explore the APOE-4 effects on whole-brain networks. Brown et al. (2011) utilized DTI tractography methods to investigate the relationship between the age and the topology of human brain structural network in normal elderly people. They found that only in the APOE-4 group the cost, Cp and σ showed significant negative correlation with age, while only in the APOE-4 non-carriers group Lp showed significant positive correlation with age. The nodal Cp of APOE-4 carriers decreased more sharply along with age in the right precuneus, the left orbitofrontal cortex, the left supramarginal gyrus, and right inferior temporal gyrus. This study demonstrated that APOE mediated the topological organization of human brain structural connectome in aging. Further studies would be important to combine different imaging modalities to systematically explore how the APOE-4 and other genetic risk factors of AD affect the topology of human connectome in health and AD.
Future Perspectives
Despite the abundance of findings already obtained from the method, graph theoretical analysis of the AD network is only in its infancy and still has some problems. Future studies should take in account a number of considerations, which will be discussed in this section.
First, the existing works on AD brain networks are at the macroscale. The interplay between macroscale network property alterations associated with AD and the biological and pathological mechanisms of AD have not been studied thoroughly. AD could cause neuron loss and white matter aberrance, which may be account for the gray matter atrophy revealed by volume loss or cortical thinning and the white matter fiber changes found in diffusion studies. Also, one study demonstrated that Aβ deposition locations corresponded with hub regions of healthy brain networks (Buckner et al., 2009). However, the relationship between these pathological changes and network abnormalities still needs further exploration. Empirical studies of AD pathology and neuroimaging would be helpful in clarifying this issue.
Second, multi-modal analysis represents one potential avenue for future research on connectomics. Data from sMRI, diffusion MRI, fMRI, and EEG/MEG all reveal meaningful information about human brain connectome from different perspectives, the combination of datasets from various modalities would thus give us a full view of human connectome in health and disease. For example, Villain et al. (2008) showed that the hippocampal atrophy in patients with AD was specifically related to cingulum bundle atrophy, which is in turn highly correlated to hypometabolism of the posterior cingulate cortex, suggesting the hypometabolism might result from hippocampal atrophy via cingulum bundle disruption.
Third, little is known about the dynamic progress of AD. Most of the AD network studies focus on comparing indices to those of normal controls and demonstrating significant differences between the two groups. We have little knowledge of longitudinal changes in brain connectomics. An R-fMRI study (Zhang et al., 2010) compared the posterior cingulate cortex connectivity of healthy controls to those of patients with mild, moderate ad severe AD. The researchers suggested that the patients with AD had abnormal posterior cingulate cortex connectivity patterns and that the disruption intensified with disease progression. This study demonstrated the dynamic changes in brain connectivity in AD, but the relationship between the network and the disease progression remains unclear. Continuous longitudinal observations of AD development are needed to characterize the developmental changes.
Fourth, further studies are necessary to determine whether the abnormalities found in network studies are specific to AD. De Haan et al. (2009) demonstrated that network property alterations such as decreases in Cp and Lp were not observed in patients with frontotemporal lobar degeneration and that degree correlation decreased in AD but increased in frontotemporal lobar degeneration. Still many more studies are needed to compare the disruptions of brain connectivity patterns between AD and other dementia, such as dementia with Lewy bodies.
Fifth, the reliability of network property changes as a biomarker of AD needs to be examined, given that controversial results were obtained from different studies mentioned in this review (Table 2). Several studies on the reliability of network topological metrics of healthy people have been done in MEG (Deuker et al., 2009), fMRI (Telesford et al., 2010; Wang et al., 2011), and diffusion MRI (Vaessen et al., 2010; Bassett et al., 2011), but little is known about using these indices to diagnose AD. This is an important issue in establishing a topological biomarker for diagnosing and monitoring AD.
Finally, some individuals are at high risk of developing AD, such as those with the APOE-4 genotype and patients with amnesia MCI. Sorg et al. (2007) demonstrated that patients with amnesia MCI have reduced connectivity in the DMN and the executive attention network. In addition, Yao et al. (2010) discovered that the brain networks of patients with amnesia MCI and the patients with AD of both group demonstrated similar alterations compared to healthy controls, while the differences between the network topologies of the two patient groups were not significant. Some progress has been made in this field, but further studies are needed to clarify the AD-like topological alterations in people with AD risk factors.
Conclusion
To summarize, brain connectome analysis of adults with AD has provided an important methodology for studies of AD. All of the studies mentioned above demonstrated that AD brain networks are less optimally constructed and have decreased information processing efficiency. These alterations in brain connectivity patterns reveal the underlying brain structural and functional disruptions that cause the cognitive deficits of AD. Thus, these studies provide further support for the description of AD as a disconnection syndrome. The graph theory analysis methods have proved to be powerful tools for exploring the structural and functional architecture of the human brain and have provided new understanding of the biological mechanisms of AD and have uncovered potential biomarkers of early diagnosis and disease progression.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (Grant Nos. 81030028 and 30870667), Beijing Natural Science Foundation (Grant No. 7102090), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars (State Education Ministry, Yong He).
Footnotes
References
Achard, S., and Bullmore, E. (2007). Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 3, e17. doi:10.1371/journal.pcbi.0030017
Achard, S., Salvador, R., Whitcher, B., Suckling, J., and Bullmore, E. (2006). A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 26, 63–72.
Adamson, M. M., Hutchinson, J. B., Shelton, A. L., Wagner, A. D., and Taylor, J. L. (2011). Reduced hippocampal activity during encoding in cognitively normal adults carrying the APOE varepsilon4 allele. Neuropsychologia 49, 2448–2455.
Adler, G., Brassen, S., and Jajcevic, A. (2003). EEG coherence in Alzheimer’s dementia. J. Neural Transm. 110, 1051–1058.
Ahmadlou, M., Adeli, H., and Adeli, A. (2010). New diagnostic EEG markers of the Alzheimer’s disease using visibility graph. J. Neural Transm. 117, 1099–1109.
Allen, G., Barnard, H., Mccoll, R., Hester, A. L., Fields, J. A., Weiner, M. F., Ringe, W. K., Lipton, A. M., Brooker, M., Mcdonald, E., Rubin, C. D., and Cullum, C. M. (2007). Reduced hippocampal functional connectivity in Alzheimer disease. Arch. Neurol. 64, 1482–1487.
Andrews, T. J., Halpern, S. D., and Purves, D. (1997). Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J. Neurosci. 17, 2859–2868.
Bassett, D. S., Brown, J. A., Deshpande, V., Carlson, J. M., and Grafton, S. T. (2011). Conserved and variable architecture of human white matter connectivity. Neuroimage 54, 1262–1279.
Bassett, D. S., Bullmore, E., Verchinski, B. A., Mattay, V. S., Weinberger, D. R., and Meyer-Lindenberg, A. (2008). Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 28, 9239–9248.
Bassett, D. S., Meyer-Lindenberg, A., Achard, S., Duke, T., and Bullmore, E. (2006). Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl. Acad. Sci. U.S.A. 103, 19518–19523.
Behrens, T. E., Woolrich, M. W., Jenkinson, M., Johansen-Berg, H., Nunes, R. G., Clare, S., Matthews, P. M., Brady, J. M., and Smith, S. M. (2003). Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med. 50, 1077–1088.
Bekris, L. M., Yu, C. E., Bird, T. D., and Tsuang, D. W. (2010). Genetics of Alzheimer disease. J. Geriatr. Psychiatry Neurol. 23, 213–227.
Berendse, H. W., Verbunt, J. P., Scheltens, P., Van Dijk, B. W., and Jonkman, E. J. (2000). Magnetoencephalographic analysis of cortical activity in Alzheimer’s disease: a pilot study. Clin. Neurophysiol. 111, 604–612.
Bigler, E. D., Lowry, C. M., Anderson, C. V., Johnson, S. C., Terry, J., and Steed, M. (2000). Dementia, quantitative neuroimaging, and apolipoprotein E genotype. AJNR Am. J. Neuroradiol. 21, 1857–1868.
Biswal, B., Yetkin, F. Z., Haughton, V. M., and Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541.
Boccaletti, S., Latora, V., Moreno, Y., Chavez, M., and Hwang, D. U. (2006). Complex networks: structure and dynamics. Phys. Rep. 424, 175–308.
Bookheimer, S., and Burggren, A. (2009). APOE-4 genotype and neurophysiological vulnerability to Alzheimer’s and cognitive aging. Annu. Rev. Clin. Psychol. 5, 343–362.
Bookheimer, S. Y., Strojwas, M. H., Cohen, M. S., Saunders, A. M., Pericak-Vance, M. A., Mazziotta, J. C., and Small, G. W. (2000). Patterns of brain activation in people at risk for Alzheimer’s disease. N. Engl. J. Med. 343, 450–456.
Bozzali, M., Falini, A., Franceschi, M., Cercignani, M., Zuffi, M., Scotti, G., Comi, G., and Filippi, M. (2002). White matter damage in Alzheimer’s disease assessed in vivo using diffusion tensor magnetic resonance imaging. J. Neurol. Neurosurg. Psychiatr. 72, 742–746.
Brown, J. A., Terashima, K. H., Burggren, A. C., Ercoli, L. M., Miller, K. J., Small, G. W., and Bookheimer, S. Y. (2011). Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proc. Natl. Acad. Sci. U.S.A. 108, 20760–20765.
Buckner, R. L., Sepulcre, J., Talukdar, T., Krienen, F. M., Liu, H., Hedden, T., Andrews-Hanna, J. R., Sperling, R. A., and Johnson, K. A. (2009). Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J. Neurosci. 29, 1860–1873.
Buckner, R. L., Snyder, A. Z., Shannon, B. J., LaRossa, G., Sachs, R., Fotenos, A. F., Sheline, Y. I., Klunk, W. E., Mathis, C. A., Morris, J. C., and Mintun, M. A. (2005). Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 25, 7709–7717.
Bullmore, E., and Sporns, O. (2009). Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198.
Bullmore, E. T., Woodruff, P. W., Wright, I. C., Rabe-Hesketh, S., Howard, R. J., Shuriquie, N., and Murray, R. M. (1998). Does dysplasia cause anatomical dysconnectivity in schizophrenia? Schizophr. Res. 30, 127–135.
Burggren, A. C., Zeineh, M. M., Ekstrom, A. D., Braskie, M. N., Thompson, P. M., Small, G. W., and Bookheimer, S. Y. (2008). Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage 41, 1177–1183.
Busatto, G. F., Garrido, G. E., Almeida, O. P., Castro, C. C., Camargo, C. H., Cid, C. G., Buchpiguel, C. A., Furuie, S., and Bottino, C. M. (2003). A voxel-based morphometry study of temporal lobe gray matter reductions in Alzheimer’s disease. Neurobiol. Aging 24, 221–231.
Catani, M., Howard, R. J., Pajevic, S., and Jones, D. K. (2002). Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage 17, 77–94.
Celone, K. A., Calhoun, V. D., Dickerson, B. C., Atri, A., Chua, E. F., Miller, S. L., Depeau, K., Rentz, D. M., Selkoe, D. J., Blacker, D., Albert, M. S., and Sperling, R. A. (2006). Alterations in memory networks in mild cognitive impairment and Alzheimer’s disease: an independent component analysis. J. Neurosci. 26, 10222–10231.
Cherbuin, N., Anstey, K. J., Sachdev, P. S., Maller, J. J., Meslin, C., Mack, H. A., Wen, W., and Easteal, S. (2008). Total and regional gray matter volume is not related to APOE*E4 status in a community sample of middle-aged individuals. J. Gerontol. A Biol. Sci. Med. Sci. 63, 501–504.
Ciftci, K. (2011). Minimum spanning tree reflects the alterations of the default mode network during Alzheimer’s disease. Ann. Biomed. Eng. 39, 1493–1504.
De Haan, W., Pijnenburg, Y. A., Strijers, R. L., Van Der Made, Y., Van Der Flier, W. M., Scheltens, P., and Stam, C. J. (2009). Functional neural network analysis in frontotemporal dementia and Alzheimer’s disease using EEG and graph theory. BMC Neurosci. 10, 101. doi:10.1186/1471-2202-10-101
De Lacoste, M. C., and White, C. L. III. (1993). The role of cortical connectivity in Alzheimer’s disease pathogenesis: a review and model system. Neurobiol. Aging 14, 1–16.
Delbeuck, X., Van Der Linden, M., and Collette, F. (2003). Alzheimer’s disease as a disconnection syndrome? Neuropsychol. Rev. 13, 79–92.
Den Heijer, T., Oudkerk, M., Launer, L. J., Van Duijn, C. M., Hofman, A., and Breteler, M. M. (2002). Hippocampal, amygdalar, and global brain atrophy in different apolipoprotein E genotypes. Neurology 59, 746–748.
Deuker, L., Bullmore, E. T., Smith, M., Christensen, S., Nathan, P. J., Rockstroh, B., and Bassett, D. S. (2009). Reproducibility of graph metrics of human brain functional networks. Neuroimage 47, 1460–1468.
Drzezga, A., Grimmer, T., Henriksen, G., Muhlau, M., Perneczky, R., Miederer, I., Praus, C., Sorg, C., Wohlschlager, A., Riemenschneider, M., Wester, H. J., Foerstl, H., Schwaiger, M., and Kurz, A. (2009). Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology 72, 1487–1494.
Fellgiebel, A., Schermuly, I., Gerhard, A., Keller, I., Albrecht, J., Weibrich, C., Muller, M. J., and Stoeter, P. (2008). Functional relevant loss of long association fibre tracts integrity in early Alzheimer’s disease. Neuropsychologia 46, 1698–1706.
Ferri, R., Rundo, F., Bruni, O., Terzano, M. G., and Stam, C. J. (2007). Small-world network organization of functional connectivity of EEG slow-wave activity during sleep. Clin. Neurophysiol. 118, 449–456.
Filippi, M., and Agosta, F. (2011). Structural and functional network connectivity breakdown in Alzheimer’s disease studied with magnetic resonance imaging techniques. J. Alzheimers Dis. 24, 455–474.
Filippini, N., Macintosh, B. J., Hough, M. G., Goodwin, G. M., Frisoni, G. B., Smith, S. M., Matthews, P. M., Beckmann, C. F., and Mackay, C. E. (2009). Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. U.S.A. 106, 7209–7214.
Fleisher, A. S., Sherzai, A., Taylor, C., Langbaum, J. B., Chen, K., and Buxton, R. B. (2009). Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer’s disease risk groups. Neuroimage 47, 1678–1690.
Fornito, A., Zalesky, A., Bassett, D. S., Meunier, D., Ellison-Wright, I., Yucel, M., Wood, S. J., Shaw, K., O’Connor, J., Nertney, D., Mowry, B. J., Pantelis, C., and Bullmore, E. T. (2011). Genetic influences on cost-efficient organization of human cortical functional networks. J. Neurosci. 31, 3261–3270.
Frisoni, G. B., Testa, C., Zorzan, A., Sabattoli, F., Beltramello, A., Soininen, H., and Laakso, M. P. (2002). Detection of grey matter loss in mild Alzheimer’s disease with voxel based morphometry. J. Neurol. Neurosurg. Psychiatr. 73, 657–664.
Geroldi, C., Pihlajamaki, M., Laakso, M. P., Decarli, C., Beltramello, A., Bianchetti, A., Soininen, H., Trabucchi, M., and Frisoni, G. B. (1999). APOE-epsilon4 is associated with less frontal and more medial temporal lobe atrophy in AD. Neurology 53, 1825–1832.
Gong, G., He, Y., Concha, L., Lebel, C., Gross, D. W., Evans, A. C., and Beaulieu, C. (2009). Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex 19, 524–536.
Grady, C. L., Furey, M. L., Pietrini, P., Horwitz, B., and Rapoport, S. I. (2001). Altered brain functional connectivity and impaired short-term memory in Alzheimer’s disease. Brain 124, 739–756.
Greicius, M. D., Srivastava, G., Reiss, A. L., and Menon, V. (2004). Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc. Natl. Acad. Sci. U.S.A. 101, 4637–4642.
Hagmann, P., Cammoun, L., Gigandet, X., Meuli, R., Honey, C. J., Wedeen, V. J., and Sporns, O. (2008). Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159. doi:10.1371/journal.pbio.0060159
Hagmann, P., Kurant, M., Gigandet, X., Thiran, P., Wedeen, V. J., Meuli, R., and Thiran, J. P. (2007). Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE 2, e597. doi:10.1371/journal.pone.0000597
Han, S. D., Houston, W. S., Jak, A. J., Eyler, L. T., Nagel, B. J., Fleisher, A. S., Brown, G. G., Corey-Bloom, J., Salmon, D. P., Thal, L. J., and Bondi, M. W. (2007). Verbal paired-associate learning by APOE genotype in non-demented older adults: fMRI evidence of a right hemispheric compensatory response. Neurobiol. Aging 28, 238–247.
Hashimoto, M., Yasuda, M., Tanimukai, S., Matsui, M., Hirono, N., Kazui, H., and Mori, E. (2001). Apolipoprotein E epsilon 4 and the pattern of regional brain atrophy in Alzheimer’s disease. Neurology 57, 1461–1466.
He, Y., Chen, Z., and Evans, A. (2008). Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer’s disease. J. Neurosci. 28, 4756–4766.
He, Y., Chen, Z., Gong, G., and Evans, A. (2009a). Neuronal networks in Alzheimer’s disease. Neuroscientist 15, 333–350.
He, Y., Wang, J., Wang, L., Chen, Z. J., Yan, C., Yang, H., Tang, H., Zhu, C., Gong, Q., Zang, Y., and Evans, A. C. (2009b). Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS ONE 4, e5226. doi:10.1371/journal.pone.0005226
He, Y., Chen, Z. J., and Evans, A. C. (2007). Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex 17, 2407–2419.
He, Y., and Evans, A. (2010). Graph theoretical modeling of brain connectivity. Curr. Opin. Neurol. 23, 341–350.
Honea, R. A., Vidoni, E., Harsha, A., and Burns, J. M. (2009). Impact of APOE on the healthy aging brain: a voxel-based MRI and DTI study. J. Alzheimers Dis. 18, 553–564.
Humphries, M. D., Gurney, K., and Prescott, T. J. (2006). The brainstem reticular formation is a small-world, not scale-free, network. Proc. Biol. Sci. 273, 503–511.
Iturria-Medina, Y., Sotero, R. C., Canales-Rodriguez, E. J., Aleman-Gomez, Y., and Melie-Garcia, L. (2008). Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage 40, 1064–1076.
Jack, C. R. Jr., Petersen, R. C., Xu, Y. C., O’Brien, P. C., Waring, S. C., Tangalos, E. G., Smith, G. E., Ivnik, R. J., Thibodeau, S. N., and Kokmen, E. (1998). Hippocampal atrophy and apolipoprotein E genotype are independently associated with Alzheimer’s disease. Ann. Neurol. 43, 303–310.
Jak, A. J., Houston, W. S., Nagel, B. J., Corey-Bloom, J., and Bondi, M. W. (2007). Differential cross-sectional and longitudinal impact of APOE genotype on hippocampal volumes in nondemented older adults. Dement. Geriatr. Cogn. Disord. 23, 382–389.
Kiuchi, K., Morikawa, M., Taoka, T., Nagashima, T., Yamauchi, T., Makinodan, M., Norimoto, K., Hashimoto, K., Kosaka, J., Inoue, Y., Inoue, M., Kichikawa, K., and Kishimoto, T. (2009). Abnormalities of the uncinate fasciculus and posterior cingulate fasciculus in mild cognitive impairment and early Alzheimer’s disease: a diffusion tensor tractography study. Brain Res. 1287, 184–191.
Knott, V., Mohr, E., Mahoney, C., and Ilivitsky, V. (2000). Electroencephalographic coherence in Alzheimer’s disease: comparisons with a control group and population norms. J. Geriatr. Psychiatry Neurol. 13, 1–8.
Koenig, T., Prichep, L., Dierks, T., Hubl, D., Wahlund, L. O., John, E. R., and Jelic, V. (2005). Decreased EEG synchronization in Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 26, 165–171.
Lacasa, L., Luque, B., Ballesteros, F., Luque, J., and Nuno, J. C. (2008). From time series to complex networks: the visibility graph. Proc. Natl. Acad. Sci. U.S.A. 105, 4972–4975.
Latora, V., and Marchiori, M. (2001). Efficient behavior of small-world networks. Phys. Rev. Lett. 87, 198701.
Laufs, H., Krakow, K., Sterzer, P., Eger, E., Beyerle, A., Salek-Haddadi, A., and Kleinschmidt, A. (2003). Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proc. Natl. Acad. Sci. U.S.A. 100, 11053–11058.
Lehtovirta, M., Soininen, H., Laakso, M. P., Partanen, K., Helisalmi, S., Mannermaa, A., Ryynanen, M., Kuikka, J., Hartikainen, P., and Riekkinen, P. J. Sr. (1996). SPECT and MRI analysis in Alzheimer’s disease: relation to apolipoprotein E epsilon 4 allele. J. Neurol. Neurosurg. Psychiatr. 60, 644–649.
Lerch, J. P., Worsley, K., Shaw, W. P., Greenstein, D. K., Lenroot, R. K., Giedd, J., and Evans, A. C. (2006). Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage 31, 993–1003.
Lo, C. Y., Wang, P. N., Chou, K. H., Wang, J., He, Y., and Lin, C. P. (2010). Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. J. Neurosci. 30, 16876–16885.
Machulda, M. M., Jones, D. T., Vemuri, P., Mcdade, E., Avula, R., Przybelski, S., Boeve, B. F., Knopman, D. S., Petersen, R. C., and Jack, C. R. Jr. (2011). Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch. Neurol. 68, 1131–1136.
Mechelli, A., Friston, K. J., Frackowiak, R. S., and Price, C. J. (2005). Structural covariance in the human cortex. J. Neurosci. 25, 8303–8310.
Miao, X., Wu, X., Li, R., Chen, K., and Yao, L. (2011). Altered connectivity pattern of hubs in default-mode network with Alzheimer’s disease: an granger causality modeling approach. PLoS ONE 6, e25546. doi:10.1371/journal.pone.0025546
Micheloyannis, S., Pachou, E., Stam, C. J., Breakspear, M., Bitsios, P., Vourkas, M., Erimaki, S., and Zervakis, M. (2006). Small-world networks and disturbed functional connectivity in schizophrenia. Schizophr. Res. 87, 60–66.
Mori, S., Crain, B. J., Chacko, V. P., and Van Zijl, P. C. (1999). Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol. 45, 265–269.
Naggara, O., Oppenheim, C., Rieu, D., Raoux, N., Rodrigo, S., Dalla Barba, G., and Meder, J. F. (2006). Diffusion tensor imaging in early Alzheimer’s disease. Psychiatry Res. 146, 243–249.
Nierenberg, J., Pomara, N., Hoptman, M. J., Sidtis, J. J., Ardekani, B. A., and Lim, K. O. (2005). Abnormal white matter integrity in healthy apolipoprotein E epsilon4 carriers. Neuroreport 16, 1369–1372.
Persson, J., Lind, J., Larsson, A., Ingvar, M., Cruts, M., Van Broeckhoven, C., Adolfsson, R., Nilsson, L. G., and Nyberg, L. (2006). Altered brain white matter integrity in healthy carriers of the APOE epsilon4 allele: a risk for AD? Neurology 66, 1029–1033.
Persson, J., Lind, J., Larsson, A., Ingvar, M., Sleegers, K., Van Broeckhoven, C., Adolfsson, R., Nilsson, L. G., and Nyberg, L. (2008). Altered deactivation in individuals with genetic risk for Alzheimer’s disease. Neuropsychologia 46, 1679–1687.
Pihlajamaki, M., and Sperling, R. A. (2009). Functional MRI assessment of task-induced deactivation of the default mode network in Alzheimer’s disease and at-risk older individuals. Behav. Neurol. 21, 77–91.
Pijnenburg, Y. A., V D Made, Y., Van Cappellen Van Walsum, A. M., Knol, D. L., Scheltens, P., and Stam, C. J. (2004). EEG synchronization likelihood in mild cognitive impairment and Alzheimer’s disease during a working memory task. Clin. Neurophysiol. 115, 1332–1339.
Raichle, M. E., Macleod, A. M., Snyder, A. Z., Powers, W. J., Gusnard, D. A., and Shulman, G. L. (2001). A default mode of brain function. Proc. Natl. Acad. Sci. U.S.A. 98, 676–682.
Reijneveld, J. C., Ponten, S. C., Berendse, H. W., and Stam, C. J. (2007). The application of graph theoretical analysis to complex networks in the brain. Clin. Neurophysiol. 118, 2317–2331.
Reiman, E. M., Caselli, R. J., Yun, L. S., Chen, K., Bandy, D., Minoshima, S., Thibodeau, S. N., and Osborne, D. (1996). Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N. Engl. J. Med. 334, 752–758.
Reiman, E. M., Uecker, A., Caselli, R. J., Lewis, S., Bandy, D., De Leon, M. J., De Santi, S., Convit, A., Osborne, D., Weaver, A., and Thibodeau, S. N. (1998). Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann. Neurol. 44, 288–291.
Rombouts, S. A., Barkhof, F., Veltman, D. J., Machielsen, W. C., Witter, M. P., Bierlaagh, M. A., Lazeron, R. H., Valk, J., and Scheltens, P. (2000). Functional MR imaging in Alzheimer’s disease during memory encoding. AJNR Am. J. Neuroradiol. 21, 1869–1875.
Rose, S. E., Chen, F., Chalk, J. B., Zelaya, F. O., Strugnell, W. E., Benson, M., Semple, J., and Doddrell, D. M. (2000). Loss of connectivity in Alzheimer’s disease: an evaluation of white matter tract integrity with colour coded MR diffusion tensor imaging. J. Neurol. Neurosurg. Psychiatr. 69, 528–530.
Rubinov, M., and Sporns, O. (2010). Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069.
Salvador, R., Suckling, J., Coleman, M. R., Pickard, J. D., Menon, D., and Bullmore, E. (2005). Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb. Cortex 15, 1332–1342.
Sanz-Arigita, E. J., Schoonheim, M. M., Damoiseaux, J. S., Rombouts, S. A., Maris, E., Barkhof, F., Scheltens, P., and Stam, C. J. (2010). Loss of “small-world” networks in Alzheimer’s disease: graph analysis of FMRI resting-state functional connectivity. PLoS ONE 5, e13788. doi:10.1371/journal.pone.0013788
Schmitt, J. E., Lenroot, R. K., Wallace, G. L., Ordaz, S., Taylor, K. N., Kabani, N., Greenstein, D., Lerch, J. P., Kendler, K. S., Neale, M. C., and Giedd, J. N. (2008). Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb. Cortex 18, 1737–1747.
Shaw, P., Lerch, J. P., Pruessner, J. C., Taylor, K. N., Rose, A. B., Greenstein, D., Clasen, L., Evans, A., Rapoport, J. L., and Giedd, J. N. (2007). Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 6, 494–500.
Sheline, Y. I., Morris, J. C., Snyder, A. Z., Price, J. L., Yan, Z., D’Angelo, G., Liu, C., Dixit, S., Benzinger, T., Fagan, A., Goate, A., and Mintun, M. A. (2010). APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Abeta42. J. Neurosci. 30, 17035–17040.
Small, G. W., Ercoli, L. M., Silverman, D. H., Huang, S. C., Komo, S., Bookheimer, S. Y., Lavretsky, H., Miller, K., Siddarth, P., Rasgon, N. L., Mazziotta, J. C., Saxena, S., Wu, H. M., Mega, M. S., Cummings, J. L., Saunders, A. M., Pericak-Vance, M. A., Roses, A. D., Barrio, J. R., and Phelps, M. E. (2000). Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 97, 6037–6042.
Smit, D. J., Stam, C. J., Posthuma, D., Boomsma, D. I., and De Geus, E. J. (2008). Heritability of “small-world” networks in the brain: a graph theoretical analysis of resting-state EEG functional connectivity. Hum. Brain Mapp. 29, 1368–1378.
Sorg, C., Riedl, V., Muhlau, M., Calhoun, V. D., Eichele, T., Laer, L., Drzezga, A., Forstl, H., Kurz, A., Zimmer, C., and Wohlschlager, A. M. (2007). Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 104, 18760–18765.
Sperling, R. A., Bates, J. F., Chua, E. F., Cocchiarella, A. J., Rentz, D. M., Rosen, B. R., Schacter, D. L., and Albert, M. S. (2003). fMRI studies of associative encoding in young and elderly controls and mild Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatr. 74, 44–50.
Sporns, O., Tononi, G., and Kotter, R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42. doi:10.1371/journal.pcbi.0010042
Stam, C. J. (2004). Functional connectivity patterns of human magnetoencephalographic recordings: a “small-world” network? Neurosci. Lett. 355, 25–28.
Stam, C. J. (2010). Use of magnetoencephalography (MEG) to study functional brain networks in neurodegenerative disorders. J. Neurol. Sci. 289, 128–134.
Stam, C. J., De Haan, W., Daffertshofer, A., Jones, B. F., Manshanden, I., Van Cappellen Van Walsum, A. M., Montez, T., Verbunt, J. P., De Munck, J. C., Van Dijk, B. W., Berendse, H. W., and Scheltens, P. (2009). Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer’s disease. Brain 132, 213–224.
Stam, C. J., Jones, B. F., Manshanden, I., Van Cappellen Van Walsum, A. M., Montez, T., Verbunt, J. P., De Munck, J. C., Van Dijk, B. W., Berendse, H. W., and Scheltens, P. (2006). Magnetoencephalographic evaluation of resting-state functional connectivity in Alzheimer’s disease. Neuroimage 32, 1335–1344.
Stam, C. J., Jones, B. F., Nolte, G., Breakspear, M., and Scheltens, P. (2007a). Small-world networks and functional connectivity in Alzheimer’s disease. Cereb. Cortex 17, 92–99.
Stam, C. J., Nolte, G., and Daffertshofer, A. (2007b). Phase lag index: assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources. Hum. Brain Mapp. 28, 1178–1193.
Stam, C. J., and Reijneveld, J. C. (2007). Graph theoretical analysis of complex networks in the brain. Nonlinear Biomed. Phys. 1, 3.
Supekar, K., Menon, V., Rubin, D., Musen, M., and Greicius, M. D. (2008). Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput. Biol. 4, e1000100. doi:10.1371/journal.pcbi.1000100
Telesford, Q. K., Morgan, A. R., Hayasaka, S., Simpson, S. L., Barret, W., Kraft, R. A., Mozolic, J. L., and Laurienti, P. J. (2010). Reproducibility of graph metrics in FMRI networks. Front. Neuroinform. 4:117. doi:10.3389/fninf.2010.00117
Tohgi, H., Takahashi, S., Kato, E., Homma, A., Niina, R., Sasaki, K., Yonezawa, H., and Sasaki, M. (1997). Reduced size of right hippocampus in 39- to 80-year-old normal subjects carrying the apolipoprotein E epsilon4 allele. Neurosci. Lett. 236, 21–24.
Tomasi, D., and Volkow, N. D. (2010). Functional connectivity density mapping. Proc. Natl. Acad. Sci. U.S.A. 107, 9885–9890.
Ukmar, M., Makuc, E., Onor, M. L., Garbin, G., Trevisiol, M., and Cova, M. A. (2008). Evaluation of white matter damage in patients with Alzheimer’s disease and in patients with mild cognitive impairment by using diffusion tensor imaging. Radiol. Med. 113, 915–922.
Vaessen, M. J., Hofman, P. A., Tijssen, H. N., Aldenkamp, A. P., Jansen, J. F., and Backes, W. H. (2010). The effect and reproducibility of different clinical DTI gradient sets on small world brain connectivity measures. Neuroimage 51, 1106–1116.
Villain, N., Desgranges, B., Viader, F., De La Sayette, V., Mezenge, F., Landeau, B., Baron, J. C., Eustache, F., and Chetelat, G. (2008). Relationships between hippocampal atrophy, white matter disruption, and gray matter hypometabolism in Alzheimer’s disease. J. Neurosci. 28, 6174–6181.
Wada, Y., Nanbu, Y., Kikuchi, M., Koshino, Y., Hashimoto, T., and Yamaguchi, N. (1998a). Abnormal functional connectivity in Alzheimer’s disease: intrahemispheric EEG coherence during rest and photic stimulation. Eur. Arch. Psychiatry Clin. Neurosci. 248, 203–208.
Wada, Y., Nanbu, Y., Koshino, Y., Yamaguchi, N., and Hashimoto, T. (1998b). Reduced interhemispheric EEG coherence in Alzheimer disease: analysis during rest and photic stimulation. Alzheimer Dis. Assoc. Disord. 12, 175–181.
Wakana, S., Jiang, H., Nagae-Poetscher, L. M., Van Zijl, P. C., and Mori, S. (2004). Fiber tract-based atlas of human white matter anatomy. Radiology 230, 77–87.
Wang, J., Zuo, X., and He, Y. (2010). Graph-based network analysis of resting-state functional MRI. Front. Syst. Neurosci. 4:16. doi:10.3389/fnsys.2010.00016
Wang, J. H., Zuo, X. N., Gohel, S., Milham, M. P., Biswal, B. B., and He, Y. (2011). Graph theoretical analysis of functional brain networks: test-retest evaluation on short- and long-term resting-state functional MRI data. PLoS ONE 6, e21976. doi:10.1371/journal.pone.0021976
Wang, K., Liang, M., Wang, L., Tian, L., Zhang, X., Li, K., and Jiang, T. (2007). Altered functional connectivity in early Alzheimer’s disease: a resting-state fMRI study. Hum. Brain Mapp. 28, 967–978.
Wang, L., Zang, Y., He, Y., Liang, M., Zhang, X., Tian, L., Wu, T., Jiang, T., and Li, K. (2006). Changes in hippocampal connectivity in the early stages of Alzheimer’s disease: evidence from resting state fMRI. Neuroimage 31, 496–504.
Watts, D. J., and Strogatz, S. H. (1998). Collective dynamics of “small-world” networks. Nature 393, 440–442.
Wishart, H. A., Saykin, A. J., Rabin, L. A., Santulli, R. B., Flashman, L. A., Guerin, S. J., Mamourian, A. C., Belloni, D. R., Rhodes, C. H., and Mcallister, T. W. (2006). Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. Am. J. Psychiatry 163, 1603–1610.
Wolk, D. A., and Dickerson, B. C. (2010). Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 107, 10256–10261.
Wright, I. C., Sharma, T., Ellison, Z. R., Mcguire, P. K., Friston, K. J., Brammer, M. J., Murray, R. M., and Bullmore, E. T. (1999). Supra-regional brain systems and the neuropathology of schizophrenia. Cereb. Cortex 9, 366–378.
Xie, S., Xiao, J. X., Gong, G. L., Zang, Y. F., Wang, Y. H., Wu, H. K., and Jiang, X. X. (2006). Voxel-based detection of white matter abnormalities in mild Alzheimer disease. Neurology 66, 1845–1849.
Yao, Z., Zhang, Y., Lin, L., Zhou, Y., Xu, C., and Jiang, T. (2010). Abnormal cortical networks in mild cognitive impairment and Alzheimer’s disease. PLoS Comput. Biol. 6, e1001006. doi:10.1371/journal.pcbi.1001006
Zhang, H. Y., Wang, S. J., Liu, B., Ma, Z. L., Yang, M., Zhang, Z. J., and Teng, G. J. (2010). Resting brain connectivity: changes during the progress of Alzheimer disease. Radiology 256, 598–606.
Zielinski, B. A., Gennatas, E. D., Zhou, J., and Seeley, W. W. (2010). Network-level structural covariance in the developing brain. Proc. Natl. Acad. Sci. U.S.A. 107, 18191–18196.
Keywords: connectome, graph theory, small-world, cortical thickness, genetics, DTI, fMRI, EEG/MEG
Citation: Xie T and He Y (2012) Mapping the Alzheimer’s brain with connectomics. Front. Psychiatry 2:77. doi: 10.3389/fpsyt.2011.00077
Received: 27 October 2011;
Accepted: 19 December 2011;
Published online: 05 January 2012.
Edited by:
Alex Fornito, University of Melbourne, AustraliaReviewed by:
Aristotle Voineskos, Centre for Addiction and Mental Health, CanadaChristian Sorg, Klinikum rechts der Isar Technische Universität München, Germany
Copyright: © 2012 Xie and He. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yong He, State Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University, Beijing 100875, China. e-mail:yong.he@bnu.edu.cn