- 1Laboratory of Sustainable Animal Environment, Graduate School of Agricultural Science, Tohoku University, Osaki, Japan
- 2Department of Biology, Graduate School of Science, Kobe University, Kobe, Japan
- 3National Institute for Physiological Sciences, Okazaki, Japan
- 4Department of Physiological Sciences, School of Life Science, The Graduate University for Advanced Studies, Okazaki, Japan
- 5Department of Convergence Medicine, School of Medicine, Pusan National University, Yangsan, Republic of Korea
- 6Core Research Facility, Pusan National University, Yangsan, Republic of Korea
Introduction: Oxyrrhis marina is thought to have diverged from other dinoflagellates at an early stage of their evolution and is considered to show their ancestral form. As for other current dinoflagellates, the species possesses condensed chromosomes throughout the cell cycle but shows some important differences. The chromosomes of O. marina are thinner and longer than those of other dinoflagellate species and do not show the repeating arch-shaped liquid-crystal structure that is found in core dinoflagellates. These morphological features were described originally about half a century ago from cells fixed using conventional chemical methods, which are prone to producing morphological artifacts. Therefore, it is crucial to reevaluate ultrastructural features using cells fixed by other methods.
Method: In this study, O. marina was fixed with freeze-substitution, a method that is less prone to artifacts in electron microscopy, in addition to conventional chemical fixation, and the details of chromosome structure were reexamined using volume electron microscopy.
Results: In the four cells observed, the number of chromosomes was consistently nearly 400. The nucleus of O. marina has a single nucleolus at its center, to which, as in other dinoflagellates, multiple chromosomes are attached. Several nucleofilaments were observed penetrating the nucleolus. On the other hand, filamentous structures have been observed in chemically fixed chromosomes, but no such structures were observed in cells fixed by freeze-substitution. Tomographic analysis using volume electron microscopy confirmed the absence of these structural features.
Discussion: The number of chromosomes of O. marina was previously considered to be approximately 50, but this is only one-eighth of the number found in the present study (400). It is concluded that the chromosomes of O. marina are composed of tightly condensed and densely folded nucleofilaments, which are difficult to distinguish. This study revealed novel ultrastructural features in the chromosome of O. marina. These findings will help consider the evolutionary scenario through which the enigmatic dinoflagellate nucleus (dinokaryon) was established. In addition, this study indicated freeze-substitution fixation and volume electron microscopy would become a critical technique in elucidating the dinokaryon chromosome structure.
Introduction
Dinoflagellates are biflagellate protists distributed in various aquatic environments around the world. About half of the species are photosynthetic and possess plastids derived from secondary or tertiary endosymbiosis, while the others are heterotrophic or parasitic. The photosynthetic dinoflagellates play an essential role in marine ecosystems as primary producers second only to diatoms (Taylor et al., 2008). The dinoflagellates are included in the Alveolata, together with Apicomplexa such as Plasmodium (the malarial parasite), Toxoplasma, and Cryptosporidium; and the ciliates such as Tetrahymena and Paramecium (Adl et al., 2005; Adl et al., 2012, 2019).
The nucleus of dinoflagellates, called a dinokaryon, has characteristic features not found in other eukaryotes (reviewed by Fukuda and Suzaki, 2015; Soyer-Gobillard and Dolan, 2015). In the dinokaryon, chromosomes are distinctly observed even in the interphase. Therefore, the chromosomes are considered to be condensed throughout the cell cycle. The interior of the nucleus has little nucleoplasm and is occupied primarily by chromosomes, which are spindle- or rugby-ball-shaped with a cholesteric liquid crystal structure probably composed of chromatin filaments (Chow et al., 2010). Arch-like filaments can be seen in chromosome sections (Bouligand et al., 1968). Histone proteins, which provide structural support for chromosomes in eukaryotes, are barely detectable in the dinokaryon, while the nucleus contains large amounts of small basic proteins (DVNPs: Dinoflagellate/viral nucleoproteins and HCc: Histone-like proteins from Crypthecodinium cohnii) not found in other eukaryotes (Sala-Rovira et al., 1991; Gornik et al., 2012). Nucleofilaments in the dinokaryon do not exhibit the bead-and-string structure common to other eukaryotes but appear as a homogeneous thick filament slightly thicker than DNA (Rizzo and Burghardt, 1980). This structure is assumed to comprise DNA bound with small basic proteins (Irwin et al., 2018), but the specific molecular structure has not been elucidated, and so far, no evolutionary scenario has been proposed to explain how the dinokaryon arose.
Oxyrrhis marina is considered to represent an ancestral or earlier stage in the evolution of dinoflagellates because of its early divergence from most other species (Saldarriaga et al., 2003; Fukuda and Endoh, 2008; Bachvaroff et al., 2014; Janouškovec et al., 2017). It has therefore been speculated that the chromosomes of O. marina may represent an intermediate stage in the evolution of the enigmatic dinokaryon from the typical eukaryotic nucleus (Wisecaver and Hackett, 2011). Therefore, the chromosomal features of O. marina are important in elucidating how the dinokaryon was established.
Several studies have described structural features of the dinokaryon. For example, a rosette-like complex was recently found in the dinokaryon chromosome in Prorocentrum minimum (Golyshev et al., 2018), and the three-dimensional (3D) arrangement of chromosomes and nucleoli in P. cordatum has been described using focused ion beam scanning electron microscopy (FIB-SEM) (Kalvelage et al., 2023). The ultrastructure of dinoflagellate chromosomes has been investigated mostly using a selection of different ‘core’ dinoflagellates, rather than that of apparently an ancestral species such as O. marina. Furthermore, all previous studies describing ultrastructural features of O. marina have used conventional chemical fixation methods (Grell and Schwalbach, 1965; Dodge and Crawford, 1971a, 1971b, 1974; Triemer, 1982; Gao and Li, 1986; Chaoying et al., 1996), which sometimes cause morphological artifacts (Song et al., 2017; Hoshina et al., 2021). The present study attempted to reevaluate the ultrastructural features of the O. marina nucleus using freeze-substitution and volume electron microscopy.
Results
Difference in ultrastructural characteristics with freeze substitution versus conventional chemical fixation
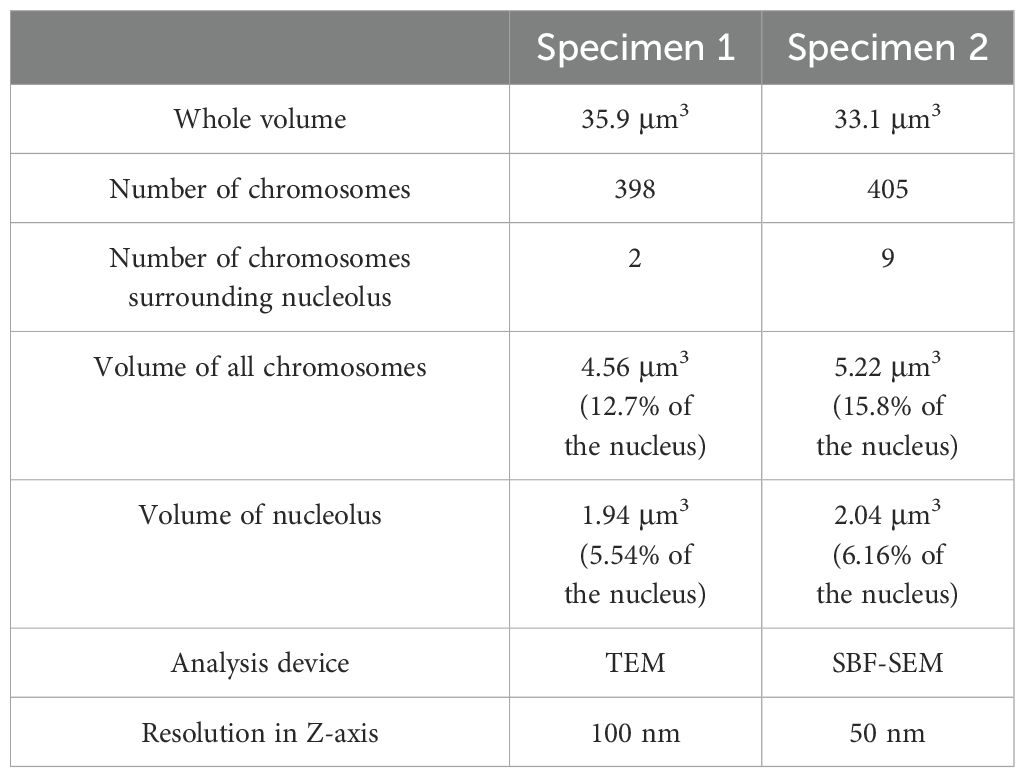
In the living cell, the nucleus of Oxyrrhis marina is located almost at the center of the cell, with a single prominent nucleolus positioned centrally within it (Figure 1A). Cells fixed with 1% glutaraldehyde were stained with DAPI and observed with confocal laser microscopy, demonstrating the presence of numerous elongated chromosomes in the nucleus except in the region of the nucleolus (Figure 1B).
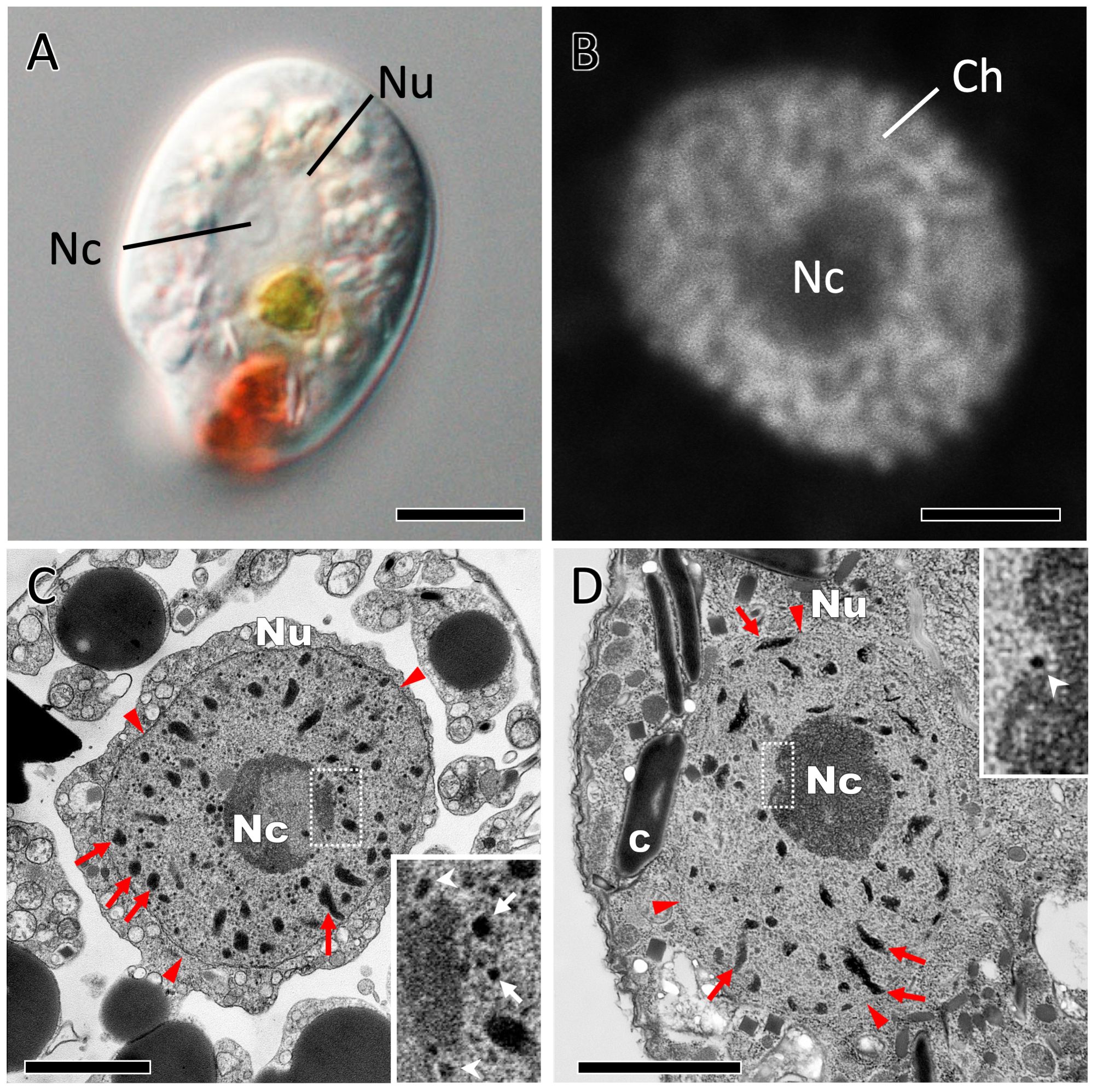
Figure 1. Representative images of Oxyrrhis marina using light and electron microscopy. (A) A light microscopic image of the living cell by differential interference contrast (DIC) microscopy. (B) A single confocal image slice of a DAPI-stained nucleus. Numerous intermingled string-like chromosomes appear within the nucleus. (C) TEM image of a cell prepared by conventional chemical fixation. Inset in (C) An enlarged image of the rectangular area of the nucleolar boundary in (C). (D) TEM image of a cell fixed by freeze substitution. Inset in (D) An enlarged image of the rectangular area of the nucleolar boundary in (D), shows an electron-dense object (white arrowhead) embedded at the surface of the nucleolus. Red arrowheads, nuclear membranes; C, crystalline structures; Ch in (B), and red arrows in (C) and (D) are Chromosomes; Nc, Nucleolus; Nu, Nucleus; White arrowheads in insets of (C) and (D), small electron-dense objects embedded at the surface of the nucleolus; White arrows in inset of (C), small electron-dense objects which presumed to be fragmented chromosomes. Scale bars: 10 µm in (A); 2 µm in (B–D).
Figure 1C is a transmission electron microscopy (TEM) image showing nuclear fine structures in O. marina by using conventional chemical fixation methods. As seen in the light microscopic observations (Figures 1A, B), a single nucleolus was present at the center of the nucleus. A number of chromosomes were observed as electron-dense granular or rod-like structures (red arrows in Figure 1C). In the nucleoplasm, there were many small objects of high electron density (white arrows in the inset image of Figure 1C), which were similar to the ones described by Dodge and Crawford (1971a) as fragmented chromosomes. Various organelles, such as mitochondria, trichocysts, and oil droplets, as well as crystal-like structures of very high electron density, were found within the cytoplasm, which were unevenly distributed around the nucleus and organelles. The remaining intracellular area could be observed as low electron density space. These morphological features are consistent with previous descriptions (Dodge and Crawford, 1971a, 1971b, 1974).
Conventional chemical fixation requires time for the fixative to diffuse through the cell membrane and form cross-linkages between molecules in the cell. Such relatively slow fixation can result in artifacts such as cytoplasmic gaps and organelle swelling. For example, Song et al. (2017) and Hoshina et al. (2021) reported that chemical fixation and subsequent dehydration led to vacuolar swelling, resulting in a gap between the perialgal vacuolar membrane and symbiotic Chlorella in ciliates. In chemically fixed O. marina, the cytoplasm was only distributed around the nucleus and organelles (Figure 1C), but this was not consistent with the images of live cells observed by differential interference contrast microscopy (Figure 1A), suggesting that conventional chemical fixation methods may produce structural artifacts in the cells. This prompted us to investigate the ultrastructure of O. marina cells using freeze-substitution fixation (which can fix cell structures much more rapidly) and compare it with conventional chemical fixation methods. In cells prepared by freeze-substitution fixation, the cytoplasm was uniformly distributed throughout the cell (Figure 1D; Supplementary Figures S1A, B). In the subcortical region of the cell, the alveoli and mitochondrial cristae that were clearly observed in the chemically fixed cells were obscured. On the other hand, cell scales were well preserved in the freeze-substituted cells. (Supplementary Figures S1C, D). The interior of the nucleus was, in outline, similar to that observed with conventional chemical fixation, with one nucleolus and numerous chromosomes. However, small electron-dense objects, such as those described as fragmented chromosomes, were not found at all in the nucleus prepared by freeze-substitution (Figure 1D; Supplementary Figures S1E, F). These findings suggest that freeze-substitution fixation, rather than conventional chemical fixation, is preferable for artifact-free ultrastructural observations of O. marina.
The 3D arrangement of chromosomes in the nucleus
The three-dimensional (3D) structure of the nucleus of O. marina fixed by freeze-substitution was reconstructed using volume electron microscopy, and morphological features of the chromosomes, including the number of chromosomes, were examined (Figures 2A–E). Figures 2A–C (Supplementary Movie S1) were reconstructed from 56 consecutive thin sections of 100 nm thickness taken by conventional TEM. Figures 2D, E were obtained from consecutive images sliced at 50 nm thickness by serial block-face scanning electron microscopy (SBF-SEM). The characteristics of the representative nuclei observed are summarized in Table 1. The estimated number of chromosomes encapsulated in one nucleus based on conventional consecutive TEM images (specimen no. 1 in Table 1 and Figures 2A–C) was 398, while that estimated from SBF-SEM images (specimen no. 2 in Table 1, and Figures 2D, E) was 405. Chromosomes were uniform in thickness (~0.1 µm) but varied in length from 0.1 to 2.5 µm (Figure 2F). The median length was 0.73 µm, and about 80% of all chromosomes were between 0.2 and 1.4 µm in length. To further confirm the number of chromosomes, two more nuclei (specimen no. 3 and 4; data not shown) were reconstructed using SBF-SEM. The results estimated 402 and 403 chromosomes, respectively, suggesting that the number of O. marina chromosomes is approximately 400.
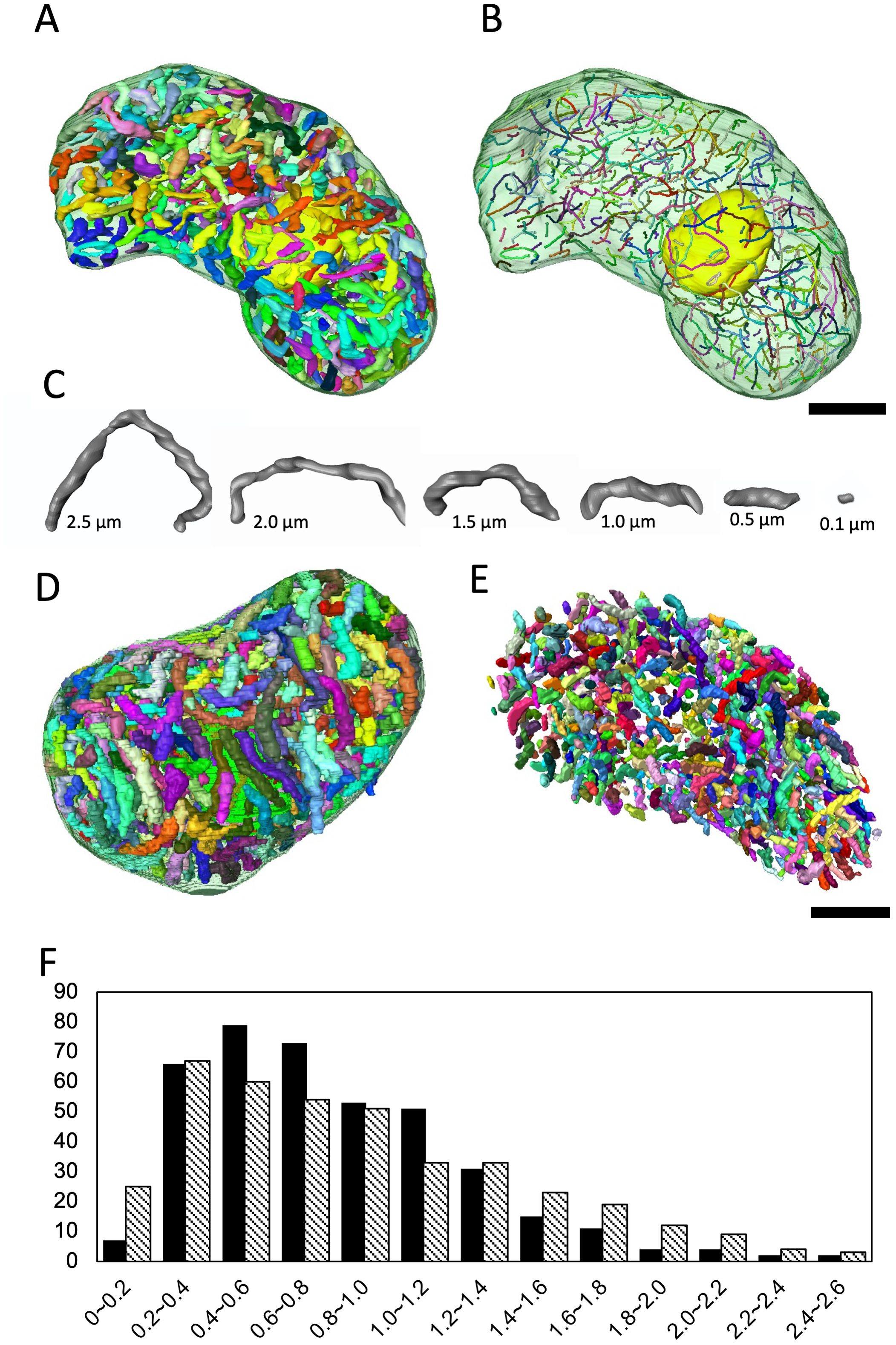
Figure 2. 3D analysis of chromosomes inside the Oxyrrhis marina nucleus. (A) Reconstructed 3D model based on consecutive conventional TEM images (specimen no. 1). The thickness of each section is 100 nm. (B) The chromosomes shown in (A) illustrated in a skeletal model, showing 398 chromosomes. The nucleolus is colored yellow. (C) Representative chromosomes of different lengths selected from those shown in (A). (D) Reconstructed 3D model based on consecutive images obtained by SBF-SEM (specimen no. 2). The z-axis resolution is 50 nm. (E) View of the 3D model in (D) excluding nuclear membrane segmentation. Scale bars: 1 µm in (B, E). (F) Comparison of the frequency distribution of chromosome lengths for consecutive TEM images (black bars) and SBF-SEM images (shaded bars). The X axis is indicated by µm, and the Y axis shows the number of chromosomes.
Structure of the nucleolus
In core dinoflagellates, the dinokaryon has multiple nucleoli (sites of ribosome biogenesis) to which many chromosomes adhere. In Prorocentrum micans, the ribosomal RNA gene locus is on chromosomes attached to the nucleolus (Géraud et al., 1991). From here, the region encoding the ribosomal RNA gene is released from the chromosome and extended to the low electron-dense central region, where active rRNA transcription occurs (Géraud et al., 1991). In O. marina, many chromosomes were observed adhering to the nucleolus, which is located in the center of the nucleus (Figures 1C, D). The structural relationship between the nucleolus and the chromosomes adhering to it was clarified by reconstructing it in 3D. Here, in addition to the four nucleoli analyzed in the nuclear fine structures in 3D (specimens no. 1 and 2 in Figure 2 and specimens no. 3 and 4), four more nucleoli (specimen no. 5–8: Figures 3, 4) were reconstructed in 3D from the other serially sectioned TEM images (eight nucleoli in total). The total number of chromosomes remained constant (approximately 400) in the four newly observed nuclei, but the number of chromosomes adhering to the nucleolus varied. Only long chromosomes adhered to the nucleolus (Figures 3, 4). While some chromosomes surrounded the surface of the nucleolus (specimen no. 5 in Figures 3A, B; Supplementary Movie S2A), chromosomes penetrating the nucleolus were also observed in some nucleoli (specimen no. 6 in Figures 3C, D; Supplementary Movie S2B).
Figure 3. 3D analysis of the Oxyrrhis marina nucleolus (specimen no. 5 and 6). (A) Three representative serial sections of a nucleolus (specimen no. 5) by TEM. Section thickness 100 nm. In the lower panels, the nucleolus is colored yellow, and each chromosome is highlighted in a different color. (B) 3D models of the nucleolus reconstituted from the consecutive images, including sections shown in (A). The nucleolus is encircled with two chromosomes colored red and green. (C) Three representative serial sections of a nucleolus (specimen no. 6). Section thickness is 100 nm. (D) 3D models of the nucleolus reconstituted from consecutive images including sections shown in (C). The nucleolus is encircled with 11 chromosomes shown in different colors. Scale bars: 200 nm.
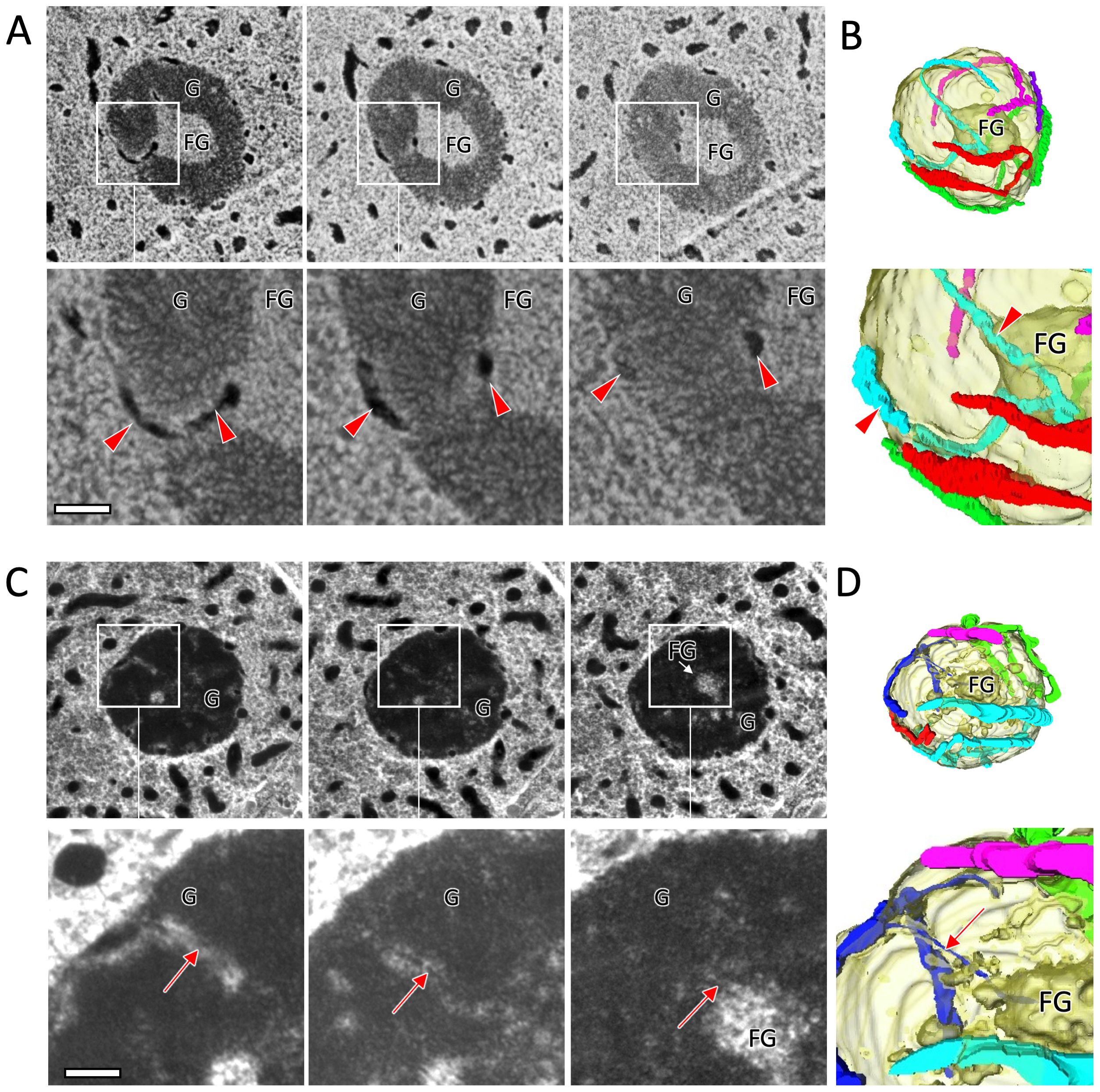
Figure 4. 3D analysis of the Oxyrrhis marina nucleolus and chromosomes/nucleofilaments penetrating the granular pre-ribosome compartment (G) region (specimen no. 7 and 8). (A) Three representative serial sections of a nucleolus (specimen no. 7) by TEM. Section thickness is 100 nm. Red arrowheads indicate chromosomes that penetrate the G region and reach the region of the fibrillogranular (FG) compartment at the center of the nucleolus. (B) Reconstructed 3D models of the nucleolus from consecutive images including sections shown in (A). The nucleolus is encircled with five chromosomes. (C) Three representative serial sections of a nucleolus (specimen no. 8) by TEM. The arrows indicate a nucleofilament branching out from a chromosome attached to the nucleolar surface. This nucleofilament extends through the G region to the FG region at the center of the nucleolus. Section thickness is 100 nm. (D) Reconstructed 3D models of the nucleolus from consecutive images including sections shown in (C) Scale bars: 200 nm.
The nucleolus of core dinoflagellates can be divided into two regions according to electron density. The high-density peripheral regions are called granular pre-ribosome compartment (G), whereas the central low-density regions are called fibrillogranular compartment (FG), where active rRNA transcription takes place (de la Espina et al., 2005). The nucleolus of O. marina may also be classified into two similar regions based on differences in electron density (Figures 4A, C), where chromosomes penetrating the G regions and reaching the low-density FG regions were also observed (specimen no. 7 in Figures 4A, B; Supplementary Movie S3A). In addition, nucleofilaments branching off from chromosomes on the surface of the nucleolus were frequently observed. These nucleofilaments were approximately 6.5 nm thick and penetrated beyond the G compartment into the FG regions at the center of the nucleolus (specimen no.8 in Figures 4C, D; Supplementary Movie S3B). Figure 5 is a schematic diagram summarizing the structure of chromosomes and nucleofilaments penetrating the nucleolus.
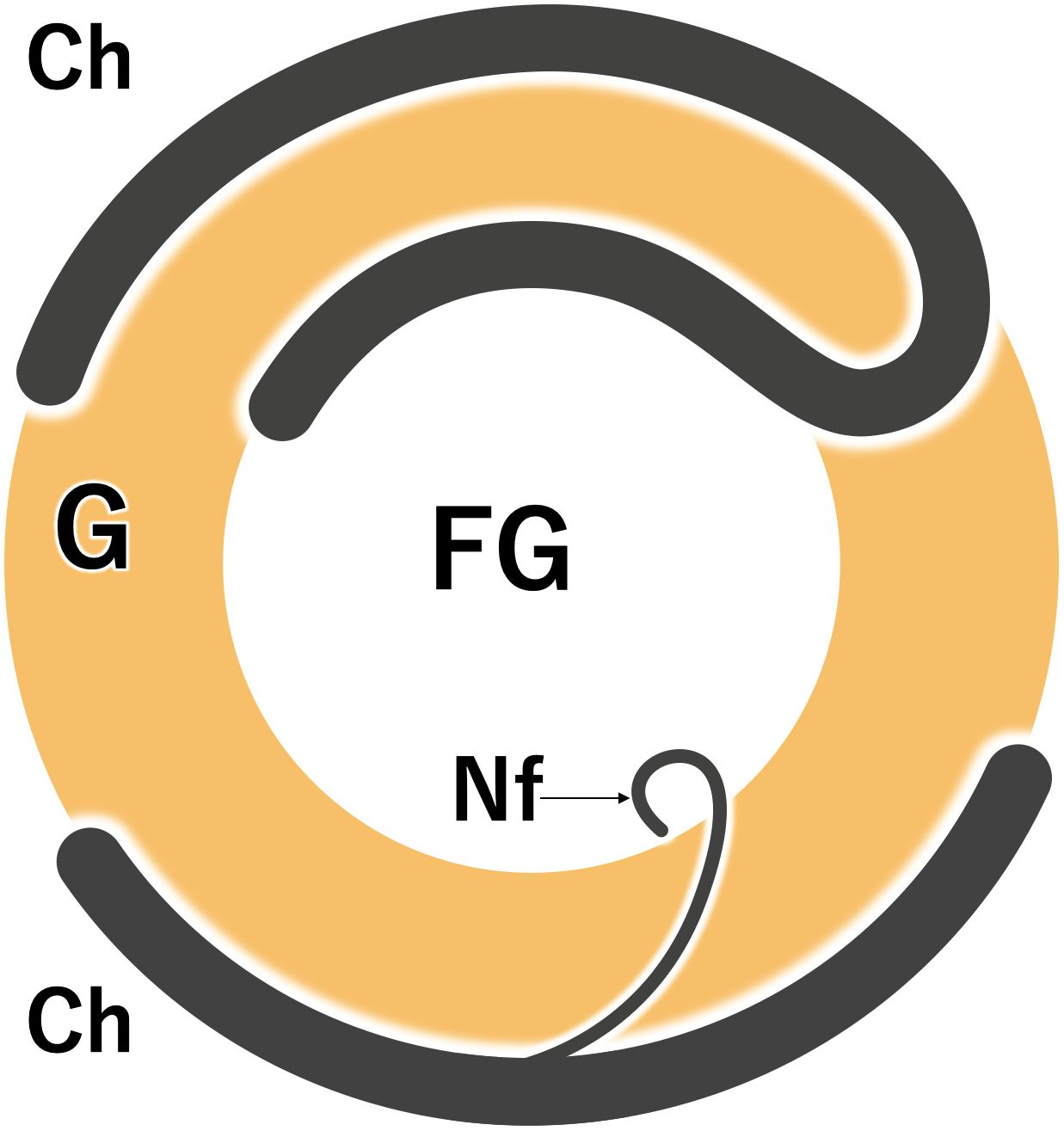
Figure 5. A schematic diagram showing a nucleolus of Oxyrrhis marina with chromosomes (Ch) and nucleofilament (Nf). FG, the fibrillogranular compartment; G, the granular pre-ribosome compartment.
Internal structure of the chromosome
The periodic arch-shaped structure is a common feature of dinokaryon chromosomes (Figure 6A). This feature has been considered to reflect the liquid cholesteric crystal structure of the dinoflagellate chromosome in which parallel nucleofilaments are repeatedly stacked in slightly different alignments (Bouligand et al., 1968). Such a periodic filament structure was not found in the O. marina chromosomes, but some disordered or randomly folded filaments appeared in the chromosome (Figure 6B). Since these nucleofilament morphologies were observed in the chemically fixed cells, an attempt was made to reevaluate the nucleofilament arrangements in O. marina chromosomes using freeze-substitution fixation.
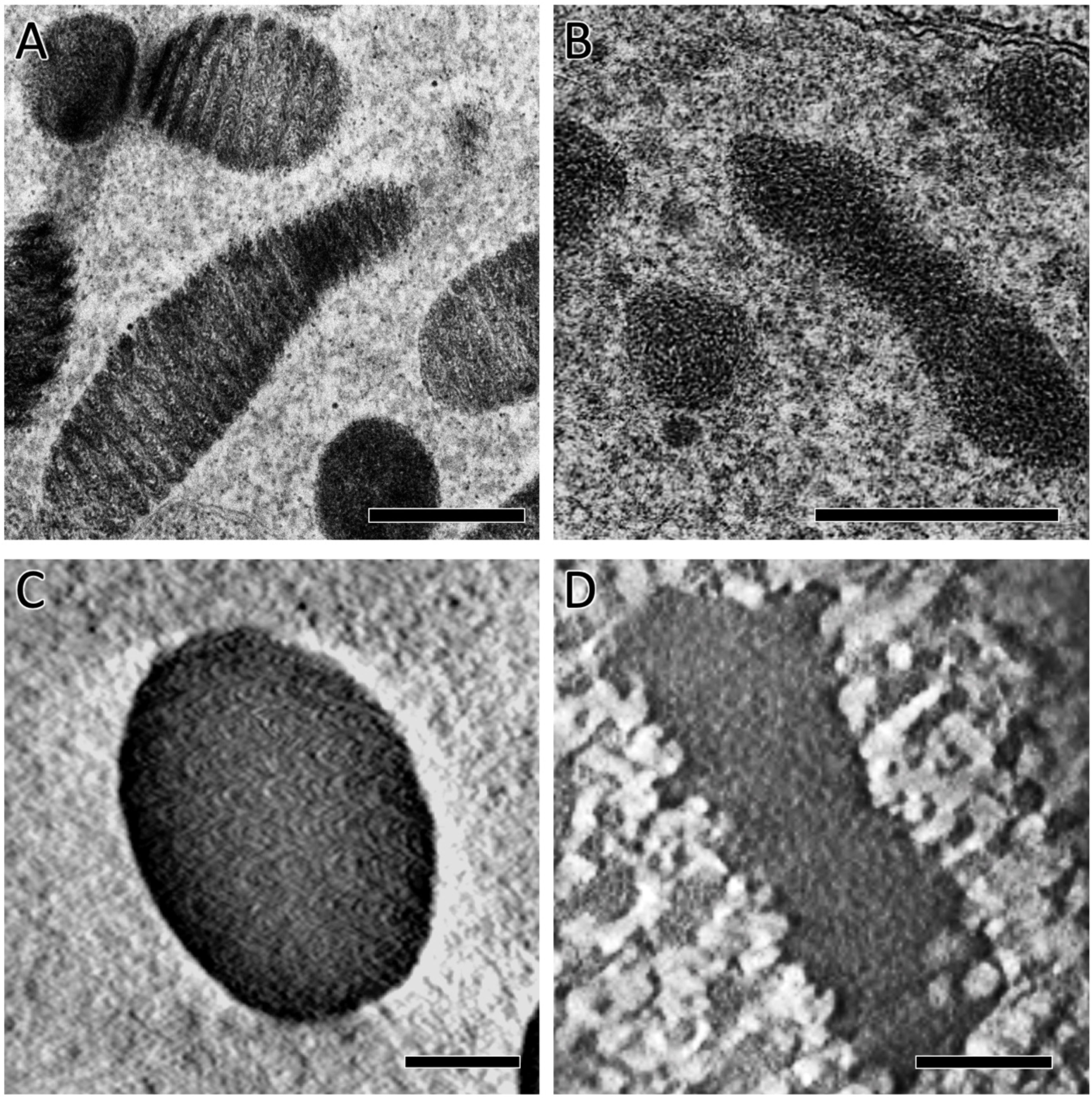
Figure 6. Comparison of chromosome ultrastructure between typical and ancestral dinoflagellate species, prepared by conventional chemical fixation (A, B) or freeze-substitution fixation and electron tomography analysis (C, D). (A) Chromosome morphology of a typical dinoflagellate species, Heterocapsa circularisquama. Periodic arch-shaped filaments, characteristic of dinokaryon chromosomes, are observed. (B) Chromosome morphology of the ancestral dinoflagellate, Oxyrrhis marina. Filamentous structures without regularity patterns appear in the chromosome. (C) Chromosome morphology of H. circularisquama shown in a tomogram slice. The chromosome shows a cholesteric liquid crystal structure. (D) Chromosome morphology of O. marina shown in a tomogram slice, in which no periodic or filamentous structures were observed. Scale bars: 500 nm in (A, B); 100 nm in (C, D).
The core dinoflagellate Heterocapsa circularisquama, which possesses a typical dinokaryon, was used for comparative analysis of chromosome structure. With freeze-substitution fixation, chromosomes in the 80-nm sections of both H. circularisquama and O. marina appeared as electron-dense images with no discernible internal structure (data not shown), suggesting the possibility of a loss of chromosomal constituents, particularly with conventional chemical fixation methods. To visualize the internal structures in the highly-dense chromosomes of the dinokaryon of H. circularisquama, we applied tomography analysis, which allowed us to find the arch-shaped structure within the chromosomes (Figure 6C; Supplementary Movie S4A). The visualized nucleofilament arcs were similar to those by conventional chemical fixation methods but were more tightly aligned (compare Figures 6A, C). In O. marina, however, no periodic filamentous structure appeared (Figure 6D; Supplementary Movie S4B), suggesting that the nucleofilaments of O. marina are either tightly packed without periodic structure or so densely folded that individual filaments are indistinguishable.
Discussion
Oxyrrhis marina appears to have diverged from most other species in the dinoflagellate phylogenetic tree at an early stage (Saldarriaga et al., 2003; Fukuda and Endoh, 2008; Bachvaroff et al., 2014; Janouškovec et al., 2017). Therefore, it may possess features that are useful for understanding the evolutionary changes that gave rise to the dinokaryon, the unusual eukaryotic nucleus characteristic of dinoflagellates. Previous studies describing morphological features of the O. marina nucleus used conventional chemical fixation methods (Grell and Schwalbach, 1965; Dodge and Crawford, 1971a, 1971b, 1974; Triemer, 1982; Gao and Li, 1986; Chaoying et al., 1996) but fixatives such as glutaraldehyde diffuse relatively slowly through the cell and may produce artifacts such as cell swelling and gaps in the cytoplasm (Song et al., 2017; Hoshina et al., 2021). In the present study, cells were fixed using freeze-substitution, and their ultrastructural morphology was compared with that obtained by conventional chemical fixation. A comparison of the TEM images obtained (Figures 1C, D) strongly suggested that freeze-substitution fixation is preferable for observing non-artifactual ultrastructural morphologies in O. marina.
In this study, the number of O. marina chromosomes was estimated to be nearly 400. Dodge (1963) reported the chromosome number of O. marina to be 55, and an unpublished estimate of approximately 100 was based on serially sectioned TEM images (Kato, personal communication). These estimates are significantly less than those obtained from the reconstructed 3D structure of O. marina nuclei using freeze-substitution fixation and volume electron microscopy. Ultrathin sections obtained from conventional chemical fixation exhibited the presence of many electron-dense granules in the nuclei (Figure 1C). Dodge and Crawford (1971a) described these granules as small fragmented chromosomes. However, such small electron-dense granules were not found in the ultrathin sections using the freeze-substitution fixation technique (Figure 1D). The reconstructed 3D structure of the nucleus reveals that the length of chromosomes in O. marina differs widely, with many chromosomes shorter than 600 nm (Figure 2F). These observations suggest that in previous studies, short chromosomes were probably not maintained and were disintegrated into small fragments, and only long chromosomes that retained their morphology were counted.
Ribosome biogenesis takes place in the nucleolus. In the core dinoflagellate Prorocentrum micans, the rRNA coding gene is located on the chromosomes adhering to the nucleolus, and the nucleofilament carrying the rRNA gene loci extends to the FG of the nucleolus. rRNA is transcribed in the FG compartment, and the transcribed rRNA precursor is transported to the highly electron-dense G compartment, where the rRNA precursors become mature ribosomes (Géraud et al., 1991). In O. marina, several chromosomes adhere to the nucleolus, and some penetrate and extend into the FG (Figures 4A, B). Some nucleofilaments branching off from chromosomes also extend into the FG (Figures 4C, D). The thickness of the nucleofilaments was 6.5 nm, which is consistent with the reported thickness of the dinoflagellate nucleofilament consisting of small basic proteins and DNA (Rizzo and Burghardt, 1980). These results suggest that the model of ribosome biogenesis proposed by Géraud et al. (1991) also applies to this ancestral dinoflagellate. In O. marina, the ribosomal genes may also be encoded on chromosomes adhering to the surface of the nucleolus, and the rRNA genes-coding nucleofilaments may perhaps also extend to the nucleolus FG, where the rRNAs are actively transcribed.
In most eukaryotes, the nucleosome shows the bead-and-string appearance consisting of DNA wrapped around a core histone octamer, which is considered to be the fundamental unit of chromatin. Histone proteins undergo modifications such as acetylation and methylation, which alter the local chromatin conformation, resulting in gene expression regulation (Strahl and Allis, 2000; Allis and Jenuwein, 2016). Dinoflagellates are the only eukaryotes that lack histone proteins (e.g., Rizzo and Noodén, 1974; Jing-Yan, 1984). Several structural models have been proposed to explain the liquid crystalline nature of dinoflagellate chromosomes, but the actual structure is still obscure (reviewed by Fukuda and Suzaki, 2015; Soyer-Gobillard and Dolan, 2015). The dinoflagellate O. marina is considered to be the most ‘ancestral’ or ‘primitive’ dinoflagellate, so it has been proposed that its chromosome structure may be intermediate between the typical histone-dependent eukaryotic chromosome structure and the histone-independent structure of the dinokaryon (Wisecaver and Hackett, 2011). The chemically fixed cell showed disordered or randomly folded nucleofilaments in the chromosome (Figure 6B), which Grell and Schwalbach (1965) considered to be folded into chromosomes without a periodic structure. However, such structures were not found in cells fixed by freeze-substitution and observed by electron tomography. In the past decade, cryo-electron microscopy revealed that nucleosomes are folded irregularly without forming 30-nm fibers within the mitotic chromosomes (Maeshima et al., 2019). The absence of obvious filamentous structures in the chromosomes of the ancestral dinoflagellate O. marina may indicate that its nucleofilaments are tightly and irregularly packed, as in other typical eukaryotic chromosomes. These findings suggest that cryo-electron microscopy will be required in the future to investigate the relationship among O. marina, core dinoflagellates and the rest of the eukaryotic chromosome structures.
Methods
Cell culture
Oxyrrhis marina was obtained from a publicly available algal culture stock center (the Culture Collection of Algae at Göttingen University; international acronym SAG), Strain No. 21.89. It was maintained in natural seawater sterilized by filtration and fed on the green alga Dunaliella tertiolecta (SAG Strain No. 13.86), which was grown in a modified f/2 medium under a 12:12 h light-dark (LD) cycle at 25°C (Fukuda and Endoh, 2006). The O. marina cells were also maintained under 12:12 LD at 25°C.
Heterocapsa circularisquama was sourced from the National Research Institute of Fisheries and Environment, Hiroshima, Japan, and cultured in a modified SWM3 medium under a 12:12 LD cycle (Kang et al., 2015).
Fluorescence microscopy
To visualize nuclear morphology, O. marina cells were fixed with 1% glutaraldehyde for 30 min and washed several times with filtered natural seawater. Cells were then resuspended in filtered natural seawater containing 1 μg/mL DAPI (Nacalai-Tesque, Kyoto, Japan), and mounted on a glass slide under a coverslip. Microscopic observation was performed with a confocal laser scanning microscope (Olympus FV-1000; Evident Corporation, Tokyo, Japan), equipped with a x100 objective lens.
Conventional chemical fixation procedure for transmission electron microscopy
Cells were prefixed with 2% glutaraldehyde in filtered natural seawater. Cell suspension was mixed 1:1 with double-strength fixative solution at room temperature, and then the suspension was left at 4°C for 60 min. Fixed cells were washed three times with cooled filtered natural seawater and then transferred to 0.5% OsO4 in filtered natural seawater for post-fixation at 4°C for 30 min. The fixed cells were then washed several times with filtered water, to eliminate fixative and salts, dehydrated through a graded ethanol series. The dehydrated sample was subjected to centrifugation, after which half of the supernatant was discarded, and an equal amount of Spurr resin (Polysciences Inc., Warrington, PA, USA) was added, followed by mixing. This process was repeated twice. Subsequently, the entire supernatant was replaced with Spurr resin and mixed again, repeating this process twice (30 minutes each). The sample embedded in resin was transferred to the Beem® capsule, and the cells were concentrated at the bottom of the capsule through centrifugation. Finally, the capsulated sample was placed in a pre-heated polymerization oven and polymerized at 70°C for 8 hours. Ultrathin sections (approximately 80 nm thick) were prepared using an ultramicrotome (EM UC7; Leica Microsystems GmbH, Wetzlar, Germany), collected onto Formvar-coated grids (Cu, one slot), stained with 3% uranyl acetate and lead citrate, and examined using a H-7100 transmission electron microscope (Hitachi High-Tech, Tokyo, Japan) operating at 75 kV.
Freeze-substitution fixation technique
Cells were collected by low-speed centrifugation, and suspensions at high cell densities were cryofixed by slamming them onto a liquid nitrogen-cooled (-196°C) copper block using a metal-contact quick-freezing device (VFZ-101, Japan Vacuum Device Inc., Mito, Ibaraki, Japan). The frozen materials were transferred to cold (-80°C) acetone containing 1% OsO4, and substitution was performed by incubation at -80°C for 72 h. The temperature was then manually elevated stepwise (-20°C for 1 h, 4–10°C for 0.5 h, and then room temperature for 0.5 h). The materials were washed with 100% acetone at room temperature and embedded in Spurr’s resin.
Volume electron microscopy
Two methods were used to obtain consecutive image series to create three-dimensional (3D) models. One was to collect consecutive sections and then take sequential images with a TEM; and the other was to automatically acquire sequential images using serial block face-scanning electron microscopy (SBF-SEM).
For the first method, consecutive ultrathin sections (approximately 100 nm thick) were prepared using an ultramicrotome and 56 consecutive sections were collected onto Formvar-coated grids (Cu, one slot) and stained with 3% uranyl acetate and lead citrate. They were then examined using TEM operating at 75 kV. The images taken at a nominal magnification of 17k were recorded in a 1k × 1k CCD camera (C4741-95; Hamamatsu Photonics, Hamamatsu, Japan) at a pixel size of 5.43 nm.
For SBF-SEM observation, the resin block containing the samples was trimmed and glued onto an aluminum rivet with conductive epoxy resin (SPI Conductive Silver Epoxy; Structure Probe Inc., West Chester, PA, USA) and coated with gold using an ion coater. A SBF-SEM system (Sigma VP, Carl Zeiss Microscopy, Jena, Germany) equipped with a back-scattered electron detector (3View; Gatan Inc., Pleasanton, CA, USA) was used to slice and image the specimen block surface. The SEM was operated at a low accelerating voltage of 1.0 kV to reduce charging. Consecutive image series were automatically acquired using Gatan Digital Micrograph software. All images were taken at an image size of 8192 × 8192 pixels (pixel size = 3 nm). Once an SEM image had been acquired, a 50-nm-thick layer was removed from the block face by the diamond knife, and the next freshly exposed surface was imaged by SEM in the same manner. This image acquisition cycle was repeated until images of a complete dinokaryon had been obtained. Two dinokaryon samples were obtained, one with 127 images and the other with 145 images. The consecutive images were aligned using the IMOD software package (Kremer et al., 1996). Segmentation in the 3D reconstruction was performed with Amira version 5.4.5 (FEI Visualization Science Group, Burlington, MA, USA).
Electron tomography
Specimens of O. marina and H. circularisquama prepared by the freeze-substitution fixation method were also subjected to electron tomography. A 80 nm-thick section on a single-slot grid was stained with 3% uranyl acetate and lead citrate. A 200 kV electron microscope (JEM-2100F; JEOL, Tokyo, Japan) was used for data acquisition. Tilt series were recorded with a Gatan Tridium imaging filter with a 2k × 2k CCD camera at 2° increments in a tilt range from -60° to +60°. Automated data acquisition for electron tomography was performed using the Recorder module in the TEMography suite (System in Frontier Inc., Tokyo, Japan). After image alignment, the 3D reconstruction was performed by weighted back-projection using IMOD (Kremer et al., 1996).
An overview of sample preparation and imaging conditions
Sample preparation methods and imaging conditions are listed in a Supplementary Table S1.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YF: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. TS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. KM: Formal analysis, Methodology, Writing – review & editing. CS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by JSPS KAKENHI Grant No. 21K06009 to YF, and by the National Research Foundation of Korea Grant No. 2022R1A5A2027161 to CS. Use of cryo-EM facilities of NEXUS consortium was supported by a National Research Foundation of Korea grant RS-2024-00440289. This study was supported by the joint research program at National Institute for Physiological Sciences, Okazaki, Japan.
Acknowledgments
The authors sincerely acknowledge Dr. Koichi Kato (Emeritus professor, Nagoya City University, Japan) for providing unpublished results of the chromosome number of O. marina based on serially sectioned TEM images. The authors also sincerely acknowledge Dr. Ian G. Gleadall (AiCeph LLC) for providing valuable suggestions and indications during this manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frpro.2025.1512258/full#supplementary-material
Supplementary Figure 1 | A comparison of subcellular ultrastructures obtained by conventional chemical fixation and freeze-substitution fixation methods. (A) A whole-cell image of a longitudinal section obtained by conventional chemical fixation. The cytoplasm is unevenly distributed, with cytoplasmic gaps (*) present. (B) A whole-cell image of a longitudinal section obtained by the freeze-substitution fixation method. No cytoplasmic gaps are observed. The electron density is uniformly high throughout the cell, making it difficult to distinguish organelles such as mitochondria. (C) A TEM image near the cell surface region obtained by conventional chemical fixation. Trichocysts and mitochondria with tubular cristae are clearly visible. (D) A TEM image near the cell surface region obtained by the freeze-substitution fixation method. On the cell surface, the cell scale (Sc), which is not observed in conventional chemical fixation, is preserved. The alveolus is observed as a thin, flattened vacuolar structure located just beneath the plasma membrane. (E) A TEM image of the nucleus by conventional chemical fixation. In addition to numerous thick chromosomes, small electron-dense granules (magenta arrows), described as fragmented chromosomes, are observed within the nucleus. (F) A TEM image of the nucleus by freeze-substitution fixation. Due to the fixation method, the membrane structures appear obscure, rendering the nuclear membrane (magenta triangle) difficult to recognize. Numerous thick chromosomes are present in the nucleus, while the fragmented chromosomes observed in cells fixed by the conventional chemical fixation method are absent. *: Cytoplasmic gap; Black arrow: trichocyst; FL: flagellum; Cr: crystal structure; Mt: mitochondria; Od: oil drop; Av: alveolus; Sc: cell scale; Magenta arrow: nuclear membrane; Green triangle: chromosome; Green arrow: fragmented chromosome.
References
Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119. doi: 10.1111/jeu.12691
Adl S. M., Simpson A. G. B., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451. doi: 10.1111/j.1550-7408.2005.00053.x
Adl S. M., Simpson A. G. B., Lane C. E., Lukeš J., Bass D., Bowser S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493. doi: 10.1111/j.1550-7408.2012.00644.x
Allis C. D., Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Bachvaroff T. R., Gornik S. G., Concepcion G. T., Waller R. F., Mendez G. S., Lippmeier J. C., et al. (2014). Dinoflagellate phylogeny revisited: using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogenet. Evol. 70, 314–322. doi: 10.1016/j.ympev.2013.10.007
Bouligand Y., Soyer M. O., Puiseux-Dao S. (1968). La structure fibrillaire et l’orientation des chromosomes chez les Dinoflagellés. Chromosoma 24, 251–287. doi: 10.1007/bf00336195
Chaoying Z., Congmei Z., Jingyan L. (1996). The comparative study on the chromosomes of Oxyrrhis marina and typical dinoflagellate paying special attention to the systematic position of this organism. Zoological Res. 17, 331–336.
Chow M. H., Yan K. T. H., Bennett M. J., Wong J. T. Y. (2010). Birefringence and DNA condensation of liquid crystalline chromosomes. Eukaryot. Cell 9, 1577–1587. doi: 10.1128/ec.00026-10
de la Espina S. M. D., Alverca E., Cuadrado A., Franca S. (2005). Organization of the genome and gene expression in a nuclear environment lacking histones and nucleosomes: the amazing dinoflagellates. Eur. J. Cell Biol. 84, 137–149. doi: 10.1016/j.ejcb.2005.01.002
Dodge J. D. (1963). Chromosome numbers in some marine Dinoflagellates. Bot. Mar. 5, 121–128. doi: 10.1515/botm.1963.5.4.121
Dodge J. D., Crawford R. M. (1971a). Fine structure of the dinoflagellate Oxyrrhis marina I. general structure of the cell. Protistologica 7, 295–304.
Dodge J. D., Crawford R. M. (1971b). Fine structure of the dinoflagellate Oxyrrhis marina. II. The flagellar system. Protistologica 7, 399–409.
Dodge J. D., Crawford R. M. (1974). Fine structure of the dinoflagellate Oxyrrhis marina. III. Phagotrophy. Protistologica 10, 239–244.
Fukuda Y., Endoh H. (2006). New details from the complete life cycle of the red-tide dinoflagellate Noctiluca scintillans (Ehrenberg) McCartney. Europ. J. Protistol. 42, 209–219. doi: 10.1016/j.ejop.2006.05.003
Fukuda Y., Endoh H. (2008). Phylogenetic analyses of the dinoflagellate Noctiluca scintillans based on β-tubulin and Hsp90 genes. Europ. J. Protistol. 44, 27–33. doi: 10.1016/j.ejop.2007.07.001
Fukuda Y., Suzaki T. (2015). “Unusual features of dinokaryon, the enigmatic nucleus of dinoflagellates”. In Marine Protists: Diversity and Dynamics 23–45. doi: 10.1007/978-4-431-55130-0_2
Gao X., Li J. (1986). Nuclear division in the marine dinoflagellate Oxyrrhis marina. J. Cell Sci. 85, 161–175. doi: 10.1242/jcs.85.1.161
Géraud M. L., Herzog M., Soyer-Gobillard M. O. (1991). Nucleolar localization of rRNA coding sequences in Prorocentrum micans Ehr. (dinomastigote, kingdom Protoctist) by in situ hybridization. Biosystems 26, 61–74. doi: 10.1016/0303-2647(91)90038-m
Golyshev S., Berdieva M., Musinova Y., Sheval E., Skarlato S. (2018). Ultrastructural organization of the chromatin elements in chromosomes of the dinoflagellate Prorocentrum minimum. Protistology 12:163–172. doi: 10.21685/1680-0826-2018-12-4-1
Gornik S. G., Ford K. L., Mulhern T. D., Bacic A., McFadden G. I., Waller R. F. (2012). Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 22, 2303–2312. doi: 10.1016/j.cub.2012.10.036
Grell K., Schwalbach G. (1965). Elektronenmikroskopische Untersuchungen an den Chromosomen der Dinoflagellaten. Chromosoma 17, 230–245. doi: 10.1007/bf00283600
Hoshina R., Tsukii Y., Harumoto T., Suzaki T. (2021). Characterization of a green Stentor with symbiotic algae growing in an extremely oligotrophic environment and storing large amounts of starch granules in its cytoplasm. Sci. Rep. 11, 2865. doi: 10.1038/s41598-021-82416-9
Irwin N. A. T., Martin B. J. E., Young B. P., Browne M. J. G., Flaus A., Loewen C. J. R., et al. (2018). Viral proteins as a potential driver of histone depletion in dinoflagellates. Nat. Commun. 9, 1535. doi: 10.1038/s41467-018-03993-4
Janouškovec J., Gavelis G. S., Burki F., Dinh D., Bachvaroff T. R., Gornik S. G., et al. (2017). Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. U.S.A. 114, E171–E180. doi: 10.1073/pnas.1614842114
Jing-Yan L. (1984). Studies of dinoflagellate chromosomal basic protein. Biosystems 16, 217–225. doi: 10.1016/0303-2647(83)90006-0
Kalvelage J., Wöhlbrand L., Schoon R.-A., Zink F.-M., Correll C., Senkler J., et al. (2023). The enigmatic nucleus of the marine dinoflagellate Prorocentrum cordatum. mSphere 8, e00038–e00023. doi: 10.1128/msphere.00038-23
Kang B. S., Eom C., Kim W., Kim P., Ju S. Y., Ryu J., et al. (2015). Construction of target-specific virus-like particles for the delivery of algicidal compounds to harmful algae. Environ. Microbiol. 17, 1463–1474. doi: 10.1111/1462-2920.12650
Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. doi: 10.1006/jsbi.1996.0013
Maeshima K., Ide S., Babokhov M. (2019). Dynamic chromatin organization without the 30-nm fiber. Curr. Opin. Cell Biol. 58, 95–104. doi: 10.1016/j.ceb.2019.02.003
Rizzo P. J., Burghardt R. C. (1980). Chromatin structure in the unicellular algae Olisthodiscus luteus, Crypthecodinium cohnii and Peridinium balticum. Chromosoma 76, 91–99. doi: 10.1007/BF00292229
Rizzo P. J., Noodén L. D. (1974). Partial characterization of dinoflagellate chromosomal proteins. Biochim. Biophys. Acta 349, 415–427. doi: 10.1016/0005-2787(74)90127-0
Sala-Rovira M., Geraud M., Caput D., Jacques F., Soyer-Gobillard M., Vernet G., et al. (1991). Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCc) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta). Chromosoma 100, 510–518. doi: 10.1007/BF00352201
Saldarriaga J. F., McEwan M. L., Fast N. M., Taylor F. J. R., Keeling P. J. (2003). Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int. J. Syst. Evol. Microbiol. 53, 355–365. doi: 10.1099/ijs.0.02328-0
Song C., Murata K., Suzaki T. (2017). Intracellular symbiosis of algae with possible involvement of mitochondrial dynamics. Sci. Rep. 7, 1221. doi: 10.1038/s41598-017-01331-0
Soyer-Gobillard M.-O., Dolan M. F. (2015). Chromosomes of Protists: The crucible of evolution. Int. Microbiol.: Off. J. Span. Soc Microbiol. 18, 209–216. doi: 10.2436/20.1501.01.252
Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. doi: 10.1038/47412
Taylor F. J. R., Hoppenrath M., Saldarriaga J. F. (2008). Dinoflagellate diversity and distribution. Biodiversity Conserv. 17, 407–418. doi: 10.1007/s10531-007-9258-3
Triemer R. E. (1982). A unique mitotic variation in the marine dinoflagellate Oxyrrhis marina (Pyrrophyta). J. Phycol. 18, 399–411. doi: 10.1111/j.0022-3646.1982.00399.x
Keywords: Oxyrrhis marina, dinokaryon, chromosome, freeze-substitution, 3D reconstruction, SBF-SEM
Citation: Fukuda Y, Suzaki T, Murata K and Song C (2025) Novel ultrastructural features of the nucleus of the ancestral dinoflagellate Oxyrrhis marina as revealed by freeze substitution fixation and volume electron microscopy. Front. Protistol. 3:1512258. doi: 10.3389/frpro.2025.1512258
Received: 16 October 2024; Accepted: 23 January 2025;
Published: 20 February 2025.
Edited by:
Matthew W. Parrow, University of North Carolina at Charlotte, United StatesReviewed by:
Petra Bulankova, INSERM U1024 Institut de biologie de l’Ecole Normale Supérieure, FranceAntónio José Calado, University of Aveiro, Portugal
Copyright © 2025 Fukuda, Suzaki, Murata and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chihong Song, Y2hzb25nQHB1c2FuLmFjLmty
 Yasuhiro Fukuda
Yasuhiro Fukuda Toshinobu Suzaki
Toshinobu Suzaki Kazuyoshi Murata
Kazuyoshi Murata Chihong Song
Chihong Song