
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Protistol. , 26 June 2024
Sec. Marine and Freshwater Harmful Algae and Protists
Volume 2 - 2024 | https://doi.org/10.3389/frpro.2024.1422481
 Arjun Verma1†
Arjun Verma1† Greta Gaiani1†
Greta Gaiani1† Mona Hoppenrath2
Mona Hoppenrath2 Juan Jose Dorantes-Aranda3
Juan Jose Dorantes-Aranda3 Kirsty Smith4
Kirsty Smith4 Gurjeet Singh Kohli1
Gurjeet Singh Kohli1 Clinton Wilkinson5
Clinton Wilkinson5 Gustaaf Marinus Hallegraeff3
Gustaaf Marinus Hallegraeff3 Shauna Ann Murray1*
Shauna Ann Murray1*Introduction: Harmful algal bloom (HAB)-forming species and populations exhibit substantial intraspecific functional trait variation, which can confer eco-evolutionary advantages. Phenotypic variability among populations can buffer the immediate detrimental effects of environmental fluctuations, with more diverse populations expected to survive changing conditions more efficiently than their uniform counterparts.
Methods: In February 2014, a mixed fish-killing dinoflagellate bloom occurred in the temperate waters of Coffin Bay, South Australia, causing the death of oysters and fish in the area. The bloom was dominated by Karenia mikimotoi and a cryptic species of Heterocapsa. Twenty-one monoclonal Heterocapsa isolates were established from the site and identified as H. ovata using microscopy and universal ribosomal markers (ITS/5.8S and LSU D1/D3 rDNA regions; SSU and cob were used for amplicon sequencing). These isolates were tested for ichthyotoxicity using a bioassay based on cells from the gills of rainbow trout (Oncorhynchus mykiss). Culture fraction preparations (whole cells, supernatant, and lysed cells) were analyzed to determine ichthyotoxicity levels.
Results: The highest ichthyotoxicity was observed in lysed cells, with surprisingly high inter-strain variability. This suggests that different strains of H. ovata have varying levels of toxicity.
Discussion: Results from this study expand our understanding of the adaptive strategies of HAB species and enable predictions of future population dynamics under changing climatic conditions. The substantial phenotypic variability among H. ovatastrains highlights the potential for diverse responses to environmental stressors, underscoring the importance of considering intraspecific variation in ecological and evolutionary studies of HABs.
Harmful algal blooms (HABs) are the rapid proliferation of certain microalgal species that can be related to elevated nutrients, temperature spikes, and other environmental triggers (Bruslé, 1995; Harvell et al., 1999; Van Dolah, 2000). Toxins produced by certain microalgal species during these blooms can lead to ecological consequences, such as fish kills (Kim and Oda, 2010; Dorantes-Aranda et al., 2015; Murray et al., 2015; Hallegraeff et al., 2023), marine mammal mortality (Van Dolah et al., 2002; Broadwater et al., 2018; Elorriaga-Verplancken et al., 2022), and disruptions to the normal functioning of aquatic ecosystems (Glibert, 2017; Griffin et al., 2023; Oh et al., 2023).
HAB species are characterized by high variations in the expression of specialized functional traits (Tillmann et al., 2009; John et al., 2015) that contribute to their success as populations (Fogg, 1969; Hallegraeff, 1993; Litchman et al., 2010; Carey et al., 2012). These traits include, but are not limited to, growth rate, toxin production, photosynthetic efficiency, light adaptation, and the production of allelochemicals (Alpermann et al., 2009; John et al., 2015; von Dassow et al., 2015; Brandenburg et al., 2018; Verma et al., 2020; Ajani et al., 2021). Dinoflagellates are common HAB taxa and have short generation times, meaning genotypes with beneficial traits may rapidly increase in abundance via lineage sorting, leading to rapid adaptation. Therefore, strains within the same species can exhibit variations in toxin production, with some completely lacking this capability (Murray et al., 2015; Brandenburg et al., 2018).
In Australia, frequent small-scale events of fish deaths in coastal waters can occur naturally at a seasonal frequency (Sammut et al., 1995; Roach, 1997; Hallegraeff, 2002; DPI, N.D.o.P.I.N, 2010; Adolf et al., 2015; Murray et al., 2015). Investigations into fish kills usually examine the potential of aquatic disease outbreaks, risks to human health, and environmental damage (e.g., the fish kill happens as a consequence of chemical or oil spills) (Roberts et al., 2019). Early detection of these factors is crucial to facilitating a timely and efficient response, minimizing ecological damage, and mitigating impacts on fisheries and aquaculture sectors. This proactive approach is important in supporting seafood export markets, specifically in ruling out notifiable diseases that could adversely affect international trade (Roberts et al., 2019). Over the past two decades, large-scale fish kills have occurred along the coast of South Australia in the Great Australian Bight and were caused mostly by HABs (Sammut et al., 1995; Roach, 1997; Hallegraeff, 2002; DPI, N.D.o.P.I.N, 2010; Roberts et al., 2014, 2019). The areas around Spencer Gulf and Gulf Saint Vincent (Great Australian Bight) are considered seasonally subtropical systems and present the perfect environment for temperate, sub-tropical, and tropical marine species (Rogers et al., 2003; Roberts et al., 2019). There are many key fish-killing HAB organisms that reside in South Australian waters, such as species of Alexandrium Halim, 1960, Chattonella Biecheler, 1936, Chrysochromulina Lackey, 1939/Prymnesium N. Carter, 1937, Heterocapsa F. Stein, 1883, Heterosigma Hada, 1967, Karenia G. Hansen & Moestrup, 2000, Karlodinium J. Larsen, 2000, Margalefidinium A.R. Loeblich III, 1979, and Pseudochattonella I. Iwataki, J. Daigo & T. Fukuyo, 2007 (Adolf et al., 2015; GlobalHAB, 2023).
During HABs where more than one species can co-occur (Hallegraeff, 2002; Fire et al., 2011; Adolf et al., 2015; Murray et al., 2015; Roberts et al., 2019; Rolton et al., 2022; Hallegraeff et al., 2023), management becomes a challenging task. There is a general lack of knowledge on the co-occurrence of fish-killing microalgae. This is particularly true for species of the genus Heterocapsa, a cosmopolitan genus identified in blooms associated with fish kill events, although it may not always be the dominant organism (Wang et al., 2005; Roberts et al., 2019). Some Heterocapsa species, especially in the Western Pacific region, are known to cause HABs (Horiguchi, 1995; Matsuyama, 1999; Iwataki, 2008; Choi and Kim, 2021). Blooms of H. circularisquama in Japan led to substantial mortality among cultured and wild bivalve mollusks in the late 1980s, significantly impacting the local bivalve culture industry, particularly the pearl oyster sector (Matsuyama, 1999, 2012). Of the 26 currently described Heterocapsa species, only three have been confirmed to harm shellfish mollusks (Matsuyama, 1999, 2012; Yang et al., 2015; Xiao et al., 2018; Hanifah et al., 2022). The second toxigenic species, H. bohaiensis, affected the aquaculture industries of prawns (Panaeus japonicus) and Chinese mitten-handed crabs (Eriocheir sinensis) in China (Yang et al., 2015; Xiao et al., 2018). The third one, H. borneoensis, showed a toxic effect in a cytotoxicity assay on brine shrimps (Artemia nauplii) (Hanifah et al., 2022). Other Heterocapsa species, such as H. steinii, H. rotundata, H. minima, and H. pygmaea, are known to cause extensive blooms though they have not yet been linked to the mortality of marine organisms. Many laboratory studies have focused on the toxicology and shellfish-killing mechanisms (Hansen, 1989; Lindholm and Nummelin, 1999; Litaker et al., 2002; Millette et al., 2015; Benico et al., 2021; Razali et al., 2022) of Heterocapsa species (Matsuyama, 1999; Basti and Segawa, 2010, Matsuyama, 2012; Nagai et al., 1996). Additionally, despite both H. circularisquama and H. bohaiensis showing hemolytic activity (Nishiguchi et al., 2016; Zhang et al., 2019), as yet, no tests of fish-killing activity have been performed with Heterocapsa extracts.
In February 2014, a localized high mortality event (within a 3–5-km area) of benthic marine organisms occurred in Coffin Bay (South Australia), specifically around Frenchman’s Bluff. This event was associated with a dense bloom of Karenia mikimotoi. The HAB was identified early, and active monitoring took place over the following days to assess any imminent threats to fisheries and aquaculture stakeholders. During this period, a site survey was conducted and reported high mortality among abalone species (Haliotis rubra and H. laevigata), rock lobster (Jasus edwardsii), turbo shells, various fish, and echinoderms on the seafloor. They also noted poor water visibility. Samples were collected from dying fish and water. Fish samples tested negative for abalone herpesvirus (AbHV) (Roberts et al., 2014). Two unpreserved, nonconcentrated water samples were sent by overnight courier to the University of Tasmania and tested the next morning using the rainbow trout gill cell line (RTgill W-1) bioassay (Dorantes-Aranda et al., 2011). This confirmed the high ichthyotoxic potential of natural bloom water, killing 80% of fish gill cells in two hours, with the bottom water sample (16 million Karenia mikimotoi cells L−1) being more potent than the surface water sample (5 million cells L−1).
In March 2014, a second survey was conducted in the waters between Coffin Bay and Pt. Drummond. One dead abalone was observed 1 km south of the initial mortality site. No further mortalities were observed at any sites, even with the high densities of K. mikimotoi recorded at the end of February 2014. Although water samples from the K. mikimotoi bloom tested negative for biotoxins (i.e., Brevetoxins, Gymnodimines, and Gymnocins) despite causing an ichthyotoxic reaction in fish gill cells, the high cell density of the bloom may still be implied in fish deaths. Given the co-occurrence of different species during dense blooms, these findings raised questions about whether organisms other than K. mikimotoi could be present in the same bloom, potentially sharing responsibility for fish mortality.
In this study, 21 isolates of a Heterocapsa species from Coffin Bay, South Australia, were established in March 2014 from the K. mikimotoi-dominated bloom samples and identified with light and electron microscopy and molecular analysis (ITS/5.8S and LSU D1–D3 rDNA regions). The variability of potential fish-killing activity was tested using a fish gill cell assay. Additionally, an overview of the microalgal population composition of the bloom was conducted using a metabarcoding approach.
Samples were collected from Coffin Bay (Figure 1) in February 2014, during which a high mortality of marine organisms was reported. Karenia mikimotoi was a common and abundant species present and was hypothesized to be the cause of the mortalities observed. Triplicate integrated water samples were collected from 1 m to 2 m depth. Samples were kept cool, in the dark, and returned to the laboratory (PIRSA, Port Lincoln) within 3 h. From each replicate, 1 L of sample was filtered through the stacked mesh (5 mm) and Whatman GF/F filters (pore size 0.7 mm), allowing the examination of size-fractionated phytoplankton biomass. A sample from the bloom was shipped to UTS, where single-cell isolation was performed.
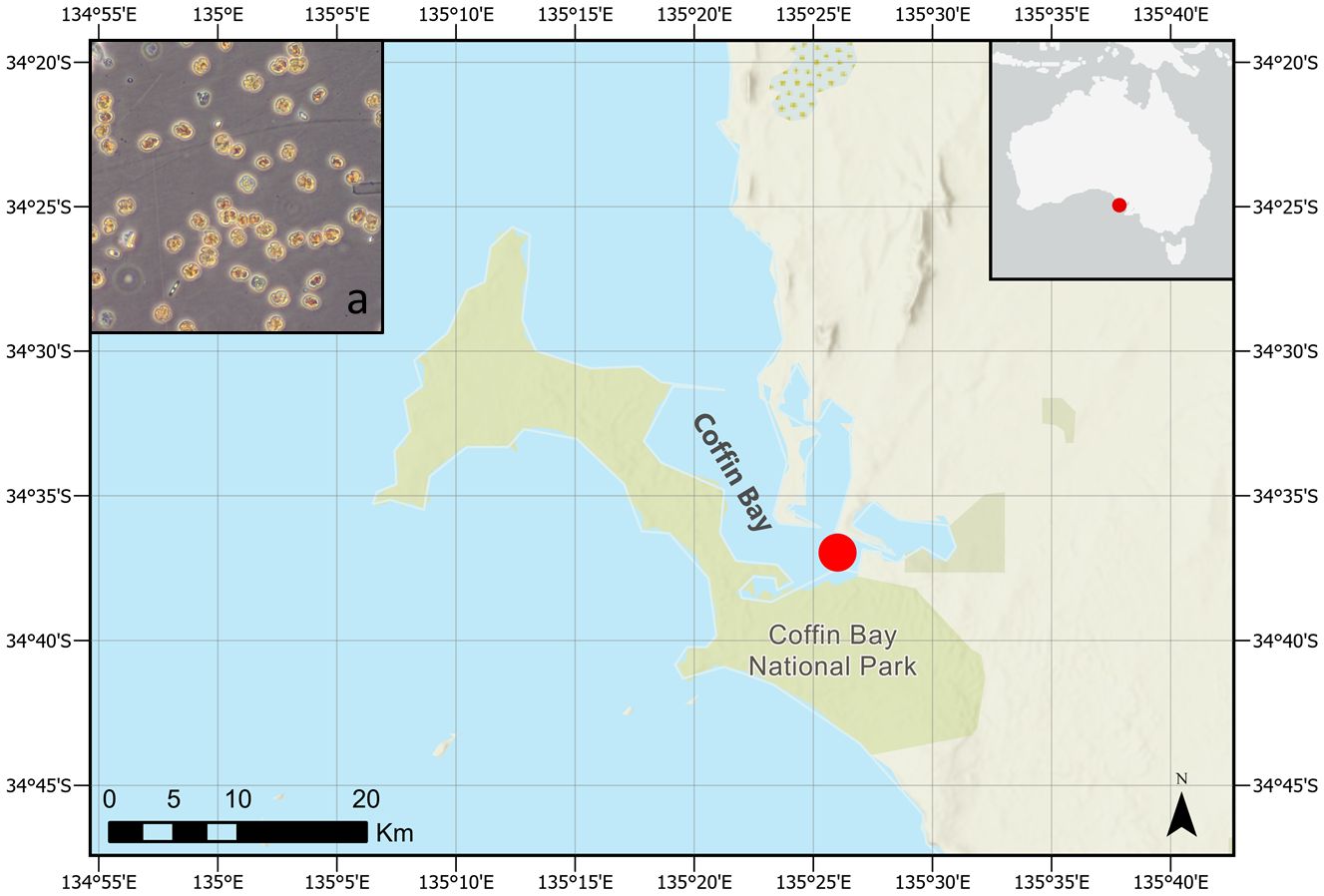
Figure 1 Coffin Bay. (a) Microscopy picture of a sample taken during the K. mikimotoi bloom (courtesy of C. Wilkinson). The map was realized with ArcGIS (2024 version).
Using a glass micropipette, Heterocapsa cells were isolated using an inverted light microscope (Nikon Eclipse TS100) and transferred to a drop of clean, filtered seawater. The transfer was repeated up to five times, and 20 non-axenic monoclonal cultures were established. The isolates were maintained in five times diluted f/2 medium (Guilard and Ryther, 1962) at 18°C and salinity of 35 under the photoillumination of 60–80 μmol m−2 s−1 and a 12:12-h light–dark cycle in 24 multi-well culture plate (Corning Inc., Durham, NC, USA) with 1 mL of medium. After 4–6 weeks, they were transferred to 50 mL of sterile tissue culture flasks (Becton Dickinson, Macquarie Park, Sydney, Australia) and maintained in the same conditions. Cultures were subcultured in five times diluted f/2 medium every 4–5 weeks thereafter.
Living cells of strain SA2 were picked using a Leica DMIL inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany), placed on an object slide, and observed with a Leica DMRB (Leica Microsystems GmbH, Wetzlar, Germany) equipped with differential interference contrast and epifluorescence optics at ×400 and ×630 magnification with oil immersion objectives. Digital photos were taken using a Leica DFC420C camera (Leica Microsystems GmbH, Wetzlar, Germany). Fixed cells were stained with Solophenyl Flavine 7GFE 500 (Ciba Specialty Chemicals, High Point, NC, USA) as described by Chomérat et al. (2017), and thecal plates visualized by epifluorescence microscopy.
Cell dimensions of mid-log phase cultures were measured under ×400 using a calibrated eyepiece of an Eclipse TS100 inverted microscope with bright field optics (Nikon, Hilton, Australia). Cells were harvested from the culture medium phase 8–10 days after subculturing and fixed in 4% acidic Lugol’s solution to measure the length and width using dorsoventral diameter (DV) and transdiameter width (W) using ImageJ v1.48 (Rasband 1997–2013).
For SEM, the culture SA2 was fixed with Lugol’s solution and stored in the dark. Cells were placed on a 5-µm Millipore filter of mixed cellulose esters (MCE), rinsed in distilled water, and dehydrated in a series of increasing ethanol concentrations (30%, 50%, 70%, 85%, 90%, and 100%), followed by chemical drying with hexamethyldisilazane at room temperature for 20 min, and finally at 50°C in a drying oven for 5 min. The sample/filter was mounted on a stub and sputter coated with gold–palladium (Bal-Tec SCD 050; BAL-TEC Präparations-gerätevertrieb, Wallof, Germany). Cells were observed using a Tescan VEGA3 microscope (Elekronen-Optik-Service GmbH, Dortmund, Germany) at 10 kV using the SE detector.
For transmission electron microscopy (TEM), cells were concentrated in a microfuge tube by slow centrifugation (8×g for 1.5 min). The pellet was prefixed with 2.5% glutaraldehyde in filtered seawater at 4°C for 30 min. Cells were washed twice in filtered seawater before postfixation with 1% OsO4 in filtered seawater at room temperature for 40 min. Fixed cells were dehydrated through a graded series of ethanol (30%, 50%, 70%, 85%, 90%, 95%, 2× 100%; 10 min each), followed by 2× 100% propylene oxide, infiltrated with propylene oxide-resin mixtures (2:1, 1:1, 1:2), and embedded in EMBed-812 resin (Science Services, Munich, Germany). The block was polymerized at 60°C for 22 h and sectioned with a diamond knife on a Reichert Ultracut microtome (Reichert-Jung, Vienna, Austria). Thin sections were directly viewed under an EM 902A TEM (Zeiss, Oberkochen, Germany) operated at 80 kV. Digitized images were taken with a 1-k Proscan High-Speed SSCCD camera (Proscan, Lagerlechfeld, Germany) operated by the iTEM Five software (Olympus, Munster, Germany).
For negative staining TEM, 50 µL of an old culture was used. The detached body scales were allowed to adsorb onto Formvar-coated grids for 5 min. The grids were stained with 1% (w/v) uranyl acetate for 1 min, washed in two drops of distilled water, and air-dried. The sample was also investigated in the EM 902A TEM (Zeiss) operated at 80 kV.
DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, USA), according to the manufacturer’s protocol. The extracted DNA was visualized on a 1% agarose gel and quantified using a Nanodrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA). The partial nuclear D1/D3 region of the large subunit ribosomal RNA (LSU rRNA) gene and the internal transcribed spacer regions and 5.8S rRNA gene (ITS1/5.8S) were amplified and sequenced as described in Verma et al. (2016). All PCR reactions were performed in 25 μL reaction volumes containing 12.5 μL 2× Immomix (Bioline, Sydney, Australia), 7.5 pmol of each primer, 1 μg μL−1 of BSA (Biolabs, Arundel, Australia), 1 μL of template DNA, and PCR-grade water to give the final volume. Thermocycling conditions consisted of an initial denaturing step of 95°C for 10 min, followed by 30 cycles of 95°C for 20 s, 30 s of annealing, 72°C for 1 min, and a final extension of 72°C for 7 min. PCR products were purified with DNA Clean and Concentrator (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. The PCR products were sequenced using a commercial service (Macrogen Inc., Seoul, Korea).
Analyses on the ITS1/5.8S and D1/D3 regions of LSU rDNA were conducted as described in Verma et al. (2016). In summary, multiple sequence alignments, postmanual inspection, were performed using ClustalW v1.6 as implemented in MEGA v6 (Tamura et al., 2013). Substitution models were selected for each dataset based on the lowest Bayesian information criterion (BiC). Phylogenetic analyses were performed using both maximum likelihood (ML) and Bayesian inference (BI) approaches. ML trees were produced in MEGA v6 using Tamura 3 (GRT) +G +I with five gamma categories substitution model for all analyses with a bootstrap of 1,000 replications. Bayesian analyses were performed using MrBayes v3.2.2 (Ronquist and Huelsenbeck, 2003) as implemented in Geneious v6 (Kearse et al., 2012), using general time reversible model (GTR) + the gamma model for all analyses.
Genetic distance (pairwise uncorrected p-distance) was estimated from the ITS/5.8S and D1/D3 LSU rDNA sequences using the p-distance model and bootstrap procedure (1,000 replicates) in MEGA v6 (Tamura et al., 2013). All positions containing gaps and missing data were eliminated for the analyses.
Approximately 1 L of water collected from the bloom was filtered, and DNA was extracted as described above and used for metabarcoding analyses. Three primer pairs, targeting three different gene regions (18S SSU rDNA, SSU556F 3′-CGCGGTAATTCCAGCTYC-5′ and SSU911R 3′-ATYCAAGAATTTCACCTCTGAC-5 primers (Smith et al., 2017); 28S LSU rDNA, D1R 3′-ACCCGCTGAATTTAAGCATA-5′ (Scholin et al., 1994) and 305R 3′-TTTAAYTCTCTTTYCAAAGTCC-5′ (Smith et al., 2017); and the mitochondrial COB gene dinocob4f 3′-AGCATTTATGGGTTATGTNTTACCTTT-5′ COB 440 (Lin et al., 2009) and dinocob6r 3′-ATTGGCATAGGAAATACCATTCAGG-5′ (Lin et al., 2009), were used. DNA extractions (> 20 ng total) were sent to New Zealand Genomics Limited (NZGL, Massey Genome Service, Massey University, Palmerston North, New Zealand) for further processing. Subsequent processing involved normalizing the samples to a concentration of 5 ng μL−1. Library preparation was carried out using the Illumina™ two-step PCR amplicon library preparation method (using the primers above), and the sequencing was performed utilizing an Illumina™ MiSeq sequencer, generating 2× 250 base paired-end reads.
All data generated were quality-checked with the following processes: FastQC, FastQ screen, and SolexaQA (Cox et al., 2010). Further analyses were carried out in Mothur v1.34.0 (Schloss et al., 2009). Paired-end reads were assembled and the resulting contigs screened; reads shorter than 200 bases, homopolymers longer than six bases, and those with more than eight ambiguous bases were removed. Unique reads were assessed for chimeras using a de novo approach in UCHIME (Edgar et al., 2011) implemented in Mothur. To facilitate alignment and classification, a custom dinoflagellate reference sequence database was established for each gene region. Dinoflagellate sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) into Geneious® v6.0.4 (Drummond, 2010). Alignments were created for each gene region using ClustalW (Thompson et al., 1994) implemented in Geneious. Next, the reads were clustered using the average neighbor method. Afterward, reads were classified using the dinoflagellate-specific gene alignments as reference files and the classify.seqs command in Mothur. Taxa with fewer than 10 reads across all samples were subsequently excluded from the analysis. The LSU rDNA and COB reads were aligned and classified with the respective custom sequence databases. SILVA database was used to furtherly align the SSU reads (release 119: SSU Ref NR 99) (Quast et al., 2012), which were then classified using the Protist Ribosomal Reference database (PR2; GenBank release 203) (Guillou et al., 2012).
The fish gill cell line RTgill-W1 was obtained from the American Type Culture Collection (CRL-2523). Primary cells were originally initiated from the gill filaments of rainbow trout, Oncorhynchus mykiss (Bols et al., 1994). Fish gill cells were cultured in 25-cm2 culture-treated flasks with Leibovitz medium (L1518, Sigma, Sydney, Australia) supplemented with 10% fetal bovine serum (v/v) (12003C, Sigma, Sydney, Australia) and an antibiotic–antimycotic solution (A5955, Sigma, Sydney, Australia) at 20°C in the dark. Confluent gill cells were detached with a 0.25% trypsin-0.02% EDTA solution (59428C, Sigma, Sydney, Australia) for subculturing and seeding purposes (Dorantes-Aranda et al., 2011). After 18 days (stationary phase), pellets of microalgal cultures were obtained from all 21 strains included in this study and were subsequently used to create three culture preparations for each strain.: whole cells (untreated), supernatant (centrifuged and cell pellet discarded), and lysed cells (prepared by sonication). A culture medium obtained by filtration of each algal culture was used in order to adjust the same cell concentrations for all algal experimental treatments (whole and lysed cell preparations to 40 × 104 cells mL−1). Algal culture aliquots were used without further preparation after adjusting the cell concentration and keeping the algae alive (whole cell preparation); other aliquots were sonicated for 5 min to rupture the cells (lysed cell preparation); finally, the third group of aliquots was centrifuged (3,000 rpm; 10 min), and the supernatant was transferred into new tubes (supernatant preparation). Gill cells were detached, counted, and seeded at a concentration of 1.5 × 105 cells mL−1 in quadruplicate in Greiner Bio-One 96-well microplates (655180, Interpath Services Pty Ltd., Heidelberg West, Australia) 48 h prior to the experiments (Dorantes-Aranda et al., 2011). Gill cells were exposed to the algal culture preparations extracts and control treatments (GSe culture medium) for 2 h. After completion of the 2-h exposure period, experimental solutions were discarded, and gill cells were rinsed with phosphate buffer saline (PBS). Next, alamarBlue (Sigma, Sydney, Australia) was added to all wells and incubated for 2 h in the dark to measure the viability of gill cells (Dorantes-Aranda et al., 2015). Experiments were performed in triplicate. Fluorescence of the oxidated alamarBlue was measured with a microplate reader (FLUOstar OPTIMA, BMG Labtech, Melbourne VIC 413–3350), using excitation and emission filters of 540 nm and 590 nm, respectively. The methodology involved comparing cell counts between treatment and control groups within each biological replicate to obtain percentages of cell viability. Subsequently, we conducted a one-way analysis of variance (ANOVA) on these percentages using Statistica v13 software (StatSoft, Dell, Australia). Additionally, a post-hoc Tukey’s test for multiple comparisons was employed. We maintained a significance level of 95% (α = 0.05) for all analyses to determine statistical significance among the experimental groups.
Cells of strain SA2 were oval in shape with epi- and hyposome of about equal size and a median, slightly descending cingulum (about half a cingulum width) (Figures 2, 3A, E). The large nucleus was located centrally in the episome and one pyrenoid with starch sheath was in the upper part of the hyposome (Figures 2A, 4A, B). A reticulated chloroplast was visible in the cell periphery (Figures 2C, D, 4A, B). The thecal plate pattern was APC (Po cp X h) 5′ 3a 7″ 6c 5s 5′″ 0p 2″″ (Figures 2E, F, 3) with the apical pore complex (APC) composed of the outer/apical pore plate (Po), a cover plate (cp), a canal plate (X), and a hinge structure (h) between X and cp (Figures 3C, D). The apical pore looked horseshoe-shaped from the outside and was centrally bordered by a strong rim of the cp plate (Figures 3C, D). The second anterior intercalary plate (2a) was seven-sided (Figures 3E, F). Thecal plates were smooth, with pores surrounded by a rim (Figures 3C, D). Body scales were visible on the cell surface (Figures 5A–C). The rounded triangular scales had a reticulated basal plate with six ridges, a central spine, six peripheral uprights, and nine peripheral bars (Figures 5C–I). Typical dinoflagellate organelles were visible in TEM sections (Figure 4), like dinokaryon (Figures 4A, B), trichocyst (Figure 4G), mitochondrion (Figure 4H), and peripheral chloroplast parts (Figures 4A, B, F). The pyrenoid contained some tubular cytoplasmic invaginations and was surrounded by starch (Figures 4C–E). Cells were 22–39 µm (28 µm ± 6.87 µm) long and 11–24 µm (15 µm ± 4.31 µm) wide (see Supplementary Table S1).
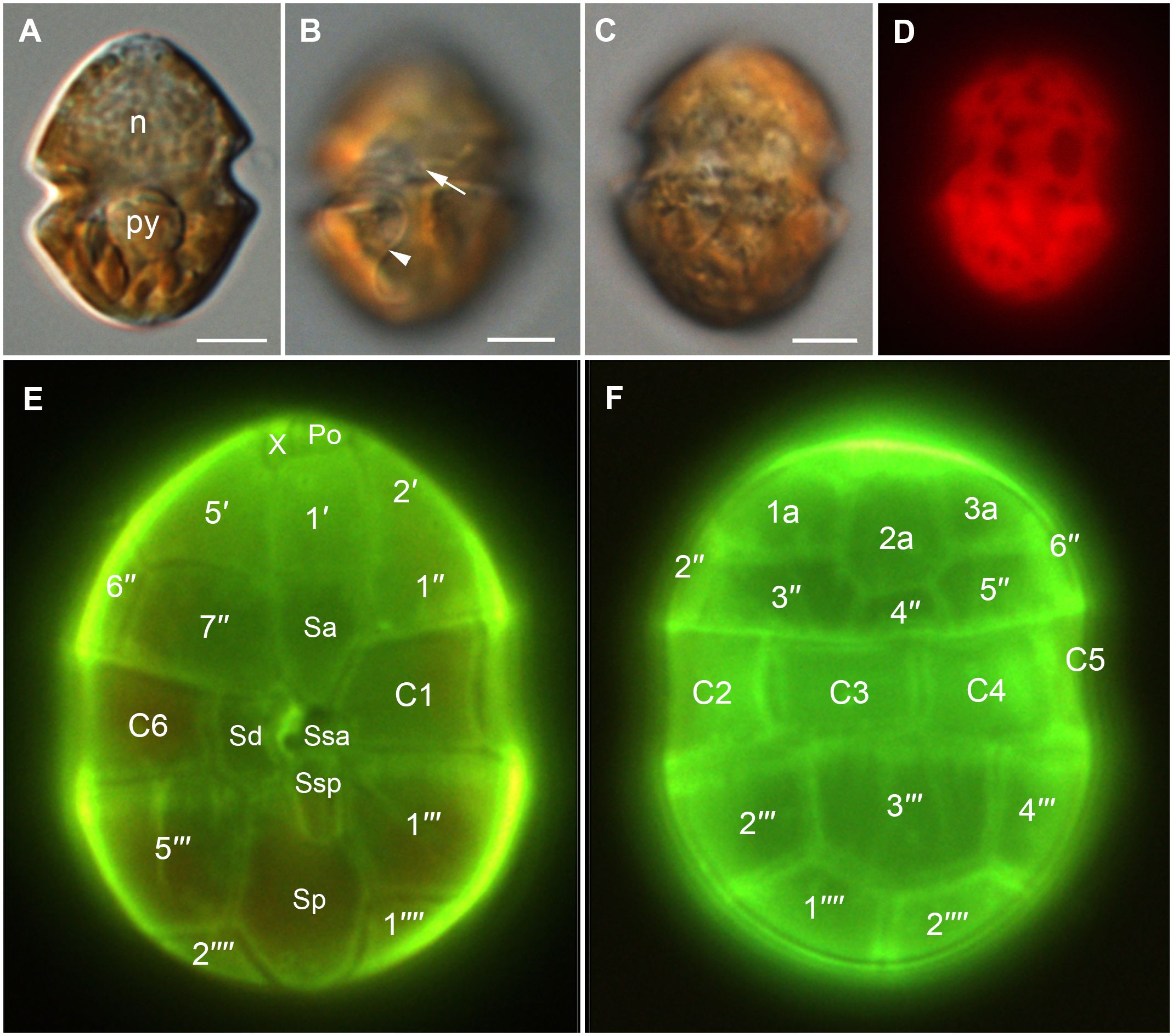
Figure 2 Light micrographs of Heterocapsa ovata (strain SA2). (A–C) DIC LM of living cells. (A) General cell shape. Cell in about mid-cell focus showing the nucleus (n) in the episome and the pyrenoid (py) surrounded by starch in the hyposome. (B) Ventral view with a focus on the central sulcal area showing the insertion of the transverse flagellum (arrow) and part of the longitudinal flagellum (arrowhead). (C) Dorsal view with a focus on the peripheral chloroplast. (D) The same cell as in (C) observed with epifluorescence showing the chloroplast autofluorescence. (E, F) Cells stained with solophenyl flavine showing thecal plates. (E) Ventral view. (F) Dorsal view. Scale bars = 5 µm.
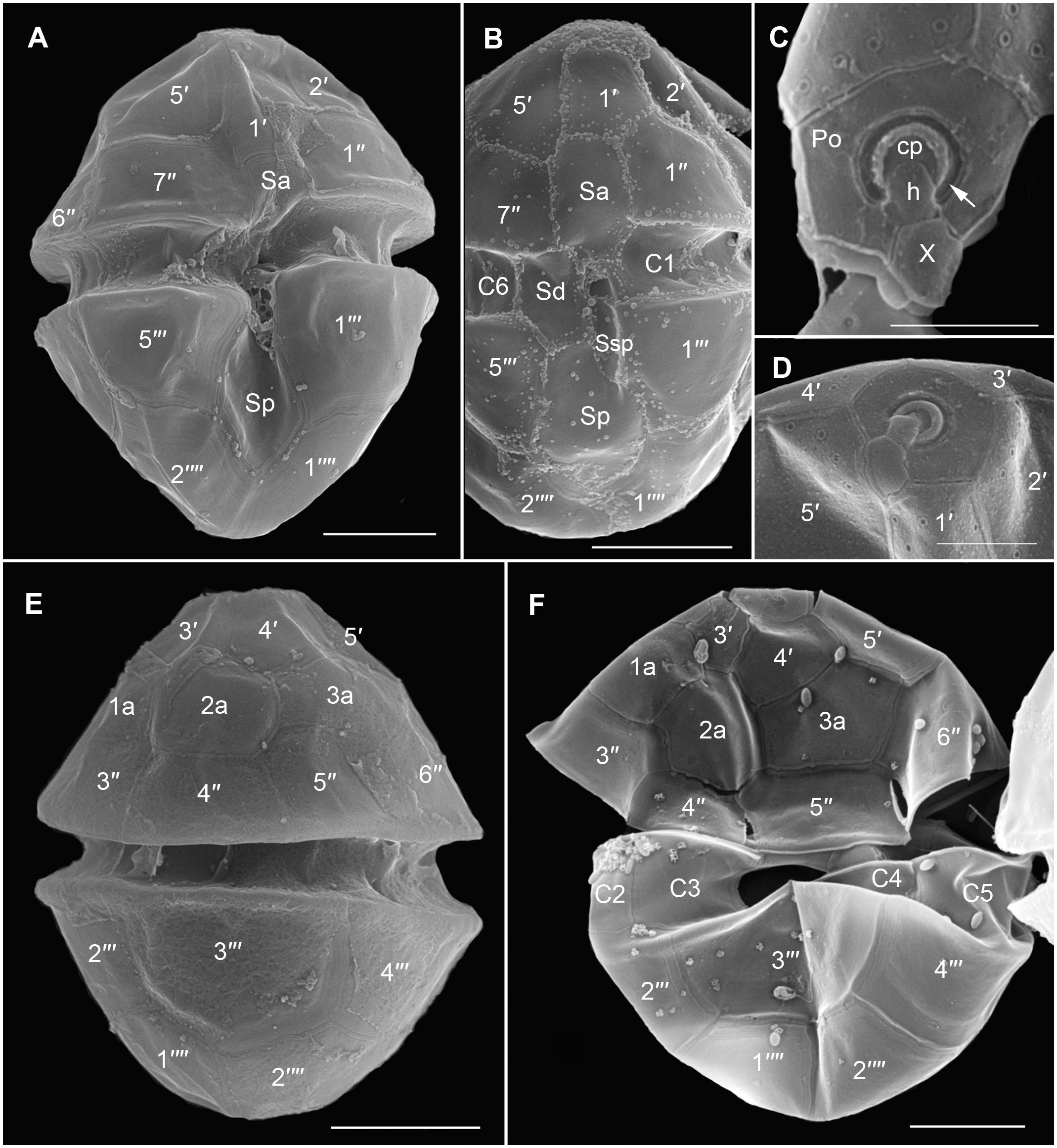
Figure 3 Scanning electron micrographs of Heterocapsa ovata (strain SA2) showing the thecal tabulation. (A) Ventral view. (B) Ventral view showing sulcal plates. (C, D) Apical pore complex (APC). (C) The APC consists of the outer/apical pore plate (Po), a cover plate (cp), a canal plate (X), and a hinge structure (h). Recognize the horseshoe-shaped apical pore area (arrow). (E, F) Dorsal view. Scale bars = 5 µm, except (C, D) 2 µm.
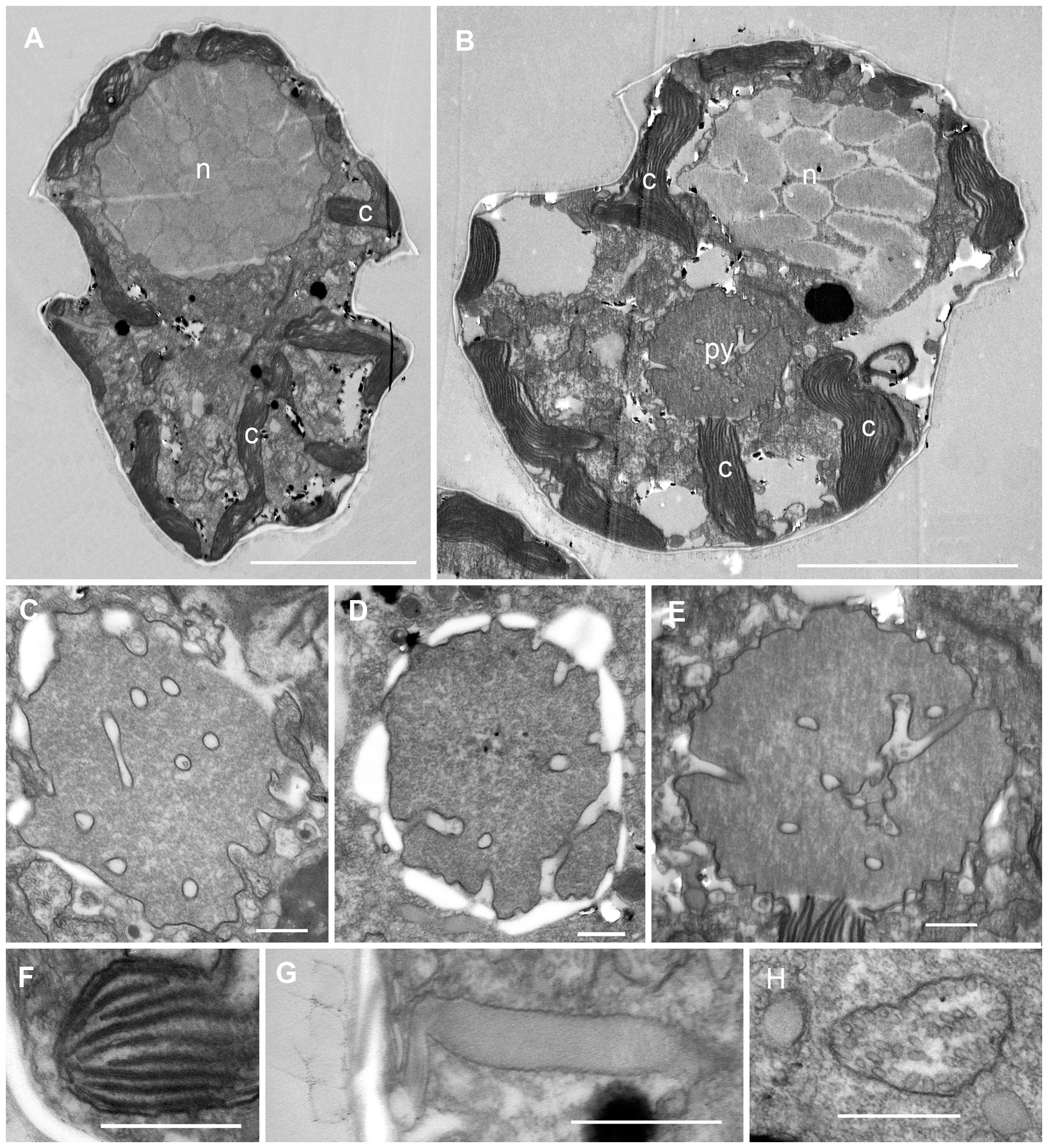
Figure 4 Transmission electron micrographs of Heterocapsa ovata (strain SA2). (A, B) Longitudinal section through a whole cell. Note the nucleus (n) in the episome, the peripheral parts of the chloroplast (c), and the central pyrenoid (py) in the hyposome. (C–E) Pyrenoids with tubular cytoplasmic invaginations and surrounding starch. (F) Chloroplast detail showing stacks of thylakoids. (G) Trichocyst in longitudinal section. (H) Mitochondrion with tubular christae. Scale bars = 5 µm (A, B) and 500 nm (C–H).
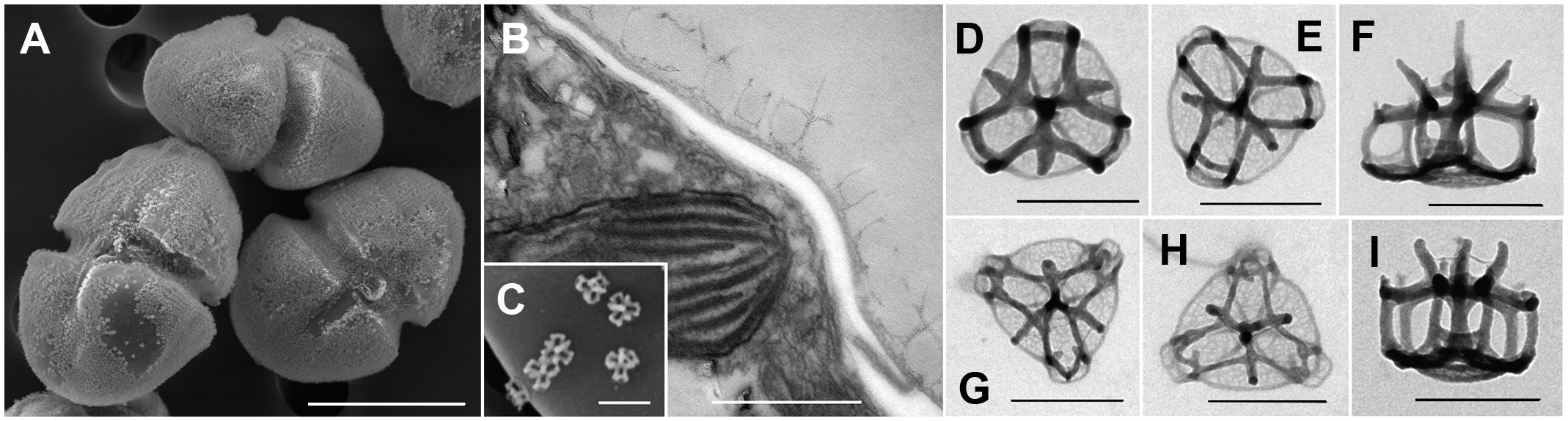
Figure 5 Scanning and transmission electron micrographs of Heterocapsa ovata (strain SA2) showing the body scales. (A) Cells with a cover of scales on the cell surface. (B) Section of a part of the cell surface showing the scales outside of the cell on top of the thecal plates. (C) Single scales on a thecal plate. (D–I) Ultrastructure of the scales. Scale bars = 10 µm (A), 500 nm (B, C), and 200 nm (D–I).
The ITS/5.8S rDNA analysis performed on the 21 Heterocapsa strains indicated that they belong to H. ovata species. The ML tree inferred from 83 sequences and employing the GTR+ G + I model is shown in Figure 6. The tree obtained from the analysis of the ITS region showed about 26 clades (Figure 6), and the 21 strains from this study clustered together with the unique GenBank entry available for H. ovata (AB084098). The monophyletic clade of H. ovata taxa shows high bootstrapping support and diverges from the H. bohaiensis and H. cf. pygmaea lineages. Sequences obtained from the H. ovata strains in this study showed little to no variability in the primary sequence (Supplementary Table S2).
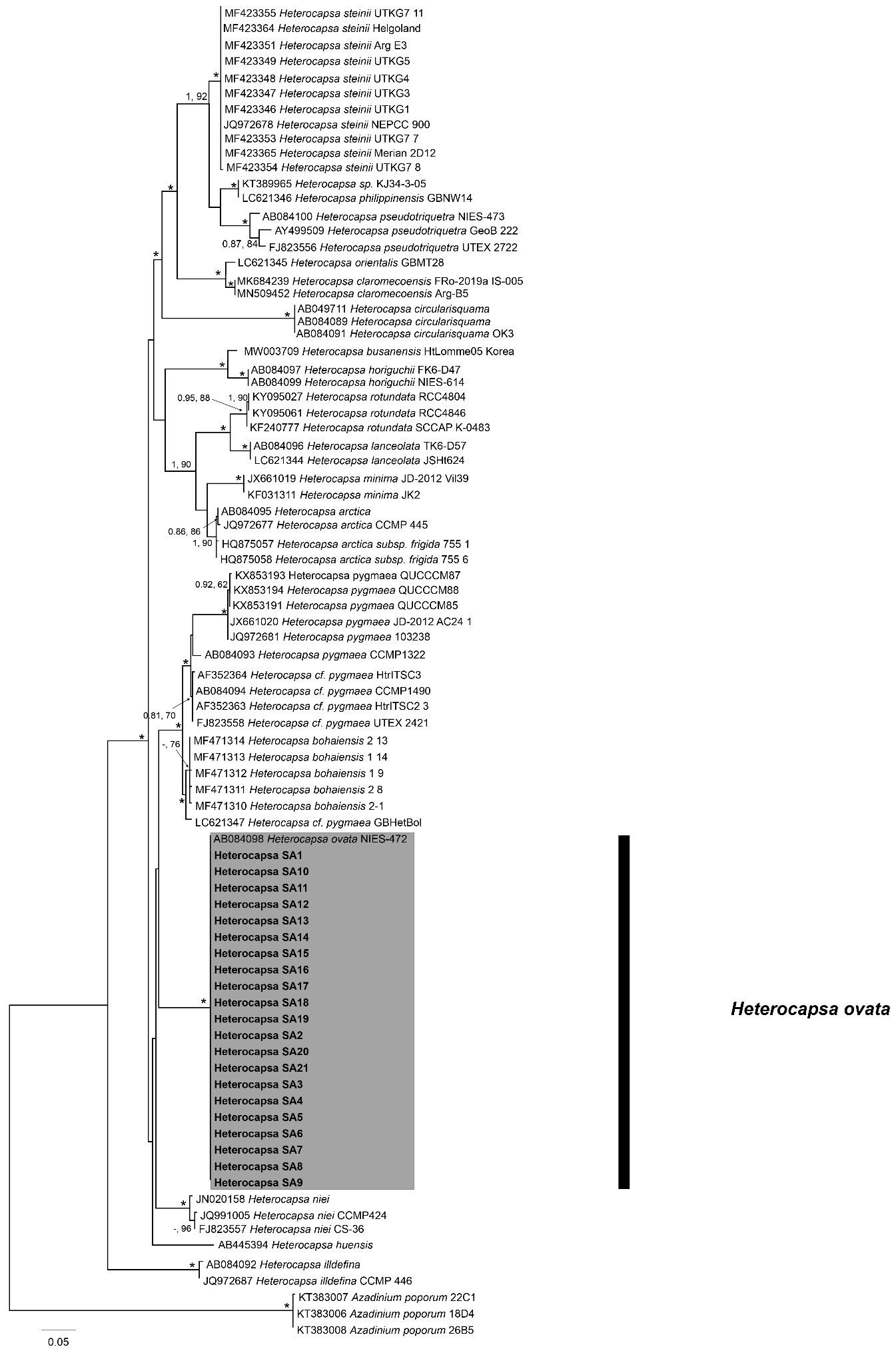
Figure 6 Maximum Likelihood (ML) phylogenetic trees of various Heterocapsa ovata strains using primer sets for ITS/5.8S. External black vertical bars show each distinct Heterocapsa clade. Numbers at nodes represent posterior probabilities from Bayesian Inferences (BI) and bootstrap support values from ML based on 1,000 pseudo-replicates. Asterisk represents 1,100 support values for BI and ML, respectively.
The analysis performed with the D1–D3 marker confirmed the species assignation to H. ovata of the 21 strains. The morphology of the tree shows about 18 clades (Figure 7). Again, the obtained tree (Figure 7) shows the 21 strains clustered together, though the unavailability of H. ovata LSU sequences in GenBank prevented the comparison with other existing strains. However, the monophyletic nature of this cluster is supported by a high bootstrap value (0.88). This is the first report of H. ovata in Australian waters.
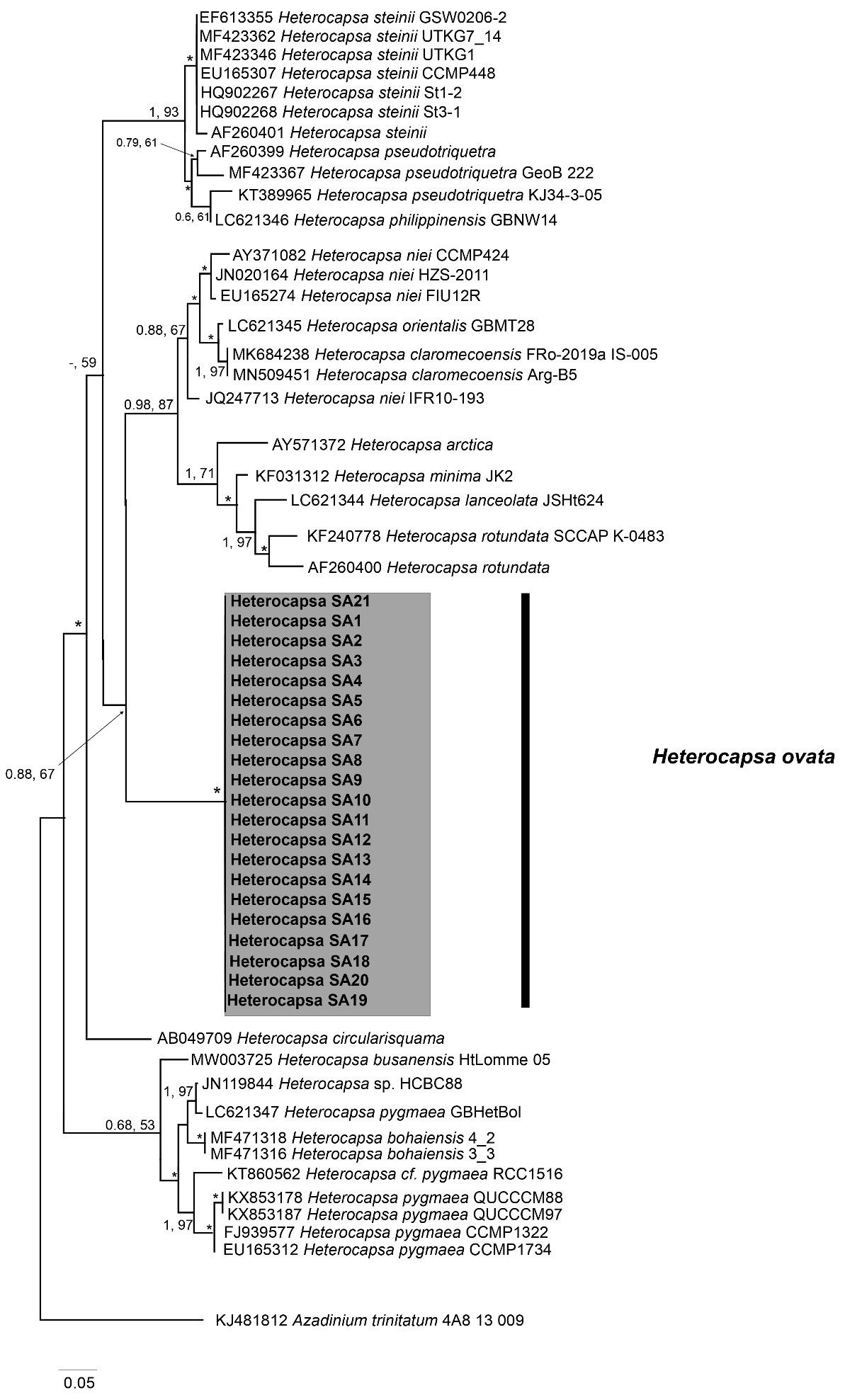
Figure 7 Maximum Likelihood (ML) phylogenetic trees of various Heterocapsa ovata strains using primer sets for D1/D3 LSU rDNA. External black vertical bars show each distinct Heterocapsa clade. Numbers at nodes represent posterior probabilities from Bayesian Inferences (BI) and bootstrap support values from ML based on 1,000 pseudo-replicates. Asterisk represents 1,100 support values for BI and ML, respectively.
The metabarcoding analysis of COB, 18S, and 28S markers revealed that the majority of the operational taxonomic units (OTUs) belonged to Karenia spp., as expected considering that K. mikimotoi had been identified as the most abundant taxon. However, other HAB-forming genera appear alongside Karenia and Heterocapsa, such as Azadinium and Karlodinium. Table 1 reports the most prevalent genera identified. Interestingly, many species of the reported genera can produce toxins that have ichthyotoxic activity (i.e., Gonyaulax, Karenia, Karlodinium, Margalefidinium, and Pfiesteria) (Hallegraeff et al., 2023; Oh et al., 2023). In particular, only analyses using the COB marker revealed the presence of Azadinium and Margalefidinium, while only the 18S marker identified five genera, including Coolia, Noctiluca, and Protoperidinium. The 28S marker identified the most genera, including Heterocapsa (Table 1).
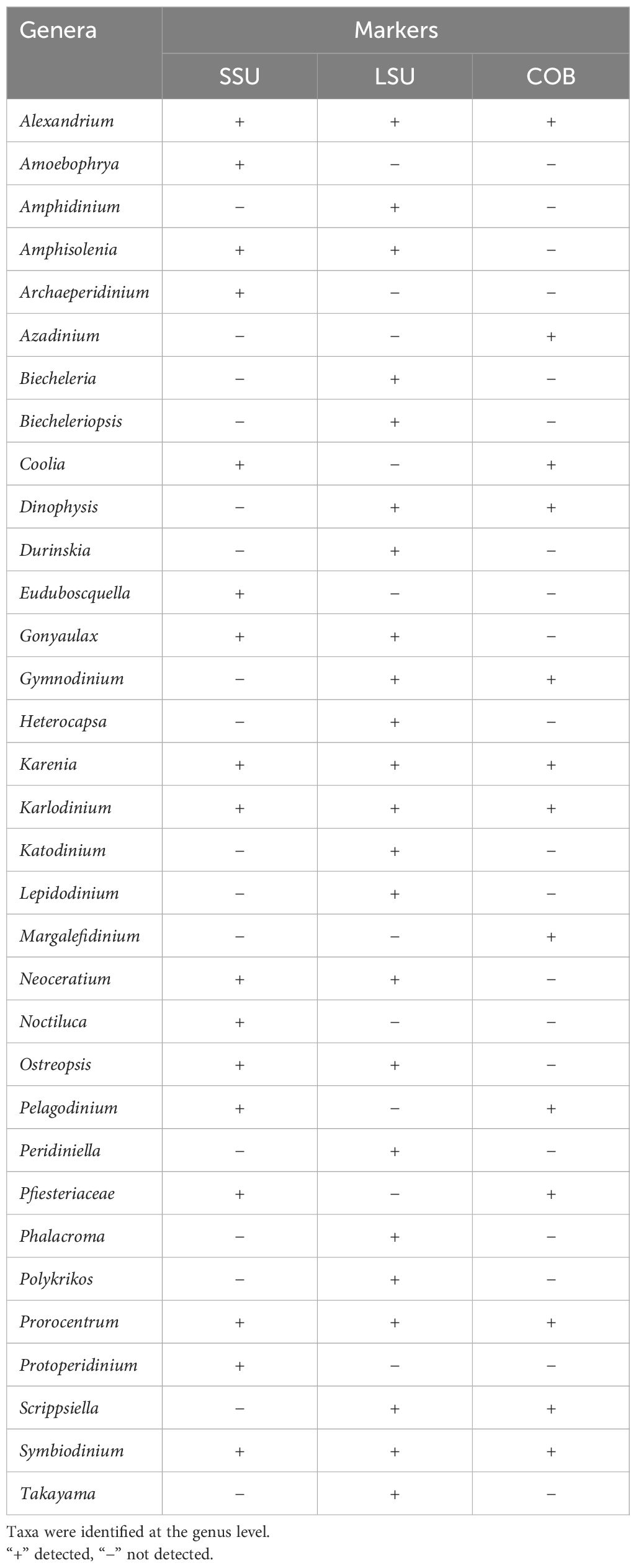
Table 1 Dinoflagellate taxon diversity from water samples, Coffin Bay, South Australia, using three molecular genetic markers (SSU, LSU, and COB).
The toxicity analysis showed that different strains have different effects on fish gill cell viability. In general terms, among the treatments performed, the one with lysed cells affected cell viability the most, with a recorded viability of ≥ 80% only in four out of 21 samples (strains 2, 5, 7, and 16, Figure 8). The other treatments did not result in a decrease in the viability of > 50%, apart from strain 14, in which all the treatments resulted in a viability of about 20%, indicating the highly toxic nature of this strain. Moreover, even if both the treatments with supernatant and whole cells had similar effects on cell viability, the latter seemed to be more toxic for strains 1, 6, 11, 12, and 20 (Figure 8). The supernatant treatment showed higher toxicity than the other two only in strain 17, indicating that the toxins are mostly preserved in the cells or on their external walls rather than released in the medium.
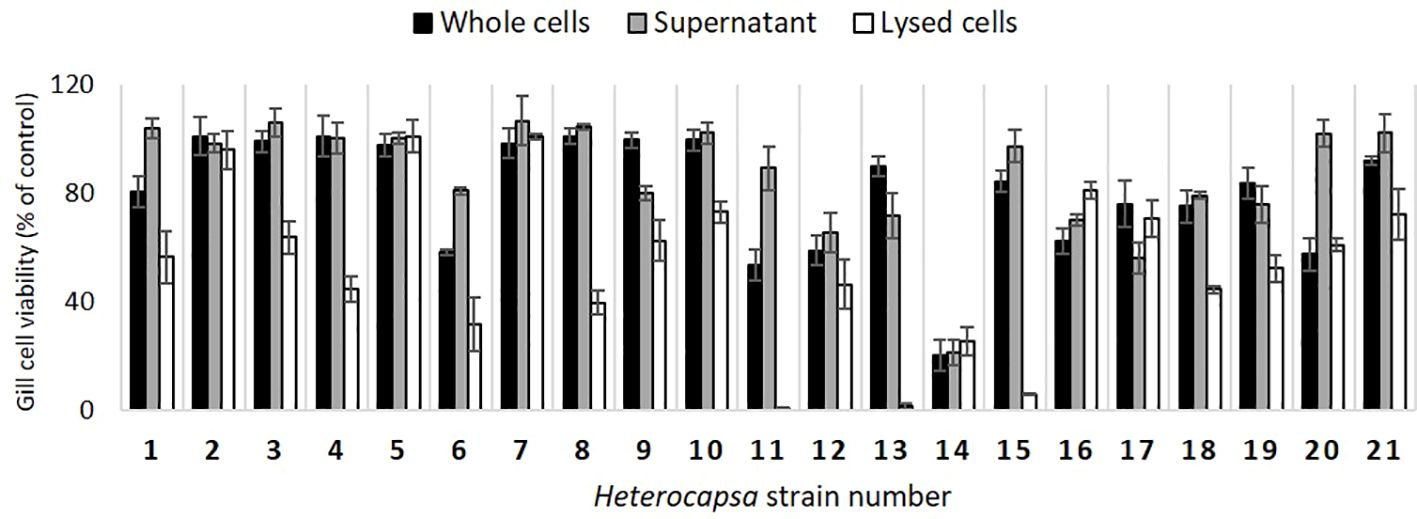
Figure 8 Viability (% of control) of the gill cells exposed to the H. ovata strains of this study. Whole H. ovata cells (black), supernatant (grey), and lysed H. ovata cells (white) were tested for each strain (the culture collection codes correspond to “SA” followed by the respective strain numbers indicated in the graph).
Molecular genetic techniques can allow for rapid characterization of the diversity of HABs, identifying both the most abundant species and less abundant taxa that may nonetheless contribute to harmful impacts. However, as observed in this study, results can vary according to the molecular marker used (Smith et al., 2017).
To identify Heterocapsa species, generally two or three molecular markers are used: ITS and SSU/LSU gene sequences, even though ITS is considered the most suitable marker to identify dinoflagellate species (Litaker et al., 2007; Stern et al., 2012). In this study, the phylogenetic trees obtained from the sequencing of ITS and LSU clustered all the sequences that were identified as H. ovata together (Figures 6, 7).
To confirm the molecular identification of this morpho-species and to add to our morphological knowledge of Heterocapsa species and populations from different geographical regions, a detailed microscopic study of one strain was conducted. The described general cell morphology (relative sizes and shapes of the epi- and hypostome, cingulum displacement, position and shape of the nucleus, number, and position of the pyrenoid) and also the ultrastructural detail of the pyrenoid (tubular invaginations) and the micromorphological details of the body scales (size, shape, and morphology of the basal plate, central spine, and the number of peripheral uprights and bars) conform with the original description of H. ovata (Iwataki et al., 2003). Details of the APC of this species are shown for the first time. Cells of strains isolated in this study in Australian plankton samples were on average narrower than in Japan, 28 µm ± 6.87 µm long compared to 26.9 µm and 15 µm ± 4.31 µm wide compared to 21.4 µm (Iwataki et al., 2003).
The use of various gene markers for the metabarcoding analyses provided insights into the taxon diversity of the analyzed bloom samples. The SSU marker allowed the identification of 16 out of the 33 total taxa detected, with seven taxa exclusively identified through this marker and three taxa (Alexandrium, Gonyaulax, and Karlodinium). The coverage provided by this marker is better than that obtained with the COB gene, as also observed by Smith et al. (2017). The LSU marker detected 20 taxa, the highest coverage of the three markers. In particular, 11 taxa were exclusively identified by this marker, while only two taxa were identified using all markers. Only with LSU primers was it possible to identify the genus Heterocapsa. These findings underscore the importance of employing multiple markers to ensure the reliability of results in the analysis of bloom samples. Moreover, SSU and LSU reference databases are more extensive, allowing the classification of OTUs to the genus level. The COB marker allowed the classification of a lower proportion of sequences probably because there are a smaller number of sequences available for this gene (Kohli et al., 2014). Notably, many genera have been identified as potential toxin producers with ichthyotoxic activity. For instance, Alexandrium, Karenia, Karlodinium, Margalefidinium, and Pfiesteria have been documented to produce ichthyotoxins that can adversely affect fish populations (Hallegraeff et al., 2023; Oh et al., 2023). This highlights the complex and varied nature of HABs, emphasizing the importance of continued research and monitoring efforts to better understand and mitigate their ecological impacts.
Toxicological analyses will help in understanding the way in which species of Heterocapsa, which were present at a comparatively low abundance, may have contributed to fish-killing activities during the K. mikimotoi bloom event. This study presented the screening of 21 Heterocapsa ovata strains for ichthyotoxicity on fish gills. Unexpectedly, strains showed substantial variability in their fish gill cell activity, with some causing substantial cell lysis for four out of 21 strains. High ichthyotoxicity has been previously reported due to the following species: Chattonella marina, Karlodinium veneficum (Dorantes-Aranda et al., 2011; Mooney et al., 2011; Dorantes-Aranda et al., 2013) Heterosigma akashiwo, Fibrocapsa japonica, Karenia mikimotoi, Alexandrium catenella, and Prymnesium parvum (Dorantes-Aranda et al., 2015). In these studies, higher ichthyotoxicity was consistently observed after cell lysis, apart from Heterosigma akashiwo, which appears to require living cells for ichthyotoxicity (GlobalHAB, 2023). In the present study, a similar action was observed for the majority of the strains. The whole cells rarely had an effect on cell viability of more than 40% (only for strain 14, Figure 8), meaning that the ichthyotoxicity is expressed only under conditions that cause cell lysis, similar to what was observed for Karlodinium veneficum (Mooney et al., 2010; Dorantes-Aranda et al., 2015). In the same study, the authors observed that this species was the most toxic after extraction and toxic purification in methanol, while other species, such as C. marina, affected the gill cell viability in a higher percentage, indicating that different species can have different ichthyotoxic mechanisms. The results obtained in this study strengthen this hypothesis and make evident that inter-strain toxicity variability is present. Such variability for Heterocapsa strains has been observed by Wu et al. (2022). In that study, three species of Heterocapsa (H. cf. niei, H. horiguchii, and H. cf. pygmaea) were identified as ichthyotoxic since they were hemolytic to rabbit erythrocytes and lethal to brine shrimps. However, Wu and colleagues pointed out the challenges of connecting lab-based toxicity results with fish and shellfish mortalities during HAB events, and the co-occurrence of many species likely makes this correlation even more difficult. In conclusion, despite toxicity being observed and reported in several studies, the mechanism is still unclear, but worth further investigation.
Large fish mortality events are a cause for concern in Australia, in relation to both their environmental and economic impacts. During an HAB event, more than one species can co-occur, which can make it difficult to assign the causes of observed impacts. In this study, samples were collected from Coffin Bay (South Australia) in February 2014 during which high mortality of marine organisms was reported. Karenia mikimotoi was a common and abundant species present and was hypothesized to be the cause of the mortalities observed. However, it was not known whether taxa other than K. mikimotoi contributed to the observed fish mortality. Thus, 21 isolates were established and identified as Heterocapsa ovata by light and electron microscopy, as well as phylogenetic analyses. Little or no genetic variability was found between strains at the level of rRNA gene regions. Toxicological analyses were performed and demonstrated substantial variability in ichthyotoxicity, with most of the H. ovata strains having a negative effect on the viability of fish gill cells. This study shows that substantial differences may exist within a harmful dinoflagellate species in its toxicological impacts, even when molecular genetic differences in barcoding marker regions such as rRNA are comparatively low. Future research on harmful dinoflagellates causing fish kills should be mindful of the substantial inter-strain variability likely in phenotypic traits such as the production of compounds with impacts on marine life.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
AV: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. GG: Data curation, Visualization, Writing – original draft, Writing – review & editing. MH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. JD-A: Formal analysis, Investigation, Methodology, Writing – review & editing. KS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. GK: Conceptualization, Methodology, Writing – review & editing. CW: Investigation, Methodology, Writing – review & editing. GH: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Writing – review & editing. SM: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Erhard Rhiel (Carl-von-Ossietzky-Universitaat, Oldenburg, Germany) is thanked for help with negative staining TEM and Janis Ortgies (Senckenberg am Meer, DZMB) for help with TEM work. Ms. Dora Vig is thanked for her help with map realization.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frpro.2024.1422481/full#supplementary-material
Adolf J. E., Bachvaroff T. R., Deeds J. R., Place A. R. (2015). Ichthyotoxic Karlodinium veneficum (Ballantine) J Larsen in the upper Swan River estuary (Western Australia): ecological conditions leading to a fish kill. Harmful Algae 48, 83–93. doi: 10.1016/j.hal.2015.07.006
Ajani P. A., Petrou K., Larsson M. E., Nielsen D. A., Burke J., Murray S. A. (2021). Phenotypic trait variability as an indication of adaptive capacity in a cosmopolitan marine diatom. Environ. Microbiol. 23, 207–223. doi: 10.1111/1462-2920.15294
Alpermann T. J., Beszteri B., John U., Tillmann U., Cembella A. D. (2009). Implications of life-history transitions on the population genetic structure of the toxigenic marine dinoflagellate Alexandrium tamarense. Mol. Ecol. 18, 2122–2133. doi: 10.1111/j.1365-294X.2009.04165.x
Basti L., Segawa S. (2010). Mortality of the short-neck clam Ruditapes philippinarum induced by the toxic dinoflagellate Heterocapsa circularisquama. Fisheries Sci. 76, 625–631. doi: 10.1007/s12562-010-0252-4
Benico G., Lum W. M., Takahashi K., Yñiguez A. T., Iwataki M. (2021). Thecal tabulation, body scale morphology and phylogeny of Heterocapsa philippinensis sp. nov. (Peridiniales, Dinophyceae) from the Philippines. Eur. J. Protistology 80, 125811. doi: 10.1016/j.ejop.2021.125811
Bols N., Barlian A., Chirino-Trejo M., Caldwell S., Goegan P., Lee L. (1994). Development of a cell line from primary cultures of rainbow trout, Oncorhynchus mykiss (Walbaum), gills. J. Fish Dis. 17, 601–611. doi: 10.1111/j.1365-2761.1994.tb00258.x
Brandenburg K. M., Wohlrab S., John U., Kremp A., Jerney J., Krock B., et al. (2018). Intraspecific trait variation and trade-offs within and across populations of a toxic dinoflagellate. Ecol. Lett. 21, 1561–1571. doi: 10.1111/ele.13138
Broadwater M. H., Van Dolah F. M., Fire S. E. (2018). “Vulnerabilities of Marine Mammals to Harmful Algal Blooms,” in Harmful algal blooms: a compendium desk reference (Wyley), 191–222. doi: 10.1002/9781118994672
Bruslé J. (1995). The impact of harmful algal blooms on finfish-mortality, pathology and toxicology. Mortality Pathol. toxicology. Repères Oceans 10, 75.
Carey C. C., Ibelings B. W., Hoffmann E. P., Hamilton D. P., Brookes J. D. (2012). Eco-physiological adaptations that favour freshwater cyanobacteria in a changing climate. Water Res. 46, 1394–1407. doi: 10.1016/j.watres.2011.12.016
Choi H., Kim S. (2021). Heterocapsa busanensis sp. nov.(Peridiniales, Dinophyceae): A new marine thecate dinoflagellate from Korean coastal waters. Eur. J. Protistology 79, 125797. doi: 10.1016/j.ejop.2021.125797
Chomérat N., Gatti C. M., Nézan É., Chinain M. (2017). Studies on the benthic genus Sinophysis (Dinophysales, Dinophyceae) II. S. canaliculata from Rapa Island (French Polynesia). Phycologia 56, 193–203. doi: 10.2216/16-96.1
Cox M. P., Peterson D. A., Biggs P. J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinf. 11, 1–6. doi: 10.1186/1471-2105-11-485
Dorantes-Aranda J. J., Nichols P. D., David Waite T., Hallegraeff G. M. (2013). Strain variability in fatty acid composition of Chattonella marina (Raphidophyceae) and its relation to differing ichthyotoxicity toward rainbow trout gill cells. J. Phycology 49, 427–438. doi: 10.1111/jpy.12053
Dorantes-Aranda J. J., Seger A., Mardones J. I., Nichols P. D., Hallegraeff G. M. (2015). Progress in understanding algal bloom-mediated fish kills: the role of superoxide radicals, phycotoxins and fatty acids. PloS One 10, e0133549. doi: 10.1371/journal.pone.0133549
Dorantes-Aranda J. J., Waite T. D., Godrant A., Rose A. L., Tovar C. D., Woods G. M., et al. (2011). Novel application of a fish gill cell line assay to assess ichthyotoxicity of harmful marine microalgae. Harmful Algae 10, 366–373. doi: 10.1016/j.hal.2011.01.002
Drummond A. (2010). Geneious v5. 3. Available online at: http://www.geneious.com.
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Elorriaga-Verplancken F. R., Hernandez-Camacho C. J., Alvarez-Santamaria L., Paniagua-Mendoza A., Robles-Hernandez R., Rebolledo-Villa F., et al. (2022). Largest mortality event to date of California Sea Lions in Mexico might be linked to a harmful algal bloom. Aquat. Mammals 48, 59–67. doi: 10.1578/AM.48.1.2022.59
Fire S. E., Wang Z., Byrd M., Whitehead H. R., Paternoster J., Morton S. L. (2011). Co-occurrence of multiple classes of harmful algal toxins in bottlenose dolphins (Tursiops truncatus) stranding during an unusual mortality event in Texas, USA. Harmful Algae 10, 330–336. doi: 10.1016/j.hal.2010.12.001
Fogg G. E. (1969). The Leeuwenhoek Lecture 1968-The physiology of an algal nuisance. Proc. R. Soc. London. Ser. B. Biol. Sci. 173, 175–189. doi: 10.1098/rspb.1969.0045
Glibert P. M. (2017). Eutrophication, harmful algae and biodiversity—Challenging paradigms in a world of complex nutrient changes. Mar. pollut. Bull. 124, 591–606. doi: 10.1016/j.marpolbul.2017.04.027
GlobalHAB. (2023). “Fish-Killing Marine Algal Blooms: Causative Organisms, Ichthyotoxic Mechanisms, Impacts and Mitigation,” in IOC Manuals and Guides (UNESCO-IOC/SCOR, Paris), 96.
Griffin L. P., Friess C., Bakenhaster M. D., Bassos-Hull K., Burnsed S. W., Brownscombe J. W., et al. (2023). Assessing the potential for red tide (Karenia brevis) algal bloom impacts on Atlantic tarpon (Megalops atlanticus) along the southwestern coast of Florida. Environ. Biol. Fishes 106, 255–273. doi: 10.1007/s10641-022-01324-7
Guilard R., Ryther J. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Gran Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., et al. (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Hallegraeff G. M. (1993). A review of harmful algal blooms and their apparent global increase. Phycologia 32, 79–99. doi: 10.2216/i0031-8884-32-2-79.1
Hallegraeff G. (2002). Aquaculturists' guide to harmful Australian microalgae (Hobart, Tasmania: University of Tasmania).
Hallegraeff G. M., Anderson D. M., Davidson K., Gianella F., Hansen P. (2023). “Fish-killing marine algal blooms: causative organisms, ichthyotoxic mechanisms, impacts and mitigation,” UNESCO. IOC Manuals and guides. (Paris, France: UNESCO-IOC/SCOR), 93.
Hanifah A. H., Teng S. T., Law I. K., Abdullah N., Uncha S., Chiba A., et al. (2022). Six marine thecate Heterocapsa (Dinophyceae) from Malaysia, including the description of three novel species and their cytotoxicity potential. Harmful Algae 120, 102338. doi: 10.1016/j.hal.2022.102338
Hansen G. (1989). Ultrastructure and morphogenesis of scales in Katodinium rotundatum (Lohmann) Loeblich (Dinophyceae). Phycologia 28, 385–394. doi: 10.2216/i0031-8884-28-3-385.1
Harvell C. D., Kim K., Burkholder J., Colwell R. R., Epstein P. R., Grimes D. J., et al. (1999). Emerging marine diseases–climate links and anthropogenic factors. Science 285, 1505–1510. doi: 10.1126/science.285.5433.1505
Horiguchi T. (1995). Heterocapsa circularisquama sp. nov.(Peridiniales, Dinophyceae): a new marine dinoflagellate causing mass mortality of bivalves in Japan. Phycological Res. 43, 129–136. doi: 10.1111/j.1440-1835.1995.tb00016.x
Iwataki M. (2008). Taxonomy and identification of the armored dinoflagellate genus Heterocapsa (Peridiniales, Dinophyceae). Plankton Benthos Res. 3, 135–142. doi: 10.3800/pbr.3.135
Iwataki M., Botes L., Sawaguchi T., Sekiguchi K., Fukuyo Y. (2003). Cellular and body scale structure of Heterocapsa ovata sp. nov. and Heterocapsa orientalis sp. nov. (Peridiniales, Dinophyceae). Phycologia 42, 629. doi: 10.2216/i0031-8884-42-6-629.1
John U., Tillmann U., Hülskötter J., Alpermann T. J., Wohlrab S., Van de Waal D. B. (2015). Intraspecific facilitation by allelochemical mediated grazing protection within a toxigenic dinoflagellate population. Proc. R. Soc. B: Biol. Sci. 282, 20141268. doi: 10.1098/rspb.2014.1268
Kearse M., Sturrock S., Meintjes P. (2012). The Geneious 6.0. 3 read mapper (Auckland, New Zealand: Biomatters, Ltd).
Kim D., Oda T. (2010). “Possible factors responsible for the fish-killing mechanisms of the red tide phytoplankton, Chattonella marina and Cochlodinium polykrikoides,” Coastal environmental and ecosystem issues of the East China sea, eds. Ishimatsu A., Lie H.-J.. (Terrapub and Nagasaki University), 245–268.
Kohli G. S., Neilan B. A., Brown M. V., Hoppenrath M., Murray S. A. (2014). Cob gene pyrosequencing enables characterization of benthic dinoflagellate diversity and biogeography. Environ. Microbiol. 16, 467–485. doi: 10.1111/1462-2920.12275
Lin S., Zhang H., Hou Y., Zhuang Y., Miranda L. (2009). High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob genes for dinoflagellate DNA barcoding. Appl. Environ. Microbiol. 75, 1279–1290. doi: 10.1128/AEM.01578-08
Lindholm T., Nummelin C. (1999). “Red tide of the dinoflagellate Heterocapsa triquetra (Dinophyta) in a ferry-mixed coastal inlet,” in Biological, Physical and Geochemical Features of Enclosed and Semi-enclosed Marine Systems, Åland Islands, Finland; 9–13 June 1997 (Netherlands: Kluwer Academics Publishers), 393, 245–251.
Litaker R., Tester P., Duke C., Kenney B., Pinckney J. L., Ramus J. (2002). Seasonal niche strategy of the bloom-forming dinoflagellate Heterocapsa triquetra. Mar. Ecol. Prog. Ser. 232, 45–62. doi: 10.3354/meps232045
Litaker R. W., Vandersea M. W., Kibler S. R., Reece K. S., Stokes N. A., Lutzoni F. M., et al. (2007). Recognizing dinoflagellate species using its rDNA sequences 1. J. phycology 43, 344–355. doi: 10.1111/j.1529-8817.2007.00320.x
Litchman E., de Tezanos Pinto P., Klausmeier C. A., Thomas M. K., Yoshiyama K. (2010). Linking traits to species diversity and community structure in phytoplankton (Fifty years after the ‘‘Homage to Santa Rosalia’’: Old and new paradigms on biodiversity in aquatic ecosystems). (Hydrobiologia), 15–28.
Matsuyama Y. (1999). Harmful effect of dinoflagellate Heterocapsa circularisquama on shellfish aquaculture in Japan. Japan Agric. Res. Q. 33, 283–294.
Matsuyama Y. (2012). Impacts of the harmful dinoflagellate Heterocapsa circularisquama bloom on shellfish aquaculture in Japan and some experimental studies on invertebrates. Harmful Algae 14, 144–155. doi: 10.1016/j.hal.2011.10.019
Millette N. C., Stoecker D. K., Pierson J. (2015). Top-down control by micro-and mesozooplankton on winter dinoflagellate blooms of Heterocapsa rotundata. Aquat. Microbial Ecol. 76, 15–25. doi: 10.3354/ame01763
Mooney B. D., Hallegraeff G. M., Place A. R.. (2010). Ichthyotoxicity of four species of gymnodinioid dinoflagellates (Kareniaceae, Dinophyta) and purified karlotoxins to larval sheepshead minnow. Harmful Algae 9, 557–563. doi: 10.1016/j.hal.2010.04.005
Mooney B. D., Dorantes-Aranda J. J., Place A. R., Hallegraeff G. M. (2011). Ichthyotoxicity of gymnodinioid dinoflagellates: PUFA and superoxide effects in sheepshead minnow larvae and rainbow trout gill cells. Mar. Ecol. Prog. Ser. 426, 213–224. doi: 10.3354/meps09036
Murray S. A., Kohli G. S., Farrell H., Spiers Z. B., Place A. R., Dorantes-Aranda J. J., et al. (2015). A fish kill associated with a bloom of Amphidinium carterae in a coastal lagoon in Sydney, Australia. Harmful Algae 49, 19–28. doi: 10.1016/j.hal.2015.08.003
Nagai K., Matsuyama Y., Uchida T., Yamaguchi M., Ishimura M., Nishimura A., et al. (1996). Toxicity and LD50 levels of the red tide dinoflagellate Heterocapsa circularisquama on juvenile pearl oysters. Aquaculture 144, 149–154. doi: 10.1016/S0044-8486(96)01307-5
Nishiguchi T., Cho K., Yasutomi M., Ueno M., Yamaguchi K., Basti L., et al. (2016). Intracellular haemolytic agents of Heterocapsa circularisquama exhibit toxic effects on H. circularisquama cells themselves and suppress both cell-mediated haemolytic activity and toxicity to rotifers (Brachionus plicatilis). Aquat. Toxicol. 179, 95–102. doi: 10.1016/j.aquatox.2016.08.019
Oh J. W., Pushparaj S. S. C., Muthu M., Gopal J. (2023). Review of harmful algal blooms (HABs) causing marine fish kills: toxicity and mitigation. Plants (Basel) 12, 3936. doi: 10.3390/plants12233936
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Razali R. M., Mustapa N. I., Yaacob K. K. K., Yusof F., Teng S. T., Hanafiah A. H., et al. (2022). Diversity of Heterocapsa (Dinophyceae) and the algal bloom event in the mariculture areas of Johor Strait, Malaysia. Plankton Benthos Res. 17, 290–300. doi: 10.3800/pbr.17.290
Roach A. (1997). The effect of acid water inflow on estuarine benthic and fish communities in the Richmond River, N. S. W. Australia. Australas. J. Ecotoxicology 3, 25–56.
Roberts S. D., Van Ruth P. D., Wilkinson C., Bastianello S. S., Bansemer M. S. (2019). Marine heatwave, harmful algae blooms and an extensive fish kill event during 2013 in South Australia. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00610
Roberts S. D., Wilkinson C., Stobart B., Doubell M., van Ruth P., Gilliland J. (2014). Fish Kill Investigation: Coffin Bay harmful algal (Karenia mikimotoi) bloom February 2014., Primary Industries and Regions South Australia. PIRSA Fisheries Aquaculture Division Adelaide.
Rogers P., Geddes M., Ward T. (2003). Blue sprat Spratelloides robustus (Clupeidae: Dussumieriinae): a temperate clupeoid with a tropical life history strategy? Mar. Biol. 142, 809–824. doi: 10.1007/s00227-002-0973-8
Rolton A., Rhodes L., Hutson K. S., Biessy L., Bui T., MacKenzie L., et al. (2022). Effects of harmful algal blooms on fish and shellfish species: A case study of New Zealand in a changing environment. Toxins 14, 341. doi: 10.3390/toxins14050341
Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sammut J., Melville M. D., Callinan R. B., Fraser G. C. (1995). Estuarine acidification: impacts on aquatic biota of draining acid sulphate soils. Aust. Geographical Stud. 33, 89–100. doi: 10.1111/j.1467-8470.1995.tb00687.x
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Scholin C. A., Herzog M., Sogin M., Anderson D. M. (1994). Identification of group-and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). ii. sequence analysis of a fragment of the LSU rRNA gene 1. J. phycology 30, 999–1011. doi: 10.1111/j.0022-3646.1994.00999.x
Smith K. F., Kohli G. S., Murray S. A., Rhodes L. L. (2017). Assessment of the metabarcoding approach for community analysis of benthic-epiphytic dinoflagellates using mock communities. New Z. J. Mar. Freshw. Res. 51, 555–576. doi: 10.1080/00288330.2017.1298632
Stern R. F., Andersen R. A., Jameson I., Küpper F. C., Coffroth M.-A., Vaulot D., et al. (2012). Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. Plos ONE. 7 (8), e42780. doi: 10.1371/journal.pone.0042780
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tillmann U., Alpermann T. L., da Purificação R. C., Krock B., Cembella A. (2009). Intra-population clonal variability in allelochemical potency of the toxigenic dinoflagellate Alexandrium tamarense. Harmful Algae 8, 759–769. doi: 10.1016/j.hal.2009.03.005
Van Dolah F. M. (2000). Marine algal toxins: origins, health effects, and their increased occurrence. Environ. Health Perspect. 108, 133–141. doi: 10.1289/ehp.00108s1133
Van Dolah F. M., Doucette G. J., Gulland F. M., Bossart T.L.R.G.D. (2002). “Impacts of algal toxins on marine mammals,” in Toxicology of marine mammals. eds. Vos J. G., Bossart G., Fournier M., O’Shea T.. (CRC Press). doi: 10.1201/9780203165577
Verma A., Hoppenrath M., Dorantes-Aranda J. J., Harwood D. T., Murray S. A. (2016). Molecular and phylogenetic characterization of Ostreopsis (Dinophyceae) and the description of a new species, Ostreopsis rhodesae sp. nov., from a subtropical Australian lagoon. Harmful Algae 60, 116–130. doi: 10.1016/j.hal.2016.11.004
Verma A., Hughes D. J., Harwood D. T., Suggett D. J., Ralph P. J., Murray S. A. (2020). Functional significance of phylogeographic structure in a toxic benthic marine microbial eukaryote over a latitudinal gradient along the East Australian Current. Ecol. Evol. 10, 6257–6273. doi: 10.1002/ece3.6358
von Dassow P., John U., Ogata H., Probert I., Bendif E. M., Kegel J. U., et al. (2015). Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 9, 1365–1377. doi: 10.1038/ismej.2014.221
Wang Q., Deeds J. R., Place A. R., Belas R. (2005). Dinoflagellate community analysis of a fish kill using denaturing gradient gel electrophoresis. Harmful Algae 4, 151–162. doi: 10.1016/j.hal.2004.01.002
Wu X., Liu Y., Weng Y., Li L., Lin S. (2022). Isolation, identification and toxicity of three strains of Heterocapsa (Dinophyceae) in a harmful event in Fujian, China. Harmful Algae 120, 102355. doi: 10.1016/j.hal.2022.102355
Xiao J., Sun N., Zhang Y., Sun P., Li Y., Pang M., et al. (2018). Heterocapsa bohaiensis sp. nov. (Peridiniales: Dinophyceae): a novel marine dinoflagellate from the Liaodong Bay of Bohai Sea, China. Acta Oceanologica Sin. 37, 18–27. doi: 10.1007/s13131-018-1296-z
Yang X., Sun N., Li Y., Li X. (2015). Effects of two harmful species algae on reproduction of rotifer Brachionus plicatilis and metamorphosis of zoea of Chinese mitten handed crab Eriocheir sinensis. J. Dalian Ocean Univ. 30, 351–356. doi: 10.16535/jcnki.dlhyxb.2015.04.002
Keywords: Heterocapsa, inter-strain variability, fish gill cell line bioassay, harmful algal blooms, fish kill event, morphology
Citation: Verma A, Gaiani G, Hoppenrath M, Dorantes-Aranda JJ, Smith K, Kohli GS, Wilkinson C, Hallegraeff GM and Murray SA (2024) Characterization and inter-strain variability in ichthyotoxicity of Heterocapsa ovata (Peridiniales, Dinophyceae) from temperate waters of South Australia. Front. Protistol. 2:1422481. doi: 10.3389/frpro.2024.1422481
Received: 24 April 2024; Accepted: 30 May 2024;
Published: 26 June 2024.
Edited by:
An Suk Lim, Gyeongsang National University, Republic of KoreaReviewed by:
Margarita Fernández Tejedor, Institute of Agrifood Research and Technology (IRTA), SpainCopyright © 2024 Verma, Gaiani, Hoppenrath, Dorantes-Aranda, Smith, Kohli, Wilkinson, Hallegraeff and Murray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shauna Ann Murray, c2hhdW5hLm11cnJheUB1dHMuZWR1LmF1
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.