
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 March 2025
Sec. Plant Metabolism and Chemodiversity
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1570758
This article is part of the Research Topic In-Depth Interpretation of Critical Genomic Information Related to the Biosynthesis of Key Specialized (Secondary) Metabolism in Medicinal Plants View all 9 articles
Phenylphenalenone is an important phytoalexin for banana plant protection, yet the mechanisms governing its biosynthesis and regulation remain unclear in plant. WRKY transcription factors play essential roles in modulating plant growth, development, and the biosynthesis of secondary metabolites. In this study, we identified 158 WRKY genes (MlWRKYs) from a phenylphenalenone-rich plant species Musella lasiocarpa. Phylogenetic analysis classified the MlWRKY genes into three distinct subfamilies: type I, type II, and type III. Chromosomal distribution revealed that the MlWRKY genes are clustered on nine respective chromosomes. Additionally, synteny analysis between M. lasiocarpa and Musa balbisiana uncovered highly conserved collinear regions. MIWRKY15, MIWRKY111, MIWRKY122 were identified as candidate genes for regulating PhPNs biosynthesis by integration of multi-omics approaches. We further investigated the expression pattern of MIWRKY15, MIWRKY111, MIWRKY122 genes, as well as their putative target genes MlOMT22 and MlOMT27, the known phenylphenalenone biosynthesis genes in various tissues, including leaves, stems, roots, and seeds. MlWRKY15 and MlOMT22 showed similar expression patterns across tissues. MlWRKY122 and MlOMT27 also displayed consistent expression patterns, suggesting MlWRKY122 may regulate MlOMT27. Additionally, MlWRKY111’s expression was inversely correlated with MlOMT27, indicating a potential negative regulation of MlOMT27 by MlWRKY111. This study provides valuable insights into the WRKY family in M. lasiocarpa and will serve as a useful genetic resource for elucidating the regulatory mechanisms of phenylphenalenone biosynthesis.
Phenylphenalenones (PhPNs) are predominantly found in monocot families, such as Strelitziaceae and Musaceae (Norman et al., 2019). These compounds have been identified as significant phytoalexins in wild banana species, making them a valuable resource for developing disease-resistant banana varieties (Flors and Nonell, 2006; Chen et al., 2018). However, cultivated bananas typically exhibit low concentrations and limited structural diversity of PhPNs. Genetic modification techniques represent a promising strategy to address this issue through the targeted manipulation of biosynthetic pathways and enzymes associated with PhPN production, thereby conferring enhanced disease resistance in banana plants. Research has indicated that PhPNs are synthesized via the phenylpropanoid biosynthetic pathway, with their linear precursors undergoing intramolecular cyclization (Norman et al., 2019). For example, a chalcone synthase named WtPKS1, which catalyzes the initial step in diarylheptanoid biosynthesis, was characterized from Wachendorfia thyrsiflora (Brand et al., 2006). Our research group has previously characterized three O-methyltransferases (OMT) involved in the phenylphenalenone biosynthetic pathway from Chinese dwarf banana Musella lasiocarpa (Zhao et al., 2024). However, the mechanisms governing PhPN biosynthesis and regulation remain unknown. M. lasiocarpa, an endemic species in China and the sole representative of the genus Musella, is primarily distributed in southwestern regions, particularly in Yunnan Province (Liu et al., 2003; Ma et al., 2019). Traditionally, the flowers and bracts of this plant have been utilized in folk medicine for their hemostatic and anti-inflammatory properties (Liu et al., 2003). Recent phytochemical studies have revealed that M. lasiocarpa contains a variety of PhPNs as well as linear diarylheptanoids, which are believed to be precursors in the biosynthesis of PhPNs (Dong et al., 2011). Consequently, M. lasiocarpa exhibits considerable potential as a model organism for elucidating the biosynthetic pathways of PhPN.
WRKY transcription factors constitute a vital class of plant genes that play an indispensable role in various physiological processes, including stress responses and the biosynthesis of secondary metabolites (Jiang et al., 2017; Viana et al., 2018; Liu et al., 2020; Wang et al., 2022; Zhang et al., 2023; Ma et al., 2024b). These transcription factors regulate the expression of target genes by specifically binding to W-box cis-elements [(T)TGAC(C/T)] in promoter regions. Additionally, they interact with a diverse array of proteins to execute their functions across multiple signaling pathways (Chen et al., 2017; Javed and Gao, 2023). WRKY transcription factors orchestrate the biosynthesis of plant secondary metabolites through transcriptional regulation of rate-limiting enzymes in secondary metabolic pathways (Schluttenhofer and Yuan, 2015). PeWRKY30, a crucial transcription factor that is co-expressed alongside flavonoid accumulation in yellow-fruited Passiflora edulis, may contribute to enhancing resistance against both biotic and abiotic stresses (Ma et al., 2024a). The PsWRKY transcription factor in Papaver somniferum binds to the W-box in the promoter regions of benzylisoquinoline alkaloid pathway genes, thus stimulating transcriptional activity from the tyrosine/DOPA decarboxylase promoter (Mishra et al., 2013). Several members of the WRKY family play pivotal roles in regulating terpenoid biosynthesis pathways. Specifically, the Artemisia annua transcription factor AaWRKY1 enhances ADS expression by directly binding to the promoter region of the artemisinin biosynthesis gene ADS, thereby modulating artemisinin biosynthesis (Han et al., 2014; Jiang et al., 2016). Similarly, overexpression of TcWRKY8 and TcWRKY47 in Taxus chinensis significantly increases the expression levels of paclitaxel-related synthase genes, thereby enhancing the biosynthetic pathway of paclitaxel (Han et al., 2014; Jiang et al., 2016). Similarly, overexpression of TcWRKY8 and TcWRKY47 in Taxus chinensis significantly upregulates the expression levels of paclitaxel-related synthase genes, leading to enhanced biosynthesis of paclitaxel (Zhang et al., 2018). Furthermore, the homodimer of GhWRKY41 from Gossypium hirsutum directly enhances the expression of GhC4H and Gh4CL, which in turn regulates the accumulation of lignin and flavonoids (Xiao et al., 2023). To our konwledge, the regulatory role of WRKY transcription factors in PhPN biosynthesis remains unexplored, highlighting a critical gap in our understanding of this metabolic pathway.
In this study, we aimed to comprehensively characterize the WRKY gene family in M. lasiocarpa through genome-wide identification and analysis, focusing on gene classification, chromosomal distribution, and phylogenetic relationships, to provide a foundation for future functional studies. Through integration of multi-omics approaches, MIWRKY15, MIWRKY111, and MIWRKY122 were identified as candidate genes potentially involved in regulating PhPNs biosynthesis. These findings will provide a robust foundation for molecular studies and genetic engineering initiatives aimed at enhancing disease resistance in Musaceae.
The M. lasiocarpa was gathered from Nanhua County, Yunnan Province, China (coordinates: 118°50’38”E, 32°3’44”N) and subsequently relocated to an experimental field in Nanjing, China (coordinates: 101°1’2”E, 25°9’54”N). Species identification of the experimental materials was conducted by Professor Yu Chen from the Institute of Botany Jiangsu Province, Chinese Academy of Sciences. For total RNA extraction experiments, leaf, stem, and seed samples were harvested from the same plant individual at three developmental stages (yellow seed, S2; brown seed, S4; and black seed, S6).
The genome sequence of M. lasiocarpa (GenBank number: PRJNA1009687) was retrieved from our research group. The AtWRKY protein sequences were retrieved from the Arabidopsis Information Resource database available at https://www.arabidopsis.org. Potential MlWRKY proteins were initially identified through homology searches using BLAST and Hidden Markov Model (HMM) algorithms. The predicted MlWRKY annotations were then validated by cross-referencing with the Swiss-Prot database, followed by further analysis using the NCBI Conserved Domain Database (CDD) at https://www.ncbi.nlm.nih.gov/cdd. The presence of conserved domains in these candidate proteins was verified using the Pfam database. Various physicochemical properties, including coding sequence (CDS) length, isoelectric point, and molecular weight, were predicted for the identified MlWRKY proteins using the EXPASy-ProtParam tool.
Phylogenetic analysis was performed using the Maximum Likelihood approach, supported by 1,000 bootstrap replicates, utilizing MEGA version 5.05. This analysis utilized the Jones-Taylor-Thornton substitution model with a stringent requirement of minimum 95% site coverage. Conserved domains were identified through Batch-Search and TBtools software (Chen et al., 2020). To better understand the functional characteristics of MlWRKY proteins, their conserved domains were analyzed via the MEME program (https://meme-suite.org/meme/). Furthermore, TBtools was used to create a distribution map that visualizes the organization of MlWRKY genes. The Dual Synteny Plotter feature in TBtools was employed to analyze the syntenic relationships between MlWRKYs and WRKY genes from various species. Genomic data for collinearity analysis were obtained from the following sources: Arabidopsis thaliana (https://www.arabidopsis.org), Oryza sativa (https://plants.ensembl.org/info/data/ftp/index.html), and Musa balbisiana (https://www.ncbi.nlm.nih.gov/datasets/genome/). To investigate the tissue-specific expression patterns of MlWRKYs in M. lasiocarpa, we utilized an RNA-seq dataset (accession number PRJNA100968) deposited by our team in NCBI.
To isolate total RNA, we employed the FastPure Universal Plant Total RNA Isolation Kit (RC411) from Vazyme Biotech Co., Ltd. (Nanjing, China), following the protocol specified by the manufacturer. For qRT-PCR purposes, the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme) was used to synthesize cDNA from the extracted total RNA. The qRT-PCR experiments were performed on the qTOWER 2.2 system, manufactured by Analytik Jena in Germany. The PCR amplification conditions consisted of 40 cycles with denaturation at 95°C for 10 seconds and annealing/extension at 60°C for 30 seconds. To assess relative expression levels, the EF-α gene from M. lasiocarpa served as an internal control, and the comparative cycle threshold (2−ΔΔCt) method was applied, incorporating t values for analysis. Each sample included both biological and technical replicates. Primer sequences for all target transcripts were designed using Primer Premier software (Supplementary Table S1).
To demonstrate the significant differences between the two groups, a two-tailed unpaired Student’s t-test was employed. The thresholds for significance were established as *p < 0.05, **p < 0.01, and ***p < 0.001. Results are presented as mean ± SD. All statistical analyses were conducted using GraphPad Prism 9. Additionally, TBtools was utilized to generate a distribution map that illustrates the arrangement of MlWRKY genes.
We identified MlWRKY genes in the M. lasiocarpa genome by screening for conserved WRKY domains, followed by manual validation of gene structures and functional motifs. A total of 158 MlWRKY proteins, each containing more than 100 amino acids, were identified after filtering out incomplete sequences and shorter variants. The 158 genes were designated as MlWRKY1 to MlWRKY158 according to their chromosomal localization (Table 1). To better understand the molecular characteristics of these MlWRKY members, their physiological and biochemical properties were examined. The proteins varied in length from 102 to 2004 amino acids (aa), with corresponding molecular weights (MW) between 11,294.9 and 223,829.29 Daltons. Their isoelectric points also differed, ranging from 4.41 to 10.45. Eighteen distinct domains were identified through the analysis of conserved motifs using the MEME program. The results demonstrated that all MlWRKY genes contain at least one WRKY conserved domain, thereby highlighting the conservation these genes (Supplementary Figure S1).
A chromosomal mapping approach was developed to investigate the genetic variation and duplication events within the WRKY family in M. lasiocarpa. 158 MlWRKY genes were unevenly distributed across nine M. lasiocarpa chromosomes (as shown in Figure 1). Chromosome 7 (chr7) harbored the highest number of MlWRKY genes (27), followed by chr9 (26), chr1 (21), chr8 (16), chr3 (15), chr4 (14), chr5 (14), and chr2 (13). Whereas only 12 genes were found on chr6.
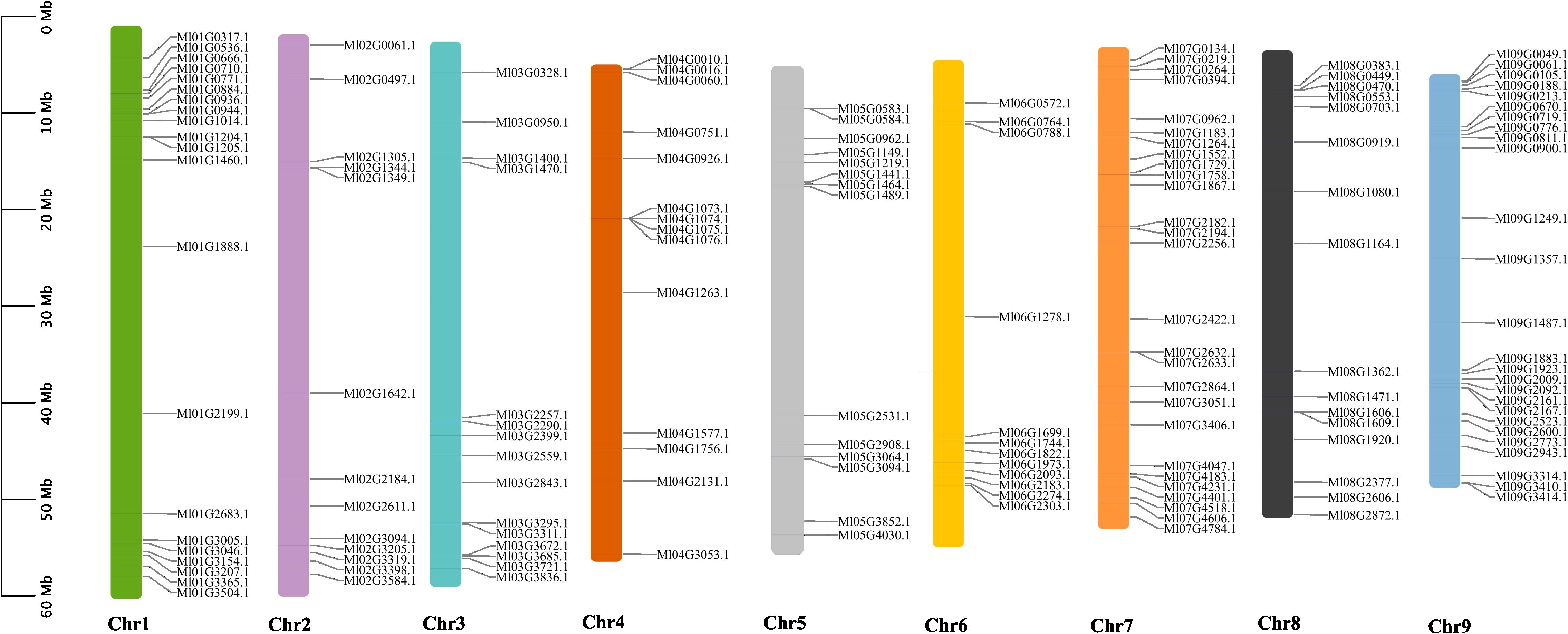
Figure 1. Chromosomal distribution of MlWRKY genes within the M. lasiocarpa genome. A total of 158 MlWRKY genes were localized across nine chromosomes in M. lasiocarpa. The left side of the figure indicates the scale in megabases (Mb). Each chromosome is depicted as a vertical line, with its name labeled beneath the corresponding line.
To examine the evolutionary relationship between MlWRKYs in M. lasiocarpa and the A. thaliana, a maximum likelihood phylogenetic tree was constructed. The MlWRKY family was classified into three major groups (Types I–III), in accordance with the classification of AtWRKYs from A. thaliana (Figure 2). In M. lasiocarpa, Type I included 25 members, compared to 14 in A. thaliana. Type II represented the largest group, with 116 MlWRKY proteins, and was subdivided into five subgroups: IIa, IIb, IIc, IId, and IIe. Furthermore, Type III contained 17 MlWRKY members, exceeding the 13 found in A. thaliana.
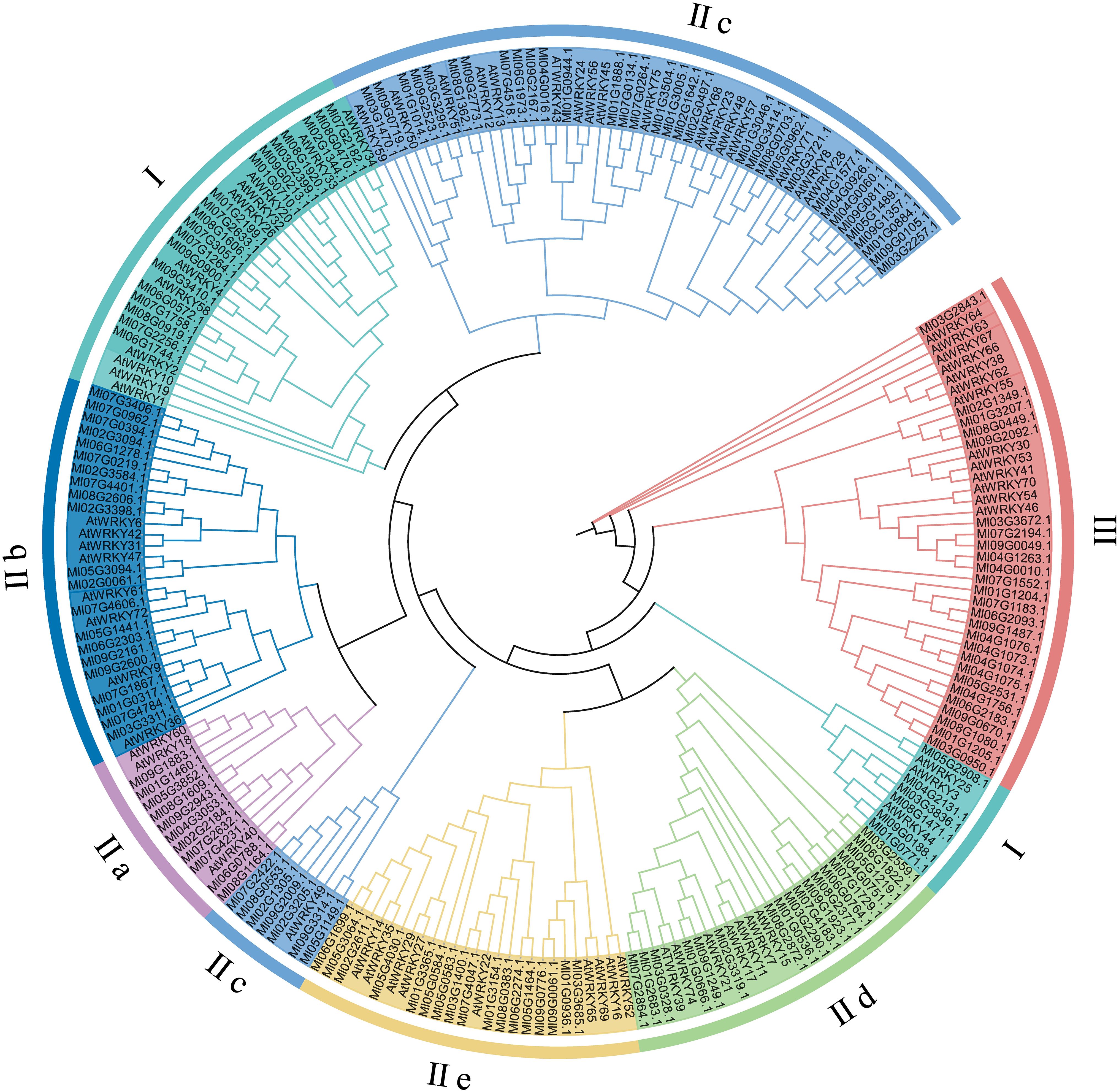
Figure 2. Phylogenetic relationships of WRKY gene families in M. lasiocarpa and A. thaliana were reconstructed using the maximum likelihood (ML) method in MEGA5. Multiple sequence alignment was performed with MUSCLE, and the best-fit substitution model (JTT+G) was selected. Branch support was evaluated with 1000 bootstrap replicates.
Collinearity analyses among M. lasiocarpa, Musa balbisiana, A. thaliana, and Oryza sativa were conducted to investigate the evolutionary relationships among these model species, including dicotyledonous and monocotyledons. The interspecies collinearity analysis revealed that MlWRKYs exhibited syntenic relationships with genes on all chromosomes of A. thaliana (Figure 3). Notably, the number of collinear gene pairs between M. lasiocarpa and O. sativa exceeded that between M. lasiocarpa and A. thaliana, likely due to both M. lasiocarpa and O. sativa being monocotyledons. Similarly, a higher number of syntenic gene pairs were observed between M. lasiocarpa and M. balbisiana, attributable to their closer evolutionary relationship within the Musaceae family.
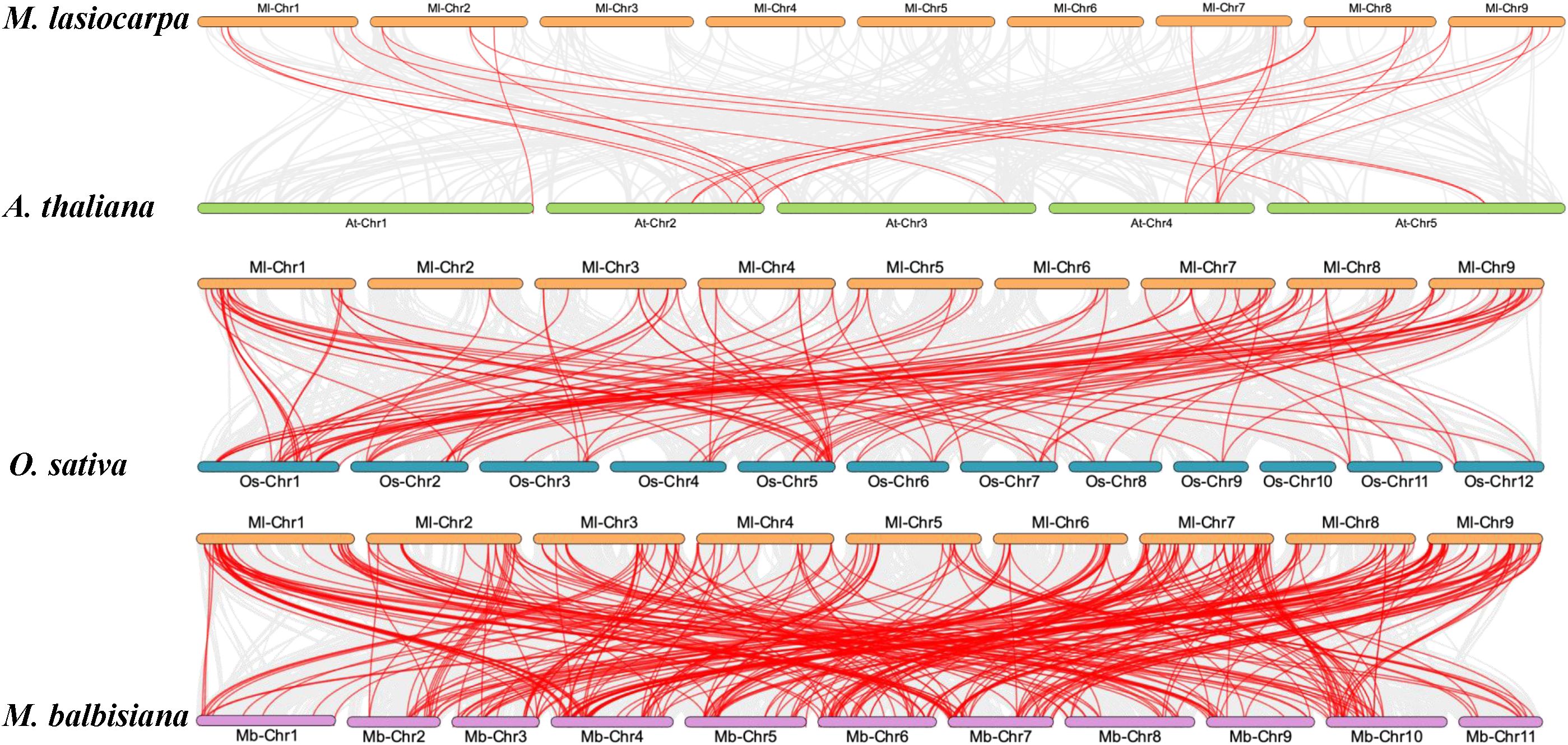
Figure 3. The collinearity gene pair analysis of WRKY among M. lasiocarpa (Ml), M. balbisiana (Mb), A. thaliana, (At) and O. sativa (Os). The M. lasiocarpa, A. thaliana, O. sativa, and M. balbisiana were represented by saffron yellow, lawn green, lake blue, and lilac, respectively. The collinearity gene pairs were indicated by red lines.
We previously reported PhPNs content in yellow seed (stage S2), brown seed (S4), and block seed (S6) of M. lasiocarpa. The results demonstrated a gradual increase in PhPN content, following the order S2 < S4 < S6 (Zhao et al., 2024). To investigate the expression patterns of MlWRKYs in M. lasiocarpa seeds across different developmental stages, RNA-Seq data from previous studies (accession PRJNA1009687) were analyzed. A heat map generated using FPKM values illustrated the expression profiles of MlWRKYs in seeds at S2, S4, and S6 stages (Figure 4). The majority of genes exhibited either no expression or low expression levels in the three developmental stages of M. lasiocarpa seeds. Only 11 out of 158 MlWRKY genes had an FPKM value exceeding 20 in least one of the tested samples. Among these 11 genes, MlWRKY15, MlWRKY84, MlWRKY109, and MlWRKY122 showed expression patterns consistent with the growth trend of PhPNs content. While MlWRKY6, MlWRKY50, MlWRKY59, MlWRKY148, and MlWRKY149 showed opposite expression pattern with the growth trend of PhPNs content. Previous research has demonstrated that CsWRKY57like from Camellia sinensis influences the biosynthesis of methylated epigallocatechin gallate (EGCG) by regulating the CCoAOMT gene (Luo et al., 2022). A homology search using the amino acid sequence of CsWRKY57like as a query against MlWRKYs revealed higher similarity with MlWRKY15, MlWRKY111, and MlWRKY122. Based on these findings, we hypothesize that MlWRKY15, MlWRKY111, and MlWRKY122 are likely to regulate PhPNs biosynthesis through the modulation of MlOMT genes expression. Furthermore, our previous studies demonstrated that two MlOMT genes, Ml04G2958 (MlOMT22), and Ml08G0855 (MlOMT27), are involved in PhPNs biosynthesis (Zhao et al., 2024). The expression pattern of MlOMT22 was consistent with those of MlWRKY15 and MlWRKY122, whereas the expression pattern of MlOMT27 was consistent with that of MlWRKY111 (Figure 4). Furthermore, the identification of W-box motifs in the promoters of MlOMT22 and MlOMT27 indicates their potential regulation by MlWRKY15, MlWRKY111, and MlWRKY122, providing insights into the transcriptional network underlying secondary metabolism in M. lasiocarpa.
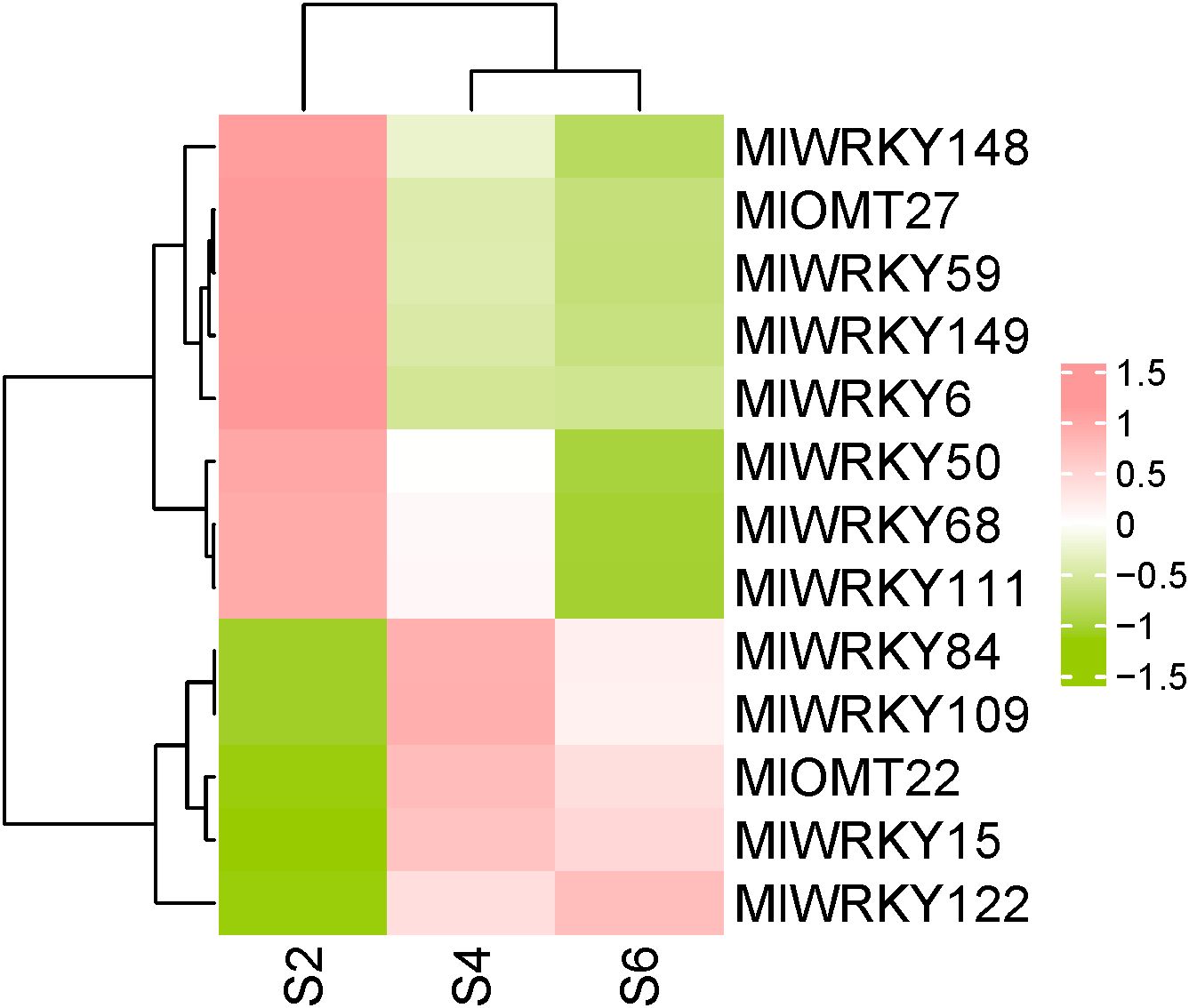
Figure 4. Heat map representing the expression levels of MlWRKY and MlOMT genes. The heat map illustrating the expression profiles of MlWRKY and MlOMT genes across in M. lasiocarpa seeds during different developmental stages, measured by FPKM. Eleven MlWRKY genes (MlWRKY6, MlWRKY15, MlWRKY50, MlWRKY59, MlWRKY68, MlWRKY84, MlWRKY109, MlWRKY111, MlWRKY122, MlWRKY148, and MlWRKY149) had an FPKM value exceeding 20 in least one of the tested samples. The color bar indicates gene expression levels, with expression increasing from green to red.
We analyzed the transcriptional levels of MlWRKY genes, MlWRKY15, MlWRKY111, and MlWRKY122 and their candidate target genes MlOMT22 and MlOMT27 in four different tissues of M. lasiocarpa by qRT-PCR. It demonstrated that each gene could be detected in all four tested tissues (Figure 5). MlWRKY15 and MlOMT22 are predominantly expressed in the root, exhibiting similar expression patterns across different tissues. This suggests that MlWRKY15 may positively regulate MlOMT22 expression in various tissues. Similarly, MlWRKY122 and MlOMT27 are primarily expressed in seeds (black seeds), with consistent expression patterns across different tissues, indicating that MlWRKY122 is likely to regulate MlOMT27 expression in these tissues. Moreover, the expression pattern of MlWRKY111 is inversely correlated with that of MlOMT27, suggesting a potential negative regulatory role of MlWRKY111 on MlOMT27. However, further experimental validation is required to confirm these specific regulatory mechanisms.
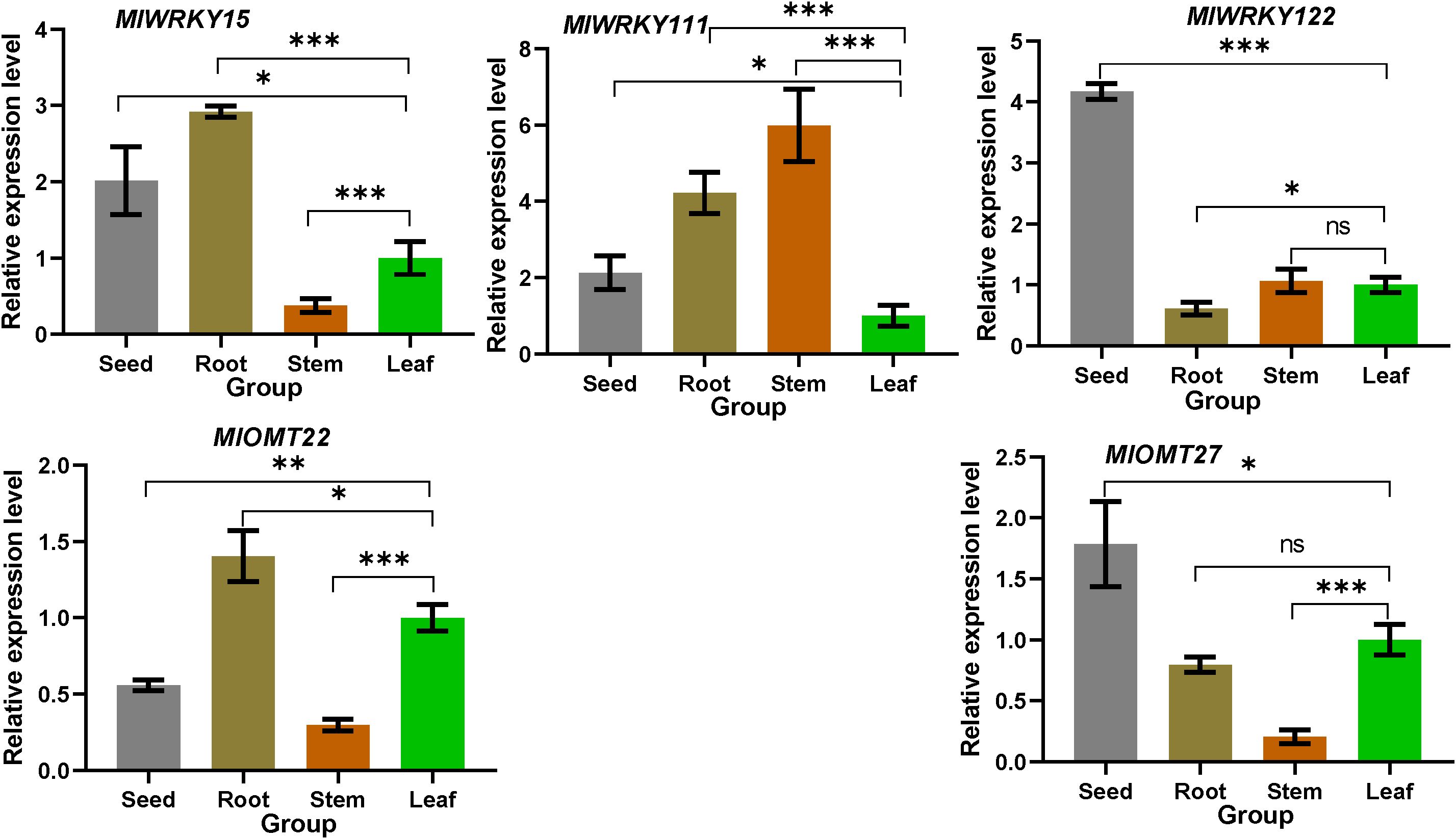
Figure 5. Expression patterns of three selected MlWRKY genes and their candidate target MlOMT genes in different tissues of M. lasiocarpa. All experiments were performed in triplicate, with relative expression levels normalized to the EF-α gene of M. lasiocarpa. Data are presented as mean ± SD. *, **, and *** denote that the genes from other tissues exhibit significant differences compared to leaf (with P values less than 0.05, 0.01, and 0.001 respectively). “ns” indicates no significant difference (P ≥ 0.05).
WRKY transcription factors exert essential regulatory functions in the biosynthesis of plant secondary metabolites (Yang et al., 2012; Kumar et al., 2023). They modulate the equilibrium between plant defense responses and metabolic pathways via transcriptional and signaling regulatory mechanisms. As our understanding of WRKY transcription factors deepens, their regulatory roles in plant secondary metabolism and their potential for industrial applications—such as metabolic engineering and biopharmaceutical production—will be increasingly clarified. Future studies should focus on the interactions of WRKY transcription factors with other transcription factors and signaling pathways, as well as their applications in plant metabolic engineering. For instance, transgenic or gene editing technologies can be employed to enhance WRKY transcription factor expression, thereby improving the yield and quality of plant secondary metabolites (Das et al., 2024). Banana is one of the most important fruits globally, yet its production faces significant threats from various diseases, particularly those caused by fungi, bacteria, and viruses (Drenth and Kema, 2021). While traditional chemical pesticides are widely employed to protect bananas from these pathogens, concerns about their potential environmental and health risks have spurred research into natural, efficient, and less toxic alternatives (Ferreyra-Suarez et al., 2024). PhPN, a type of natural secondary metabolite, plays a crucial role in plant self-protection, demonstrating significant biological activity in disease resistance, antibacterial, and antifungal properties (Krishnamurthy et al., 2023). Additionally, ketenes exhibit substantial potential in antioxidant, anti-inflammatory, antibacterial, and anticancer activities, with broad applications in drug development, food, fragrance, and cosmetics industries. PhPN, derived from phenylpropanoid compounds, typically features a phenyl-ketene structure, comprising a benzene ring and an unsaturated ketene moiety. This structural configuration enables PhPN to engage in diverse biochemical reactions and interact with various biomolecules. Notably, PhPN content in bananas is very low; it primarily exists in certain plants of the Musaceae family, such as M. lasiocarpa, which is rich in PhPN and serves as an ideal material for studying its biosynthesis and regulation (Zhao et al., 2024).
Plants regulate their responses to various stresses through transcription factors such as WRKY (Liu et al., 2020) (Li et al., 2024; Ma et al., 2024a), with an important mechanism being the regulation of secondary metabolite biosynthesis (Jiang et al., 2017). In recent years, the role of WRKY transcription factors in regulating secondary metabolite biosynthesis has garnered significant attention (Li et al., 2025; Zhao et al., 2025b). WRKY transcription factors recognize and bind to the W-box cis-element (TTGACC/T) in the promoters of target genes. For instance, SlWRKY35 from tomatoes directly activates the expression of the SlDXS1 gene, thereby enhancing carotenoid biosynthesis and accumulation (Yuan et al., 2022). Conversely, ElWRKY48 negatively regulates ingenol biosynthesis by modulating the expression of genes involved in diterpenoid biosynthesis (Zhao et al., 2025a). We identified similar binding motifs (W-box) in the promoters of MlOMT22 and MlOMT27 genes in M. lasiocarpa, suggesting a conserved regulatory mechanism. Our co-expression analysis revealed that MlWRKY15, MlWRKY111 and MlWRKY122 similar expression patterns with MlOMT22 and MlOMT27, implying potential protein-protein interactions that may synergistically activate PhPN biosynthetic genes. However, it remains unclear how MlWRKYs regulate the biosynthesis of PhPNs by modulating the expression of MlOMT genes, which will be the focus of our future research. We selected MlWRKY15, MlWRKY111, and MlWRKY122 for tissue expression analysis based on the following evidence: 1) Expression Patterns: As shown in Figure 4, MlWRKY15, MlWRKY111, and MlWRKY122 exhibited expression patterns consistent with the growth trend of PhPNs content in M. lasiocarpa seeds (S2 < S4 < S6). This correlation suggests a potential regulatory role in PhPNs biosynthesis. 2) Homology Analysis: A homology search using the amino acid sequence of CsWRKY57like (a known regulator of methylated EGCG biosynthesis in Camellia sinensis) revealed higher similarity with MlWRKY15, MlWRKY111, and MlWRKY122. This finding further supports their potential involvement in the regulation of methylated compounds, such as PhPNs. 3) Functional Relevance: Previous studies have demonstrated that WRKY transcription factors often regulate secondary metabolite biosynthesis by modulating the expression of key enzymes (e.g., O-methyltransferases). Given the homology and expression patterns, we hypothesize that MlWRKY15, MlWRKY111, and MlWRKY122 may regulate PhPNs biosynthesis through the modulation of MlOMT genes.
In this study, we performed the first comprehensive analysis of the WRKY gene family in M. lasiocarpa. In sum, 158 MlWRKY genes were identified in the genome, showing an uneven distribution across all nine chromosomes. We systematically examined the evolution, chromosomal localization, protein-protein interactions, and expression patterns of these MlWRKY genes. Specifically, MIWRKY15, MIWRKY111, and MIWRKY122 were pinpointed as candidate genes potentially involved in the regulation of PhPN biosynthesis. These genes represent promising targets for future functional studies aimed at elucidating their roles in PhPN biosynthesis in M. lasiocarpa. This study lays a solid foundation for future research into the functions of MlWRKY genes and the molecular mechanisms governing PhPN biosynthesis. While this study has identified potential WRKY genes that may regulate PhPN biosynthesis, the specific regulatory mechanisms remain to be elucidated, which will be a focus of our subsequent research. Overall, this is the first identification of the WRKY gene family in M. lasiocarpa, providing a basis for understanding the biosynthetic mechanism of PhPNs and offering insights into other potential functions of WRKY genes in plants.
In summary, this study represents the first comprehensive analysis of the WRKY gene family in M. lasiocarpa, identifying 158 MlWRKY genes unevenly distributed across nine chromosomes. We characterized their evolutionary relationships, chromosomal distribution, protein-protein interactions, and expression patterns. MIWRKY15, MIWRKY111, MIWRKY122 were identified as candidate genes potentially involved in regulating PhPNs biosynthesis by integration of multi-omics approaches. These genes serve as promising candidates for future functional studies aimed at elucidating their roles in PhPNs biosynthesis regulation in M. lasiocarpa. This research provides a foundation for further investigation into the functions of MIWRKY genes and the molecular mechanisms underlying PhPNs biosynthesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LH: Investigation, Writing – review & editing. PL: Investigation, Writing – review & editing. MT: Writing – review & editing, Data curation. XF: Writing – review & editing. YC: Writing – review & editing, Funding acquisition. BF: Writing – review & editing, Methodology. WZ: Conceptualization, Writing – original draft.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Natural Science Foundation of Jiangsu Province (grant number: BK20220752 and BK20220749), the National Natural Science Foundation of China (22207047), and the Talent Fund of the Jiangsu Institute of Botany (JIBTF202304).
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1570758/full#supplementary-material
Brand, S., Hölscher, D., Schierhorn, A., Svatos, A., Schröder, J., Schneider, B. (2006). A type III polyketide synthase from Wachendorfia thyrsiflora and its role in diarylheptanoid and phenylphenalenone biosynthesis. Planta 224, 413–428. doi: 10.1007/s00425-006-0228-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, Y., Paetz, C., Schneider, B. (2018). Precursor-directed biosynthesis of phenylbenzoisoquinolindione alkaloids and the discovery of a phenylphenalenone-based plant defense mechanism. J. Nat. Prod. 81, 879–884. doi: 10.1021/acs.jnatprod.7b00885
Das, S., Kwon, M., Kim, J.-Y. (2024). Enhancement of specialized metabolites using CRISPR/Cas gene editing technology in medicinal plants. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1279738
Dong, L.-B., He, J., Li, X.-Y., Wu, X.-D., Deng, X., Xu, G., et al. (2011). Chemical constituents from the aerial parts of Musella lasiocarpa. Natural Products Bioprospecting 1, 41–47. doi: 10.1007/s13659-011-0007-7
Drenth, A., Kema, G. (2021). The vulnerability of bananas to globally emerging disease threats. Phytopathology 111, 2146–2161. doi: 10.1094/PHYTO-07-20-0311-RVW
Ferreyra-Suarez, D., García-Depraect, O., Castro-Muñoz, R. (2024). A review on fungal-based biopesticides and biofertilizers production. Ecotoxicol. Environ. Saf. 283, 116945. doi: 10.1016/j.ecoenv.2024.116945
Flors, C., Nonell, S. (2006). Light and singlet oxygen in plant defense against pathogens: phototoxic phenalenone phytoalexins. Acc Chem. Res. 39, 293–300. doi: 10.1021/ar0402863
Han, J., Wang, H., Lundgren, A., Brodelius, P. E. (2014). Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102, 89–96. doi: 10.1016/j.phytochem.2014.02.011
Javed, T., Gao, S.-J. (2023). WRKY transcription factors in plant defense. Trends Genet. 39, 787–801. doi: 10.1016/j.tig.2023.07.001
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Jiang, W., Fu, X., Pan, Q., Tang, Y., Shen, Q., Lv, Z., et al. (2016). Overexpression of aaWRKY1 leads to an enhanced content of artemisinin in artemisia annua. BioMed. Res. Int. 2016, 7314971. doi: 10.1155/2016/7314971
Krishnamurthy, P., Ravikumar, M. J., Arumugam Palanivelu, S., Pothiraj, R., Suthanthiram, B., Subbaraya, U., et al. (2023). Phenylphenalenone-type phytoalexins in banana (Musa species): a comprehensive review for new research directions. Phytochem. Rev. 22, 187–210. doi: 10.1007/s11101-022-09839-8
Kumar, S., Korra, T., Thakur, R., Arutselvan, R., Kashyap, A. S., Nehela, Y., et al. (2023). Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress 8, 100154. doi: 10.1016/j.stress.2023.100154
Li, M., Shao, Y., Pan, B., Liu, C., Tan, H. (2025). Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J. Adv. Res. doi: 10.1016/j.jare.2025.01.009
Li, J., Wang, X., Lu, J., Song, H., Lei, H., Niu, T., et al. (2024). Genome-wide identification and expression analysis of the WRKY gene family in Sophora flavescens during tissue development and salt stress. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1520786
Liu, A.-Z., Kress, W. J., Long, C.-L. (2003). The ethnobotany of Musella lasiocarpa (Musaceae), an endemic plant of southwest China. Economic Bot. 57, 279–281. doi: 10.1663/0013-0001(2003)057[0279:TEOMLM]2.0.CO;2
Liu, A., Liu, C., Lei, H., Wang, Z., Zhang, M., Yan, X., et al. (2020). Phylogenetic analysis and transcriptional profiling of WRKY genes in sunflower (Helianthus annuus L.): Genetic diversity and their responses to different biotic and abiotic stresses. Ind. Crops Products 148, 112268. doi: 10.1016/j.indcrop.2020.112268
Luo, Y., Huang, X.-X., Song, X.-F., Wen, B.-B., Xie, N.-C., Wang, K.-B., et al. (2022). Identification of a WRKY transcriptional activator from Camellia sinensis that regulates methylated EGCG biosynthesis. Horticulture Res. 9, uhac024. doi: 10.1093/hr/uhac024
Ma, J., Wang, Y., Hong, Y., Zhao, M., Ma, X., Liu, J., et al. (2024b). SlWRKY55 coordinately acts with SlVQ11 to enhance tomato thermotolerance by activating SlHsfA2. Plant J. 119, 2904–2918. doi: 10.1111/tpj.16960
Ma, H., Wang, D.-X., Li, T.-Q., Li, Z.-H. (2019). Phylogeographic study of Musella lasiocarpa (Musaceae): providing insight into the historical river capture events. Pakistan J. Bot. 51, 191–199. doi: 10.30848/PJB2019-1(17
Ma, F., Zhou, H., Yang, H., Huang, D., Xing, W., Wu, B., et al. (2024a). WRKY transcription factors in passion fruit analysis reveals key PeWRKYs involved in abiotic stress and flavonoid biosynthesis. Int. J. Biol. Macromolecules 256, 128063. doi: 10.1016/j.ijbiomac.2023.128063
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M., Shanker, K., et al. (2013). Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PloS One 8, e52784. doi: 10.1371/journal.pone.0052784
Norman, E. O., Lever, J., Brkljača, R., Urban, S. (2019). Distribution, biosynthesis, and biological activity of phenylphenalenone-type compounds derived from the family of plants, Haemodoraceae. Nat. Prod. Rep. 36, 753–768. doi: 10.1039/c8np00067k
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Viana, V., Marini, N., Busanello, C., Pegoraro, C., Fernando, J., Da Maia, L., et al. (2018). Regulation of rice responses to submergence by WRKY transcription factors. Biol. plantarum 62, 551–560. doi: 10.1007/s10535-018-0806-3
Wang, C., Hao, X., Wang, Y., Maoz, I., Zhou, W., Zhou, Z., et al. (2022). Identification of WRKY transcription factors involved in regulating the biosynthesis of the anti-cancer drug camptothecin in Ophiorrhiza pumila. Horticulture Res. 9, uhac099. doi: 10.1093/hr/uhac099
Xiao, S., Ming, Y., Hu, Q., Ye, Z., Si, H., Liu, S., et al. (2023). GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol. J. 21, 961–978. doi: 10.1111/pbi.14008
Yang, C. Q., Fang, X., Wu, X. M., Mao, Y. B., Wang, L. J., Chen, X. Y. (2012). Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 54, 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yuan, Y., Ren, S., Liu, X., Su, L., Wu, Y., Zhang, W., et al. (2022). SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 234, 164–178. doi: 10.1111/nph.17977
Zhang, J., Zhao, H., Chen, L., Lin, J., Wang, Z., Pan, J., et al. (2023). Multifaceted roles of WRKY transcription factors in abiotic stress and flavonoid biosynthesis. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1303667
Zhang, M., Chen, Y., Nie, L., Jin, X., Liao, W., Zhao, S., et al. (2018). Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 8 (1), 5197. doi: 10.1038/s41598-018-23558-1
Zhao, W., Li, P., Huang, L., Wang, R., Tian, M., Xu, S., et al. (2025a). Genome-wide identification of WRKY transcription factor genes in Euphorbia lathyris reveals ElWRKY48 as a negative regulator of phosphate uptake and ingenol biosynthesis. Int. J. Biol. Macromolecules 302 (2025), 139859. doi: 10.1016/j.ijbiomac.2025.139859
Zhao, W., Liu, J., Chen, Y. (2025b). Advances in the biosynthesis of naturally occurring benzylisoquinoline alkaloids. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1548471
Keywords: WRKY, Musella lasiocarpa, regulation, phenylphenalenone biosynthesis, O-methyl transferase
Citation: Huang L, Li P, Tian M, Feng X, Chen Y, Feng B and Zhao W (2025) Comprehensive characterization of the WRKY gene family and their potential roles in regulation phenylphenalenone biosynthesis in Musella lasiocarpa. Front. Plant Sci. 16:1570758. doi: 10.3389/fpls.2025.1570758
Received: 04 February 2025; Accepted: 25 February 2025;
Published: 12 March 2025.
Edited by:
Xinyi Guo, Central European Institute of Technology (CEITEC), CzechiaReviewed by:
Linlin Yang, Henan University of Chinese Medicine, ChinaCopyright © 2025 Huang, Li, Tian, Feng, Chen, Feng and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanli Zhao, emhhb3dhbmxpdGNtQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.