
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 March 2025
Sec. Technical Advances in Plant Science
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1564079
This article is part of the Research Topic Machine Vision and Machine Learning for Plant Phenotyping and Precision Agriculture, Volume II View all 20 articles
Accurate segmentation of leaf diseases is crucial for crop health management and disease prevention. However, existing studies fall short in addressing issues such as blurred disease spot boundaries and complex feature distributions in disease images. Although the vision foundation model, Segment Anything Model (SAM), performs well in general segmentation tasks within natural scenes, it does not exhibit good performance in plant disease segmentation. To achieve fine-grained segmentation of leaf disease images, this study proposes an advanced model: Enhanced Multi-Scale SAM (EMSAM). EMSAM employs the Local Feature Extraction Module (LFEM) and the Global Feature Extraction Module (GFEM) to extract local and global features from images respectively. The LFEM utilizes multiple convolutional layers to capture lesion boundaries and detailed characteristics, while the GFEM fine-tunes ViT blocks using a Multi-Scale Adaptive Adapter (MAA) to obtain multi-scale global information. Both outputs of LFEM and GFEM are then effectively fused in the Feature Fusion Module (FFM), which is optimized with cross-branch and channel attention mechanisms, significantly enhancing the model’s ability to handle blurred boundaries and complex shapes. EMSAM integrates lightweight linear layers as classification heads and employs a joint loss function for both classification and segmentation tasks. Experimental results on the PlantVillage dataset demonstrate that EMSAM outperforms the second-best state-of-the-art semantic segmentation model by 2.45% in Dice Coefficient and 6.91% in IoU score, and surpasses the baseline method by 21.40% and 22.57%, respectively. Particularly, for images with moderate and severe disease levels, EMSAM achieved Dice Coefficients of 0.8354 and 0.8178, respectively, significantly outperforming other semantic segmentation algorithms. Additionally, the model achieved a classification accuracy of 87.86% across the entire dataset, highlighting EMSAM’s effectiveness and superiority in plant disease segmentation and classification tasks.
Accurate segmentation of leaf lesions is essential for the early diagnosis and precise management of crop diseases. The area, shape, and distribution of lesions reflect disease severity and guide subsequent prevention and control measures (Shoaib et al., 2022). Manual segmentation of disease spots by plant pathology experts is time-consuming, labor-intensive, and inefficient. Moreover, this approach is prone to bias and requires significant investment in human and material resources (Singh and Misra, 2017). Computer vision-based methods generally offer better efficiency, consistency, and automation than manual segmentation. They can capture complex feature details, facilitating large-scale processing and analysis. Traditional methods for plant disease segmentation employ edge detection, thresholding, and region growing (Pang et al., 2011; Revathi and Hemalatha, 2012; Wang et al., 2013). These methods are effective for images with simple backgrounds and distinct disease spots. However, they lack robustness when dealing with lesions that have blurred boundaries, complex shapes, and varying sizes.
Deep learning models can automatically learn complex features, showing strong robustness against noise and complex backgrounds, making them efficient for lesion segmentation (Giménez-Gallego et al., 2020). Models based on deep learning are primarily divided into two categories: Convolutional Neural Network (CNN) and Vision Transformer (ViT). CNN-based segmentation models, such as U-Net (Ronneberger et al., 2015) and the DeepLab series (Chen et al., 2018a), achieve precise segmentation of target regions. Liu et al. (2022) used a U-Net model with DenseNet as the backbone to segment three common rice leaf diseases, achieving a mean Dice coefficient (mDice) of 0.86. Yuan et al. (2022) employed an improved DeepLab+ model to segment grape leaf black rot, introducing channel attention mechanisms and pyramid feature fusion networks, resulting in a mean Intersection over Union (mIoU) score of 0.85 on a custom orchard dataset. Pal and Kumar (2023) proposed an AgriDet framework, which integrates the Inception-Visual Geometry Group Network with a Kohonen-based deep learning network to classify the severity of plant diseases. Within this framework, a multi-variate Grab-Cut algorithm is employed to achieve effective image segmentation under complex background occlusion, significantly mitigating the issue of background interference. Divyanth et al. (2023) proposed a two-stage model combining U-Net and DeepLab+. It first extracts leaves from the background, followed by disease spot extraction, achieving an mIoU of 0.74 for corn leaf diseases. Although these works have made progress in leaf disease image detection, CNNs inherently lack global information perception. Pal et al. (2024) proposed a novel framework for plant disease recognition, which initially employs DeepLabV3 to accurately segment the diseased plant regions. Additionally, the framework utilizes Bayesian Task Augmentation-Model Agnostic Meta-Learning with multi-scale spatial attention to optimize the network, enabling the model to achieve exceptional performance even with limited datasets. Experimental results on two datasets demonstrated outstanding performance, with an accuracy rate of 99.1%, a sensitivity of 99.5%, and a specificity of 98.7%. This limits their performance when dealing with leaf disease spots that exhibit distributional differences. Transformer architectures excel at global modeling, which improves segmentation accuracy and helps handle complex disease regions (Dosovitskiy et al., 2021). For instance, Jiang et al. (2023) employed the Trans-Unet model to segment pine nematode disease, employing a novel loss function based on precision and recall, achieving an mDice Coefficient of 0.87. Yang et al. (2024) utilized the Swin-Unet model, optimized with SENet modules to focus on global target features, achieving an mDice Coefficient of 0.85 in corn leaf disease segmentation tasks. However, these methods require extensive annotated data and are less effective in handling blurred boundaries. Additionally, task-specific models require targeted training, limiting their adaptability and generalization to diverse leaf disease segmentation scenarios.
Traditional models designed for specific tasks often have limited adaptability and generalization when applied to diverse leaf disease segmentation scenarios. In contrast, the Segment Anything Model (SAM), pre-trained on extensive image datasets, shows exceptional generalization performance. This enables SAM to adapt more effectively and generalize across various leaf disease segmentation tasks. Its efficiency in segmenting targets with both sparse and dense prompts makes it a promising solution for overcoming the limitations of task-specific models, thereby enhancing the versatility and performance in agricultural applications (Kirillov et al., 2023). SAM excels in generalization, performing well in simple natural image segmentation tasks without the need for retraining. However, for specialized tasks such as plant disease segmentation, SAM typically requires fine-tuning to achieve optimal performance (Zhang and Jiao, 2023). Since its release, SAM has garnered significant attention, with applications in medical imaging (Cheng et al., 2023; Wu et al., 2023; Ma et al., 2024), remote sensing (Osco et al., 2023; Wang et al., 2023; Gui et al., 2024), and plant disease segmentation. In the field of plant disease segmentation, Zhang et al. (2023) fine-tuned SAM on a tobacco leaf dataset, achieving an mIoU of 0.84 across different growth stages. Moupojou et al. (2024) employed a two-stage framework where SAM first segments all recognizable objects in an image, and then a self-constructed classification network performs image classification, resulting in a 10% improvement in classification accuracy compared to traditional classification networks. Balasundaram et al. (2025) used SAM to segment diseased tea leaf areas. These segments were processed by a custom CNN for feature extraction, followed by classification, achieving 95.06% accuracy. The above studies use SAM to segment diseases in private datasets, not open datasets, making it difficult to comprehensively evaluate SAM’s segmentation performance on plant diseases. Moreover, Transformer-based SAM faces challenges in capturing fine details, showing limitations in tasks that require fine-grained segmentation, such as blurred boundaries, camouflaged objects, and fragmented features (Zhang et al., 2024). Plant disease images often exhibit these traits, with lesions of low severity showing blurred boundaries and fragmented, fine-grained features. Directly applying SAM to leaf disease segmentation yields unsatisfactory results, as shown in Figure 1. Figure 1A shows segmentation using point prompts, where foreground points mark disease spots, and background points mark leaves and other areas. However, achieving better results requires precise point settings, significantly reducing efficiency. Figure 1B illustrates box prompts, using a bounding box to segment leaves. While box prompts segment the leaf, they fail to isolate disease spots. Figure 1C shows automatic segmentation results, distinguishing foreground from background but missing detailed disease regions. resulting masks lack any semantic information. As a result, to enhance SAM’s performance in the leaf disease monitoring field, it often requires fine-tuning with high-quality plant disease segmentation images. This highlights the need for fine-tuning large vision models to adapt them for specific segmentation tasks.
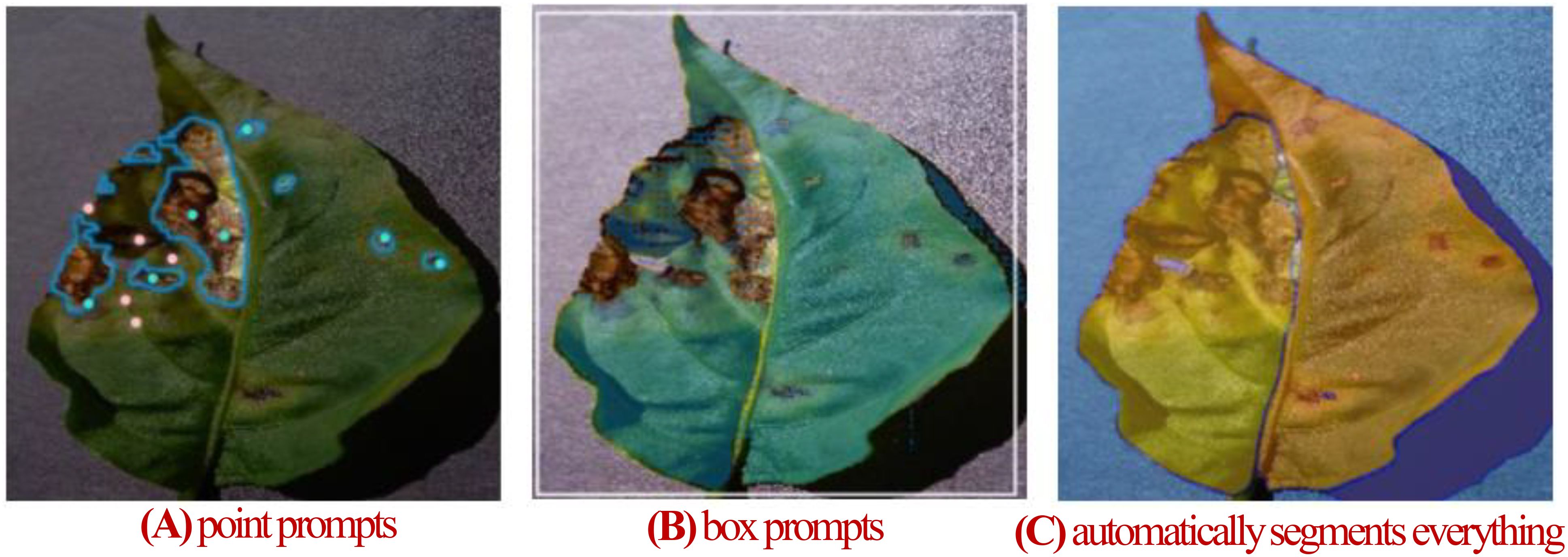
Figure 1. (A) Segmentation with SAM’s point prompts. (B) Segmentation with SAM’s box prompts. (C) SAM automatically segments everything.
To enable SAM to better learn domain-specific knowledge, parameter-efficient fine-tuning techniques are considered the most effective solutions (Xu et al., 2023). Chen et al. (2023b) proposed SAM-Adapter, integrating domain-specific information or prompts into SAM via efficient adapters. This approach facilitates the adaptation to downstream tasks. Zhang and Liu (2023) proposed SAMed, which fine-tunes SAM’s image encoder using Low-Rank Adaptation (LoRA). This method achieves performance comparable to state-of-the-art semantic segmentation techniques in various medical image segmentation tasks. Li et al. (2023) developed an agriculture-specific SAM adapter, improving the Dice Coefficient by 41.48%. However, these fine-tuning methods still fail to allow SAM to perform effectively with complex features. The main reason for these issues is that plant disease images differ significantly in feature details and distribution from SAM’s training images. The unclear boundary and irregular shape of a lesion in a plant disease image pose significant challenges for SAM. These challenges can be summarized as follows: (1) Leaves with mild disease severity often have indistinct boundaries, resulting in minimal differences between the foreground and background. (2) Lesion areas vary widely, ranging from large clusters to fragmented distributions, or a combination of both. (3) The absence of label information in SAM’s training data prevents the use of individual small lesions for disease type classification. These challenges significantly constrain the model’s ability to effectively segment leaf disease regions with complex features.
In response to these challenges, we propose the Enhanced Multi-Scale SAM (EMSAM), a framework designed to improve SAM’s performance in complex disease segmentation tasks. EMSAM achieves fine-grained segmentation of leaf disease images by focusing on the following key objectives: (1) Adapting the base model specifically for plant disease segmentation. (2) Integrating multi-scale feature modeling to effectively capture lesion characteristics. (3) Combining global feature extraction with local detail capture to improve sensitivity to disease-specific features. (4) Jointly optimizing segmentation and classification in the decoding stage using a unified loss function. The main contributions of this study include:
1. Development of the EMSAM framework, which integrates more efficient adapter tuning techniques to enhance its performance in plant disease segmentation. The framework improves SAM’s ability to handle disease images with blurred boundaries and complex shapes.
2. Design of the Multi-Scale Adaptive Adapter (MAA): The MAA module captures multi-scale pyramid features to extract disease features at various scales, improving the model’s segmentation precision and robustness. This is achieved with minimal trainable parameters, enabling efficient parameter tuning.
3. Introduction of an efficient feature fusion mechanism: Combining a Local Feature Extraction Module (LFEM) with the Feature Fusion Module (FFM), EMSAM incorporates cross-branch and channel attention mechanisms to optimize the integration of CNN and ViT features, achieving a balance between global and local feature representation.
4. Incorporation of a lightweight classification head and a joint loss function: This enables EMSAM to accurately segment lesion regions while predicting disease categories, significantly enhancing the practical utility of the model.
5. We conduct the first comprehensive evaluation of SAM and EMSAM on the PlantVillage dataset, which is the well-known open plant disease image dataset, establishing a comparable baseline for future research in plant disease segmentation.
The remainder of this paper is organized as follows: Section 2 provides a detailed description of the dataset construction and EMSAM architecture. Section 3 describes the experimental setup and analyzes the results. Section 4 discusses the implications and significance of the study and summarizes the overall research.
The dataset employed in this study is the widely recognized PlantVillage dataset, which is extensively utilized in the field of plant disease segmentation (Hughes and Salathe, 2016). Comprising a total of 54,306 plant leaf images, the dataset is organized into 12 categories of healthy leaves and 26 categories of diseased leaves. Notably, all disease types have been diagnosed by plant pathology experts, ensuring the accuracy and reliability of the data. The images are in RGB format, with a uniform resolution of 256x256 pixels, facilitating consistent preprocessing and analysis.
We employ the “EISeg” tool (Hao et al., 2022) from Baidu’s PaddlePaddle framework for pixel-level annotations of disease lesion areas, saving the annotated data in PNG format. For our study, we annotated 200 images per category across 26 diseased leaf categories, following these criteria:
1. Diversity of Lesion Characteristics: Images were selected to capture variations in lesion shapes, sizes, color patterns, and spatial distributions, ensuring representation of early to late disease stages.
2. Class Balance: Each category was strictly limited to 200 images to prevent model bias toward overrepresented classes.
The resulting dataset, named the PlantVillage Segmentation Dataset (PSD), contains 5,200 images (26 classes × 200 images) split into a training set (4,160 images) and a test set (1,040 images) with an 8:2 ratio. Table 1 details the category distribution and annotation statistics.
The PSD includes images displaying various lesion distributions: some with small, fragmented lesions; others with large, concentrated lesions; and some exhibiting a combination of both characteristics. These complex shapes reflect the irregularity of lesion area distributions on leaves, posing higher demands on models tasked with leaf disease segmentation. Figure 2 illustrates selected annotated images from the PSD, showcasing original images alongside their corresponding grayscale ground truth masks. The first-row features images with fragmented disease spot distributions, the second row shows images with mixed fragmented and concentrated distributions, and the third row displays images with concentrated disease spots.
To demonstrate the model’s capability in segmenting leaves with varying levels of disease severity, we adopt the leaf disease severity classification method proposed by Ji and Wu (2022), utilizing the Percentage of Infections () as a baseline metric. The infection percentage is calculated using Equation 1:
Where and denote the mutilated leaf area and the total leaf area, respectively, and denote the pixel size of the diseased spot area and the pixel size of the total leaf area, respectively. Based on this metric, disease severity is categorized into three levels: light (0< ≤ 0.2), moderate (0.2< ≤ 0.5), and severe (0.5< ≤ 1). Utilizing these three severity levels, we further subdivide the test set to facilitate more detailed experimental validation.
For the task of plant leaf disease segmentation and classification, we propose the Enhanced Multi-scale SAM (EMSAM), as illustrated in Figure 3. The EMSAM architecture comprises three primary components: an image encoder, SAM’s prompt encoder, and a hybrid decoder. The image encoder is responsible for fusing global and local features, SAM’s prompt encoder is responsible for generating the prompt embedding, and the hybrid decoder concurrently handles classification and segmentation tasks. The image encoder consists of two branches: a ViT branch forming the Global Feature Extraction Module (GFEM) and a CNN branch comprising the Local Feature Extraction Module (LFEM). The GFEM employs stacked ViT blocks to extract global features, with most parameters frozen during training. Only the parameters related to the MAA integrated into the ViT blocks are fine-tuned. The LFEM leverages convolutional modules and multi-scale feature extraction techniques to capture local features, thereby enhancing sensitivity to edges and textures. The global and local features are subsequently fed into the FFM. The FFM employs cross-branch attention mechanisms and progressively integrated Squeeze-and-Excitation (SE) blocks (Hu et al., 2018) to facilitate effective interaction and weighted fusion, resulting in comprehensive feature representations. In the decoding phase, the Mask-Class Hybrid Decoder integrates the fused features from the image encoder with the prompt embeddings from SAM’s Prompt Encoder. A lightweight linear layer acts as the classification head, responsible for predicting mask confidence, Intersection over Union (IoU) tokens, and label identifiers.
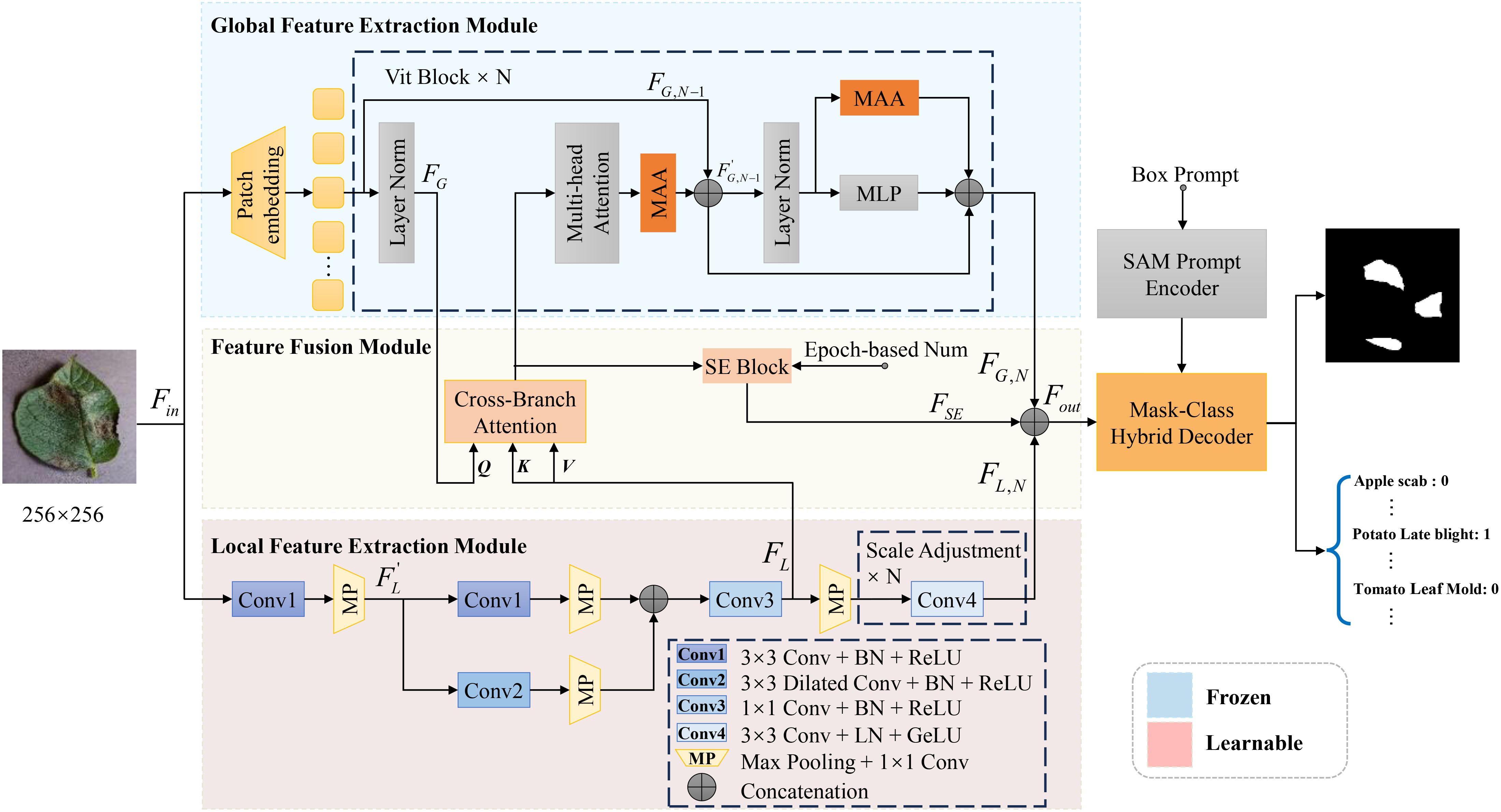
Figure 3. Overall architecture of the proposed EMSAM. It enhances the multi-scale fine-grained image processing capability of SAM through four novel modules: MAA, LFEM, FFM, and mask-category hybrid decoder.
Although SAM excels in natural scene segmentation, it necessitates fine-tuning for specific downstream tasks. The image encoder of SAM, which employs stacked ViT blocks, results in a large number of trainable parameters, thereby limiting performance and increasing the risk of overfitting, particularly when training data are scarce. To address these challenges, adapter tuning offers an efficient and cost-effective approach to adapt pre-trained models (Sung et al., 2022). Consequently, we design the MAA to efficiently adapt SAM for the leaf disease image domain. The detailed architecture of MAA is illustrated in Figure 4.
Within the GFEM, each ViT Block comprises a multi-head attention mechanism and an MLP layer, with layer normalization applied before each sublayer. In our architecture, we insert two MAAs into each ViT Block as trainable parameters, while freezing the remaining components to preserve the pre-trained weights. These MAAs incorporate depthwise separable convolutions, enabling a lightweight design that significantly reduces the training cost while enhancing the model’s adaptability to the specific task of leaf disease segmentation.
Traditional Adapter structures include a down-sampling linear layer (Down), a ReLU activation function, and an up-sampling linear layer (Up). In our design, to obtain multi-scale features and optimize adapter-tuning, we add a Multi-Scale Pyramid Feature Module (MSPM) after the ReLU activation function. Thus, the adapter tuning process can be represented by Equation 2:
Where denotes the input features, which are sequentially processed through a linear down-sampling layer and a ReLU activation function. The processed features are then transformed into within the Multiscale Pyramid Feature Module (MSPM) for subsequent multi-scale spatial information processing, while represents a reduction factor introduced to decrease the dimensionality of the input features. denotes the reshaping operation applied to the input features, where the global features are decomposed into multiple sub-features to enable parallel processing of information at different scales or across distinct regions.
In the MSPM, to enhance the utilization efficiency of multi-scale features, we employ four global average pooling layers (AP) to extract multi-scale features, denoted as . Subsequently, simple 1×1 convolution layers are designed to generate dynamic weights . These dynamic weights allow each input feature to dynamically adjust the importance of each scale based on its content, effectively avoiding the fixed contribution of features at each scale. This process is formally represented by Equations 3, 4, and 5:
Where the dynamic weights are constrained within a reasonable range using the Sigmoid activation function. defines a convolutional layer with a kernel of 1×1 and a kernel of 3×3 depthwise separable convolution with GELU activation function. is a bilinear interpolation-based upsampling method that restores downsampled features to their original resolution.
Next, we concatenate the multi-scale features , representing the processed multi-scale spatial information in Equation 6:
Where defines a 3×3 depthwise separable convolutional layer with a GELU activation function, optimizing the overall feature information extraction capability. represents the process of channel-wise concatenation of features from different scales. defines a 1×1 convolutional layer, which is applied to impose weight control on the channel dimension of the output feature, acting on the concatenated feature map and facilitating the fusion of global channel weights.
Finally, the entire MSPM output is mathematically represented by Equation 7:
Where similarly represents the operation of reshaping the features to match the input feature dimensions. This design enables EMSAM to fine-tune the ViT blocks efficiently, minimizing the training cost.
Traditional segmentation models relying exclusively on CNN or ViT often face challenges in effectively capturing both global and local information, particularly when dealing with the complex and irregular feature distributions characteristic of leaf disease images (Ngo et al., 2024). To address these limitations, we introduce the Enhanced Detail Feature Image Encoder, as depicted in Figure 5. This encoder consists of two key components: the GFEM, designed to capture global contextual information, and the LFEM, focused on extracting boundary and detailed features. By integrating these modules, the encoder significantly enhances the model’s feature extraction capabilities, thereby improving segmentation performance.
In the GFEM, we inherit the ViT framework from SAM and incorporate the MAA in each ViT Block. The MAA module, through depthwise separable convolutions and the multi-scale pyramid feature module (MSPM), optimizes the ViT’s ability to capture global information specific to leaf disease images. This process can be expressed by Equation 8:
Additionally, the GFEM leverages multi-head self-attention mechanisms to effectively aggregate critical information globally, enabling the model to capture the overall characteristics of diseased leaves. In the N-th ViT Block, the entire process is given in Equations 9, 10 and 11:
Where denotes the input to the , and represent the output features of the N-th and (N-1)-th ViT Blocks, respectively, represents the intermediate parameters within the ViT Block, and denotes the feature fusion module.
In the LFEM, a multi-layer convolutional structure comprising four convolutional groups and a max-pooling convolution group is employed to precisely capture detailed features. The input features first undergo a 3×3 convolution to extract core features while maintaining computational stability. A parallel branch incorporates a dilated convolution to expand the receptive field and capture larger-scale local features. Post addition, a 1×1 convolution reduces computational load through channel compression and performs nonlinear feature mapping. Each convolutional layer is followed by a Batch Normalization layer to normalize outputs, thereby accelerating the training process and enhancing model performance and stability. ReLU activation functions are applied to all convolutional layers. To match the spatial resolution of the GFEM’s output feature maps, the final stage employs N 3×3 convolutions with Layer Normalization and GELU activation. This process is illustrated in Equations 12, 13, and 14:
Where represents the portion of the LFEM features input to the FFM. denotes the intermediate features during the processing stage. refers to the max-pooling operation, which is used to downsample feature maps while retaining prominent local features. Notably, and maintain identical spatial resolutions. Thus, the combined output from the image encoder can be expressed by Equation 15:
To effectively integrate global and local features from the image encoder, we designed the Feature Fusion Module (FFM), as depicted in Figure 6. This module balances global and local features, enabling the model to learn the diverse distributions of disease spots in leaf images. To enhance feature flexibility, we incorporated a SE Block into the FFM. The SE Block adjusts channel-wise feature weights, emphasizing key features. The weight adjustment progressively increases over training epochs, guided by an epoch-based scaling factor, ensuring the model adapts to the influence of the SE Block.
In the FFM, the Cross-Branch Attention (CBA) replaces the Multi-head Attention in the original transformer block. These two are essentially the same in nature, except that the keys (K) and values (V) input to the CBA are derived from the local feature extraction module. To emphasize the interaction between different branches, we refer to this module as Cross-Branch Attention. We first apply CBA to interact between global features and local features . Using query (Q), key, and value, CBA uncovers effective complementary information across different branches, given in Equations 16 and 17:
Where , , and are the learnable weight matrices, while represents the feature dimension used to scale the inner product.
For the initially interacted features, we employ the SE Block to implement dynamic weight adjustment. The SE Block dynamically adjusts the importance of global and local features based on training epochs, enhancing the adaptability and robustness of the model’s information processing. The operations of the SE Block can be expressed by Equations 18 and 19:
Where represents the global average pooling result of the feature vector. and denote the Sigmoid and ReLU activation functions, respectively. and correspond to the weights of fully connected layers. is a dynamic coefficient associated with the training epochs, controlled by an incremental function in the form of exponential growth, expressed as follows:
In the above equation, represents the current training epoch, and is a hyperparameter controlling the growth rate.
Although the SAM mask decoder effectively generates segmentation masks through multi-layer cross-attention mechanisms, it lacks the ability to provide class-specific predictions. To enhance disease segmentation efficiency, which requires both precise lesion delineation and accurate disease type classification, we introduce the Mask-Class Hybrid Decoder. This architecture integrates classification functionalities into the SAM mask decoder, as shown in Figure 7.
The SAM mask decoder utilizes self-attention and bidirectional cross-attention to extract interactive features between image and prompt embeddings. These features are processed by an upsampling convolution module to produce binary segmentation masks. However, this approach does not offer class-specific predictions. To address this, we propose adding a lightweight classification head to the existing mask decoder architecture. This head consists of two fully connected layers preceded by a global average pooling layer for dimensionality reduction. By projecting the image embeddings through these layers, we obtain class probability distributions corresponding to the generated segmentation masks. The lightweight design ensures minimal additional training overhead while enabling concurrent classification tasks effectively.
To simultaneously optimize the performance of segmentation and classification tasks, we define a joint loss function , which comprises three components: segmentation loss , IoU loss , and classification loss . These components are linearly combined as follows:
Here, , , and are the weights assigned to each loss component.
The segmentation loss serves as the primary objective function, focusing on the precision of boundary delineation and the consistency of segmented regions. We adopt a combination of Dice Loss and Binary Cross-Entropy (BCE) Loss for the segmentation loss, optimizing both the overlap rate of target regions and pixel-level classification accuracy, as illustrated in Equation 22:
Dice Loss emphasizes the overlap between the target region and the prediction, addressing the imbalance between foreground and background areas, which is suitable for segmentation tasks like lesion segmentation where the foreground area is relatively small. BCE Loss measures the correctness of each pixel classification, suitable for scenarios where the foreground and background are relatively balanced. Dice Loss and BCE Loss are defined below, respectively, as shown in Equations 23 and 24:
Where and represent the predicted value and the ground truth value of the i-th pixel, respectively.
IoU loss aims to further enhance the model’s prediction of the overlap ratio between the predicted and ground truth regions, effectively focusing on boundary information. This process is illustrated in Equation 25:
Where represents the number of pixels in the predicted region, denotes the number of pixels in the ground truth region, and correspond to the number of pixels in the intersection and union of the predicted and ground truth regions, respectively.
The classification loss extends the capability of the segmentation task by optimizing the prediction of class labels through cross-entropy loss, as shown in Equation 26:
Where is the number of classes, and are the true label and the predicted label for class c, respectively.
To evaluate the performance of EMSAM in leaf disease image segmentation and classification tasks, we employ three categories of evaluation metrics: Dice Coefficient (Dice), Intersection over Union (IoU), and Accuracy (Acc).
The Dice is a metric used to measure the overlap between the predicted segmentation and the ground truth, reflecting the model’s ability to accurately predict the foreground regions. The expression is given by Equation 27:
The IoU is a metric that measures the ratio of the intersection to the union between the predicted results and the ground truth segmentation. Compared to the Dice Coefficient, IoU places greater emphasis on strict matching of boundary regions, making it suitable for evaluating the model’s precision in delineating the boundaries of diseased areas. The equation is given by Equation 28:
Acc refers to the proportion of correctly classified labels for a given instance averaged over the total number of labels (including both predicted and actual values). The calculation equation is given by Equation 29:
Where TP (True Positives) and TN (True Negatives) represent correctly predicted positive and negative instances, respectively, while FP (False Positives) and FN (False Negatives) represent incorrectly predicted instances.
By employing these metrics, we can comprehensively assess both the segmentation and classification performance of the EMSAM model.
The hardware configuration for our experiments is as follows: CPU - Intel® Core™ i7-13700KF @ 5.4GHz, GPU - NVIDIA GeForce RTX 3090 (24GB), and memory - 32GB Samsung DDR5 5600MHz (16GB×2). The experimental environment is set up on an Ubuntu 20.04.6 LTS 64-bit operating system, using Python 3.11 as the programming language, PyTorch 2.0.1 as the deep learning framework, CUDA Toolkit version 12.2, and cuDNN version 8.9.0.
During training, the primary parameter settings are as follows: we select the ViT-b image encoder as the pre-trained model, with an input image size of 256×256 pixels. The patch size for the patch embedding block is set to 16, and the windowed attention size is 14×14. The batch size is set to 4, and the total number of training epochs, including those for comparative experiments, is 200. The training process utilizes the AdamW optimizer with an initial learning rate of 0.0005, adopting an exponential decay learning rate scheduling strategy. The learning rate at the -th iteration is defined as: . Where refers to the learning rate at the -th iteration, is the initial learning rate at the start of training, is the decay rate constant, and represents the current training iteration number. This setup facilitates rapid convergence in the early stages of training, helping the model quickly locate a favorable region in the parameter space and avoid over-reliance on local minima. As training progresses, the learning rate gradually decreases, reducing the step size of model updates, which aids in fine-tuning model parameters and effectively prevents oscillations and overfitting in the later stages of training.
We compare EMSAM with six other models to evaluate its effectiveness in segmenting diseased leaf regions. The models include SAMUS (Lin et al., 2024) and MedSAM (Ma et al., 2024) (SAM extensions), Swin-Unet (Cao et al., 2023) and Trans-Unet (Chen et al., 2021) (ViT-based), DeepLabv3+ (Chen et al., 2018b) (ResNet-based), and HRNet-48 (Wang et al., 2021) (HRNet-based). The comparison focuses on three key metrics: Dice coefficient, IoU score, and the backbone networks utilized by each model. These metrics collectively assess the feature extraction and segmentation effect of each model. The quantitative experimental results are presented in Table 2.
From Table 2, it is evident that EMSAM outperforms all other models on the PSD. Leveraging deep CNN-ViT feature fusion, EMSAM effectively captures both global and local features, highlighting the efficacy of combining CNN and ViT for plant disease segmentation. Models relying solely on ViT, such as MedSAM and Trans-Unet, exhibit lower Dice and IoU scores due to insufficient attention to local features. While DeepLabv3+ achieves an IoU of 0.6504, its ResNet-50 backbone struggles with fine-grained boundary information. Swin-Unet and Trans-Unet show relatively lower performance, indicating room for improvement in segmenting complex disease images. HRNet-48, with Dice and IoU scores of 0.6911 and 0.5643 respectively, demonstrates advantages in multi-scale feature extraction but still falls short of EMSAM’s overall performance. Notably, EMSAM achieves the highest Dice coefficient of 0.7925, surpassing the second-best model, DeepLabv3+, by 2.45%, and outperforming the lowest-performing Swin-Unet by 31.92%. This underscores EMSAM’s superior overall match quality for segmented regions, particularly in boundary detection and small region segmentation. Additionally, its IoU of 0.6987 surpasses DeepLabv3+ and Swin-Unet by 6.91% and 35.84%, respectively.
This study compares EMSAM with SOTA methods in terms of total parameters, learnable parameters, and floating-point operations (FLOPs), as detailed in Table 3. EMSAM has a total of 589.6M parameters and 133.9M learnable parameters, demonstrating its ability to capture complex features. Its computational cost of 322.5G FLOPs is higher than that of smaller models such as DeepLabv3+ and Swin-Unet but significantly lower than larger models like MedSAM and Trans-Unet. As a transformer-based model, EMSAM has the highest overall parameter count among these models. However, the adoption of parameter-efficient fine-tuning techniques and lightweight module designs helps keep the number of learnable parameters at a moderate level. Overall, EMSAM achieves a balance between accuracy and model complexity by maintaining sufficient representational capacity while optimizing parameter efficiency and computational cost. This makes it particularly suitable for challenging segmentation tasks involving ambiguous lesion boundaries and diverse morphological variations.
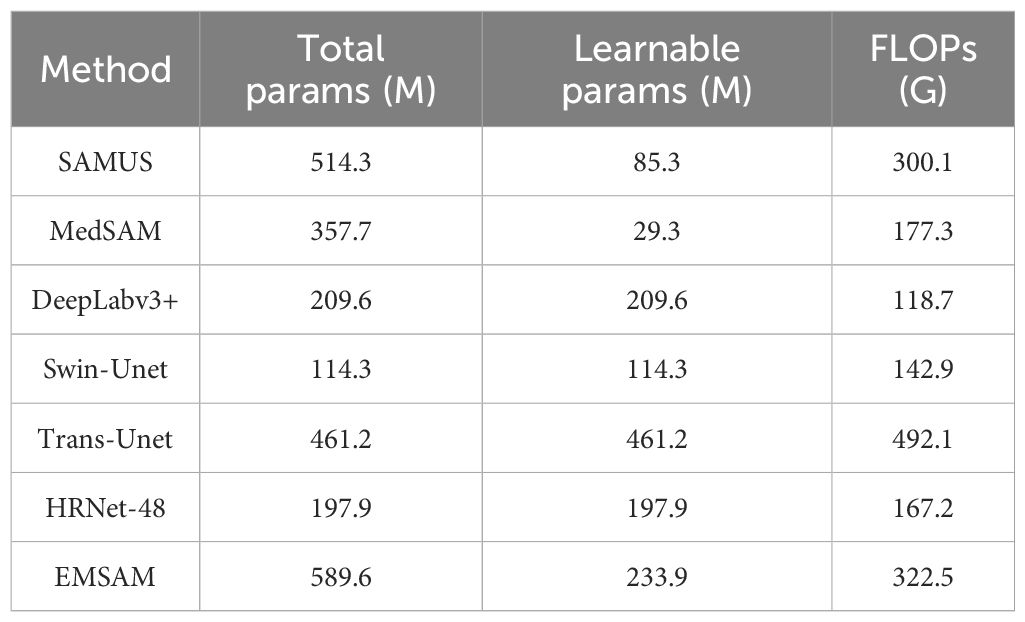
Table 3. Comparison of EMSAM and SOTA methods in terms of model parameters, learnable parameters, and training resource consumption.
Figure 8 demonstrates that EMSAM achieves refined pixel-level segmentation, effectively capturing both global and local features while maintaining the integrity of disease regions and preserving superior boundary details. Compared to EMSAM, SAMUS exhibits difficulties in detecting small lesions, while MedSAM’s sensitivity to noise results in false boundary delineations. In the first row, representing small and fragmented lesions, EMSAM outperforms the other models with accurate segmentation. In the second row, depicting large and concentrated lesions, EMSAM excels in preserving boundary details, while DeepLabv3+, although robust in segmenting large areas, struggles with fine-grained precision. In the third row, depicting mixed feature distributions, EMSAM accurately segments disease regions while avoiding noise, unlike HRNet-48, which exhibits noticeable lesion adhesion.
To validate EMSAM’s adaptability in segmenting leaf diseases across different severity levels, we reclassified the test set based on the disease severity classification method outlined in Section 2.1, categorizing the leaves into light, moderate, and severe severity levels. A detailed analysis of each model’s performance under these conditions is presented in Table 4, showing the Dice and IoU scores for different models across the three severity levels.
Under light disease conditions, DeepLabv3+ achieves the highest Dice score of 0.7564 and IoU of 0.6372, significantly outperforming other models and demonstrating superior fine feature capture. EMSAM follows closely, exhibiting high stability, while Swin-Unet records the lowest performance with an IoU of 0.3195, indicating challenges in handling small lesion areas. In medium disease conditions, EMSAM achieves Dice and IoU scores of 0.8354 and 0.7365, respectively, outperforming MedSAM, which shows a relatively lower IoU of 0.5213, highlighting limited robustness in moderately complex lesions. For severe disease conditions, EMSAM maintains its leading performance with Dice and IoU scores of 0.8187 and 0.7342, respectively, showcasing strong adaptability and generalization in complex scenarios Figure 9 presents visual segmentation outcomes across varying disease severity levels, revealing model performance disparities. MedSAM and Swin-Unet, sensitive to foreground-background discrepancies, exhibit poor performance on leaves with ambiguous boundaries. In light disease scenarios, CNN-based models such as DeepLabv3+ and HRNet-48 excel at capturing small lesion areas, while EMSAM demonstrates a more balanced performance. For medium severity cases, EMSAM performs notably well, whereas MedSAM struggles with indistinct lesion boundaries. Under severe disease conditions, Trans-Unet and SAMUS experience over-segmentation due to heightened sensitivity to complex backgrounds, hindering performance improvement.
In Section 2.2.3, after introducing the SE Block, we design a dynamic coefficient to control the growth process of Epoch-based Num, as expressed in Equation 20. Additionally, in Section 2.2.1, we introduce a joint loss function to simultaneously cater to segmentation and classification tasks, as defined in Equation 21. These equations introduce two new hyperparameters to EMSAM: the growth rate control parameter and the weights for the loss component. To investigate the impact of dynamic and fixed parameter settings on model performance, we designed relevant experiments and visualized key training indicators, as shown in Figure 10. The left plot shows the Dice score variation over epochs, while the right plot shows the loss variation over epochs. In this experiment, we selected three values (0.1, 0.5, and 0.9) to control the influence of the SE block on the training process. When =0.1, the SE block has a relatively minor effect on the overall model training, allowing the Dice score to increase rapidly in the early training phase while maintaining relatively high segmentation accuracy in later stages. Moreover, the overall training process remains stable. In contrast, when =0.9, the SE block exerts a much stronger influence on the model training, leading to significantly lower Dice scores compared to other settings. This suggests that an excessively strong influence from the SE block during the early training phase can cause the model to become overly sensitive to its effects, making the optimization process too slow to adapt to later-stage loss variations. Consequently, the model fails to fully leverage the feature representations learned in the early stages, which ultimately compromises performance.
The experimental results in Figure 10 indicate that dynamically adjusting the influence of the SE block during training is essential for achieving a better balance between convergence speed and final accuracy. To observe the effects of the dynamic parameter on model performance during training, we experiment with different values for each and conduct training sessions to identify the appropriate hyperparameter settings. Figure 11 illustrates the impact of varying values on the model’s loss function and Dice Coefficient.
As shown in the figure above, different values of have a significant impact on the model’s early convergence speed and the final stable value. When , the convergence speed is the fastest, but the final loss value is higher than those of other hyperparameter settings, which suggests that the model may converge too quickly, leading to underfitting. In contrast, and exhibit lower final loss values. Considering the effect of different values on the Dice, contributes more effectively to optimizing the model’s performance, striking a good balance between training efficiency and segmentation performance. Table 5 presents the effects of different weight settings for the three components of the joint loss function on model performance.
From Table 5, it is evident that the weights and positively influence segmentation performance, while the Acc metric does not exhibit a clear monotonic trend, instead fluctuating based on the weight combinations. Setting , , effectively balances the weight distribution between segmentation and classification tasks, resulting in notable performance improvements for the model.
In EMSAM, the core components are the proposed MAA, LFEM, and FFM. Table 6 presents ablation experiment results on the PSD, analyzing the contributions of these components. The baseline model, shown in the first row, employs traditional adapter tuning for SAM transfer (Chen et al., 2023a). The second row incorporates MAA for adapter tuning, validating its effectiveness in extracting multi-scale information. The third row introduces LFEM as an efficient detail feature supplement, utilizing standard cross-modal attention for information fusion. The fourth row further incorporates an SE-based attention mechanism into the information fusion process to prevent detail loss during feature integration.
To illustrate the contributions of EMSAM’s core components, we present a visual analysis of segmentation performance across different model configurations. Figure 12 provides a visual comparison of segmentation outcomes for EMSAM’s core components: MAA, LFEM, and FFM. The baseline model shows notable limitations in segmenting complex lesion regions, particularly in capturing boundary details and detecting small lesions. Incorporating the MAA module substantially improves segmentation performance, enabling more effective extraction of multi-scale features compared to traditional adapter tuning methods. Adding the LFEM further enhances the model’s ability to process details and boundaries, leading to more precise local feature extraction, particularly in small lesion areas and at lesion edges. Incorporating the FFM module effectively integrates features from the CNN and ViT branches, resulting in the best overall segmentation performance for both lesion area segmentation and boundary detail capture.
The ablation study demonstrates that the introduced components (MAA, LFEM, and FFM) each significantly improve segmentation performance. MAA enhances multi-scale feature extraction in complex lesion regions, LFEM boosts local feature extraction and boundary processing, and FFM facilitates efficient channel-level feature fusion between CNN and ViT branches. Collectively, these modules synergize to elevate the model’s segmentation capabilities, particularly in handling complex and detailed disease regions.
In this study, we propose EMSAM to address the challenges of blurred boundaries and complex shapes in leaf disease images. Built upon the SAM architecture, EMSAM achieves superior performance over state-of-the-art segmentation algorithms on the PSD, demonstrating robust generalization across varying disease severities and diverse scenarios. The MAA, specifically designed for leaf disease image processing, efficiently captures blurred boundary features through a multi-scale information extraction module. The LFEM employs lightweight convolutional groups to extract fine-grained features, complementing the global information captured by ViT blocks and enabling balanced attention to both global and local features. The FFM uses SE blocks to dynamically balance the weights between CNN and ViT branches, effectively reducing redundancy. Lastly, a lightweight classification head in the decoder integrates segmentation and classification tasks for efficient multi-task learning.
Despite the promising performance of EMSAM in leaf disease segmentation, several limitations need to be acknowledged: (1) The study primarily relies on the PlantVillage dataset, which, while widely used in plant disease research, consists of images captured in controlled environments with simple and uniform backgrounds. This limits the model’s ability to generalize to real-world agricultural settings where lighting variations, occlusions, and complex backgrounds pose additional challenges. Future work should evaluate EMSAM on diverse field datasets to enhance its robustness and applicability. (2) Although EMSAM integrates parameter-efficient tuning techniques, the inclusion of MAA, FFM, and attention mechanisms increases computational overhead compared to standard CNN-based approaches. The additional FLOPs and memory requirements may hinder real-time deployment on edge devices with limited resources. Future optimizations, such as knowledge distillation or pruning, could help reduce inference costs while maintaining performance. To further enhance the capabilities of EMSAM, future research could explore two primary avenues. Firstly, expanding the training dataset to encompass a broader range of plant species and environmental conditions would likely bolster the model’s generalization across diverse scenarios. Secondly, exploring more efficient multi-task learning paradigms could optimize performance on auxiliary tasks without increasing model complexity, thereby enhancing both primary and auxiliary task outcomes. These directions hold the potential to significantly advance the effectiveness and applicability of EMSAM in real-world settings. Overall, EMSAM presents innovative research directions and technical frameworks to enhance image processing in leaf disease segmentation, advancing the development of more efficient and accurate disease detection solutions.
In addition to its demonstrated superiority in leaf disease segmentation, the proposed EMSAM framework exhibits considerable potential for broader real-world applications. The integration of multi-scale adaptive modules and hybrid feature extraction—combining CNN-based local detail capture with ViT-based global context modeling—enables EMSAM to effectively handle images characterized by blurred boundaries and complex object shapes. Consequently, this methodology is not only well-suited for plant disease detection in precision agriculture but can also be readily adapted to other challenging segmentation tasks. For instance, its robust performance in delineating fine structures makes it a promising candidate for medical image analysis (e.g., tumor or lesion segmentation), remote sensing applications (e.g., land cover mapping), and industrial quality control (e.g., defect detection). The modular design and parameter-efficient tuning strategy further facilitate customization for domain-specific requirements, even in scenarios with limited annotated data.
The datasets presented in this article are not readily available because the dataset is part of ongoing research efforts and cannot be shared publicly until these studies are completed. Requests to access the datasets should be directed to Li Junlong, bGlqdW5sb25nMTMyMUAxNjMuY29t.
JL: Writing – original draft, Conceptualization, Formal Analysis, Investigation, Software. QF: Conceptualization, Formal Analysis, Methodology, Software, Writing – original draft, Writing – review & editing. JZ: Conceptualization, Methodology, Writing – review & editing. SY: Conceptualization, Formal Analysis, Investigation, Validation, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No.32160421).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Balasundaram, A., Sundaresan, P., Bhavsar, A., Mattu, M., Kavitha, M. S., Shaik, A. (2025). Tea leaf disease detection using segment anything model and deep convolutional neural networks. Results Eng. 25, 103784. doi: 10.1016/j.rineng.2024.103784
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., et al. (2023). “Swin-unet: Unet-like pure transformer for medical image segmentation,” in Computer Vision – ECCV 2022 Workshops. Eds. Karlinsky, L., Michaeli, T., Nishino, K. (Springer Nature Switzerland, Cham), 205–218. doi: 10.1007/978-3-031-25066-8_9
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., et al. (2021). ). TransUNet: Transformers make strong encoders for medical image segmentation. doi: 10.48550/arXiv.2102.04306
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A. L. (2018a). DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40, 834–848. doi: 10.1109/TPAMI.2017.2699184
Chen, T., Zhu, L., Deng, C., Cao, R., Wang, Y., Zhang, S., et al. (2023a). SAM-adapter: Adapting segment anything in underperformed scenes. Available online at: https://openaccess.thecvf.com/content/ICCV2023W/VCL/html/Chen_SAM-Adapter_Adapting_Segment_Anything_in_Underperformed_Scenes_ICCVW_2023_paper.html (Accessed January 12, 2025).
Chen, T., Zhu, L., Ding, C., Cao, R., Wang, Y., Li, Z., et al. (2023b). SAM fails to segment anything? – SAM-adapter: Adapting SAM in underperformed scenes: camouflage, shadow, medical image segmentation, and more. doi: 10.48550/arXiv.2304.09148
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H. (2018b). Encoder-decoder with atrous separable convolution for semantic image segmentation. Available online at: https://openaccess.thecvf.com/content_ECCV_2018/html/Liang-Chieh_Chen_Encoder-Decoder_with_Atrous_ECCV_2018_paper.html (Accessed January 12, 2025).
Cheng, J., Ye, J., Deng, Z., Chen, J., Li, T., Wang, H., et al. (2023). SAM-med2D. doi: 10.48550/arXiv.2308.16184
Divyanth, L. G., Ahmad, A., Saraswat, D. (2023). A two-stage deep-learning based segmentation model for crop disease quantification based on corn field imagery. Smart Agric. Technol. 3, 100108. doi: 10.1016/j.atech.2022.100108
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al. (2021). An image is worth 16x16 words: Transformers for image recognition at scale. doi: 10.48550/arXiv.2010.11929
Giménez-Gallego, J., González-Teruel, J. D., Jiménez-Buendía, M., Toledo-Moreo, A. B., Soto-Valles, F., Torres-Sánchez, R. (2020). Segmentation of multiple tree leaves pictures with natural backgrounds using deep learning for image-based agriculture applications. Appl. Sci. 10, 202. doi: 10.3390/app10010202
Gui, B., Bhardwaj, A., Sam, L. (2024). Evaluating the efficacy of segment anything model for delineating agriculture and urban green spaces in multiresolution aerial and spaceborne remote sensing images. Remote Sens. 16, 414. doi: 10.3390/rs16020414
Hao, Y., Liu, Y., Chen, Y., Han, L., Peng, J., Tang, S., et al. (2022). EISeg: An efficient interactive segmentation tool based on PaddlePaddle. doi: 10.48550/arXiv.2210.08788
Hu, J., Shen, L., Sun, G. (2018).Squeeze-and-excitation networks. Available online at: https://openaccess.thecvf.com/content_cvpr_2018/html/Hu_Squeeze-and-Excitation_Networks_CVPR_2018_paper.html (Accessed January 12, 2025).
Hughes, D. P., Salathe, M. (2016). An open access repository of images on plant health to enable the development of mobile disease diagnostics. doi: 10.48550/arXiv.1511.08060
Ji, M., Wu, Z. (2022). Automatic detection and severity analysis of grape black measles disease based on deep learning and fuzzy logic. Comput. Electron. Agric. 193, 106718. doi: 10.1016/j.compag.2022.106718
Jiang, Y., Chen, Y., Du, C., Zhao, Y., Zhuang, S., Yin, Z. (2023). “Automatic image screening of pine wilt disease based on TransUNet,” in 2023 9th International Conference on Control Science and Systems Engineering (ICCSSE). USA: IEEE. 281–286. doi: 10.1109/ICCSSE59359.2023.10245235
Kirillov, A., Mintun, E., Ravi, N., Mao, H., Rolland, C., Gustafson, L., et al. (2023). Segment anything. Available online at: https://openaccess.thecvf.com/content/ICCV2023/html/Kirillov_Segment_Anything_ICCV_2023_paper.html (Accessed January 12, 2025).
Li, Y., Wang, D., Yuan, C., Li, H., Hu, J. (2023). Enhancing agricultural image segmentation with an agricultural segment anything model adapter. Sensors 23, 7884. doi: 10.3390/s23187884
Lin, X., Xiang, Y., Yu, L., Yan, Z., et al. (2024). “Beyond adapting SAM: Towards end-to-end ultrasound image segmentation via auto prompting,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. Eds. Linguraru, M. G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K. (Springer Nature Switzerland, Cham), 24–34. doi: 10.1007/978-3-031-72111-3_3
Liu, W., Yu, L., Luo, J. (2022). A hybrid attention-enhanced DenseNet neural network model based on improved U-net for rice leaf disease identification. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.922809
Ma, J., He, Y., Li, F., Han, L., You, C., Wang, B. (2024). Segment anything in medical images. Nat. Commun. 15, 654. doi: 10.1038/s41467-024-44824-z
Moupojou, E., Retraint, F., Tapamo, H., Nkenlifack, M., Kacfah, C., Tagne, A. (2024). Segment anything model and fully convolutional data description for plant multi-disease detection on field images. IEEE Access 12, 102592–102605. doi: 10.1109/ACCESS.2024.3433495
Ngo, B. H., Do-Tran, N.-T., Nguyen, T.-N., Jeon, H.-G., Choi, T. J. (2024). Learning CNN on ViT: A hybrid model to explicitly class-specific boundaries for domain adaptation. Available online at: https://openaccess.thecvf.com/content/CVPR2024/html/Ngo_Learning_CNN_on_ViT_A_Hybrid_Model_to_Explicitly_Class-specific_CVPR_2024_paper.html (Accessed January 12, 2025).
Osco, L. P., Wu, Q., de Lemos, E. L., Gonçalves, W. N., Ramos, A. P. M., Li, J., et al. (2023). The segment anything model (SAM) for remote sensing applications: From zero to one shot. Int. J. Appl. Earth Observation Geoinformation 124, 103540. doi: 10.1016/j.jag.2023.103540
Pal, A., Kumar, V. (2023). AgriDet: Plant leaf disease severity classification using agriculture detection framework. Eng. Appl. Artif. Intell. 119, 105754. doi: 10.1016/j.engappai.2022.105754
Pal, A., Kumar, V., Hassan, K. L., Singh, B. K. (2024). A framework for leaf disease analysis and estimation using MAML with DeepLabV3. Microsyst Technol. doi: 10.1007/s00542-024-05686-z
Pang, J., Bai, Z., Lai, J., Li, S. (2011). “Automatic segmentation of crop leaf spot disease images by integrating local threshold and seeded region growing,” in 2011 International Conference on Image Analysis and Signal Processing. USA: IEEE. 590–594. doi: 10.1109/IASP.2011.6109113
Revathi, P., Hemalatha, M. (2012). “Classification of cotton leaf spot diseases using image processing edge detection techniques,” in 2012 International Conference on Emerging Trends in Science, Engineering and Technology (INCOSET). USA: IEEE. 169–173. doi: 10.1109/INCOSET.2012.6513900
Ronneberger, O., Fischer, P., Brox, T. (2015). “U-net: Convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Eds. Navab, N., Hornegger, J., Wells, W. M., Frangi, A. F. (Springer International Publishing, Cham), 234–241. doi: 10.1007/978-3-319-24574-4_28
Shoaib, M., Hussain, T., Shah, B., Ullah, I., Shah, S. M., Ali, F., et al. (2022). Deep learning-based segmentation and classification of leaf images for detection of tomato plant disease. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1031748
Singh, V., Misra, A. K. (2017). Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 4, 41–49. doi: 10.1016/j.inpa.2016.10.005
Sung, Y.-L., Cho, J., Bansal, M. (2022). VL-adapter: Parameter-efficient transfer learning for vision-and-language tasks. Available online at: https://openaccess.thecvf.com/content/CVPR2022/html/Sung_VL-Adapter_Parameter-Efficient_Transfer_Learning_for_Vision-and-Language_Tasks_CVPR_2022_paper.html (Accessed January 12, 2025).
Wang, J., He, J., Han, Y., Ouyang, C., Li, D. (2013). An adaptive thresholding algorithm of field leaf image. Comput. Electron. Agric. 96, 23–39. doi: 10.1016/j.compag.2013.04.014
Wang, J., Sun, K., Cheng, T., Jiang, B., Deng, C., Zhao, Y., et al. (2021). Deep high-resolution representation learning for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 43, 3349–3364. doi: 10.1109/TPAMI.2020.2983686
Wang, D., Zhang, J., Du, B., Xu, M., Liu, L., Tao, D., et al. (2023). SAMRS: Scaling-up remote sensing segmentation dataset with segment anything model. doi: 10.48550/arXiv.2305.02034
Wu, J., Ji, W., Liu, Y., Fu, H., Xu, M., Xu, Y., et al. (2023). Medical SAM adapter: Adapting segment anything model for medical image segmentation. doi: 10.48550/arXiv.2304.12620
Xu, L., Xie, H., Qin, S.-Z. J., Tao, X., Wang, F. L. (2023). Parameter-efficient fine-tuning methods for pretrained language models: A critical review and assessment. doi: 10.48550/arXiv.2312.12148
Yang, Y., Wang, C., Zhao, Q., Li, G., Zang, H. (2024). SE-SWIN UNET FOR IMAGE SEGMENTATION OF MAJOR MAIZE FOLIAR DISEASES. Eng. Agríc. 44, e20230097. doi: 10.1590/1809-4430-eng.agric.v44e20230097/2024
Yuan, H., Zhu, J., Wang, Q., Cheng, M., Cai, Z. (2022). An improved deepLab v3+ Deep learning network applied to the segmentation of grape leaf black rot spots. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.795410
Zhang, C., Cho, J., Puspitasari, F. D., Zheng, S., Li, C., Qiao, Y., et al. (2024). A survey on segment anything model (SAM): Vision foundation model meets prompt engineering. doi: 10.48550/arXiv.2306.06211
Zhang, Y., Jiao, R. (2023). Towards segment anything model (SAM) for medical image segmentation: A survey. doi: 10.48550/arXiv.2305.03678
Zhang, K., Liu, D. (2023). Customized segment anything model for medical image segmentation. doi: 10.48550/arXiv.2304.13785
Keywords: segment anything model, parameter efficient fine-tuning, adapter tuning, leaf disease segmentation, multi-task learning
Citation: Li J, Feng Q, Zhang J and Yang S (2025) EMSAM: enhanced multi-scale segment anything model for leaf disease segmentation. Front. Plant Sci. 16:1564079. doi: 10.3389/fpls.2025.1564079
Received: 21 January 2025; Accepted: 26 February 2025;
Published: 14 March 2025.
Edited by:
Huajian Liu, University of Adelaide, AustraliaReviewed by:
Arunangshu Pal, Tripura, IndiaCopyright © 2025 Li, Feng, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Feng, ZnF1YW5AZ3NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.