- 1Hunan Engineering & Technology Research Center for Agricultural Big Data Analysis & Decision-Making, College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, Food Crop Institute, Hubei Academy of Agricultural Sciences, Wuhan, China
- 3Crop Phenomics Research Center, Huazhi Biotechnology Co., Ltd, Changsha, China
Introduction: Thioredoxins (TRX) are redox-active proteins critical for plant stress adaptation. As a TRX family member, nucleoredoxin (NRX) maintains drought-induced redox homeostasis, yet its genome-wide characterization in rice remains uninvestigated.
Methods: Using HMMER3.0 (E-value <1e-5) and TBtools, we identified OsNRX genes across three rice varieties (Minghui63, Nipponbare, 9311). Conserved domains were verified by SMART/CDD, while promoter cis-elements were systematically predicted with PlantCARE. Tissue-specific expression patterns were analyzed using RiceXPro data, and drought responses were quantified via qRT-PCR in drought-tolerant (Jiangnong Zao 1B) versus sensitive (TAISEN GLUTINOUS YU 1157) varieties under PEG6000 stress.
Results: Ten OsNRX genes were classified into three subfamilies (NRX1/NRX2/NRX3) exhibiting conserved domain architectures. Promoter analysis identified abundant stress-responsive elements (ABRE, MBS) and phytohormone signals (ABA/JA/SA). Tissue-specific expression profiles revealed NRX1a dominance in roots/hulls, versus NRX1b/NRX2 enrichment in endosperm. Drought stress triggered rapid OsNRX upregulation (20-53-fold within 3-6h), with tolerant varieties showing earlier NRX2 activation than sensitive counterparts.
Discussion: OsNRX genes exhibit dynamic drought-responsive regulation, while their spatiotemporal expression in glumes, embryos, and endosperm suggests potential dual roles in stress adaptation and grain development. These results provide molecular targets for improving drought resilience in rice breeding.
Introduction
Nucleoredoxin (NRX), initially discovered by Kurooka et al. (1997) in the nucleus of mouse (Mus musculus) cells, is named for its ability to reduce oxygen and is classified within the thioredoxin (TRX) superfamily due to its TRX-like domain. The TRX superfamily is involved in a series of physiological and biochemical processes within the cell that are associated with redox reactions, by reducing disulfide bonds in their target proteins (Piaz et al., 2010). Laughner et al. (1998) were the first to isolate plant nucleoredoxin from maize (Zea mays), confirming that NRX proteins possess significant insulin-degrading activity and the capacity to catalyze redox reactions. Marchal et al. (2014) categorized NRX into three subgroups based on the number of TRX-like domains: Type I NRX contains three TRX-like domains, with the first and third domains harboring typical WC[G/P]PC redox sites, while the second domain is an atypical TRX domain with weaker activity (Cheng et al., 2021a); Type II NRX contains two TRX domains with atypical active sites WY[P/A][K/P]C and [W/R/H]C[L/A/V/R]P[C/G]; Type III NRX contains three TRX domains, with the first domain having a highly conserved WCRPC redox site, and the second domain typically containing a classic Trx active site WCPP[C/F/S].
NRX plays a crucial role in plants’ response to drought stress. Zhang et al. (2021) constructed overexpression and RNA interference wheat lines for TaNRX1 to investigate its mechanism in regulating wheat drought tolerance. The results indicated that the overexpression lines had higher drought tolerance indicators such as survival rate, leaf chlorophyll, proline, soluble sugar content, and antioxidant enzyme activity compared to the wild type, while the RNAi lines showed the opposite, demonstrating that TaNRX1 positively regulates wheat’s tolerance to drought stress. Tian et al. (2019) demonstrated that TaNRX1 interacts with TaPDI, TaTRX-h, and TaPP2Ac proteins in the nucleus and cell membrane, and that the overexpression of TaPDI, TaTRX-h, and TaPP2Ac alleviated the inhibitory effects of drought stress on yeast growth. In addition, Zhang et al. (2022) showed that overexpression of SiNRX1 in foxtail millet (Setaria italica) enhanced Arabidopsis tolerance to drought and salt stress. Jedmowski et al. (2014) conducted drought stress experiments on two sorghum (Sorghum bicolor) varieties with differing drought tolerance and found that SbNRX expression significantly increased during both stress and recovery phases in the drought-tolerant variety, whereas in the drought-sensitive variety, SbNRX expression increased only during the stress phase.
NRX is also involved in regulating plant growth and development and responding to other stresses. In rice (Oryza sativa), NRX regulates cytokinin signaling by inhibiting the dimerization of the Oryza sativa histidine kinase 4 (OHK4), thereby influencing regeneration capacity and leaf senescence (Yao et al., 2023). In Arabidopsis, AtNRX1 has been shown to integrate signals from maternal tissues and guide pollen tube growth toward the ovule, thereby regulating pollen tube growth and pollen fertility (Qin et al., 2009). AtNRX2 is specifically induced by jasmonic acid (JA) and promotes trichome formation by regulating the expression of genes related to trichome formation (Lee et al., 2020). In tomato (Lycopersicon esculentum), SLNRX1 is a positive regulator of heat stress tolerance, protecting cells from oxidative damage and protein denaturation induced by heat stress by enhancing antioxidant capacity (Cha et al., 2022). In grape (Vitis vinifera), Gauthier et al. (2014) found that after inoculation with grape downy mildew Plasmopara viticola, the expression of NRX1 was specifically induced by salicylic acid (SA), thereby improving grape resistance to downy mildew.
Rice is a major staple crop worldwide and serves as the primary energy source for more than half of the global population (Hassan et al., 2023). Due to its high water consumption during production, rice yield is highly susceptible to drought (Panda et al., 2021). To date, studies on the NRX gene family have primarily been derived from research on the TRX family, but a systematic investigation of the NRX gene family in rice remains lacking. Therefore, this study aims to identify the OsNRX gene family and investigate its role in rice’s response to drought stress, providing a foundation for in-depth functional analysis of rice NRX genes and offering scientific support for molecular breeding to enhance drought resistance in rice. Using bioinformatics methods, this study identified the NRX genes in the rice genome and conducted a comprehensive analysis of their protein physicochemical properties, subcellular localization, protein structure, conserved domains, phylogenetic tree and cis-acting elements. Furthermore, the expression patterns of the OsNRX gene family under drought stress were examined to elucidate the regulatory effects of drought stress on OsNRX gene expression.
Materials and methods
Data sources
Genomic data and annotation files for the rice varieties Nipponbare and 9311 were downloaded from the Ensembl Plants database (http://plants.ensembl.org/info/data/ftp/index.html) (Yates et al., 2022), accessed on 2023.7.27. The whole-genome data and its annotation files for the rice variety Minghui63 were obtained from the Rice Information Network RIGW (http://rice.hzau.edu.cn/cgi-bin/rice_rs1/download_ext) (Song et al., 2018), accessed on 2023.7.27. The HMM file for the NRX gene (PF13905) was downloaded from the InterPro database (https://www.ebi.ac.uk/interpro) (Paysan-Lafosse et al., 2023), accessed on 2023.7.27. The genotype data used for SNP Analysis were obtained from Meng et al. (2022).
Identification of OsNRX gene family members
The hmmsearch program of HMMER3.0 and the Blast Compare Two Seqs program of TBtools are employed to search for proteins similar to the NRX domain within the total protein sequences of rice (Eddy, 2011; Chen et al., 2020), with an E-value of 1×10-5 used to filter the search results, yielding candidate rice NRX gene family members. The intersection of the two results is the final set of candidate gene family members. The protein sequences of the final set of candidate gene family members were submitted to the SMART (http://smart.embl.de) (Letunic et al., 2021) and CDD (https://www.ncbi.nlm.nih.gov/cdd) (Wang et al., 2023) databases, and sequences containing the NRX domain were confirmed as rice NRX gene family members.
Physicochemical property analysis and subcellular localization of OsNRX proteins
The online software ExPASy (http://web.expasy.org/protparam) (Duvaud et al., 2021) was utilized to analyze the protein sequences encoded by the rice NRX genes, predicting their relative molecular weight, isoelectric point, instability index, grand average of hydropathy, and hydrophobicity. The online tool Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi) (Chou and Shen, 2010) was used for subcellular localization prediction.
Secondary and 3D structure prediction of OsNRX proteins
The SOPMA website (https://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) (Geourjon and Deléage, 1995) was used to predict the secondary structure of proteins encoded by the rice NRX gene family; the SWISS-MODEL website (https://swissmodel.expasy.org/interactive) (Waterhouse et al., 2018) was used for 3D structure prediction of the OsNRX protein family.
Conserved domain and motif analysis of OsNRX proteins
TBTools and the online software MEME (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2015) were used to predict conserved domains and motifs of the OsNRX proteins. MUSCLE in MEGAX was used to align the rice NRX protein sequences to obtain the evolutionary relationships between NRX proteins (Kumar et al., 2018). MEME was used to identify conserved motifs (Motif) in the NRX protein sequences, with a maximum of 10 conserved structural domains and motif width set to 6-20 bp. TBtools were then used to analyze and visualize the conserved motif information.
Phylogenetic analysis of the OsNRX family
Genomic data and annotation files for wheat, foxtail millet, Arabidopsis, soybean, and maize were downloaded from the Ensembl Plants database (http://plants.ensembl.org/info/data/ftp/index.html), accessed on 2023.8.2. The method described above for identifying rice NRX gene family members was used to identify NRX gene family members in wheat, foxtail millet, Arabidopsis, soybean, and maize. ClustalW in MEGAX was used to align the NRX protein sequences of these species with the rice NRX protein sequences, and a phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with a bootstrap value set to 1000 and other parameters default. The iTOL website (https://itol.embl.de) (Letunic and Bork, 2024) was used to beautify the phylogenetic tree.
Prediction of cis-acting elements in the OsNRX family
To identify cis-acting elements related to the NRX genes, 2000 bp of genomic sequence upstream of the transcription start site were extracted and submitted to the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Lescot, 2002) for prediction of cis-acting elements. Common cis-acting elements, light response elements, and elements of unknown function were removed, and a diagram of the cis-acting elements of the OsNRX family was drawn.
Expression analysis of the OsNRX family at different developmental stages
The expression data of OsNRX genes across various tissues and developmental stages of Nipponbare rice were downloaded from the RiceXPro V3 database (https://ricexpro.dna.afrc.go.jp/index.html) (Sato et al., 2013). The data were processed as follows: (1) Cy3 raw signals were normalized to the 75th percentile using Agilent Feature Extraction software (version 9.5.3.1); (2) log2 transformation was applied. The analyzed tissues included leaf, sheath, root, stem, panicle, anther, pistil, lemma, palea, ovary, embryo, and endosperm, while the developmental stages covered the vegetative, reproductive, and mature phases. The R heatmap package was used to visualize the expression patterns of OsNRX genes across these tissues and developmental stages.
Plant materials and stress treatments
In this study, we used two varieties obtained from the Institute of Grain Crops, Hubei Academy of Agricultural Sciences: Jiangnong Zao 1B, which is characterized by drought resistance during the seedling stage, and TAISEN GLUTINOUS YU 1157, which is known for its drought sensitivity at the seedling stage. Cultivation was conducted in an artificial climate chamber (16 h of light at 28°C, 8 h of darkness at 26°C, 70% relative humidity, and light intensity of 200 μmol/m2/s). Cultivate the plants with tap water for the first 4 days, then with half-strength Yoshida nutrient solution (Coolaber, Beijing, China) for the next 3 days, followed by full-strength Yoshida nutrient solution for 7 days, changing the solution every two days. At 14 days old, subject the seedlings to drought stress using full-strength Yoshida nutrient solution supplemented with 20% PEG6000. Samples were collected at 0, 3, 6, 12, and 24 h post-stress. For each species, three plants with uniform growth conditions were selected, and 0.1 g of fresh young leaves were quickly placed in 1.5 mL centrifuge tubes, tightly capped, and stored in liquid nitrogen and then stored in an ultra-low temperature refrigerator at -80°C.
Real-time quantitative PCR (qRT-PCR)
Fluorescence quantitative PCR primers were designed using Primer Premier 6 (Supplementary Table S5), with OsACT selected as the reference gene. The melting temperature is 55-60°C, the primer length is 19-23bp, the GC content is 47-57%, and the amplicon size is 179-222bp. The total RNA of the sample was extracted using the RNA Easy Fast Plant Tissue Kit (Tiangen, China). cDNA was synthesized using RevertAid Reverse Transcriptase (Thermo Fisher Scientific, USA; Cat# EP0441), following the manufacturer’s protocol. qRT-PCR was then performed using the PerfectStart Green qPCR SuperMix (TransGen Biotech, China) on a QuantStudio 1 Plus Real-Time PCR System (Thermo Fisher Scientific, USA). Related experimental parameters are provided in Supplementary Table S6.
Statistical analysis
Each sample was repeated for 3 times, and the data were processed by 2-△△Ct method (Livak and Schmittgen, 2001). Statistical analysis was performed using R (R Core Team, 2024). Data were analyzed using the Student T-test, and least significant difference post hoc tests were used to determine significant differences between means (*, statistically significant at P < 0.1; **, statistically significant at P < 0.05; ***, statistically significant at P < 0.01).
SNP analysis
The reference genome used for SNP calling was the Os-Nipponbare-Reference-IRGSP-1.0 (Li and Durbin, 2010). Variants were identified using the GATK (DePristo et al., 2011), with single-nucleotide polymorphisms (SNPs) retained if they met the following criteria: minor allele frequency (MAF) ≥ 5% and missing rate ≤ 20%. Missing genotypes were imputed using IMPUTE2 (Howie et al., 2009), and SNPs specific to Nipponbare, Minghui63, 9311, Jiangnong Zao 1B, and TAISEN GLUTINOUS YU were extracted using Plink (Purcell et al., 2007).
Results
Identification and basic characteristics of OsNRX gene family members
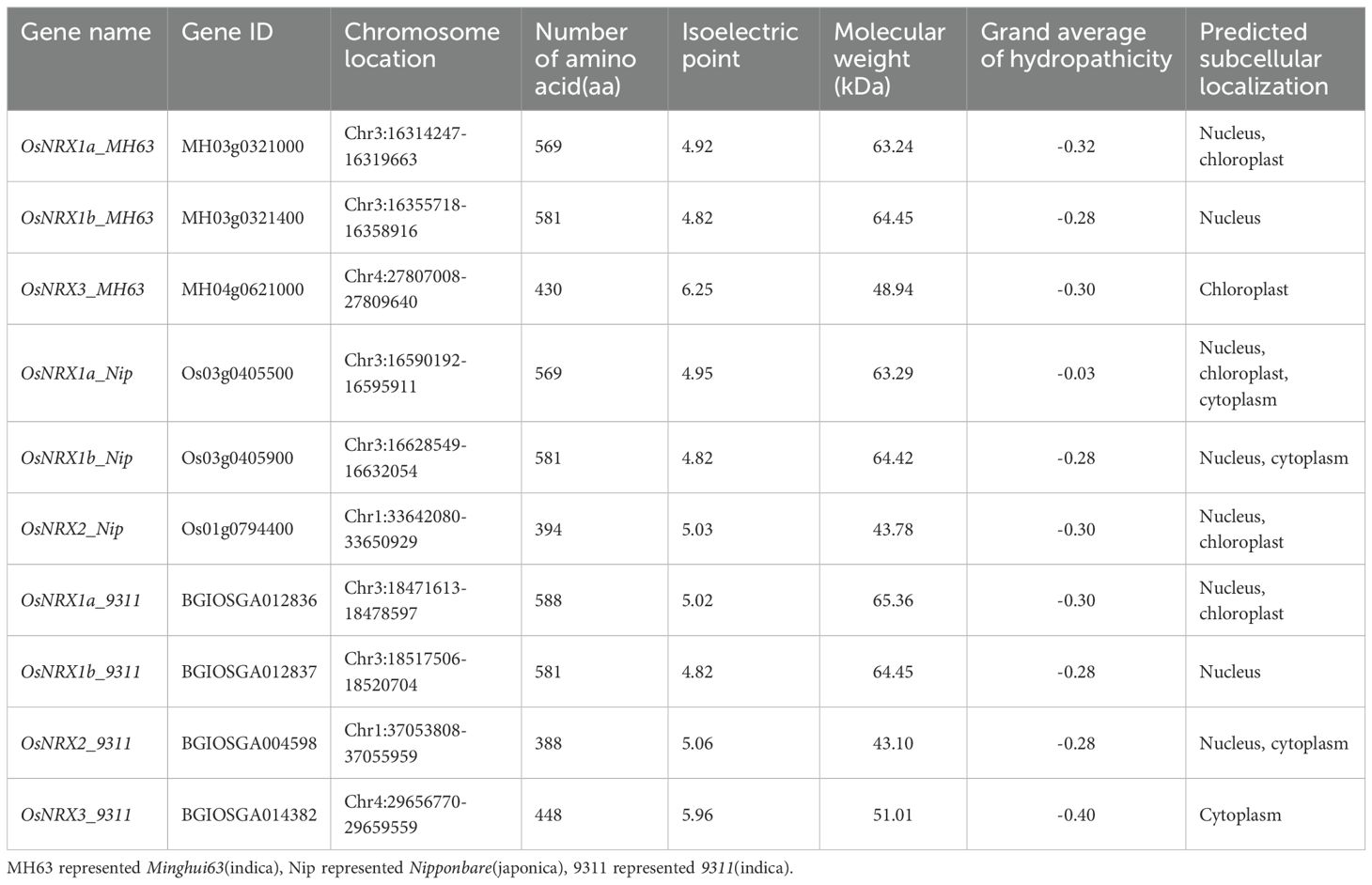
A total of 10 candidate NRX genes were identified in the rice varieties Minghui63 (indica), Nipponbare (japonica), and 9311 (indica) using HMMER3.0 and TBtools software (Supplementary Table S1). The protein sequences of these candidate gene family members were analyzed using SMART and CDD, and the results showed that all candidates contained the NRX domain. Following the classification method of NRX family by Marchal et al. (2014) (Figure 1a), the NRX genes in Minghui63 were named OsNRX1a_MH63, OsNRX1b_MH63, and OsNRX3_MH63; in Nipponbare, they were named OsNRX1a_Nip, OsNRX1b_Nip, and OsNRX2_Nip; and in 9311, they were named OsNRX1a_9311, OsNRX1b_9311, OsNRX2_9311, and OsNRX3_9311.
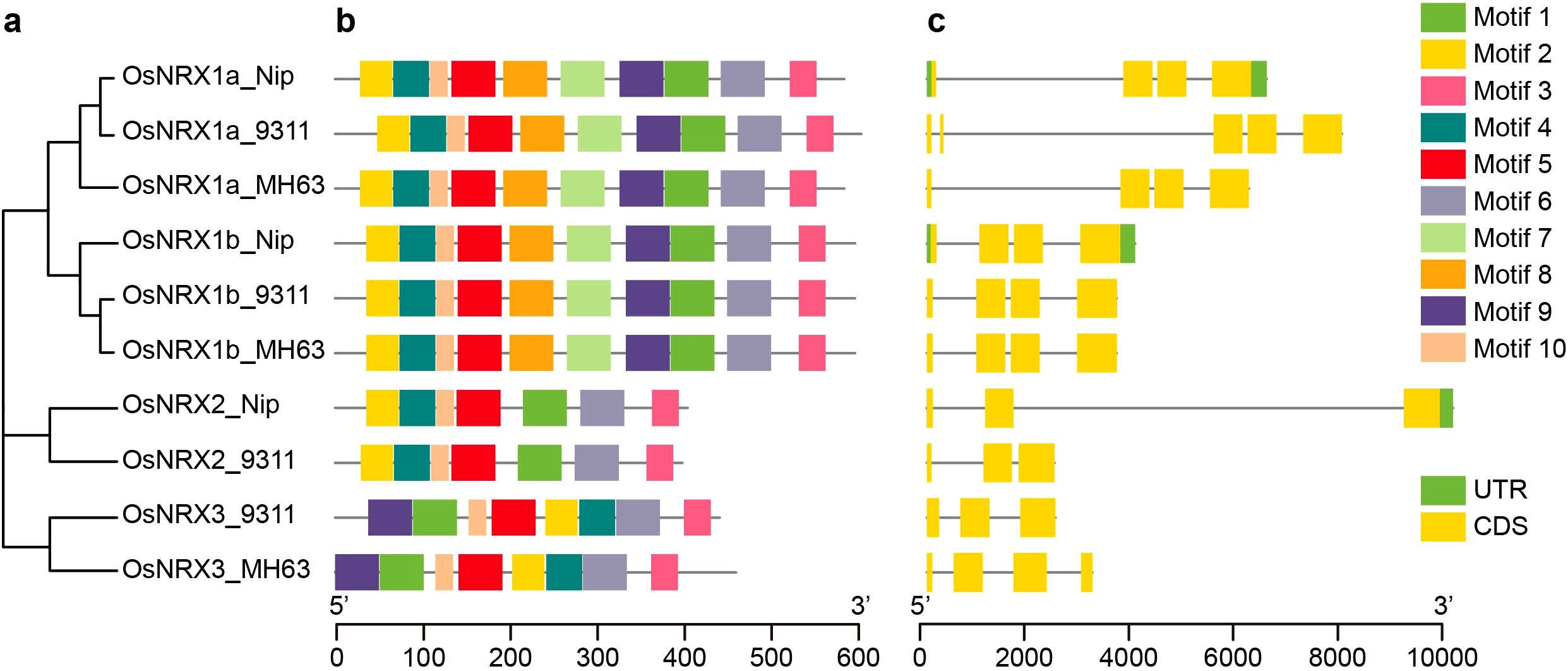
Figure 1. Phylogenetic relationship, conserved motif structure and gene structure of OsNRX protein. (a) The phylogenetic tree of OsNRX. MH63 represented Minghui63(indica), Nip represented Nipponbare(japonica), 9311 represented 9311(indica). (b) The distribution of conserved motifs. Boxes with different colors represented different conserved motifs (Motif 1-10). (c) The gene structure of OsNRX. Green boxes represented the UTRs, yellow boxes represented exons.
The physicochemical properties and subcellular localization of the 10 NRX protein sequences in rice were analyzed using the online software ExPASy and Plant-mPLoc, predicting their isoelectric point, molecular weight, grand average of hydropathicity (Table 1). The size of OsNRX proteins ranges from 388 to 581 amino acids; their molecular weights range from 43.10 to 65.36 kDa; and their isoelectric points are all below 7, indicating acidic properties; the average hydrophilicity index is less than 0, suggesting that the NRX gene family proteins are hydrophilic. Subcellular localization predictions suggest that OsNRX family members may be distributed across different organelles, such as the nucleus, chloroplast, or cytoplasm. These details imply that OsNRX members may perform distinct functions in different parts of the cell.
Gene structure analysis and protein conserved motif analysis of OsNRX
To explore the sequence information of the OsNRX gene family members, the gene structure and conserved motifs were analyzed using MEME and TBtools software. Regarding the protein conserved motifs, the number and distribution of motifs within the OsNRX subfamily members were largely consistent (Figure 1b). All OsNRX family members contain Motif1, Motif2, Motif3, and Motif6. Notably, Motif1 and Motif2 harbor the characteristic “WCGPC” sequence and are located within the TRX domain, whereas Motif3 and Motif6 are found in the cysteine-rich C-terminal domain, the function of which remains unclear. The gene structures of OsNRX genes show some variation (Figure 1c), with OsNRX2-Nip being the longest at 8.8 kb and OsNRX3_MH63 the shortest at only 2.1 kb. All OsNRX genes contained at least three CDS segments, with only OsNRX1a-Nip, OsNRX1b-Nip, and OsNRX2-Nip containing UTR fragments.
Secondary and 3D structure prediction of OsNRX proteins
Secondary structures of OsNRX proteins were analyzed by the SOPMA online website.OsNRX1a OsNRX1b and OsNRX2 proteins have similar composition and structure in three rice varieties, while the OsNRX3 protein shows some variation among different varieties (Figure 2). All OsNRX proteins were consisted of four secondary structures: alpha helix, extended helix, random coil, and extended strand, and the proportions of alpha helix > extended strand > random coil> beta turn.
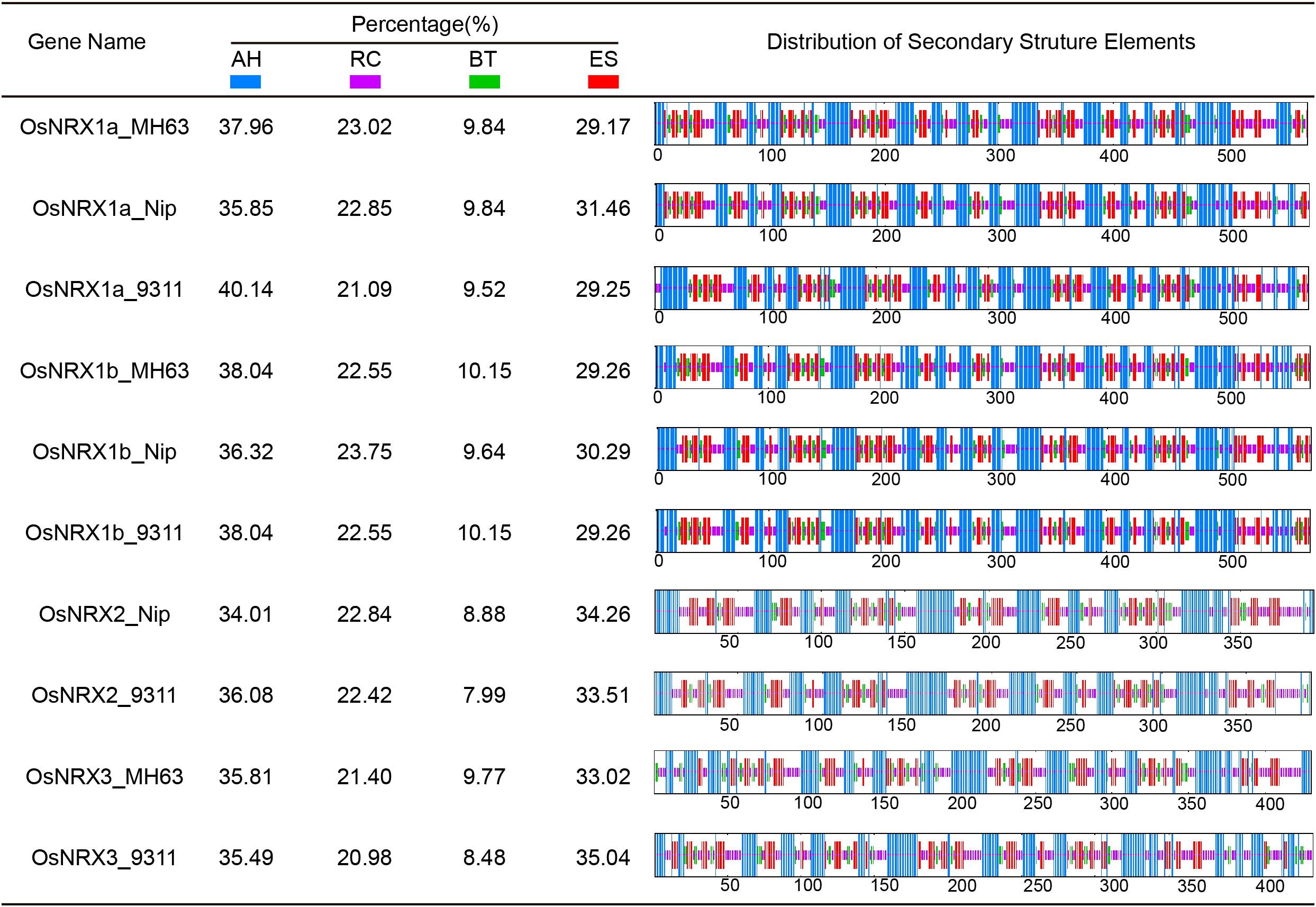
Figure 2. Secondary structure analysis of OsNRX proteins. MH63 represented Minghui63(indica), Nip represented Nipponbare(japonica), 9311 represented 9311(indica). The blue color represents alpha helix (AH), the purple color represents random coil (RC), the green color represents beta turn (BT) and the red color represents extended strand (ES).
The 3D structures of proteins encoded by the OsNRX gene family were predicted using the online SWISS-MODEL platform. The results revealed that the 3D structures of NRX1a and NRX1b are highly similar across the three rice varieties. Similarly, the tertiary structures of NRX2 show high similarity between Nipponbare and 9311, whereas the 3D structures of NRX3 exhibit lower similarity between Minghui 63 and 9311 (Figure 3). The GMQE values for all these predicted structures exceed 0.8, indicating relatively high reliability of the predictions (Biasini et al., 2014).
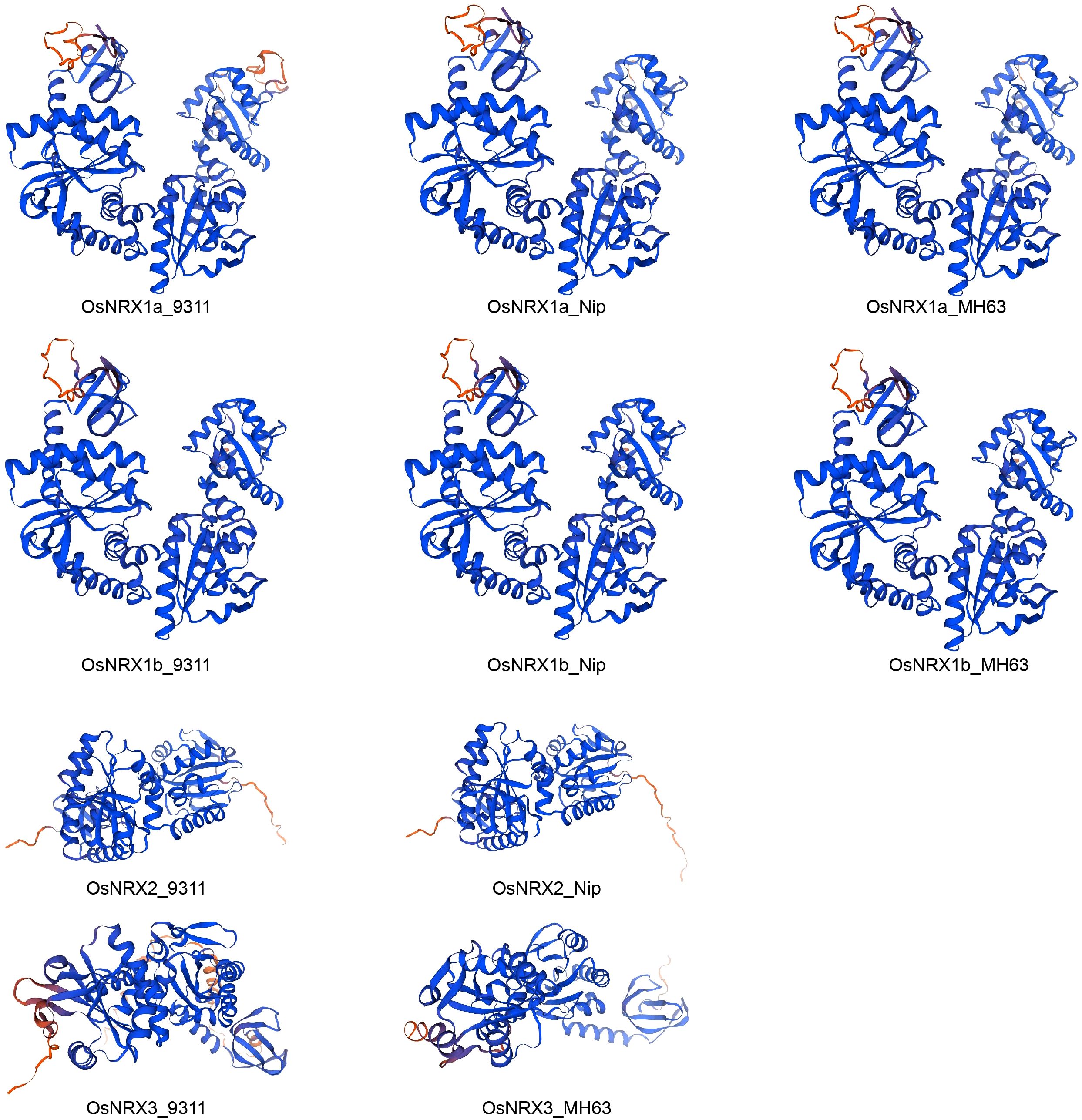
Figure 3. Protein 3D structure prediction model of OsNRX gene families. MH63 represented Minghui63(indica), Nip represented Nipponbare(japonica), 9311 represented 9311(indica).
Phylogenetic tree analysis of the OsNRX gene family
To investigate the evolutionary relationships of the OsNRX genes, NRX gene family members were identified in wheat, foxtail millet, Arabidopsis, soybean, and maize. The phylogenetic tree of these NRX family members was constructed (Figure 4, Supplementary Table S1). This comprehensive analysis encompasses a diverse range of species, such as rice with three distinct varieties—Minghui63, Nipponbare, and 9311—as well as wheat, foxtail millet, Arabidopsis, soybean, and maize. The phylogenetic tree of NRX gene members is divided into three subfamilies, with 12 genes assigned to the NRX1 subfamily, 10 genes to the NRX2 subfamily, and 6 genes to the NRX3 subfamily. In conjunction with the protein sequences of the NRX genes, the results of the phylogenetic tree are fully consistent with the NRX subfamily classification by Marchal et al. (2014). Consistent with their evolutionary relationships, the NRX genes of the three rice varieties show the closest phylogenetic relationships, followed by wheat, foxtail millet and maize, while exhibiting the most distant relationships with Arabidopsis and soybean. All 6 species have at least one NRX1 gene, and the three rice varieties each contain at least two NRX1 genes. The study by Hazra et al. (2024) demonstrated that NRX1 genes are present in 15 species, and that Oryza sativa, Populus trichocarpa, and Vitis vinifera each contain multiple NRX1 members. Our findings are consistent with their results, further suggesting that NRX1 is a conserved gene that may play a critical role in plants. The NRX1 genes of wheat, foxtail millet and maizet are more closely related to those of rice in the phylogenetic tree. NRX1 in foxtail millet was able to balances the excess reactive oxygen species (ROS) due to drought and salt stress, reduces oxidative damage in plants, and maintains normal growth and development (Zhang et al., 2022). Combined with the involvement of the TaNRX1-D gene in regulating the response to drought stress in wheat (Cheng et al., 2021b), we speculate that the NRX family in rice may have similar functions.
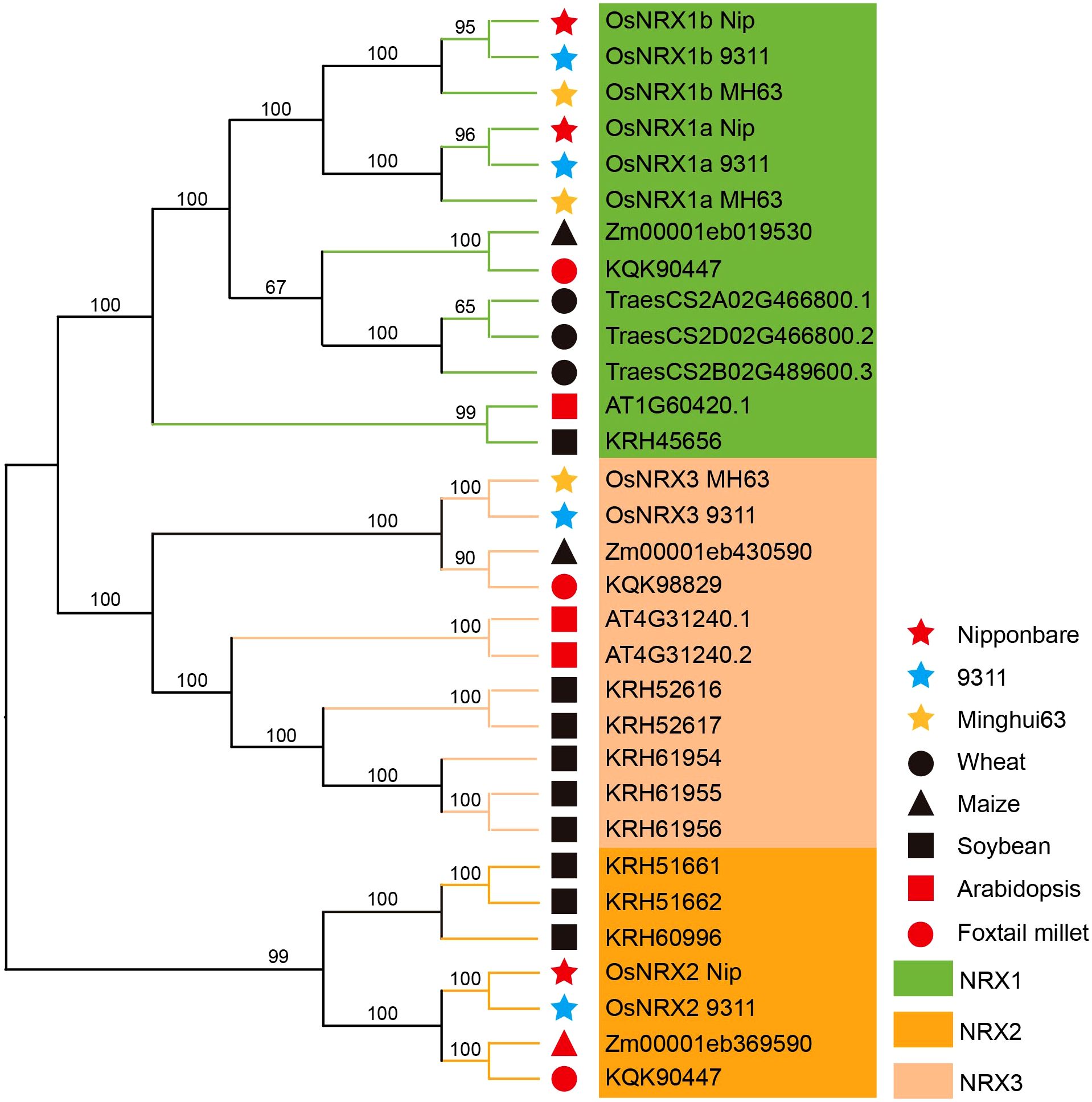
Figure 4. Phylogenetic tree of NRX gene family proteins in rice and five species. Phylogenetic trees were constructed using the Neighbor-Joining (NJ) method of the MEGAX software with the bootstrap value set to 1000.
Cis-acting elements of the OsNRX family
To predict the biological functions of the OsNRX genes, we conducted a cis-acting element analysis on the 2000 bp promoter region upstream of the start codon of the OsNRX genes using the PlantCARE database (Figure 5, Supplementary Table S2). The results indicated that the promoter regions of OsNRX-related genes contain a rich array of cis-acting elements that may significantly influence their biological functions. After initial screening, a variety of cis-acting elements were discovered, some of which are associated with known biological functions, while others are of unknown function. After removing elements of unknown function, a total of 36 different types of cis-acting elements were predicted, summing up to 877 elements. The number and types of cis-acting elements in the promoter region of OsNRX gene are obviously different. The analysis revealed significant differences in the number and types of cis-acting elements in the promoter regions of different OsNRX genes (Figure 5). In terms of quantity, OsNRX1 has the fewest cis-acting elements, with an average of 9 elements per promoter region; OsNRX2 has the most, with an average of 45 elements per promoter region; and OsNRX3 has an average of 18 elements. In terms of types, in addition to common cis-acting elements such as CAAT-box, TATA-box, and light response elements, specific cis-acting elements related to plant response to environmental stress and growth and development were also identified. Among these, abscisic acid response elements (ABRE) were the most abundant, accounting for about 45% of the uncommon cis-acting elements. Additionally, the OsNRX family includes elements responsive to JA (CGTCA-motif, TGACG-motif), SA (TCA-element), IAA (TGA-element), GA (P-box), growth and development (CAT-box, GCN4_motif, RY-element), and drought induction (ARE, MBS). The presence of these elements suggests that OsNRX genes may be involved in the response and adaptation mechanisms to drought stress by regulating plant water absorption and utilization.
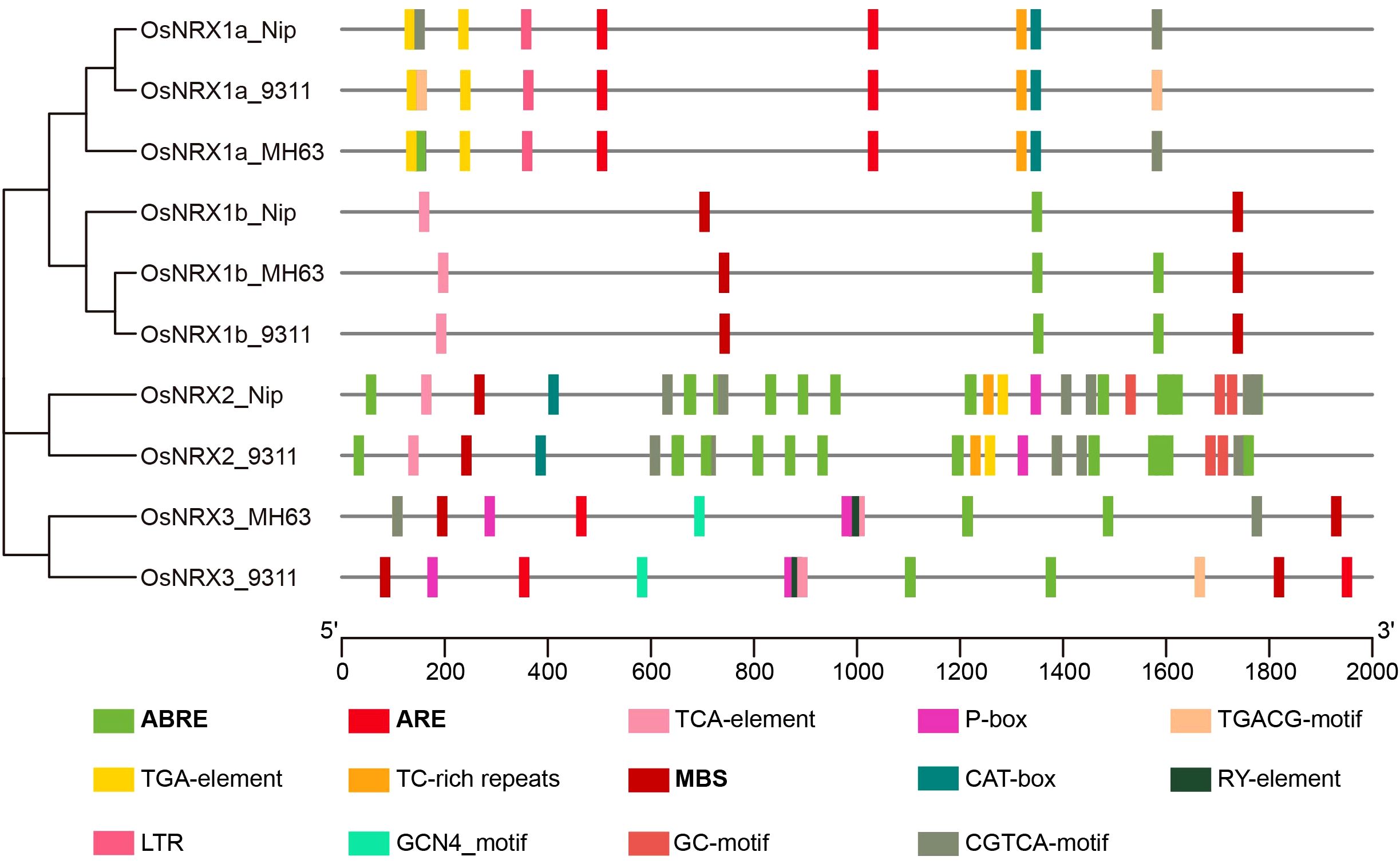
Figure 5. Distribution of cis-acting elements in promoter regions of OsNRX genes. MH63 represented Minghui63 (indica), Nip represented Nipponbare (japonica), 9311 represented 9311 (indica). Boxes with different colors represent cis-regulatory elements, with bold fonts indicating cis-regulatory elements related to drought induction, and the scale at the bottom represents the length of the promoter sequence.
Expression patterns of the OsNRX family at different developmental stages
Both phylogenetic tree analysis and cis-acting element prediction suggested that the OsNRX gene family might be involved in drought stress response. To further investigate the function of OsNRX genes at different developmental stages and the possibility of their involvement in response to drought stress, we downloaded gene expression data from the RiceXpro database (https://ricexpro.dna.afrc.go.jp/index.html) for different tissues of Nipponbare at vegetative and reproductive growth stages. The RiceXpro database employs a single microarray system for data collection, ensuring consistency across experiments and providing a reliable platform for comparing gene expression across tissues/organs, growth stages, and experimental conditions (Sato et al., 2013). Expression data indicated that OsNRX genes are expressed in all tissues, with particularly significant differences in expression in roots, lemma, palea, embryo, and endosperm (Figure 6, Supplementary Table S3). OsNRX1a has the highest expression level in roots, with expression decreasing as roots, embryos, and endosperm develop, while increasing as the length of the glumes (lemma and palea) increases. The expression level of OsNRX1b increases with the development of the endosperm. In other tissues, the expression pattern of OsNRX1b is similar to that of OsNRX1a. OsNRX2 has the highest expression level in endosperm, with expression increasing as roots, glumes, embryos, and endosperm develop. The expression patterns of OsNRX1a, OsNRX1b, and OsNRX2 in different tissues are complementary, and whether there is interaction between OsNRX gene families needs further study.
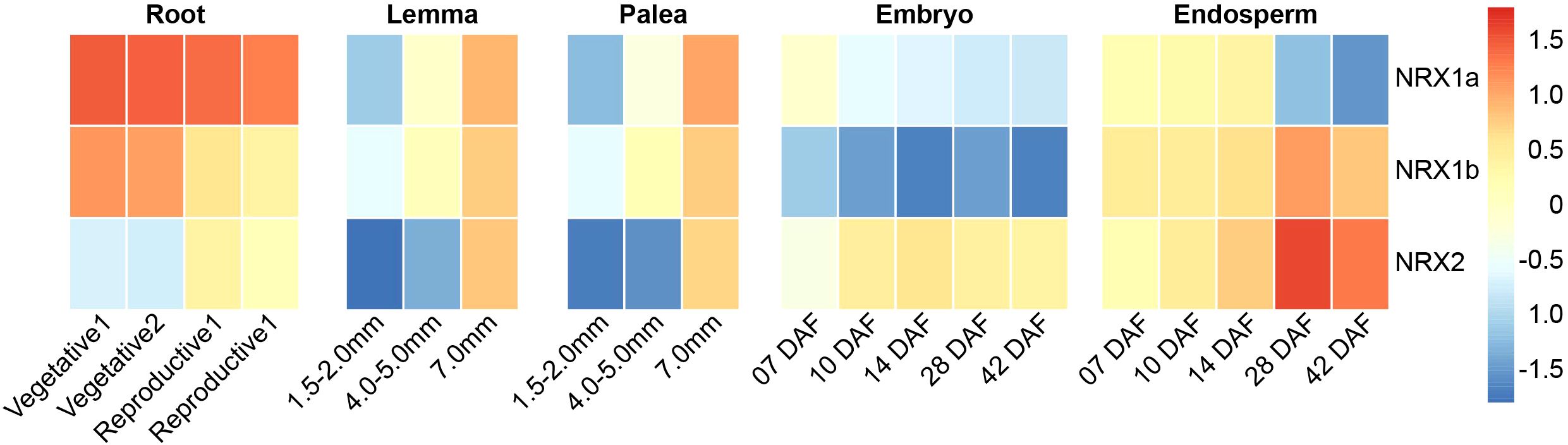
Figure 6. Expression patterns of the OsNRX gene family in rice. The color scale represents the relative expression levels, ranging from low (blue) to high (red). “mm” represents millimeters, and “DAF” represents days after flowering.
Expression patterns of the OsNRX family under drought stress
Studies have indicated that the expression of the NRX family increases in response to drought stress in plants such as wheat, sorghum, foxtail millet (Zhang et al., 2021; Jedmowski et al., 2014; Zhang et al., 2022). These findings suggest that the NRX gene family may play a significant role in plants’ responses to drought stress. In this study, by analyzing the cis-acting elements of the OsNRX gene family, we found that the OsNRX family contains abscisic acid-responsive elements (ABRE) associated with drought response. The expression of OsNRX1a and OsNRX1b in rice roots was significantly higher than that in other tissues or organs. Additionally, OsNRX1b, OsNRX2, and OsNRX3 were found to have MYB binding sites (MBS) and CCAAT-box involved in drought induction capabilities. Therefore, we hypothesize that the NRX family in rice also responds to drought stress.
To validate whether the expression of the OsNRX family is associated with drought stress, two rice varieties, Jiangnong Zao 1B (drought-tolerant, DT) and TAISEN GLUTINOUS YU 1157 (drought-sensitive, DS), were selected for qRT-PCR analysis. There were significant differences in drought tolerance between DT and DS. DT exhibited partial leaf rolling after 24 h of drought stress and nearly full recovery after rehydration, while DS showed complete leaf rolling after 24 h and no recovery after rehydration (Supplementary Figure S1). The gene expression level at 0h after PEG6000 treatment was used as a control, and the expression levels of OsNRX genes at different time points under drought treatment were analyzed (Figure 7, Supplementary Table S4). The relative expression level of OsNRX1a exhibited the most substantial variation in DS, with a 7.5-fold increase at 6h compared to 0h, and significant differences between DT and DS were observed at 6h and 24h. Similarly, OsNRX1b displayed a significant change in DS, with a 20-fold increase at 6h compared to 0h after PEG6000 treatment, and a notable difference in expression levels between DT and DS. For OsNRX2, the relative expression level increased by more than 20-fold after 3h of PEG6000 treatment compared to untreated samples. Specifically, DS showed a 26.1-fold increase at 3h, while DT reached a 53.2-fold increase at 6h, with significant differences in relative expression levels between DT and DS at both 3h and 6h. For OsNRX3, the relative expression level in DT increased 7.7-fold after 3h of PEG6000 treatment and then gradually decreased, whereas in DS, the expression level increased 7-fold after 6h, followed by a decrease and then an increase. Significant differences in relative expression levels between DT and DS were observed after 3h of treatment. In conclusion, the OsNRX gene family exhibited significant expression changes under drought stress, typically showing an initial increase followed by a decrease. The expression levels peaked around 6h post-stress, then decreased, but remained higher than those before the stress. Moreover, there were significant differences in the expression levels of the OsNRX gene family between DT and DS.
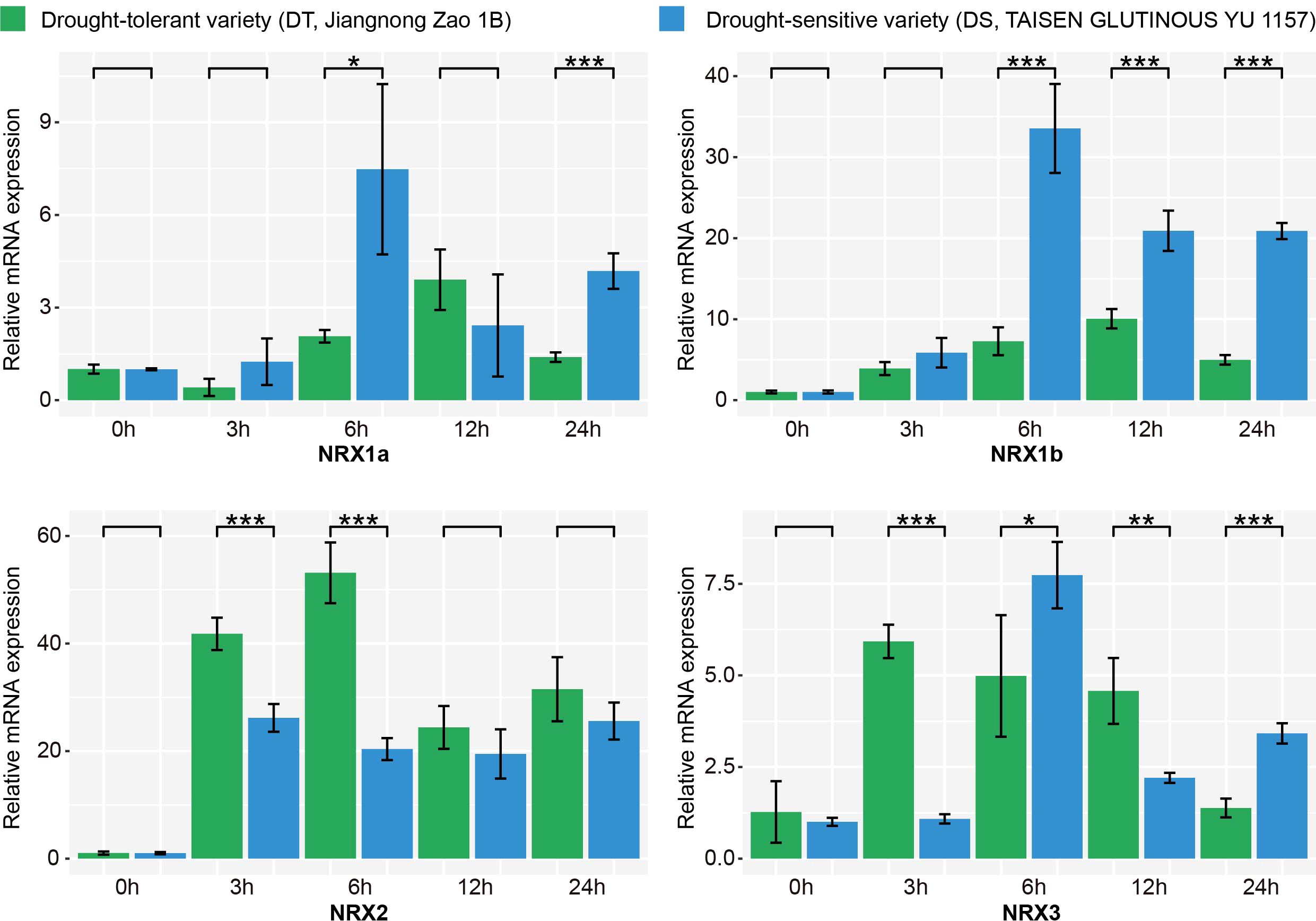
Figure 7. Expression changes of OsNRX gene family under PEG6000 treatment in Drought-tolerant and Drought-sensitive varieties. Response of drought-tolerant (DT, green) and drought-sensitive (DS, blue) varieties to PEG6000 treatment over time (0, 3, 6, 12, and 24 hours). Asterisks (*) indicate significant differences between DT and DS at each time point, as determined by Student’s t-test: *p < 0.1, **p < 0.05, and ***p < 0.01. The results were the mean of three independent biological replicates using quantitative real-time PCR (qRT-PCR) technology.
Discussion
Identification and characterization of the OsNRX gene family
The NRX gene family, which contains TRX-like domains, is classified within the TRX family. Research on the NRX gene family primarily stems from studies related to the TRX family. Zhou et al. (2022) identified 48 TRX genes in wheat, including 3 NRX gene family members; similarly, Zhang et al. (2022) identified 25 TRX gene family members in foxtail millet, with 3 being NRX gene family members. In this study, we used bioinformatics methods to identify 10 NRX family members in the genomes of three rice varieties (Minghui 63, Nipponbare, and 9311). These members can be divided into three subgroups: NRX1, NRX2, and NRX3, and each variety contains NRX1a and NRX1b members. The physicochemical property analysis of the OsNRX gene family proteins revealed that these proteins are acidic and hydrophilic. In rice, the subcellular localization of both OsNRX1 and OsNRX2 genes includes the nucleus, which is consistent with the initial discovery of NRX in the nucleus (Kurooka et al., 1997; Laughner et al., 1998). However, in different rice varieties, the OsNRX gene family is also localized in various cellular structures. According to the study by Mata-Pérez and Spoel (2019), biotic and abiotic stresses recruit members of the TRX family to specific sites, which may be related to the differences in subcellular localization of NRX in various rice varieties. In Minghui 63, Nipponbare, and 9311, the protein structures and conserved motifs of OsNRX1a, OsNRX1b, and OsNRX2 are similar, which may imply similar functions. Phylogenetic tree analysis indicates that the NRX gene family is highly conserved in plants, particularly NRX1, with at least one NRX1 gene present in every plant species.
Impact of drought stress on OsNRX gene family expression
ROS are byproducts of aerobic metabolism and act as secondary messengers under various environmental stresses. When plants experience abiotic stresses such as drought, heavy metals, and salinity, excessive ROS are produced, causing plant cell damage and even cell death (Gill and Tuteja, 2010; Panda et al., 2021). To maintain the dynamic balance of ROS, plants rely on antioxidant defense mechanisms, which are activated by antioxidant enzymes such as catalase (CAT) and superoxide dismutase (SOD), as well as non-enzymatic antioxidant components (Das and Roychoudhury, 2014). CAT requires interaction with its PEX5 site on the peroxisomal membrane to enter the peroxisome and eliminate ROS. However, excessive ROS can lead to mono-ubiquitination of PEX5, preventing CAT from maintaining its reduced state and entering the peroxisome. NRX1 can bind to CAT, maintaining its reduced state and thus protecting plant cells from oxidative stress induced by adverse environmental conditions (Kneeshaw et al., 2017; Calderón et al., 2018). Davoudi et al. (2022) found that under drought stress, the expression of NRX and CAT in grafted cucumber and pumpkin significantly increased, enhancing resistance to drought stress. In foxtail millet, Zhang et al. (2022) constructed transgenic Arabidopsis plants expressing SiNRX1, and the results showed that the overexpression lines had significantly higher antioxidant capacity and ROS scavenging ability under drought stress than the wild type, enhancing their drought and salt tolerance.
By analyzing the cis-acting elements of the OsNRX family, we found that the OsNRX family contains a large number of elements associated with drought stress, particularly the abscisic acid-responsive element (ABRE), which accounts for about 45% of all uncommon cis-acting elements. Abscisic acid is an important signaling hormone in plants’ response to drought stress, and the ABRE cis-acting element is a key component in the abscisic acid signaling pathway. Cheng et al. (2021b) studied the promoter of TaNRX1 and found that a 36 bp fragment containing two ABRE cis-acting elements is a key sequence for TaNRX1 gene response to polyethylene glycol (PEG) and abscisic acid (ABA). In plants, PEG is commonly used to simulate drought stress because it can induce physiological responses similar to drought conditions by reducing osmotic potential (Pereira et al., 2020; Fan et al., 2021; Wang et al., 2022). ABA, as an important hormone regulating plant stress resistance, can enhance plants’ tolerance to drought stress by inducing the expression of a series of stress-related genes (Liu et al., 2022). The expression patterns of the OsNRX family at different developmental stages indicate that OsNRX1a and OsNRX1b have significantly higher expression in rice roots than in other tissues or organs, and as the roots develop, the expression levels of OsNRX1a and OsNRX1b gradually decrease, while the expression level of OsNRX2 gradually increases. Therefore, we speculate that the function of the OsNRX gene family is related to drought stress. To verify this hypothesis, we conducted drought simulation stress on two rice varieties with different seedling drought resistance. The results showed that the expression of the OsNRX gene family changed significantly, and the expression patterns were not the same in rice varieties with different drought resistance capabilities, indicating that the OsNRX family may have a regulatory function in drought resistance in rice.
SNP analysis of the OsNRX gene family (Supplementary Table S7) revealed one SNP in NRX1a and five SNPs in NRX2 among Nipponbare, Minghui63, 9311, Jiangnong Zao 1B, and TAISEN GLUTINOUS YU 1157, while no SNPs were detected in NRX1b or NRX3. Integrated with drought-responsive expression patterns, NRX2 exhibited significant upregulation at 3 h post-stress and differential expression between drought-tolerant (DT) and drought-sensitive (DS) varieties (Figure 7), potentially influenced by genetic variation. NRX1a showed differential expression between DT and DS at 24 h post-stress, likely due to its single SNP. Despite the absence of SNPs, NRX1b was rapidly upregulated at 6 h and exhibited differential expression between DT and DS, possibly regulated by upstream transcription factors or epigenetic modifications. Similarly, NRX3, despite lacking SNPs, showed differential expression between DT and DS, potentially mediated by post-transcriptional or epigenetic regulation. These hypotheses require further experimental validation.
Potential of OsNRX gene family for drought resistance breeding
Among the OsNRX gene family, NRX2 and NRX1b emerge as the most promising candidates for drought resistance breeding. NRX2 exhibited the most dramatic response to drought stress, with relative expression levels increasing by over 20-fold within 3-12h post-PEG6000 treatment (Figure 7, Supplementary Table S4). Notably, the drought-tolerant (DT) variety showed a 53.2-fold increase at 6 h, significantly higher than the drought-sensitive (DS) variety (26.1-fold at 3 h), suggesting its critical role in rapid drought adaptation. 5 SNPs within the NRX2 between DT and DS varieties may underlie the observed expression differences and contribute to drought tolerance. Despite lacking SNPs, NRX1b displayed a 20-fold upregulation in DS at 6 hours post-stress, with significant differences between DT and DS (Figure 7). Studies in wheat and foxtail millet have demonstrated that NRX1 homologs (TaNRX1, SiNRX1) directly enhance drought tolerance by modulating antioxidant pathways and stress-related gene networks (Zhang et al., 2022; Zhou et al., 2022).
Although NRX1a showed a 7.5-fold increase in DS at 6 h and NRX3 showed a 7-fold increase in DS at 6 h, the magnitude of change in their relative expression in the wake of drought stress was not as large as that of NRX2 and NRX1b. The strong drought-inducible expression of NRX2 and NRX1b, coupled with their genetic/epigenetic plasticity, positions them as prime targets for marker-assisted selection or CRISPR-based editing to enhance drought tolerance. Future studies should prioritize functional characterization of NRX2 and NRX1b to unravel their mechanistic roles and translational potential.
OsNRX gene family may regulate rice grain size
In addition to environmental influences, the size of plant seeds depends on the genotype of the embryo and endosperm and is limited by the pericarp (Li and Li, 2016). In Arabidopsis, Wang et al. (2010) found that the IKU1 mutant leads to reduced endosperm growth and smaller grain size; IKU1 is expressed in early endosperm, and restoring IKU1 function in the endosperm can increase seed size. In rice, Ren et al. (2018) knocked out fzp and found that rice cell proliferation and expansion were affected, the pericarp was smaller than the wild type, and ultimately the rice grain size was reduced. By analyzing the expression patterns of OsNRX at different developmental stages, we found that as the length of the pericarp increases, the expression level of OsNRX also tends to rise. As the embryo and endosperm develop, the expression level of OsNRX1a gradually decreases, while the expression level of OsNRX2 gradually increases, and the expression level of OsNRX1b changes little. Therefore, we speculate that the OsNRX gene family may have a function in regulating the size of rice grains in rice, which needs further verification by experiments.
Conclusion
Using bioinformatics approaches, we identified a total of 10 OsNRX genes across three rice varieties, with Minghui63 harboring 3, Nipponbare 3, and 9311 4. All identified OsNRX proteins are hydrophilic. The expression of OsNRX genes is influenced by drought stress and varies among rice varieties with differing drought resistance, suggesting a potential role for OsNRX in regulating drought tolerance. The expression levels of OsNRX change with the development of the glumes, and the presence of cis-acting elements related to growth and development within 2000 bp upstream of their promoters, which suggests that OsNRX may be involved in regulating rice grain size.
Data availability statement
Genomic data and annotation files for the rice varieties Nipponbare and 9311 were downloaded from the Ensembl Plants database (http://plants.ensembl.org/info/data/ftp/index.html) (Yates et al., 2022), accessed on 2023.7.27. The whole-genome data and its annotation files for the rice variety Minghui63 were obtained from the Rice Information Network RIGW (http://rice.hzau.edu.cn/cgi-bin/rice_rs1/download_ext) (Song et al., 2018), accessed on 2023.7.27. The HMM file for the NRX gene (PF13905) was downloaded from the InterPro database (https://www.ebi.ac.uk/interpro) (Paysan-Lafosse et al., 2023), accessed on 2023.7.27. The genotype data used for SNP Analysis were obtained from Meng et al. (2022).
Author contributions
ZL: Investigation, Software, Visualization, Writing – original draft, Writing – review & editing. XZ: Resources, Supervision, Validation, Writing – review & editing, Investigation. DY: Resources, Supervision, Writing – review & editing. LL: Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing, Conceptualization. HY: Visualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the science and technology innovation Program of Hunan Province, China (2023NK2001), the Changsha Natural Science Foundation, China (kq2402109), the Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, China (2022LZJJ08), the Special Funds for Construction of Innovative Provinces in Hunan Province, China (2021NK1011).
Conflict of interest
Author HY was employed by Huazhi Biotechnology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1562718/full#supplementary-material.
References
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Calderón, A., Sevilla, F., Jiménez, A. (2018). “Redox protein thioredoxins: function under salinity, drought and extreme temperature conditions,” in Antioxidants and antioxidant enzymes in higher plants. Eds. Gupta, D. K., Palma, J. M., Corpas, F. J. (Springer International Publishing, Cham), 123–162. doi: 10.1007/978-3-319-75088-0_7
Cha, J.-Y., Ahn, G., Jeong, S. Y., Shin, G.-I., Ali, I., Ji, M. G., et al. (2022). Nucleoredoxin 1 positively regulates heat stress tolerance by enhancing the transcription of antioxidants and heat-shock proteins in tomato. Biochem. Biophys. Res. Commun. 635, 12–18. doi: 10.1016/j.bbrc.2022.10.033
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, J., Song, T., Wei, F., Li, R., Yu, M., Hao, Z., et al. (2021a). Effects of different domains on activity of wheat (Triticum aestivum) taNRX1-D. J. Agric. Biotechnol. 29, 443–452. doi: 10.3969/j.issn.1674-7968.2021.03.004
Cheng, J., Wei, F., Zhang, M., Li, N., Song, T., Wang, Y., et al. (2021b). Identification of a 193 bp promoter region of TaNRX1-D gene from common wheat that contributes to osmotic or ABA stress inducibility in transgenic Arabidopsis. Genes Genom 43, 1035–1048. doi: 10.1007/s13258-021-01115-x
Chou, K.-C., Shen, H.-B. (2010). Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335. doi: 10.1371/journal.pone.0011335
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2. doi: 10.3389/fenvs.2014.00053
Davoudi, M., Song, M., Zhang, M., Chen, J., Lou, Q. (2022). Long-distance control of the scion by the rootstock under drought stress as revealed by transcriptome sequencing and mobile mRNA identification. Horticulture Res. 9, uhab033. doi: 10.1093/hr/uhab033
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., Durinx, C. (2021). Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49, W216–W227. doi: 10.1093/nar/gkab225
Eddy, S. R. (2011). Accelerated profile HMM searches. PloS Comput. Biol. 7, e1002195. doi: 10.1371/journal.pcbi.1002195
Fan, T., Lv, T., Xie, C., Zhou, Y., Tian, C. (2021). Genome-wide analysis of the IQM gene family in rice (Oryza sativa L.). Plants 10, 1949. doi: 10.3390/plants10091949
Gauthier, A., Trouvelot, S., Kelloniemi, J., Frettinger, P., Wendehenne, D., Daire, X., et al. (2014). The Sulfated Laminarin Triggers a Stress Transcriptome before Priming the SA- and ROS-Dependent Defenses during Grapevine’s Induced Resistance against Plasmopara viticola. PloS One 9, e88145. doi: 10.1371/journal.pone.0088145
Geourjon, C., Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hassan, M. A., Dahu, N., Hongning, T., Qian, Z., Yueming, Y., Yiru, L., et al. (2023). Drought stress in rice: morpho-physiological and molecular responses and marker-assisted breeding. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1215371
Hazra, S., Chatterjee, A., Bhattacharyya, S., Sen, P. (2024). Nucleoredoxin vis-à-vis a novel thioredoxin in regulating oxidative stress in plants: A review. Agric. Res. 13, 400–418. doi: 10.1007/s40003-024-00737-3
Howie, B. N., Donnelly, P., Marchini, J. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PloS Genet. 5, e1000529. doi: 10.1371/journal.pgen.1000529
Jedmowski, C., Ashoub, A., Beckhaus, T., Berberich, T., Karas, M., Brüggemann, W. (2014). Comparative Analysis of Sorghum bicolor Proteome in Response to Drought Stress and following Recovery. Int. J. Proteomics 2014, 1–10. doi: 10.1155/2014/395905
Kneeshaw, S., Keyani, R., Delorme-Hinoux, V., Imrie, L., Loake, G. J., Le Bihan, T., et al. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc. Natl. Acad. Sci. 114, 8414–8419. doi: 10.1073/pnas.1703344114
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurooka, H., Kato, K., Minoguchi, S., Takahashi, Y., Ikeda, J., Habu, S., et al. (1997). Cloning and characterization of the nucleoredoxin gene that encodes a novel nuclear protein related to thioredoxin. Genomics 39, 331–339. doi: 10.1006/geno.1996.4493
Laughner, B. J., Sehnke, P. C., Ferl, R. J. (1998). A novel nuclear member of the thioredoxin superfamily. Plant Physiol. 118, 987–996. doi: 10.1104/pp.118.3.987
Lee, E. S., Park, J. H., Hwang, G. Y., Chi, Y. H., Kang, C. H., Chae, H. B., et al. (2020). Nucleoredoxin2 (NRX2) promotes jasmonate-mediated trichome formation in arabidopsis. J. Plant Biol. 63, 495–503. doi: 10.1007/s12374-020-09277-z
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2024). Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, H., Durbin, R. (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, N., Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Liu, H., Song, S., Zhang, H., Li, Y., Niu, L., Zhang, J., et al. (2022). Signaling transduction of ABA, ROS, and ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 23, 14824. doi: 10.3390/ijms232314824
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marchal, C., Delorme-Hinoux, V., Bariat, L., Siala, W., Belin, C., Saez-Vasquez, J., et al. (2014). NTR/NRX define a new thioredoxin system in the nucleus of arabidopsis thaliana cells. Mol. Plant 7, 30–44. doi: 10.1093/mp/sst162
Mata-Pérez, C., Spoel, S. H. (2019). Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 279, 27–33. doi: 10.1016/j.plantsci.2018.05.001
Meng, B., Wang, T., Luo, Y., Guo, Y., Xu, D., Liu, C., et al. (2022). Identification and allele combination analysis of rice grain shape-related genes by genome-wide association study. IJMS 23, 1065. doi: 10.3390/ijms23031065
Panda, D., Mishra, S. S., Behera, P. K. (2021). Drought tolerance in rice: focus on recent mechanisms and approaches. Rice Sci. 28, 119–132. doi: 10.1016/j.rsci.2021.01.002
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. doi: 10.1093/nar/gkac993
Pereira, W. J., Melo, A. T. D. O., Coelho, A. S. G., Rodrigues, F. A., Mamidi, S., Alencar, S. A. D., et al. (2020). Genome-wide analysis of the transcriptional response to drought stress in root and leaf of common bean. Genet. Mol. Biol. 43, e20180259. doi: 10.1590/1678-4685-gmb-2018-0259
Piaz, F. D., Braca, A., Belisario, M. A., De Tommasi, N. (2010). Thioredoxin system modulation by plant and fungal secondary metabolites. Curr. Medicinal Chem. 17, 479–494. doi: 10.2174/092986710790226165
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Qin, Y., Leydon, A. R., Manziello, A., Pandey, R., Mount, D., Denic, S., et al. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PloS Genet. 5, e1000621. doi: 10.1371/journal.pgen.1000621
R Core Team (2024). R: A language and environment for statistical computing. MSOR Connections 1, 23–25. doi: 10.11120/msor.2001.01010023
Ren, D., Hu, J., Xu, Q., Cui, Y., Zhang, Y., Zhou, T., et al. (2018). FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69, 4853–4866. doi: 10.1093/jxb/ery264
Sato, Y., Takehisa, H., Kamatsuki, K., Minami, H., Namiki, N., Ikawa, H., et al. (2013). RiceXPro Version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, D1206–D1213. doi: 10.1093/nar/gks1125
Song, J.-M., Lei, Y., Shu, C.-C., Ding, Y., Xing, F., Liu, H., et al. (2018). Rice information gateWay: A comprehensive bioinformatics platform for indica rice genomes. Mol. Plant 11, 505–507. doi: 10.1016/j.molp.2017.10.003
Tian, S., Cheng, J., Yang, J., Li, R., Zhang, Y., Hao, X., et al. (2019). Interaction analysis of taNRX1 with taPDI, taTRX-h and taPP2Ac and drought resistance study. J. Agric. Biotechnol. 27, 1901–1911. doi: 10.3969/j.issn.1674-7968.2019.11.001
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Wang, X., Pan, C., Long, J., Bai, S., Yao, M., Chen, J., et al. (2022). Genome-wide identification of the jumonji C domain- containing histone demethylase gene family in wheat and their expression analysis under drought stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.987257
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Yao, Y., Xiang, D., Wu, N., Wang, Y., Chen, Y., Yuan, Y., et al. (2023). Control of rice ratooning ability by a nucleoredoxin that inhibits histidine kinase dimerization to attenuate cytokinin signaling in axillary buds. Mol. Plant 16, 1911–1926. doi: 10.1016/j.molp.2023.10.009
Yates, A. D., Allen, J., Amode, R. M., Azov, A. G., Barba, M., Becerra, A., et al. (2022). Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50, D996–D1003. doi: 10.1093/nar/gkab1007
Zhang, S., Yu, Y., Song, T., Zhang, M., Li, N., Yu, M., et al. (2022). Genome-wide identification of foxtail millet’s TRX family and a functional analysis of SiNRX1 in response to drought and salt stresses in transgenic Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946037
Zhang, Y., Zhou, J., Wei, F., Song, T., Yu, Y., Yu, M., et al. (2021). Nucleoredoxin gene taNRX1 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756338
Keywords: rice, nucleoredoxin gene family, drought stress, genome-wide identification, expression pattern
Citation: Liu Z, Zheng X, Yang D, Li L and Yin H (2025) Genome-wide identification of the nuclear redox protein gene family revealed its potential role in drought stress tolerance in rice. Front. Plant Sci. 16:1562718. doi: 10.3389/fpls.2025.1562718
Received: 18 January 2025; Accepted: 28 March 2025;
Published: 22 April 2025.
Edited by:
Dev Mani Pandey, Birla Institute of Technology, Mesra, IndiaReviewed by:
Klára Kosová, Crop Research Institute (CRI), CzechiaPrachi Pandey, National Institute of Plant Genome Research (NIPGR), India
Elsayed Nishawy, Desert Research Center, Egypt
Copyright © 2025 Liu, Zheng, Yang, Li and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanzhi Li, bGlsYW56aGlAaHVuYXUuZWR1LmNu; Hexing Yin, aGV4aW5nLnlpbkBoaWdlbnRlYy5jb20=
 Zijie Liu
Zijie Liu Xingfei Zheng2
Xingfei Zheng2 Dabing Yang
Dabing Yang Lanzhi Li
Lanzhi Li Hexing Yin
Hexing Yin