
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 February 2025
Sec. Plant Metabolism and Chemodiversity
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1557544
This article is part of the Research TopicMetabolomics in Crop Research – Current and Emerging Methodologies, Volume IIIView all 13 articles
The oil palm (Elaeis guineensis Jacq.) is a perennial oilseed crop whose mesocarp produces palm oil rich in the unsaturated fatty acid oleic acid, known for its oxidative stability and cardiovascular health benefits. However, the regulatory mechanisms and pathways responsible for variations in oleic acid biosynthesis during fruit development remain inadequately elucidated. The study examined the mesocarp of oil palm fruits from three developmental stages in seedless and Tenera varieties to evaluate oleic acid content. Fruits from Seedless (MS) and Tenera (MT) oil palms, pollinated for 95 days (MS1 and MT1), 125 days (MS2 and MT2), and 185 days (MS3 and MT3), were analyzed using metabolomics via liquid chromatography-tandem mass spectrometry (LC-MS/MS). RNA sequencing was conducted to profile gene expression associated to oleic acid biosynthesis and accumulation. Differential genes and metabolites were mapped and functionally enriched through KEGG pathway analysis. The result revealed that SAD, FabD, LACS6, BC, FabB, and FabI were positively associated with oleic acid content, whereas LACS9 exhibited either a negative or strongly negative correlation. By integrating metabolomic and transcriptomic techniques, this study elucidates the distinct mechanisms of oleic acid biosynthesis in seedless and thin-shelled oil palm varieties. These findings provide a scientific foundation for enhancing oleic acid content and improving the quality of oil palm-derived products.
The oil palm (Elaeis guineensis Jacq.) is one of the most important oil crops globally, renowned for its superior productivity compared to other oilseed crops. Its oil yield is approximately eight to ten times greater than that of soybean and rapeseed, respectively, making it the most productive oil crop worldwide (Papilo et al., 2022). The mesocarp of oil palm fruit is the primary oil-producing tissue, containing predominantly 38–45% palmitic acid (C16:0) and 38–44% oleic acid (C18:1) (Dussert et al., 2013; Morcillo et al., 2021).
Oleic acid, a monounsaturated fatty acid, is known for its exceptional oxidative stability, which supports immune regulation and helps prevent cardiovascular diseases (Katariya et al., 2021). After ingestion, oleic acid undergoes hydrolysis in the stomach and enters the bloodstream intact as glycerol monoesters (Voon et al., 2019). Moreover, studies suggest that high-oleic oil palm can generate an estimated economic value of $1,500 per hectare annually when oleic acid content exceeds 65% (Sambanthamurthi et al., 2002). In comparison, high-oleic acid rapeseed oil typically contains 75–84% oleic acid (Chang et al., 2022). As a result, oleic acid holds significant nutritional and economic value. Consequently, the developing oil palm varieties with elevated oleic acid content through genetic enhancement has become a critical objective in breeding programs (Bahariah et al., 2021).
Previous research has identified key enzyme genes involved in oleic acid synthesis in oil palm. Sun et al. (2016) successfully cloned Δ12-desaturase, a gene associated with the biosynthesis of long-chain polyunsaturated fatty acids (LC-PUFAs), and demonstrated that egFAD12 exhibited peak activity in oil palm fruits 120–140 days after pollination. This enzyme facilitates the conversion of oleic acid to linoleic acid. Ben Ayed et al. (2022) performed a comparative analysis of 24 FAD2 sequences from different species and identified two single nucleotide polymorphisms (SNPs), SNP373 and SNP718, associated with oleic and linoleic acid content. These SNPs were shown to regulate FAD2 activity, either reducing or increasing oleic acid levels. Additionally, Bahariah et al. (2023) utilized a multiplex CRISPR/Cas9 platform to successfully induce mutations in the EgFAD2 gene, located on chromosome 8, and the EgPAT gene (also known as EgFATB-1), found on chromosomes 3 and 7. This approach aims to reduce the activity of the PAT and FAD2 enzymes, thereby enhancing oleic acid content. To achieve high levels of 18:1 for improved oil stability and human health, FAD2 has become a prime target for disruption using the powerful genome editing tool CRISPR/Cas9 (Okuzaki et al., 2018; Al Amin et al., 2019; Do et al., 2019). Wei et al. (2024) further identified six enzyme genes—BC, ACC, FabB, FabI, FabG, and FabD—that promote oleic acid accumulation during oil palm fruit development.
An AGAMOUS-like MADS-box transcription factor, EgAGL9, was identified through expression profiling at various developmental stages of the oil palm pericarp. Research by Zhang et al. (2022) demonstrated that EgAGL9 interacts with the enzyme genes EgSAD, EgTSA, and EgSDH in the fatty acid synthesis pathway, leading to a substantial reduction in the proportion of unsaturated fatty acids, including oleic, linoleic, and linolenic acids. Additionally, EgMADS2 has been shown to regulate EgDGAT expression, significantly decreasing linoleic acid content while increasing oleic acid levels in transgenic embryos (Li et al., 2020). A full-length cDNA encoding EgLACS9 was also cloned from oil palm pericarp. Overexpression of EgLACS9 in the LACS-deficient Saccharomyces cerevisiae strain YB525 resulted in reduced levels of C18:1 (oleic acid) and C18:2 (linoleic acid) (Wang et al., 2021). Moreover, downregulation of the transcription factor EgGRP2A resulted in decreased expression of EgFATA and a concomitant reduction in oleic acid content (Luo et al., 2024). A seed-like fruit-specific complex network has been established in the mesocarp of oil palm.NF-YA3, NF-YC2, and ABI5 directly activate WRI1-1 and a subset of FA synthesis genes. NF-YA3 also physically interacts with NF-YC2, ABI5 and WRI1-1 to form transcription complexes that regulate gene expression. WRKY40 cooperates with WRKY2 to repress ABI5, resulting in the synthesis of oils and oils (Yeap et al., 2017). Additionally, researchers have explored the biosynthesis, transport, key enzymes and their regulation of C18 UFAs, especially the emerging regulatory network involving transcription factors and upstream signaling pathways (He et al., 2020). The findings collectively contribute to understanding the genetic regulation of oleic acid synthesis and highlight potential targets for genetic improvement to enhance oil quality.
Although initial investigations into oleic acid in oil palm have been conducted, there is a lack of comprehensive studies examining the variation in oleic acid content across different oil palm varieties and developmental stages. This study utilized high-throughput sequencing technology for transcriptomics and liquid chromatography-tandem mass spectrometry (LC-MS/MS) for metabolomics to examine the changing patterns of oleic acid during oil palm growth and development. By analyzing the differences in oleic acid content across various oil palm varieties, this research elucidates the dynamic characteristics of oleic acid at different developmental stages and provides a scientific foundation for enhancing the quality of oil palm.
The experimental samples were collected from oil palm fruits of seedless and thin-shelled varieties, harvested at different developmental stages from the Coconut Research Institute of the Chinese Academy of Tropical Agricultural Sciences (19°33′N, 110°47′E), located at the institute’s oil palm test site. The samples consisted of mesocarp tissue obtained from oil palm fruits at three distinct developmental stages: 95 days (early developmental stage: MS1 and MT1), 125 days (mid-developmental stage: MS2 and MT2), and 185 days (late developmental stage: MS3 and MT3) post-pollination. For each developmental stage, three biological replicates were taken, ensuring a robust representation of the variation within each stage. Following collection, all samples were immediately stored in liquid nitrogen at -80°C to preserve metabolomic and transcriptomic integrity for subsequent analysis. These stored samples were then subjected to high-throughput sequencing and liquid chromatography-tandem mass spectrometry (LC-MS/MS) for in-depth investigation of the metabolic and transcriptional profiles.
Mesocarp samples were prepared for metabolomic analysis using an ExionLC ultra-high-performance liquid chromatography (UHPLC) system coupled with a SCIEX QTRAP 6500+ tandem mass spectrometer. Metabolite extraction involved tissue homogenization, solvent extraction, and filtration to ensure sample integrity and reproducibility. Metabolite separation was conducted under optimized chromatographic conditions, and detection was performed in multiple reaction monitoring (MRM) mode for high sensitivity and specificity. Raw mass spectrometry data were processed with Analyst 1.6.3 software, including peak detection, alignment, and integration. Metabolite identification was achieved by matching spectral data with local lipid databases, while spectral peaks across samples were aligned and corrected for variability. Absolute metabolite concentrations were determined from integral peak areas. Differentially abundant metabolites were identified through orthogonal partial least squares discriminant analysis (OPLS-DA), with selection criteria including a variable importance in projection (VIP) score of ≥ 1, a fold change of ≥ 2 for upregulation, or ≤ 0.5 for downregulation. Results from MRM mode were visualized in Supplementary Figure S2, and identified metabolites were further analyzed to provide insights into oleic acid biosynthesis, elucidating developmental and varietal differences in oil palm.
Total RNA was extracted from mesocarp tissues collected at 95, 125, and 185 days after pollination using a commercial RNA isolation kit (Tiangen Biotech, China), following the manufacturer’s protocol, with homogenization performed under liquid nitrogen to preserve RNA integrity. RNA quality and integrity were assessed using an Agilent 2100 Bioanalyzer, with samples achieving RNA integrity number (RIN) values ≥ 7.0 deemed suitable for downstream applications. Purity was evaluated using A260/A280 and A260/A230 ratios measured on a NanoDrop spectrophotometer, while RNA concentrations were quantified with a Qubit RNA assay kit. The cDNA fragments underwent end repair, A-tailing, adapter ligation, and PCR amplification, with library quality assessed using an Agilent 2100 Bioanalyzer and concentrations determined via qPCR. After passing the library check, different libraries are pooled according to the effective concentration and the target downstream data volume required for Illumina sequencing, and 150bp paired-end reads are generated. Four types of fluorescently labeled dNTP, DNA polymerase, and junction primers are added to the sequencing flow cell for amplification. When extending the complementary strand of each sequencing cluster, each fluorescently labeled dNTP added releases corresponding fluorescence, which is then captured by the sequencer, and converted into sequencing peaks by the computer software, thus obtaining the sequence information of the fragment to be sequenced.
Raw data were filtered using fastp v 0.19.3, mainly removing reads with adapters; when the N content of any sequencing read exceeded 10% of the number of bases in the read, the paired reads were removed; when the number of low-quality (Q<=20) bases in any sequencing read exceeded 50% of the number of bases in the read, the paired reads were removed. All subsequent analyses were based on clean reads. reference genomes and their annotation files were downloaded from the indicated websites, indexes were constructed using HISAT v2.1.0, and clean reads were aligned to the reference genomes. Differentially expressed genes (DEGs) were identified using DESeq2 with stringent criteria: false discovery rate (FDR) < 0.05 and |log2FoldChange| ≥ 1. Identified DEGs underwent further analysis, including Nr functional annotation, enrichment analysis, and other bioinformatics approaches, to explore their roles in metabolic and regulatory pathways.
Data analysis was performed using Excel 2019 and SPSS software. For graphical representation, Origin 2022 and GraphPad Prism 9.5 were utilized. Venn diagrams and heatmaps were generated using the online tool available at https://www.omicshare.com/tools/. Statistical significance was determined based on the appropriate tests for each data set, with p-values < 0.05 considered statistically significant.
Studies on oleic acid metabolites in the pulp of seedless and thin-shelled oil palm varieties at various developmental stages revealed that the free fatty acid composition in the pulp consisted of 13 unsaturated and 17 saturated fatty acids, with oleic acid being the dominant fatty acid. In the seedless oil palm variety, oleic acid content gradually increased from 277.30 nmol/g at the MS1 stage to 31,536.14 nmol/g at the MS2 stage, reaching a peak of 32,992.80 nmol/g at the MS3 stage. This increase represented 15.64%, 58.81%, and 56.66% of the total free fatty acid content at each respective stage. Furthermore, oleic acid accounted for 64.27%, 75.44%, and 69.31% of the total unsaturated fatty acids at these stages (Figures 1A–C).

Figure 1. Dynamic changes of oleic acid content in Seedless and Tenera oil palm at different developmental stages. (A) Dynamics of oleic acid, unsaturated fatty acids, and total free fatty acids (B) Oleic acid as a percentage of total free fatty acids (C) Oleic acid as a percentage of total unsaturated fatty acids (D) Dynamic distribution of substances in all substance groups. Each point in the graph represents a lipid molecule. The vertical coordinate represents the corresponding content (log10 conversion) of each lipid molecule, and the lipid molecules with the highest contents are labelled. Different color curves represent different groupings.
In the thin-shelled oil palm variety, oleic acid content increased from 4,549.25 nmol/g at the MT1 stage to 26,021.06 nmol/g at the MT2 stage, reaching a maximum of 30,396.86 nmol/g at the MT3 stage. This corresponded to 48.85%, 56.88%, and 50.51% of the total free fatty acid content, respectively. Oleic acid also made up 68.87%, 78.2%, and 73.31% of the total unsaturated fatty acids at the respective stages (Figures 1A–C).
The increase in oleic acid content from the MT1 to MT2 stage was greater in seedless oil palm (31,258.84 nmol/g) than in thin-shelled oil palm (21,471.81 nmol/g). From mid-development onwards, the oleic acid content of seedless oil palm exceeded that of thin-shelled oil palm, and the oleic acid content of the two varieties of oil palm reached a maximum at a later stage of development, a trend reflecting the pattern of unsaturated fatty acid content of the two varieties. The contribution of lipids in the pulp of the two varieties of oil palm at different developmental stages is as follows: at the early stage of development of the seedless species (MS1), the highest content of phosphatidylethanolamine (18:2_16:0), at the middle stage of development of the fruit (MS2), the highest content of fatty acids in the fruit pulp is said to be oleic acid (18:1), and at the late stage of development (MS3), the highest content of glycerol diesters (18:1_18:1). MS3), the highest content in the pulp was triglycerides (18:1_18:1); in the early stage of development of thin-shelled species of oil palm (MT1), the highest content in the pulp was phosphatidic acid (16:0_18:2), and in the middle stage of development of thin-shelled species of oil palm (MT2) and the late stage of development (MT3), the highest content in the pulp was triglycerides (16:0_18:1) (Figure 1D).
Differential metabolite analysis was conducted across different developmental periods of oil palm, using criteria of fold-change ≥ 2 or fold-change ≤ 0.5 and VIP ≥ 1. A total of 19 differential metabolites were identified through comparisons of three developmental stages of oil palm (Table 1). In the comparison between the MS1 vs. MT1 stage, 12 differential metabolites were found, consisting of 11 up-regulated metabolites and 1 down-regulated metabolite (Figure 2A). At the MS2 vs. MT2 stage, 7 differential metabolites were identified, with 1 up-regulated metabolite and 6 down-regulated metabolites (Figure 2B). Finally, at the MS3 vs. MT3 stage, 9 differential metabolites were observed, with 4 up-regulated and 5 down-regulated metabolites (Figure 2C).
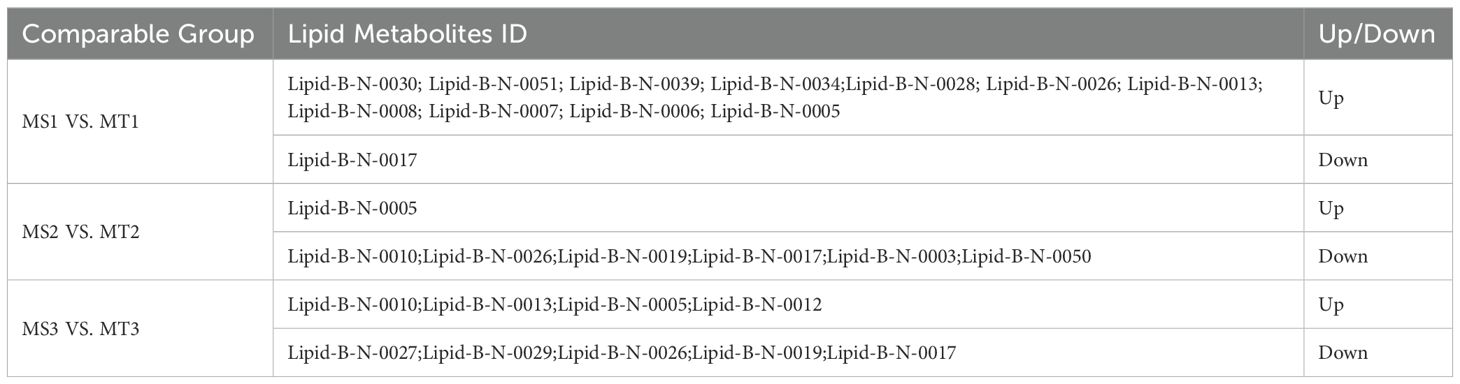
Table 1. Differential metabolites in different comparison groups of oil palm mesocarp in Seedless and Tenera oil palm.
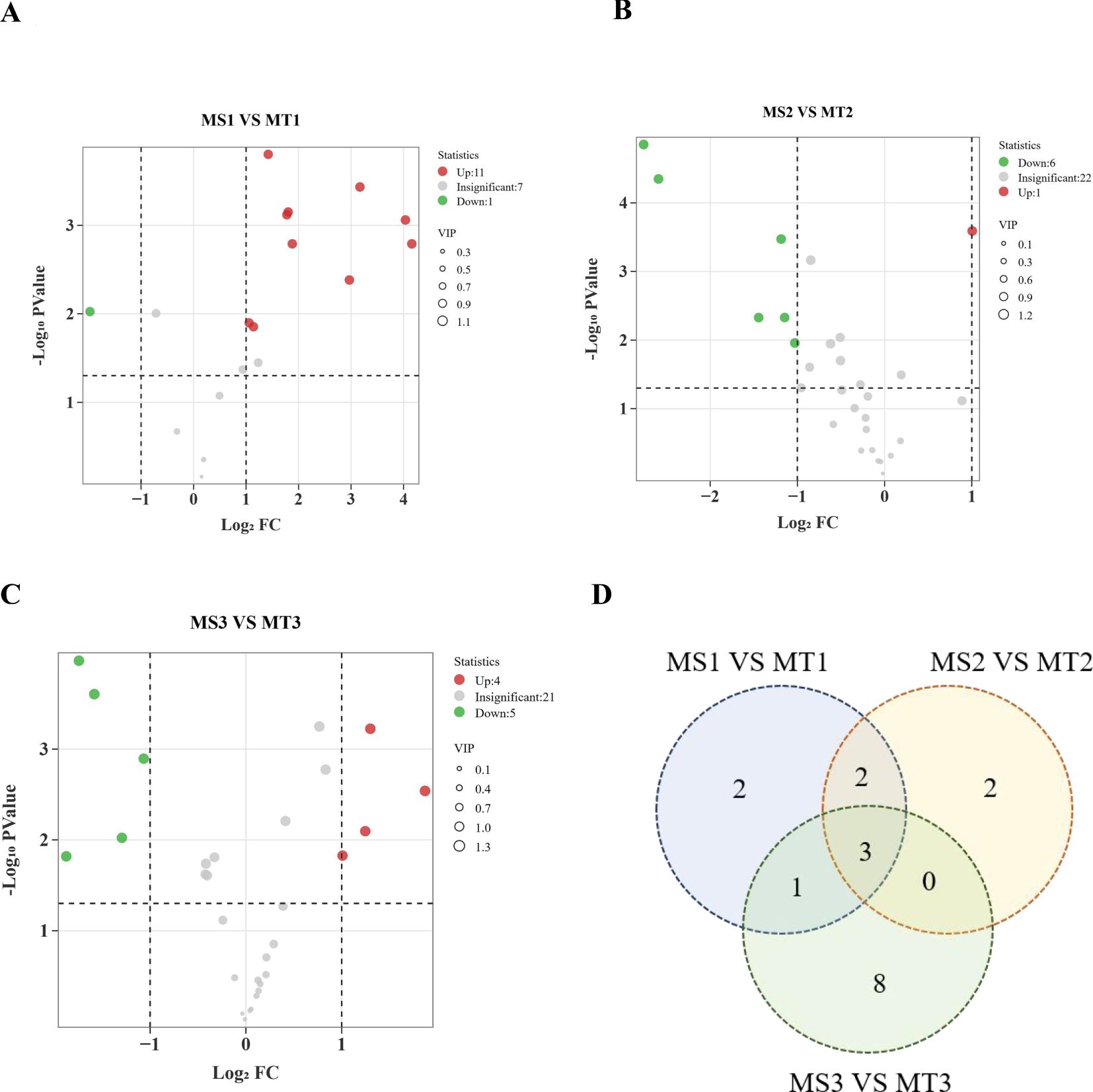
Figure 2. Statistics of different metabolites of oil palm in Seedless and Tenera (A-C). (A-C) are volcano plots of MS1 VS MT1, MS2 VS MT2 and MS3 VS MT3 differential metabolites, respectively (D) Venn map. Each point represents a metabolite, with the horizontal axis showing the logarithm of the fold change between the two sample varieties (Log2 fold change) and the vertical axis representing the significance of the variable (VIP). Larger absolute values on the horizontal axis indicate greater significance of the difference. Green dots denote down-regulated metabolites, red dots represent up-regulated metabolites, and gray dots indicate metabolites with significant changes.
Three metabolites were commonly differential across all three comparison groups (Figure 2D): tridecanoic acid (Lipid-B-N-0017), myristic acid (Lipid-B-N-0005), and palmitoleic acid (Lipid-B-N-0026) (Table 2). Among these, only myristic acid was consistently up-regulated across all stages, while tridecanoic acid was the only metabolite consistently down-regulated. Palmitoleic acid was up-regulated in the early developmental stages but down-regulated in the mid- and late stages. Additionally, oleic acid (Lipid-B-N-0028) was up-regulated during the early stages of development. These findings offer valuable insights into the dynamic changes of fatty acid metabolites in the pulp of oil palm across different developmental stages.
Analysis of differentially expressed genes (DEGs) across developmental periods in oil palm revealed significant up- and down-regulation of gene expression. A comparison of transcriptome data between the two oil palm varieties at each growth stage during three developmental periods (MS1 vs. MT1, MS2 vs. MT2, and MS3 vs. MT3) identified 7140, 4881, and 4686 DEGs, respectively. Of these, 3159, 2395, and 2567 were up-regulated, while 3991, 2491, and 2129 were down-regulated (Figure 3A). Across all three comparative groups, a total of 1401 genes were significantly differentially expressed (Figure 3B). Additionally, 454 up-regulated and 626 down-regulated genes were common across both oil palm varieties in the three developmental periods (Figures 3C, D).
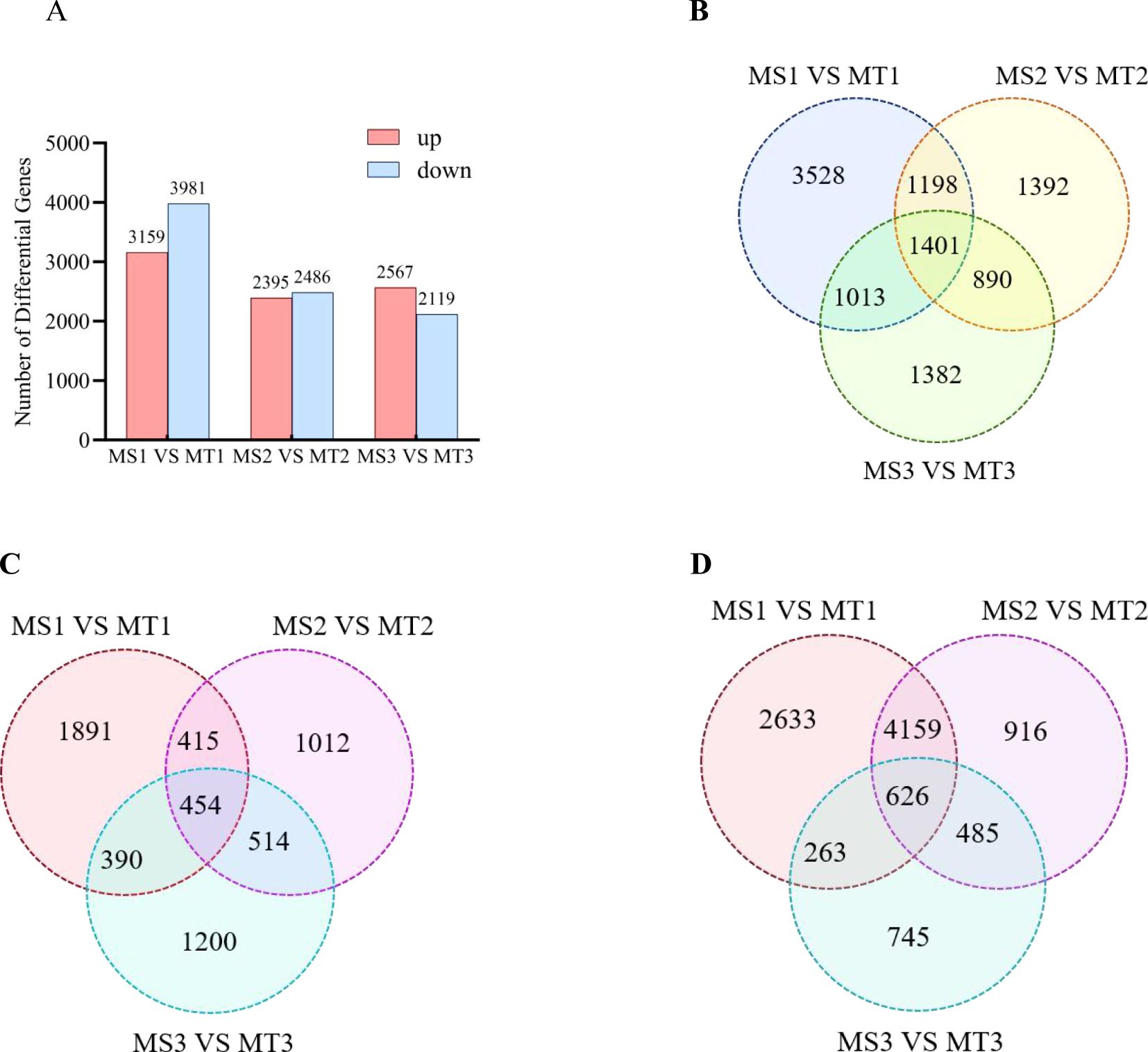
Figure 3. Statistics of different genes of oil palm in Seedless and Tenera (A) Histogram (B) Venn map of up/down-regulated differential genes (C) Venn map of up-regulated differential genes (D) Venn map of down-regulated differential genes.
Enrichment analysis of significantly differentially expressed genes (DEGs) in oil palm fruits across different developmental periods, integrating metabolome data with transcriptome data and referencing KEGG metabolic pathway information, revealed that free fatty acids were enriched in five key lipid metabolism pathways of oil palm (Figures 4A–C). The five fatty acids with significant differences in the fatty acid biosynthesis pathways of oil palm, including those from Seedless and Tenera species, were two unsaturated fatty acids—oleic acid and palmitoleic acid—and three saturated fatty acids—palmitic acid, lauric acid, and myristic acid. These fatty acids corresponded to 37 differential genes (Table 3).
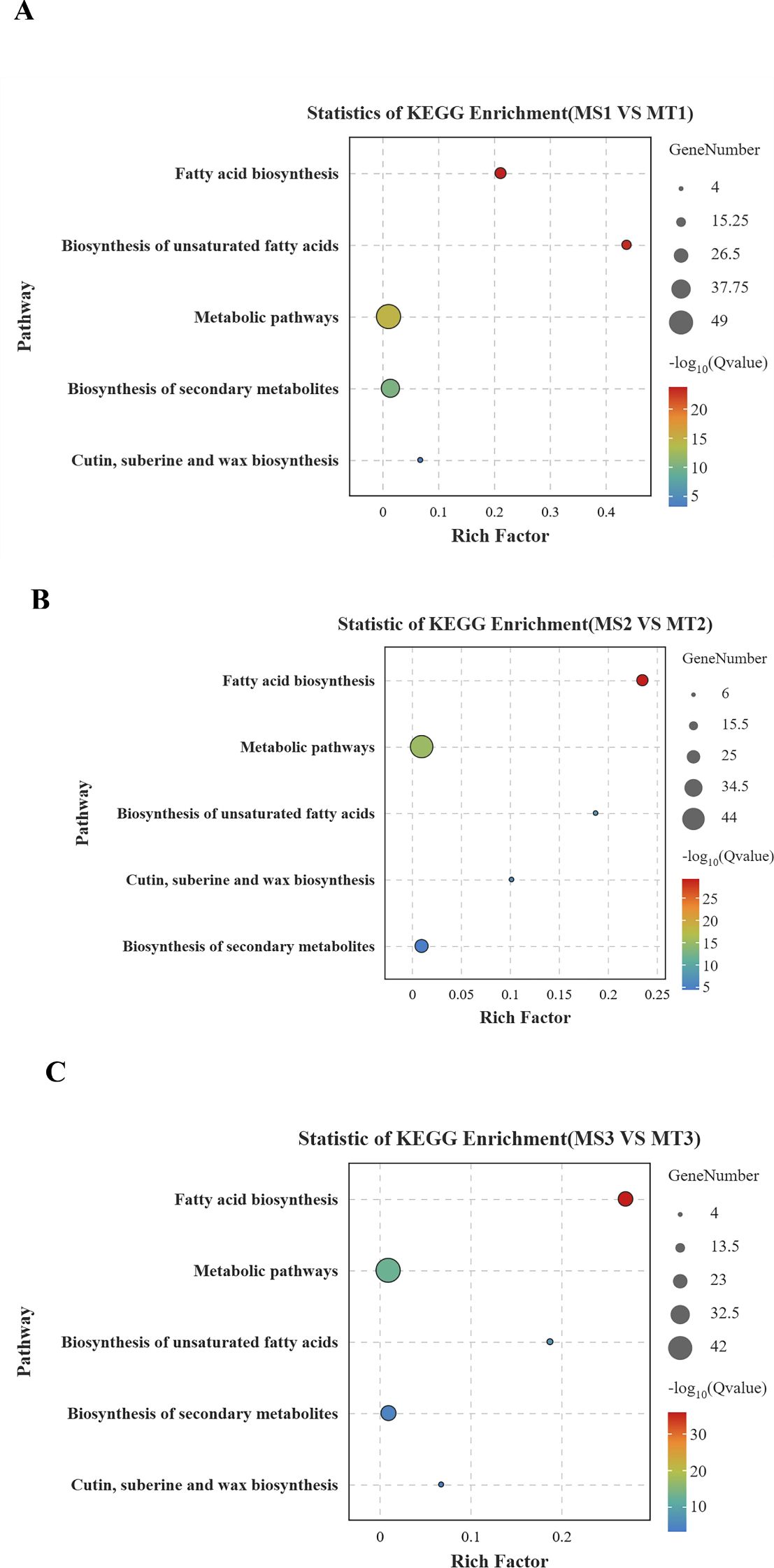
Figure 4. Bubble map of KEGG enrichment in oil palm fruit at different developmental periods. (A) differential gene KEGG results for MS1 vs MT1; (B) differential gene KEGG results for MS2 vs MT2; (C) differential gene KEGG results for MS3 vs MT3. The size of the circle indicates the number of different genes, and the larger the circle, the more genes. q value is a p-value that has been verified by multiple factors.
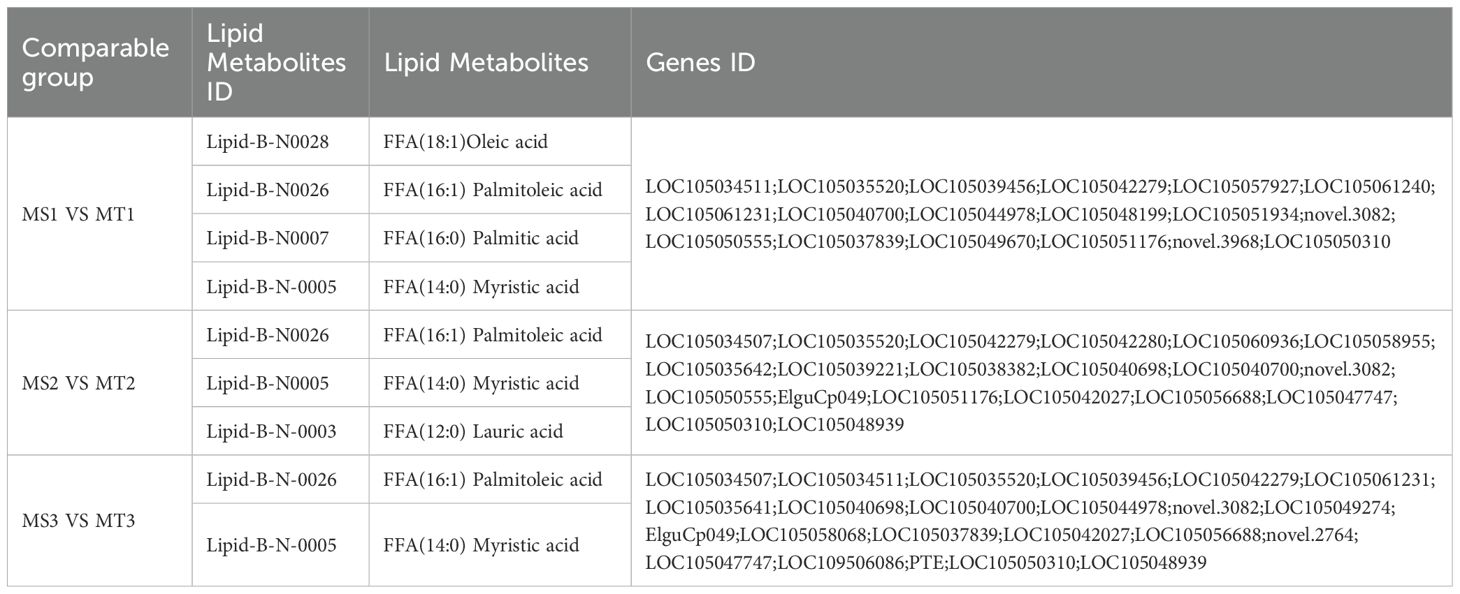
Table 3. Differential lipid metabolites and gene statistics of fatty acid biosynthesis (ko00061) pathway at different developmental periods of Seedless and Tenera oil palm mesocarp.
NR annotation of the 37 significantly differentially expressed genes in Seedless and Tenera oil palm species highlighted that genes such as SAD、 FabD, LACS6, LACS9, FabB, BC, and FabI were highly expressed during oil palm fruit development. Regarding the dynamic changes in the expression of these enzyme genes, SAD and BC exhibited a trend of increasing and then decreasing expression during fruit development in both seedless and thin-shelled oil palms, peaking at the middle developmental stage (Figures 5A, B). LACS6 expression increased and peaked at the late developmental stage, while LACS9 expression patterns differed between the two varieties: in seedless oil palm, LACS9 expression was highest at MS1 and then decreased, whereas in thin-shelled oil palm, LACS9 expression was lowest at MT1, subsequently increased, and peaked at MT3 (Figure 5C). FabB expression increased with development in thin-shelled oil palm and peaked at MT3 (Figure 5D), and the expression patterns of FabD and FabI increased and then decreased, reaching a maximum during the middle developmental stage (Figures 5E, F).
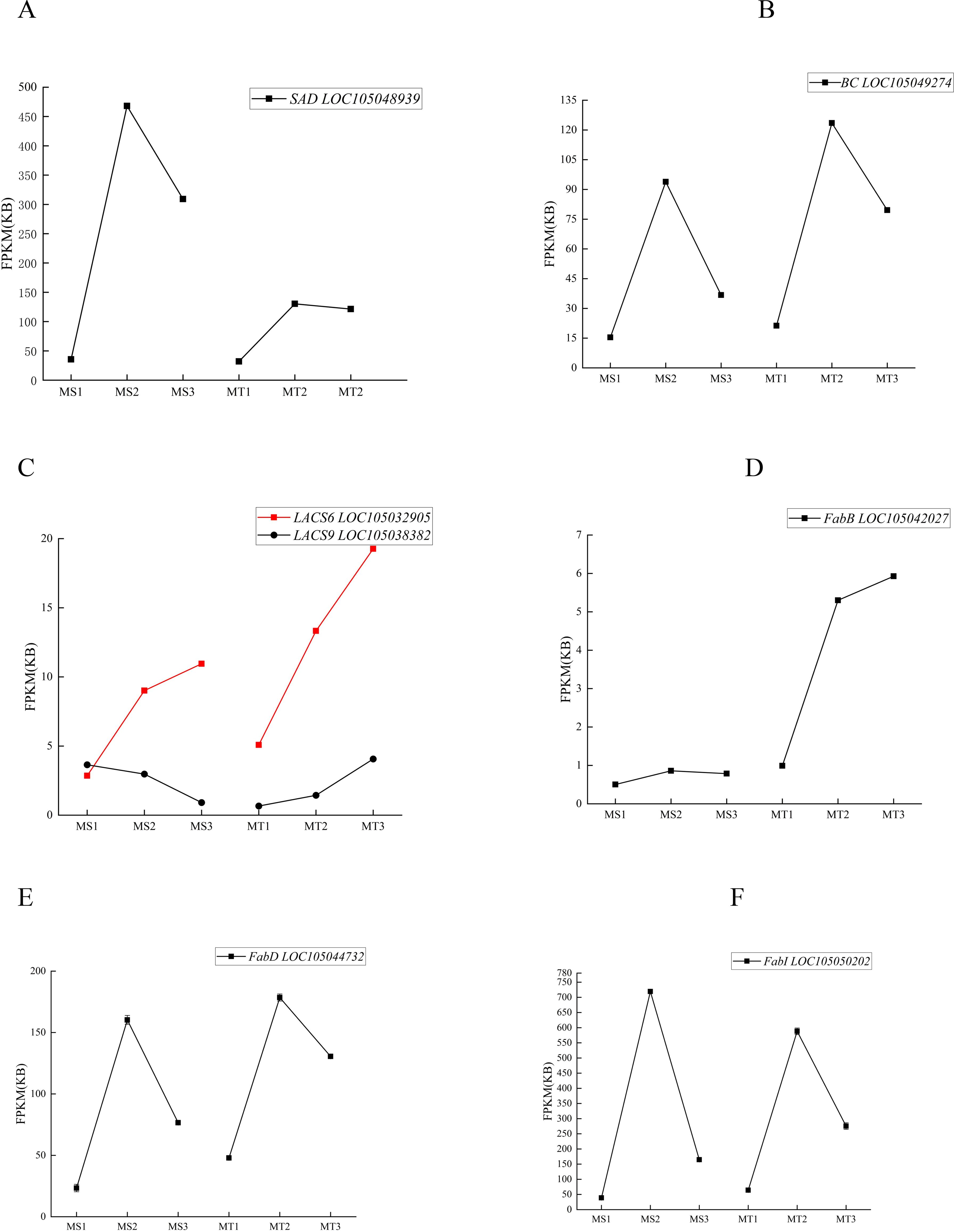
Figure 5. The expression levels of key enzyme genes changed dynamically in different developmental stages of seedless and Tenera oil palm (A-F).
The dynamic changes in the content of the five differential metabolites were correlated with the expression changes of these seven enzyme genes (Figure 5; Tables 4, 5). The results indicated that SAD, FabD, LACS6, BC, FabB, and FabI were positively correlated with oleic acid content in both oil palm varieties, while LACS9 exhibited a negative correlation with these five free fatty acids (Tables 4, 5). This suggests that the expression of these enzyme genes (SAD, FabD, LACS6, BC, FabB, and FabI) promotes oleic acid accumulation in the pulp, while LACS9 expression inhibits this accumulation.
The study revealed a significant increase in oleic acid levels during the developmental stages of oil palm fruits in both seedless and thin-shelled varieties, with pronounced accumulation occurring in the middle stage. By the late stage, the oleic acid content stabilized. In the late developmental stage, oleic acid content stabilized. Throughout the developmental process, the seedless species consistently exhibited higher oleic acid levels compared to the thin-shelled species. These results provide a foundational basis for the development of high-oleic-acid planting materials and strategies to enhance oleic acid content.
Previous research supports these findings, highlighting the conserved roles of LACS family genes across different plant species. In Arabidopsis thaliana, most of the LACS family genes (comprising nine members) have been well characterized, with distinct functions in lipid metabolism (Zhang et al., 2018). Notably, LACS6 in Arabidopsis has been implicated in the activation of fatty acids for storage lipid synthesis, aligning with our observations in oil palm. Additionally, studies in other crops have revealed similar patterns. For instance, two LACS6 enzyme genes were found to exhibit strong correlations between their expression levels and metabolite content in oil-producing tissues. Specifically, LACS6 showed high expression in the pulp, with peak expression occurring during the mid-to-late developmental stages (MT3 period), which is consistent with our findings in oil palm. Similar trends were also reported in stalked lentils (Bao et al., 2021), where LACS6 expression was closely associated with increased lipid accumulation during key developmental phases. Zhong et al. (2023) reported that the expression of nearly all LACS genes was significantly higher in high-seeded oilseed cotton compared to low-seeded varieties, highlighting their importance in seed oil production. Similarly, in kale-type oilseed rape, BnLACS2 plays a crucial role in seed oil biosynthesis, exhibiting substrate preferences for fatty acids such as 14:0, 16:0, 18:0, 18:1, and 22:1 (Ding et al., 2020). Furthermore, LACS genes have been shown to exhibit distinct expression patterns across different tissues and developmental stages (Ayaz et al., 2021). For instance, earlier research found that overexpression of EgLACS9 in oil palm led to a significant reduction in palmitic, oleic, and linoleic acids (Wang et al., 2021), which is consistent with our observation that LACS9 may inhibit oleic acid accumulation in seedless oil palms. In contrast, LACS6 has been identified as key enzyme in activating long-chain fatty acids for cellular lipid synthesis and degradation through β-oxidation (Shockey et al., 2002). In the present study, the LACS6 exhibited a positive correlation with the five major free fatty acids in seedless oil palm (Table 4), indicating its role in enhancing the synthesis of these fatty acids in this variety. However, in thin-shelled oil palm, LACS6 showed a negative correlation with lauric acid but positive correlation with palmitic acid, oleic acid, palmitoleic acid, and myristic acid (Table 5). This suggests that LACS6 may promote the synthesis of these four fatty acids while inhibiting the lauric acid production in the mesocarp of thin-shelled oil palm.
The BC gene plays an important role in fatty acid biosynthesis by catalyzing the biotin carboxylation reaction (Laseke et al., 2023). In the present study, BC exhibited a positive correlation with the levels of five free fatty acids in the pulp of seedless oil palm (Table 4), suggesting that BC may facilitate the synthesis of these fatty acids in this variety. Interestingly, in thin-shelled oil palm, BC showed a contrasting pattern- it was negatively correlated with lauric acid but positively correlated with palmitic acid, palmitoleic acid, oleic acid, and myristic acid (Table 5). This indicates that the expression of the BC enzyme gene may enhance the synthesis of the four fatty acids while simultaneously inhibiting the production of lauric acid in thin-shelled oil palm.
SAD genes play a pivotal role in fatty acid biosynthesis by catalyzing the desaturation of stearic acid (C18:0) to oleic acid (C18:1), a key step in the production of unsaturated fatty acids (Liu et al., 2020). In hybrid oil palm, the expression of OeSAD1, OeSAD2, and OeSAD3 has been observed to increase during early developmental stages before declining in later stages, highlighting their dynamic role in lipid metabolism (Wang et al., 2022). Similarly, in peony seeds, SAD1 and SAD2 have been shown to catalyze the desaturation of C18:0 to C18:1, underscoring their importance in the biosynthesis of unsaturated fatty acids (Li et al., 2021). In the present study, SAD genes exhibited a positive correlation with palmitic acid, palmitoleic acid, oleic acid, myristic acid, and lauric acid in the pulp of seedless oil palm (Table 4). This suggests that SAD may promote the synthesis of these fatty acids during fruit development in seedless varieties. However, in thin-shelled oil palm, SAD genes showed a positive correlation with palmitic acid, palmitoleic acid, oleic acid, and myristic acid, but a negative correlation with lauric acid (Table 5). This indicates that SAD may enhance the synthesis of the former four fatty acids while suppressing lauric acid production in thin-shelled varieties. The result suggest that the dual role of SAD genes in regulating fatty acid composition, depending on the oil palm variety and developmental stage. The positive correlation between SAD and oleic acid in both seedless and thin-shelled oil palms underscores its critical role in promoting oleic acid biosynthesis. At the same time, the contrasting effects on lauric acid production suggest that SAD may act as a regulatory switch, modulating the balance between saturated and unsaturated fatty acids.
The FabB, FabD, and FabI genes play critical roles in the fatty acid biosynthesis pathway, contributing significantly to the accumulation of oleic acid in the mesocarp of oil palm. FabB acts as an activator of fatty acid synthase (FAS) and functions as a β-oxoacyl-ACP synthase, catalyzing the addition of acyl-ACP to malonyl-ACP to form β-oxoacyl-ACP, a key step in fatty acid synthesis II pathway (Lee et al., 2013). Enzymes encoded by the FabA and FabB genes are known to introduce double bonds in decane precursors, which are elongated into unsaturated fatty acyl chain such as the 16:1Δ9 and 18:1Δ11 essential for functional membrane phospholipids (Wei et al., 2024). In microalgae fabB/F has been identified as playing a key role in the elongation of medium fatty acids (C < 18) and the conversion of cis-16:1 to cis-18:1 (Gong and Miao, 2019). In this study, FabB was positively correlated with palmitic acid and myristic acid, oleic acid, and lauric acid in the pulp of seedless oil palm (Table 4). In thin-shelled oil palm, FabB showed positive correlation with palmitic acid, myristic acid, oleic acid, and palmitoleic acid but a negative correlation with lauric acid (Table 5). The results suggest that FabB supports the production of palmitic acid, myristic acid, and oleic acid in both seedless and thin-shelled oil palm varieties, while also enhancing lauric acid synthesis in seedless species and inhibiting its formation in thin-shelled species during fruit development.
FabD is a crucial enzyme in the type II fatty acid synthesis pathway, responsible for transferring malonyl groups from malonyl coenzyme A to acyl carrier protein (ACP) to form malonyl-ACP and free coenzyme A-SH (Magnuson et al., 1993). In this study, FabD was positively correlated with the levels of five free fatty acids in the pulp of seedless oil palm (Table 4), and with oleic acid content in the pulp of Tenera oil palm (Table 5). The results suggest that the FabD likely facilitates oleic acid synthesis in both seedless and thin-shelled oil palm species.
FabI catalyzes the final step in type II fatty acid biosynthesis and is widely utilized for the production of pharmacologically relevant chiral intermediates, such as palmitic and oleic acids, due to its unique properties in reduction and asymmetric enantioselective catalysis (Zhou et al., 2023). In this study, the FabI was positively correlated with palmitic and oleic acid content in the pulp of seedless oil palm (Table 4). Additionally, FabI showed a strong positive correlation with oleic acid in the pulp of thin-shelled oil palm, with a notably high expression levels during the middle stage of development (Table 5). These findings suggest that the FabI plays a significant role in promoting oleic acid synthesis.
Significant differences in oleic acid content were observed between seedless and thin-shelled oil palms at various developmental stages. During the early developmental period, oleic acid content was higher in thin-shelled oil palms compared to seedless species. This difference may be attributed to the high expression of LACS9 in seedless oil palms, which could inhibit oleic acid accumulation. In contrast, the elevated expression of FabB and LACS6 genes in the thin-shelled oil palms appeared to promote oleic acid accumulation in thin-shelled oil palms. The mid-developmental stage emerged as crucial for oleic acid synthesis, with genes such as FabD, LACS6, BC, SAD, FabB, and FabI exhibiting high expression, levels, lead to a significant increase in oleic acid content during this period. By the late developmental stage, oleic acid levels stabilized, with comparable levels observed in both seedless and thin-shelled varieties, This stabilization likely resulted from distinct regulatory mechanisms involving the expression of LACS9 and LACS6. These findings provide a theoretical foundation for selecting germplasm with high oleic acid content and breeding oil palm varieties enriched in unsaturated fatty acids. Future research incorporating proteomic analyses will be essential to identify differentially accumulated enzymes and establish stronger connections between transcriptomic changes and metabolite profiles. Such approaches will offer deeper insights into the regulatory mechanisms governing oleic acid biosynthesis, enabling more precise strategies for metabolic engineering and crop improvement.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WX: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. JJ: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. XL (3rd Author): Data curation, Investigation, Writing – original draft, Writing – review & editing. XL (4th Author): Validation, Visualization, Writing – original draft, Writing – review & editing. SC: Formal analysis, Supervision, Writing – original draft, Writing – review & editing. HC: Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research work was financially supported by the National Key R&D program of China (2023YFD2200700), China Agriculture Research System (CARS-14-2-31), Central Public Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630152022001)and Protection of species and variety resources on tropical palm of Ministry of Agriculture and Rural Affair (No. 1820003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1557544/full#supplementary-material
Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max. L). BMC Biotechnol. 19, 9. doi: 10.1186/s12896-019-0501-2
Ayaz, A., Saqib, S., Huang, H., Zaman, W., Lü, S., Zhao, H. (2021). Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 161, 1–11. doi: 10.1016/j.plaphy.2021.01.042
Bahariah, B., Masani, M. Y. A., Fizree, M.P.M.A.A., Rasid, O. A., Parveez, G. K. A. (2023). Multiplex CRISPR/Cas9 gene-editing platform in oil palm targeting mutations in EgFAD2 and EgPAT genes. J. Genet. Eng. Biotechnol. 21, 3. doi: 10.1186/s43141-022-00459-5
Bahariah, B., Masani, M. Y. A., Rasid, O. A., Parveez, G. K. A. (2021). Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm. J. Genet. Eng. Biotechnol. 19, 86. doi: 10.1186/s43141-021-00185-4
Bao, W. Q., Ao, D., Wang, L., Ling, Z. H., Chen, M. S., Bai, Y., et al. (2021). Dynamic transcriptome analysis identifies genes related to fatty acid biosynthesis in the seeds of Prunus pedunculata Pall. BMC. Plant Biol. 21, 152. doi: 10.1186/s12870-021-02921-x
Ben Ayed, R., Chirmade, T., Hanana, M., Khamassi, K., Ercisli, S., Choudhary, R., et al. (2022). Comparative analysis and structural modeling of elaeis oleifera FAD2, a fatty acid desaturase involved in unsaturated fatty acid composition of american oil palm. Biol. (Basel) 11, 529. doi: 10.3390/biology11040529
Chang, T., Wu, J., Wu, X., Yao, M., Zhao, D., Guan, C., et al. (2022). Comprehensive evaluation of high-oleic rapeseed (Brassica napus) based on quality, resistance, and yield traits: A new method for rapid identification of high-oleic acid rapeseed germplasm. PloS One 17, e0272798. doi: 10.1371/journal.pone.0272798
Ding, L. N., Gu, S. L., Zhu, F. G., Ma, Z. Y., Li, J., Li, M., et al. (2020). Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus. BMC Plant Biol. 20, 21. doi: 10.1186/s12870-020-2240-x
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19, 311. doi: 10.1186/s12870-019-1906-8
Dussert, S., Guerin, C., Andersson, M., Joët, T., Tranbarger, T. J., Pizot, M., et al. (2013). Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 162, 1337–1358. doi: 10.1104/pp.113.220525
Gong, Y., Miao, X. (2019). Short chain fatty acid biosynthesis in microalgae synechococcus sp. PCC 7942. Mar. Drugs 17, 255. doi: 10.3390/md17050255
He, M., Qin, C. X., Wang, X., Ding, N. Z. (2020). Plant unsaturated fatty acids: biosynthesis and regulation. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00390
Katariya, A., Gajera, V., Lambole, V., Shah, D., Joshi, H. (2021). Review on: role of oleic acid in various disease. Int. J. Pharm. Sci. Rev. Res. 197–205. doi: 10.47583/ijpsrr.2021.v67i02.031
Laseke, A. J., Boram, T. J., Schneider, N. O., Lohman, J. R., St Maurice, M. (2023). Allosteric Site at the Biotin Carboxylase Dimer Interface Mediates Activation and Inhibition in Staphylococcus aureus Pyruvate Carboxylase. Biochemistry 62, 2632–2644. doi: 10.1021/acs.biochem.3c00280
Lee, S., Lee, S., Yoon, Y. J., Lee, J. (2013). Enhancement of Long-Chain Fatty Acid Production in Escherichia coli by Coexpressing Genes, Including fabF, Involved in the Elongation Cycle of Fatty Acid Biosynthesis. Appl. Biochem. Biotechnol. 169, 462–476. doi: 10.1007/s12010-012-9987-y
Li, C., Hu, L., Que, B., Hu, Y., Guo, Y., Zhang, M., et al. (2021). Expression profiles of genes involved in fatty acid and lipid biosynthesis in developing seeds of Paeonia ostii. Genes Genomics 43, 885–896. doi: 10.1007/s13258-021-01102-2
Li, S. Y., Zhang, Q., Jin, Y. H., Zou, J. X., Zheng, Y. S., Li, D. D. (2020). A MADS-box gene, EgMADS21, negatively regulates EgDGAT2 expression and decreases polyunsaturated fatty acid accumulation in oil palm (Elaeis guineensis Jacq.). Plant Cell Rep. 39, 1505–1516. doi: 10.1007/s00299-020-02579-z
Liu, G., Wu, Z., Peng, Y., Shang, X., Xie, Y., Arnold, R. J. (2020). Transcriptome analyses reveals the dynamic nature of oil accumulation during seed development of Plukenetia volubilis L. Sci. Rep. 10, 20467. doi: 10.1038/s41598-020-77177-w
Luo, S., Huang, J., Jin, L., Zou, J., Zheng, Y., Li, D. (2024). Transcription factor EgGRP2A regulates EgFATA expression and promotes oleic acid accumulation in oil palm (Elaeis guineensis). J. Plant Physiol. 299, 154263. doi: 10.1016/j.jplph.2024.154263
Magnuson, K., Jackowski, S., Rock, C. O., Cronan, J. E., Jr. (1993). Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542. doi: 10.1128/mr.57.3.522-542.1993
Morcillo, F., Vaissayre, V., Serret, J., Avallone, S., Domonhédo, H., Jacob, F., et al. (2021). Natural diversity in the carotene, tocochromanol and fatty acid composition of crude palm oil. Food Chem. 365, 130638. doi: 10.1016/j.foodchem.2021.130638
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica Napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Papilo, P., Marimin, M., Hambali, E., Machfud, M., Yani, M., Asrol, M., et al. (2022). Palm oil-based bioenergy sustainability and policy in Indonesia and Malaysia: A systematic review and future agendas. Heliyon 8, e10919. doi: 10.1016/j.heliyon.2022.e10919
Sambanthamurthi, R., Akmar, A. S. N., Parveez, G. K. A. (2002). Genetic manipulation of the oil palm - challenges and prospects. Planter 78, 547–562.
Shockey, J. M., Fulda, M. S., Browse, J. A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129, 1710–1722. doi: 10.1104/pp.003269
Sun, R., Gao, L., Yu, X., Zheng, Y., Li, D., Wang, X. (2016). Identification of a Δ12 fatty acid desaturase from oil palm (Elaeis guineensis Jacq.) involved in the biosynthesis of linoleic acid by heterologous expression in Saccharomyces cerevisiae. Gene 591, 21–26. doi: 10.1016/j.gene.2016.06.039
Voon, P. T., Lee, S. T., Ng, T. K. W., Ng, Y. T., Yong, X. S., Lee, V. K. M., et al. (2019). Intake of palm olein and lipid status in healthy adults: A meta-analysis. Adv. Nutr. 10, 647–659. doi: 10.1093/advances/nmy122
Wang, M., Li, R., Cao, H., Jin, L., Li, X. (2022). Cloning and expression analysis of the oil palm Δ9 stearoyl-ACP dehydrogenase EgSAD gene. J. Trop. Crops 43, 235–243. doi: 10.3969/j.issn.1000-2561.2022.02.002
Wang, Y., Zhao, J., Chen, H., Zhang, Q., Zheng, Y., Li, D. (2021). Molecular cloning and characterization of long-chain acyl-CoA synthetase 9 from the mesocarp of African oil palm (Elaeis guineensis Jacq.). Scientia Hortic. 276, 109751. doi: 10.1016/j.scienta.2020.109751
Wei, L., Yang, C., John Martin, J. J., Li, R., Zhou, L., Cheng, S., et al. (2024). Metabonomics and transcriptomic analysis of free fatty acid synthesis in seedless and tenera oil palm. Int. J. Mol. Sci. 25, 1686. doi: 10.3390/ijms25031686
Yeap, W. C., Lee, F. C., Shabari Shan, D. K., Musa, H., Appleton, D. R., Kulaveerasingam, H. (2017). WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 91, 97–113. doi: 10.1111/tpj.13549
Zhang, Q., Jin, Y.-h., Zou, J.-x., Zheng, Y.-s., Li, D.-d. (2022). Characterization and functional analysis of the MADS-box EgAGL9 transcription factor from the mesocarp of oil palm (Elaeis guineensis Jacq.). Plant Sci. 321, 111317. doi: 10.1016/j.plantsci.2022.111317
Zhang, C., Maddelein, M.-L., Wai-Yin-Sun, R., Gornitzka, H., Cuvillier, O., Hemmert, C. (2018). Pharmacomodulation on Gold-NHC complexes for anticancer applications – is lipophilicity the key point? Eur. J. Medicinal Chem. 157, 320–332. doi: 10.1016/j.ejmech.2018.07.070
Zhong, Y., Wang, Y., Li, P., Gong, W., Wang, X., Yan, H., et al. (2023). Genome-wide analysis and functional characterization of LACS gene family associated with lipid synthesis in cotton (Gossypium spp.). Int. J. Mol. Sci. 24, 8530. doi: 10.3390/ijms24108530
Zhou, J., Zhang, L., Wang, Y., Song, W., Huang, Y., Mu, Y., et al. (2023). The molecular basis of catalysis by SDR family members ketoacyl-ACP reductase fabG and enoyl-ACP reductase fabI in type-II fatty acid biosynthesis. Angewandte Chemie-international Edition 62, e202313109. doi: 10.1002/anie.202313109
Keywords: oil palm, oleic acid biosynthesis, transcriptome, metabolome, lipid metabolism
Citation: Xu W, John Martin JJ, Li X, Liu X, Cheng S and Cao H (2025) Transcriptional and metabolic analysis of oleic acid synthesis in seedless and tenera oil palm species. Front. Plant Sci. 16:1557544. doi: 10.3389/fpls.2025.1557544
Received: 08 January 2025; Accepted: 05 February 2025;
Published: 25 February 2025.
Edited by:
Marta Sousa Silva, University of Lisbon, PortugalReviewed by:
Michał Rurek, Adam Mickiewicz University, Poznań, PolandCopyright © 2025 Xu, John Martin, Li, Liu, Cheng and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shunghong Cheng, MTk5OTAzM0B5bmF1LmVkdS5jbg==; Hongxing Cao, Y2FvaHhAY2F0YXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.