- State Key Laboratory of Tree Genetics and Breeding, Co-Innovation Center for Sustainable Forestry in Southern China, Jiangsu Key Laboratory for Poplar Germplasm Enhancement and Variety Improvement, Nanjing Forestry University, Nanjing, China
In order to survive harsh winter conditions, perennial trees in the temperate and frigid regions enter a dormant state and cease growth in late summer after vigorous growth in spring and summer. After experiencing prolonged cold temperature and short days in winter, trees release their dormancy, and they resume growth to produce new buds in the following spring, a process known as bud break. The establishment/release of bud dormancy and bud break are crucial for the adaptations of woody plants and their survival in the natural environment. Photoperiod and temperature are key regulators in the bud dormancy and break cycle. In recent years, significant progress has been made in understanding the molecular mechanism for how photoperiod and temperature regulate seasonal growth and dormancy. Here, we summarized the regulatory network and mechanisms underlying the seasonal growth of perennial woody plants in the temperate and frigid regions, focusing on several molecular modules including the photoperiod, circadian clock, EARLY BUD BREAK 1 (EBB1) - SHORT VEGETATIVE PHASE Like (SVL) - EARLY BUD BREAK 3 (EBB3) module and hormone regulation. Through these modules, we will summarize how perennial trees release dormancy and bud break in order to better understand their differences and connections. By elucidating the interactions among these factors, we also point out the questions and challenges need to be addressed in understanding the bud dormancy and break cycle of perennial plants.
1 Introduction
Unlike annual plants, perennial woody plants undergo a repeated cycle of growth, dormancy, and recovery. The cycle of plant dormancy and active growth has been proposed to include the transitions between endodormancy and ecodormancy (Espinosa-Ruiz et al., 2004; Welling and Palva, 2006). In late summer, buds of perennial plants cease their growth and enter a state known as bud set which is induced by disadvantageous environment such as short daylength. And this state is also called ecodormancy which could be translated to active growth by promotive conditions (Lang et al., 1987). With the transition from autumn to winter, trees enter endodormancy under the influence of low temperature and endogenous factors. Once endodormancy is established, active growth cannot be restored by promotive conditions unless endodormancy state is broken by long-term low temperature rather than merely the presence of low temperature which is similar to seed dormancy (Maurya and Bhalerao, 2017).
After experiencing a prolonged cold period during winter, trees release dormancy and reenter ecodormancy at the end of winter. Furthermore, upon exposure to promotive conditions, trees will initiate bud break and subsequently engage in active growth.
In recent years, significant progress has been made in understanding the molecular mechanisms underlying the seasonal growth of trees. The molecular regulation mechanism of bud break in woody plants is highly correlated with the regulation of flowering in terms of signaling pathways. In this process, the FLOWERING LOCUS T (FT) plays an important role in Populus L (Böhlenius et al., 2006). Photoreceptors, e.g., Phytochrome B (PHYB), work as an upstream element to transmit light signals and affect downstream flowering key genes such as FT2 to regulate the active growth of tree buds (Ding et al., 2021). Low temperature promotes the expression of the transcription factor EBB1 from the AP2/ERF family, which inhibits the expression of the MADS-box gene family SVL to promote bud break (Azeez et al., 2021). Hormones, as important endogenous signals in plants, also play a crucial role in the dormancy release and bud break processes of perennial woody plants. In the previous reviews, significant attention has been paid to highlight the mechanisms of tree dormancy (Horvath et al., 2003; Rohde and Bhalerao, 2007; Cooke et al., 2012). This article aims to summarize the regulatory modules and mechanisms involved in the regulation of dormancy release and bud break in perennial plants, especially the genes and their characteristics that participate in bud break regulation. We hope that this exposition can provide a better understanding of the dormancy release and bud break mechanism in the process of tree adaptation to the environment, and raise some questions and potential research directions for the future.
2 Physiological and biochemical changes during the transition during dormancy release and bud break
Under low-temperature conditions prevalent in winter, the expression of fatty acid desaturase genes in trees is induced to reduce the saturation of membrane fatty acids, thereby maintaining membrane fluidity (Welling and Palva, 2006). Additionally, the expression of sucrose, raffinose synthase, and starch-degrading enzymes are upregulated in the cambial region of poplar, with an increase in sugar abundance suggesting a potential positive role in cold adaptation (Welling and Palva, 2006), although this upregulation has already occurred under short-day conditions (Druart et al., 2007). Some proteins, such as antifreeze proteins (AFPs), small heat shock proteins (sHSP) and dehydrins, begin accumulating to response to low temperature to enhance plant tolerance for successful overwintering (Druart et al., 2007; Chang et al., 2021). In addition to these changes, reactive oxygen species accumulate during the endodormancy phase to promote release from dormancy, and an increase in oxidative phosphorylation efficiency during the ecodormancy phase facilitates germination (Welling and Palva, 2006; Saito et al., 2017; Beauvieux et al., 2018).
It was indicated that starch content cannot influence the bud set and bud break in poplar (Wang et al., 2022). Prior to leaf abscission in autumn, leaf proteins are hydrolyzed, and the yielded amino acids are translocated to overwintering organs to produce bark storage protein (BSP). In spring, auxin can be synthesized normally and translocated to the phloem, promoting the hydrolysis of BSP. The poplars with BSP-RNAi exhibit a significant delay in bud break during spring (Li et al., 2020).
In addition to changes in the composition of the contents, the microstructure within tree buds also undergoes changes during the transition from dormancy to bud break. A model has been proposed to facilitate our understanding towards dormancy and its release. Plasmodesmata (PD) are intercellular channels that connect and transport molecules between adjacent cells (Maule, 2008). In Arabidopsis, PD closing is induced by callose deposition, which is catalyzed by callose synthases gene CALS1. On the other hand, PD opening is induced by the endocellular activity of callose-degrading endoglucanases (Levy et al., 2007; Simpson et al., 2009). When PD is closed, nutrients and some large molecules are hindered from reaching the shoot apical meristem (SAM), resulting in cellular isolation and dormancy. PD also plays a similar important role in trees, extracellular ring of protein and callose form dormancy sphincter complexes (DSCs) to close PD and reject growth-promotive components (Rinne and Van Der, 1998; Rinne and van der Schoot, 2003).
In the recent study, samples were collected at six time points (December to March: Dec, Jan1, Jan2, Feb, Mar1, Mar2) during the dormancy release phase of poplar, with the Mar2 stage corresponding to bud break and the others representing the dormancy release phase. The ultrastructure and physiological state of samples from Jan1, Feb, Mar1, and Mar2 were observed. It was found that buds in Jan1 contained darkly stained material, a large number of lipid bodies, and plasmodesmata blocked by callose. As dormancy release progressed, the number of lipid bodies and starch granules gradually decreased, the cell walls thinned, and the number of plasmodesmata sphincters diminished (Hu et al., 2024).
The apical bud of a tree is composed of the central zone, peripheral zone, rib zone, leaf primordia, and the subapical meristem located beneath the bud tip (Liu et al., 2018). The longitudinal micrographs of Picea glauca depicting the transition from active growth to dormancy clearly illustrate the cessation of cell division and elongation in the subapical meristem, as well as the formation of bud scales (resulting from the inhibition of internode elongation above the bud scales, leading to the formation of bud set) (Cooke et al., 2012).
Morphologically, autumn buds consist SAM and leaf primordia enclosed by protective bud scales (Cooke et al., 2012). The buds transform from a reddish-brown hue to a tender green color and undergo significant enlargement during the Mar2 stage (Hu et al., 2024). Due to the lack of detailed differentiation between bud break and active growth in many research papers, the emergence of new leaf growth visible to the naked eye is generally presented as the results of bud break. Therefore, in this study, the process of more rapid production of tender buds after prolonged exposure to low temperature is considered as an indication that dormancy release or bud break has been promoted.
3 Molecular basis for bud dormancy and dormancy release
During bud dormancy, the transition from the G1 phase to the M phase is generally inhibited (Velappan et al., 2017), so the upregulation of D-CYCLIN expression which regulates the transition from G1 to S phase, is particularly important for bud growth during dormancy release and bud break (Shimizu-Sato and Mori, 2001). In hybrid poplar (Populus tremula × tremuloides), cytokinin treatment enhances the expression of CYCD3 (Randall et al., 2015). Moreover, the short-day induced decrease of PttCYCD3, and PttCYCD6 expression is necessary for bud set (Karlberg et al., 2011). It means that D-type CYCLINs may have an important function in poplar bud growth.
In Arabidopsis, the FT is partially regulated by CO, which is modulated by the circadian clock and diurnal rhythms. Under long day conditions, CO protein reaches its expression peak and it remains stable under light, thus activating the expression of downstream FT and thereby promoting flowering (Kobayashi et al., 1999). In the economically significant crop soybean, GmFT5 (Arabidopsis FT orthologs) also promotes flowering under long-day conditions (Su et al., 2024). In poplar, it was discovered that FT not only promotes flowering but also inhibits growth cessation under short-day conditions (Böhlenius et al., 2006). Overexpression of FT1 in poplar prevents bud set and allows continuous growth under short-day conditions. On the contrary, bud set of the FT1-RNAi lines occur earlier than wild type plants under long-day conditions, indicating that downregulation of FT expression is necessary for bud dormancy. What’s more, the expression of FT exhibits diurnal rhythm when the day length exceeds the critical day length for poplar. However, experiments indicated that FT1 does not show a clear diurnal rhythm throughout the day (Hsu et al., 2011). Other studies showed that FT1 is expressed in buds during winter and is induced by cold, while FT2 is highly expressed in leaves and is induced by warm long-day conditions in poplar (André et al., 2022). Further studies revealed the functional differentiation of the two homologous FT genes, of which FT1 primarily regulates bud dormancy release, and FT2 primarily promotes active growth after bud dormancy release (André et al., 2022).
As the day length increases from winter to spring, the expression of FT2 is induced. How does the expression of FTs reactivate bud growth in poplar? AP2 Family AINTEGUMENTA-Like 1 (AIL1) is expressed in shoot apical meristem and leaf primordia. Downregulation of AIL1 expression is necessary for growth cessation in poplar, although there is no direct interaction between AIL1 and FT (Karlberg et al., 2011). The APETALA1 (AP1) in Arabidopsis contains a MADS domain and is expressed in floral meristems. It subsequently localizes to petals and sepals as the flower develops, playing a role in determining the identity of the floral meristem (Abe et al., 2005). The discovery of Like-AP1 (LAP1), a poplar homolog of Arabidopsis AP1, established the regulatory link between FT2 and AIL1. PttLAP1-OE lines delay bud set compared to wild type and SD-induced downregulation of AIL1 expression is significantly suppressed in PttLAP1-OE lines, which indicates that AIL1 functions upstream of FT2 and downstream of LAP1 (Azeez et al., 2014). In Arabidopsis, the FD (a kind of bZIP transcription factor) protein is primarily expressed in the shoot apex and forms a complex with FT to regulate flowering (Abe et al., 2005). A similar complex exists in poplar. FD-Like 1/2 (FDL1/2) are FD homologs in poplar, and only FDL1 participates in light-mediated growth regulation. Additionally, BRANCHED1 (BRC1), which is the homologue of Arabidopsis BRC1, controls branching, functions in light-mediated bud growth cessation. BRC1 acts downstream of LAP1 and AIL1, and its expression is suppressed by LAP1. Thus, the suppression of FT2 expression induced by short-day promotes the expression of BRC1, which in turn inhibits FT2 expression and accelerates growth cessation (Maurya et al., 2020; Cubas, 2020). In long-day conditions, the inhibition of BRC1 is crucial for bud outgrowth. The regulatory mechanisms by which FT2 in poplar influences bud growth through direct downstream factors have been elucidated. FT2 and FDL physically interact to form a protein complex, which promotes the expression of AIL1 in buds through LAP1. Consequently, AIL1 directly binds to the promoter of D-type cyclin genes and promotes its expression, it accelerates the transition from the G1 phase to the S phase of the cell cycle in buds (Randall et al., 2015; Karlberg et al., 2011) (Figure 1). In Vitis vinifera, short-day conditions suppress VvFT-VvAP1-VvAIL2 pathway (Vergara et al., 2016). The FT gene has also undergone some functional differentiation across different species. For instance, in Norway Spruce (Picea abies), PaFT1 is predominantly expressed in summer while PaFT2 is mainly expressed in autumn, and both promote bud set (Karlgren et al., 2013).
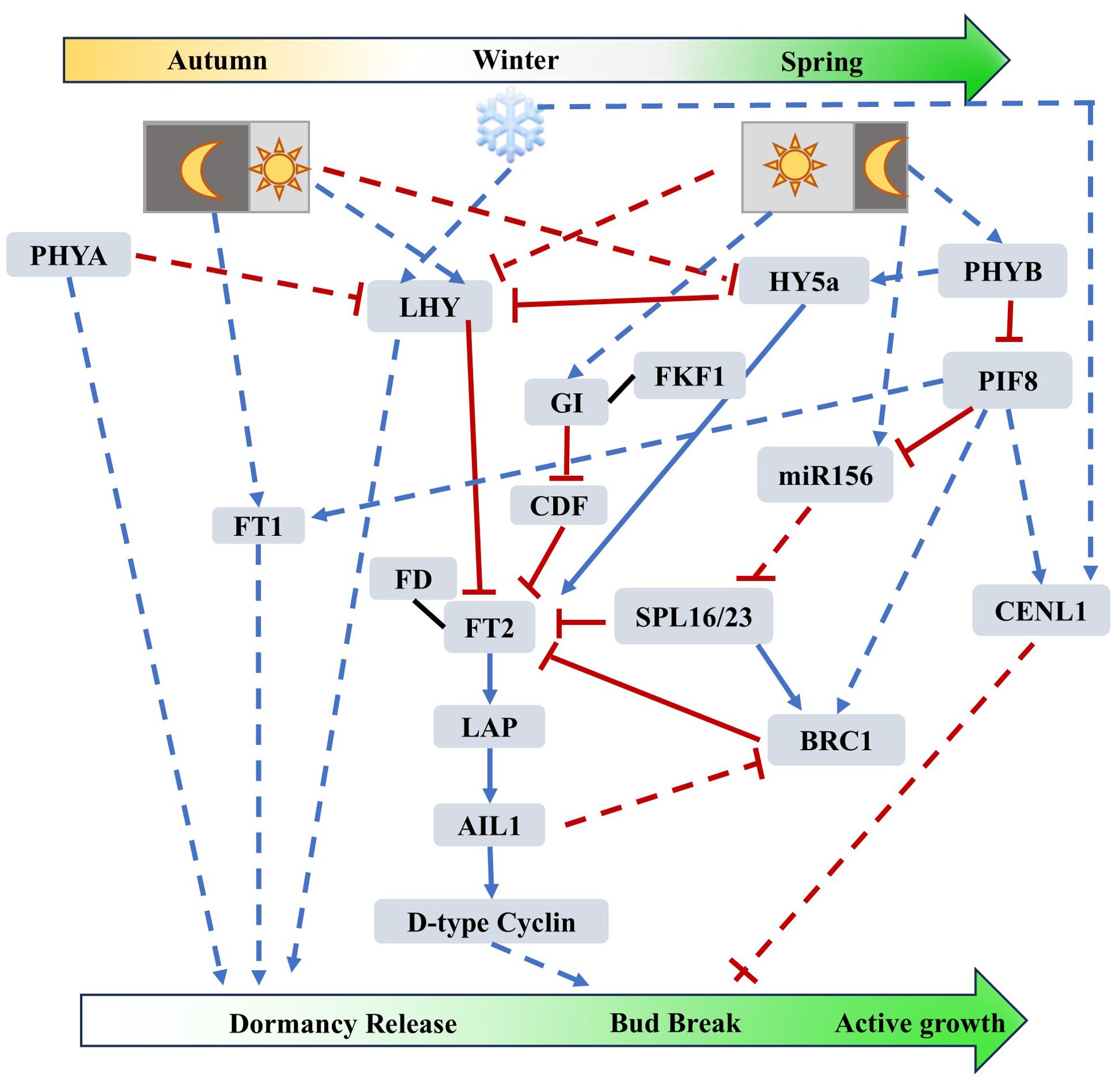
Figure 1. Photoperiodic- and Circadian Clock-mediated regulatory networks of tree dormancy release, bud break and active growth (based on studies from poplar). Prolonged exposure to low temperatures induces the accumulation of LHY, which suppresses germination in plants under cold conditions, while FT1 also accumulates to facilitate dormancy release. Under long-day conditions, FT2 accumulates in a GI-dependent and GI-independent manner, interacts with FD to exert its function, and promotes downstream D-type CYCLIN to facilitate cell division, thereby promoting bud break. Trees primarily respond to day length through phytochromes, and the expression of PHYB is upregulated under long-day conditions. On one hand, PHYB positively regulates FT2 to promote dormancy release and bud break by inhibiting the expression of SPL16/23 (an inhibitor of FT2) through PIF8 and miR156; on the other hand, PHYB positively regulates the expression of HY5a under long-day conditions to enhance the expression of FT2. PIF8 negatively regulates bud break through the entire pathway by modulating CENL1 and BRC1. Blue arrows indicate positive regulation, while red bars indicate negative regulation. The black solid lines indicate protein-protein interactions. The solid lines represent direct interactions between two elements. Dash lines indicate indirect regulation.
4 Molecular modules regulating dormancy release and bud break
Perennial plants primarily regulate their growth by perceiving changes in temperature and day length (Singh et al., 2017). The cessation of growth and establishment of dormancy in trees are mainly induced by short day length (Druart et al., 2007; Olsen, 1997). Although rapid growth in spring is induced by warmer temperature and longer day length, the reactivation of plant growth is primarily triggered by prolonged periods of low temperature, so long-term low temperature is like a signal that prompt trees spring is approaching (Hsu et al., 2011; Espinosa-Ruiz et al., 2004; Fadón et al., 2020). Previous sections have primarily elucidated the role of FTs in regulating dormancy release and bud break in perennial plants. How do perennial plants perceive environmental signals and modulate downstream signaling to control dormancy release and bud break? This section will elaborate on the molecular modules of dormancy release and bud break regulation from the following several aspects.
4.1 Photoreceptors - circadian clock - FT pathway
Seed plants typically contain 3 - 5 phytochrome genes, which play important roles in light perception. Most plants possess PHYA/B/C, while poplar only have PHYA and PHYB (Howe et al., 1998). In Arabidopsis, there are five phytochrome genes, namely PHYA-E. In Arabidopsis, PHYB primarily senses red and far-red light. PHYTOCHROME-INTARACTING FACTORS (PIFs), primarily PIF4 and PIF7, positively regulates the elongation of the hypocotyl in Arabidopsis. Under shading conditions, far-red light increases, the ratio of red light to far-red light decreases. Hence PHYB transforms from active form to inactive form and releases the repression of PIFs which leads to an increase in plant height and enhanced light capture, a behavior known as shade avoidance syndrome (SAS) (Lorrain et al., 2008; Mizuno et al., 2015). In hybrid poplar, the overexpression of PttPHYB1/2 (Populus tremula × tremuloides) is able to shorten the time required for bud break after dormancy release, while PHYB2 is more effective than PHYB1 and primarily regulates PIF8 for seasonal growth. Further experiments have revealed that FT1 and CENTRORADIALIS-LIKE1 (CENL1) maintain high expression levels even after transferred from short-day low-temperature conditions to warm conditions in PHYB-RNAi and PIF8-OE genotypes, suggesting that CENL1 and FT1 in poplar are positively regulated by PIF8. In addition, the expression level of PIF8 also exerts a negative regulation on FT2 and a positive regulation on BRC1, thereby fulfilling its function in indirect negative regulation of bud break (Ding et al., 2021). Recent research has elucidated the mechanism by which PIF8 negatively influences poplar bud break by regulating FT2 and BRC1. The findings indicate that short photoperiods suppress miR156a/c expression while miR156a/c inhibits the expression of SPL16/23 in Populus tomentosa Carr. SPL16 and SPL23 directly repress FT2 and activate BRC1 by binding to their promoters. The study explains the function of the important components between PHYB-PIF8 and FT2, BRC1 (Wei et al., 2024a). Additionally, PHYB2 has been linked to bud set timing in Populus trichocarpa × Populus deltoides (Frewen et al., 2000). Another study indicates that overexpression of barley PHYA in poplar can prevent bud set induced by short days (Olsen et al., 1997). However, bud set can occur when oat PHYA-overexpressing poplar is subjected to a 6-h light/6-h dark cycle. This experiment demonstrates the importance of the consistency between the endogenous biological clock and environmental photoperiod for the growth of poplar buds (Kozarewa et al., 2010). In addition to phytochromes, there is another class of blue light receptors in plants, cryptochromes. In Arabidopsis, CRY1 and CRY2 primarily regulate blue light-induced photomorphogenesis and flowering control (Bouveret et al., 1998; Lin et al., 1998; Mockler et al., 2002). In poplar, there are three cryptochrome genes: CRY1a, CRY1b, and CRY2. Among them, CRY1s negatively regulate poplar height and biomass, while CRY2 does not affect plant height but significantly enhance biomass. CRY2-OE lines significantly delay their bud set time induced by short-day conditions, indicating that CRY2 has a positive regulatory effect on bud active growth (Wei et al., 2024b).
GIGANTEA (GI) is another important gene in the Photoreceptors-FT pathway. In Arabidopsis, GI functions in the central oscillator of LATE ELONGATED HYPOCOTYL (LHY)/CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and TIMING OF CAB2 EXPRESSION1 (TOC1), which acts upstream of CO/FT (Cockram et al., 2007). At the same time, Arabidopsis GI can also regulate flowering through microRNA172 but independently of CO (Jung et al., 2007). Ding’s study revealed that overexpression of GI inhibits poplar growth while promoting bud break, resembling the phenotype of FT2 overexpression. Interestingly, in GI-overexpressed poplar, the expression of FT2 significantly increases under long-day conditions, while the expression of CO1 and CO2 showed minimal changes (Ding et al., 2018). What’s more, overexpression of CO1/CO2 don’t influence poplar (Populus alba × Populus tremula) bud set, it indicates that the influence of CO on FT2 may be minimal (Hsu et al., 2012). These findings suggest that PttGI may mainly regulate FT2 expression independent of CO during bud break although PttGI/PttGIL can also bind to the promoter region of CO2 (Ding et al., 2018). In Arabidopsis, CYCLING DOF FACTOR (CDF) can bind to the promoter region of CO/FT to suppress their transcription (Sawa et al., 2007). In poplar, GI and GI-Like (GIL) acts as a protein complex with FLAVIN-BINDING KELCH DOMAIN F-BOX PROTEIN 1 (FKF1) to inhibit CDF expression, which releases the inhibition of FT2 expression, thereby promoting bud break (Ding et al., 2018).
LHY/CCA1 and PSEUDO-RESPONSE REGULATOR1 (PRR1)/TOC1 are morning and evening components of the central oscillator of the circadian clock in Arabidopsis, respectively. Their transcription levels reach the peak during the morning and dawn, respectively. LHY/CCA1 are functionally redundant homologs in Arabidopsis, and their functions are conserved in both monocots and dicots. During daytime, AtLHY/AtCCA1 binds to the promoter regions of TOC1 and PRR1 to suppress their expression (Alabadí et al., 2001; Gendron et al., 2012). The expression of LHY1, LHY2, and TOC1 in poplar buds change as bud break progresses from 8°C short days (16-h light/8-h dark) to 18°C long days (6-h light/18-h dark), suggesting their potential roles during seasonal transitions. The expression of LHY2 in poplar is induced by darkness and low temperature and reaches its peak at dawn (Ramos-Sánchez et al., 2019; Ibáñez et al., 2010). Additionally, the bud break is delayed in poplar lhy mutants which is advanced in toc1 mutants, indicating the positive function of LHY and negative function of TOC1 in dormancy release regulation (Ibáñez et al., 2010). The bud set is delayed in lhy mutants upon transition from long day to short day. It indicates that LHY positively regulate poplar bud break (Ibáñez et al., 2010). Under short-day conditions, the PttFT2 expression level in the GI-overexpression remained similar to the wild type. It differs from Arabidopsis that GI overexpression can induce CO during short days (Mizoguchi et al., 2005; Ding et al., 2018). Moreover, the expression of CO2 shows no significant difference under short-day and long-day conditions, whereas CO1 expression is slightly induced under short-day conditions and overexpression of COs cannot change the bud set time of poplar (Hsu et al., 2012). These results indicate poplar can regulate their growth cessation through other means but not CO under short day conditions (Hsu et al., 2012). Interestingly, the expression of FT2 is directly inhibited by LHY2, which is regulated by night length, it shows that how upstream factors regulate bud break through FT2 under short day conditions (Ramos-Sánchez et al., 2019).
The recent research results indicate that PtoHY5a can directly bind to the FT2 promoter to activate its expression and bind to the LHY2 promoter to suppress its expression, thereby delaying the growth cessation induced by short-day conditions in poplar. After long-term low-temperature conditions, the active gibberellic acid (GA) content rises to promote bud break. When transitioning from low-temperature short-day to warm long-day conditions, overexpression of HY5 represses the expression of GA biosynthesis-related genes and promotes the expression of GA deactivation-related genes, thereby inhibiting bud break but promoting the active growth of the bud (Gao et al., 2024) (Figure 1).
4.2 EBB1-SVL-EBB3 regulatory module
The MADS-box genes FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) play important roles in regulating flowering in Arabidopsis. They form dimers to inhibit the expression of FT, thereby suppressing flowering in Arabidopsis (Hartmann et al., 2000; Mateos et al., 2015). The epigenetic modifications of FLC, specifically DNA methylation, are important for the regulation of FLC expression during vernalization in Arabidopsis (Bastow et al., 2004; Zhu et al., 2021). The discovery of dormancy-related MADS-box genes in evergreen peach raises the possibility that the release of bud dormancy in tree species may also be regulated by MADS-box genes. Studies have reported a decrease in the expression of DORMANCY ASSOCIATED MADS-BOX (DAMS) during dormancy release in peach (Prunus persica) (Leida et al., 2012). Overexpression of the DAMS in apple (Malus × domestica ‘Royal Gala’) leads to delayed bud break (Wu et al., 2017a). These findings suggest that MADS-box genes may play important regulatory roles in bud break in woody plants. As bud break is temperature-regulated, the effect of temperature on SVL (PpMADS) expression was investigated. The results showed that low temperature negatively regulates SVL expression (Leida et al., 2012; Saito et al., 2015). Furthermore, further experiments showed that PttSVL can directly interact with the CArG motif of FT1 promoter region to inhibit its expression, thus repressing dormancy release and bud break (Singh et al., 2018).
At the same time, EBB1, an AP2/ERF transcription factor, had been identified to regulate bud break through screening a poplar activation tagging population (Yordanov et al., 2014). EBB1 is primarily expressed in bud tissues of poplar and its expression is rapidly increased before dormancy release. EBB1 acts as a positive regulator of bud break, as its overexpression transgenic lines show significant bud break delay. Afterward, genetic screening for early bud break mutants identified EBB3 (Azeez et al., 2021). Building on previous studies, Azeez et al. investigated the relationships between EBB1, EBB3, and SVL. They demonstrated that EBB1 directly binds to the GCCGCCA motif of the SVL promoter to inhibit its expression. Meanwhile, SVL inhibits the expression of EBB3, and EBB3 promotes the expression of D-CYCLIN to facilitate cell division (Azeez et al., 2021). On the other hand, SVL is involved in accumulated low temperature promoted bud break through downregulating expression of TCP18 (TEOSINTE BRANCHED1, CYCLOIDEA, PCF, a transcription factor that regulates axillary bud outgrowth and controls abscisic acid (ABA) signaling)/BRC1 (Singh et al., 2018). They also show that low temperature induces EBB1 expression, and such temperature-dependent expression regulation of EBB3 is controlled by histone modifications. H3 lysine 27 trimethylation (H3K27me3) is a typical histone modification and has been studied in peach and pear (Leida et al., 2012; Saito et al., 2015). The levels of H3K27me3 at the EBB3 locus are significantly reduced following low-temperature induction, thereby promoting dormancy release. Taken together, EBB1, SVL, and the recently identified EBB3 act together as a regulatory loop in bud break. The discovery of this regulatory module leads us to a better understanding of the molecular mechanism of bud break.
In addition to the findings in poplar, the EBB1-SVL module has also been extensively studied in other perennial plants. The expression patterns of EBBs in peach (Prunus persica var. nectarina cultivar Zhongyou 4), pear (Pyrus pyrifolia Nakai), and apple are similar to that in poplar (Zhao et al., 2020; Anh Tuan et al., 2016). Additionally, EBB1 in pear and peach both promote bud break. Interestingly, overexpression of peach CBF in apple (“Malling 26” rootstock) leads to increased expression of apple EBB1which may be the reason for the delayed bud break (Wisniewski et al., 2015). Overexpression of peach EBB1 in poplar leads to increased branching and enrichment of differentially expressed genes related to growth and development (Zhao et al., 2021b). PpEBB1 was transiently transformed into peach buds, resulting in early bud break. PpEBB1 also regulates auxin biosynthesis by binding to the promoter of some related genes including STYLISH1 (STY1), SHI RELATED SEQUENCE 5 (SRS5), and YUCCA1 (YUC1) (Zhao et al., 2021a). In addition, the expression pattern of SVP or SVP-Like in other woody plants such as apple, cherry and kiwifruit are similar to that in poplar which indicate their possible functions in bud break regulation (Wu et al., 2017a; Wang et al., 2021; Wu et al., 2012). And AcSVP in kiwifruit (Actinidia deliciosa, ‘Hayward’), MdSVPa and MdDAMb (a homolog of SVP in MADS-box family) in apple (Malus × domestica ‘Royal Gala’) negatively regulate the bud break (Wu et al., 2017a, Wu et al., 2017b). Interestingly, SVP-Like genes undergo some functional differentiations during evolution. For example, in kiwifruit, SVP3 differs from the other three SVP-Like genes. Overexpressing of SVP3 in kiwifruit does not affect bud break or flowering time but affect flower color and petal development (Wu et al., 2014). In plums, SVP does not play a role in dormancy but regulate floral bud differentiation along with DAMS (Zhao et al., 2022). In addition to SVP and FLC, several other genes originating from the DAM gene family can also regulate dormancy and bud break in perennial plants. For instance, overexpression of the Prunus DAM6 gene in apple (Malus domestica) results in delayed bud break (Yamane et al., 2019). Furthermore, overexpression of the apricot (Prunus mume) DAM6 in poplar delays bud break (Sasaki et al., 2011). Overexpression of the peach DAM6 in apple (Malus domestica) inhibits the outgrowth of apical vegetative buds and advances bud set (Zhao et al., 2023).
Similar to SVP, FLC works as a flowering regulator in annual plants. Does FLC regulate bud break in perennial plants as well? Four FLC genes (PtFLC2-5) have been identified in Populus tremula. PtFLC4 is predominantly expressed during the dormancy stage and high temperature downregulates its expression, which is similar to the expression pattern of MdFLC in apple (Malus × domestica Borkh.), VvFLC2 in grape (Vitis vinifera L.), and CsFLC in tea (Camellia sinensis) (Nishiyama et al., 2021; Díaz-Riquelme et al., 2012; Liu et al., 2022). In kiwifruit (Actinidia chinensis’Hort16A’), AcFLCL (FLC-Like) shows high expression during the dormancy period, and overexpression of AcFLCL promotes bud break (Voogd et al., 2022). Since FT1 is mainly induced by cold and downregulated under warm temperature, while FT2 is induced by warm temperature (Hsu et al., 2011). It could be speculated that some FLCs may regulate dormancy release by acting on FT1 in woody plants. In apple (Malus × domestica Borkh.), MdFLC may have a growth-inhibiting function during the end of dormancy to protect buds when the temperature is still low (Nishiyama et al., 2021). Similar to FLC expression in Arabidopsis, PtFLC2 in poplar, VvFLC1 in grape, and PEP1 in perennial Brassicaceae showed low expression in winter and increased expression after dormancy release, it means there may also be some differentiation in FLC (Wang et al., 2009).
In summary, after the transition from autumn to winter, low temperature promotes the expression of EBB1. EBB1 inhibits the expression of SVL and relieves the expression inhibition of FT1, thus accelerating dormancy release. EBB3 can be induced by temperature-dependent histone modifications at low temperature. Meanwhile, the inhibition of EBB3 expression by SVL is released, leading to increased expression of CYCLIN in poplar (Figure 2).
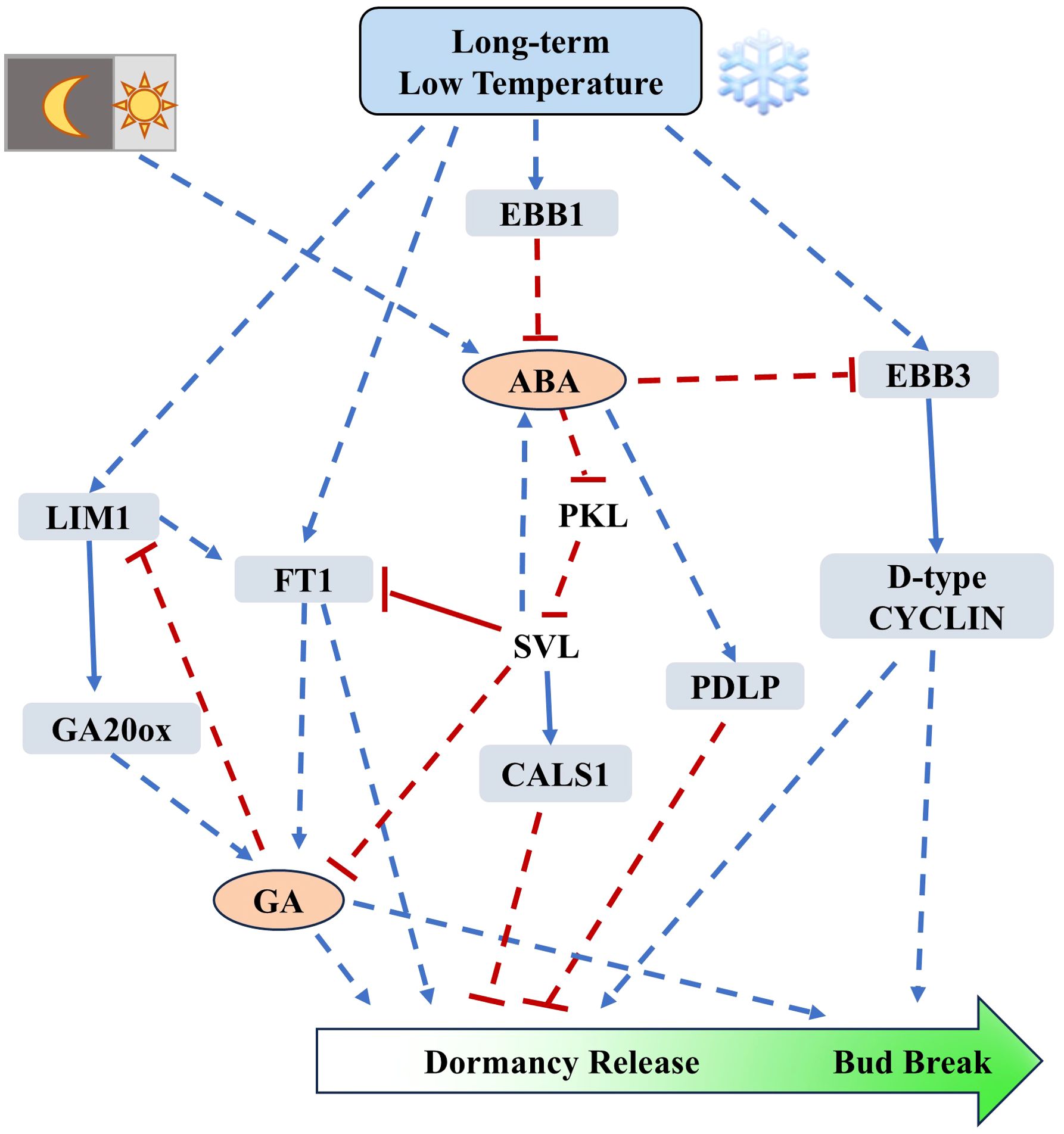
Figure 2. Regulation of dormancy release and bud break mediated by EBB1-SVL-EBB3 module and hormones (based on studies from poplar). PttEBB1 is induced under low-temperature conditions, where it represses ABA to promote the expression of EBB3. Additionally, EBB3 can be induced by temperature-dependent histone modifications at low temperatures. EBB3 facilitates cell division by promoting the expression of D-type CYCLIN. Hormonal regulation of dormancy release primarily involves the modulation of callose deposition; ABA positively regulates the expression of CALS1, which promotes callose deposition and plasmodesmata occlusion, thereby inhibiting dormancy release and bud break by blocking cell-to-cell communication. In contrast, GA can promote dormancy release by removing callose and opening plasmodesmata. The synthesis of ABA is suppressed under low temperatures, while the synthesis of bioactive GA is positively regulated by LIM and FT1 under cold conditions. Blue arrows indicate positive regulation, while red bars indicate negative regulation. The solid lines represent direct interactions between two elements. Dash lines indicate indirect regulation.
4.3 Hormonal regulations
As important endogenous factors regulating plant growth and development, phytohormones also play crucial roles in the dormancy release and bud break processes of perennial plants. Ethylene synthesis and signaling are triggered by a 2-week short-day treatment, while ABA signaling reaches its peak at 3-4 weeks under short-day treatment (Cooke et al., 2012). The content of GA in trees is synthesized and downregulated in response to short day conditions, and the cessation of cell division in the subapical meristem under short day conditions can be restored by applying GA, indicating that GA may regulate the release of tree dormancy (Eriksson and Moritz, 2002; Olsen, 2010). Additionally, jasmonic acid (JA) and brassinosteroids (BR) crosstalk can positively regulate dormancy release in pears (Pyrus pyrifolia) (Wang et al., 2024).
Ethylene may be involved in dormancy induction and dormancy release in response to the change of day length. Studies in grape have shown that ethylene content increases during the dormancy period and gradually decreases during dormancy release. This may be due to the anaerobic conditions within buds wrapped in scale leaves for an extended period, which boost ethylene synthesis. At the same time, protein and lipid degradation is activated to cope with starvation, which serves as a mandatory switch for meristematic tissue growth (Shi et al., 2018, Shi et al., 2020). In Betula pendula, ethylene promotes the growth cessation and formation of bud set induced by short-day conditions. Additionally, the buds of ethylene-insensitive birch trees do not accumulate ABA under short-day conditions, indicating crosstalk between ethylene and ABA signals (Zhao et al., 2023). The application of GA to poplar can promote bud break while the application of ABA to birch delays bud break (Rinne et al., 1994, Rinne et al., 2011). Long-term exposure to low temperature induces the accumulation of GA in the stem apex of trees (Rinne et al., 2011). Research indicates that GA3 and GA4 treatments induce different 1,3-β-glucanase genes (glucan hydrolase family 17, GH17) expression while GA3 induces dormancy release by enabling poplar bypass the cold accumulation stage and GA4 promote bud break by opening the blocked PD (Rinne et al., 2011). What is the molecular mechanism underlying the regulation of low temperature on GA content? One of the explanations is SVL protein. SVL has an inhibitory effect on the expression of a key enzyme in GA biosynthesis, GA20 oxidase (GA20ox). The reduced expression of SVL after prolonged low temperature accelerates GA synthesis, but the in-depth regulatory mechanism between SVL and GA20ox remains to be elucidated (Singh et al., 2018). In addition, SVL also functions with genes involved in ABA synthesis and signaling, such as 9-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3, encoding a key enzyme in ABA biosynthesis), REGULATOR COMPONENT OF ABA RECRPTOR (RCAR)/PYRABACTIN RESISTANCE 1 (PYL1, homolog of ABA receptors), and TCP18 (Singh et al., 2018). Meanwhile, ABA can induce SVL expression and the levels of ABA decrease following dormancy release in Betula pubescens (Rinne et al., 1994; Lebedev et al., 2023).
The physiological mechanisms underlying bud dormancy, dormancy release, and bud break in trees are primarily regulated by the modulation of plasmodesmata trafficking through the deposition and removal of callose (Sankoh and Burch-Smith, 2021). And do ABA and GA regulate dormancy release and bud break by modulating the callose in trees?
Researches sampled buds of Populus tremula × tremuloides after cold treatment and analyzed transcription factors related to PD opening that were co-expressed with FT1 and GA20ox, successfully identifying the MADs-box family gene Low-temperature-Induced MADS-box 1 (LIM1) whose expression increased with cold treatment. Under low-temperature conditions, the expression of LIM1 in SVL-RNAi lines and ft1 mutants show no significant difference compared to wild type, indicating that the regulation of LIM1 by low temperature is independent of SVL and FT1 (Pandey et al., 2024). Overexpression of LIM1 leads to earlier bud break, while LIM1-RNAi lines exhibited a significant delay in bud break after transition from short to long day conditions, suggesting that LIM1 positively regulates bud break in poplar. In contrast to WT, overexpression of LIM1 shows direct bud break and active growth upon transition from short to long day conditions, indicating that LIM1 promotes dormancy release in poplar. The callose content in buds of OE lines was significantly lower than that of RNAi lines, it indicates that LIM1 negatively regulates callose deposition (Pandey et al., 2024). LIM1 overexpression promoted GA synthesis and Yeast two-hybrid experiments proved the interaction between them, and grafting experiments confirmed that LIM1 overexpression facilitated PD opening (Pandey et al., 2024). In addition, the expression of FT1 was also significantly increased in LIM1-OE lines. Further studies showed that mutating FT1 alone or reducing active GA content did not delay bud break in LIM1-OE lines, unless paclobutrazol (GA biosynthesis inhibitor) was applied to LIM1-OE/ft1 poplar, suggesting functional redundancy between FT1 and LIM1 in regulating dormancy release and bud break (Pandey et al., 2024). In Arabidopsis, PACLOBUTRAZOL RESISTANCE 1 (PRE1), a bHLH transcription factor, integrates signals from BR, GA, and light pathways. Additionally, overexpression of PRE1 promotes early flowering in Arabidopsis. In apple, MdoPRE1 plays a crucial role in bud break under warm conditions, possibly by interacting with GA signaling (Porto et al., 2015; Miotto et al., 2019).
Another study further elucidated the role of ABA in dormancy release. The ABA-insensitive genotype abi1-1 was able to undergo bud break without long-term cold treatment when transferred from short to long day conditions, while the wild type could not. This suggests that ABA negatively regulates poplar dormancy release. Additionally, transcriptomic results indicated that short day conditions upregulated genes associated with plasmodesmata function and callose deposition-related CALS1, while downregulating GH17. The abi1/PDLP-OE lines (PDLP, PLASMODESMATA-LOCATED PROTEIN 1) were unable to undergo bud break after transferred from short to long day conditions, it indicates that ABA suppresses dormancy release by regulating the closure of plasmodesmata. PICKLE (PKL) is an antagonist of polycomb repression complex 2 which promote seed dormancy by positively regulating GA signaling and negatively regulating ABA signaling in Arabidopsis (Aichinger et al., 2009; Bouyer et al., 2011). And PKL is downregulated in wild type poplar under short day conditions, but upregulated in the abi1 lines which indicates its possible function in dormancy release. The abi1/PKL-RNAi lines exhibit impaired bud break after transferred from short to long day conditions and show a higher degree of plasmodesmata blockage, indicating that ABA regulates the closure of plasmodesmata through PKL. Furthermore, grafting ten-week short-day treated abi1 scions, rather than wild type, onto FT1-OE poplar allowed bud break after seven weeks of short days, further confirming that ABA positively regulates dormancy through plasmodesmata status (Tylewicz et al., 2018). The previous discussion has already addressed the positive regulatory relationship between SVL and ABA, and the role of ABA in callose deposition is known in poplar (Tylewicz et al., 2018). The question arises whether there are additional components downstream of ABA that participate in the dormancy release and break of poplar. It has been demonstrated that short-day conditions cannot induce the expression of SVL in the abi1 lines, indicating that the induction of SVL by short-day photoperiod requires ABA. Previous studies have shown that ABA positively regulates poplar dormancy by inhibiting PKL, thus prompting an investigation into the relationship between PKL, SVL, and ABA (Tylewicz et al., 2018). Under short-day conditions, the expression of SVL in PKL-RNAi/abi1 lines returned to wild type levels, suggesting that ABA’s induction of SVL under short-day conditions requires the suppression of PKL. After transferred from short-day to long-day conditions, abi1 lines are able to undergo bud break, while the SVL-OE/abi1 lines could not, indicating that the promotion of dormancy release by abi1 lines need the downregulation of SVL (Singh et al., 2019). In the subsequent study, the authors investigated the regulation of plasmodesmata-related genes and GA-related genes by SVL, and the results indicated that SVL can directly target CALS1 and GA2ox (GA synthesis negative regulatory gene) to promote their transcription. Further experiments using SVL-RNAi, abi1, GA2ox-OE/SVL-RNAi, and GA2ox-OE/abi1 lines showed that after transferred from short-day to long-day conditions, bud break occurred in SVL-RNAi and abi1 lines, but not in GA2ox-OE/SVL-RNAi and GA2ox-OE/abi1 lines, indicating that GA and ABA regulate dormancy release together (Singh et al., 2019) (Figure 2).
4.4 Other genes involved in the regulation of bud break in trees
CENL1 is predominantly expressed in shoot tip, axillary vegetative buds, terminal buds, and flowers, while CENL2 is primarily expressed in stems, leaves, floral buds in poplar. And CENL1 reaches its peak expression level in April after bud break. RNAi lines of CENL1/2 leads to earlier bud outgrowth, whereas overexpression of CENL1/2 results in noticeable bud break delay. Although the underlying mechanism remains unclear, the downregulation of CENL1/2 is crucial for dormancy release (Ruonala et al., 2008; Mohamed et al., 2010; Sheng et al., 2023).
In a recent study, samples were collected from poplar buds during the bud break stage, and their transcriptomes, methylomes, and proteomes were analyzed. A lncRNA named Phenology Responsive Intergenic lncRNA 1 (PRIR1) was identified. The experimental results indicated that PRIR1 can promote bud break by activating EXORDIUM LIKE 5 (PtEXL5), and the Arabidopsis EXORDIUM which is the homolog of PtEXL5 is known to facilitate cell division (Hu et al., 2024; Schröder et al., 2009).
In apple, the overexpression of PpCBF in apple has been shown to induce growth cessation and delay bud break, which may be related to the fact that apple dormancy is induced by low temperature rather than short-day photoperiods, as CBF is also induced by low temperature (Wisniewski et al., 2015; Heide and Prestrud, 2005). We have summarized the relevant information of some genes in Table 1.
5 Conclusion and perspective
The cessation of growth, dormancy induction, and dormancy release form a seasonal dormancy cycle of perennial plants. Such cycles enable perennial trees to adapt to seasonal changes, ensuring that their growth patterns align with environment changes. Current research articles predominantly focus on the issues of dormancy and bud break, with experimental results often depicting trees that have already undergone bud break and are in active growth. The distinction between dormancy release and bud break is challenging due to the gradual nature transition. Therefore, establishing a quantitative criterion, potentially based on gene expression, to determine the onset of these two phases would be highly valuable. However, these two processes are by no means entirely distinct. Some genes simultaneously regulate both dormancy release and bud break. For instance, in the lhy mutant, after the transition from long-day to short-day conditions, bud set and growth cessation are delayed compared to the wild type (Ibáñez et al., 2010). Additionally, considering the fact that LHY can bind to and repress the transcription of FT2 (Ramos-Sánchez et al., 2019). This indicates that LHY inhibits bud growth in poplar by repressing the expression of FT2. In the lhy mutants, after the transition from short-day conditions to low-temperature conditions and then to long-day conditions, bud break is delayed compared to the wild type (Ibáñez et al., 2010). This appears to be contrary to the phenotype where LHY inhibits bud growth through FT2. This result is consistent with the fact that LHY is induced by low temperatures. Additionally, FT1 is also induced by low temperatures and can promote dormancy release. After low-temperature induction, LHY may facilitate the process of dormancy release. However, this hypothesis requires further experimental verification.
Day length and temperature are crucial factors influencing dormancy states in trees. Growth cessation in autumn is primarily triggered by short-day, establishing reversible environmentally induced dormancy. Of course, there are exceptions to this pattern, such as in the case of apples, where dormancy establishment is not dependent on short-day photoperiods but rather on low temperatures (Heide and Prestrud, 2005). One perplexing gene is CO. In CO1/CO2-overexpressed poplar, the timing of bud set and bud break remains unchanged. However, PttGI can directly bind to the promoter of CO2. In GI-overexpressed lines, CO expression is only upregulated slightly at night. CO likely plays only a minimal role in regulating dormancy release and bud break in poplar, and its potential function requires further investigation.
Prolonged duration of low temperature serves as the inducing condition for dormancy release, during which a series of signal transductions promote the accumulation of FT1. Following the transition to warm spring, FT1 expression is rapidly downregulated, while FT2 expression increases. This expression regulation appears reasonable, as FT1 functions more like a switch of the sufficient chilling units to induce bud break, aligning with the impending warm environment. FT2 primarily regulates cell division and is responsible for bud break and as well as the rapid growth following dormancy release (Hsu et al., 2011; André et al., 2022). Additionally, while FT2 accelerates cell division, the potential existence of other genes that may promote bud cell division or differentiation represents a direction worthy of future exploration. Thus, FT1 and FT2 act as pivotal regulatory nodes in the annual growth cycle of trees.
As sensors of environmental factors, mainly photoperiod and temperature, photoreceptors/clock genes and EBB1 work cooperatively upstream of FT1 and FT2 to regulate the seasonal dormancy cycle. The EBB1 represents an intrinsic molecular mechanism for temperature sensing, where low temperature enhances the activity of EBB1 to suppress the signal pathway of ABA and promote the expression of EBB3, thereby promoting cell division. And expression of EBB3 increases after low-temperature because of H2K27me3 modification (histone modifications). H3K4me3 and H3K27me3 are well-studied epigenetic modifications that is influenced by temperature, and DAM/SVP are primary targets of epigenetic regulation. Research on the epigenetic regulation of dormancy release and bud break in perennial trees is currently mainly focused on temperate fruit trees, with other tree species being less studied. In addition to histone modifications, plants possess other significant thermosensing mechanisms. For instance, in Arabidopsis, ELF3 responds to environmental temperature through phase separation. It is also worth investigating other temperature-regulated genes in trees that control dormancy release and bud break. Dormancy and bud break in different temperate tree species have their own critical photoperiod, timing regulations and cold accumulation, and related studies contribute to a deeper understanding of bud break in trees. Photoreceptors function as dual signal sensors for both light and temperature in Arabidopsis, potentially providing valuable insights in tree research (Bianchetti et al., 2020). The potential for phytochrome chromophores to respond to temperature variations represents one of the directions for future exploration.
EBB1, along with SVL, not only regulates FT1 expression but also participates in inhibiting GA synthesis and promoting ABA synthesis (Azeez et al., 2021; Singh et al., 2018). Moreover, both ABA and GA primarily influence the pore size of plasmodesmata by affecting callose synthesis. The latest findings indicate that LIM1 promotes dormancy release by positively regulating GA20ox and FT1. Interestingly, FT1 ultimately modulates dormancy release by regulating GA synthesis, which in turn affects callose synthesis. However, the specific active GA that downstream of FT1 remains to be elucidated. Additionally, it is unclear whether LIM1 has direct upstream regulators and is subject to temperature-regulated protein modifications.
Researches on mechanisms of dormancy release and bud break hold important implications for tree protection and introduction under the backdrop of global warming, as well as for the productive application in both timber and non-timber production. What’s more, warming winters shorten the dormancy period of trees, potentially weakening their cold hardiness and leading to extended cold accumulation periods. And rising temperatures in spring cause trees to break dormancy prematurely and begin bud break. This increases the risk of late frost damage, which can harm young tissues and affect tree health and growth.
Author contributions
YZ: Writing – original draft. YM: Writing – review & editing. HQ: Writing – review & editing. LZ: Writing – review & editing. KH: Writing – review & editing. YY: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study is supported by the grants from the National Natural Science Foundation of China (32200213 to Y.J.Y, 32300249 to L.J.Z), the Natural Science Foundation of Jiangsu Province (BK20220417 to Y.J.Y, BK20220418 to L.J.Z), and the STI 2030 Major Projects (grant
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 309, 1052–1056. doi: 10.1126/science.1115983
Aichinger, E., Villar, C. B. R., Farrona, S., Reyes, J. C., Hennig, L., Köhler, C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PloS Genet. 5, e1000605. doi: 10.1371/journal.pgen.1000605
Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 293, 880–883. doi: 10.1126/science.1061320
André, D., Marcon, A., Lee, K. C., Goretti, D., Zhang, B., Delhomme, N., et al. (2022). FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees. Curr. Biol. 32, 2988–2996.e4. doi: 10.1016/j.cub.2022.05.023
Anh Tuan, P., Bai, S., Saito, T., Imai, T., Ito, A., Moriguchi, T. (2016). Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia nakai) lateral flower buds: expression, histone modifications and possible target genes. Plant Cell Physiol. 57, 1038–1047. doi: 10.1093/pcp/pcw041
Azeez, A., Miskolczi, P., Tylewicz, S., Bhalerao, R. P. (2014). A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr. Biol. 24, 717–724. doi: 10.1016/j.cub.2014.02.037
Azeez, A., Zhao, Y. C., Singh, R. K., Yordanov, Y. S., Dash, M., Miskolczi, P., et al. (2021). EARLY BUD-BREAK 1 and EARLY BUD-BREAK 3 control resumption of poplar growth after winter dormancy. Nat. Commun. 12, 1–12. doi: 10.1038/s41467-021-21449-0
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 427, 164–167. doi: 10.1038/nature02269
Beauvieux, R., Wenden, B., Dirlewanger, E. (2018). Bud dormancy in perennial fruit tree species: A pivotal role for oxidative cues. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00657
Bianchetti, R., De Luca, B., De Haro, L. A., Rosado, D., Demarco, D., Conte, M., et al. (2020). Phytochrome-dependent temperature perception modulates isoprenoid metabolism. Plant Physiol. 183, 869–882. doi: 10.1104/pp.20.00019
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 312, 1040–1043. doi: 10.1126/science.1126038
Bouveret, R., Schönrock, N., Gruissem, W., Hennig, L. (1998). Regulation of flowering time by Arabidopsis Photoreceptors. Development. 133, 1693–1702. doi: 10.1242/dev.02340
Bouyer, D., Roudier, F., Heese, M., Andersen, E. D., Gey, D., Nowack, M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PloS Genet. 7, e1002014. doi: 10.1371/journal.pgen.1002014
Chang, C. Y. Y., Bräutigam, K., Hüner, N. P. A., Ensminger, I. (2021). Champions of winter survival: cold acclimation and molecular regulation of cold hardiness in evergreen conifers. New Phytol. 229, 675–691. doi: 10.1111/nph.16904
Cockram, J., Jones, H., Leigh, F. J., O’Sullivan, D., Powell, W., Laurie, D. A., et al. (2007). Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244. doi: 10.1093/jxb/erm042
Cooke, J. E. K., Eriksson, M. E., Junttila, O. (2012). The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Cubas, P. (2020). Plant seasonal growth: how perennial plants sense that winter is coming. Curr. Biol. 30, R21–R23. doi: 10.1016/j.cub.2019.11.044
Díaz-Riquelme, J., Grimplet, J., Martínez-Zapater, J. M., Carmona, M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 12, 181. doi: 10.1186/1471-2229-12-181
Ding, J., Böhlenius, H., Rühl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). GIGANTEA-like genes control seasonal growth cessation in Populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Ding, J., Zhang, B., Li, Y., André, D., Nilsson, O. (2021). Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 modulate seasonal growth in trees. New Phytol. 232, 2339–2352. doi: 10.1111/nph.17350
Druart, N., Johansson, A., Baba, K., Schrader, J., Sjödin, A., Bhalerao, R. R., et al. (2007). Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J. 50, 557–573. doi: 10.1111/j.1365-313X.2007.03077.x
Eriksson, M. E., Moritz, T. (2002). Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (Populus tremula L. x P. tremuloides Michx.). Planta. 214, 920–930. doi: 10.1007/s00425-001-0703-3
Espinosa-Ruiz, A., Saxena, S., Schmidt, J., Mellerowicz, E., Miskolczi, P., Bakó, L., et al. (2004). Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant J. 38, 603–615. doi: 10.1111/j.1365-313X.2004.02070.x
Fadón, E., Fernandez, E., Behn, H., Luedeling, E. (2020). A conceptual framework for winter dormancy in deciduous trees. Agronomy. 10, 241. doi: 10.3390/agronomy10020241
Frewen, B. E., Chen, T. H. H., Howe, G. T., Davis, J., Rohde, A., Boerjan, W., et al. (2000). Quantitative trait loci and candidate gene mapping of bud set and bud flush in populus. Genetics. 154, 837–845. doi: 10.1093/genetics/154.2.837
Gao, Y., Chen, Z., Feng, Q., Long, T., Ding, J., Shu, P., et al. (2024). ELONGATED HYPOCOTYL 5a modulates FLOWERING LOCUS T2 and gibberellin levels to control dormancy and bud break in poplar. Plant Cell. 36, 1963–1984. doi: 10.1093/plcell/koae022
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U. S. A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., Huijser, P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313X.2000.00682.x
Heide, O. M., Prestrud, A. K. (2005). Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 25, 109–114. doi: 10.1093/treephys/25.1.109
Horvath, D. P., Anderson, J. V., Chao, W. S., Foley, M. E. (2003). Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci. 8, 534–540. doi: 10.1016/j.tplants.2003.09.013
Howe, G. T., Bucciaglia, P. A., Hackett, W. P., Furnier, G. R., Cordonnier-Pratt, M. M., Gardner, G. (1998). Evidence that the phytochrome gene family in black cottonwood has one PHYA locus and two PHYB loci but lacks members of the PHYC/F and PHYE subfamilies. Mol. Biol. Evol. 15, 160–175. doi: 10.1093/oxfordjournals.molbev.a025912
Hsu, C. Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U. S. A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Hsu, C. Y., Adams, J. P., No, K., Liang, H., Meilan, R., Pechanova, O., et al. (2012). Overexpression of constans homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PloS One 7, e45448. doi: 10.1371/journal.pone.0045448
Hu, Z., Wu, Z., Zhu, Q., Ma, M., Li, Y., Dai, X., et al. (2024). Multilayer regulatory landscape and new regulators identification for bud dormancy release and bud break in Populus. Plant Cell Environ. 47, 3181–3197. doi: 10.1111/pce.14938
Ibáñez, C., Kozarewa, I., Johansson, M., Ögren, E., Rohde, A., Eriksson, M. E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153, 1823–1833. doi: 10.1104/pp.110.158220
Jung, J. H., Seo, Y. H., Pil, J. S., Reyes, J. L., Yun, J., Chua, N. H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 19, 2736–2748. doi: 10.1105/tpc.107.054528
Karlberg, A., Bako, L., Bhalerao, R. P. (2011). Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PloS Genet. 7, e1002361. doi: 10.1371/journal.pgen.1002361
Karlgren, A., Gyllenstrand, N., Clapham, D., Lagercrantz, U. (2013). FLOWERING LOCUS T/TERMINAL FLOWER1-like genes affect growth rhythm and bud set in Norway spruce. Plant Physiol. 163, 792–803. doi: 10.1104/pp.113.224139
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science. 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kozarewa, I., Ibáñez, C., Johansson, M., Ögren, E., Mozley, D., Nylander, E., et al. (2010). Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Mol. Biol. 73, 143–156. doi: 10.1007/s11103-010-9619-2
Lang, G. A., Early, J. D., Martin, G. C., Darnell, R. L. (1987). Endo-, para-, and ecodormancy: ph ysiological terminology and classification for dormancy research. 22, 371-377. Horti. Sci. 22, 371–377. doi: 10.21273/HORTSCI.22.3.371
Lebedev, V. G., Korobova, A. V., Shendel, G. V., Shestibratov, K. A. (2023). Hormonal status of transgenic birch with a pine glutamine synthetase gene during rooting in vitro and budburst outdoors. Biomolecules 13. doi: 10.3390/biom13121734
Leida, C., Conesa, A., Llácer, G., Badenes, M. L., Ríos, G. (2012). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193, 67–80. doi: 10.1111/j.1469-8137.2011.03863.x
Levy, A., Erlanger, M., Rosenthal, M., Epel, B. L. (2007). A plasmodesmata-associated β-1,3-glucanase in arabidopsis. Plant J. 49, 669–682. doi: 10.1111/j.1365-313X.2006.02986.x
Li, G., Lin, R., Egekwu, C., Blakeslee, J., Lin, J., Pettengill, E., et al. (2020). Seasonal nitrogen remobilization and the role of auxin transport in poplar trees. J. Exp. Bot. 71, 4512–4530. doi: 10.1093/jxb/eraa130
Liu, X., Chen, J., Zhang, X. (2018). Genetic regulation of shoot architecture in cucumber. Hortic. Res. 8, 437–468. doi: 10.1038/s41438-021-00577-0
Liu, Y., Dreni, L., Zhang, H., Zhang, X., Li, N., Zhang, K., et al. (2022). A tea plant (Camellia sinensis) FLOWERING LOCUS C-like gene, csFLC1, is correlated to bud dormancy and triggers early flowering in arabidopsis. Int. J. Mol. Sci. 23, e15711. doi: 10.3390/ijms232415711
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Mateos, J. L., Madrigal, P., Tsuda, K., Rawat, V., Richter, R., Romera-Branchat, M., et al. (2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16, 1–23. doi: 10.1186/s13059-015-0597-1
Maule, A. J. (2008). Plasmodesmata: structure, function and biogenesis. Curr. Opin. Plant Biol. 11, 680–686. doi: 10.1016/j.pbi.2008.08.002
Maurya, J. P., Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: A molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Maurya, J. P., Singh, R. K., Miskolczi, P. C., Prasad, A. N., Jonsson, K., Wu, F., et al. (2020). Branching regulator BRC1 mediates photoperiodic control of seasonal growth in hybrid aspen. Curr. Biol. 30, 122–126.e2. doi: 10.1016/j.cub.2019.11.001
Miotto, Y. E., Tessele, C., Czermainski, A. B. C., Revers, L. F. (2019). Spring is coming : genetic analyses of the bud break date locus reveal candidate genes from the cold perception pathway to dormancy release in apple (Malus × domestica borkh.). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00033
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 17, 2255–2270. doi: 10.1105/tpc.105.033464
Mizuno, T., Oka, H., Yoshimura, F., Ishida, K., Yamashino, T. (2015). Insight into the mechanism of end-of-day far-red light (EODFR)-induced Shade avoidance responses in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 79, 1987–1994. doi: 10.1080/09168451.2015.1065171
Mockler, T., Yang, H., Yu, X. H., Parikh, D., Cheng, Y., Dolan, S., et al. (2002). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 100, 2140–2145. doi: 10.1073/pnas.0437826100
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Nishiyama, S., Matsushita, M. C., Yamane, H., Honda, C., Okada, K., Tamada, Y., et al. (2021). Functional and expressional analyses of apple FLC-like in relation to dormancy progress and flower bud development. Tree Physiol. 41, 562–570. doi: 10.1093/treephys/tpz111
Olsen, J. E. (1997). Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J. 12, 1339–1350. doi: 10.1046/j.1365-313x.1997.12061339.x
Olsen, J. E. (2010). Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Mol. Biol. 73, 37–47. doi: 10.1007/s11103-010-9620-9
Pandey, S. K., Maurya, J. P., Aryal, B., Drynda, K., Nair, A., Miskolczi, P., et al. (2024). A regulatory module mediating temperature control of cell-cell communication facilitates tree bud dormancy release. EMBO J. 43, 5793–5812. doi: 10.1038/s44318-024-00256-5
Porto, D. D., Bruneau, M., Perini, P., Anzanello, R., Renou, J. P., Dos Santos, H. P., et al. (2015). Transcription profiling of the chilling requirement for bud break in apples: A putative role for FLC-like genes. J. Exp. Bot. 66, 2659–2672. doi: 10.1093/jxb/erv061
Ramos-Sánchez, J. M., Triozzi, P. M., Alique, D., Geng, F., Gao, M., Jaeger, K. E., et al. (2019). LHY2 integrates night-length information to determine timing of poplar photoperiodic growth. Curr. Biol. 29, 2402–2406.e4. doi: 10.1016/j.cub.2019.06.003
Randall, R. S., Miyashima, S., Blomster, T., Zhang, J., Elo, A., Karlberg, A., et al. (2015). AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open 4, 1229–1236. doi: 10.1242/bio.013128
Rinne, P., Tuominen, H., Junttila, O. (1994). Seasonal changes in bud dormancy in relation to bud morphology, water and starch content, and abscisic acid concentration in adult trees of Betula pubescens. Tree Physiol. 14, 549–561. doi: 10.1093/treephys/14.6.549
Rinne, P. L. H., Van Der, S. C. (1998). Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1485, 1477–1485. doi: 10.1242/dev.125.8.1477
Rinne, P. L. H., van der Schoot, C. (2003). Plasmodesmata at the crossroads between development, dormancy, and defense. Can. J. Bot. 81, 1182–1197. doi: 10.1139/b03-123
Rinne, P. L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjärvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell. 23, 130–146. doi: 10.1105/tpc.110.081307
Rohde, A., Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Ruonala, R., Rinne, P. L. H., Kangasjärvi, J., van der Schoot, C. (2008). CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell. 20, 59–74. doi: 10.1105/tpc.107.056721
Saito, T., Bai, S., Imai, T., Ito, A., Nakajima, I., Moriguchi, T. (2015). Histone modification and signalling cascade of the dormancy-associatedMADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38, 1157–1166. doi: 10.1111/pce.12469
Saito, T., Wang, S., Ohkawa, K., Ohara, H., Ikeura, H., Ogawa, Y., et al. (2017). Lipid droplet-associated gene expression and chromatin remodelling in LIPASE 5′-upstream region from beginning- to mid-endodormant bud in ‘Fuji’ apple. Plant Mol. Biol. 95, 441–449. doi: 10.1007/s11103-017-0662-0
Sankoh, A. F., Burch-Smith, T. M. (2021). Plasmodesmata and hormones: pathways for plant development. Am. J. Bot. 108, 1580–1583. doi: 10.1002/ajb2.1733
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. doi: 10.1104/pp.111.181982
Sawa, M., Nusinow, D., Kay, A., Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 318, 261–265. doi: 10.1126/science.1146994
Schröder, F., Lisso, J., Lange, P., Müssig, C. (2009). The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 9, 1–12. doi: 10.1186/1471-2229-9-20
Sheng, X., Mahendra, R. A., Wang, C.-T., Brunner, A. M. (2023). CRISPR/Cas9 mutants delineate roles of Populus FT and TFL1/CEN/BFT family members in growth, dormancy release and flowering. Tree Physiol. 43, 1042–1054. doi: 10.1093/treephys/tpad027
Shi, Z., Halaly-Basha, T., Zheng, C., Sharabi-Schwager, M., Wang, C., Galbraith, D. W., et al. (2020). Identification of potential post-ethylene events in the signaling cascade induced by stimuli of bud dormancy release in grapevine. Plant J. 104, 1251–1268. doi: 10.1111/tpj.14997
Shi, Z., Halaly-Basha, T., Zheng, C., Weissberg, M., Ophir, R., Galbraith, D. W., et al. (2018). Transient induction of a subset of ethylene biosynthesis genes is potentially involved in regulation of grapevine bud dormancy release. Plant Mol. Biol. 98, 507–523. doi: 10.1007/s11103-018-0793-y
Shimizu-Sato, S., Mori, H. (2001). Control of outgrowth and dormancy in axillary buds. Plant Physiol. 127, 1405–1413. doi: 10.1104/pp.010841
Simpson, C., Thomas, C., Findlay, K., Bayer, E., Maule, A. J. (2009). An arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell. 21, 581–594. doi: 10.1105/tpc.108.060145
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkovič, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Miskolczi, P., Maurya, J. P., Bhalerao, R. P. (2019). A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 29, 128–133.e2. doi: 10.1016/j.cub.2018.11.006
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Su, Q., Chen, L., Cai, Y., Wang, L., Chen, Y., Zhang, J., et al. (2024). The FLOWERING LOCUS T 5b positively regulates photoperiodic flowering and improves the geographical adaptation of soybean. Plant Cell Environ. 47, 246–258. doi: 10.1111/pce.14739
Tylewicz, S., Petterle, A., Marttila, S., Miskolczi, P., Azeez, A., Singh, R. K., et al. (2018). Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science. 360, 212–215. doi: 10.1126/science.aan8576
Velappan, Y., Signorelli, S., Considine, M. J. (2017). Cell cycle arrest in plants: What distinguishes quiescence, dormancy and differentiated G1? Ann. Bot. 120, 495–509. doi: 10.1093/aob/mcx082
Vergara, R., Noriega, X., Parada, F., Dantas, D., Pérez, F. J. (2016). Relationship between endodormancy, FLOWERING LOCUS T and cell cycle genes in Vitis vinifera. Planta 243, 411–419. doi: 10.1007/s00425-015-2415-0
Voogd, C., Brian, L. A., Wu, R., Wang, T., Allan, A. C., Varkonyi-Gasic, E. (2022). A MADS-box gene with similarity to FLC is induced by cold and correlated with epigenetic changes to control budbreak in kiwifruit. New Phytol. 233, 2111–2126. doi: 10.1111/nph.17916
Wang, R., Farrona, S., Vincent, C., Joecker, A., Schoof, H., Turck, F., et al. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature. 459, 423–427. doi: 10.1038/nature07988
Wang, J., Jiu, S., Xu, Y., Sabir, I. A., Wang, L., Ma, C., et al. (2021). SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium L.). Plant Physiol. Biochem. 159, 277–284. doi: 10.1016/j.plaphy.2020.12.013
Wang, W., Talide, L., Viljamaa, S., Niittylä, T. (2022). Aspen growth is not limited by starch reserves. Curr. Biol. 32, 3619–3627.e4. doi: 10.1016/j.cub.2022.06.056
Wang, X., Wei, J., Wu, J., Shi, B., Wang, P., Alabd, A., et al. (2024). Transcription factors BZR2/MYC2 modulate brassinosteroid and jasmonic acid crosstalk during pear dormancy. Plant Physiol. 194, 1794–1814. doi: 10.1093/plphys/kiad633
Wei, H., Luo, M., Deng, J., Xiao, Y., Yan, H., Liu, H., et al. (2024a). SPL16 and SPL23 mediate photoperiodic control of seasonal growth in Populus trees. New Phytol. 241, 1646–1661. doi: 10.1111/nph.19485
Wei, H., Sun, F., Mo, J., Hu, B., Luo, K. (2024b). Overexpression of CRYPTOCHROME 2 enhances shoot growth and wood formation in poplar under growth-restrictive short days. J. Genet. Genomics 51, 1–4. doi: 10.1016/j.jgg.2024.08.003
Welling, A., Palva, E. T. (2006). Molecular control of cold acclimation in trees. Physiol. Plant 127, 167–181. doi: 10.1111/j.1399-3054.2006.00672.x
Wisniewski, M., Norelli, J., Artlip, T. (2015). Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00085
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017a). SVP-Like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00477
Wu, R. M., Walton, E. F., Richardson, A. C., Wood, M., Hellens, R. P., Varkonyi-Gasic, E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63, 797–807. doi: 10.1093/jxb/err304
Wu, R., Wang, T., McGie, T., Voogd, C., Allan, A. C., Hellens, R. P., et al. (2014). Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 65, 4985–4995. doi: 10.1093/jxb/eru264
Wu, R., Wang, T., Warren, B. A. W., Allan, A. C., Macknight, R. C., Varkonyi-Gasic, E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68, 1071–1082. doi: 10.1093/jxb/erx014
Yamane, H., Wada, M., Honda, C., Matsuura, T., Ikeda, Y., Hirayama, T., et al. (2019). Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS One 14, 1–24. doi: 10.1371/journal.pone.0214788
Yordanov, Y. S., Ma, C., Strauss, S. H., Busov, V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U. S. A. 111, 10001–10006. doi: 10.1073/pnas.1405621111
Zhao, X., Han, X., Wang, Q., Wang, X., Chen, X., Li, L., et al. (2020). EARLY BUD BREAK 1 triggers bud break in peach trees by regulating hormone metabolism, the cell cycle, and cell wall modifications. J. Exp. Bot. 71, 3512–3523. doi: 10.1093/jxb/eraa119
Zhao, X., Wen, B., Li, C., Liu, L., Chen, X., Li, D., et al. (2021a). PpEBB1 directly binds to the GCC box-like element of auxin biosynthesis related genes. Plant Sci. 306, 110874. doi: 10.1016/j.plantsci.2021.110874
Zhao, X., Wen, B., Li, C., Tan, Q., Liu, L., Chen, X., et al. (2021b). Overexpression of the peach transcription factor early bud-break 1 leads to more branches in poplar. Front. Plant Sci. 12, e681283. doi: 10.3389/fpls.2021.681283
Zhao, K., Zhou, Y., Zheng, Y., Zheng, R. Y., Hu, M., Tong, Y., et al. (2022). The collaborative mode by PmSVPs and PmDAMs reveals neofunctionalization in the switch of the flower bud development and dormancy for Prunus mume. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1023628
Zhao, Y. L., Li, Y., Cao, K., Yao, J. L., Bie, H. L., Khan, I. A., et al. (2023). MADS-box protein PpDAM6 regulates chilling requirement-mediated dormancy and bud break in peach. Plant Physiol. 193, 448–465. doi: 10.1093/plphys/kiad291
Keywords: perennial plants, environment perception, regulatory modules, dormancy release, bud break
Citation: Zhao Y, Ma Y, Qiu H, Zhou L, He K and Ye Y (2025) Wake up: the regulation of dormancy release and bud break in perennial plants. Front. Plant Sci. 16:1553953. doi: 10.3389/fpls.2025.1553953
Received: 31 December 2024; Accepted: 17 February 2025;
Published: 06 March 2025.
Edited by:
Marta Joanna Monder, Warsaw University of Life Sciences, PolandReviewed by:
Ioannis-Dimosthenis S. Adamakis, National and Kapodistrian University of Athens, GreeceDurga Prasad Biswal, National Institute of Science Education and Research (NISER), India
David Lázaro Gimeno, University of Helsinki, Finland
Copyright © 2025 Zhao, Ma, Qiu, Zhou, He and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yajin Ye, eWFqaW55ZUBuamZ1LmVkdS5jbg==
 Yue Zhao
Yue Zhao Yahui Ma
Yahui Ma Lijuan Zhou
Lijuan Zhou Yajin Ye
Yajin Ye