
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 30 January 2025
Sec. Plant Metabolism and Chemodiversity
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1548471
This article is part of the Research Topic In-Depth Interpretation of Critical Genomic Information Related to the Biosynthesis of Key Specialized (Secondary) Metabolism in Medicinal Plants View all 9 articles
Benzylisoquinoline alkaloids (BIAs) are a prominent class of plant metabolites with significant pharmaceutical and industrial significance that have garnered substantial attention from researchers worldwide. BIAs exhibit several pharmacological activities and have been used extensively. Examples include analgesics such as morphine, tetrahydropalmatine, antimicrobials such as berberine, and antineoplastic agents including cepharanthine. Most BIAs are derived and isolated from medicinal plants; however, these plants are predominantly wild resources that are scarce. Their high environmental impact, slow growth rate, scarcity of resources, and expensive direct extraction costs pose a significant challenge. Certain BIAs are present in trace amounts in medicinal plants; moreover, they have complex chemical structures and unstable properties. Designing chemical synthesis routes and processes is challenging. Thus, a major obstacle in developing and utilizing these natural products in the pharmaceutical industry lies in their low abundance in nature. Consequently, the limited supply of these molecules fails to meet high research and market demands. In recent years, biosynthesis approaches have emerged as a novel and efficient method to obtain BIAs. In this review, recent progress in the field of enzymes related to the elucidation of biosynthetic pathways and the biosynthesis of BIAs are discussed, and future perspectives for designing viable strategies for their targeted manipulation are presented.
Benzylisoquinoline alkaloids (BIAs) constitute a major group of alkaloids, wherein approximately 2,500 compounds have been identified (Facchini and De Luca, 2008; Minami et al., 2008; Tian et al., 2024). BIAs exhibit numerous pharmacological properties and have been used extensively. Notable examples include morphine, codeine, and tetrahydropalmatine as analgesics; berberine as an antiseptic; and cepharanthine as an antineoplastic and antiviral agent (Figure 1) (Hagel and Facchini, 2013; Fan et al., 2022). Currently, BIAs are predominantly found in plant families such as Galleriferaceae, Tetranyaceae, and Ranunculaceae. Most of these plants are wild resources and their growth is highly influenced by environmental factors. Moreover, they grow slowly, have limited availability, and are associated with high extraction costs. Certain BIAs are present as trace compounds in medicinal plants. They have complex chemical structures and are unstable molecules; moreover, developing synthetic processes for these compounds is challenging. In recent years, researchers have explored novel approaches to obtain naturally occurring bioactive compounds that show therapeutic potential. According to a report in the United States, approximately 25% of pharmaceuticals originate from plant-derived natural chemicals (Li et al., 2018a). However, a major obstacle in harnessing and exploiting natural products to their full potential for pharmaceutical use is their scarcity in nature, leading to an insufficient supply that fails to meet the high research and market demands. Furthermore, the complete elucidation of biosynthetic pathways for secondary plant metabolites is poorly understood. Natural products are currently isolated from plant cells or using tissue culture; however, the high costs associated with plant cell culture, the challenges in developing plant cell lines, and limited yields are some drawbacks that limit the widespread commercial application of this technique (Atanasov et al., 2015). However, advances in new generation sequencing technology, genomics, metabolomics, and bioinformatics have led to the emergence of synthetic biology as a promising approach in addressing sourcing issues related to several natural products that are of high value (Voigt, 2020; Jiang et al., 2024). The heterologous biosynthesis of naturally occurring phytochemicals in bacteria, yeast, and other host plants shows immense potential. The use of synthetic biology techniques to obtain terpenoids, alkaloids, flavonoids, and polyphenols has met with considerable success. Its efficient and environmentally friendly production chain is widely recognized by both the scientific community and the pharmaceutical industry (Paddon et al., 2013; Yao et al., 2013; Nett et al., 2023; Jiang et al., 2024). A well-defined biosynthetic pathway and the elucidation of key enzymes in these pathways serve as the foundation for BIA biosynthesis. The objective of this review was to conduct a comprehensive and systematic bibliometric analysis of the biosynthetic pathways of BIAs, elucidating key enzymes and bridging the knowledge gap in existing bibliometric reviews. We hope that this review will provide novel insights into the biosynthesis of BIAs.
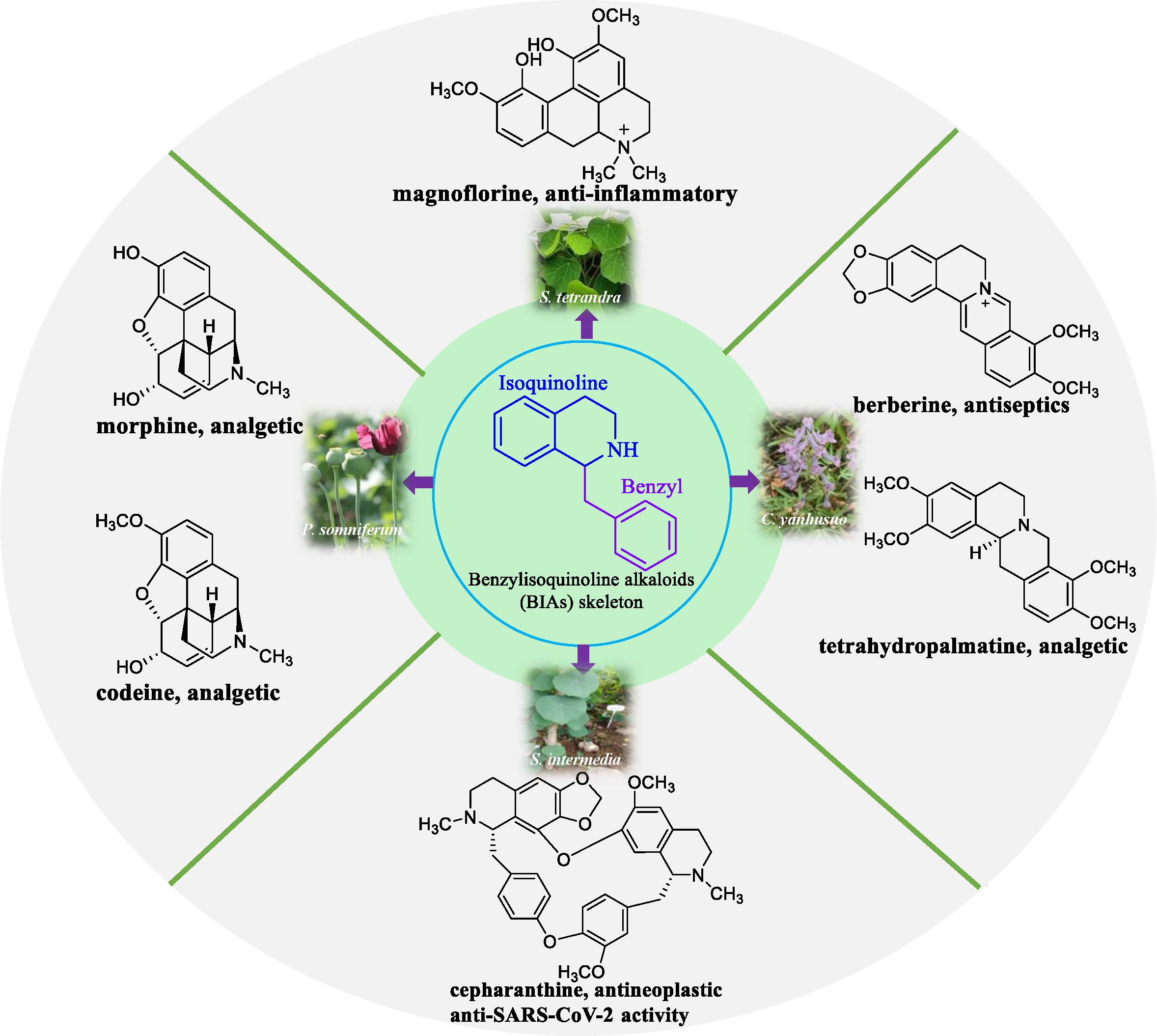
Figure 1. Representative compounds of BIAs and their source plants. The structure skeleton of BIA consists of an isoquinoline ring and a benzyl group. Representative BIA compounds include the analgesics morphine and codeine derived from Papaver somniferum, the antitumor agent cepharanthine from Stephania intermedia, the analgesic tetrahydropalmatine and the antibacterial agent berberine from Corydalis yanhusuo, and the anti-inflammatory compound magnoflorine from Stephania tetrandra.
A detailed analysis of biosynthetic pathways and the study of key enzymes involved in BIA biosynthesis are essential prerequisites when using synthetic biology techniques. Furthermore, these efficient enzymes should be assembled into a biosynthetic module to construct BIA-producing engineering strains. Additionally, the screening of key enzymes and their regulatory genes in biosynthetic pathways is crucial in understanding secondary metabolic pathways. Although high-throughput sequencing enables the rapid acquisition of genome information, selecting candidate genes involved in the biosynthesis of specific natural products from numerous genes poses a challenge. Transcriptomics, metabonomics, and genomics are the primary methods currently used to screen genes that encode key enzymes in biosynthetic pathway (Zhan et al., 2022; He et al., 2023). Most BIAs share a well-defined upstream biosynthetic pathway, thereby necessitating the analysis of their downstream metabolic pathways when elucidating the biosynthetic pathway of a specific component or class. The biosynthetic pathway of BIAs utilizes L-tyrosine as its precursor, which undergoes catalysis via multiple enzymatic reaction steps to yield a series of compounds (Tan et al., 2020; Wang et al., 2022). For example, for berberine, the biosynthetic precursor tyrosine undergoes enzymatic catalysis to yield tyrosine derivatives, including dopamine and 4- hydroxyphenylacetaldehyde (Figure 2) (Samanani et al., 2004; Minami et al., 2007; Vimolmangkang et al., 2016). Then, dopamine and 4-hydroxyphenylacetaldehyde reacted to form norcoclaurine catalyzed by norcoclaurine synthase (NCS). The subsequent conversion of these derivatives involves norcoclaurine 6-O-methyltransferase (6OMT), 4′-O-methyltransferase (4′OMT), coclaurine N-methyltransferase (CNMT), and cytochrome P450 oxidoreductase (NMCH), leading to the production of (S)-reticuline (Ikezawa et al., 2003; Liscombe and Facchini, 2007; Khodorova et al., 2013). The next reaction is catalyzed by berberine bridge enzyme (BBE), wherein (S)-reticuline forms tetrahydroproberberine (S)-scoulerine. (S)-scoulerine is then catalyzed by (S)-scoulerine 9-O-methyltransferase (9OMT) to form the intermediate (S)-tetrahydrocolumbamine (Nakagawa et al., 2012). (S)-tetrahydrocolumbamine is acted upon by canadine synthase (CAS) and (S)-tetrahydroprotoberberine oxidase (STOX) to yield berberine (Diaz Chavez et al., 2011; Gesell et al., 2011; Hagel et al., 2012; Dang and Facchini, 2014). Reticuline serves as a pivotal intermediate and a crucial node in the biosynthetic pathways of morphinan type and aporphine type. While the upstream biosynthetic pathway of BIAs and the key enzymes that are involved are relatively well understood, there is a lack of clarity related to the specific downstream biosynthetic pathway for a particular compound. Furthermore, it is worth noting that the catalytic efficiency of a particular enzyme can vary across different plant sources. Therefore, it is crucial to analyze and elucidate the downstream biosynthetic pathways of BIAs, identify the pivotal enzymes involved in the pathway, and utilize these highly efficient synthetases for the optimal biosynthesis of these compounds.
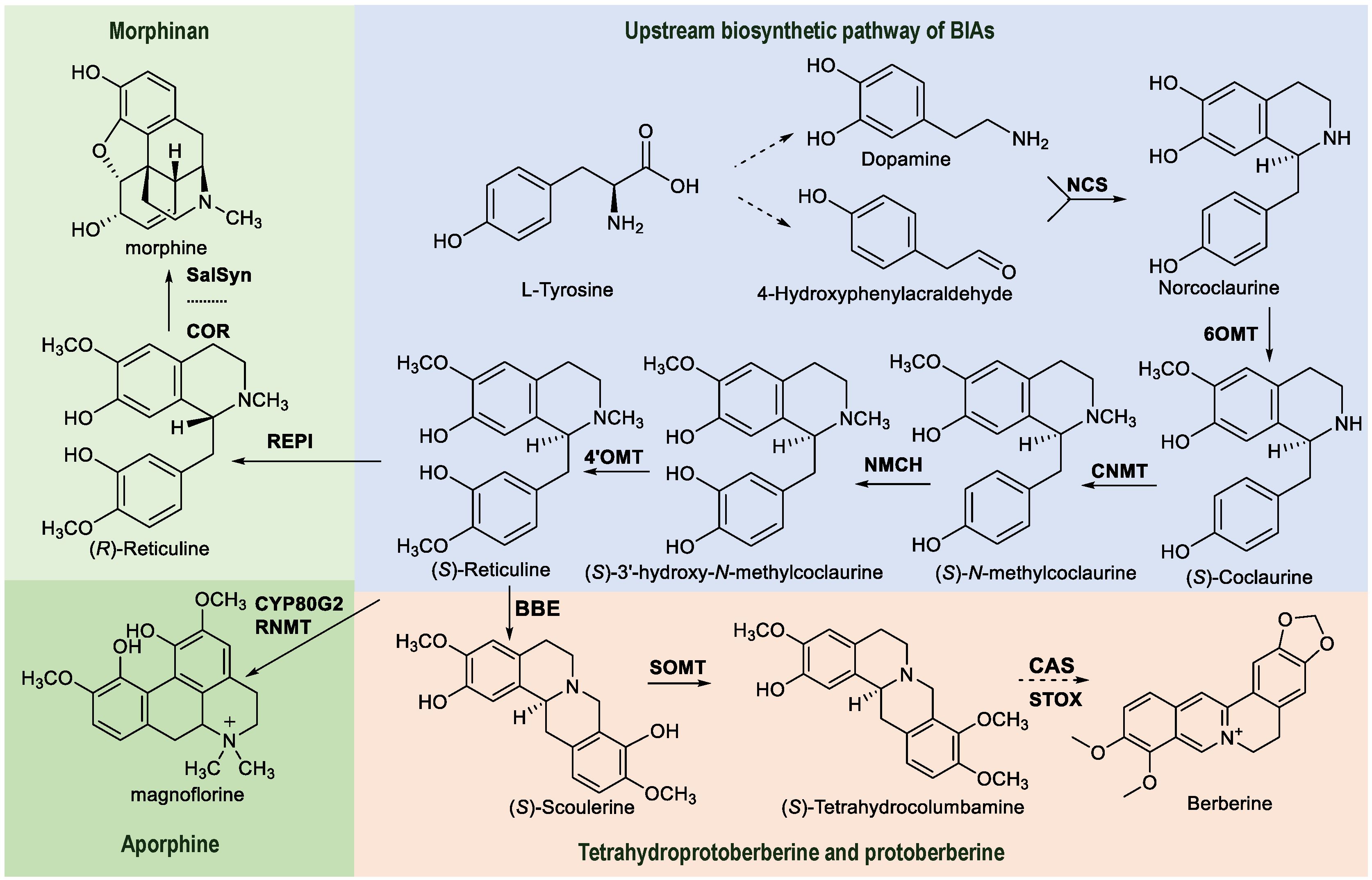
Figure 2. Biosynthetic pathways for the major types of BIAs, including the associated enzymes. The powder blue box represents the shared upstream biosynthetic pathway and key branch points for the synthesis of BIAs. The flesh-colored box represents the tetrahydroprotoberberine and protoberberine types; the green apple-colored box represents the morphinan type; the Maldivian green box represents the aporphine type. Norcoclaurine synthase (NCS); norcoclaurine 6-O-methyltransferase (6OMT); Coclaurine N-methyltransferase (CNMT); N-methylcoclaurine 3’-hydroxylase (NMCH); 4’-O-methyltransferase (4’OMT); Berberine bridge enzyme (BBE); (S)-scoulerine 9-O-methyltransferase (SOMT); Canadine synthase (CAS); (S)-tetrahydroprotoberberine oxidase (STOX); Salutaridine synthase (SalSyn); Reticuline N-methyltransferase (RNMT); Reticuline epimerase (REPI).
The identification and characterization of candidate enzyme genes involved in the biosynthesis of secondary metabolic products is crucial in comprehending the molecular mechanisms underlying these biosynthetic pathways (He et al., 2018). BIAs are present in various families of plants, including Papaveraceae, Ranunculaceae, Berberidaceae, and Caprophyllaceae. The biosynthetic pathways of BIAs have been extensively studied in medicinal plants such as Papaver somniferum, Eschscholzia californica, and Coptis japonica (Hara et al., 1994; Liang et al., 2008; Liscombe et al., 2009; Inui et al., 2012; Tekleyohans et al., 2013; Son et al., 2014; Boke et al., 2015; Agarwal et al., 2016; Purwanto et al., 2017; Oh et al., 2018). (S)-reticuline is a pivotal intermediate in the biosynthesis of various BIAs, facilitating the biosynthesis of diverse BIAs via the action of numerous enzymes (Farrow et al., 2012). Another crucial intermediate, (S)-scoulerine, is produced from (S)-reticuline via BBE catalysis (Figure 2). This key enzyme has been identified in numerous plant species (Facchini et al., 1996; Samanani et al., 2005). Furthermore, the gene for the enzyme SOMT, a crucial component in the biosynthetic pathway of BIAs, has been successfully cloned and expressed from P. somniferum, Glaucium flavum, and C. japonica consecutively (Morishige et al., 2010; Dang and Facchini, 2012; Chang et al., 2015). Subsequently, (S)-scoulerine undergoes methylation or cyclization to generate a series of BIAs. The research teams of Jillian and Facchini conducted metabolomic analysis of more than 20 BIA-producing plants using a multiplatform approach and integrating analytical methods to identify 20 nonmodel plant metabolites (Hagel et al., 2015). They also established a comprehensive compound library of related metabolites. These valuable metabolomics resources, in conjunction with genomic resources, provide favorable conditions to investigate the biosynthesis of BIAs in nonmodel plant species. CYP80 genes involved in the biosynthesis of BIAs and aporphine were identified based on the transcriptome sequencing of lotus (Nelumbo nucifera) (Deng et al., 2018). The main active components of the traditional Chinese medicine Fenfangji (Stephania tetrandra) are BIAs. A total of 42 candidate genes involved in the 15 steps of BIA biosynthesis was discovered through metabolite and transcriptome analyses of the stems, leaves, xylem, and epidermis of Stephania tetrandra. Furthermore, a novel (S)-norcoclurine-6OMT gene was characterized for its role as a catalyst in the formation of (S)-coclaurine through the methylation of (S)-norcoclaurine in vitro (Li et al., 2022). Fourteen cytochrome P450s (CYP450s) and 33 methyltransferases associated with BIA biosynthesis were identified using the weighted gene co-expression network analysis of the transcriptional and metabolic profiles from 18 different tissues of Phellodendron amurens. Notably, a PR10/Bet v1 protein was characterized as norcoclaurine synthase, which was responsible for catalyzing the conversion of dopamine and 4-hydroxyphenylacetaldehyde into (S)-norcoclaurine (Liu et al., 2024a). Yang et al. identified candidate genes involved in the biosynthetic pathways of BIAs, such as sinomenine, magnoflorine, and tetrahydropalmatine from Sinomenium acutum through comprehensive full-length transcriptome sequencing and metabolite analysis (Yang et al., 2022). Zhao et al. elucidated the biosynthetic pathway of BIAs and identified candidate genes, including 6OMT, CNMT, NMCH, BBE, SOMT1, CFS, SPS, STOX, MSH, TNMT, and P6H in C. yanhusuo through widely targeted metabolomic and transcriptomic analyses (Zhao et al., 2024). Furthermore, methyltransferases and cytochrome P450 family genes involved in BIA biosynthetic pathway were identified based on the comparative transcriptomics of Berberis koreana, B. thunbergii and B. amurensis (Roy et al., 2022). A combination of full-length transcriptomics and targeted metabolomics was used to identify the BIA biosynthetic genes in Corydalis corydalis. Ten candidate genes of columbamine-O-methyltransferase were screened to lay a foundation for the identification of the last key enzyme of tetrahydropalmatine in C. corydalis (Xu et al., 2021). The combination of full-length transcriptome sequencing and metabolomics to elucidate the biosynthetic pathways of plant secondary metabolites has attracted increasing attention due to its high efficiency and relatively low cost.
The integration of genomics, metabolomics, and transcriptomics enables determining the correlation between changes in secondary metabolites and the expression of related enzyme genes, facilitating the identification of biosynthetic genes involved in the biosynthesis of secondary metabolites and the analysis of secondary metabolic pathways. This advancement also contributes to an enhancement in the development and utilization of medicinal plant resources. Ye Kai et al. investigated the complete genome sequence of opium poppy, elucidating significant rearrangement events and duplication events that occurred during its history of evolution. Furthermore, they provided insights into the evolutionary history of gene clusters involved in the biosynthesis of morphine alkaloids in poppy, laying a crucial foundation for the further exploration of its medicinal value and unveiling the evolutionary history of Papaveraceae (Guo et al., 2018). The biosynthetic pathway genes of sanguinarine (a BIA) were successfully identified and validated by Zeng Jianguo et al. Using whole-genome sequencing of medicinal plants and construction of the Macleaya cordata genome map, significantly advancing the industrial production of sanguinarine (Liu et al., 2017). Cepharanthine, a class of BIAs primarily found in plants in the Stephania genus, has been approved by the Japanese Medicines and Medical Devices Agency to treat cancer and inflammation. It exerts anti-coronavirus effect against severe acute respiratory syndrome coronavirus-2. The genomes of S. yunnanensis, S. cepharantha, S. japonica, Corydalis tomentella, Tinospora sagittata, Corydalis yanhusuo, Aristolochia contorta, and Menispermum dauricum have been reported (Cui et al., 2022; Xu et al., 2022b; Alami et al., 2024; An et al., 2024; Leng et al., 2024; Liu et al., 2024b; Xu et al., 2024a). These genome reports provide valuable genetic resources to analyze the biosynthetic pathways of BIAs. The use of genomics has expedited the discovery of novel enzyme genes, whereas, the integration of genomics, metabolomics, and transcriptomics has provided initial insights into the regulation of BIAs. Structural analysis of key enzymes involved in BIA biosynthesis offers valuable resources for the development of biomimetic biosynthesis. However, challenges still exist in elucidating the biosynthetic pathway of BIAs, warranting further investigation to elucidate the regulatory mechanisms in plants. Due to the complexity and high costs associated with plant genome sequencing, most BIA-containing medicinal plants remain currently unexplored. With advances in sequencing technology and cost reduction, substantial genomic information on medicinal plants can be unveiled via gene sequencing.
The types and contents of BIAs from different medicinal plants exhibit significant variation. Researchers have investigated the formation mechanisms underlying the structural diversity of BIAs. A whole-genome duplication event occurred in the Papaver somniferum genome approximately 7.8 million years ago, while a segmental genome duplication event took place at least 110 million years ago (Guo et al., 2018). Additionally, 15 genes involved in the biosynthesis of antitussives noscapine and analgesic morphine from P. somniferum were identified to form supergene clusters on chromosome 11. Thus, gene duplication, rearrangement, and fusion events lead to the formation of supergene clusters that can cooperatively and efficiently synthesize specialized metabolic products, such as morphine and noscapine, in P. somniferum. Xu and his colleagues investigated the species evolution, whole genome duplication events, and chromosomal evolution in Menispermum dauricum. By comparing genomes, they identified a significant expansion of the CYP80 gene family in M. dauricum. This expansion, along with tissue-specific expression patterns, contributes to the diversity of BIAs. Notably, they discovered a novel enzyme MdCYP80G10 to catalyze the C2′-C4a phenol coupling of (S)-reticuline into sinoacutine, which is the enantiomer of morphinan compounds, with stereospecificity (An et al., 2024). Additionally, the team observed that genes encoding berberine biosynthesis in Coptis chinensis and other plants are dispersed across different genomic locations, whereas in Phellodendron amurense, enzymes involved in berberine biosynthesis, such as CYP71BG and OMT, form gene clusters (Xu et al., 2024b). These clusters underwent two species-specific whole genome duplication events, leading to the replication and neofunctionalization of key genes. Their work elucidates the convergent evolutionary mechanism of berberine biosynthesis mediated by non-homologous enzymes, offering a new paradigm for understanding how genome evolution drives plant metabolic diversity.
Enzymes involved in the biosynthesis of BIAs include lyases, transferases, and redox enzymes. Lyase primarily refers to NCS, which catalyzes the condensation of dopamine and 4-hydroxyphenylacetaldehyde to generate a precursor of alkaloid biosynthesis. Methyltransferases, including O-methyltransferase and N-methyltransferase, play a crucial role in the biosynthetic pathway of BIAs. Additionally, BBE and cytochrome P450 are the main oxidoreductase enzymes that are involved.
The methyltransferases involved in BIA biosynthesis consist of O- and N- methyltransferases. Methyltransferases play a pivotal role in secondary metabolism in plants, significantly diversifying the range of secondary metabolites through substrate methylation. Furthermore, methyltransferases guide intermediates into specific biosynthetic pathways, thereby exerting key regulatory control over the generation of secondary metabolites in plants (Morishige et al., 2010). Three crucial O-methyltransferases, namely 4′-O-methyltransferase (4′OMT), (S)-normonine 6OMT, and (S)-scoulerine 9OMT, were successively isolated and characterized in the biosynthetic pathway of BIAs (Figure 2) (Rueffer et al., 1983; Takeshita et al., 1995; Morishige et al., 2000; Sato et al., 2010). O-methyltransferases typically catalyze the transfer of a methyl group from S -adenosylmethionine (SAM) to their respective substrates. Although the substrate structures of different O-methyltransferases are similar, they exhibit strict substrate specificity. Comparative analysis of O-methyltransferase sequences from various sources reveals a highly conserved C-terminal region responsible for SAM binding, whereas the N-terminal region tends to vary. The N-terminal domain plays a pivotal role in substrate recognition, and modification of this domain can potentially alter the substrate specificity of the enzyme (Joshi and Chiang, 1998; Zubieta et al., 2001). Reticuline N-methyltransferase, pavine N-methyltransferase, and tetrahydroproberberine N-methyltransferase were characterized during the biosynthesis of BIAs (Kum-Boo et al., 2002; Liscombe and Facchini, 2007; Morris and Facchini, 2016; Torres et al., 2016). Currently, the successful heterologous expression of these N-methyltransferases serves as a useful resource for the heterologous reconstruction of BIA biosynthetic pathways.
Although more than 30 components of BIAs have been isolated from lotus, the biosynthetic enzymes associated with their production are poorly understood. In 2020, Menendez-Perdomo and Facchini reported the identification of 2 O-methyltransferases from N. nucifera, namely NnOMT1 and NnOMT5 (Menéndez-Perdomo and Facchini, 2020). The functional characterization of these recombinant proteins revealed NnOMT1 as a regiospecific 6OMT that was capable of accepting both R and S substrates. Conversely, NnOMT5 primarily acts as a 7-O-methyltransferase with minor activity toward 6OMT, exhibiting a strong stereospecific preference for S-enantiomers. Given the limited understanding of the mechanism of formation of (R)-enantiospecific BIAs, the protein crystal structure of NnNCS1 was determined to serve as an invaluable enzymatic tool for synthetic biology studies related to the biosynthesis of (R)-BIAs (Zhang et al., 2023). Multiple crystal structures of the 2 variants of scoulerine 9OMT (S9OMT) from Thalictrum flavum were resolved, enabling comparative analysis with the crystal structure of TfS9OMT and T. flavum norcoclaurine 6OMT. This analysis identified a crucial residue responsible for regional specificity and was further validated based on mutagenesis and in vitro experiments. Subsequently, several mutants of TfS9OMT were generated to expand the substrate range for multiple BIAs, whereas strategically designed mutants with region-specific changes were analyzed in scoulerine-producing yeast chassis cells. This led to the successful facilitation of the production of tetrahydropalmatrubine and tetrahydropalmatine (Valentic et al., 2020). Aporphine alkaloids, an important class of BIAs, are natural compounds that have a broad spectrum of pharmacological effects. Recently, the involvement of the CYP80 enzymes AcCYP80G7 and AcCYP80Q8 in the formation of the aporphine alkaloid skeleton in Aristolochia contorta has been reported (Meng et al., 2024).
BBE plays a pivotal role in the biosynthetic pathway of tetrahydroproberberine alkaloids (belonging to BIAs). It catalyzes the N-methyl cyclization on (S)-reticuline, leading to the formation of the “C” ring and the production of (S)-scoulerine, which constitutes the fundamental framework of tetrahydroproberberine alkaloids (Figure 2). This enzyme has been found in plants including Eschscholzia californica, P. somniferum, and T. flavum (Facchini et al., 1996; Winkler et al., 2006; Zubi Liu et al., 2013). The molecular structure of BBE consists of the following 2 domains: the FAD-binding domain and the α/β-binding domain (Winkler et al., 2008). Its N-terminal region contains a signal peptide consisting of more than 20 amino acids. It localizes within vacuoles as a nontransmembrane protein. BBE targets the endoplasmic reticulum through signal peptide cleavage and is subsequently transported to vacuoles as intracavicular proteins within the intimal system. Due to the acidic conditions in vacuoles (pH lower than the optimal alkaline pH for BBE activity), BBE is inactivated upon entering the vacuoles (Bird and Facchini, 2001). Researchers have successfully achieved the functional expression of BBE in Saccharomyces cerevisiae and Pichia pastoris (Silvia et al., 2012; Galanie and Smolke, 2015). No detectable functional expression of BBE was noted in Escherichia coli (E. coli), impeding the development of the BIA biosynthetic pathways from heterologous constructs in this bacterial host (Kuroki, 1994). Although co-culturing has been used by some researchers, the upstream pathway of BBE utilizes E.coli, whereas BBE requires yeast for the biosynthesis of (S)-scoulerine. However, due to the inconvenience in cell-culturing conditions as well as the entry and exit of intermediate cells, the co-culture method is unsuitable for large-scale cultivation. The main challenge in biosynthesizing this alkaloid using a prokaryotic chassis organism lies in achieving the prokaryotic activity expression of BBE. Our team was the first to successfully achieve the active expression of the key enzyme BBE in the tetrahydroproberberine alkaloid biosynthetic pathway in a prokaryotic host by optimizing gene sources, expression tags, and vectors. This breakthrough effectively addressed the bottleneck associated with reconstructing the entire tetrahydroproberberine alkaloid biosynthetic pathway in E. coli (Zhao et al., 2021). This breakthrough accomplishment has laid a solid foundation for the heterobiosynthesis of BIAs using E. coli. Furthermore, the researchers discovered that the berberine pontozyme-like gene, which undergoes tandem replication in the Corydalis tomentella genome, appear to be involved in the biosynthesis of cavitine (a BIAs compound) (Xu et al., 2022b). Recently, an endoplasmic reticulum compartmentalization strategy has been developed to enhance the activity of the vacuolar protein BBE, leading to a greater than 200% increase in the production of the key intermediate (S)-scoulerine (Jiao et al., 2024).
Cytochrome P450 is mainly hydroxylated in the BIA biosynthetic pathway by NMCH (Figure 2) (Pauli and Kutchan, 1998; Ikezawa et al., 2003; Yang et al., 2017), cheilanthifoline synthase (Takemura et al., 2013; Yahyazadeh et al., 2017), salutaridine reductase (Geissler et al., 2007; Ziegler et al., 2009; Higashi et al., 2010, 2011), and codeinone reductase (Alcantara et al., 2005; Sharafi et al., 2013; Dastmalchi et al., 2018). When plant cytochrome P450s are heterologously expressed in prokaryotes, the absence of matching endoplasmic reticulum limits the formation of functions. Cytochrome P450 is predominantly expressed in eukaryotic host cells; however, recent studies have demonstrated the successful expression of numerous plant-derived P450 enzymes in E. coli by modifying these enzymes, including those involved in the BIA biosynthetic pathway (Nakagawa et al., 2011; Lin and Yan, 2012). In 2016, Minami successfully expressed the P450 enzymes STORR and SaLSN in E. coli by removing the transmembrane domain. They subsequently synthesized opioid alkaloids based on the step-by-step fermentation of engineered E. coli (Nakagawa et al., 2016). This study provides valuable insights into the heterologous prokaryotic expression of P450 enzymes in the BIA biosynthetic pathway, enabling BIA biosynthesis using prokaryotic chassis cells. The successful expression of plant-derived P450 enzymes in prokaryotic hosts therefore paves the way for the heterologous reengineering of secondary metabolic product biosynthetic pathways in prokaryotic chassis organisms. Traditionally, FAD was believed to rely on oxidase to catalyze the classical berberine-bridging activity; however, recent studies have proposed that cytochrome oxidase CYP71BG29 functions as a berberine-bridge enzyme in Phellodendron amurense (Xu et al., 2024b).
The biosynthesis of plant secondary metabolites involves not only enzyme genes but also transcription factor regulation, with the latter playing a crucial role in this process (Schluttenhofer and Yuan, 2015). By modulating the expression of target genes, transcription factors can effectively promote the biosynthesis of secondary metabolites, thereby enhancing resistance to both biotic and abiotic stresses (Broun, 2004; Cao et al., 2018; Lai et al., 2019; Xu et al., 2022a). Transcription factors can modulate the biosynthesis of secondary metabolites by orchestrating complex metabolic pathways in plants from a holistic perspective (Yuan and Grotewold, 2015). To date, transcription factors have been identified as regulators of the biosynthesis of BIAs in multiple plant species (Deng et al., 2018; Huang et al., 2023; Hong et al., 2025). The PsWRKY transcription factor in P. somniferum interacts with the W-box, a consensus cis-element found in the promoters of BIAs pathway genes, thereby activating transcription from the tyrosine/DOPA decarboxylase (TYDC) promoter (Mishra et al., 2013). StWRKY8 regulates the BIAs biosynthetic pathway in potato, enhancing resistance to late blight (Yogendra et al., 2017). Overexpression of the CjWRKY1 gene from C. japonica led to a significant increase in the accumulation of BIAs in the culture medium of E. californica cells (Yamada et al., 2017). CcbHLH001 and CcbHLH0002 from C. chinensis were found to interact with the promoters of the berberine biosynthesis pathway genes CcBBE and CcCAS, indicating their potential regulatory roles in BIA biosynthesis. In addition, researchers have also identified that the AP2/ERF transcription factors play significant roles in the biosynthetic regulation of BIAs (Zhang et al., 2024).
BIAs are mainly extracted from medicinal plants and have a wide range of pharmacological activities. With the exception of some BIAs such as berberine in Coptis coptis and tetrahydropalmatine in Stephania intermedia, the content of most BIAs in medicinal plants is relatively low (Zhao et al., 2020). Therefore, several scholars have studied them after their biosynthesis. Galanie et al. introduced 7 BIA biosynthesis–related genes from different plants into Saccharomyces cerevisiae and achieved berberine biosynthesis through enzyme-mutation screening, gene copy number optimization, intermediate addition, and culture condition optimization. Furthermore, the biosynthesis of opioid (thebaine and hydrocodone) alkaloids has now been achieved (Galanie et al., 2015).
The genetic background of E. coli is well established and encompasses a diverse range of suitable strains and various vectors. Moreover, E. coli has been successfully used to express numerous eukaryotic genes at high levels. Additionally, the simplicity of operating and culturing E. coli, along with its cost-effectiveness and lower requirements, further contribute to its advantages (Lee and Lee, 2003). Recently, there have been numerous reports on the biosynthesis of BIAs using E. coli. Fumihiko Sato employed the E. coli fermentation system to biosynthesize (S)-reticuline (55 mg/L), a crucial intermediate for BIAs, using dopamine as a substrate. The co-culture of genetically engineered strains of E. coli and S. cerevisiae resulted in the production of magnoflorine and (S)-scoulerine at yields of 7.2 mg/L and 8.3 mg/L, respectively (Minami et al., 2008). Furthermore, using E. coli as the base organism, this research group initiated the biosynthesis of the key BIA intermediate (S)-reticuline starting from L-tyrosine, resulting in a good yield of 46 mg/L (Nakagawa et al., 2012). Next, the biosynthesis of (R,S)-tetrahydropapaveroline was achieved in E. coli using glycerol as a starting material (Nakagawa et al., 2014). Subsequently, this team accomplished the step-by-step fermentation synthesis of thebaine in E. coli using glycerol as a starting material, resulting in a yield of 2.1 mg/L. However, there are challenges associated with the transmembrane transport of intermediates between these 2 microorganisms and scaling up their production via fermentation using the co-culture approach. Our team identified the biosynthesis-related genes of tetrahydroproberberine from plants and microorganisms. Subsequently, E. coli was used as a host to reconstruct the entire biosynthetic pathway, overcoming the bottleneck in the biosynthesis of tetrahydroproberberine using E. coli. By implementing a modular biosynthesis strategy, 5 synthetic modules for tetrahydroproberberine alkaloids were successfully constructed and introduced into E. coli to generate multiple engineered strains that were capable of producing tetrahydroproberberine, and corydamine (Zhao et al., 2021). Subsequently, directed methylation biosynthesis of tetrahydroproberberine compounds was achieved by combining different O-methyltransferases (Zhao et al., 2023).
The yeast system has been used extensively for the heterologous reconstruction of secondary biosynthetic pathways of plant metabolites. Yeast systems have well-defined metabolites and genetic backgrounds (Borodina and Nielsen, 2014; Yunzi et al., 2015). Moreover, compared with bacterial systems, yeast exhibits a microenvironment more akin to plant metabolism, enabling the posttranslational processing and modification of proteins that contribute to the expression of plant-derived membrane proteins. There are numerous reports on the expression of single enzymes or multiple synthases using yeast systems (Ikezawa et al., 2007; Deloache et al., 2015; Trenchard et al., 2015; Hori et al., 2016; Chen et al., 2018; Farrow et al., 2018; Dastmalchi et al., 2019; Ping et al., 2019; Srinivasan and Smolke, 2019). Although E. coli is routinely used to express plant-derived P450 enzymes, it is generally acknowledged that yeast facilitates the relatively facile expression of plant-derived membrane proteins. Hawkins and Smolke introduced various enzymes including 6OMT, CNMT, 4′OMT, SMT, and BBE from plants and CYP2D6 from humans into the chassis organism S. cerevisiae through different combinations. Using (R,S)-norrhodanine as a substrate, the team achieved yields of 32.9 mg/L of (R,S)-reticuline, 60 mg/L of (S)-tetrahydrococlumbamine, 30 mg/L of (S)-tetrahydroberberine, and 20 mg/L of salutaridine (Hawkins and Smolke, 2008). Furthermore, the biosynthesis of stylopine, cis-N-methylstylopine, protoopine, and sanguinarine was achieved using (R,S)-norlordanine as a substrate (Trenchard and Smolke, 2015). In the same year, the research team expressed the enzymes involved in multiple BIA biosynthetic pathways in yeast, and successfully biosynthesized thebaine (expressing 21 enzymes) and hydrocodone (expressing 23 enzymes) using glucose (Galanie et al., 2015). In 2018, Smolke et al. successfully integrated more than 30 genes sourced from diverse organisms including plants, animals, bacteria, and yeast into yeast chassis cells. They successfully achieved the de novo biosynthesis of noscapine, an opioid derivative, via highly optimized (S)-reticuline biosynthesis. By optimizing key enzymes and using suitable strategic approaches, a high yield of 2.2 mg/L of noscapine was obtained when this fermentation process was used (Li et al., 2018b). Jamil et al. successfully biosynthesized tetrahydropapaverine in yeast and obtained the drug papaverine using semisynthetic methods, indicating a novel and alternative synthetic approach (Jamil et al., 2022). Qishuang Li et al. successfully achieved the de novo biosynthesis of magnoflorine in yeast using glucose as a substrate, resulting in a yield of 75.8 mg/L (Li et al., 2024). However, the heterologous biosynthetic yield of alkaloids is relatively low and can be attributed to the complex, lengthy, biosynthetic pathways that involve numerous enzymatic reactions and enzymes. For example, the biosynthetic pathway of noscapine involves more than 30 enzymatic reactions, posing a challenge to optimize its expression and culture conditions. Recent studies have demonstrated biocatalytic cascades to biosynthesize (S)-norcoclaurine (Zhao et al., 2022) and methylated tetrahydroprotoberberine and protoberberine alkaloids (Roddan et al., 2021), highlighting the suitability of alternative routes for BIA biosynthesis.
The biosynthetic pathways of most BIAs are highly intricate and involve membrane-binding proteins, thereby necessitating specialized intracellular infrastructure. Transferring such mechanisms for component biosynthesis to other organisms is therefore challenging. Compared with microbial hosts, exogenous plant hosts offer microenvironments and core metabolic pathways that closely resemble those of the original host plants in terms of exogenous protein expression. Therefore, the latter serve as natural chemical precursors for downstream biosynthesis pathways. A limited number of model plants are currently being used to engineer chassis organisms to synthesize natural products. The Tobacco BY-2 cell line, derived from tobacco seedlings, has been extensively utilized in cell suspension culture and serves as a model organism to study the molecular biology and physiology of plants. Moreover, this cell line is susceptible to several plant viruses, rendering it an ideal model for studying plant–pathogen interactions. Additionally, the utilization of tobacco has facilitated the reprogramming of various biosynthetic pathways involving compounds such as sesquiterpenes, diterpenes, monoterpenes, lignans, and glucosinolates via the incorporation of up to 10 enzymes (Wu et al., 2006; Lau and Sattely, 2015; Kotopka et al., 2018). Moreover, nonmodel plants can be selected as foreign hosts for the biosynthesis of natural products. A primary reason for utilizing a nonmodel plant host is the presence of a specific upstream substrate in the chosen organism. For instance, Artemisia annua has been used to biosynthesize taxadiene owing to its abundant terpene precursors (Li et al., 2015). A. annua grows rapidly and has an efficient genetic transformation system that can be converted into functional expression heterologous plant enzymes. The establishment of hairy root cultures and genetic transformation provides a robust foundation for the production of BIAs using plant chassis (Zhang et al., 2023). However, allogenic plant systems, such as native plants, have slower growth rates compared to microbial hosts and often lack convenient and stable genome engineering systems. Moreover, they demonstrate a complex metabolite background that may contain compounds similar to the target compounds, thereby introducing challenges in isolating the required components.
Reconstruction of biosynthetic pathways is met with numerous challenges due to the complex nature of certain natural products and their associated biosynthetic pathways. Synthetic biology approaches have been developed to overcome these obstacles. Some such approaches include precise regulation of foreign gene expression, enhancement of the functional expression of foreign enzymes, and modification of central metabolism to augment the entry routes of precursor compounds. These strategies have been used to promote pathway-reconstruction approaches for plant-based natural products. Furthermore, elucidation of biosynthetic pathways in plants that are not currently well understood is being attempted via enzyme mining or engineering by using both natural and nonnatural hosts. By enhancing efficiency, selectivity, expression levels, or electron-transfer capabilities, the activity of plant-derived enzymes—particularly that of cytochrome P450—is bolstered. The overall reaction efficiency in multienzyme pathways can be enhanced via dynamic control by partitioning or optimization of host metabolism (Li et al., 2018a). To enhance the efficiency of multienzyme co-expression in a pathway, the synthesis of natural product components using microbial chassis involves a diverse array of endogenous metabolic networks that orchestrate intracellular reactions. Consequently, pathway efficiency is influenced by both intra- and extracellular interactions between metabolites and enzymes. To optimize pathway efficiency, researchers have devised numerous strategies such as the dynamic modulation of enzyme expression levels in the pathway, compartmentalization of enzymes to facilitate interactions with metabolites in the pathway, and the global optimization of heterologous biosynthesis pathways utilizing endogenous microbial metabolic networks.
Recent advances in technology have increased the feasibility of utilizing microorganisms to biosynthesize secondary plant metabolites having complex structures and high value. Heterologous biosynthesis platforms reported to date demonstrate the possibility of achieving high yields and support the expression of numerous heterologous enzymes involved in complex biosynthetic pathways. The integration of genomics, bioinformatics, and synthetic biology has significantly facilitated the discovery of secondary plant metabolites and their potential biosynthetic routes. Despite rapid progress in sequencing data analysis in the fields of plant genomics and functional genomics, inherent trade-offs between data quality, scale, and measurement speed persist. A systematic interface between computational research and experimental science is therefore essential to bridge the gap between the transitioning of raw data to valuable knowledge. Future developments encompassing annotated information related to plant genomics, transcriptome analysis associated with plant secondary metabolism, and microbial and plant biosynthetic pathway reconstruction can be integrated into a comprehensive pipeline for microbial biosynthesis platforms that will advance microbial engineering toward synthesizing complex secondary plant metabolites while enhancing our understanding of the intricate protocols that govern natural product biosynthesis.
BIAs are crucial in advancing medical and societal domains; however, their availability is often limited by the scarcity of plant resources. Recent advances in synthetic biology have elucidated the biosynthetic pathways and enzymes involved in the biosynthesis processes of several alkaloids. Heterologous reconstruction of BIA biosynthetic pathways in bacteria and yeast enables a cost-effective production of complex active ingredients or drugs using inexpensive starting materials. Nevertheless, the biosynthesis of BIAs is met with several challenges. First, most biosynthetic pathways for BIAs, particularly those involving complex structures, are poorly understood, posing a challenge in identifying and transforming highly efficient synthases within these metabolic routes. Second, although various enzymes involved in BIA biosynthetic pathways have been successfully expressed in heterologous hosts (including membrane-binding proteins), the current levels of production and demands cannot meet the high industrial requirements due to factors such as low enzyme expression and difficulties in optimizing metabolism using engineered strains. Therefore, further developments in synthetic biology are warranted for the discovery of innovative solutions—particularly through cross-disciplinary integration with multiomics technologies—to enable the large-scale biosynthesis of trace natural products.
WZ: Data curation, Funding acquisition, Writing – original draft. JL: Conceptualization, Funding acquisition, Writing – review & editing. YC: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Natural Science Foundation of Jiangsu Province (BK20220752), the Open Fund of Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (JSPKLB202309), the Jiangsu Institute of Botany Talent Fund (JIBTF202304), Nanjing Postdoctoral Science Foundation (032105302), and the Agricultural Science and Technology Independent Innovation Fund Project of Jiangsu Province (CX (22)3068).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agarwal, P., Pathak, S., Lakhwani, D., Gupta, P., Asif, M. H., Trivedi, P. K. (2016). Comparative analysis of transcription factor gene families from Papaver somniferum: identification of regulatory factors involved in benzylisoquinoline alkaloid biosynthesis. Protoplasma 253, 857–871. doi: 10.1007/s00709-015-0848-8
Alami, M. M., Shu, S., Liu, S., Ouyang, Z., Zhang, Y., Lv, M., et al. (2024). Chromosome-scale genome assembly of medicinal plant Tinospora sagittata (Oliv.) Gagnep. from the Menispermaceae family. Sci. Data 11, 610. doi: 10.1038/s41597-024-03315-y
Alcantara, J., Bird, D. A., Franceschi, V. R., Facchini, P. J. (2005). Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol. 138, 173–183. doi: 10.1104/pp.105.059287
An, Z., Gao, R., Chen, S., Tian, Y., Li, Q., Tian, L., et al. (2024). Lineage-specific CYP80 expansion and benzylisoquinoline alkaloid diversity in early-diverging eudicots. Adv. Sci. 11, 2309990. doi: 10.1002/advs.202309990
Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E. M., Linder, T., Wawrosch, C., Uhrin, P., et al. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614. doi: 10.1016/j.bioteChadv.2015.08.001
Bird, D. A., Facchini, P. J. (2001). Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213, 888–897. doi: 10.1007/s004250100582
Boke, H., Ozhuner, E., Turktas, M., Parmaksiz, I., Ozcan, S., Unver, T. (2015). Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol. J. 13, 409–420. doi: 10.1111/pbi.12346
Borodina, I., Nielsen, J. (2014). Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 9, 609–620. doi: 10.1002/biot.201300445
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 202–209. doi: 10.1016/j.pbi.2004.01.013
Cao, W., Wang, Y., Shi, M., Hao, X., Zhao, W., Wang, Y., et al. (2018). Transcription factor smWRKY1 positively promotes the biosynthesis of tanshinones in salvia miltiorrhiza. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00554
Chang, L., Hagel, J. M., Facchini, P. J. (2015). Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in glaucium flavum. Plant Physiol. 169, 1127–1140. doi: 10.1104/pp.15.01240
Chen, X., Hagel, J. M., Chang, L., Tucker, J. E., Shiigi, S. A., Yelpaala, Y., et al. (2018). A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nat. Chem. Biol. 14, 738–743. doi: 10.1038/s41589-018-0059-7
Cui, X., Meng, F., Pan, X., Qiu, X., Zhang, S., Li, C., et al. (2022). Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Hortic. Res. 9, uhac005. doi: 10.1093/hr/uhac005
Dang, T. T., Facchini, P. J. (2012). Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631. doi: 10.1104/pp.112.194886
Dang, T. T., Facchini, P. J. (2014). Cloning and characterization of canadine synthase involved in noscapine biosynthesis in opium poppy. FEBS Lett. 588, 198–204. doi: 10.1016/j.febslet.2013.11.037
Dastmalchi, M., Chang, L., Torres, M. A., Ng, K. K. S., Facchini, P. J. (2018). Codeinone reductase isoforms with differential stability, efficiency and product selectivity in opium poppy. Plant J. 4), 631–647. doi: 10.1111/tpj.13975
Dastmalchi, M., Chen, X., Hagel, J. M., Chang, L., Chen, R., Ramasamy, S., et al. (2019). Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat. Chem. Biol. 15, 384–390. doi: 10.1038/s41589-019-0247-0
Deloache, W. C., Russ, Z. N., Narcross, L., Gonzales, A. M., Martin, V. J., Dueber, J. E. (2015). An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 11, 465–471. doi: 10.1038/nchembio.1816
Deng, X., Zhao, L., Fang, T., Xiong, Y., Ogutu, C., Yang, D., et al. (2018). Investigation of benzylisoquinoline alkaloid biosynthetic pathway and its transcriptional regulation in lotus. Hortic. Res. 5, 29. doi: 10.1038/s41438-018-0035-0
Diaz Chavez, M. L., Rolf, M., Gesell, A., Kutchan, T. M. (2011). Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 507, 186–193. doi: 10.1016/j.abb.2010.11.016
Facchini, P. J., De Luca, V. (2008). Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 54, 763–784. doi: 10.1111/j.1365-313X.2008.03438.x
Facchini, P. J., Penzes, C., Johnson, A. G., Bull, D. (1996). Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 112, 1669–1677. doi: 10.1104/pp.112.4.1669
Fan, H., He, S. T., Han, P., Hong, B., Liu, K., Li, M., et al. (2022). Cepharanthine: A promising old drug against SARS-coV-2. Adv. Biol. 6, 2200148. doi: 10.1002/adbi.202200148
Farrow, S. C., Hagel, J. M., Facchini, P. J. (2012). Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochem. 77, 79–88. doi: 10.1016/j.phytochem.2012.02.014
Farrow, S. C., Kamileen, M. O., Meades, J., Ameyaw, B., Xiao, Y., O’connor, S. E. (2018). Cytochrome P450 and O-methyltransferase catalyze the final steps in the biosynthesis of the anti-addictive alkaloid ibogaine from Tabernanthe iboga. J. Biol. Chem. 293, 13821–13833. doi: 10.1074/jbc.RA118.004060
Galanie, S., Smolke, C. D. (2015). Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb. Cell Fact. 14, 144. doi: 10.1186/s12934-015-0332-3
Galanie, S., Thodey, K., Trenchard, I. J., Filsinger Interrante, M., Smolke, C. D. (2015). Complete biosynthesis of opioids in yeast. Science 349, 1095–1100. doi: 10.1126/science.aac9373
Geissler, R., Brandt, W., Ziegler, J. (2007). Molecular modeling and site-directed mutagenesis reveal the benzylisoquinoline binding site of the short-chain dehydrogenase/reductase salutaridine reductase. Plant Physiol. 143, 1493–1503. doi: 10.1104/pp.106.095166
Gesell, A., Chavez, M. L., Kramell, R., Piotrowski, M., Macheroux, P., Kutchan, T. M. (2011). Heterologous expression of two FAD-dependent oxidases with (S)-tetrahydroprotoberberine oxidase activity from Arge mone mexicana and Berberis wilsoniae in insect cells. Planta 233, 1185–1197. doi: 10.1007/s00425-011-1357-4
Guo, L., Winzer, T., Yang, X., Li, Y., Ning, Z., He, Z., et al. (2018). The opium poppy genome and morphinan production. Science 362, 343–347. doi: 10.1126/science.aat4096
Hagel, J. M., Beaudoin, G. A., Fossati, E., Ekins, A., Martin, V. J., Facchini, P. J. (2012). Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J. Biol. Chem. 287, 42972–42983. doi: 10.1074/jbc.M112.420414
Hagel, J. M., Facchini, P. J. (2013). Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672. doi: 10.1093/pcp/pct020
Hagel, J. M., Mandal, R., Han, B., Han, J., Dinsmore, D. R., Borchers, C. H., et al. (2015). Metabolome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 15, 220. doi: 10.1186/s12870-015-0594-2
Hara, M., Tanaka, S., Tabata, M. Induction of a specific methyltransferase activity regulating berberine biosynthesis by cytokinin in Thalictrum minus cell cultures. Phytochemistry (1994) 36, 327–332. doi: 10.1016/S0031-9422(00)97070-5
Hawkins, K., Smolke, C. (2008). Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 4, 564–573. doi: 10.1038/nchembio.105
He, S. M., Liang, Y. L., Cong, K., Chen, G., Zhao, X., Zhao, Q. M., et al. (2018). Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in coptis species. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00731
He, B., Qian, K., Han, X., Li, J., Zhou, Q., Xu, L.-A., et al. (2023). Novel mechanisms for the synthesis of important secondary metabolites in Ginkgo biloba seed revealed by multi-omics data. Front. Plant Sci. 14. doi: 10.3389/fps.2023.1196609
Higashi, Y., Kutchan, T. M., Smith, T. J. (2011). Atomic structure of salutaridine reductase from the opium poppy (Papaver somniferum). J. Biol. Chem. 286, 6532–6541. doi: 10.1074/jbc.M110.168633
Higashi, Y., Smith, T. J., Jez, J. M., Kutchan, T. M. (2010). Crystallization and preliminary X-ray diffraction analysis of salutaridine reductase from the opium poppy Papaver somniferum. Acta Crystallogr. Sect F Struct. Biol. Cryst Commun. 66, 163–166. doi: 10.1107/s174430910904932x
Hong, U. V. T., Tamiru-Oli, M., Hurgobin, B., Lewsey, M. G. (2025). Genomic and cell-specific regulation of benzylisoquinoline alkaloid biosynthesis in opium poppy. J. Exp. Bot. 76, 35–51. doi: 10.1093/jxb/erae317
Hori, K., Okano, S., Sato, F. (2016). Efficient microbial production of stylopine using a Pichia pastoris expression system. Sci. Rep. 6, 22201. doi: 10.1038/srep22201
Huang, X., Jia, A., Huang, T., Wang, L., Yang, G., Zhao, W. (2023). Genomic profiling of WRKY transcription factors and functional analysis of CcWRKY7, CcWRKY29, and CcWRKY32 related to protoberberine alkaloids biosynthesis in Coptis chinensis Franch. Front. Genet. 14. doi: 10.3389/fgene.2023.1151645
Ikezawa, N., Iwasa, K., Sato, F. (2007). Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274, 1019–1035. doi: 10.1111/j.1742-4658.2007.05652.x
Ikezawa, N., Tanaka, M., Nagayoshi, M., Shinkyo, R., Sakaki, T., Inouye, K., et al. (2003). Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 278, 38557–38565. doi: 10.1074/jbc.M302470200
Inui, T., Kawano, N., Shitan, N., Yazaki, K., Kiuchi, F., Kawahara, N., et al. (2012). Improvement of benzylisoquinoline alkaloid productivity by overexpression of 3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase in transgenic Coptis japonica plants. Biol. Pharm. Bull. 35, 650–659. doi: 10.1248/bpb.35.650
Jamil, O. K., Cravens, A., Payne, J. T., Kim, C. Y., Smolke, C. D. (2022). Biosynthesis of tetrahydropapaverine and semisynthesis of papaverine in yeast. Proc. Natl. Acad. Sci. U. S. A 119, e2205848119. doi: 10.1073/pnas.2205848119
Jiang, B., Gao, L., Wang, H., Sun, Y., Zhang, X., Ke, H., et al. (2024). Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science 383, 622–629. doi: 10.1126/science.adj3484
Jiao, X., Fu, X., Li, Q., Bu, J., Liu, X., Savolainen, O., et al. (2024). De novo production of protoberberine and benzophenanthridine alkaloids through metabolic engineering of yeast. Nat. Commun. 15, 8759. doi: 10.1038/s41467-024-53045-3
Joshi, C. P., Chiang, V. L. (1998). Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol.Biol. 37, 663–674. doi: 10.1023/A:1006035210889
Khodorova, N. V., Shavarda, A. L., Lequart-Pillon, M., Laberche, J. C., Voitsekhovskaja, O. V., Boitel-Conti, M. (2013). Biosynthesis of benzylisoquinoline alkaloids in Corydalis bracteata: compartmentation and seasonal dynamics. Phytochemistry 92, 60–70. doi: 10.1016/j.phytochem.2013.04.008
Kotopka, B. J., Li, Y., Smolke, C. D. (2018). Synthetic biology strategies toward heterologous phytochemical production. Nat. Prod Rep. 35, 902–920. doi: 10.1039/c8np00028j
Kum-Boo, C., Takashi, M., Nobukazu, S., Kazufumi, Y., Fumihiko, S. (2002). Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277, 830–835. doi: 10.1074/jbc.M106405200
Lai, X., Stigliani, A., Vachon, G., Carles, C., Smaczniak, C., Zubieta, C., et al. (2019). Building transcription factor binding site models to understand gene regulation in plants. Mol. Plant 12, 743–763. doi: 10.1016/j.molp.2018.10.010
Lau, W., Sattely, E. S. (2015). Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349, 1224–1228. doi: 10.1126/science.aac7202
Lee, P. S., Lee, K. H. (2003). Escherichia coli–a model system that benefits from and contributes to the evolution of proteomics. Biotechnol. Bioeng 84, 801–814. doi: 10.1002/bit.10848
Leng, L., Xu, Z., Hong, B., Zhao, B., Tian, Y., Wang, C., et al. (2024). Cepharanthine analogs mining and genomes of Stephania accelerate anti-coronavirus drug discovery. Nat. Commun. 15, 1537. doi: 10.1038/s41467-024-45690-5
Li, K., Chen, X., Zhang, J., Wang, C., Xu, Q., Hu, J., et al. (2022). Transcriptome analysis of stephania tetrandra and characterization of norcoclaurine-6-O-methyltransferase involved in benzylisoquinoline alkaloid biosynthesis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.874583
Li, M., Jiang, F., Yu, X., Miao, Z. (2015). Engineering isoprenoid biosynthesis in Artemisia annua L. for the production of taxadiene: a key intermediate of taxol. BioMed. Res. Int. 2015, 504932. doi: 10.1155/2015/504932
Li, Q., Jiao, X., Li, X., Shi, W., Ma, Y., Tan, X., et al. (2024). Identification of the cytochrome P450s responsible for the biosynthesis of two types of aporphine alkaloids and their de novo biosynthesis in yeast. J. Integr. Plant Biol. 66, 1703–1717. doi: 10.1111/jipb.13724
Li, S., Li, Y., Smolke, C. D. (2018a). Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 10, 395–404. doi: 10.1038/s41557-018-0013-z
Li, Y., Li, S., Thodey, K., Trenchard, I., Cravens, A., Smolke, C. D. (2018b). Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E3922–E3931. doi: 10.1073/pnas.1721469115
Liang, Y., Minami, H., Sato, F. (2008). Isolation of herbicide-resistant 4-hydroxyphenylpyruvate dioxygenase from cultured Coptis japonica cells. Biosci. Biotechnol. Biochem. 72, 3059–3062. doi: 10.1271/bbb.80466
Lin, Y., Yan, Y. (2012). Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Fact 11, 42. doi: 10.1186/1475-2859-11-42
Liscombe, D. K., Facchini, P. J. (2007). Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282, 14741–14751. doi: 10.1074/jbc.M611908200
Liscombe, D. K., Ziegler, J., Schmidt, J., Ammer, C., Facchini, P. J. (2009). Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 60, 729–743. doi: 10.1111/j.1365-313X.2009.03980.x
Liu, X., Liu, Y., Huang, P., Ma, Y., Qing, Z., Tang, Q., et al. (2017). The Genome of Medicinal Plant Macleaya cordata Provides New Insights into Benzylisoquinoline Alkaloids Metabolism. Mol. Plant 10, 975–989. doi: 10.1016/j.molp.2017.05.007
Liu, Z., Shen, S., Wang, Y., Sun, S., Yu, T., Fu, Y., et al. (2024b). The genome of Stephania japonica provides insights into the biosynthesis of cepharanthine. Cell Rep. 43(3). doi: 10.1016/j.molp.2017.05.007
Liu, T., Zhang, W., Wang, S., Tian, Y., Wang, Y., Gao, R., et al. (2024a). Metabolome and transcriptome association study reveals biosynthesis of specialized benzylisoquinoline alkaloids in Phellodendron amurense. Chin. Herb. Med. 17(1), 178–188. doi: 10.1016/j.chmed.2024.11.003
Menéndez-Perdomo, I. M., Facchini, P. J. (2020). Isolation and characterization of two O-methyltransferases involved in benzylisoquinoline alkaloid biosynthesis in sacred lotus (Nelumbo nucifera). J. Biol. Chem. 295, 1598–1612. doi: 10.1074/jbc.RA119.011547
Meng, F., Zhang, S., Su, J., Zhu, B., Pan, X., Qiu, X., et al. (2024). Characterization of two CYP80 enzymes provides insights into aporphine alkaloid skeleton formation in Aristolochia contorta. Plant J. 118(5), 1439–1454 doi: 10.1111/tpj.16686
Minami, H., Dubouzet, E., Iwasa, K., Sato, F. (2007). Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 282, 6274–6282. doi: 10.1074/jbc.M608933200
Minami, H., Kim, J. S., Ikezawa, N., Takemura, T., Katayama, T., Kumagai, H., et al. (2008). Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U.S.A. 105, 7393–7398. doi: 10.1073/pnas.0802981105
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M., Shanker, K., et al. (2013). Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PloS One 8, e52784. doi: 10.1371/journal.pone.0052784
Morishige, T., Tamakoshi, M., Takemura, T., Sato, F. (2010). Molecular characterization of O-methyltransferases involved in isoquinoline alkaloid biosynthesis in Coptis japonica. Proc. Jpn. Acad. Ser. B 86, 757–768. doi: 10.2183/pjab.86.757
Morishige, T., Tsujita, T., Yamada, Y., Sato, F. (2000). Molecular characterization of the S-adenosyl-L-methionine:3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275, 23398–23405. doi: 10.1074/jbc.M002439200
Morris, J. S., Facchini, P. J. (2016). Isolation and characterization of reticuline N-methyltransferase involved in biosynthesis of the aporphine alkaloid magnoflorine in opium poppy. J. Biol. Chem. 291, 23416–23427. doi: 10.1074/jbc.M116.750893
Nakagawa, A., Matsumura, E., Koyanagi, T., Katayama, T., Kawano, N., Yoshimatsu, K., et al. (2016). Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 7, 10390. doi: 10.1038/ncomms10390
Nakagawa, A., Matsuzaki, C., Matsumura, E., Koyanagi, T., Katayama, T., Yamamoto, K., et al. (2014). (R,S)-tetrahydropapaveroline production by stepwise fermentation using engineered Escherichia coli. Sci. Rep. 4, 6695. doi: 10.1038/srep06695
Nakagawa, A., Minami, H., Kim, J. S., Koyanagi, T., Katayama, T., Sato, F., et al. (2011). A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2, 326. doi: 10.1038/ncomms1327
Nakagawa, A., Minami, H., Kim, J. S., Koyanagi, T., Katayama, T., Sato, F., et al. (2012). Bench-top fermentative production of plant benzylisoquinoline alkaloids using a bacterial platform. Bioeng Bugs 3, 49–53. doi: 10.4161/bbug.3.1.18446
Nett, R. S., Dho, Y., Tsai, C., Passow, D., Martinez Grundman, J., Low, Y.-Y., et al. (2023). Plant carbonic anhydrase-like enzymes in neuroactive alkaloid biosynthesis. Nature 624, 182–191. doi: 10.1038/s41586-023-06716-y
Oh, J., Shin, Y., Ha, I. J., Lee, M. Y., Lee, S. G., Kang, B. C., et al. (2018). Transcriptome profiling of two ornamental and medicinal papaver herbs. Int. J. Mol. Sci. 19, 1–20. doi: 10.3390/ijms19103192
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Pauli, H. H., Kutchan, T. M. (1998). Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3’-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 13, 793–801. doi: 10.1046/j.1365-313x.1998.00085.x
Ping, Y., Li, X., You, W., Li, G., Yang, M., Wei, W., et al. (2019). De novo production of the plant-derived tropine and pseudotropine in yeast. ACS Synth Biol. 8, 1257–1262. doi: 10.1021/acssynbio.9b00152
Purwanto, R., Hori, K., Yamada, Y., Sato, F. (2017). Unraveling additional O-methylation steps in benzylisoquinoline alkaloid biosynthesis in california poppy (Eschscholzia californica). Plant Cell Physiol. 58, 1528–1540. doi: 10.1093/pcp/pcx093
Roddan, R., Subrizi, F., Broomfield, J., Ward, J. M., Keep, N. H., Hailes, H. C. (2021). Chemoenzymatic cascades toward methylated tetrahydroprotoberberine and protoberberine alkaloids. Org Lett. 23, 6342–6347. doi: 10.1021/acs.orglett.1c02110
Roy, N. S., Park, N.-I., Kim, N.-S., Park, Y., Kim, B.-Y., Kim, Y.-D., et al. (2022). Comparative transcriptomics for genes related to berberine and berbamine biosynthesis in Berberidaceae. Plants 11, 2676. doi: 10.3390/plants11202676
Rueffer, M., Nagakura, N., Zenk, M. H. (1983). ). Partial purification and properties of S-adenosylmethionine: (R), (S)-norlaudanosoline-6-O-methyltransferase from argemone platyceras cell cultures. Planta Med. 49, 131–137. doi: 10.1055/s-2007-969833
Samanani, N., Liscombe, D. K., Facchini, P. J. (2004). Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40, 302–313. doi: 10.1111/j.1365-313X.2004.02210.x
Samanani, N., Park, S. U., Facchini, P. J. (2005). Cell type-specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis. Plant Cell 17, 915–926. doi: 10.1105/tpc.104.028654
Sato, F., Tsujita, T., Katagiri, Y., Yoshida, S., Yamada, Y. (2010). Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 225, 125–131. doi: 10.1111/j.1432-1033.1994.00125.x
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Sharafi, A., Sohi, H. H., Mousavi, A., Azadi, P., Khalifani, B. H., Razavi, K. (2013). Metabolic engineering of morphinan alkaloids by over-expression of codeinone reductase in transgenic hairy roots of Papaver bracteatum, the Iranian poppy. Biotechnol. Lett. 35, 445–453. doi: 10.1007/s10529-012-1080-7
Silvia, W., Andreas, W., Sabrina, R., Corinna, D., Stefanie, H., Karl, G., et al. (2012). Catalytic and structural role of a conserved active site histidine in berberine bridge enzyme. Biochemistry 51, 6139–6147. doi: 10.1021/bi300411n
Son, S. Y., Rhee, H. S., Lee, M. W., Park, J. M. (2014). Analysis of benzo[c]phenanthridine alkaloids in Eschscholtzia californica cell culture using HPLC-DAD and HPLC-ESI-MS/MS. Biosci. Biotechnol. Biochem. 78, 1103–1111. doi: 10.1080/09168451.2014.917264
Srinivasan, P., Smolke, C. D. (2019). Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nat. Commun. 10, 3634. doi: 10.1038/s41467-019-11588-w
Takemura, T., Ikezawa, N., Iwasa, K., Sato, F. (2013). Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry 91, 100–108. doi: 10.1016/j.phytochem.2012.02.013
Takeshita, N., Fujiwara, H., Mimura, H., Fitchen, J. H., Yamada, Y., Sato, F. (1995). Molecular cloning and characterization of S-adenosyl-L-methionine : scoulerine-9-O-methyltransferase from cultured cells of coptis japonica. Plant Cell Physiol. 36, 29–36. doi: 10.1093/oxfordjournals.pcp.a078741
Tan, X., Song, W., Chen, X., Liu, L., Wu, J. (2020). Recent advances in biocatalytic derivatization of L-tyrosine. Appl. Microbiol. Biotechnol. 104, 9907–9920. doi: 10.1007/s00253-020-10949-6
Tekleyohans, D. G., Lange, S., Becker, A. (2013). Virus-induced gene silencing of the alkaloid-producing basal eudicot model plant Eschscholzia californica (California Poppy). Methods Mol. Biol. 975, 83–98. doi: 10.1007/978-1-62703-278-0_7
Tian, Y., Kong, L., Li, Q., Wang, Y., Wang, Y., An, Z., et al. (2024). Structural diversity, evolutionary origin, and metabolic engineering of plant specialized benzylisoquinoline alkaloids. Nat. Prod. Rep. 41, 1787–1810. doi: 10.1039/D4NP00029C
Torres, M. A., Hoffarth, E., Eugenio, L., Savtchouk, J., Chen, X., Morris, J. S., et al. (2016). Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism. J. Biol. Chem. 291, 23403–23415. doi: 10.1074/jbc.M116.747261
Trenchard, I. J., Siddiqui, M. S., Thodey, K., Smolke, C. D. (2015). De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 31, 74–83. doi: 10.1016/j.ymben.2015.06.010
Trenchard, I. J., Smolke, C. D. (2015). Engineering strategies for the fermentative production of plant alkaloids in yeast. Metab. Eng. 30, 96–104. doi: 10.1016/j.ymben.2015.05.001
Valentic, T. R., Payne, J. T., Smolke, C. D. (2020). Structure-guided engineering of a scoulerine 9-O-methyltransferase enables the biosynthesis of tetrahydropalmatrubine and tetrahydropalmatine in yeast. ACS Catal. 10, 4497–4509. doi: 10.1021/acscatal.9b05417
Vimolmangkang, S., Deng, X., Owiti, A., Meelaph, T., Ogutu, C., Han, Y. (2016). Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants. Sci. Rep. 6, 26323. doi: 10.1038/srep26323
Voigt, C. A. (2020). Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 11, 1–6. doi: 10.1038/s41467-020-20122-2
Wang, Y., Subrizi, F., Carter, E. M., Sheppard, T. D., Ward, J. M., Hailes, H. C. (2022). Enzymatic synthesis of benzylisoquinoline alkaloids using a parallel cascade strategy and tyrosinase variants. Nat. Commun. 13, 5436. doi: 10.1038/s41467-022-33122-1
Winkler, A., Hartner, F., Kutchan, T. M., Glieder, A., Macheroux, P. (2006). Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 281, 21276–21285. doi: 10.1074/jbc.M603267200
Winkler, A., Lyskowski, A., Riedl, S., Puhl, M., Kutchan, T. M., Macheroux, P., et al. (2008). A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol. 4, 739–741. doi: 10.1038/nchembio.123
Wu, S., Schalk, M., Clark, A., Miles, R. B., Coates, R., Chappell, J. (2006). Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 24, 1441–1447. doi: 10.1038/nbt1251
Xu, Z., Li, Z., Ren, F., Gao, R., Wang, Z., Zhang, J., et al. (2022b). The genome of Corydalis reveals the evolution of benzylisoquinoline alkaloid biosynthesis in Ranunculales. Plant J. 111, 217–230. doi: 10.1111/tpj.15788
Xu, D., Lin, H., Tang, Y., Huang, L., Xu, J., Nian, S., et al. (2021). Integration of full-length transcriptomics and targeted metabolomics to identify benzylisoquinoline alkaloid biosynthetic genes in Corydalis yanhusuo. Hortic. Res. 8, 16. doi: 10.1038/s41438-020-00450-6
Xu, Z., Tian, Y., Wang, J., Ma, Y., Li, Q., Zhou, Y., et al. (2024b). Convergent evolution of berberine biosynthesis. Sci. Adv. 10, eads3596. doi: 10.1126/sciadv.ads3596
Xu, Y., Wang, Y., Du, J., Pei, S., Guo, S., Hao, R., et al. (2022a). A DE1 BINDING FACTOR 1-GLABRA2 module regulates rhamnogalacturonan I biosynthesis in Arabidopsis seed coat mucilage. Plant Cell 34 (4), 1396–1414. doi: 10.1093/plcell/koac011
Xu, D., Ye, Z., Huang, Y., Zhu, K., Xu, H., Yu, J., et al. (2024a). Haplotype-resolved genome assembly of Corydalis yanhusuo, a traditional Chinese medicine with unusual telomere motif. Hortic. Res. 11, uhad296. doi: 10.1093/hr/uhad296
Yahyazadeh, M., Ratmoyo, P., Bittner, F., Sato, F., Selmar, D. (2017). Cloning and characterization of cheilanthifoline and stylopine synthase genes from chelidonium majus. Plant Cell Physiol. 58, 1421–1430. doi: 10.1093/pcp/pcx077
Yamada, Y., Shimada, T., Motomura, Y., Sato, F. (2017). Modulation of benzylisoquinoline alkaloid biosynthesis by heterologous expression of CjWRKY1 in Eschscholzia californica cells. PloS One 12, e0186953. doi: 10.1371/journal.pone.0186953
Yang, Y., Sun, Y., Wang, Z., Yin, M., Sun, R., Xue, L., et al. (2022). Full-length transcriptome and metabolite analysis reveal reticuline epimerase-independent pathways for benzylisoquinoline alkaloids biosynthesis in Sinomenium acutum. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1086335
Yang, M., Zhu, L., Li, L., Li, J., Xu, L., Feng, J., et al. (2017). Digital Gene Expression Analysis Provides Insight into the Transcript Profile of the Genes Involved in Aporphine Alkaloid Biosynthesis in Lotus (Nelumbo nucifera). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00080
Yao, Y. F., Wang, C. S., Qiao, J., Zhao, G. R. (2013). Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab. Eng. 19, 79–87. doi: 10.1016/j.ymben.2013.06.001
Yogendra, K. N., Dhokane, D., Kushalappa, A. C., Sarmiento, F., Rodriguez, E., Mosquera, T. (2017). StWRKY8 transcription factor regulates benzylisoquinoline alkaloid pathway in potato conferring resistance to late blight. Plant Sci. 256, 208–216. doi: 10.1016/j.plantsci.2016.12.014
Yuan, L., Grotewold, E. (2015). Metabolic engineering to enhance the value of plants as green factories. Metab. Eng. 27, 83–91. doi: 10.1016/j.ymben.2014.11.005
Yunzi, L., Bing-Zhi, L., Duo, L., Lu, Z., Yan, C., Bin, J., et al. (2015). Engineered biosynthesis of natural products in heterologous hosts. Chem. Soc Rev. 46, 5265–5290. doi: 10.1039/C5CS00025D
Zhan, C., Shen, S., Yang, C., Liu, Z., Fernie, A. R., Graham, I. A., et al. (2022). Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 27, 981–1001. doi: 10.1016/j.tplants.2022.03.002
Zhang, X., Bu, J., Zhao, Y., Li, Q., Li, X., Ma, Y., et al. (2023). Establishment of hairy root culture and its genetic transformation of Stephania tetrandra S. Moore for production of BIAs. Med. Plant Biol. 2022, 2028. doi: 10.48130/MPB-2023-0008
Zhang, M., Lu, P., Zheng, Y., Huang, X., Liu, J., Yan, H., et al. (2024). Genome-wide identification of AP2/ERF gene family in Coptis Chinensis Franch reveals its role in tissue-specific accumulation of benzylisoquinoline alkaloids. BMC Genomics 25, 972. doi: 10.1186/s12864-024-10883-1
Zhao, W., Liu, M., Liu, K., Liu, H., Liu, X., Liu, J. (2023). An enzymatic strategy for the selective methylation of high-value-added tetrahydroprotoberberine alkaloids. Int. J. Mol. Sci. 24, 15214. doi: 10.3390/ijms25020782
Zhao, W., Liu, M., Shen, C., Liu, K., Liu, H., Ou, C., et al. (2021). Biosynthesis of plant-specific alkaloids tetrahydroprotoberberines in engineered Escherichia coli. Green Chem. 23, 5944–5955. doi: 10.1039/d1gc01670a
Zhao, W., Liu, M., Shen, C., Liu, H., Zhang, Z., Dai, W., et al. (2020). Differentiation, chemical profiles and quality evaluation of five medicinal Stephania species (Menispermaceae) through integrated DNA barcoding, HPLC-QTOF-MS/MS and UHPLC-DAD. Fitoterapia 141, 104453. doi: 10.1016/j.fitote.2019.104453
Zhao, X., Pan, Y., Tan, J., Lv, H., Wang, Y., Chen, D.-X. (2024). Metabolomics and transcriptomics reveal the mechanism of alkaloid synthesis in Corydalis yanhusuo bulbs. PloS One 19, e0304258. doi: 10.1371/journal.pone.0304258
Zhao, M., Qin, Z., Abdullah, A., Xiao, Y. (2022). Construction of biocatalytic cascades for the synthesis of benzylisoquinoline alkaloids from p-coumaric acid derivatives and dopamine. Green Chem. 24, 3225–3234. doi: 10.1039/D1GC04759K
Ziegler, J., Brandt, W., Geissler, R., Facchini, P. J. (2009). Removal of substrate inhibition and increase in maximal velocity in the short chain dehydrogenase/reductase salutaridine reductase involved in morphine biosynthesis. J. Biol. Chem. 284, 26758–26767. doi: 10.1074/jbc.M109.030957
Zubieta, C., He, X. Z., Dixon, R. A., Noel, J. P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8, 271–279. doi: 10.1038/85029
Keywords: benzylisoquinoline alkaloids, biosynthesis, secondary metabolite, biosynthetic pathway, berberine bridge enzyme, cytochrome P450, methyltransferase
Citation: Zhao W, Liu J and Chen Y (2025) Advances in the biosynthesis of naturally occurring benzylisoquinoline alkaloids. Front. Plant Sci. 16:1548471. doi: 10.3389/fpls.2025.1548471
Received: 19 December 2024; Accepted: 14 January 2025;
Published: 30 January 2025.
Edited by:
Zefu Wang, Nanjing Forestry University, ChinaReviewed by:
Cheng Song, West Anhui University, ChinaCopyright © 2025 Zhao, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Chen, eWNoZW5AamliLmFjLmNu; Jihua Liu, bGl1amlodWFAY3B1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.