
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 March 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1546660
Introduction: Vascular cambium proliferates and differentiates into the secondary xylem (wood), enabling the perennial increase in stem diameter for wood formation. In our previous study, we identified 95 vascular-cambium-specific (VCS) transcription factors (TFs) in Populus trichocarpa.
Methods: In this study, we characterized the function of the highly vascular cambium-expressed heat shock TF among these VCSs, PtrSCZ1, using PtrSCZ1-overexpressing transgenic lines and gene-edited mutants in P. trichocarpa.
Results: Overexpressing PtrSCZ1 or its homolog PtrSCZ3 (OE-PtrSCZ1, OE-PtrSCZ3) led to enhanced cambium activity, increased stem diameter, and a larger xylem proportion. CRISPR-based mutants of PtrSCZ1 and PtrSCZ3 exhibited phenotypes opposite to the OE-PtrSCZ1 and OE-PtrSCZ3 plants. This suggests that PtrSCZ1 and PtrSCZ3 redundantly promote cambium activity and secondary growth, leading to increased radial growth in P. trichocarpa. Overexpression and knockout of PtrSCZ1 and PtrSCZ3 significantly affected the expression of key regulatory factors of cambium (PtrWOX4a, PtrWOX4b, PtrWOX13a, PtrPXYa, PtrVCM1, and PtrVCM2) and disrupted cell wall-related gene expression. This demonstrates that PtrSCZ1 and PtrSCZ3 may function in cambium division activity by regulating these key cambium-associated transcription factors for wood formation.
Discussion: Our work identifies PtrSCZ1 and PtrSCZ3 as promising target genes for enhancing wood yield through molecular breeding, and illustrates the role of vascular cambium systems in understanding lateral meristem development.
Vascular tissue formation in forest trees initiates from the procambium within the shoot apical meristems (SAM). This tissue differentiates into vascular bundles while maintaining a fascicular cambium layer that retains meristematic potential. The fascicular cambium, together with the interfascicular cambium derived from parenchyma cells, forms a continuous ring of secondary meristematic tissue, collectively referred to as the vascular cambium (Mazur et al., 2014; Zhu et al., 2018, 2019). The vascular cambium continuously produces secondary xylem inward and secondary phloem outward during secondary growth (Timell, 1980; Włoch, 1981; Włoch et al., 2023; Li et al., 2024). Fusiform initials differentiate into vessels and fibers, contributing to increased stem diameter and wood formation (Esau, 1965; Evert, 2006). Therefore, the activity of the vascular cambium directly affects the secondary growth of trees and, consequently, wood formation. Cambium activity is tightly regulated by transcription factors (TFs), plant hormones, signaling peptides, small RNAs, long non-coding RNAs, and enzymes. These elements contribute to transcriptional, post-transcriptional, and epigenetic regulation according to developmental programs (Ortega-Martínez et al., 2007; Matsumoto-Kitano et al., 2008; Suer et al., 2011; Nieminen et al., 2015; Johnsson and Fischer, 2016; Bhalerao and Fischer, 2017; Fischer et al., 2019; Zhang et al., 2019b; Hu et al., 2024). However, the mechanisms governing this complex regulatory process remain largely unknown.
Several TFs expressed in the stem cambium have been identified in tree species. These include WOX4a/b, PXY, PtrVCM1 and PtrVCM2, PtrWOX13a, PtrVCS2, PtrHB4, PtrHB7, popREVOLUTA (PRE), ARK2, POPCORONA, PtoTCP20, PagGRF15, PagMYB31, and PagJAZ5. They have been shown to directly or indirectly regulate cambium activity (Du et al., 2009, 2011; Robischon et al., 2011; Zhu et al., 2013; Kucukoglu et al., 2017; Zhu et al., 2018; Hou et al., 2020; Zheng et al., 2021; Dai et al., 2023; Hu et al., 2024; Zhang et al., 2024; Zhou et al., 2024). Disturbances in the expression levels of these genes affect the number of cambium cell layers, subsequently influencing xylem development. This indicates their crucial roles in regulating vascular cambium activity and xylem development.
Heat shock transcription factors (HSFs) are crucial transcriptional regulators involved in early heat shock responses (Li et al., 2010; Nishizawa-Yokoi et al., 2011; Ikeda et al., 2011) and disease resistance (Kumar et al., 2009). Based on structural characteristics, such as the adjacent hydrophobic heptad repeat oligomerization domain (HR-A/B), plant HSFs can be grouped into three conserved classes (Nover et al., 2001; Scharf et al., 2012). AtHSFB4/SCHIZORIZA (SCZ), a B-class heat-shock TF, is one of the 21 TFs in the Arabidopsis HSF family (Nover et al., 2001; Pernas et al., 2010). The expression of SCZ is barely affected by heat treatment, unlike typical HSF genes such as HSFA1s, HSFA2, HSFB1, and HSFB2b (Miller and Mittler, 2006; von-Koskull-Döring et al., 2007; Scharf et al., 2012; Pardal et al., 2023). This suggests that SCZ has evolved toward functions beyond the stress response typical of the HSF family (Pardal et al., 2023). During plant development, SCZ is essential for the early establishment of stem cells that generate ground tissue in the embryonic root meristem (Pernas et al., 2010). This transcription factor also plays a key role in regulating segregation of different root tissue fates in Arabidopsis (Pardal et al., 2023). Overexpressing AtHSFB4 causes abnormal growth in root meristematic region (Begum et al., 2013), while the AtHsfB4 mutant exhibits aberrant periclinal division of the ground tissue during embryogenesis, disturbed asymmetric cell division, abnormal root hair development, and misexpression of tissue identity markers in root development (Mylona et al., 2002; ten Hove et al., 2010; Pardal et al., 2023).
Thirty HSF members have been identified in P. trichocarpa (Wang et al., 2011; Scharf et al., 2012; Zhang et al., 2015; Liu et al., 2019; Guo et al., 2023). PtrHSFB3-1 is a tension wood-responsive TF that directly regulates cell-wall genes, including PAL2, PAL4, PAL5, C4H1, C3H3, CSE1, CCoAOMT2, and CCR2, to reprogram wood formation (Liu et al., 2021). While some HSF genes are involved in secondary growth, most play crucial roles in stress response processes in trees. For instance, PuHSFA4a regulates abiotic stress responses and root development-related genes, enhancing tolerance to excess Zn in P. ussuriensis roots (Zhang et al., 2019a). Additionally, PsnHSF21 in poplar contributes to salt resistance (Guo et al., 2023).
In our previous study, we identified 95 vascular-cambium-specific (VCS) TFs in P. trichocarpa (Dai et al., 2023). Among the 95 VCS TFs, three belonged to the HSF family: PtrVCS9, PtrVCS38, and PtrVCS80. Notably, PtrVCS9 exhibited higher transcript abundance in the vascular cambium compared to xylem and phloem, and its expression level ranked 9th among the 95 TFs (Dai et al., 2023). In this study, we investigated the function of PtrVCS9 (renamed as PtrSCZ1) in regulating vascular cambium activity during wood formation in P. trichocarpa. We demonstrated that PtrVCS9 and its homologous PtrVCS80 (renamed as PtrSCZ3) are crucial for regulating vascular cambium proliferation, which in turn influences xylem cell division during secondary stem growth in P. trichocarpa. The two TFs regulate the expression of key genes in the stem vascular cambium, thereby modulating wood formation.
Populus trichocarpa (Nisqually-1) was cultivated in a phytotron under a 16-hour light/8-hour dark photoperiod at 25°C and utilized for gene cloning and expression analysis in this study. Following generation, screening, and verification of transgenic plants, they were maintained in the phytotron for 4 months before undergoing detailed characterization.
Based on the P. trichocarpa genome (https://phytozome-next.jgi.doe.gov), the full-length sequences of PtrSCZ1 and PtrSCZ3 were amplified from cDNA derived from cambium–phloem tissue of P. trichocarpa plants. The specific primers used for amplification are listed in Supplementary Table S1. The resulting amplicons were cloned into pENTR/D-TOPO vector (Invitrogen), and their sequences were subsequently verified by sequencing. Sequence alignment was performed using Clustal W, and the corresponding figure was generated using ESPript 3.0 (Robert and Gouet, 2014). Phylogenetic analysis of SCZs was conducted using MEGA 5 software, employing the neighbor-joining method with 1000 bootstrap replicates.
Well-developed plants were selected for bark removal. Stem-differentiating xylem (SDX) from the debarked stems and cambium–phloem tissues from the bark side were separately collected using single-edged razors and immediately frozen in liquid nitrogen. Total RNA was isolated from SDX tissues using the RNeasy Plant Mini Kit (Qiagen, 74904) for RT-qPCR analysis of lignin biosynthesis-related genes. For cambium–phloem tissues, RNA extraction was performed using the CTAB method (Lorenz et al., 2010) to determine the relative expression levels of PtrSCZ1, PtrSCZ3, and other cambium-related genes. RNA concentration was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific).
Reverse transcription (RT) reactions were performed using TaqMan Reverse Transcription Reagents (Invitrogen, N8080234) according to the manufacturer’s protocol. All quantitative PCR (qPCR) analyses were conducted on the Agilent M×3000P Real-Time PCR System with FastStart Universal SYBR Green Master Mix (Roche, 4913914001) following the standard protocol. Gene expression levels were normalized to PtrACTIN. The specific primers used for RT-qPCR analysis are listed in Supplementary Table S1.
The eighth stem internodes of P. trichocarpa were collected and fixed in FAA solution (50% [v/v] ethanol, 5% [v/v] acetic acid, and 3.7% [v/v] formaldehyde). Following dehydration, the fixed tissues were embedded in paraffin (Sigma) and sectioned to a thickness of 10 µm using a rotary microtome (Leica RM2245). A 172-bp region of PtrSCZ1 was used as a specific probe for in situ hybridization. Both antisense and sense probes were synthesized using T7 and SP6 RNA polymerases, respectively. Probe labeling was performed using a digoxigenin RNA labeling kit (Roche). After pretreatment, the slide-mounted sections were hybridized with the digoxigenin-labeled PtrSCZ1 antisense or sense RNA probes in hybridization solution. Hybridized signals were detected by incubation with alkaline phosphatase-conjugated anti-digoxigenin antibodies according to the manufacturer’s protocol provided with the digoxigenin nucleic acid detection kit (Roche). Color development was achieved using alkaline phosphatase substrates. Microscopic images were captured using a Leica DM6B microscope. The Primer sequences used for probe amplification are summarized in Supplementary Table S1.
The 35S::PtrSCZ1-FLAG and 35S::PtrSCZ3-FLAG constructs were generated using the pBI121 vector. Double-knockout mutants of PtrSCZ1 and PtrSCZ3 genes were created using the CRISPR-Cas9 system. The single guide RNAs (sgRNAs) were designed using CRISPR-P 2.0 (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR). The sgRNA sequences were synthesized and subsequently cloned into the pEgP237-2A GFP vector (Ueta et al., 2017). All vectors were introduced into P. trichocarpa plants through Agrobacterium tumefaciens-mediated transformation, following a previously described protocol (Li et al., 2017). The expression levels of PtrSCZ1 and PtrSCZ3 in transgenic plants were analyzed by RT-qPCR as described above. To verify the genome editing in knockout mutants, specific primers flanking the sgRNA target sites were designed for PCR amplification. The resulting PCR products were cloned into the pMD18-T vector, and a minimum of 20 colonies were selected for sequencing analysis. All primer sequence are listed in Supplementary Table S1.
Stem internodes of P. trichocarpa were sectioned into 2-mm fragments and fixed in FAA solution (composition as described above). The fixed stem segments were subsequently processed through a graded ethanol series (50%, 60%, 70%, 85%, and 100% [v/v]) at 4°C for dehydration, followed by incubated in ethanol:xylene solutions (75:25, 50:50, and 25:75 [v/v]) and finally in 100% xylene at room temperature. The dehydrated samples were then immersed in 75:25 (v/v) xylene:paraffin solution at 42°C overnight and embedded in pure paraffin (Sigma). The paraffin-embedded stem segments were sectioned into 12-µm thickness using a rotary microtome (Leica RM2245) and stained with Toluidine Blue. Digital images were acquired using a M8 microscope scanner (Percipient), and the quantitative measurements of cambium cells, stem area, and xylem area were performed using LASv4.8 and LASXv2.0 software (Leica).
Stem segments from OE-PtrSCZ1-L14, OE-PtrSCZ3-L13, ptrscz1/ptrscz3-L1, and wild-type P. trichocarpa plants were ground to 40–60 mesh size. Extraction was carried out using a benzene:ethanol mixture (2:1 v/v) for 24 hours. The air-dried wood samples from different transgenic lines and wild-type plants were pooled, and their lignin and sugar contents were analyzed according to established procedures (Abraham et al., 2013; Wang et al., 2018).
All statistical analyses were performed using two-tailed Student’s t-tests to determine significance, with significance levels set at *P < 0.05 and **P < 0.01. The RNA-seq data for PtrSCZ1 (PtrVCS9) and PtrSCZ3 (PtrVCS80) presented in this study were obtained from previously characterized vascular-cambium-specific TFs. These TFs were identified through RNA-seq analysis of laser capture microdissection (LCM)-isolated cambium, differentiating xylem, and phloem tissues (Supplementary Table 1 in Dai et al., 2023), integrated with RNA-seq data from leaves, roots, and shoots (Shi et al., 2017).
Based on vascular-cambium-specific transcriptome data acquired by LCM (Dai et al., 2023) and tissue-specific datasets (Shi et al., 2017) from P. trichocarpa, we identified that the HSF gene PtrVCS9 shows preferential expression in the vascular cambium compared to other tissues including xylem, phloem, leaf, root, and shoot (Figure 1A). Comparative analysis with Arabidopsis HSF family genes revealed that PtrVCS9 shares sequence similarity with HSFB4 (Supplementary Figure S1), also known as SCZ (SCHIZORIZA) (Pernas et al., 2010; ten Hove et al., 2010). Consequently, we designated PtrVCS9 as PtrSCZ1. RT-qPCR analysis demonstrated that PtrSCZ1 expression in the cambium becomes evident beginning at the 4th internode, coinciding with the initiation of secondary growth (Figure 1B). In situ RNA hybridization using a PtrSCZ1-specific probe on 4-month-old P. trichocarpa 8th internodes showed strong signal accumulation in the vascular cambium zone and weaker signals in xylem cells (Figure 1C). The expression pattern suggests PtrSCZ1’s involvement in vascular cambium development. To explore PtrSCZ1’s functions in cambium activity, we generated CRISPR-mediated knockout and overexpressing lines in P. trichocarpa for functional characterizations.

Figure 1. PtrSCZ1 was highly expressed in vascular cambium. (A) Tissue-specific expression patterns of PtrSCZ1 analyzed by RNA-seq across six P. trichocarpa tissues (cambium, differentiating xylem, phloem, leaf, root, and shoot). FPKM values represent fragments per kilobase of transcript per million fragments mapped reads. (B) Transcript abundance of PtrSCZ1, as determined by RT-qPCR in the shoot apices (containing the apical meristem, leaf primordia, developing leaves and early vascular tissues), and cambium–phloem tissues from 1st–4th, 20th, and 40th internodes of P. trichocarpa stems. (C) Spatial expression pattern of PtrSCZ1 revealed by in situ hybridization in 8th stem internode of P. trichocarpa. Stem cross-sections were hybridized with digoxigenin-labeled antisense RNA probes for PtrSCZ1 (left) or sense RNA probes as negative control (right). ca, cambium; pf, phloem fiber; ph, phloem; xy, xylem. Bar, 100 µm. Error bars in (A, B) represent standard errors (SE) of three biological replicates from independent pools of P. trichocarpa tissues.
We generated loss-of-function mutants of PtrSCZ1 in P. trichocarpa using CRISPR-based genome editing with Streptococcus pyogenes Cas9 (Deltcheva et al., 2011; Jiang et al., 2013; Heler et al., 2015). The single-knockout ptrscz1 lines showed comparable plant height, stem diameter, and cambium cell layer numbers to wild-type (WT) plants (Supplementary Figure S2), suggesting potential functional redundancy among PtrSCZ family members. We therefore investigated PtrSCZ1’s homologue, PtrSCZ3, which shares 93.3% amino acid sequence identity with PtrSCZ1 (Supplementary Figure S1) and displays high expression levels in cambium, root, and shoot tissues (Supplementary Figure S3A) (Shi et al., 2017; Dai et al., 2023), though lacking cambium-specific expression across different internodes (Supplementary Figure S3B). In situ hybridization demonstrated that PtrSCZ3 exhibits a comparable expression pattern to PtrSCZ1 in stem tissues (Supplementary Figures S3C, D). We subsequently created two independent biallelic double mutants, ptrscz1/ptrscz3-L1 and ptrscz1/ptrscz3-L10, containing insertions and/or deletions that introduce frameshifts and premature stop codons (Figures 2A, B). The ptrscz1/ptrscz3 lines displayed significantly reduced stem diameters compared to WT plants (Figures 2C, D; Supplementary Figure S4). Stem cross-sectional analysis revealed that ptrscz1/ptrscz3 internodes consistently lacked a fixed number (one to three) of cambium cell layers compared to the WT (5th to 8th internodes shown in Figures 2E, F). These results indicate that the loss-of-function in PtrSCZ1 and PtrSCZ3 intrinsically reduces cambium cell layer formation during stem elongation.
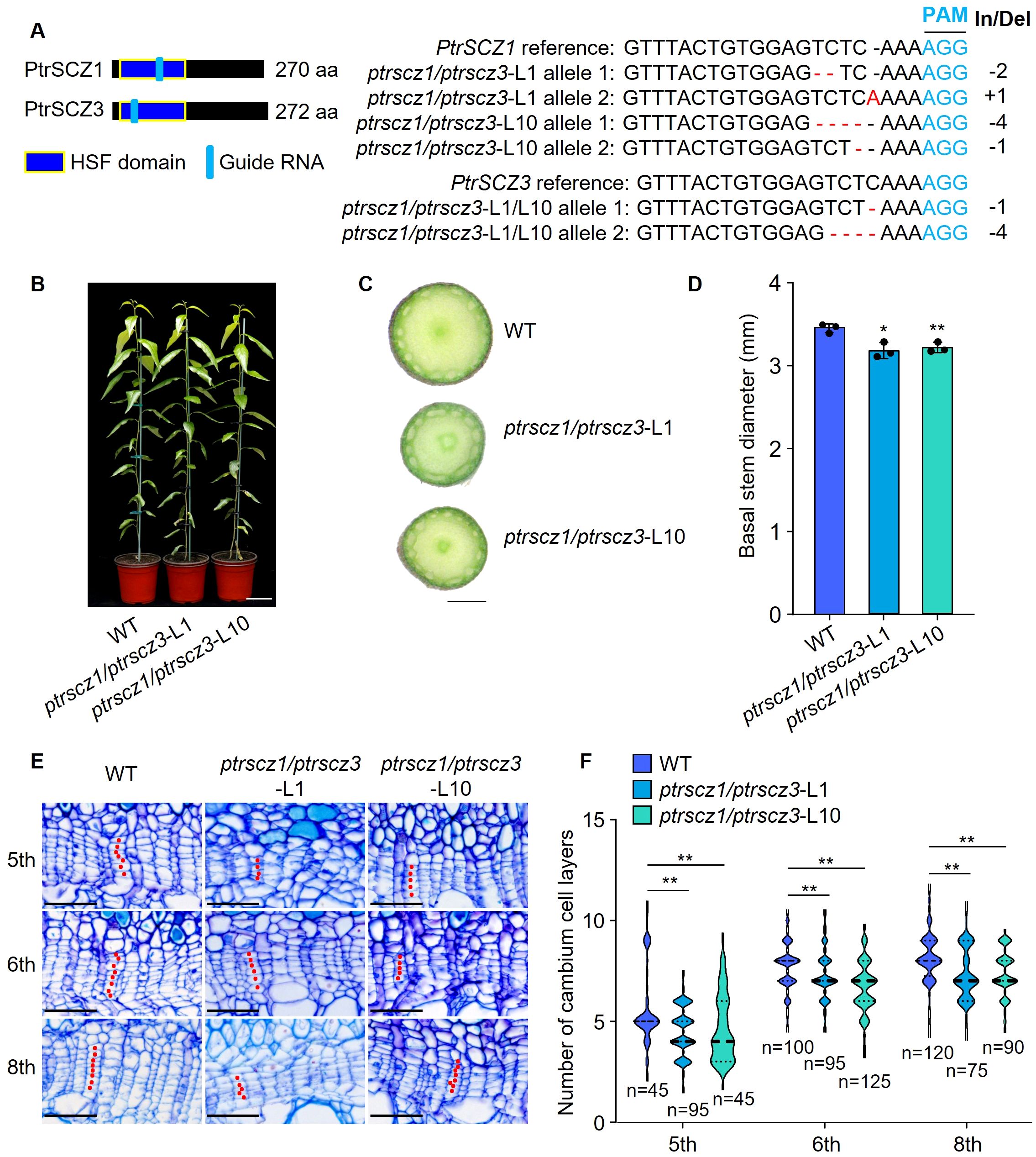
Figure 2. Effects of knocking out PtrSCZ1 and PtrSCZ3 on vascular cambium proliferation. (A) Mutations at the sgRNA target sites in PtrSCZ1 and PtrSCZ3 from two independent ptrscz1/ptrscz3 mutant lines. Deletions are indicated by red dashes, while nucleotide substitutions and insertions are highlighted in red. The protospacer-adjacent motif (PAM) is shown in blue, with insertion/deletion (In/Del) lengths indicated on the right. (B) Growth phenotypes of 4-month-old WT, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 plants. Bar, 10 cm. (C) Basal stems morphology of WT, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 mutant plants. Bar, 1 mm. (D) Quantitative analysis of stem diameters of WT, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 plants. Data represent means ± SE from three biological replicates (two-tailed Student’s t-test, *P< 0.05, **P< 0.01). (E) Histochemical and histological characterization of stem sections from WT, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 plants. Bars, 50 µm. (F) Cambium cell layer quantification in stem vascular tissues of WT, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 plants. For each biological replicate, at least ten radial cell files were analyzed per cross-section. Data from three biological replicates are presented (two-tailed Student’s t-test, **P< 0.01). n, total number of samples per dataset. The width of the violin represents the distribution shape of the data. The center line represents the median, and the upper and lower dotted lines correspond to the 25th and 75th percentiles, respectively.
We overexpressed PtrSCZ1 in P. trichocarpa under the control of a cauliflower mosaic virus 35S promoter. Three independent overexpression transgenic lines of OE-PtrSCZ1 (-L1, -L7 and -L14) were generated (Figures 3A, B). These transgenic lines displayed stunted growth (Figure 3B) but showed increased stem diameter during secondary development compared to WT (Figures 3C, D; Supplementary Figure S5). To examine PtrSCZ1’s role in cambium development during wood formation, we selected OE-PtrSCZ1-L14, which showed the highest transgene overexpression level, for detailed phenotypic analysis. Four-month-old transgenic plants were evaluated under greenhouse conditions. Histological examination revealed that OE-PtrSCZ1-L14 developed 2–3 additional cambium cell layers in the 5th to 8th stem internodes compared to WT (Figures 3E, F). Comparable phenotypic alterations were observed in OE-PtrSCZ1-L13 (Figures 3E, F). The opposing cambium cell layer phenotypes between gain- and loss-of-function transgenics indicate that PtrSCZ1 and PtrSCZ3 play specific roles in regulating cambium cell proliferation.
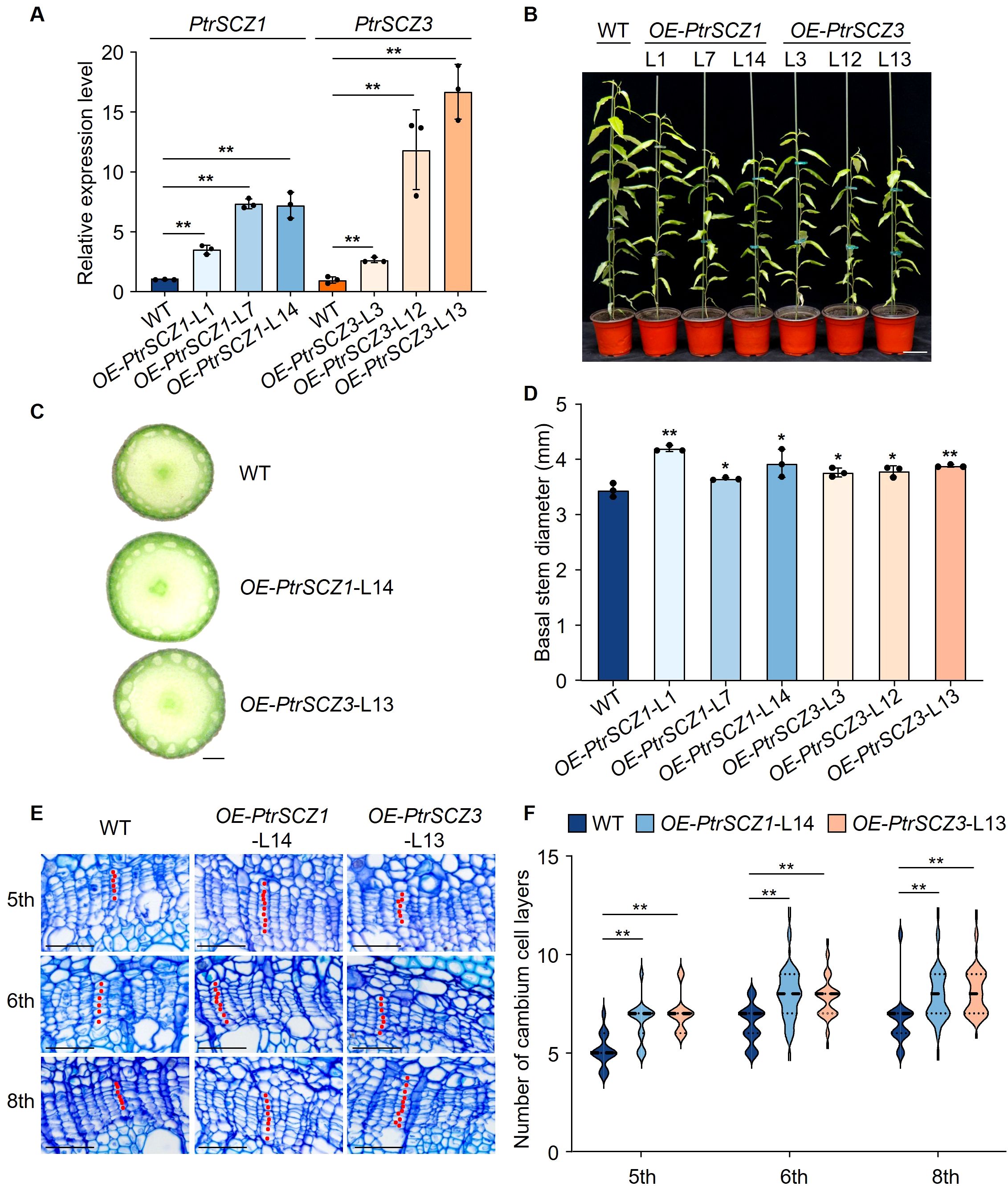
Figure 3. Growth phenotype and statistical analysis of PtrSCZ1 and PtrSCZ3 overexpression transgenic lines. (A) Expression profiles of PtrSCZ1 and PtrSCZ3 in cambium–phloem tissues from transgenic lines. (B) Growth phenotypes of 4-month-old WT, OE-PtrSCZ1 (-L1, -L7, -L14), and OE-PtrSCZ3 (-L3, -L12, -L13) plants. Bar, 10 cm. (C) Basal stem morphology of WT, OE-PtrSCZ1-L14, and OE-PtrSCZ3-L13 transgenic plants. Bar, 1 mm. (D) Quantitative analysis of stem diameters in WT, OE-PtrSCZ1 (-L1, -L7, -L14), and OE-PtrSCZ3 (-L3, -L12, -L13) plants. In (A, D), error bars represent mean ± SE from three biological replicates, with three P. trichocarpa plants per genotype per replicate (two-tailed Student’s t-test, *P< 0.05, **P<0.01). (E) Histochemical and histological characterization of stem sections from WT, OE-PtrSCZ1-L14, and OE-PtrSCZ3-L13 transgenic plants. Bars, 50 µm. (F) Cambium cell layer quantification in stem vascular tissues of WT, OE-PtrSCZ1-L14, and OE-PtrSCZ3-L13 transgenic plants. For each biological replicate, at least ten radial cell files were counted per cross-section. Data from three biological replicates are shown (two-tailed Student’s t-test, **P< 0.01). n = 30 in (F). The width of the violin represents the distribution shape of the data. The center line represents the median, and the upper and lower dotted lines correspond to the 25th and 75th percentiles, respectively.
The ptrscz1/ptrscz3 mutants displayed reduced cambium cell layer numbers compared to WT plants, whereas overexpression transgenic plants showed increased cambial cell layers (Figures 2, 3). These results indicate that PtrSCZ1 and PtrSCZ3 positively regulate cambial activity. To investigate how alterations in PtrSCZ1 and PtrSCZ3 affect cambial development, we analyzed the expression levels of key cambium regulators, including positive regulators of cell division (PtrWOX4a, PtrWOX4b, PtrWOX13a, and PtrPXYa) and negative regulators of vascular cambium proliferation (PtrVCM1 and PtrVCM2) in OE-PtrSCZ1, OE-PtrSCZ3, and ptrscz1/ptrscz3 plants (Etchells et al., 2013; Kucukoglu et al., 2017; Zheng et al., 2021; Dai et al., 2023). Transcript levels of PtrWOX4a, PtrWOX4b, PtrWOX13a, and PtrPXYa were consistently upregulated in OE-PtrSCZ1 and OE-PtrSCZ3 plants but downregulated in ptrscz1/ptrscz3 mutants compared to WT (Figures 4A–D). PtrVCM2 expression was significantly reduced in OE-PtrSCZ1 plants, while PtrVCM1 levels remained unchanged in both OE-PtrSCZ1 and OE-PtrSCZ3 lines (Figures 4E, F). Most key TFs associated with cambium activity showed altered expression patterns in response to PtrSCZ1 and PtrSCZ3 manipulation. These findings suggest that PtrSCZ1 and PtrSCZ3 may promote cambium cell division by modulating these key TFs.
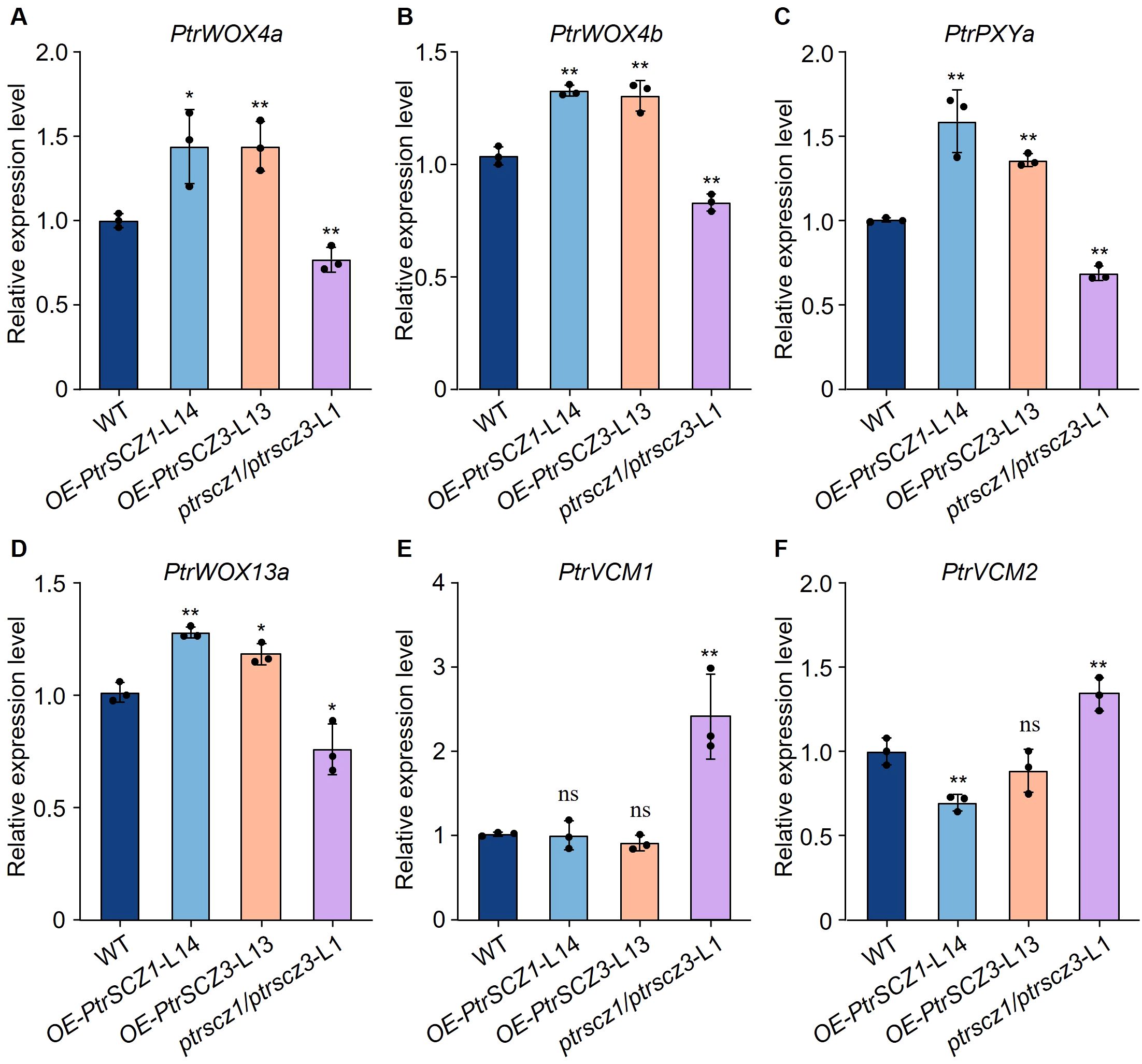
Figure 4. RT-qPCR analysis of key TFs in cambium development in transgenic plants. (A–F) Expression levels of PtrWOX4a (A), PtrWOX4b (B), PtrPXYa (C), PtrWOX13a (D), PtrVCM1 (E), and PtrVCM2 (F) were quantified by RT-qPCR in cambium–phloem tissues from 4-month-old WT, OE-PtrSCZ1-L14, OE-PtrSCZ3-L13, and ptrscz1/scz3-L1 transgenic plants. Error bars represent mean ± SE from three biological replicates using independent pools of P. trichocarpa cambium–phloem tissues (two-tailed Student’s t-test, *P< 0.05, **P< 0.01). ns, not significant differences.
To determine whether cambium cell layer alterations affect xylem development, we compared xylem zones between transgenic and WT plants. Stem cross-sectional analysis revealed that OE-PtrSCZ1-L14 and OE-PtrSCZ3-L13 transgenic lines displayed increased xylem-to-stem area ratios compared to WT (Figures 5A, C; Supplementary Figures S6A, B), demonstrating enhanced xylem development in the overexpression plants. In contrast, ptrscz1/ptrscz3 loss-of-function mutants showed significantly reduced xylem area proportions relative to total stem area (Figures 5B, D; Supplementary Figures S6C, D), indicating inhibited xylem development. These findings demonstrate that PtrSCZ1 and PtrSCZ3 positively regulates secondary growth. We subsequently characterized and quantified the three primary components of wood cell walls.
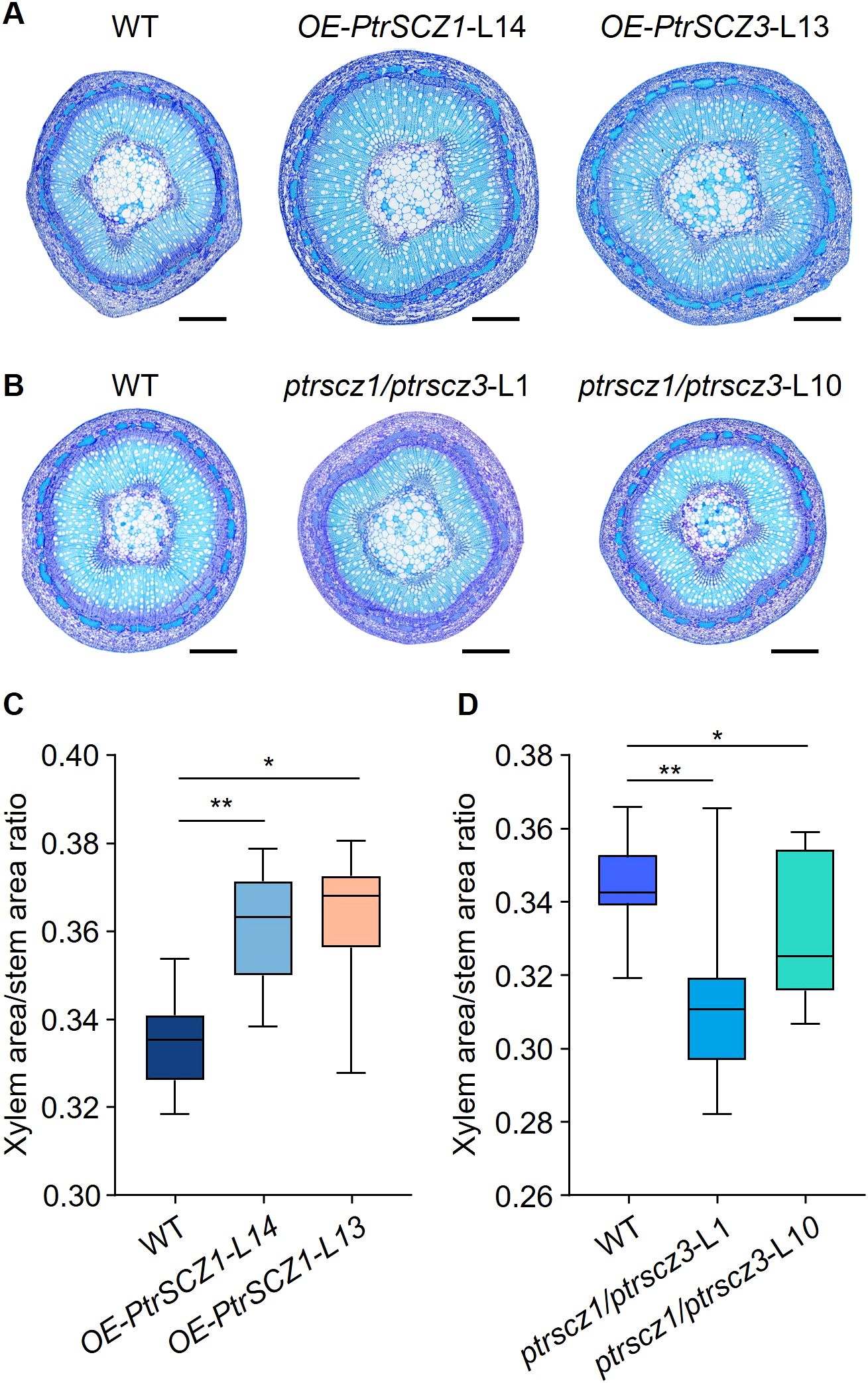
Figure 5. PtrSCZ1 and PtrSCZ3 affect the xylem development. (A, B) Histological observations of stem cross-sections from 8th internodes of WT, OE-PtrSCZ1-L14, OE-PtrSCZ3-L13, ptrscz1/ptrscz3-L1, and ptrscz1/ptrscz3-L10 plants. Bars, 500 µm. (C, D) Quantitative analysis of xylem area proportion in stems among OE-PtrSCZ1-L14, OE-PtrSCZ3-L13, and WT (C), ptrscz1/ptrscz3-L1, ptrscz1/ptrscz3-L10, and WT (D). Xylem and stem areas were measured from five cross-sections per stem internode for each biological replicate. Data from three biological replicates are shown (two-tailed Student’s t-test, *P< 0.05, **P < 0.01). n=15. Box plots display median and interquartile ranges, with whiskers representing data ranges excluding outliers.
Wood composition analysis showed a notable increase in lignin content (13%–15%) in OE-PtrSCZ1-L14 and OE-PtrSCZ3-L13 (Table 1), with no significant change in total carbohydrate content in these transgenic plants. Notably, monolignol composition in the lignin of overexpression transgenics showed a 10% reduction in S-monomers and a 20% increase in G-monomers, leading to S:G ratios of 1.48–1.62 compared to 2.00 in WT (Table 2). Analytical results showed reduced lignin content (approximately 9%) and the carbohydrate-to-lignin ratio (C:L) in ptrscz1/ptrscz3-L1 mutants, contrasting with the increases observed in OE-PtrSCZ1-L14 and OE-PtrSCZ3-L13 (Table 1). S:G ratios in ptrscz1/ptrscz3-L1 mutants showed minimal changes, remaining statistically indistinguishable from WT values (Table 2). These findings suggest that PtrSCZ1 and PtrSCZ3 regulate lignin biosynthesis without substantially affecting lignin monomer composition. We subsequently examined the effect of PtrSCZ1 and PtrSCZ3 overexpression or mutation on the transcriptional levels of genes related to monolignol biosynthesis.
Given that altering PtrSCZ1 and PtrSCZ3 functions resulted in changes in lignin content and S/G ratio, we use RT-qPCR to examine the expression of monolignol biosynthesis enzyme genes in transgenic plants. Enzyme genes associated with the G-lignin biosynthesis (Zhang et al., 2020), including PtrCCoAOMT1, as well as those regulating total lignin content (PtrPAL1, PtrCCR2, and PtrCAD1), showed upregulated expression in both OE-PtrSCZ1 and OE-PtrSCZ3 (Figure 6). Conversely, PtrCAld5H1 expression was downregulated (Figure 6), consistent with the reduced S-monomer levels observed in OE-PtrSCZ1-L14 and OE-PtrSCZ3-L13 (Table 2). In ptrscz1/ptrscz3-L1 mutants, disruption of PtrSCZ1 and PtrSCZ3 did not significantly alter the expression of most lignin biosynthesis genes, except for PtrCCoAOMT1 and PtrCAld5H1 (Figure 6; Supplementary Figure S7). Compared to overexpressed plants, these two genes displayed opposite expression patterns in the mutant (Figure 6). These results suggest that PtrSCZ1 and PtrSCZ3 likely function as transcriptional activators regulating lignin biosynthesis during secondary wall formation in P. trichocarpa.
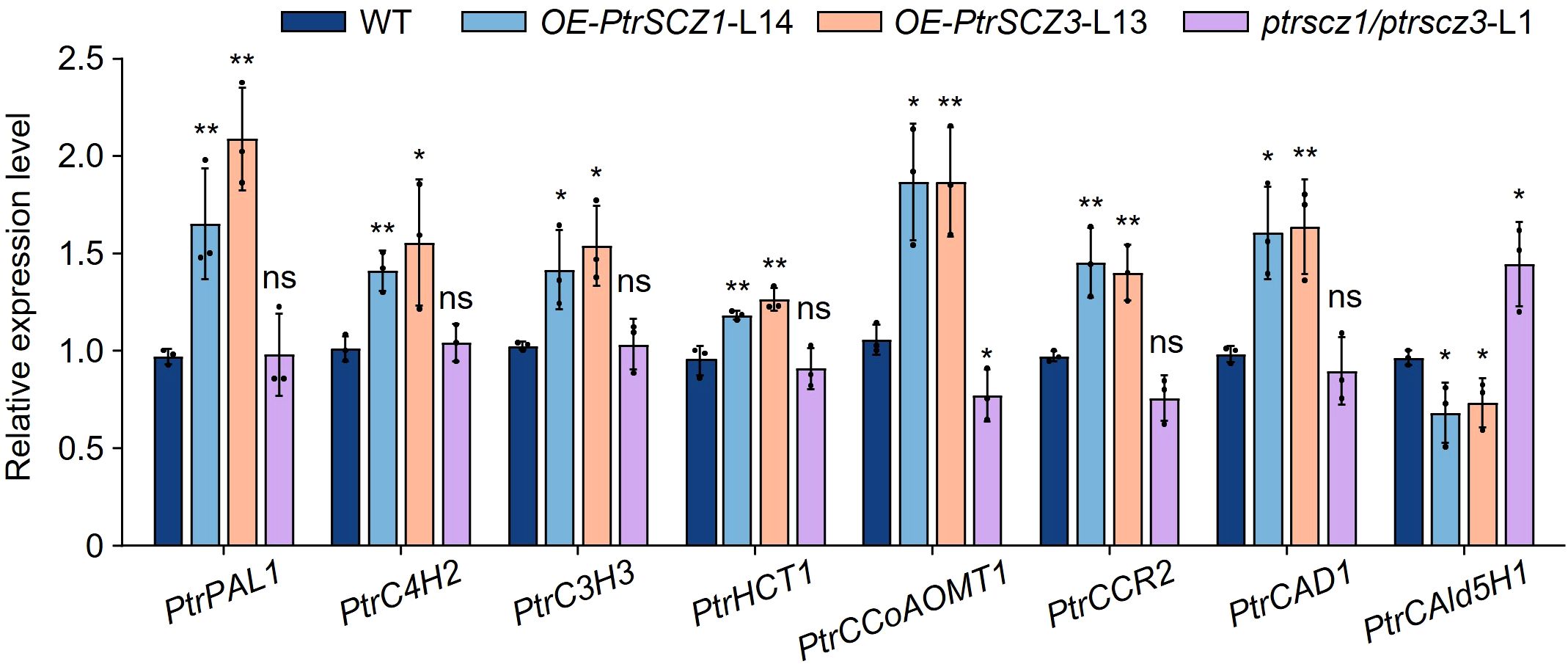
Figure 6. Expression levels of cell wall component genes in transgenic plants. The SDX tissues from WT, OE-PtrSCZ1-L14, OE-PtrSCZ3-L13, and ptrscz1/ptrscz3-L1 plants were analyzed by RT-qPCR to examine expression patterns of monolignol biosynthesis genes. Genes are arranged according to their enzymatic order in the lignin biosynthesis pathway. Error bars represent mean ± SE from three biological replicates using independent pools of P. trichocarpa SDX tissues (two-tailed Student’s t-test, *P< 0.05, **P< 0.01). ns, not significant differences.
Collectively, transcriptional analysis of PtrSCZ1 and PtrSCZ3 knockout and overexpressed plants reveals their active role in secondary growth through directly or indirectly regulating the vascular cambium and monolignol genes, which are essential for the cambium and xylem development.
Division and differentiation of vascular cambium cells result in secondary xylem formation, a sequential developmental process that includes secondary cell wall (SCW) deposition and programmed cell death (Du and Groover, 2010; Luo and Li, 2022). In this study, we revealed the homologous gene pair PtrSCZ1 and PtrSCZ3 function redundantly in the regulation of vascular cambium activity and secondary xylem tissue formation in P. trichocarpa stems. We speculate that PtrSCZ1 and PtrSCZ3 may control wood formation by direct or indirect regulating specific vascular cambium and monolignol genes during plant secondary growth.
A key characteristic of perennial trees is the secondary growth of stems, which depends on vascular cambium proliferation activity. The expression level of PtrSCZ1 is significantly upregulated in the vascular cambium during the secondary growth stage (Figure 1B). Overexpression of PtrSCZ1 and its homologous PtrSCZ3 promoted vascular cambium proliferation and enhanced the secondary growth of stems. The double mutants of PtrSCZ1 and PtrSCZ3 exhibit decreased stem diameter (Figures 2C, D), reduced number of vascular cambium cell layers (Figures 2E, F), and a reduction in the proportion of xylem (Figures 5B, D). The observed transgenic phenotypes demonstrate that PtrSCZ1 and its homologs positively regulate cambium activity and xylem development. There are 4 homologous HSFB4 genes in P. trichocarpa including PtrSCZ1, PtrSCZ2, PtrSCZ3, and PtrSCZ4 (Supplementary Figure S1). Both PtrSCZ2 (PtrVCS38) and PtrSCZ4 show expression in multiple tissues, including cambium, xylem, phloem, leaves, roots, and shoots (Shi et al., 2017; Dai et al., 2023). Notably, PtrSCZ2 has been identified as a cambium-specific TF gene (Dai et al., 2023). The potential roles of PtrSCZ2 and PtrSCZ4 in regulating cambium activity and affecting xylem development remain to be investigated. Generating and analyzing a quadruple mutant (ptrscz1/ptrscz2/ptrscz3/ptrscz4) could provide further insights into the collective functions of PtrSCZ genes in regulating secondary growth in woody trees.
The activity of the vascular cambium is indicated by the number of cambium cell layers, which is controlled by the expression levels of key cambium-regulated TFs. Overexpression of PttPXY and PttCLE41 in hybrid aspen resulted in vascular tissue abnormalities and poor plant growth (Etchells et al., 2015; Kucukoglu et al., 2017). The PttWOX4 genes have been shown to function downstream of PXY and control cell division activity in the vascular cambium, thereby influencing growth in stem girth (Kucukoglu et al., 2017). PtrWOX13a directly regulates the transcription of PtrWOX4a (Dai et al., 2023). The hierarchical regulation of cambium development is precisely controlled by these TFs. Changes in the expression levels of PtrSCZ1 and PtrSCZ3 were positively correlated with those of four genes (PtrWOX4a, PtrWOX4b, PtrWOX13a, and PtrPXYa) (Figure 4), indicating that PtrSCZ1 and PtrSCZ3 might operate within the same regulatory pathway, directly or indirectly influencing their expression. Knockdown of two vascular cambium-related MADS-box genes, VCM1 and VCM2, enhanced cambium proliferation and subsequent xylem differentiation in Populus (Zheng et al., 2021). Changes in the expression of PtrSCZ1 and PtrSCZ3 also affect the expression of these two genes (Figure 4). The previous report indicated that the complex PtrWOX13a–PtrVCS2–PtrGCN5-1–PtrADA2b-3 binds directly to PtrWOX4a promoter through PtrWOX13a, where PtrVCS2 prevented the interaction between PtrGCN5-1 and PtrADA2b-3. This prevention resulted in PtrWOX4a promoter hypoacetylation, leading to fewer cambium cell layers (Dai et al., 2023). Overexpressing PtrSCZ1 or PtrSCZ3 elevates PtrWOX4a levels, yet their impact on the acetylation of the PtrWOX13a–PtrVCS2–PtrGCN5-1–PtrADA2b-3 complex remains unexplored. These findings underscore the crucial role of PtrSCZ1 and PtrSCZ3 in cambium activity and suggest they might establish a complex regulatory network with key TFs for cambium regulation. High levels of PtrSCZ1 and PtrSCZ3 may signal a regulatory cascade impacting xylem radial width, consistent with their expression in developing xylem (Figure 1C; Supplementary Figure S3C).
RT-qPCR assays showed that the monolignol genes PtrPAL1, PtrC4H2, PtrC3H3, PtrHCT1, PtrCCoAOMT1, PtrCCR2, and PtrCAD1 were upregulated in the differentiating xylem of OE-PtrSCZ1-L14 and OE-PtrSCZ3-L13 plants (Figure 6). However, mutations in PtrSCZ1 and PtrSCZ3 did not significantly alter the expression of most monolignol genes, except for PtrCCoAOMT1 and PtrCAld5H1 (Figure 6; Supplementary Figure S7). This might be due to PtrSCZ1 and PtrSCZ3 functioning as direct regulators of cambium development rather than as controllers of cell wall components. The observed increase in total lignin content in OE-PtrSCZ1 and OE-PtrSCZ3 (Table 1) might be attributed to an indirect effect of the larger xylem area (Figures 5A, C; Supplementary Figure S6A). Another possibility is that changes in lignin biosynthesis gene expression in the overexpressed plants (Figure 6) could be due to the ectopic expression of PtrSCZ1 and PtrSCZ3.
Similarly, plants with overexpressed PtrSCZ1 and PtrSCZ3 were dwarfed (Figure 3), while those with loss-of-function did not exhibit changes in height (Figure 2), likely due to ectopic overexpression of these genes. Considering the crucial roles of phytohormones in plant growth and development, regulations may be altered in transgenics with ectopic expression of PtrSCZ1 and PtrSCZ3 (Vanstraelen and Benková, 2012; Vargas et al., 2016; Hu et al., 2024). PtrSCZ1 and PtrSCZ3 might interact with a receptor protein perceived with phytohormones, disrupting a signaling pathway essential for releasing hormones that support normal plant growth and development. High levels of PtrSCZ1 and PtrSCZ3 may inhibit the hormone release, resulting in impeded growth.
In this study, we characterized PtrSCZ1, a previously uncharacterized HSF gene exhibiting specific expression in the vascular cambium. Overexpression of PtrSCZ1 and its homolog PtrSCZ3 significantly enhanced vascular cambial proliferation, resulting in increased stem radial growth during secondary growth. The ptrscz1/ptrscz3 double mutants displayed phenotypes opposite to those observed in the overexpression plants. Subsequent RT-qPCR analysis of key TFs regulating cambium development in transgenic plants demonstrated that PtrSCZ1 and PtrSCZ3 promote cambium activity through directly or indirectly regulation of these factors, establishing a complex regulatory network controlling cambial division and xylem development. These findings identify PtrSCZ1 and PtrSCZ3 as promising target genes for molecular breeding strategies aimed at enhancing wood production. Moreover, this work establishes vascular cambium systems as valuable models for investigating lateral meristem development, particularly wood formation mechanisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. JJ: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. QZ: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. JZ: Data curation, Investigation, Methodology, Writing – original draft. HM: Formal Analysis, Methodology, Writing – original draft. DL: Formal Analysis, Methodology, Writing – original draft. SL: Conceptualization, Supervision, Writing – review & editing. CZ: Conceptualization, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. WL: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Key Research and Development Program of China (no. 2021YFD2200700 to WL), the Fundamental Research Funds for the Central Universities of China (grant no. 2572022DQ01 to WL), and the 111 Project (B16010).
The authors thank Prof. Vincent Chiang for his support and encouragement throughout the project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1546660/full#supplementary-material
Abraham, P., Giannone, R. J., Adams, R. M., Kalluri, U., Tuskan, G. A., Hettich, R. L. (2013). Putting the pieces together: High-performance LC-MS/MS provides network-, pathway-, and protein-level perspectives in Populus. Mol. Cell. Proteomics 12, 106–119. doi: 10.1074/mcp.M112.022996
Begum, T., Reuter, R., Schöffl, F. (2013). Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 130, 54–60. doi: 10.1016/j.mod.2012.05.008
Bhalerao, R. P., Fischer, U. (2017). Environmental and hormonal control of cambial stem cell dynamics. J. Exp. Bot. 68, 79–87. doi: 10.1093/jxb/erw466
Dai, X., Zhai, R., Lin, J., Wang, Z., Meng, D., Li, M., et al. (2023). Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa. Nat. Plants 9, 96–111. doi: 10.1038/s41477-022-01315-7
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. doi: 10.1038/nature09886
Du, J., Groover, A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52, 17–27. doi: 10.1111/j.1744-7909.2010.00901.x
Du, J., Mansfield, S. D., Groover, A. T. (2009). The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J. 60, 1000–1014. doi: 10.1111/j.1365-313X.2009.04017.x
Du, J., Miura, E., Robischon, M., Martinez, C., Groover, A. (2011). The Populus class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PloS One 6, e17458. doi: 10.1371/journal.pone.0017458
Etchells, J. P., Laxmi, S., Kumar, M., Campbell, L., Simon, R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055. doi: 10.1016/j.cub.2015.02.023
Etchells, J. P., Provost, C. M., Mishra, L., Turner, S. R. (2013). WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–2234. doi: 10.1242/dev.091314
Evert, R. F. E. (2006). Esau’s Plant Anatomy, Meristems, Cells, and Tissues of the Plant Body: their Structure, Function, and Development. 3rd edn (Hoboken, NJ, USA: John Wiley & Sons).
Fischer, U., Kucukoglu, M., Helariutta, Y., Bhalerao, R. P. (2019). The dynamics of cambial stem cell activity. Annu. Rev. Plant Biol. 70, 293–319. doi: 10.1146/annurev-arplant-050718-100402
Guo, Q., Wei, R., Xu, M., Yao, W., Jiang, J., Ma, X., et al. (2023). Genome-wide analysis of HSF family and overexpression of PsnHSF21 confers salt tolerance in Populus simonii × P. nigra. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1160102
Heler, R., Samai, P., Modell, J. W., Weiner, C., Goldberg, G. W., Bikard, D., et al. (2015). Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202. doi: 10.1038/nature14245
Hou, J., Xu, H., Fan, D., Ran, L., Li, J., Wu, S., et al. (2020). MiR319a-targeted PtoTCP20 regulates secondary growth via interactions with PtoWOX4 and PtoWND6 in Populus tomentosa. New Phytol. 228, 1354–1368. doi: 10.1111/nph.16782
Hu, M. X., Guo, W., Song, X. Q., Liu, Y. L., Xue, Y., Cao, Y., et al. (2024). PagJAZ5 regulates cambium activity through coordinately modulating cytokinin concentration and signaling in poplar. New Phytol. 243, 1455–1471. doi: 10.1111/nph.19912
Ikeda, M., Mitsuda, N., Ohme-Takagi, M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. doi: 10.1104/pp.111.179036
Jiang, W., Bikard, D., Cox, D., Zhang, F., Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi: 10.1038/nbt.2508
Johnsson, C., Fischer, U. (2016). Cambial stem cells and their niche. Plant Sci. 252, 239–245. doi: 10.1016/j.plantsci.2016.08.002
Kucukoglu, M., Nilsson, J., Zheng, B., Chaabouni, S., Nilsson, O. (2017). WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 215, 642–657. doi: 10.1111/nph.14631
Kumar, M., Busch, W., Birke, H., Kemmerling, B., Nürnberger, T., Schöffl, F. (2009). Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol. Plant 2, 152–165. doi: 10.1093/mp/ssn095
Li, M., Doll, J., Weckermann, K., Oecking, C., Berendzen, K. W., Schöffl, F. (2010). Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur. J. Cell Biol. 89, 126–132. doi: 10.1016/j.ejcb.2009.10.012
Li, W., Lin, Y. J., Chen, Y. L., Zhou, C., Li, S., De Ridder, N., et al. (2024). Woody plant cell walls: Fundamentals and utilization. Mol. Plant 17, 112–140. doi: 10.1016/j.molp.2023.12.008
Li, S., Zhen, C., Xu, W., Wang, C., Cheng, Y. (2017). Simple, rapid and efficient transformation of genotype Nisqually-1: a basic tool for the first sequenced model tree. Sci. Rep. 7, 2638. doi: 10.1038/s41598-017-02651-x
Liu, B., Hu, J., Zhang, J. (2019). Evolutionary divergence of duplicated Hsf genes in Populus. Cells 8, 438. doi: 10.3390/cells8050438
Liu, B., Liu, J., Yu, J., Wang, Z., Sun, Y., Li, S., et al. (2021). Transcriptional reprogramming of xylem cell wall biosynthesis in tension wood. Plant Physiol. 186, 250–269. doi: 10.1093/plphys/kiab038
Lorenz, W. W., Yu, Y.-S., Dean, J. F. D. (2010). An improved method of RNA isolation from loblolly Pine (P. taeda L.) and other conifer species. J. Visualized Experiments 36), 1751. doi: 10.3791/1751
Luo, L., Li, L. (2022). Molecular understanding of wood formation in trees. Forestry Res. 2, 5. doi: 10.48130/fr-2022-0005
Matsumoto-Kitano, M., Kusumoto, T., Tarkowski, P., Kinoshita-Tsujimura, K., Václavíková, K., Miyawaki, K., et al. (2008). Cytokinins are central regulators of cambial activity. PNAS 105, 20027–20031. doi: 10.1073/pnas.0805619105
Mazur, E., Kurczyńska, E. U., Friml, J. (2014). Cellular events during interfascicular cambium ontogenesis in inflorescence stems of Arabidopsis. Protoplasma 251, 1125–1139. doi: 10.1007/s00709-014-0620-5
Miller, G., Mittler, R. (2006). Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 98, 279–288. doi: 10.1093/aob/mcl107
Mylona, P., Linstead, P., Martienssen, R., Dolan, R. (2002). SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129, 4327–4334. doi: 10.1242/dev.129.18.4327
Nieminen, K., Blomster, T., Helariutta, Y., Mähönen, A. P. (2015). Vascular cambium development. Arabidopsis Book 13, e0177. doi: 10.1199/tab.0177
Nishizawa-Yokoi, A., Nosaka, R., Hayashi, H., Tainaka, H., Maruta, T., Tamoi, M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. doi: 10.1093/pcp/pcr045
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., Scharf, K.-D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ortega-Martínez, O., Pernas, M., Carol, R. J., Dolan, L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510. doi: 10.1126/science.1143409
Pardal, R., Scheres, B., Heidstra, R. (2023). SCHIZORIZA domain–function analysis identifies requirements for its specific role in cell fate segregation. Plant Physiol. 193, 1866–1879. doi: 10.1093/plphys/kiad456
Pernas, M., Ryan, E., Dolan, L. (2010). SCHIZORIZA controls tissue system complexity in plants. Curr. Biol. 20, 818–823. doi: 10.1016/j.cub.2010.02.062
Robert, X., Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Robischon, M., Du, J., Miura, E., Groover, A. (2011). The Populus Class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 155, 1214–1225. doi: 10.1104/pp.110.167007
Scharf, K.-D., Berberich, T., Ebersberger, I., Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Shi, R., Wang, J. P., Lin, Y.-C., Li, Q., Sun, Y.-H., Chen, H., et al. (2017). Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 245, 927–938. doi: 10.1007/s00425-016-2640-1
Suer, S., Agusti, J., Sanchez, P., Schwarz, M., Greb, T. (2011). WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23, 3247–3259. doi: 10.1105/tpc.111.087874
ten Hove, C. A., Willemsen, V., de Vries, W. J., van Dijken, A., Scheres, B., Heidstra, R. (2010). SCHIZORIZA encodes a nuclear factor regulating asymmetry of sem cell divisions in the Arabidopsis root. Curr. Biol. 20, 452–457. doi: 10.1016/j.cub.2010.01.018
Timell, T. E. (1980). Organization and ultrastructure of the dormant cambial zone in compression wood of Picea abies. Woocl Sci. Technol. 14, 161–179. doi: 10.1007/BF00350566
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507. doi: 10.1038/s41598-017-00501-4
Vanstraelen, M., Benková, E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487. doi: 10.1146/annurev-cellbio-101011-155741
Vargas, L., Cesarino, I., Vanholme, R., Voorend, W., de Lyra Soriano Saleme, M., Morreel, K., et al. (2016). Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol. Biofuels 9, 139. doi: 10.1186/s13068-016-0551-9
von-Koskull-Döring, P., Scharf, K. D., Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wang, F., Dong, Q., Jiang, H., Zhu, S., Chen, B., Xiang, Y. (2011). Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 39, 1877–1886. doi: 10.1007/s11033-011-0933-9
Wang, J. P., Matthews, M. L., Williams, C. M., Shi, R., Yang, C. M., Tunlaya-Anukit, S., et al. (2018). Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 9, 1579. doi: 10.1038/s41467-018-03863-z
Włoch, W. (1981). Nonparallelism of cambium cells in neighboring row. Acta Societatis Botanicorum Poloniae 50, 625–636. doi: 10.5586/ASBP.1981.084
Włoch, W., Iqbal, M., Jura-Morawiec, J. (2023). Calculating the growth of vascular cambium in woody plants as the cylindrical surface. Botanical Rev. 89, 237–249. doi: 10.1007/s12229-023-09291-z
Zhang, Y., Chen, S., Xu, L., Chu, S., Yan, X., Lin, L., et al. (2024). Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar. Plant Cell 36, 1806–1828. doi: 10.1093/plcell/koae040
Zhang, J., Eswaran, G., Alonso-Serra, J., Kucukoglu, M., Xiang, J. L., Yang, W. B., et al. (2019b). Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants 5, 1033–1042. doi: 10.1038/s41477-019-0522-9
Zhang, J., Liu, B., Li, J., Zhang, L., Wang, Y., Zheng, H., et al. (2015). Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics 16, 181. doi: 10.1186/s12864-015-1398-3
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., Chen, J.-G. (2020). Transcriptional and post-transcriptional regulation of lignin biosynthesis pathway genes in Populus. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00652
Zhang, H., Yang, J., Li, W., Chen, Y., Lu, H., Zhao, S., et al. (2019a). PuHSFA4a enhances tolerance to excess zinc by regulating reactive oxygen species production and root development in Populus. Plant Physiol. 180, 2254–2271. doi: 10.1104/pp.18.01495
Zheng, S., He, J., Lin, Z., Zhu, Y., Sun, J., Li, L. (2021). Two MADS-box genes regulate vascular cambium activity and secondary growth by modulating auxin homeostasis in Populus. Plant Commun. 2, 100134. doi: 10.1016/j.xplc.2020.100134
Zhou, H., Song, X., Lu, M.-Z. (2024). Growth-regulating factor 15-mediated vascular cambium differentiation positively regulates wood formation in hybrid poplar (Populus alba × P. glandulosa). Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1343312
Zhu, Y., Song, D., Sun, J., Wang, X., Li, L. (2013). PtrHB7, a class III HD-Zip gene, plays a critical tole in regulation of vascular cambium differentiation in Populus. Mol. Plant 6, 1331–1343. doi: 10.1093/mp/sss164
Zhu, Y., Song, D., Xu, P., Sun, J., Li, L. (2018). A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 16, 808–817. doi: 10.1111/pbi.12830
Keywords: vascular cambium, xylem development, wood formation, transcription factor, PtrSCZs, Populus trichocarpa
Citation: Sun Y, Jiang J, Zhang Q, Zhao J, Ma H, Li D, Li S, Zhou C and Li W (2025) The vascular-cambium-specific transcription factor PtrSCZ1 and its homologue regulate cambium activity and affect xylem development in Populus trichocarpa. Front. Plant Sci. 16:1546660. doi: 10.3389/fpls.2025.1546660
Received: 17 December 2024; Accepted: 21 February 2025;
Published: 11 March 2025.
Edited by:
Rebecca Grumet, Michigan State University, United StatesReviewed by:
Ana Campilho, University of Porto, PortugalCopyright © 2025 Sun, Jiang, Zhang, Zhao, Ma, Li, Li, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Li, d2VpbGkyMDE1QG5lZnUuZWR1LmNu; Chenguang Zhou, emhvdWNoZW5ndWFuZ0BuZWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.