
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 March 2025
Sec. Plant Development and EvoDevo
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1546092
 ZhengFeng Fan1,2
ZhengFeng Fan1,2 Li Zhang3,4
Li Zhang3,4 SiQi Li1,2
SiQi Li1,2 ShengQun Pang1,2
ShengQun Pang1,2 YiBing Zhang3,4
YiBing Zhang3,4 ChuanQiang Xu5
ChuanQiang Xu5 YuDong Liu1,2*
YuDong Liu1,2* MingFang Qi5*
MingFang Qi5*AP2/ERF transcription factors regulate plants’ growth, development, and stress responses. In this study, the seed germination rate and seedling growth were reduced in the tomato slerf4-9 mutant. The fresh weight, drought weight, number of primary lateral roots (LRs), average root diameter, and number of root tips were also decreased in the mutant. The findings suggest that SlERF4-9 plays a significant role in root growth and development. The results of RNA-seq analysis of young roots indicated that the mutation of SlERF4-9 did not affect the expression of genes related to auxin biosynthesis or signal transduction, but it did reduce the expression of the auxin efflux carrier genes SlAEC2 and SlPIN5. Moreover, the mutation of SlERF4-9 affected the distribution of auxin in the roots of DR5 × WT and DR5 × slerf4-9 hybrid tomato seedlings. However, the promoters of SlAEC2 and SlPIN5 do not possess the GCC-box or DRE elements, suggesting that SlERF4-9 does not directly regulate their transcription. In addition, the expression levels of the two Cycling DOF Factors (CDFs) SlCDF1 and SlCDF3 decreased in the roots of the slerf4-9 mutant. Moreover, the GCC-box was present in the promoters of SlCDF1 and SlCDF3. Therefore, exploring the regulatory relationships between SlERF4-9, SlCDF1/3, and SlAEC2/SlPIN5 will further our understanding of the molecular mechanisms of tomato root growth and development.
Roots are responsible for transporting water and nutrients during plant growth and development, and they can take in water and nutrients from the soil to meet the demands of the plant and enhance its tolerance to abiotic stresses. In many plants, the number of lateral roots (LRs) often determines the efficiency of water and nutrient utilization (Casimiro et al., 2003). Therefore, cultivating new varieties with enhanced and better-developed root systems has become an important goal in crop breeding.
The growth and development of plant roots cannot be separated from auxin regulation. Auxin is an important phytohormone that is involved in almost all stages of plant growth and development (Overvoorde et al., 2010; Yu et al., 2022). This phytohormone is synthesized in source tissues such as the stem tip, young leaves, cotyledons, and root tips (Ljung et al., 2001; Peret et al., 2009), and it is transported to other tissues through polar or phloem transport (Benková et al., 2003). Phloem transport relies on the concentration gradient of organic matter, while polar transport is a short-distance and unidirectional transport mode between the parenchymal cells of the coleoptile, young stem, and young roots (Blakeslee et al., 2005). During polar auxin transport, auxin efflux (PIN), and influx (AUX/LAX) carrier proteins are involved in the regulation of plant growth and development (Bennett et al., 1996; Petrasek et al., 2006; Swarup et al., 2001). In roots, AUX/LAX and PIN synergistically regulate cell differentiation by controlling the cellular level of free auxin. In Arabidopsis, AtAUX1 regulates cell differentiation in the lateral root cap and epidermis (De Smet et al., 2007), whereas AtLAX3 regulates lateral root emergence (Swarup et al., 2008). The polar transport of auxin has been found to be significantly reduced in the atpin1 mutant (Okada et al., 1991). Moreover, PIN1 can transport auxin via PIN1–PIN1 dimers (Teale et al., 2021).
Ethylene, as a plant-specific hormone, is involved in fruit ripening and softening, organ senescence, flower organ development, root development, and the response to abiotic stresses (Jiroutova et al., 2018; Pieterse et al., 2009). During root development, treatment with the ethylene inhibitor 1-MCP was found to improve root elongation and increase the number of LRs in tomatoes while simultaneously repressing root penetration into the soil (Santisree et al., 2011). In the Arabidopsis aco1 mutant, ethylene production in the root tips is reduced, and the number of LRs is increased (Park et al., 2018). These results suggest that ethylene positively regulates root gravitropism while inhibiting root elongation and lateral root development. The AP2/ERF regulatory genes encode specific ethylene-responsive transcription in plants, playing important roles in plant growth, development, and stress response (Jia et al., 2021; Xu et al., 2024; Yuste-Lisbona et al., 2020). In Arabidopsis, AtERF070 negatively regulates the number and length of LRs (Ramaiah et al., 2014). However, in Populus, PtaERF003 had been found to promote lateral root formation (Trupiano et al., 2013). Tomato SlERF4-9 (SlERF.H12/SlDREB3) overexpression improves the root growth of tobacco and cold tolerance in tomatoes (Upadhyay et al., 2017; Wang et al., 2019; Zhang et al., 2022). However, the mechanism by which SlERF4-9 regulates root development remains unclear.
In addition to the AP2/ERF family, the DNA binding with One Finger (DOF) family is a unique transcription factor in plants, and its members are involved in various aspects of plant growth and development, including seed germination, the photoperiodic control of flowering, vascular development, and shoot branching (Fornara et al., 2009; Gabriele et al., 2010; Konishi and Yanagisawa, 2007; Zou et al., 2013). In bananas, MaDof23 regulates fruit ripening by interacting with MaERF9 (Feng et al., 2016). In Arabidopsis, the AtDof5.1 mutation reduces the expression levels of the auxin-responsive genes IAA6 and IAA19 and alters auxin homeostasis in the root tips (Kim et al., 2010). Therefore, there may be a relationship between AP2/ERF, DOF, and auxin during plant root development.
In this study, we found that root growth was significantly reduced in the slerf4-9 mutants. RNA-seq analysis of young roots revealed that the expression levels of genes involved in auxin biosynthesis and signal transduction were not significantly altered. However, the expression of the auxin efflux carriers SlAEC2 and SlPIN5 was significantly decreased in slerf4-9 roots, suggesting that SlERF4-9 may regulate root growth and development by modulating their transcription.
Interestingly, while the SlAEC2 and SlPIN5 promoters lack the GCC-box or DRE elements, they contain multiple DOF protein binding elements, indicating that their reduced expression may be an indirect consequence of the SlERF4-9 function. Furthermore, we observed the significant downregulation of Cycling DOF Factors (SlCDF1 and SlCDF3) in the roots of slerf4-9, whose promoters bear GCC-box elements, suggesting a possible regulatory link between SlERF4-9 and SlCDF1/3 in the control of SlAEC2 and SlPIN5.
These findings suggest a regulatory module, where SlERF4-9 may influence root development through the modulation of SlCDF1/3, which in turn could affect auxin transport via SlAEC2 and SlPIN5 gene regulation. Further investigations into these regulatory interactions will shed light on the role of SlERF4-9 on auxin transport during root growth.
All tomato plants were derived from the Ailsa Craig (AC) strain. The slerf4-9 knockout mutants were created using CRISPR/CAS9 technology with AC as the background material. The AC wild-type (WT) and slerf4-9 mutant lines #2 and #31 were used in this study. The tomato plants were grown for 15 and 30 days under 25°C 16 h/15°C 8 h day/night conditions. The roots of the 30-day seedlings were scanned and analyzed using WinRHIZO Pro root analysis software (Epson Perfection V850 Pro), and the soil volume analysis parameters were set to 1.0 cm3. To investigate auxin transport in tomato roots, the pollen of tomato DR5 was used to pollinate the stigma of the WT and slerf4-9 mutant to generate DR5 × WT and DR5 × slerf4-9 hybrid seeds. These seeds were then used in a GUS (β-glucuronidase) staining assay.
Thirty seeds of the WT and slerf4-9 mutant lines #2 and #31 were heated at 55°C for 15 min, placed in a culture dish with moist filter paper, and germinated at 25°C for 7 days. In addition, 100 mg of the young roots of seeds germinated for 7 days was gathered for transcriptomics analysis. Three biological replicates were set for each treatment.
A total of 100 mg of the fresh leaves of the WT tomatoes was used to extract DNA using a Plant Genomic DNA Kit (DP305-03; Tiangen, Beijing, China) to subsequently amplify the promoters of SlCDF1 (Solyc03g115940), SlCDF3 (Solyc06g069760), SlAEC2 (Solyc02g082450), and SlPIN5 (Solyc01g068410). The young roots of the WT and slerf4-9 mutants were used to extract total RNA using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). The RNA was used for transcriptomics sequencing, quantitative real-time PCR (qRT-PCR), and the amplification of the Coding sequence (CDS) fragments of SlERF4-9, SlCDF1, and SlCDF3.
To determine whether SlERF4-9 had potential nuclear regulatory functions, the CDS of SlERF4-9 was amplified using the primers ERF4-9 F/R (the primers are listed in Supplementary Table S1) and inserted between the cleavage sites BamHI and SalI in the pCAMBIA1300 vector using an In-Fusion PCR Cloning Kit to form a 35S::SlERF4-9-GFP recombinant vector. The recombinant vector was then transformed into Agrobacterium strain GV3101. Subsequently, 15 mL of a suspension of Agrobacterium cells with the 35S::SlERF4-9-GFP vector (OD 1.0) was centrifuged at 4°C and 5,000 × g for 5 min; after this, the supernatant was removed, and the cells were resuspended in 5 mL of solution (10 mM·mL−1 MES, 10 mM·mL−1 MgCl2, and 20 μM·mL−1 AS, pH 5.6) and placed in the dark at 25°C for 3 h. Finally, the above-mentioned solution was injected into tobacco leaves, and the tobacco plants were placed in the dark at 25°C for 2 days. Agrobacterium cells carrying the pCAMBIA1300 vector were used as a control in this assay.
The young roots (200 mg) of the WT and slerf4-9 mutant lines #2 and #31 from seeds germinated for 7 days were collected for transcriptomics analysis. The WT roots (young roots of AC, named WT) were analyzed as a control group, and the E2 roots (young roots of the slerf4-9 mutant line #2, named E2) and E31 roots (young roots of the slerf4-9 mutant line #31, named E31) comprised the experimental groups. The experiment adopted a paired design, in which the young root samples of WT, E2, and E31 were synchronously processed under the same culture conditions, with three independent biological replicates set for each genotype. When conducting paired analysis, the WT was matched one-to-one with the three biological replicates of each mutant to eliminate batch effects. Total RNA was extracted using TRIzol Reagent (Invitrogen) and employed to construct RNA-seq libraries. The sequences were read using the Illumina HiSeq 2000 platform. Agilent 2100 Bioanalyzer was used for quality control of libraries. The raw data were evaluated for quality using FastQC (v0.11.9), and low-quality bases were removed using Trimmomatic. Clean reads were aligned to the tomato reference genome (https://solgenomics.net/ftp/tomato_genome/annotation/ITAG4.0_release/) using HISAT2 (v2.2.1) and filter reads with unique alignment rate ≥90% using SAMtools (v1.12). The fragments per kilobase of exon model per million mapped fragments (FPKM) values of each gene were computed using the HiSeq (ver. 0.6.1) and DESeq Bioconductor packages. Differentially expressed genes (DEGs) were analyzed by DESeq2 (v1.34.0), and the screening criteria were |log2FoldChange| > 1 and false discovery rate (FDR) corrected p-value < 0.05. FDR calculation adopted the Benjamini–Hochberg method. Gene Ontology (GO) TermFinder (v0.4) was used to analyze the GO enrichment by a hypergeometric test based on the ITAG4.0 annotated tomato GO database, with filtering criteria of corrected p-value < 0.05 and exclusion of entries containing “unknown function”. Finally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using KOBAS (v3.0), with a pathway significance threshold of FDR < 0.05.
Total RNA was extracted using TRIzol Reagent (Invitrogen), and cDNA was synthesized using a PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Kyoto, Japan). qRT-PCR was conducted using TB Green® Premix Ex Taq™ II FAST qPCR (TaKaRa, Kyoto, Japan). The reaction conditions were as follows: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 10 s. The primers used in this assay are listed in Supplementary Table S1. A dissociation curve was used to validate the specificity of the primer pair. Three biological replicates were set up for this assay. The cycle threshold value of each gene was used to calculate the relative expression level of each gene using the 2−ΔΔCT method (Nolan et al., 2006). In addition, the differences in expression levels were analyzed using t-tests.
To obtain the SlERF4-9 protein, the CDS of SlERF4-9 was amplified using the primers 6P-ERF4-9 F/R (the primers are listed in Supplementary Table S1) and inserted between the cleavage sites BamHI and XhoI of the pGEX6P-1 (GST-tag) vector using an In-Fusion PCR Cloning Kit to form a 35S::SlERF4-9-GST recombinant vector. The recombinant vector was then transformed into Escherichia coli competent BL21 cells (DE3). The E. coli cells with the 35S::SlERF4-9-GST vector were cultivated in 50 mL of lysogeny broth liquid medium with 50 μg·mL−1 ampicillin at 37°C for 200 × g for 12 h, then supplied with 50 μL of 1 M isopropyl-beta-d-thiogalactopyranoside, and finally cultivated at 37°C for 200 × g for 12 h. Next, the above cells were centrifuged at 5,000 × g for 10 min at 4°C, and the supernatant was removed. Then, 10 mL of phosphate buffered saline (PBS) with lysozyme (1 mg·mL−1) was used to break down the cell walls using an ultrasonic crusher in an ice bath for 30 min. The bacterial fluid was centrifuged at 10,000 × g for 10 min at 4°C to separate the supernatant and precipitate. The supernatant was used to purify the SlERF4-9-GST fusion proteins using a GST-tag Protein Purification Kit (Beyotime, Shanghai, China). Finally, 20 μL of the purified protein was detected using Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE).
The purified SlERF4-9-GST fusion protein was used to detect the binding ability of SlERF4-9 with the GCC-box of the SlCDF1 and SlCDF3 promoters. The biotin probes (named prob-Bio), cold probes (named prob-Cold), and mutated probes (named Mu-prob-Bio) of the P1 and P2 sites were synthesized (the probe sequences are listed in Supplementary Table S1). The specific binding of the protein with DNA was validated by competition with the prob-Cold (100×) and Mu-prob-Bio probes. The electrophoretic mobility shift assay (EMSA) was carried out using a Chemiluminescent EMSA Kit (Beyotime, Shanghai, China).
The promoters of SlCDF1 (−1 to −1,500 bp upstream of ATG), SlCDF3 (−1 to −1,487 bp upstream of ATG), SlAEC2 (+33 to −1,502 bp upstream of ATG), and SlPIN5 (+15 to −1,489 bp upstream of ATG) were amplified and inserted into a pGreenII-0800-Luc vector using an In-Fusion PCR Cloning Kit to form the proSlCDF1-Luc, proSlCDF3-Luc, proSlAEC2-Luc, and proSlPIN5-Luc recombinant vectors. The CDS fragments of SlERF4-9, SlCDF1, and SlCDF3 were cloned and inserted into the pGreenII-62-SK vector to construct the SK-SlERF4-9, SK-SlCDF1, and SK-SlCDF3 recombinant vectors, respectively. The primers used for amplification are listed in Supplementary Table S1. The above-mentioned recombinant vectors, alongside pGreenII-0800-Luc and pGreenII-62-SK empty vectors, were then transformed into Agrobacterium GV3101 competent cells. The cells (OD600 value = 0.8) with SK-SlERF4-9 were combined with those of proSlCDF1-Luc and proSlCDF3-Luc; the cells with proSlAEC2-Luc and proSlPIN5-Luc were combined with those of SK-SlCDF1 and SK-SlCDF3; and the cells of pGreenII-62-SK and pGreenII-0800-Luc replaced those of the corresponding recombinant vectors as negative controls. The mixtures of cells were placed in the dark at 28°C for 3 h, injected into tobacco leaves, and cultured in the dark at 25°C for 24 h. d-Luciferin potassium salt was then sprayed on the injected tobacco leaves. The fluorescent signal of the injected tobacco leaves was detected using a plant multispectral fluorescence imaging system. The luciferase was extracted using a dual-luciferase reporter assay system (Promega, Madison, WI, USA; E1910), and the activity analysis of LUC and REN was conducted utilizing a SpectraMax iD3 instrument (Molecular Devices, San Jose, CA, USA).
The germinated 7-day seedlings of the DR5 × WT and DR5 × slerf4-9 hybrid seeds in sterile water were placed in a 15-mL centrifuge tube containing 10 mL of GUS staining solution (500 mg·L−1 X-Gluc, 5-bromo-4-chloro-3-indolyl glucuronide) and stained at 37°C for 12 h. The staining solution was removed, and the seedlings were destained at 37°C using 70% ethanol until the green color of the cotyledons and hypocotyl had faded.
Transcriptomics data were analyzed using R (http://www.r-project.org/). All heatmaps in this study were produced using the Pvclust package (Suzuki and Shimodaira, 2006) and the pcaMethods Bioconductor package (Stacklies et al., 2007). The phylogenetic tree of the CDF proteins of tomato and Arabidopsis was constructed using MEGA v7.0 and CLUSTAL W. The parameters for the ML method were a + JTT matrix model with a site coverage cutoff of 50%, Poisson correction, and self-expansion value of 1,000. Conserved motifs of the CDF proteins in tomato and Arabidopsis were analyzed using the MEME (Multiple Em for Motif Elicitation) online software v.5.4 (https://meme-suite.org/meme/tools/meme). All data were analyzed as the mean ± standard error of three biological repeats. Statistically significant differences (p < 0.05) for the mean of each sample (Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001) were detected using a post-hoc test following an analysis of variance (ANOVA).
The subcellular localization results indicated that the SlERF4-9 protein was only detected in the nucleus (Figure 1A). Therefore, SlERF4-9, as a nuclear transcription factor, may have a regulatory role in tomato growth and development. To explore the function of SlERF4-9, an slerf4-9 mutant was created using CRISPR/Cas9 technology. In the slerf4-9 mutants, the gRNA site of slerf4-9#2 had “AGCTCTA” sequences deleted, while that of slerf4-9#31 had an “A” inserted (Figure 1B). The results suggested that both slerf4-9#2 and slerf4-9#31 would be unable to be correctly translated to the SlERF4-9 protein. After seed germination at 25°C for 3 days, 43.33%, 32.22%, and 20% of the seeds germinated in the WT, slerf4-9#2, and slerf4-9#31, respectively. After germination for 4 days, 90%, 68.89%, and 37.78% of the seeds germinated in the WT, slerf4-9#2, and slerf4-9#31, respectively. After germination for 7 days, the germination rates of the WT, slerf4-9#2, and slerf4-9#31 seeds were 93.3%, 72.2%, and 41.1%, respectively (Figures 1C, D). The results suggest that the SlERF4-9 mutation reduces the germination ability of tomato seeds at 25°C.
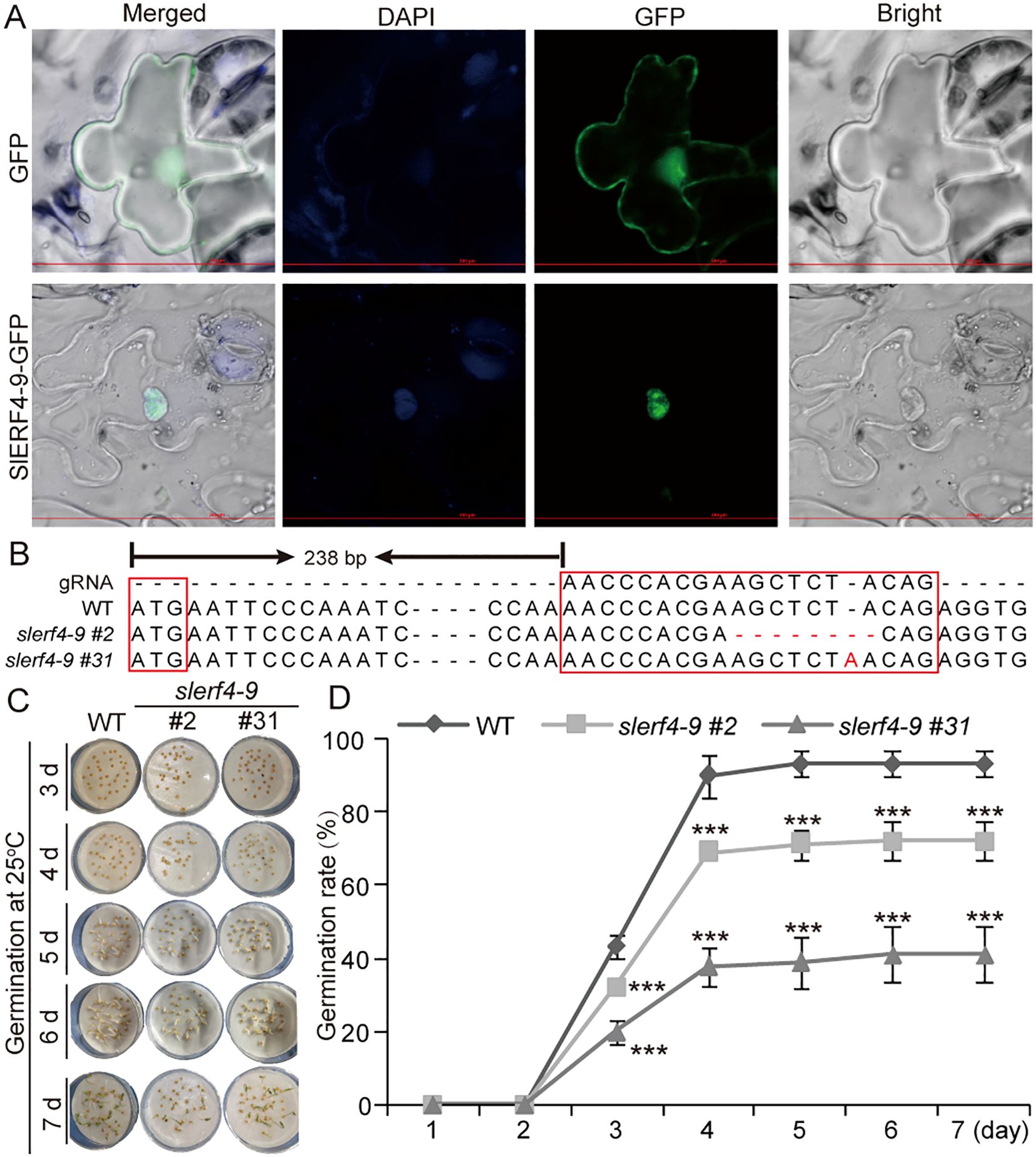
Figure 1. Seed germination analysis of the slerf4-9 mutants. (A) Subcellular localization of SlERF4-9. (B) Mutation site analysis of SlERF4-9. (C, D) Germination rate analysis of slerf4-9#2 and slerf4-9#31 mutants. WT, tomato Ailsa Craig (AC). Asterisks denote significance relative to WT (Student’s t-test; ***p < 0.001).
After seed germination at 25°C for 16 h/16°C for 8 h (day/night) for 15 days, the WT seedlings grew faster than the slerf4-9#2 and slerf4-9#31 mutants (Figure 2A). In addition, the WT 30-day seedlings showed numerous and stronger lateral roots than the slerf4-9 mutants (Figures 2B, C). The fresh weight, dry weight, primary lateral root number, average diameter, and the number of tips of the roots in the mutants were reduced compared with those of the WT (Figures 2D–I). However, the total length and surface area of the roots showed an opposite trend (Figures 2J, K). The reason for this may be that the LRs of the slerf4-9 mutants were slender and elongated, leading to an increase in the root surface area. The results indicate that SlERF4-9 regulates tomato lateral root development.
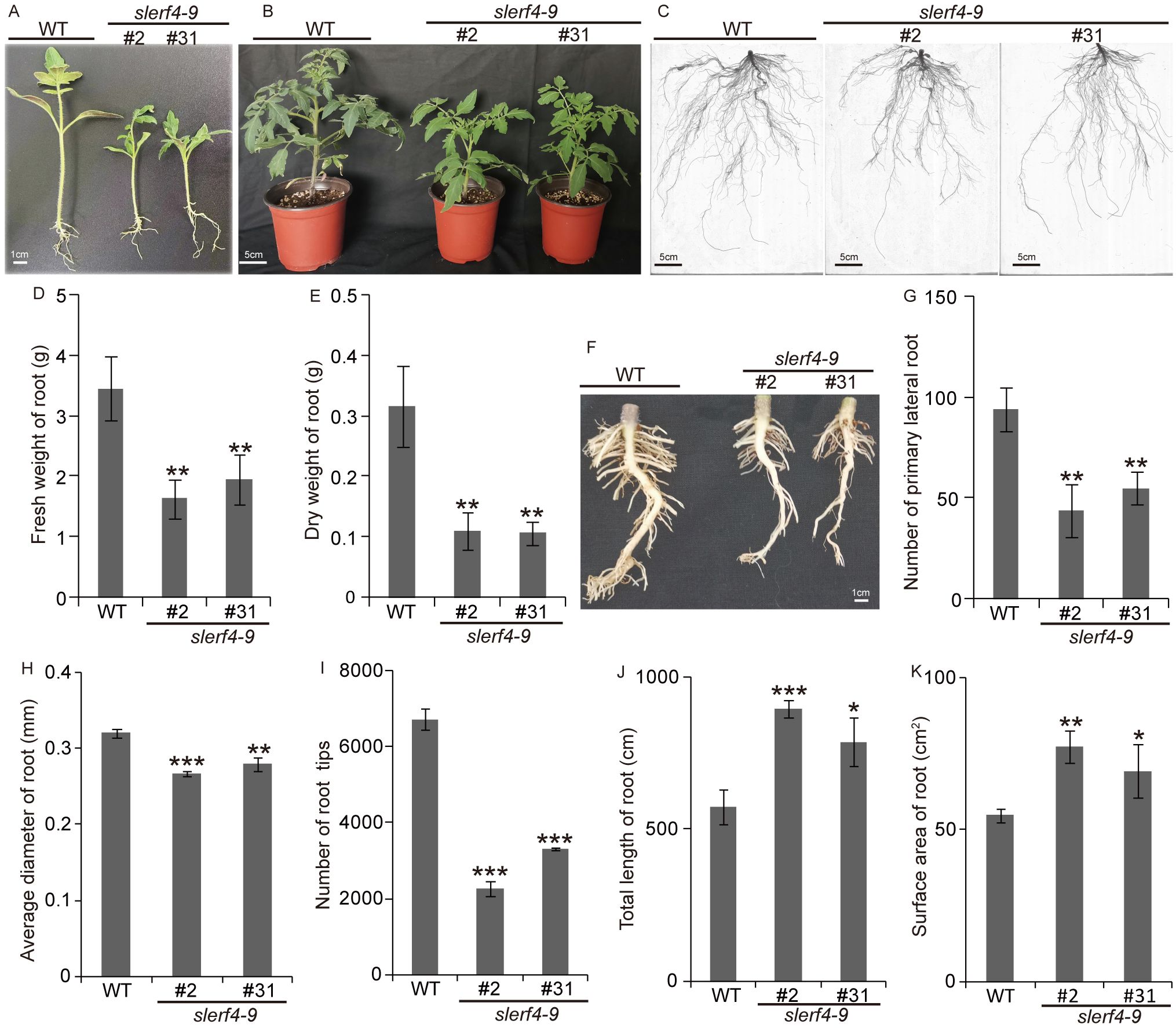
Figure 2. Phenotype analysis of slerf4-9 mutants. (A, B) Phenotype of the growth 15- and 30-day mutant seedlings. (C) Complete roots of the 30-day mutant seedlings using WinRHIZo Pro root analysis software. (D, E) Comparison of fresh weight and drought weight of the 30-day mutant roots. (F, G) Comparison of primary LR number of the 30-day mutant roots. (H) The average diameter. (I) Tip number. (J) Total length. (K) Surface area of root. WT, tomato Ailsa Craig (AC). Asterisks denote significance relative to WT (Student’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001). LR, lateral root.
To understand the regulatory network of SlERF4-9 in tomato root development, the young roots of WT, slerf4-9#2, and slerf4-9#31 germinated for 7 days were collected and subjected to RNA-seq analysis. The transcriptome sequencing raw data were uploaded to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (PRJNA1091043). The FPKM values of 34,075 genes in the transcriptome data were analyzed (Supplementary Table S2). The DEG analysis identified 479 and 617 DEGs in the WT_vs_E2 and WT_vs_E31 comparisons, respectively. There were 255 DEGs common to the WT_vs_E2 and WT_vs_E31 comparisons (Figure 3A). In addition, there were 331 upregulated and 148 downregulated DEGs in the WT_vs_E2 comparison and 419 upregulated and 198 downregulated DEGs in the WT_vs_E31 comparison (Figure 3B; Supplementary Tables S3-S6). Several downregulated DEGs were enriched in the response to stimulus and antioxidant systems in the GO analysis; plant hormone signal transduction and phenylpropanoid biosynthesis were identified by the KEGG pathway enrichment analysis of WT_vs_E2 and WT_vs_E31 (Figure 3C). The results suggest that the SlERF4-9 mutation causes a decrease in the expression of genes related to hormone signal transduction and antioxidant pathways. In addition, some upregulated DEGs were enriched in catalytic activity, oxidation–reduction process, and oxidoreductase activity in the GO analysis, and in substance biosynthesis and metabolism in the KEGG pathway enrichment analysis, of WT_vs_E2 and WT_vs_E31 (Figure 3D). The results indicate that SlERF4-9 may negatively regulate the expression of genes related to biosynthesis and metabolism during tomato root development.
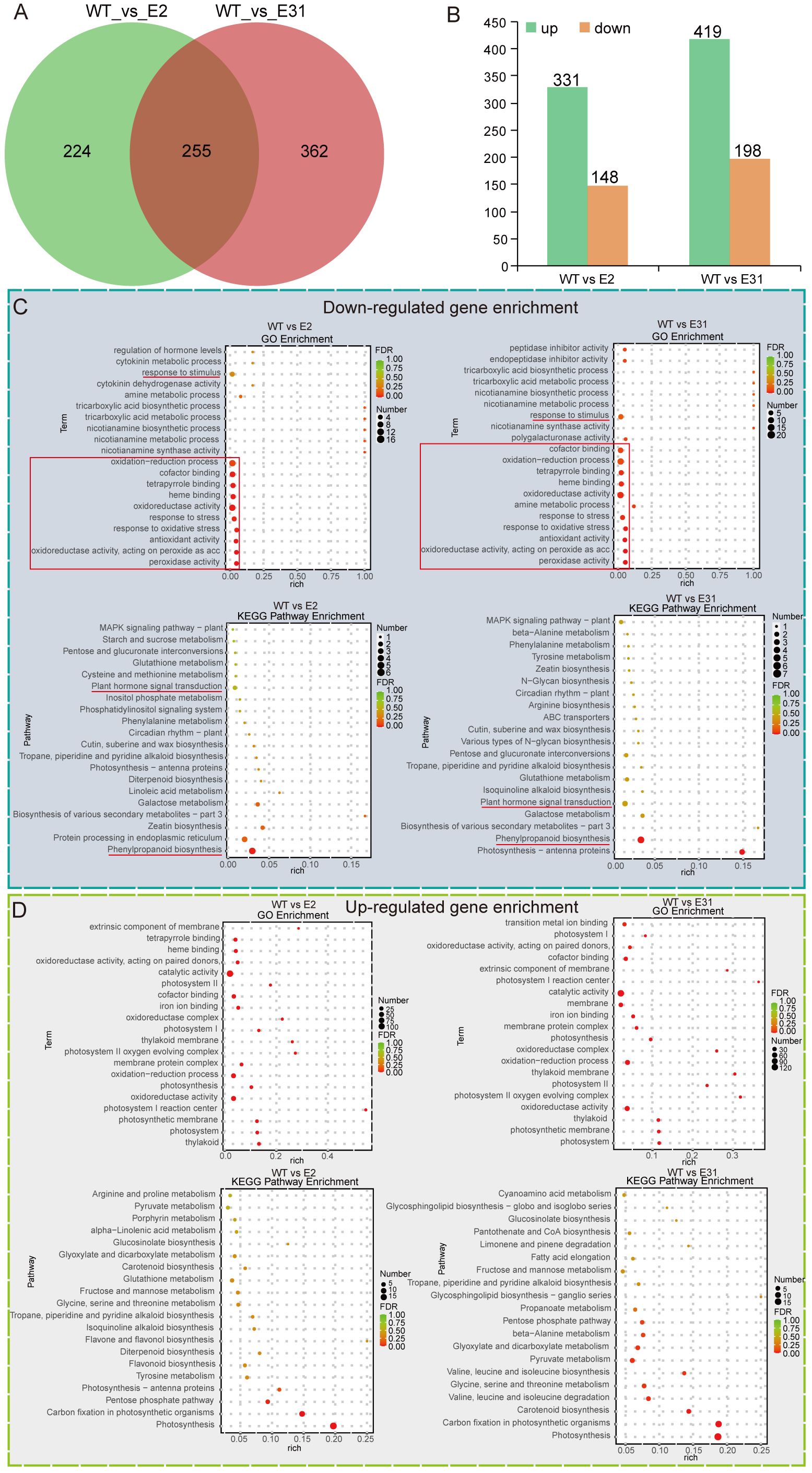
Figure 3. The Venn diagram, GO, and KEGG enrichment analysis. (A) Comparison of DEGs between WT_vs_E2 and WT_vs_E31. (B) Statistics on the number of upregulated and downregulated DEGs. (C, B) The GO and KEGG enrichment analyses of upregulated and downregulated DEGs, respectively. WT, E2, and E31, respectively indicate the 7-day seedling roots of AC, slerf4-9#2, and slerf4-9#31. GO, Gene Ontology; DEGs, differentially expressed genes.
The transcriptome data demonstrated that the FPKM values of 10 SlYUCCA and three SlTAA genes in two slerf4-9 mutants did not differ from those in the WT (Supplementary Figures S1A, B). This indicated that SlERF4-9 was not involved in auxin biosynthesis. In addition, the FPKM values of 22 SlARF and 25 SlIAA genes in two slerf4-9 mutants did not differ from those of the WT (Supplementary Figures S1C, D). However, we found that among eight auxin efflux carrier (AEC) family members, only the FPKM value of SlAEC2 (Solyc02g082450) significantly decreased in two slerf4-9 mutants, while those of SlAEC8 (Solyc12g095750) and SlAEC1 (Solyc02g037550) were zero; those of SlAEC3 (Solyc02g091240), SlAEC5 (Solyc03g032075), and SlAEC6 (Solyc03g032080) did not show significant changes; and those of SlAEC4 (Solyc03g031990) and SlAEC7 (Solyc04g082830) showed slight changes between the WT and the mutants (Figure 4A). In addition, only the FPKM value of SlPIN5 was downregulated in the two slerf4-9 mutants among the 10 SlPIN genes (Figure 4B). The qRT-PCR results showed the same patterns (Figure 4C).
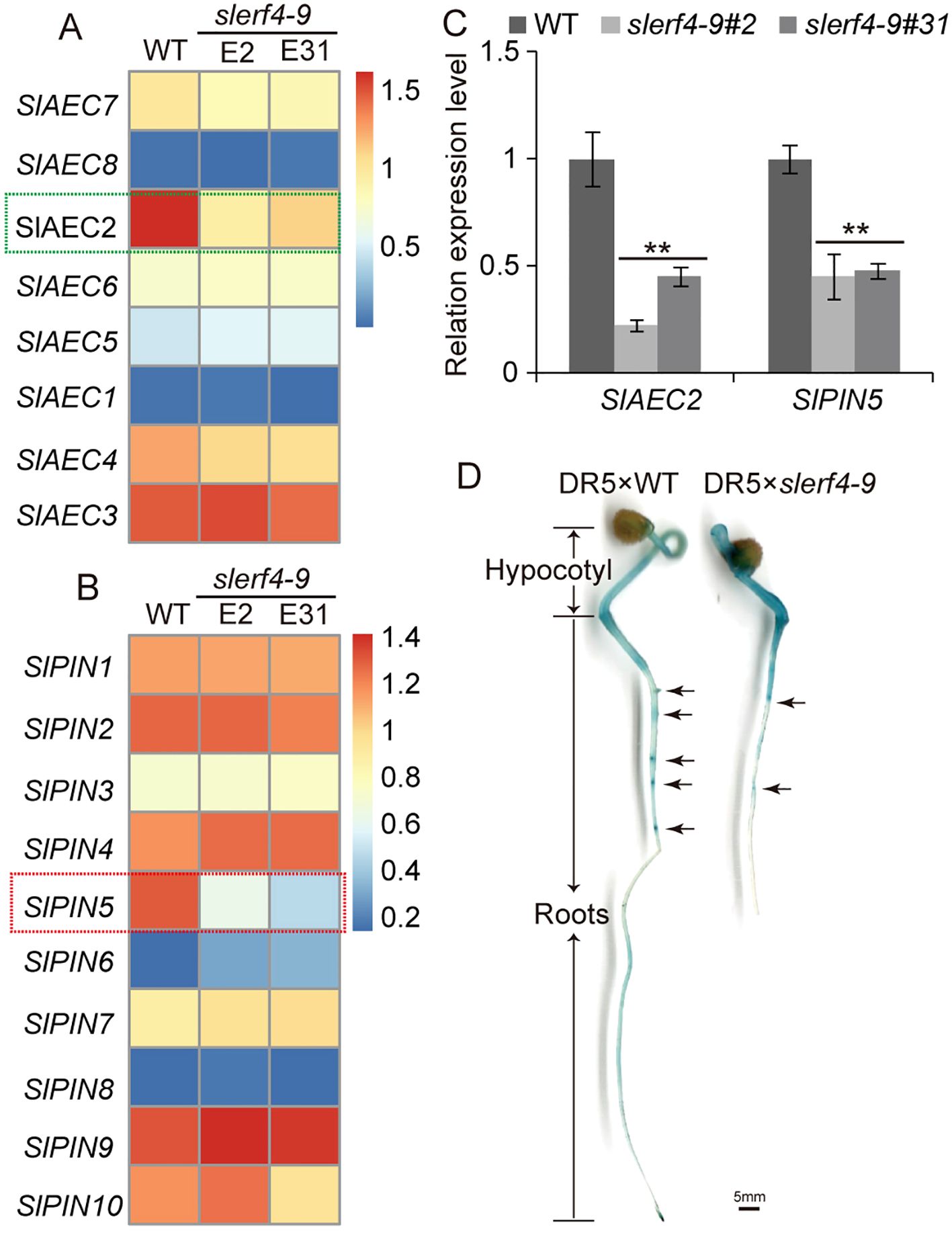
Figure 4. Auxin transport analysis. The FPKM value comparison of (A) SlAEC and (B) SlPIN family members. (C) Expression analysis of SlAEC2 and SlPIN5. (D) GUS staining analysis of the DR5 × WT and DR5 × slerf4-9 hybrid seed germinated seedlings. Asterisks denote significance relative to WT (Student’s t-test; **p < 0.01).
To investigate auxin transport in tomato roots, the DR5 strain was hybridized with the WT and the slerf4-9 mutant. The results of the GUS staining assay suggested that the mutation of SlERF4-9 led to the reduced accumulation of auxin in LR primordia (Figure 4D). However, there were no GCC-box or DRE elements in the 5,000-bp promoters of SlAEC2 and SlPIN5. Thus, SlERF4-9 may affect the efflux of auxin from intracellular to extracellular by indirectly regulating SlAEC2 and SlPIN5 expression and thereby improving LR development in tomatoes.
In the downregulated DEG analysis, two DOF zinc finger proteins, SlCDF1 (Solyc03g115940) and SlCDF3 (Solyc06g069760), were identified in the slerf4-9 mutants. The phylogenetic analysis indicated that the motif distributions of SlCDF1 and SlCDF3 proteins were similar to those of AtCDF1 and AtCDF3; moreover, SlCDF4 and SlCDF5 were similar to AtCDF2, and SlCDF2 was similar to AtTDDF1 (Figure 5A). The results suggest that CDFs with a similar motif distribution may have similar functions. In the transcriptome results, the FPKM values of SlCDF1 and SlCDF3 in two slerf4-9 mutants were lower than those in the WT, and those of SlCDF2, SlCDF4, and SlCDF5 were not significantly different between the WT and the mutants (Figure 5B). Moreover, the qRT-PCR results indicated that the SlERF4-9 mutation reduced the expression of SlCDF1 and SlCDF3 in the young tomato roots (Figure 5C). In addition, the promoters of SlCDF1 and SlCDF3 had three and one GCC-boxes, respectively (Figure 5D). Therefore, we speculated that SlERF4-9 may directly regulate the transcription of SlCDF1 and SlCDF3. In the EMSA, the SlERF4-9-GST fusion protein could bind with the GCC-boxes of the SlCDF1 and SlCDF3 promoters (Figure 5E), especially the SlCDF1 promoter. The dual-luciferase transient expression assay showed that SlERF4-9 could positively activate the transcription of SlCDF1 and SlCDF3 (Figures 5F, G). In addition, the luciferase fold analysis showed that the SlERF4-9 protein enhanced LUC expression via the promoters of SlCDF1 and SlCDF3 (Figure 5H). Therefore, the results suggest that SlERF4-9 can positively regulate SlCDF1 and SlCDF3 expression by directly binding the GCC-boxes of their promoters.
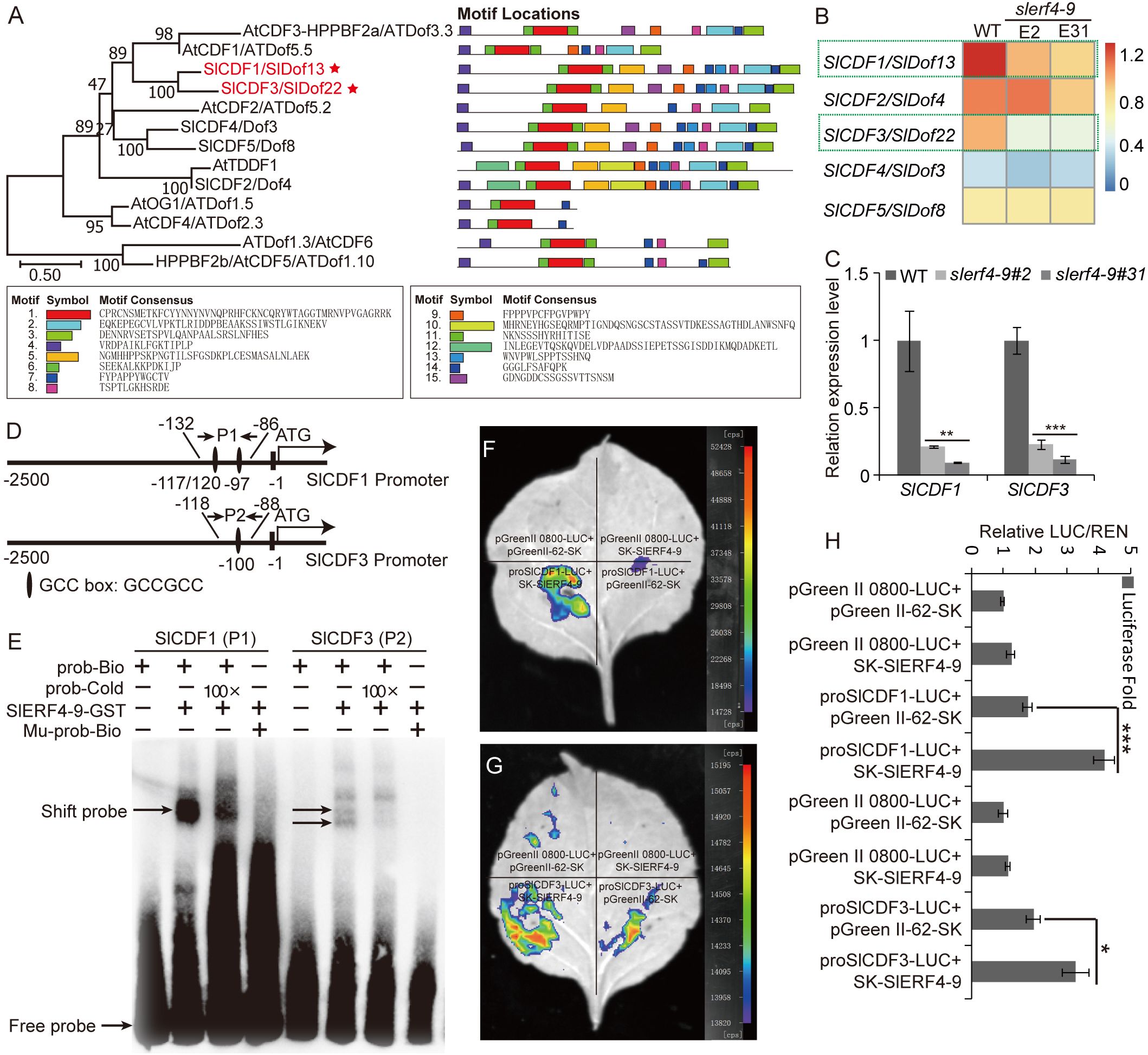
Figure 5. Regulatory relationship of SlERF4-9 and SlCDF1/3. (A) Phylogenetic and motif analyses of SlCDF1 and SlCDF3 proteins, (B) Heatmap analysis using the FPKM values of SlCDF1 and SlCDF3. (C) qRT-PCR analysis of SlCDF1 and SlCDF3. (D) GCC-box site analysis of SlCDF1 and SlCDF3 promoters. (E) The binding assays of SlERF4-9-GST protein with the P1 site of SlCDF1 promoter and the P2 site of SlCDF3 promoter. (F, G) Dual-luciferase transient expression assays of SlERF4-9 protein bind with the promoter of SlCDF1 and SlCDF3. (H) Luciferase fold analysis of LUC/REN using the leaves injected in the previous step. Asterisks denote significance relative to WT (Student’s t-test; *P < 0.05, **p < 0.01 and ***p < 0.001).
There are eight SlAEC and 10 SlPIN proteins in tomatoes, and there are eight AtPIN-like and seven AtPIN proteins in Arabidopsis. These proteins were subjected to a phylogenetic analysis. The results indicated that all PIN proteins were clustered in the I subgroup, while all SlAEC and AtPIN-like proteins were in the II subgroup (Supplementary Figure S2A). A protein sequence alignment found that all SlAEC and SlPIN proteins except SlAEC5 had both A and B domains, but SlPIN proteins had a long non-homologous sequence within the interval connecting the A and B domains (Supplementary Figure S2B). Thus, their A and B domains may be key for auxin binding and transport, while the non-homologous sequence between the A and B domains may be responsible for other functions.
There were 22 and 21 DOF transcription factor binding sites “(A/T)AAAG” on the SlAEC2 and SlPIN5 promoters, respectively (Figure 6A). We hypothesized that the transcription of SlAEC2 and SlPIN5 may be regulated by the SlCDF1 and SlCDF3 proteins. Therefore, a dual-luciferase transient expression assay was used to confirm this hypothesis. The results suggested that the transient expression of SlCDF1 and SlCDF3 activated LUC transcription through the SlAEC2 and SlPIN5 promoters (Figures 6B–E). A luciferase fold analysis yielded the same results (Figure 6F). Thus, SlCDF1 and SlCDF3 positively regulate SlAEC2 and SlPIN5 expression by binding to their promoters.
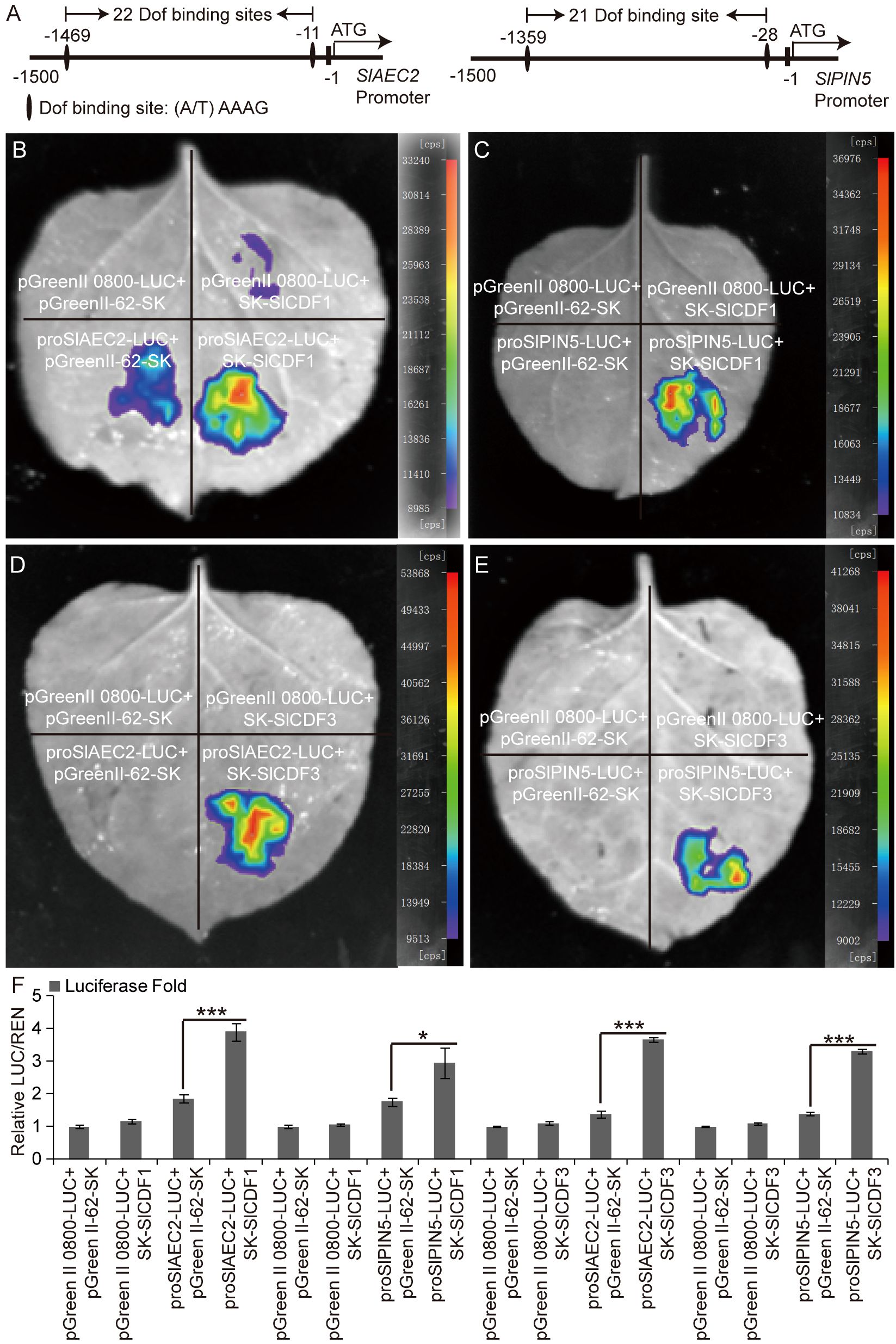
Figure 6. Regulatory relationship of SlCDF1/3 with SlAEC2 and SlPIN5. (A) DOF binding site analysis of SlAEC2 and SlPIN5 promoters. (B–E) The dual-luciferase transient expression assay of SlCDF1/3 protein binding with the promoter of SlAEC2 and SlPIN5. (F) Luciferase fold analysis of LUC/REN using the leaves injected in the previous step. Three biological replicates were conducted in the assays. Asterisks denote significance relative to WT or control (Student’s t-test; *p < 0.05 and ***p < 0.001). DOF, DNA binding with One Finger.
The roots are responsible for absorbing and transporting water and nutrients from the soil (Lal, 1979). Plants with more developed roots will have a greater ability to absorb and utilize water and nutrients. Phytohormone signal networks play important regulatory roles during root growth and development.
The biosynthesis, transport, and signal transduction of auxin involve SlYUCCA, SlTAA, SlIAA, SlARF, SlAEC, and SlPIN (Blilou et al., 2005; Cheng et al., 2006; Liscum and Reed, 2002; Stepanova et al., 2008). In this study, we found that the SlERF4-9 mutation reduced tomato seed germination and the number of LRs (Figures 1C,D, 2C,F,G,I). The transcriptome analysis of young tomato roots showed that the transcript levels of genes related to auxin biosynthesis and signal transduction such as SlYUCCA, SlTAA, SlIAA, and SlARF were unchanged (Supplementary Figure S1), while those of SlAEC2 and SlPIN5 decreased in the slerf4-9 mutant (Figures 4A–C). In Arabidopsis, lateral root initiation and root and hypocotyl growth were repressed in atpin5 mutants, and low concentrations of IAA did not improve root growth (Mravec et al., 2009). Auxin accumulation was altered in the DR5 × WT and DR5 × slerf4-9 hybrid seedling roots, especially in the LR primordia (Figure 4D). This indicated that SlERF4-9 is not involved in regulating auxin biosynthesis or signal transduction but may regulate the transcription of genes related to auxin transport and thus affect LR development. Interestingly, the AP2/ERF binding motifs GCC-box and DRE elements have not been found on the SlAEC2 and SlPIN5 promoters, but 22 and 21 DOF binding sites existed on the SlAEC2 and SlPIN5 promoters (Figure 6A). Therefore, we speculated that there may be some DOF factors between SlERF4-9 and these binding sites, regulated by SlERF4-9 and positively regulating SlAEC2 and SlPIN5 transcription to affect auxin transport in tomato roots.
The Cycling DOF Factors, a subgroup of the DOF family, regulate many plant life activities, including photoperiodic flowering, hypocotyl elongation, nitrogen assimilation, and abiotic stress responses (Corrales et al., 2017; Gao et al., 2022; Krahmer et al., 2019; Renau-Morata et al., 2020). In the DEG analysis of the transcriptome, the FPKM values of the two DOF factors SlCDF1 and SlCDF3 were reduced in slerf4-9 mutant roots (Figure 5B). The qRT-PCR analysis yielded the same result (Figure 5C). Moreover, their respective promoters showed three and one GCC-box elements (Figure 5D). This suggested that the expression of SlCDF1 and SlCDF3 may be regulated by SlERF4-9. The results of EMSA and LUC assays showed that SlERF4-9 could interact with SlCDF1 and SlCDF3 promoters to positively regulate their transcription (Figures 5E–H). Studies have demonstrated that SlCDF3 regulates flower timing, and its overexpression increases biomass production and yield (Renau-Morata et al., 2017; Xu et al., 2021). AtCDF1 overexpression not only promotes the accumulation of carbon and nitrogen metabolites in potato tubers but also increases their yield (Carrillo et al., 2023). The phylogenetic and MEME analysis results suggested that SlCDF1, SlCDF3, AtCDF1, and AtCDF3 may have similar functions (Figures 5A, B). Therefore, reduced expression of SlCDF1 and SlCDF3 will inevitably affect the carbon and nitrogen metabolism in the roots, thereby affecting their growth and development. In addition, the results of the LUC assays suggested that both SlCDF1 and SlCDF3 could improve the transcription of SlAEC2 and SlPIN5 (Figures 6B–F). Therefore, SlCDF1 and SlCDF3 are involved in auxin transport by regulating SlAEC2 and SlPIN5 expression (Figure 7).
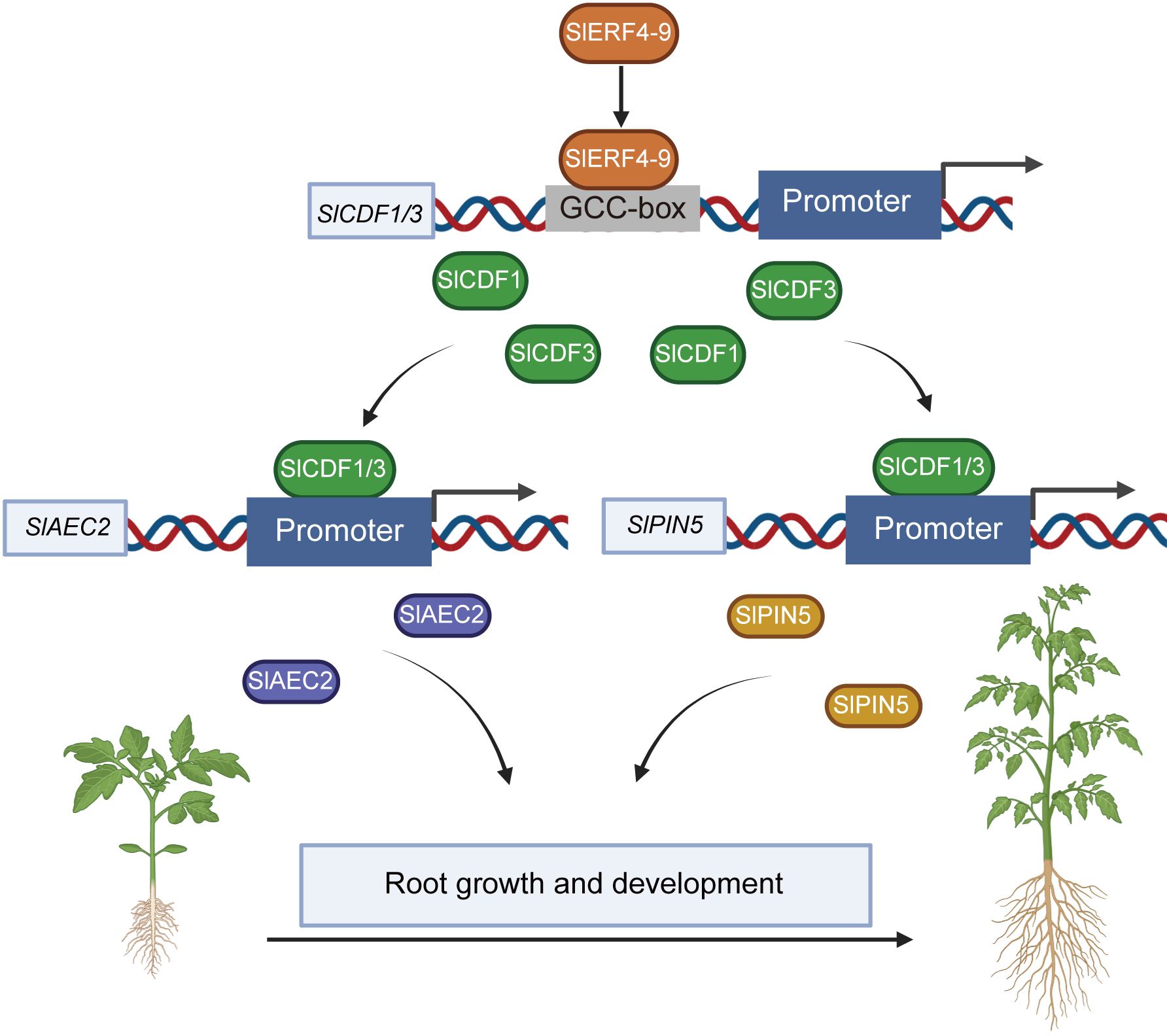
Figure 7. The regulatory network of SlERF4-9 involved in the growth and development of tomato roots.
In conclusion, SlERF4-9 positively regulates SlCDF1/3 transcription by binding to the GCC-boxes of their promoters; SlCDF1 and SlCDF3 improve the transcription of SlAEC2 and SlPIN5; and SlAEC2 and SlPIN5 coordinate auxin transport and thereby affect the growth and development of tomato roots (Figure 7). The SlERF4-9-SlCDF1/3-SlAEC2/SlPIN5 model will provide important theoretical support for cultivating new tomato varieties with highly developed root systems.
Raw data of RNA-seq was uploaded in the National Center for Biotechnology Information (NCBI) BioProject database with accession number PRJNA1091043.
YL: Funding acquisition, Project administration, Writing – original draft, Writing – review & editing. ZF: Investigation, Methodology, Writing – original draft. LZ: Data curation, Methodology, Project administration, Writing – review & editing. SL: Investigation, Software, Writing – original draft. SP: Conceptualization, Resources, Software, Writing – review & editing. YZ: Data curation, Investigation, Validation, Writing – original draft. CX: Data curation, Supervision, Validation, Writing – review & editing. MQ: Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 32160713).
We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1546092/full#supplementary-material
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as acommon module for plant organ formation. Cell 115, 591–602.
Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. doi: 10.1126/science.273.5277.948
Blakeslee, J. J., Peer, W. A., Murphy, A. S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8, 494–500. doi: 10.1016/j.pbi.2005.07.014
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 433, 39–44. doi: 10.1038/nature03184
Carrillo, L., Baroja-Fernandez, E., Renau-Morata, B., Munoz, F. J., Canales, J., Ciordia, S., et al. (2023). Ectopic expression of the AtCDF1 transcription factor in potato enhances tuber starch and amino acid contents and yield under open field conditions. Front. Plant Sci. 14, 1010669. doi: 10.3389/fpls.2023.1010669
Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., et al. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8, 165–171. doi: 10.1016/S1360-1385(03)00051-7
Cheng, Y., Dai, X., Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Corrales, A. R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40, 748–764. doi: 10.1111/pce.12894
De Smet, I., Tetsumura, T., De Rybel, B., Frei dit Frey, N., Laplaze, L., Casimiro, I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690. doi: 10.1242/dev.02753
Feng, B. H., Han, Y. C., Xiao, Y. Y., Kuang, J. F., Fan, Z. Q., Chen, J. Y., et al. (2016). The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67, 2263–2275. doi: 10.1093/jxb/erw032
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Ruhl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Gabriele, S., Rizza, A., Martone, J., Circelli, P., Costantino, P., Vittorioso, P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323. doi: 10.1111/j.1365-313X.2009.04055.x
Gao, H., Song, W., Severing, E., Vayssieres, A., Huettel, B., Franzen, R., et al. (2022). PIF4 and CDF2 co-operate to regulate cell elongation in Arabidopsis thaliana. Nat. Plants 8, 990–991. doi: 10.1038/s41477-022-01213-y
Jia, M., Li, Y., Wang, Z., Tao, S., Sun, G., Kong, X., et al. (2021). TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat. Plant J. 108, 1754–1767. doi: 10.1111/tpj.v108.6
Jiroutova, P., Oklestkova, J., Strnad, M. (2018). Crosstalk between brassinosteroids and ethylene during plant growth and under abiotic stress conditions. Int. J. Mol. Sci. 19, 3283. doi: 10.3390/ijms19103283
Kim, H. S., Kim, S. J., Abbasi, N., Bressan, R. A., Yun, D. J., Yoo, S. D., et al. (2010). The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 64, 524–535. doi: 10.1111/j.1365-313X.2010.04346.x
Konishi, M., Yanagisawa, S. (2007). Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 45 8, 623–629. doi: 10.1016/j.plaphy.2007.05.001
Krahmer, J., Goralogia, G. S., Kubota, A., Zardilis, A., Johnson, R. S., Song, Y. H., et al. (2019). Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 593, 319–338. doi: 10.1002/feb2.2019.593.issue-3
Lal, R. (1979). Plant root systems: Their function and interaction with the soil. Field Crops Res. 2, 177–179. doi: 10.1016/0378-4290(79)90020-0
Liscum, E., Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1023/A:1015255030047
Ljung, K., Bhalerao, R. P., Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474. doi: 10.1046/j.1365-313X.2001.01173.x
Mravec, J., Skupa, P., Bailly, A., Hoyerova, K., Krecek, P., Bielach, A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140. doi: 10.1038/nature08066
Nolan, T., Hands, R. E., Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabídopsis floral bud formation. Plant Cell 3, 677–684. doi: 10.2307/3869249
Overvoorde, P., Fukaki, H., Beeckman, T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2, a001537. doi: 10.1101/cshperspect.a001537
Park, C. H., Roh, J., Youn, J. H., Son, S. H., Park, J. H., Kim, S. Y., et al. (2018). Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Molecules Cells 41, 923–932. doi: 10.14348/molcells.2018.0092
Peret, B., De Rybel, B., Casimiro, I., Benková, E., Swarup, R., Laplaze, L., et al. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408. doi: 10.1016/j.tplants.2009.05.002
Petrasek, J., Mravec, J., Bouchard, R., Blakeslee, J. J., Abas, M., Seifertova, D., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918. doi: 10.1126/science.1123542
Pieterse, C. M. J., Leon-Reyes, A., van der Ent, S., Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Ramaiah, M., Jain, A., Raghothama, K. G. (2014). Ethylene response factor 070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 164, 1484–1498. doi: 10.1104/pp.113.231183
Renau-Morata, B., Carrillo, L., Cebolla-Cornejo, J., Molina, R. V., Marti, R., Dominguez-Figueroa, J., et al. (2020). The targeted overexpression of SlCDF4 in the fruit enhances tomato size and yield involving gibberellin signalling. Sci. Rep. 10, 10645. doi: 10.1038/s41598-020-67537-x
Renau-Morata, B., Molina, R. V., Carrillo, L., Cebolla-Cornejo, J., Sanchez-Perales, M., Pollmann, S., et al. (2017). Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 8, 660. doi: 10.3389/fpls.2017.00660
Santisree, P., Nongmaithem, S., Vasuki, H., Sreelakshmi, Y., Ivanchenko, M. G., Sharma, R. (2011). Tomato root penetration in soil requires a coaction between ethylene and auxin signaling. Plant Physiol. 156, 1424–1438. doi: 10.1104/pp.111.177014
Stacklies, W., Redestig, H., Scholz, M., Walther, D., Selbig, J. (2007). pcaMethods–a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167. doi: 10.1093/bioinformatics/btm069
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L., Xie, D. Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–1791. doi: 10.1016/j.cell.2008.01.047
Suzuki, R., Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Swarup, K., Benková, E., Swarup, R., Casimiro, I., Peret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., et al. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648–2653. doi: 10.1101/gad.210501
Teale, W. D., Pasternak, T., Dal Bosco, C., Dovzhenko, A., Kratzat, K., Bildl, W., et al. (2021). Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 40, e104416. doi: 10.15252/embj.2020104416
Trupiano, D., Yordanov, Y., Regan, S., Meilan, R., Tschaplinski, T., Scippa, G. S., et al. (2013). Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238, 271–282. doi: 10.1007/s00425-013-1890-4
Upadhyay, R. K., Gupta, A., Soni, D., Garg, R., Pathre, U. V., Nath, P., et al. (2017). Ectopic expression of a tomato DREB gene affects several ABA processes and influences plant growth and root architecture in an age-dependent manner. J. Plant Physiol. 214, 97–107. doi: 10.1016/j.jplph.2017.04.004
Wang, G. D., Xu, X. P., Wang, H., Liu, Q., Yang, X. T., Liao, L. X., et al. (2019). A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol. Biochem. 142, 254–262. doi: 10.1016/j.plaphy.2019.07.017
Xu, D., Li, X., Wu, X., Meng, L., Zou, Z., Bao, E., et al. (2021). Tomato SlCDF3 delays flowering time by regulating different FT-like genes under long-day and short-day conditions. Front. Plant Sci. 12, 650068. doi: 10.3389/fpls.2021.650068
Xu, L., Liu, P., Li, X., Mi, Q., Zheng, Q., Xing, J., et al. (2024). NtERF283 positively regulates water deficit tolerance in tobacco (Nicotianatabacum L.) by enhancing antioxidant capacity. Plant Physiol. Biochem. 207, 108413. doi: 10.1016/j.plaphy.2024.108413
Yu, Z., Zhang, F., Friml, J., Ding, Z. (2022). Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 64, 371–392. doi: 10.1111/jipb.13225
Yuste-Lisbona, F. J., Fernandez-Lozano, A., Pineda, B., Bretones, S., Ortiz-Atienza, A., Garcia-Sogo, B., et al. (2020). ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. U.S.A. 117, 8187–8195. doi: 10.1073/pnas.1913688117
Zhang, L., Chen, L. J., Pang, S. Q., Zheng, Q., Quan, S. W., Liu, Y. F., et al. (2022). Function analysis of the ERF and DREB subfamilies in tomato fruit development and ripening. Front. Plant Sci. 13, 849048. doi: 10.3389/fpls.2022.849048
Keywords: auxin efflux carrier, cycling DOF factors, lateral root development, SlERF4-9, tomato
Citation: Fan Z, Zhang L, Li S, Pang S, Zhang Y, Xu C, Liu Y and Qi M (2025) The SlERF4-9-SlCDF1/3-SlAEC2/SlPIN5 module regulates tomato root morphogenesis. Front. Plant Sci. 16:1546092. doi: 10.3389/fpls.2025.1546092
Received: 16 December 2024; Accepted: 10 March 2025;
Published: 28 March 2025.
Edited by:
Juan José Ripoll, Miguel Hernández University of Elche, SpainReviewed by:
Wenkun Zhou, China Agricultural University, ChinaCopyright © 2025 Fan, Zhang, Li, Pang, Zhang, Xu, Liu and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: YuDong Liu, bHlkLWZvcmV2ZXJAMTYzLmNvbQ==; MingFang Qi, cWltaW5nZmFuZ0BzeWF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.