
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Plant Sci., 07 March 2025
Sec. Plant Physiology
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1542950
MED7, a middle-module subunit of the transcriptional co-regulator Mediator complex, plays a critical role in gene regulation in Arabidopsis thaliana, where it is encoded by two paralogs, MED7A and MED7B. We present phenotypic analyses of homozygous MED7-silenced transgenic lines with significantly reduced expression of both MED7 paralogs under autotrophic conditions. Our findings demonstrate that MED7 is essential for proper cotyledon opening during de-etiolation, as the silenced lines showed a marked delay in this process. Additionally, these lines displayed distinct morphological alterations, including hyponastic cotyledons, elongated hypocotyls, and modified root architecture, such as shorter primary roots and impaired root hair development in light-grown seedlings. MED7 silencing also significantly hindered light-induced adventitious root (AR) formation on the hypocotyls of etiolated seedlings, leading to a notable reduction in AR production. Moreover, MED7 silencing impacted the timing of floral transition and shoot branching, resulting in delayed flowering and an increased number of primary cauline branches on the inflorescence stem. Together, these results underscore a central role for MED7 in orchestrating key developmental processes in plants.
Mediator is a highly conserved and essential protein complex that plays a central role in transcriptional regulation across all eukaryotes (Soutourina, 2018). As a key component of the transcriptional machinery, Mediator functions as a critical co-regulator, bridging gene-specific transcription factors (TFs) with the RNA polymerase II (Pol II) pre-initiation complex. By receiving and integrating regulatory information from promoter-bound TFs and enhancers, Mediator modulates transcription across nearly all protein-coding genes in eukaryotes (Allen and Taatjes, 2015). Beyond initiation, Mediator contributes to multiple other regulatory processes involved in gene expression, including chromatin looping, mRNA processing and export, transcriptional memory, and re-initiation (Reeves and Hahn, 2003; D’Urso et al., 2016; Chereji et al., 2017; Crawford et al., 2020; Ramasamy et al., 2023).
Mediator comprises 25 to 35 subunits, varying by organism, and is organized into distinct modules: head, middle, tail (core Mediator), and a cyclin-kinase module (CKM), which associates transiently with core Mediator (Elmlund et al., 2006). Head and middle interact with Pol II, including its C-terminal domain (CTD), as well as with general transcription factors (GTFs), while tail binds to sequence-specific TFs at promoters or enhancers (Warfield et al., 2022). Although Mediator has been extensively studied in yeast and metazoans, research on plant Mediator is more recent. Similar to its role in other eukaryotes, plant Mediator serves as a central hub for transcriptional regulation, coordinating gene expression for development, metabolism, and responses to hormones and stress (Buendía-Monreal and Gillmor, 2016; Sinha and Kumar, 2021; Blomberg et al., 2024). However, plants have evolved unique subunits, likely facilitating interactions with plant-specific TFs and enabling the integration of signals related to hormonal responses, pathogen defense, and stress adaptation (Bäckström et al., 2007; Dolan and Chapple, 2016).
In plants Mediator plays a pivotal role in regulating responses to key hormones such as auxins, gibberellins, and abscisic acid, as well as in the transcriptional activation of genes within hormonal pathways, which are essential for plant growth and development (Buendía-Monreal and Gillmor, 2016; Kumar et al., 2018). Beyond its central role in transcriptional regulation, plant Mediator acts as a crucial integrator of diverse signaling pathways, enabling plants to effectively respond and adapt to dynamic environmental conditions (Dolan and Chapple, 2016). Studies using loss-of-function mutants and silenced lines of various subunits have shed light on their distinct roles in plant development and environmental responses. For instance, a MED18 loss-of-function mutant exhibits delayed flowering and altered floral organ development (Zheng et al., 2013) while MED8 is essential for organ size regulation; mutants lacking MED8 develop smaller flowers due to reduced cell expansion (Xu and Li, 2012). MED30 is critical for early development, particularly embryogenesis, and reduced MED30 expression delays flowering (Jaskolowski et al., 2019). Furthermore, MED25 has been shown to regulate lateral root formation (Raya-González et al., 2014).
The Arabidopsis MED7 subunit is part of the Mediator middle module and is encoded by two paralogs, MED7A and MED7B. Studies in various organisms, including plants and fungi, highlight MED7’s importance in diverse biological processes such as growth, development, and responses to environmental stresses (Tebbji et al., 2014; Kumar et al., 2018). However, research on MED7’s specific functions in plants remains limited. In previous work, we demonstrated the redundant roles of MED7A and MED7B in regulating skotomorphogenic growth in Arabidopsis seedlings. Our findings showed that MED7 influences expression of genes involved in cell elongation and response to auxin and brassinosteroids processes essential for etiolated seedling growth under dark conditions. Additionally, concurrent silencing of MED7A and MED7B (hereafter referred to as med7RI lines) resulted in shorter hypocotyls and defective hook opening in dark-grown seedlings, underscoring MED7’s critical role in early seedling development during skotomorphogenesis (Kumar et al., 2018).
In this study, we investigate the effects of MED7A/MED7B silencing across multiple stages of Arabidopsis development, revealing a broader impact of MED7 on plant growth and developmental processes beyond skotomorphogenesis.
In this study, we used Arabidopsis thaliana ecotype Columbia (Col-0) as the wild-type (WT) control. Two previously characterized homozygous MED7A/MED7B silenced transgenic lines, med7RI-7 and med7RI-13, with significantly reduced expression of both MED7 paralogs, were included (Kumar et al., 2018). A homozygous empty vector (EV) line was also used as a control alongside WT plants (Kumar et al., 2018). Additionally, we employed med7RI lines conditionally complemented with epitope-tagged MED7A (FLAG-MED7AR/med7RI) and MED7B (HA-MED7BR/med7RI), each expressed individually under an estradiol-inducible promoter (cloned into the pER8 vector), along with a control complementation line (empty pER8 vector)/med7RI) as previously described (Kumar et al., 2018). Seeds from WT and transgenic lines were surface-sterilized with 70% ethanol containing 0.1% Tween 20, followed by two quick washes in 95% ethanol. After drying, seeds were plated on half-strength Murashige and Skoog (½ × MS, pH 5.7) basal salt medium (Sigma-Aldrich, Stockholm, Sweden) containing 0.8% (w/v) phytoagar (Duchefa Biochemie). The plated seeds were stratified at 4°C in darkness for three days to synchronize germination. After stratification, seeds were transferred to photoperiodic light conditions (120-130 µmol m−2s−1) with a 16-hour light/8-hour dark cycle at 22°C and grown vertically. For etiolated seedling growth, stratified seeds were exposed to light for six hours to initiate germination, then shifted to complete darkness in a vertical orientation and grown at 22°C. Conditional expression of FLAG-MED7AR and HA-MED7BR was induced by growing the respective transgenic seedlings on medium containing 1.0 μM β-estradiol (Sigma-Aldrich) (Kumar et al., 2018).
A qualitative analysis of cotyledon opening and expansion during de-etiolation was conducted using four-day-old etiolated seedlings. To induce de-etiolation, seedlings grown in darkness were exposed to continuous low-fluence white light at an intensity of 10 µmol m-2s-1 for 48 hours in a growth chamber (Percival Scientific, Iowa, USA). Cotyledon opening was monitored at regular intervals and observed under a dissecting microscope. Photographs were taken after 36 hours of light exposure to assess the progression of de-etiolation.
Seedlings were grown under photoperiodic light conditions (120–130 µmol m-2s-1, 16-hour light/8-hour dark cycle) for 10 days. They were then placed on a flat surface, and images were captured using a high-resolution camera. Hypocotyl and primary root lengths were measured using ImageJ (https://imagej.net/ij/). Hypocotyl length was measured from the base of the cotyledons to the root-hypocotyl junction, while primary root length was measured from the root-hypocotyl junction to the root tip.
Four-day-old etiolated seedlings grown in darkness on ½× MS basal salt medium (pH 5.7) containing 0.8% (w/v) phytoagar were transferred to light conditions (120–130 µmol m-2s-1). After 7 days in light, photographs were taken, and ARs on the hypocotyls were counted.
Seeds were germinated and plants were grown under long-day conditions (16-hour light/8-hour dark photoperiod; 120–130 µmol m-2s-1, 22°C) using cool fluorescent lighting. In each experiment, at least 15 plants per genotype were analyzed, with tray positions randomized to minimize positional effects within the growth chamber. Flowering time was recorded as both the total number of rosette leaves at bolting and the number of days from sowing until bolting, defined as the visible emergence of the inflorescence stem above the rosette. At the mature flowering stage, the number of primary cauline branches (arising from the main inflorescence stem) and primary rosette branches were also counted.
Total RNA was extracted from both four-day-old old dark grown etiolated seedlings and from etiolated seedlings after 6 h exposure to white light at an intensity of 120–130 µmol m-2s-1 using the Omega Bio tek plant RNA kit and treated with RNase-free ezDNase following the manufacturer’s instructions (Thermo Scientific, USA). 1 μg of total RNA was reverse transcribed using iScript reverse transcription supermix (Biorad, Solna, Sweden). RT-qPCR was performed using a CFX96 Touch qPCR instrument (BioRAD USA) and the PowerUp SYBR green master mix (Applied Biosystems, Massachusetts, USA). Gene expression levels were normalized to the AT3G18780 and AT4G36800 reference genes and displayed in relative units. Three biological and two technical repetitions were performed for each sample. The sequences of the RT-qPCR primers for HY5 were described previously (Kumar et al., 2018). The sequences for PIF4 were; Forward: 5´-CCGACCGGTTTGCTAGATACATCG-3´, Reverse: 5´-ATCTCCATCGGCTGCATCTGAGTC-3´.
Significance levels were assessed using Student´s t-test or one-way ANOVA, followed by mean comparisons with Tukey’s and Bonferroni’s post hoc tests. All statistical analyses were performed using Excel or the Origin software (https://www.originlab.com/).
Cotyledon opening in Arabidopsis is a key process in photomorphogenesis, the developmental transition triggered by light exposure. When etiolated seedlings are exposed to light, they undergo de-etiolation, which includes the expansion and opening of the cotyledons. This is crucial for shifting from heterotrophic to autotrophic growth, enabling seedlings to optimize light capture for photosynthesis. Brassinazole (BRZ), a brassinosteroid (BR) biosynthesis inhibitor, can induce photomorphogenic traits such as short hypocotyls and cotyledon opening in dark-grown Arabidopsis seedlings. In a previous study, we observed that the unfolding of cotyledons in response to BRZ treatment was significantly impaired in the med7RI lines, indicating that MED7 silencing impacts not only skotomorphogenesis but also photomorphogenesis in Arabidopsis (Kumar et al., 2018).
To further investigate the impact of MED7 silencing on cotyledon opening during de-etiolation, we monitored cotyledon opening under low-fluence continuous white light. The med7RI lines exhibited a delayed response in cotyledon opening compared to the control lines. After 36 hours of exposure to continuous low-fluence white light, control seedlings (WT and EV) showed fully opened cotyledons, while the med7RI lines remained closed (Figure 1A, Supplementary Figure 1A). However, by 48 hours, most med7RI seedlings had initiated cotyledon opening, indicating that the opening and expansion of cotyledons were delayed in the med7RI lines compared to the controls.
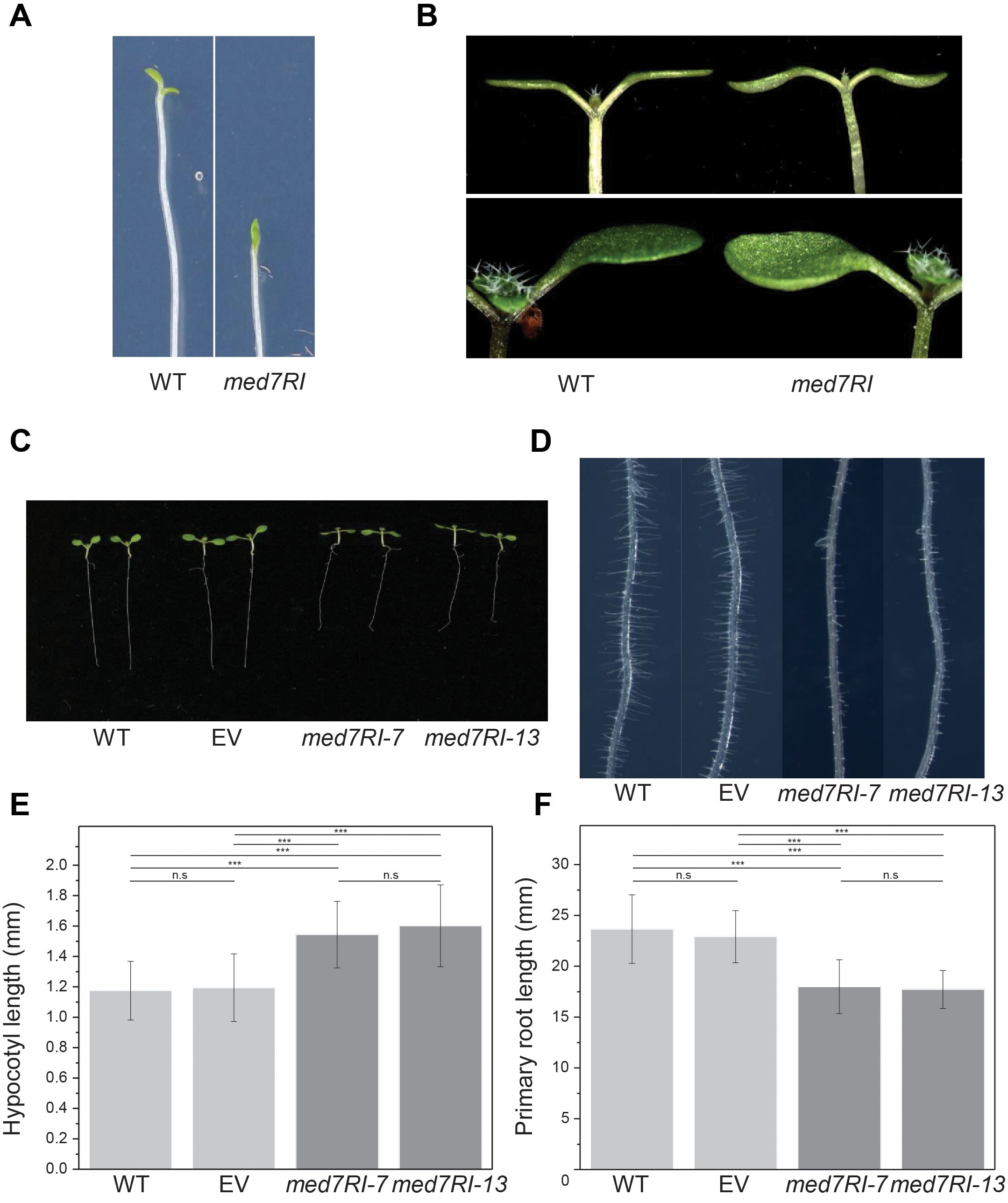
Figure 1. Effect of MED7 silencing on light-grown Arabidopsis seedlings. (A) Delayed cotyledon opening during de-etiolation. Four-day-old etiolated seedlings were exposed to 10 μE white light, and images were captured after 36 hours. Representative images of wild-type (WT) and med7RI line is shown. (B) Hyponastic cotyledon phenotype in MED7-silenced lines compared to control lines. Images of WT and med7RI seedlings are presented. (C) Ten-day-old seedlings of control (WT, EV) and med7RI (med7RI-7, med7RI-13) lines, displaying differences in growth patterns under photoperiodic light. (D) Impaired root hair development in MED7-silenced lines. Root hair growth on the primary root of ten-day-old seedlings from control (WT, EV) and med7RI lines is shown. (E) Hypocotyl length and (F) primary root length of ten-day-old light-grown seedlings from control and med7RI lines. Data represent mean ± SD, with significance indicated by asterisks (***P < 0.001). Not statistically significant differences are labelled n.s. The experiments were performed using three biological replicates, with ≥30 seedlings per line/genotype, and statistical significance was determined using one-way ANOVA, followed by Tukey’s and Bonferroni’s post hoc tests.
The delayed cotyledon opening in med7RI lines during de-etiolation highlights the importance of MED7 in regulating light signaling pathways that facilitate the transition from skotomorphogenesis to photomorphogenesis. ELONGATED HYPOCOTYL 5 (HY5) and PHYTOCHROME INTERACTING FACTORs (PIFs) are transcription factors that function as master regulators that control a large array of genes involved in light-dependent development. PIFs and HY5 are oppositely regulated by light conditions and therefore have contrasting effects on light-induced cotyledon opening in Arabidopsis (Zhang et al., 2017; Yao et al., 2024). In darkness, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) degrades HY5, relieving repression on elongation genes and enabling PIFs to accumulate and activate hypocotyl elongation pathways (Yu et al., 2013). Upon light exposure, phytochromes phosphorylate PIFs, marking them for ubiquitin-mediated degradation, which suppresses elongation and promotes photomorphogenesis (Jia et al., 2014). In light-grown seedlings, HY5 promotes inhibition of hypocotyl elongation by activating photomorphogenesis-associated genes, repressing elongation-related genes through chromatin modifications via its interaction with HDA15, and enhancing BIN2 kinase activity to suppress brassinosteroid signaling (Xu et al., 2016; Zhao et al., 2019; Li et al., 2020). We have previously reported that HY5 levels are significantly lower in etiolated med7RI seedlings grown in dark compared to wild-type (Kumar et al., 2018). We used RT-qPCR to analyze the HY5 mRNA levels in etiolated seedlings before and after 6 hours of exposure to light. We found that the expression levels of HY5 were reduced in the med7RI lines both at skotomorphogenic and during de-etiolation compared to control lines (Supplementary Figure 1B). On the other hand, PIF4 levels were relatively higher in med7RI lines compared to control lines in both dark and light conditions. This indicates that defects in expression of HY5 and PIF4 during de-etiolation contribute to the delayed cotyledon opening observed in med7Ri lines.
We observed that med7RI lines display hyponastic cotyledons, characterized by an upward bending of the cotyledons (Figure 1B). This was reversed upon expression of either Flag-MED7aR or HA-MED7bR (Supplementary Figure 2A). This morphological change appears to result from disruptions in the normal growth patterns and hormonal signaling pathways that are essential for proper cotyledon development and orientation. Importantly, hyponasty was restricted to cotyledons and was not observed in the true leaves of med7RI lines.
Auxin, a hormone essential to nearly all aspects of plant development, including postembryonic organogenesis, has been linked to similar phenotypes. For example, hyponastic cotyledons are observed in the rol1-2 mutant, where altered flavonol profiles lead to significant changes in auxin transport and distribution, marked by a basal-to-apical shift in the polar localization of PIN2, a key auxin efflux carrier (Kuhn et al., 2017). Previously, we reported that several AUX/IAA family genes and other auxin-responsive genes are differentially regulated in etiolated med7RI seedlings (Kumar et al., 2018), suggesting that disruptions in auxin homeostasis may contribute to the hyponastic cotyledons observed in med7RI lines. Interestingly, mutations in CDK8, encoding a subunit of the Mediator kinase module, reversed the hyponastic cotyledons in rol1-2 mutants to an epinastic state (Schumacher et al., 2021).
To examine the role of MED7 in light-grown conditions, we compared hypocotyl and primary root lengths in med7RI lines with WT and EV plants. In contrast to the reduced hypocotyl length seen in etiolated seedlings with MED7 silencing (Kumar et al., 2018), hypocotyls of light-grown med7RI seedlings were significantly longer than those of control lines (Figures 1C, E). Additionally, the primary root length in med7RI lines was markedly shorter than in WT (Figures 1C, F). Expression of either Flag-MED7aR or HA-MED7bR reversed both phenotypes (Supplementary Figures 2B–D). Furthermore, med7RI lines exhibited abnormal root hair development along the primary root, with predominantly short and unevenly distributed root hairs compared to controls (Figure 1D). However, no significant changes in lateral root density were observed between the control and med7RI lines (Supplementary Figure 2E).
The contrasting effects of MED7 silencing on hypocotyl elongation under dark and light conditions underscore the complexity of its role in development. In etiolated seedlings, MED7 silencing results in shorter hypocotyls due to impaired cell elongation, likely caused by altered expression of genes involved in cell wall remodeling and hormone signaling, particularly pathways involving auxin and brassinosteroids (Kumar et al., 2018). Conversely, in light-grown seedlings, MED7 silencing leads to increased hypocotyl length, potentially due to differential regulation of light-responsive genes and hormonal pathways that promote elongation in response to light (Vandenbussche et al., 2005). It is likely that the increased hypocotyl elongation in light-grown med7RI seedlings is associated with defects in light-signaling pathways mediated by the dysregulation of key regulators, such as PIF4 and HY5 whose expression levels were affected in med7RI lines (Supplementary Figures 1B, Cv ). Both HY5 and PIF4 are crucial in controlling hypocotyl growth in response to light, with HY5 and PIF protein stability being oppositely regulated by light conditions (Zhang et al., 2017).
The reduced primary root length in light-grown med7RI lines suggests that MED7 is also involved in root development under light conditions. Light signaling is known to coordinate shoot and root development, where TFs like HY5, PIFs and various hormonal pathways mediate crosstalk between organs (Chen et al., 2016; Yang and Liu, 2020). Our current and previous findings that HY5 expression is downregulated both during skotomorphogenic and de-etiolation in the med7RI lines may therefore account for their altered root growth, supporting the role of MED7 in light-dependent developmental processes (Kumar et al., 2018). These findings indicate that MED7 interacts with or regulates distinct factors in skotomorphogenic versus photomorphogenic conditions, allowing the plant to adapt root and shoot growth in response to light.
Previous studies have shown that auxin homeostasis and distribution are essential for proper root hair development, with auxin transporters like PIN proteins playing a key role in establishing the auxin gradients necessary for root hair elongation (Rigas et al., 2013). Similar root hair phenotypes have been observed in the rol1-2 mutant, where disrupted auxin homeostasis results in shorter root hairs and a reduced primary root length (Schumacher et al., 2021). Additionally, shorter root hairs have been reported in other Mediator subunit mutants, such as med12 and med13, which also affect auxin response pathways (Raya-González et al., 2014). Our findings suggest that MED7 silencing may disrupt auxin distribution or signaling, contributing to the shorter root and root hair phenotypes observed in the med7RI lines.
In Arabidopsis, AR development along the hypocotyl is a well-documented response when etiolated seedlings are exposed to light. This process is primarily regulated by auxin signaling pathways and other factors that influence hypocotyl growth during the transition from etiolation to de-etiolation. To assess the impact of MED7 silencing on AR development, we examined AR induction in med7RI seedlings. Unlike the WT and EV control lines, med7RI seedlings showed a nearly complete absence of AR formation after 7 days of light exposure (Figures 2A, B). In Arabidopsis, AR development is predominantly controlled by auxin homeostasis, transport, and signaling, along with interactions with other hormones and environmental cues, such as light (Pacurar et al., 2014; Zeng et al., 2022). Auxin-driven AR formation is mediated by AUXIN RESPONSE FACTORS (ARFs), which regulate the expression of genes involved in root initiation. ARF6 and ARF8 positively regulate AR formation, whereas ARF17 acts as a negative regulator (Gutierrez et al., 2009). Furthermore, auxin transport via PIN and AUX/LAX transporters is essential for AR development in Arabidopsis hypocotyls (Da Costa et al., 2020). The observed reduction in AR formation in med7RI lines may be linked to the previously documented defects in hypocotyl growth and auxin-responsive gene expression in these lines (Kumar et al., 2018). This suggests that the MED7 subunit is critical not only for proper etiolation and de-etiolation processes but also for maintaining auxin homeostasis—key factors for normal AR development. The absence of well-developed ARs in med7RI seedlings highlights the importance of MED7 in regulating pathways essential for AR formation. Furthermore, both MED7 paralogs successfully restored AR development in the complementation lines (Supplementary Figure 2F).
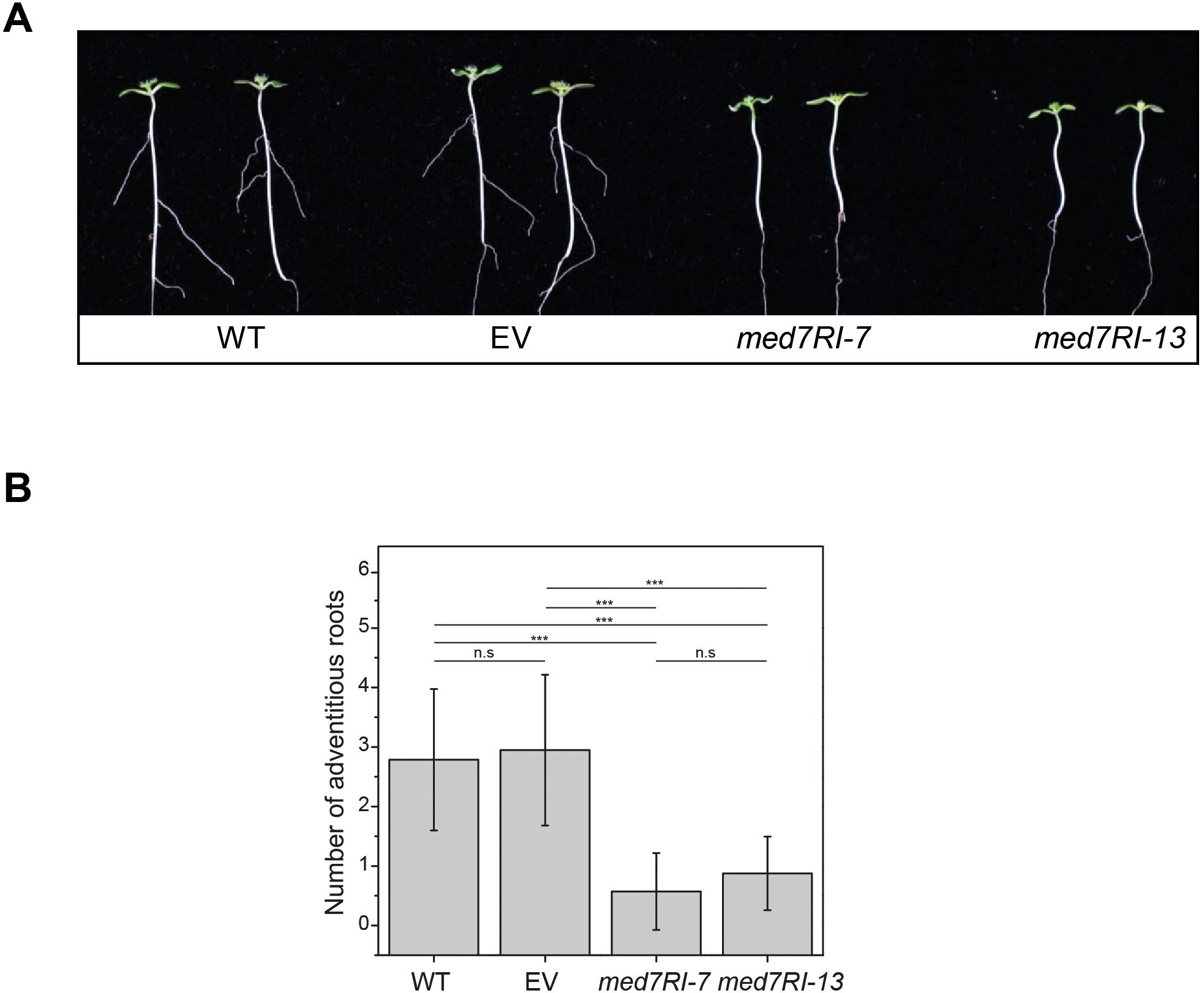
Figure 2. Effect of MED7 silencing on AR development in Arabidopsis. (A) Four-day-old etiolated seedlings grown on 1/2 x MS medium were exposed to light to induce AR formation along the hypocotyl. After 7 days of light exposure, AR development was observed, showing a marked reduction in MED7-silenced lines (med7RI-7 and med7R1-13) compared to controls (WT, EV). Representative images are shown. (B) The number of ARs was quantified after 7 days of light exposure in both control and med7RI lines. Results demonstrate a significant decrease in AR numbers in MED7-silenced lines. Data represent the mean ± SD, with significance indicated by asterisks (***P < 0.001). Not statistically significant differences are labelled n.s. The experiments were performed using three biological replicates, with ≥30 seedlings per each line/genotype, and statistical significance was determined using one-way ANOVA, followed by Tukey’s and Bonferroni’s post hoc tests.
Following our assessment of developmental phenotypes in med7RI lines during the seedling stage, we evaluated floral transition in the adult plants. In med7RI lines, flowering was significantly delayed, as indicated by an increased number of rosette leaves and a longer time to bolting (Figures 3A, B). Furthermore, med7RI plants displayed a substantial increase in the number of primary cauline branches (Figures 3C, E), while the number of rosette branches remained normal (Figure 3D).
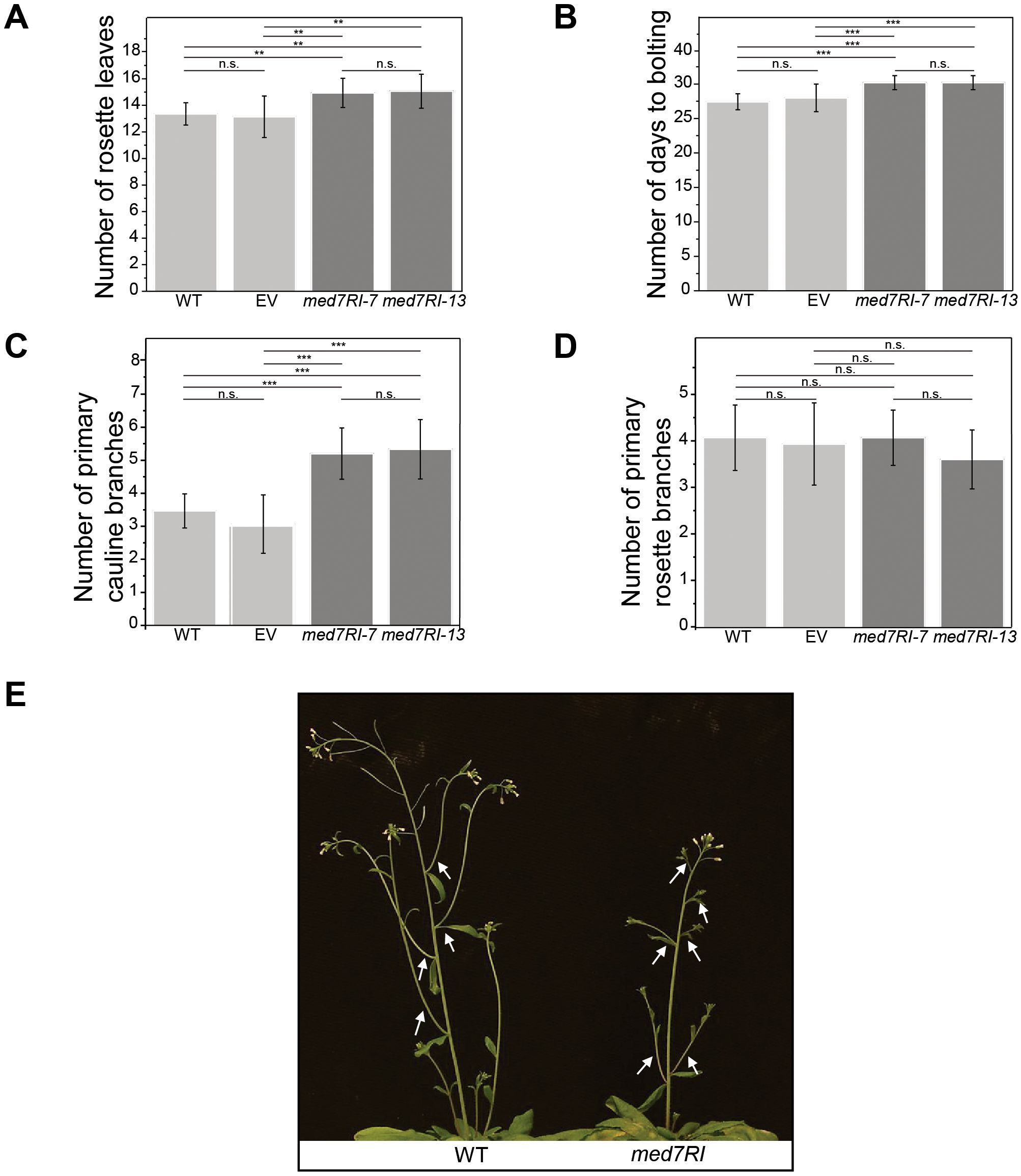
Figure 3. Effects of MED7 silencing on flowering time and branching in Arabidopsis. Plants were grown under long-day conditions (16 h light/8 h dark). (A) Number of rosette leaves: Flowering time was assessed by recording the number of rosette leaves at bolting. MED7-silenced lines (med7RI-7, med7RI-13) exhibited a greater number of rosette leaves, indicating delayed flowering compared to controls (WT, EV). (B) Days to bolting: The number of days from sowing to bolting was recorded, showing a significant delay in flowering in MED7-silenced lines compared to control lines. (C) Primary cauline branches: MED7-silenced lines displayed a significant increase in the number of primary cauline branches, suggesting altered shoot branching. (D) Primary rosette branches: The number of primary branches emerging from the rosette was analyzed, comparing control and med7 lines. (E) A comparison of WT and med7RI plants, with arrows indicating the increased number of primary cauline branches in MED7-silenced lines. Data represent the mean ± SD. Asterisks indicate the level of significance (***P < 0.001, **P < 0.01). Not statistically significant differences are labelled n.s. The experiments were performed using three biological replicates, with 15 plants per each line/genotype, and statistical significance was determined using one-way ANOVA, followed by Tukey’s and Bonferroni’s post hoc tests.
In Arabidopsis thaliana, flowering is controlled by key genes like FLOWERING LOCUS T (FT), which promotes flowering, and FLOWERING LOCUS C (FLC), which acts as a repressor. These genes integrate environmental signals, including photoperiod and vernalization, to optimize the timing of flowering (Searle et al., 2006). Additionally, multiple Mediator subunits play crucial roles in floral transition by modulating the expression of these and other flowering-related genes (Buendía-Monreal and Gillmor, 2016). Auxin also significantly influences branching by modulating levels of phytohormones like strigolactones and cytokinins, which collectively shape shoot architecture and branching patterns (Fichtner et al., 2022). In this study, the delayed flowering and increased primary cauline branching observed in med7RI lines suggest disruptions in either the regulation of flowering via FT and FLC or in the auxin and associated phytohormone pathways that control branching.
Mediator acts as a central regulator of gene expression, integrating diverse environmental and developmental signals to ensure coordinated plant growth and adaptation. The delayed cotyledon opening and impaired AR development observed with MED7 silencing highlight MED7’s key role in light- and auxin-regulated processes in Arabidopsis development. Additionally, the delays in flowering and increased cauline branching underscore MED7’s significance in orchestrating major developmental transitions. Together, these findings position MED7 as essential for multiple developmental processes. Further investigation into the precise mechanisms through which MED7 modulates these pathways will yield deeper insights into the complex regulation of plant growth and development.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
KRRK: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Resources. JB: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. SB: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by a start-up research grant from the University Grants Commission to KRRK, and research grants to SB from the Knut and Alice Wallenberg Foundation (2015-0056), the Swedish Foundation for Strategic Research (SB16-0089) and the Swedish Research Council (2016-03943).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1542950/full#supplementary-material
Allen, B. L., Taatjes, D. J. (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166. doi: 10.1038/nrm3951
Bäckström, S., Elfving, N., Nilsson, R., Wingsle, G., Björklund, S. (2007). Purification of a plant mediator from arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729. doi: 10.1016/j.molcel.2007.05.007
Blomberg, J., Tasselius, V., Vergara, A., Karamat, F., Imran, Q. M., Strand, Å., et al. (2024). Pseudomonas syringae infectivity correlates to altered transcript and metabolite levels of Arabidopsis mediator mutants. Sci. Rep. 14, 6771. doi: 10.1038/s41598-024-57192-x
Buendía-Monreal, M., Gillmor, C. S. (2016). Mediator: A key regulator of plant development. Dev. Biol. 419, 7–18. doi: 10.1016/j.ydbio.2016.06.009
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Chereji, R. V., Bharatula, V., Elfving, N., Blomberg, J., Larsson, M., Morozov, A. V., et al. (2017). Mediator binds to boundaries of chromosomal interaction domains and to proteins involved in DNA looping, RNA metabolism, chromatin remodeling, and actin assembly. Nucleic Acids Res. 45, 8806–8821. doi: 10.1093/nar/gkx491
Crawford, T., Karamat, F., Lehotai, N., Rentoft, M., Blomberg, J., Strand, Å., et al. (2020). Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 10, 5073. doi: 10.1038/s41598-020-61758-w
D’Urso, A., Takahashi, Y., Xiong, B., Marone, J., Coukos, R., Randise-Hinchliff, C., et al. (2016). Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. eLife 5, e16691. doi: 10.7554/eLife.16691
Da Costa, C. T., Offringa, R., Fett-Neto, A. G. (2020). The role of auxin transporters and receptors in adventitious rooting of Arabidopsis thaliana pre-etiolated flooded seedlings. Plant Sci. 290, 110294. doi: 10.1016/j.plantsci.2019.110294
Dolan, W. L., Chapple, C. (2016). Conservation and divergence of mediator structure and function: insights from plants. Plant Cell Physiol 58, 4–21. doi: 10.1093/pcp/pcw176
Elmlund, H., Baraznenok, V., Lindahl, M., Samuelsen, C. O., Koeck, P. J. B., Holmberg, S., et al. (2006). The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 103, 15788–15793. doi: 10.1073/pnas.0607483103
Fichtner, F., Barbier, F. F., Kerr, S. C., Dudley, C., Cubas, P., Turnbull, C., et al. (2022). Plasticity of bud outgrowth varies at cauline and rosette nodes in Arabidopsis thaliana. Plant Physiol. 188, 1586–1603. doi: 10.1093/plphys/kiab586
Gutierrez, L., Bussell, J. D., Păcurar, D. I., Schwambach, J., Păcurar, M., Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Jaskolowski, A., Iñigo, S., Arellano, S. M., Arias, L. A., Fiol, D. F., Sede, A. R., et al. (2019). The MED30 subunit of mediator complex is essential for early plant development and promotes flowering in Arabidopsis thaliana. Development 146, dev175224. doi: 10.1242/dev.175224
Jia, K.-P., Luo, Q., He, S.-B., Lu, X.-D., Yang, H.-Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in arabidopsis. Mol. Plant 7, 528–540. doi: 10.1093/mp/sst093
Kuhn, B. M., Nodzyński, T., Errafi, S., Bucher, R., Gupta, S., Aryal, B., et al. (2017). Flavonol-induced changes in PIN2 polarity and auxin transport in the Arabidopsis thaliana rol1-2 mutant require phosphatase activity. Sci. Rep. 7, 41906. doi: 10.1038/srep41906
Kumar, K. R. R., Blomberg, J., Björklund, S. (2018). The MED 7 subunit paralogs of Mediator function redundantly in development of etiolated seedlings in Arabidopsis. Plant J. 96, 578–594. doi: 10.1111/tpj.14052
Li, J., Terzaghi, W., Gong, Y., Li, C., Ling, J.-J., Fan, Y., et al. (2020). Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 11, 1592. doi: 10.1038/s41467-020-15394-7
Pacurar, D. I., Perrone, I., Bellini, C. (2014). Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiologia Plantarum 151, 83–96. doi: 10.1111/ppl.12171
Ramasamy, S., Aljahani, A., Karpinska, M. A., Cao, T. B. N., Velychko, T., Cruz, J. N., et al. (2023). The Mediator complex regulates enhancer-promoter interactions. Nat. Struct. Mol. Biol. 30, 991–1000. doi: 10.1038/s41594-023-01027-2
Raya-González, J., Ortiz-Castro, R., Ruíz-Herrera, L. F., Kazan, K., López-Bucio, J. (2014). PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in arabidopsis. Plant Physiol. 165, 880–894. doi: 10.1104/pp.114.239806
Reeves, W. M., Hahn, S. (2003). Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23, 349–358. doi: 10.1128/MCB.23.1.349-358.2003
Rigas, S., Ditengou, F. A., Ljung, K., Daras, G., Tietz, O., Palme, K., et al. (2013). Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol. 197, 1130–1141. doi: 10.1111/nph.12092
Schumacher, I., Ndinyanka Fabrice, T., Abdou, M.-T., Kuhn, B. M., Voxeur, A., Herger, A., et al. (2021). Defects in cell wall differentiation of the arabidopsis mutant rol1-2 is dependent on cyclin-dependent kinase CDK8. Cells 10, 685. doi: 10.3390/cells10030685
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sinha, S. K., Kumar, K. R. R. (2021). “Function of mediator in regulating salicylic acid mediated signaling and responses in plants,” in Jasmonates and Salicylates Signaling in Plants. Eds. Aftab, T., Yusuf, M. (Springer International Publishing, Cham), 265–279. doi: 10.1007/978-3-030-75805-9_13
Soutourina, J. (2018). Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274. doi: 10.1038/nrm.2017.115
Tebbji, F., Chen, Y., Richard Albert, J., Gunsalus, K. T. W., Kumamoto, C. A., Nantel, A., et al. (2014). A functional portrait of Med7 and the mediator complex in candida albicans. PloS Genet. 10, e1004770. doi: 10.1371/journal.pgen.1004770
Vandenbussche, F., Verbelen, J.-P., van der Straeten, D. (2005). Of light and length: Regulation of hypocotyl growth inArabidopsis. Bioessays 27, 275–284. doi: 10.1002/bies.20199
Warfield, L., Donczew, R., Mahendrawada, L., Hahn, S. (2022). Yeast Mediator facilitates transcription initiation at most promoters via a Tail-independent mechanism. Mol. Cell 82, 4033–4048.e7. doi: 10.1016/j.molcel.2022.09.016
Xu, D., Jiang, Y., Li, J., Lin, F., Holm, M., Deng, X. W. (2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 113, 7655–7660. doi: 10.1073/pnas.1607687113
Xu, R., Li, Y. (2012). The Mediator complex subunit 8 regulates organ size in Arabidopsis thaliana. Plant Signaling Behav. 7, 182–183. doi: 10.4161/psb.18803
Yang, Y., Liu, H. (2020). Coordinated shoot and root responses to light signaling in arabidopsis. Plant Commun. 1, 100026. doi: 10.1016/j.xplc.2020.100026
Yao, X., Fang, K., Qiao, K., Xiong, J., Lan, J., Chen, J., et al. (2024). Cooperative transcriptional regulation by ATAF1 and HY5 promotes light-induced cotyledon opening in Arabidopsis thaliana. Sci. Signal. 17, eadf7318. doi: 10.1126/scisignal.adf7318
Yu, Y., Wang, J., Zhang, Z., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PloS Genet. 9, e1004025. doi: 10.1371/journal.pgen.1004025
Zeng, Y., Schotte, S., Trinh, H. K., Verstraeten, I., Li, J., Van De Velde, E., et al. (2022). Genetic dissection of light-regulated adventitious root induction in arabidopsis thaliana hypocotyls. IJMS 23, 5301. doi: 10.3390/ijms23105301
Zhang, X., Huai, J., Shang, F., Xu, G., Tang, W., Jing, Y., et al. (2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174, 2487–2500. doi: 10.1104/pp.17.00418
Zhao, L., Peng, T., Chen, C.-Y., Ji, R., Gu, D., Li, T., et al. (2019). HY5 interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis. Plant Physiol. 180, 1450–1466. doi: 10.1104/pp.19.00055
Keywords: mediator, Arabidopsis thaliana, MED7, hyponastic cotyledons, elongated hypocotyls, modified root architecture
Citation: Kumar KRR, Blomberg J and Björklund S (2025) The role of mediator subunit MED7 in Arabidopsis development. Front. Plant Sci. 16:1542950. doi: 10.3389/fpls.2025.1542950
Received: 10 December 2024; Accepted: 20 February 2025;
Published: 07 March 2025.
Edited by:
Giovanna Serino, Sapienza University of Rome, ItalyReviewed by:
Javier Raya Gonzalez, Michoacana University of San Nicolás de Hidalgo, MexicoCopyright © 2025 Kumar, Blomberg and Björklund. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Björklund, c3RlZmFuLmJqb3JrbHVuZEB1bXUuc2U=
†ORCID: Stefan Björklund, orcid.org/0000-0003-1181-0415
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.