
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 February 2025
Sec. Plant Pathogen Interactions
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1540003
This article is part of the Research TopicImproving Legume Crops for Resistance to Pathogens and PestsView all 5 articles
Background: The family of membrane-bound fatty acid desaturase (FAD) genes play a vital role in plant growth, development, and stress responses. The seed-borne pathogen Fusarium fujikuroi causes seed decay disease during pre-harvest and post-harvest stages of soybean, leading to a significant reduction in yield and quality. Therefore, it is very meaningful to characterize the diversity and function of the GmFAD gene family in soybean and to elucidate their roles in seed resistance to F. fujikuroi.
Results: In this study, 30 full-length GmFAD genes were identified from the soybean genome. A range of analysis was conducted to characterize gene and protein structures, chromosomal locations, conserved motif and conserved structural domains, and results showed that GmFAD genes were clustered into seven subfamilies (FAB2, ADS, SLD, DES, FAD6, FAD2, FAD3/7/8), which is also supported by phylogenetic analysis. The diversity and expansion of the GmFAD gene family were mainly caused by segmental duplication, and their encoding proteins were observed to locate in chloroplast or endoplasmic reticulum. The promoters of GmFAD genes contained a set of cis-acting elements in response to plant hormone, defense and stress, light, and plant growth and development, indicating these genes have the complex expression regulation and diverse functions. Gene ontology (GO) and KEGG enrichment pathway analyses showed that GmFAD genes were closely related to the biosynthesis and metabolism of lipid and unsaturated fatty acids (UFAs). In addition, the expression of GmFADs was significantly changed in soybean seeds when challenged by the seed decay pathogen F. fujikuroi. Specifically, GmFAB2.1/2.2, GmFAD3.3/3-2B/7-1//8-2, and GmFAD2.3/2.5 genes displayed distinct temporal expression patterns in the resistant ND25 and susceptible CX12, highlighting their potential roles in soybean resistance against F. fujikuroi infection.
Conclusion: Our findings contribute to a deeper understanding of the GmFAD gene family and their intricate roles in soybean resistance against the seed-borne pathogen F. fujikuroi. Moreover, several distinct genes provide valuable candidates for further application in soybean resistant breeding.
Unsaturated fatty acids (UFAs) have been increasingly recognized as significant components in the plant defense against a range of biotic and abiotic stresses (He et al., 2018). Fatty acid desaturases (FADs) are key enzymes in the biosynthesis of UFAs, catalyzing the insertion of double bonds at specific sites in fatty acid chains, thereby enhancing the fluidity and structural integrity of cell membranes (Dong et al., 2016; Feng et al., 2017). Generally, plant FADs can be categorized into soluble desaturases and membrane-bound desaturases based on their solubility (Nayeri and Yarizade, 2014). Among them, stearoyl-ACP desaturase, namely FAB2, known as the typical soluble FAD located in the plastid matrix, is responsible for introducing a double bond at the Δ9 position, and facilitating the conversion of stearic acid to oleic acid (Ha et al., 2010; Xue et al., 2018). Membrane-bound FADs are further divided into four distinct subfamilies based on their functions, including omega-6 desaturases (ω6, FAD2 and FAD6), omega-3 desaturases (ω3, FAD3, FAD7 and FAD8), FAD4, and DES/ADS/SLD (Hashimoto et al., 2008; Lou et al., 2014; Saini and Kumar, 2019). Generally, FADs in the same subfamily have the highly conserved amino acid sequences (Hashimoto et al., 2008). To date, FAD genes have been characterized in many plants, such as soybean (Glycine max) (Chi et al., 2011), tobacco (Nicotiana tabacum) (Sayanova et al., 1997), banana (Cheng et al., 2022), corn (Zea mays) (Mikkilineni and Rocheford, 2003), rice (Oryza sativa) (Wang et al., 2006), etc.
Numerous studies have demonstrated that FADs play crucial roles in plant stress tolerance, such as high and low temperatures, drought, salinity, and heavy metal exposure (Xue et al., 2018). The MaFADs expression in banana are significantly activated in response to high and low temperature (Cheng et al., 2022). Similarly, a significant up-regulation of CsSLD3 and CsSLD4 is also observed under cold stress in Camellia sinensis (Jin et al., 2024). Overexpression of antisense AtFAD2 results in a decrease tolerance to drought and salt stresses in Arabidopsis (Im et al., 2002), when the transcript of the soybean homologous GmFAD2-2C was accumulated to increase in pods grown in cool conditions rather than those in warmer conditions (Li et al., 2007). AtFAD3 or AtFAD8 expressed in transgenic tobacco enhanced the resistance to drought and osmotic stresses (Zhang et al., 2005). On the other hand, since fatty acids as the main source of organic carbon can be delivered to the fungi by the host plants, thus FADs as pivotal agents in plant lipid metabolism also modulate the plant-pathogen interaction (Jiang et al., 2017; Luginbuehl et al., 2017). Researches have demonstrated that triene fatty acids originating from chloroplasts are involved in host resistance at the infection initial stage of pathogen (Chandra-Shekara et al., 2007). Transient silencing of the FAD2 homologous gene in wheat (Triticum Aestivum) increased its susceptibility to powdery mildew (Jiang et al., 2009). Conversely, the inhibition of OsFAD7 and OsFAD8 have been found to enhance the transgenic rice resistance against Magnaporthe grisea (Yara et al., 2007). In addition, the SSI2 gene encoding a stearoyl-acyl carrier protein-desaturase (SACPD) also participates in the pathogen resistance, and knockdown of OsSSI2 markedly increased accumulation of endogenous free salicylic acid (SA) and enhanced rice resistance to the fungus Magnaporthe grisea and bacterium Xanthomonas oryzae pv. oryzae (Li et al., 2011). Similarly, overexpression of the Arabidopsis ssi2 mutant TaSSI2 restored its resistance to powdery mildew fungi (Song et al., 2013). In soybean, the fatty acid composition in soybean tissues is often responsive to pathogen attack (Upchurch, 2008), and fatty acids and fatty acid-derived compounds act as signals of defense gene expression (Upchurch, 2008). Evidence suggests that the level of stearic acid and oleic acid are critical for defense against pathogens in soybean as they have been shown to be in Arabidopsis (Kachroo and Kachroo, 2009). Moreover, the oleate and linoleate content of soybean seeds appears to influence the course of seed colonization by Cerospora kikuchii and Diaporthe phaseolorum (Xue et al., 2008). Silencing of GmSACPDs confers soybean resistance to Pseudomonas syringae pv. glycinea (Kachroo et al., 2008). Thus, FADs as crucial regulatory components are capable of reacting to and being linked with various stress-induced damages in plants.
Soybean (Glycine max) is one of the largest oilseed crop worldwide and rich in high-quality vegetable protein and unsaturated fatty acids. However, soybean seed quality and yield are often affected by various seed-borne diseases (Wrather et al., 2003). Specially, seed decay emerges as one of the most damaging seed-borne diseases during the pre- and post-harvest stages of soybean, and this disease often leads to substantial yield losses, poor seed quality and nutrients, and reduced seed germination and vigor (Chang et al., 2020a; Xu et al., 2023). Some infected seeds can even become important carriers of diseases, facilitating the spread of these pathogens over extensive distances (Lv and Sun, 2007). The necrotrophic fungus Fusarium fujikuroi has previously been reported to cause soybean root rot (Chang et al., 2020b), and it was also identified as the causal agents of pod blight and seed decay (Suga et al., 2018; Chang et al., 2022). The fungus is capable of producing a variety of secondary metabolites, such as fumonisins and gibberellins, which threaten the health of humans and livestock (Zhang et al., 2021). However, there is little information on F. fujikuroi-induced seed responses in soybean. Recently, our study showed that the soybean cultivar CX12 exhibited susceptibility to F. fujikuroi, whereas the cultivar Nandou25 had high resistance to F. fujikuroi. Given the important role of FAD genes in enhancing plant resistance against various diseases, it become meaningful to explore the contribution of GmFAD to soybean seed decay.
To date, the GmFAD gene family has been reported in Glycine max through genomic and transcriptomic analyses (Chen et al., 2016; Liu et al., 2017). A total of 75 FAD genes have already reported from the genomes of different soybean species, with 23 FAD genes found in Glycine max var. Williams 82 (Chen et al., 2016). However, the diversity and function of GmFAD genes in soybean remain largely uncovered. This study aimed to analyze the functions of the soybean FAD gene and clarify the roles of GmFAD in soybean resistance to the seed-borne F. fujikuroi. By performing a genome-wide analysis using the latest genomic data from Glycine max var. Williams 82, we systematically characterized and functionally annotated the GmFAD gene family according to their physicochemical properties, gene structures, and promoter motifs. Furthermore, we analyzed the expression profiles of representative FAD genes using qRT-PCR during F. fujikuroi infection in soybean seeds. This study aids in identifying candidate genes that may improve Fusarium resistance in soybean.
Full-length genes of 30 fatty acid desaturase (GmFADs) were predicted from the genome of Glycine max var. Williams 82 (Glycine max Wm82.a4.v1) using HMM search, and were listed in Supplementary Table S1. The identified proteins corresponding to the GmFAD gene family had an amino acid sequence length from 235 aa (GmFAD6.2) to 453 aa (GmFAD7-2). The relative molecular mass of these proteins varied between 26,800.2 Da (GmFAD6.2) and 51,550.41 Da (GmSLD1.2). The isoelectric points (pI) of the proteins were distributed within a range of 5.94 (GmFAB2.1) to 9.51 (GmFAD5.1), with the majority possessing pI values above 7. The lipolysis index values ranged from 79.26 (GmSACPD) to 94.62 (GmSLD1.4). The instability coefficients of proteins were found to vary between 30.72 and 48.67, with GmFAD2.3 protein being the most stable and GmFAD6.1 protein the least stable among these members identified. Out of the 30 family members, 21 members were classified as hydrophilic proteins, whereas the others were hydrophobic (Supplementary Table S2).
The subcellular localization analysis predicted that proteins GmFAD2, GmDES, and GmSLD were localized on endoplasmic reticulum, whereas GmFAD6 and GmFAB2 proteins localized on chloroplasts. The members of GmFAD5 and GmFAD3 along with their isozymes GmFAD7/FAD8 proteins were found in both endoplasmic reticulum and chloroplasts as detailed in Supplementary Table S2. These results suggest that the subcellular location diversity of these proteins could be related to multiple functions.
To determine the evolutionary relationships of FAD proteins among A. thaliana (At), rice (Os), and soybean (Gm), a phylogenetic tree was constructed using the neighbor-joining (NJ) method with p-distance model using amino acid sequences of GmFADs (30), AtFADs (24), and OsFADs (18) (Supplementary Table S3). As depicted in Figure 1A, all FAD proteins were classified into two distinct clusters: soluble (FAB2) and membrane-bounding FAD proteins (ADS, SLD, DES, FAD6, FAD2, and FAD3/FAD7/8).
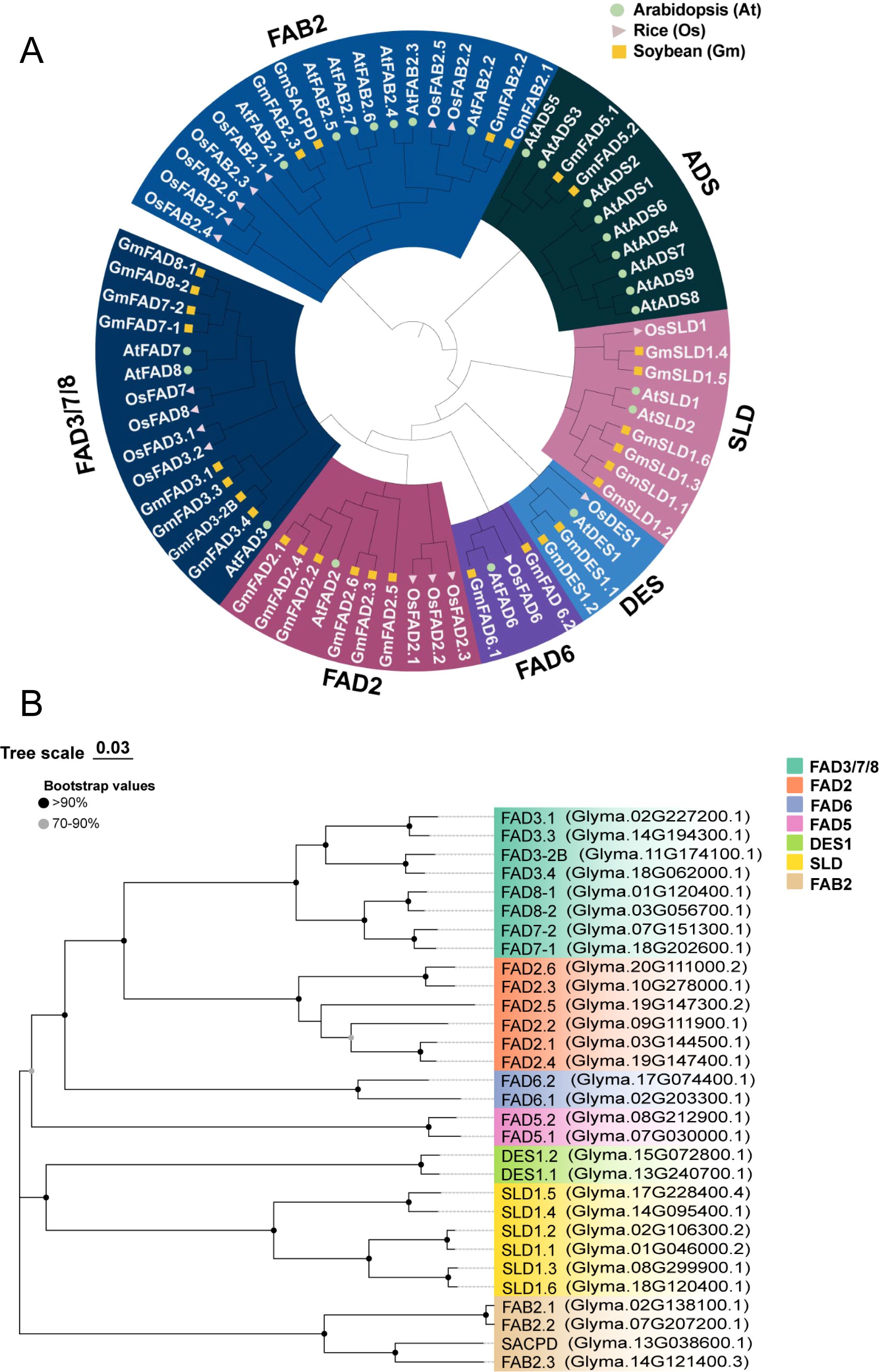
Figure 1. Phylogenetic analysis of FADs proteins. (A) Phylogenetic relationship of FAD proteins from Arabidopsis (At), rice (Os) and soybean (Gm). The amino acid sequences of GmFAD, AtFAD and OsFAD were compared using ClustalW and a phylogenetic tree was constructed using the neighbor-joining (NJ) method with p-distance model by MEGA7.0. The bootstrap support values were calculated from 1000 replicates. FAD proteins are divided into seven subfamilies as FAD3/7/8, FAD2, FAD6, DES, ADS, SLD and FAB2, which are indicated by different colors. (B) Phylogenetic relationships of GmFAD protein in soybean.
FAB2 subfamily represents the only soluble desaturase identified so far. In this study, both A. thaliana and rice were found to contain seven homologous proteins, respectively (Figure 1A). In contrast, it was observed that soybean possesses four FAB2, namely three GmFAB2 and one GmSACPD, and among them, two alleles GmFAB2.1 and GmFAB2.2 were clustered together, when GmFAB2.3 aligned with GmSACPD in a different branch (Figure 1B). Compared FAB members in three species, except for OsFAB2.1, AtFAB2.1 and OsFAB2.1, most FAB members of soybean and A. thaliana were closely clustered together but OsFAB in rice was located at the base position of this subfamily. This suggests that FAB2 in rice may have evolved earlier and could have been divided into monocotyledonous and dicotyledonous taxa.
As shown in Figure 1A, proteins encoding membrane-bound desaturases were categorized into six distinct subfamilies. Among them, the ADS subfamily was further classified into two groups, composed of nine ADS from A. thaliana along with their two homologous FAD5 from soybean. Interestingly, GmFAD5.1 and GmFAD5.2 were closely grouped with AtADS3 and AtADS5 as a small branch, indicating that GmFAD5 might evolved from AtADS. Furthermore, no members from rice were identified in the ADS subfamily, indicating that it might have undergone evolutionary loss in rice or were transmitted to dicotyledons (A. thaliana and soybean) following their divergence from the last common ancestor.
The SLD subfamily comprised six GmSLD, two AtSLD, and one OsSLD, all of which encode sphingolipid Δ7 desaturases. Except for OsSLD1 was grouped with both GmSLD1.4 and GmSLD1.5 as a separate branch, the SLD members from each plant species were distinctly clustered together, indicating a closer genetic relationship among them. The DES subfamily, which is responsible for sphingolipid Δ4 desaturases, had the members including OsDES1, AtDES1, GmDES1.1 and GmDES1.2, with OsDES1 in rice localized at the basal position of the subfamily. This suggests GmDES1 in soybean as well as AtDES1 in A. thaliana had more close relationship as the dicotyledonous plants, but they diverged from OsDES1 from the last common ancestor.
Both FAD6 and FAD2 subfamilies encode Δ12 desaturases but were differently localized in the subcellular organelles. In the plastidial FAD6 subfamily, GmFAD6.2 notably occupied at a basal position within this subfamily. In the microsomal FAD2 subfamily, three rice OsFAD2 were clustered together, while six GmFAD2 and one AtFAD2 grouped in another branch, indicating this family has diverged into monocotyledonous and dicotyledonous groups from the same ancestor. Additionally, in the FAD3/FAD7/FAD8 subfamily, eight FADs from soybean (four GmFAD3, two GmFAD7 and two GmFAD8), three from Arabidopsis, and four from rice, were clustered together and encoded their corresponding microsomal or plastidial ω3 desaturases.
As shown in Figure 2, different GmFAD genes were located in different soybean chromosomes. Except for the chromosomes 4, 5, 6, 12, and 16, the identified 30 GmFAD genes in soybean were found to distribute on the other 15 of the 20 soybean chromosomes. The maximum number of GmFAD genes were located on chromosome 2 as four genes. There were three genes located on the chromosomes 7, 14, and 18, respectively, while only one or two genes were located on other chromosomes. In addition, different gene members of each subfamily were mostly distributed in different chromosomes except that GmFAD2.4 and GmFAD2.5 was clustered in a small region of chromosome 19. This suggests that GmFAD genes are broadly dispersed throughout the soybean genome, and they might originate from diverse ancestors.
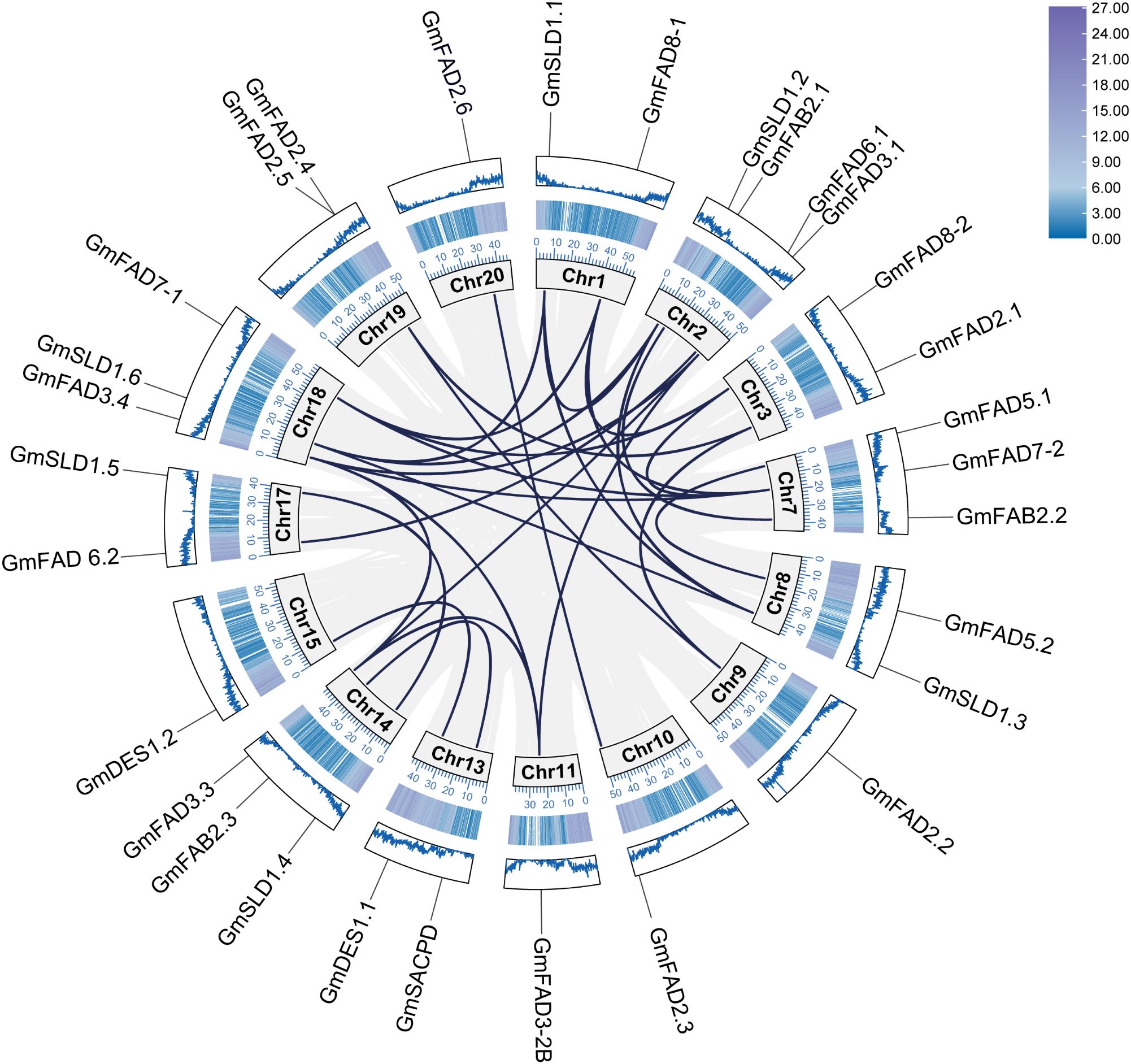
Figure 2. Covariance analysis of GmFAD gene family. Rectangles indicate chromosomes, and the location of the GmFAD genes on the chromosome is marked using the GmFAD name. Each gray and dark blue curves indicate all covariate gene pairs on the chromosome and gene duplication events of GmFAD, respectively.
The diversity and expansion of gene families often arise from crosstalk and segmental duplication events, and gene duplication serves as a key mechanism for enhancing plant genetic diversity and the generation of novel genes. In this study, covariance analysis based on MCScanX was performed to investigate gene duplication events in the GmFAD gene family, and results showed that a large number of GmFAD genes had covariance between and within soybean chromosomes, and the most GmFAD genes with covariance were localized on the chromosome 19. As shown in Table 1, both tandem and segmental duplication can be observed in the GmFAD family. Except for GmFAD2.4 and GmFAD2.5 on the chromosome 19 were caused by tandem duplication, a total of 29 segmental duplication events were identified (Table 1). Among them, the ω-3 desaturase subfamily (GmFAD3/GmFAD7/GmFAD8) had the maximum duplicated gene pairs as 13 pairs followed by the GmSLD subfamily with seven pairs, while the ω-6 desaturase subfamilies (GmFAD2 and GmFAD6) had five duplicated gene pairs. The subfamilies of GmFAB2, GmDES and GmFAD5(ADS) had less duplicated gene pairs than other subfamilies in our study. Thus, the role of segmental duplication events plays a prominent role in increasing the genetic diversity of soybean GmFAD gene families rather than tandem duplication.
To assess the selection of duplicated GmFAD gene pairs, we calculated the substitution ratios between non-synonymous and synonymous (Ka/Ks) based on a whole genome analysis of gene duplication (Table 1). In soybean, most Ka/Ks ratios for the GmFAD gene replication pairs were found to be less than 1, indicating that these genes are relatively conserved during the evolutionary process and have been subjected to purifying selection pressure to maintain gene functions and species stability. However, three gene pairs, such as GmFAD7-2/GmFAD3.4, GmFAD2.1/GmFAD2.2, and GmFAD2.2/GmFAD2.5, exhibited a Ks value greater than 1, and it indicates that a positive selection pressure occurs to produce new protein functions aiming to promote gene evolution and the adaptive changes of species.
Analysis of the exon/intron structures of 30 GmFAD genes identified in soybean (Figure 3A, Supplementary Table S4), showed that the gene structure of different GmFAD subfamily was generally variable. Both GmFAD3/FAD7/FAD8 and GmFAD6 gene subfamilies had more than 6 exons, especially GmFAD6.1 possessed ten exons. GmFAD5 subfamily had five exons as compared to GmDES and GmFAB2 subfamilies which contained two to three exons. In addition, members in the GmFAD and GmFAB subfamilies had different number of exons, for example, GmFAD6.2 and GmFAD6.1 had four and ten exons, respectively. Interestingly, GmFAD2 and GmSLD had the simplest structures with only one exon. Furthermore, most GmFAD genes within the same subfamily exhibited high similarity in their exon/intron patterns as compared to those in different subfamilies, thus the exon/intron distribution could provide strong supports for the phylogenetic classification of GmFAD proteins above.

Figure 3. Analysis of gene structure, conserved motifs and conserved structural domain interactions in soybean GmFAD proteins. (A) Exons and introns of GmFAD genes. Green boxes and blue boxex represented exons and untranslated region (UTR), respectively. Grey lines meaned introns. (B) Conserved motifs of soybean GmFAD gene family and their interacted with conserved structural domains. Motifs 1-20 represent conserved motifs. Membrane-FADS-like PLN02505, PLN02598, PLN02579, Δ6-FADS-like, Cyt-b5, PLN03198 and PLN00179 belong to conserved structural domains.
Furthermore, the gene motifs were analyzed in this study, and total 20 motifs were identified in GmFAD genes (Figure 3B). Evaluation of motifs 1-20 using the Pfam database (http://pfam.xfam.org) showed that motifs 2, 3, 6, 10, 11, 12, 16, 19, and 20 corresponded to membrane-FADS-like superfamily structural domains, while other three protein conserved structural domains PLN02505, Δ6-FADS-like, and PLN02598 were also identified as the membrane-FADS-like superfamily. Motifs 2, 3, and 10 were observed in all members of the ω-3 subfamilies (GmFAD3/GmFAD7/GmFAD8), while two unique motifs 16 and 20 were found in the GmFAD7 and GmFAD8 subfamilies. All the GmFAD2 subfamily members contained motifs 4, 6, and 11 except that GmFAD2.5 lacked motif 4. Motif 17 was present only in the GmFAD6 subfamily, but this motif is not functional. In the GmFAB2 subfamily, motifs 5, 14, 15 and 20 corresponded to the acyl-[acyl-carrier protein] desaturase structural domain (PLN00179), whereas Motif 11 was a domain of unknown function (with PLN00179 domain) and only existed in the branch of GmFAB2.3 and GmSACPD as compared to GmFAB2.1 and GmFAB2.2. The members of GmFAD5(ADS), GmDES, and GmSLD subfamilies comprised two, two, and four motifs, respectively. Notably, motif 9, as a component of the Cyt-b5 domain, was uniquely identified in the GmSLD subfamily. In short, members with similar conserved motifs and gene structures clustered together in the GmFAD gene family. The motif distribution patterns of each subfamily are similar, whereas differences between subfamilies may be related to subfamily functional convergence.
To elucidate the three-dimensional conformation of the GmFAD proteins, homology modeling method was adopted and homology modeling was performed using Swiss-Model online software. As shown in Figure 4, GmFAD primarily consisted of α-helixs, irregular coils, and β-turns, and proteins clustered into the same clade exhibited analogous 3-dimensional (3D) structures. GmFAD3.3 within the ω-3 desaturase subfamilies and GmFAD2.6 within the ω-6 desaturase subfamilies exhibited distinct characteristics compared to GmFAD proteins from other subfamilies, indicating that there exists a balance between conservation and divergence within the GmFAD proteins.
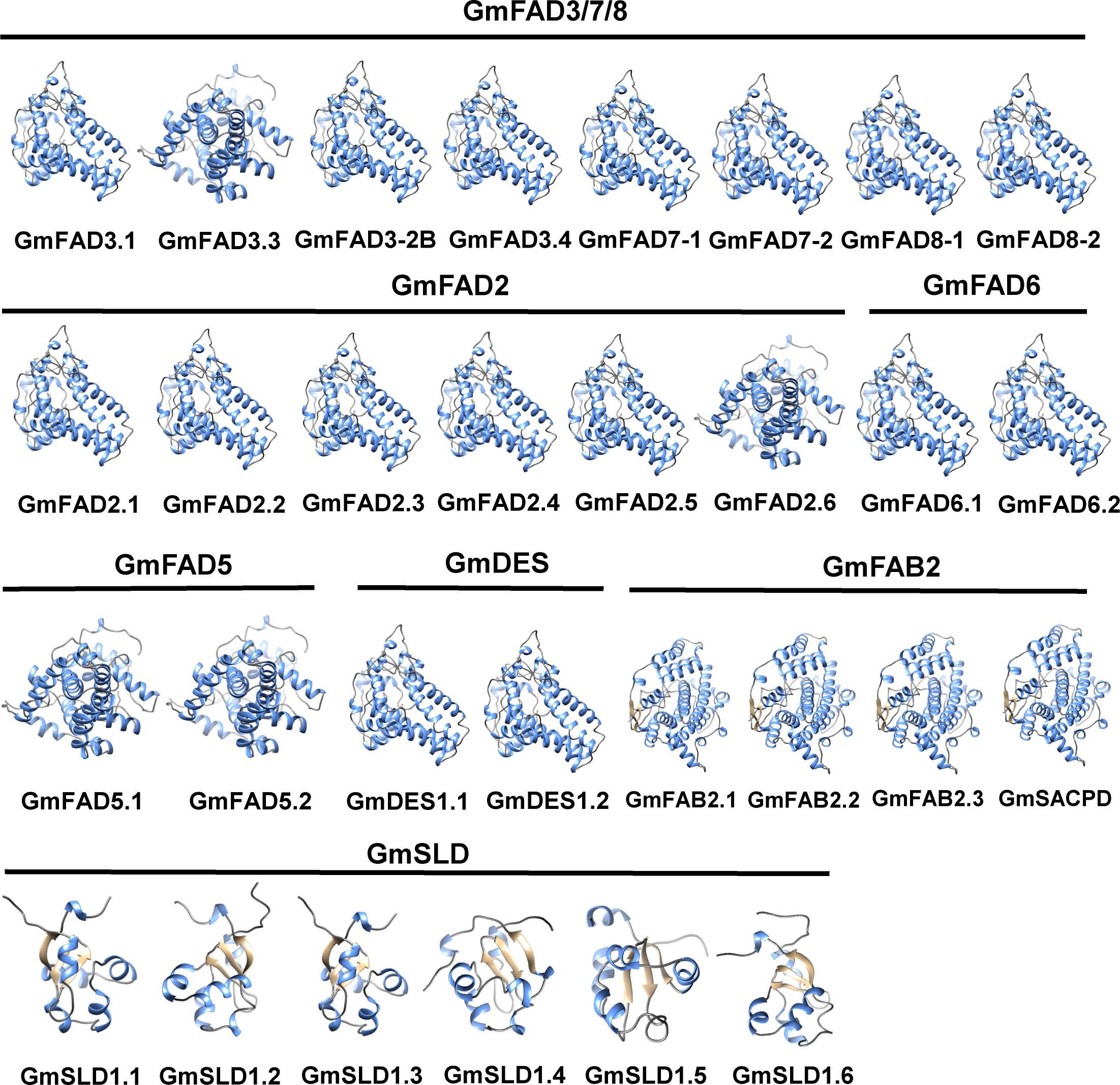
Figure 4. Three-dimensional conformation prediction of soybean GmFAD proteins. The coil, α-helix, and strand were represented in green, gray and yellow, respectively.
Promoters located in the upstream regions of genes are crucial in gene expression regulation involving in plant growth and development as well as environmental adaptation. To understand the roles of GmFAD genes and the precise regulation of gene expression, cis-acting elements within the promoter regions were analyzed (Figure 5, Supplementary Table S5). The results showed that the promoter region of the GmFAD genes contained three distinct types of cis-elements. The first type of cis-elements was related to plant growth and development, including light responsiveness, circadian control, the differentiation of fenestrated chloroplasts, and cell cycle regulation. The second type was responsible for stress responses, including those triggered by methyl jasmonate (MeJA), abscisic acid (ABA), salicylic acid (SA), GA, growth hormone, low-temperature, drought, defense and stress responsiveness. The third category is associated with specific biological processes, such as those involved in endosperm expression, zein metabolism regulation, flavonoid biosynthesis regulation, and cell cycle regulation.
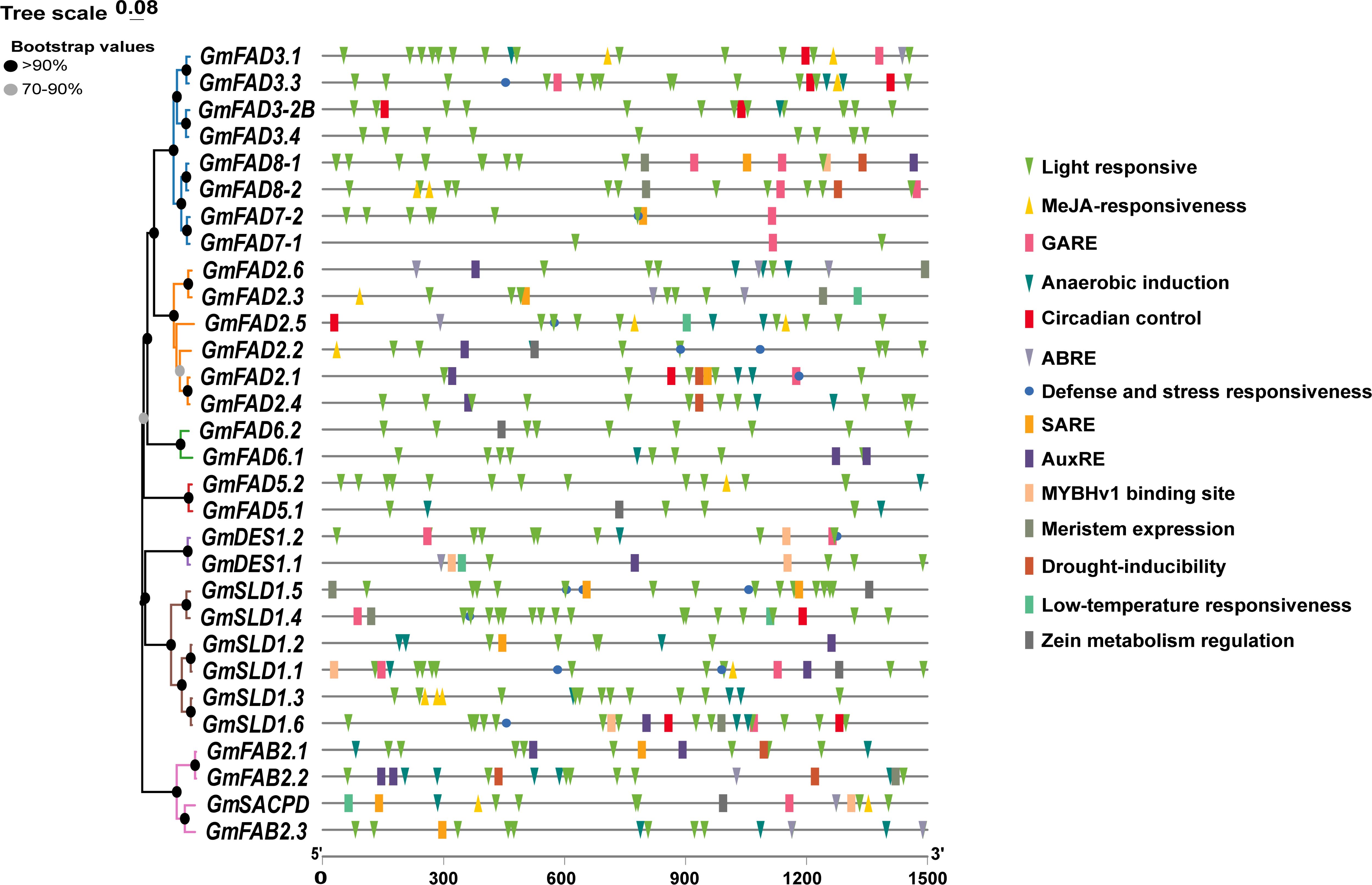
Figure 5. Map of cis-acting elements of the GmFAD gene promoter. Promoter sequences (−1500 bp) of GmFAD genes are speculated on PlantCARE. The upstream length to the translation start site can be deduced on the basis of the scale at the bottom.
As illustrated in Table 2, the promoters of most GmFAD genes contained cis-elements related to phytohormone responses, including ABRE, TGA, P-box, TATC-box, GARE-motif, CGTCA-motif, TGACG, AuxRR-core, and TGA-box, which correspond to ABA, SA, GA, MeJA, and Auxin signaling, respectively. Additionally, TC-rich repeats, which often function on the regulation of defense and stress responses, have been identified within the promoter sequences of 10 GmFAD genes (GmFAD3.3, GmFAD7-2, GmFAD2.1, GmFAD2.2, GmFAD2.5, GmDES1.2, GmSLD1.1, GmSLD1.4, GmSLD1.5, and GmSLD1.6). Moreover, the distribution patterns of TC-rich repeats within the GmFAD2 subfamily were similar to those in the GmSLD subfamily, but it is notably absent in the GmFAD6, GmFAD5, and GmFAB2 subfamilies.
All members of the GmFAD gene family possess abundant light-responsive cis-elements at the start codon upstream, with the detection of 17 such regulatory elements, such as Box4, G-Box, and GT1-Motif and others, suggesting that light signals play crucial roles in the accumulation of soybean fatty acid desaturases. Overall, these findings illuminate the potential functions of GmFAD genes in facilitating plant growth and development, enhancing stress responses, and modulating hormone signaling.
To gain a deeper understanding of the functional pathways associated with GmFAD genes, we conducted Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis to explore the potential functions of GmFAD. The results showed that GmFAD was mainly enriched in three GO terms: “molecular function”, “cellular component” and “biological process” (Figure 6A). Notably, the GmFAD gene family were significantly implicated in oxidoreductase activity, facilitating the transfer of electrons between paired donors and binding or reduction of molecular oxygen, and GmFAD genes also exhibited acyl-acyl-carrier-protein desaturase activity. Furthermore, GmFAD genes showed significant enrichment in membrane components, particularly endoplasmic reticulum membrane network, suggesting their potential involvement in the stability of membrane assembly. Additionally, both GO and KEGG annotations revealed that GmFAD genes were significantly involved in the biosynthetic and metabolic pathways of lipids and fatty acids (Figure 6B). In summary, GmFAD gene family participate in the biosynthesis and metabolism of fatty acid and lipid, and they also are responsible for redox and desaturation activities.
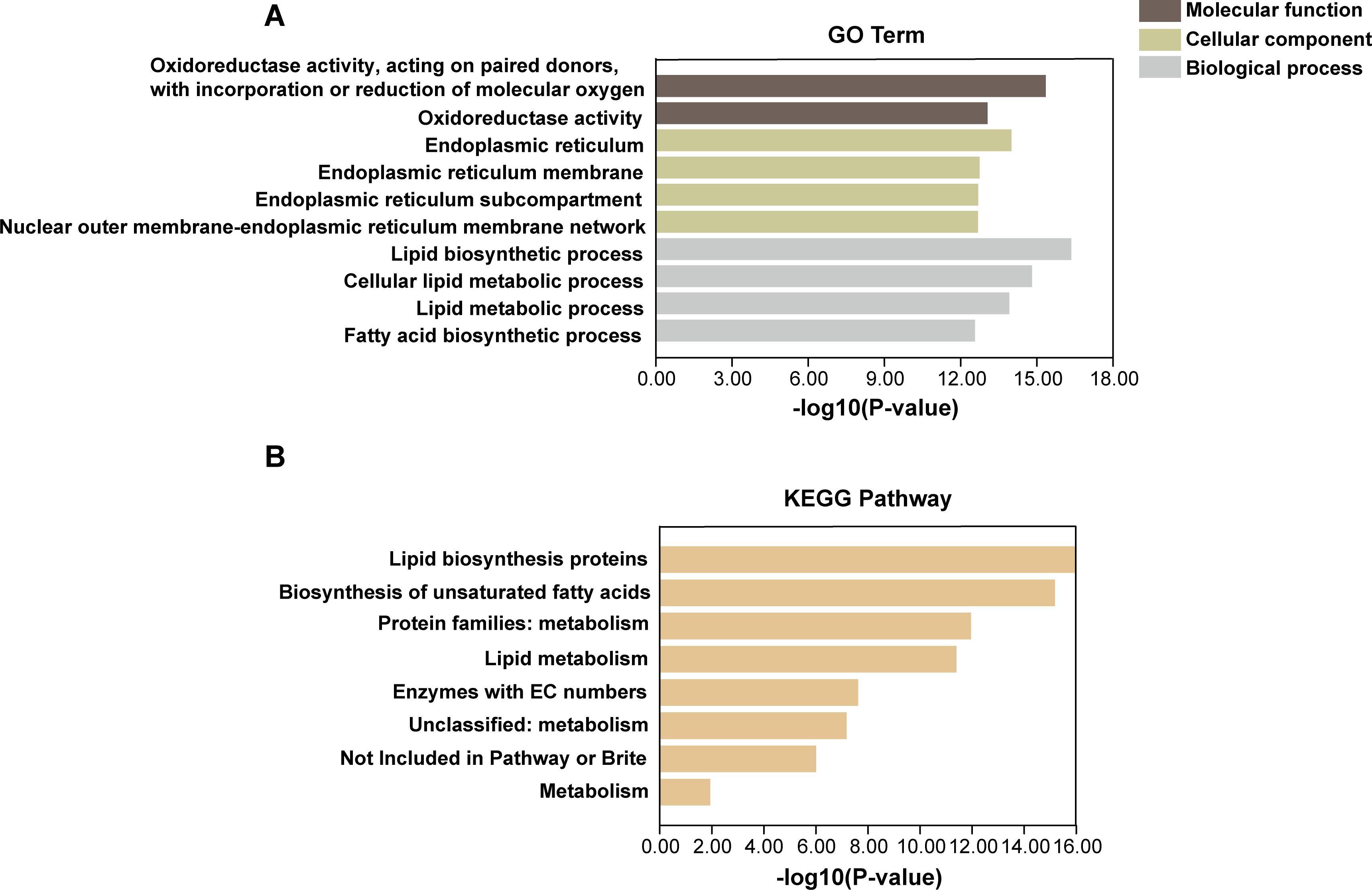
Figure 6. GO functional annotation and KEGG pathway enrichment analysis of GmFAD gene family. (A) Gene ontology (GO) analysis of the GmFAD gene family. (B) The KEGG pathway analysis of the GmFAD gene family. Both GO enrichment and KEGG pathway enrichment analysis were performed using TBtools, respectively. A total of 10 GO terms and 10 KEGG pathways with the lowest P-value were listed. The horizontal axis shows -log10(P-value), and the larger P-value means higher statistical significance.
To investigate the regulation of GmFAD genes in soybean seed resistance to F. fujikuroi, the dominant seed decay pathogen, we analyzed expression patterns of 16 GmFAD genes in soybean seeds of susceptible cultivar (CX12) and resistant cultivar (ND25) after F. fujikuroi inoculation (Figure 7). As shown in Figure 7, all representative GmFAD genes belonging to ω-3 and -6 desaturase subfamilies were significantly induced by F. fujikuroi infection but their expression patterns differed in the resistant ND25 and susceptible CX12. Most genes in the susceptible CX12 were significantly up-regulated at much earlier infection (12 hpi), whereas they were strongly induced at 48 hpi in the resistant ND25. In particular, relative expression level of five genes including GmFAD8.2, GmFAD3.3, GmFAD7-1, GmFAB2.2, GmFAD2.3 and GmFAD2.5 were as higher as 6.0 fold at 48 hpi as compared to 0 hpi. In contrast, expression of GmFAB2.3 and GmSACPD was inhibited at the early infection of F. fujikuroi (6 hpi) in both cultivars, but their expression were more rapidly recovered and reached the peak at 12 hpi in the resistant ND25 rather than those in the susceptible CX12. Additionally, GmFAD2.4 were dramatically up-regulated by F. fujikuroi at 6 hpi and then decreased after 24 hpi in the susceptible CX12, but it had different expression pattern in the resistant ND25. Expression of GmFAD2.1 and GmFAD7.2 were weakly induced in the resistant ND25 upon F. fujikuroi infection but they were up-regulated differently in the susceptible CX12. This suggests that GmFAD genes play important roles in soybean seed resistance to F. fujikuroi infection.
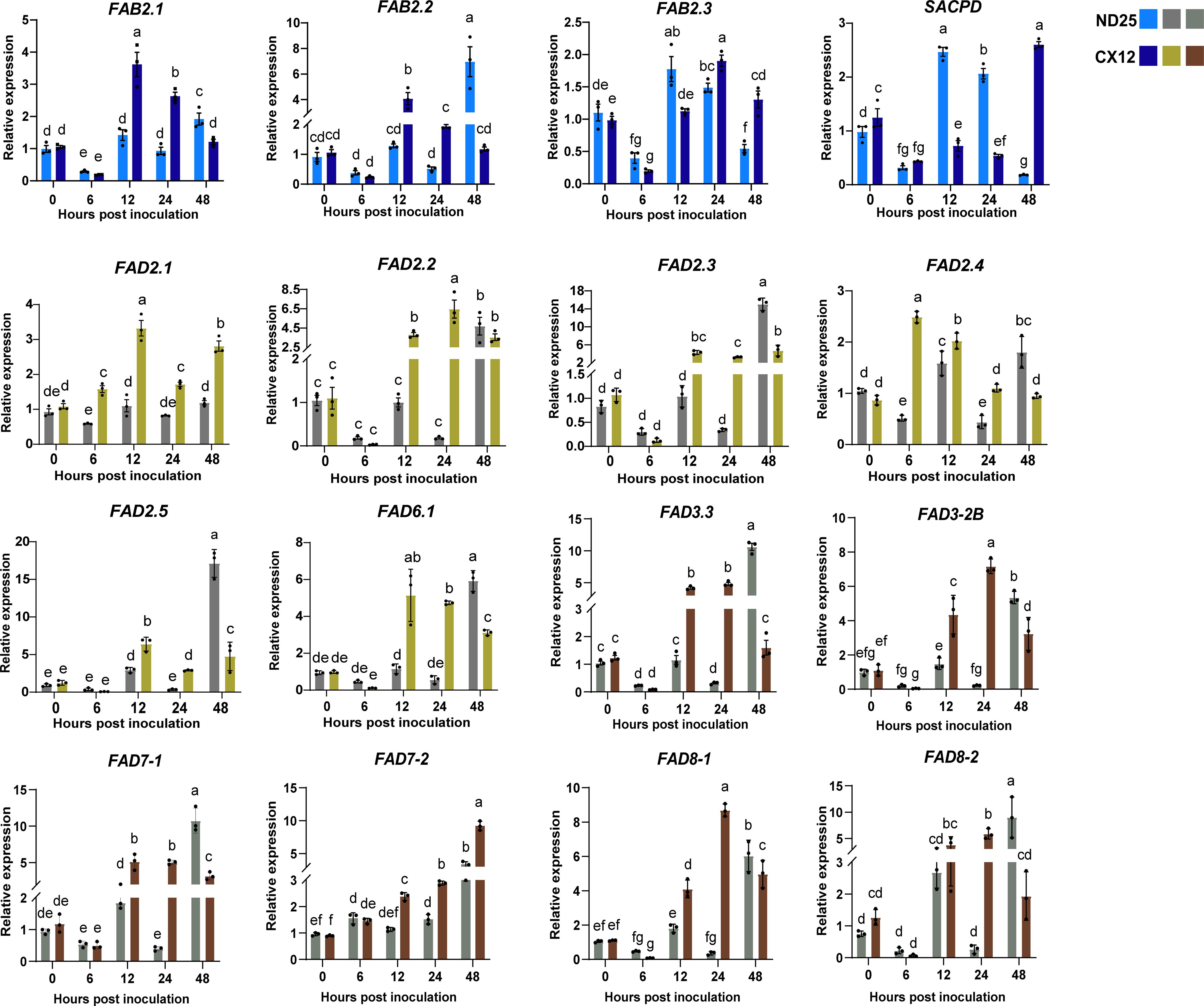
Figure 7. Expression patterns of GmFAD genes in soybean after F. fujikuroi inoculation. The expression level of GmFADs at 0 hour post inoculation was normalized as “1”. Quantitative RT-PCR was used to investigate the expression levels of each GmFAD gene upon F. fujikuroi infection from three biological and four technical replicates. Vertical bars indicated the standard errors of mean. Means denoted by the same lowercase letters when there was no significant difference at P < 0.05 as determined by Analysis of variance (ANOVA) and Duncan’s multiple range test (DMRT) using SPSS 24 software.
In recent years, the FADs gene family has been extensively studied in a wide range of plant species, and genome-wide analyses have revealed different members of FAD genes regarding plant species (Zhiguo et al., 2019). Previously, Chi et al. (2011) reported 41 GmFAD genes from an older version of the Williams 82 genome. Zhang et al. (2021) identified 23 GmFADs in Glycine max var. Williams 82 using the BLASTP method when compared to Chinese wild soybean and ancient polyploid soybean. In this study, we identified 30 full-length GmFADs genes from the soybean genome (Glycine max Wm82.a4.v1) using a Hidden Markov Model (HMM) of protein structural domains by HMM BLAST software. The difference in total gene numbers of Williams 82 may result from different genomic versions and identification criteria and parameters of the BLAST software. Phylogenetic analysis based the amino acid showed 30 GmFAD proteins in this study were categorized into seven subfamilies, including FAD3/7/8, FAD2, FAD6, FAD5(ADS), DES, and SLD, and FAB2, and except for the ADS subfamily, other six subfamilies are basically consistent with those of A. thaliana and rice (Cheng et al., 2022), indicating that those subfamilies have a common ancestor before their divergence. For the ADS family, responsible for desaturating palmitic acid to palmitoleic acid (Laureano et al., 2021), GmFAD5 was clustered with eight AtADS genes, whereas the monocotyledons rice lost this subfamily. Similar results was also reported in banana genome previously (Cheng et al., 2022), and this suggests that the ADS family might be formed after the differentiation of monocotyledons and dicotyledons. In addition, compared to A. thaliana and rice, the soybean genome possesses a larger number of genes within the ω-3 and ω-6 subfamilies as well as the SLD subfamily, and this can be predicted that these subfamilies have undergone positive selection pressures, leading to the expansion of gene families. This expansion may have granted soybean greater functional redundancy, enhancing their adaptability and diversity in lipid biosynthesis pathways. As a result, this could improve their capacity to adapt to environmental changes. Additionally, our results also demonstrated that different members within the same GmFAD gene subfamily were located on different soybean chromosomes, which is consistent with the previous findings in sunflower (Laureano et al., 2021).
Previous studies have revealed that expansion of the FAD gene family is species-specific in different plants, and this expansion is often determined by gene duplication events (Johnson and Thomas, 2007). Gene duplication plays a crucial role in generating new genes and functions, and both segmental and tandem duplication can drive the emergence of novel gene families (Cannon et al., 2004). In the wheat genome, TaFAD gene pairs were generated from tandem and segmental duplication with the pair number of 26 and 126, respectively (Hajiahmadi et al., 2020). In the poplar genome, PtFAD genes had 16 segmental repeat events and one tandem repeat event, respectively (Wei et al., 2022). However, replication of FAD family in Brassica juncea is mostly identified as segmental replications (89%) (Xue et al., 2020). In our study, there were 29 pairs of segmental repeats and one pair of tandem repeats in soybean GmFAD genes. Thus, segmental duplication in FAD family amplification is more frequently and important than tandem duplication, and it might play a vital role in increasing the genetic diversity of soybean GmFAD gene family. In addition, the selection pressure analysis showed that most GmFAD genes have undergone purifying selection to maintain the functions of GmFAD genes and species stability in soybean. For example, the FAD2 gene has experienced several duplication events, and all members of the FAD2 gene family have diverged into monopolistic and dicotyledons clusters in their evolutionary history. In some dicotyledons, the FAD2 genes is divided into two branches as constitutive and specific expression. In soybean, multiple GmFAD2 copies, such as GmFAD2.1, GmFAD2.2, GmFAD2.3, GmFAD2.4, and GmFAD2.5, have already been reported (Chi et al., 2011). Among them, GmFAD2.2 and GmFAD2.3 are constitutively expressed in both vegetative tissue and developing seeds, whereas two alleles GmFAD2-1A and GmFAD2-1B are specifically expressed in developing seeds and contributes to the polyunsaturated fatty acid contents of seed storage oil (Schlueter et al., 2007; Zhao et al., 2019). Another study on soybean genome analysis found that the transcript of GmFAD2-2C rather than GmFAD2-2A and GmFAD2-2B alleles of GmFAD2-2 was significantly accumulated in pods under cool conditions (Li et al., 2007).
The variation in gene structure is critical for the functional evolution of gene family (Cao and Shi, 2012). With the exception of GmFAD2.5 and GmFAD6.2, members within the same GmFAD subfamily exhibit similar intron/exon structures and intron patterns, and the proteins encoded by the same gene family had similar gene motif compositions and conserved protein domains. Similar results have also been found in Medicago truncatula (Zhang et al., 2018), wheat (Hajiahmadi et al., 2020), and Brassica napus (Xu et al., 2019). Our study also revealed that no common conserved sequences existed among these 30 GmFAD genes, and gene members clustered into the same clade were evolutionarily conserved. It is implied that specific conserved motifs play different roles in plant growth and development (Zhang et al., 2015). Combined with the results of the phylogenetic analyses, these results strongly support the reliability of the taxon divisions.
Previous studies have demonstrated that the FAD gene promoters contained diverse cis-elements in response to growth, development, fruiting, and defense and stresses (Soria-Garcï et al., 2019). Horiguchi et al. (1996) that the expression level of the wheat TaFAD7 gene was significantly up-regulated during leaf development stage under light and dark stresses. Liu et al. (2016) have reported that the BnFAD2-C5 promoter had SA and JA response elements, and their expression was up-regulated by SA and JA induction. In this study, we identified a range of cis-elements in the promoter region of GmFAD genes related to low temperature, drought, light, hypoxia, circadian rhythms, plant hormones (ABA, GA, SA, MeJA), and defense and stress responsive. For example, TC-rich repeats involving in the regulation of defense and stress responses were identified in the promoter sequences of 10 GmFAD genes including GmFAD3.3, GmFAD7-2, GmFAD2.5, GmFAD2.2, GmFAD2.1, GmDES1.2, GmSLD1.5, GmSLD1.4, GmSLD1.1 and GmSLD1.6, suggesting that these genes may play important roles in soybean defense and stress responses. However, expression analysis of Chinese wild soybean under salt stress demonstrated that GsDES1.1, GsDES1.2, GsFAD2.1 and GsSLD1 in leaves were not closely related to salt stress response (Zhang et al., 2021).
Growing evidence that members of the FAD gene family regulate plant defense responses to biotic stresses (Kachroo et al., 2005; Dar et al., 2017). Previous studies demonstrated AtFAD2 genes not only contribute to salt and cold tolerance in A. thaliana (Schlueter et al., 2007; Chi et al., 2011; Zhao et al., 2019), but also some alleles of GmFAD2 genes was able to respond to pathogen attack by increasing the biosynthesis of linoleic acid and palmitic-linoleic acid in soybean (Hernández et al., 2011). Li et al. (2021) showed that HaFAD3.1 and HaADS6 genes in sunflower were expressed at higher level after Orobanche cumana infection. In Brassica carinata, five genes including BolFAD6.2, BolFAD4.3, BolFAD6.3, BolFAD4.1, and BolADS17, were significantly up-regulated in response to Xanthomonas campestris infection (Shaheen et al., 2023). The Arabidopsis double mutant fad7/fad8 also exhibited reduced the accumulation of triene fatty acid in chloroplast and increased the sensitivity to Pseudomonas syringae pv. tomato DC3000 (Yaeno et al., 2004). In addition, several genes such as DES1, FAB2.3, FAB2.5, FAB2.7 and FAD3.2 were significantly down-regulated in banana when infected by the pathogenic FocTR4 causing banana wilt disease (Cheng et al., 2022). FAD genes play important roles in membrane remodeling and signaling in grapevine defense towards biotrophic pathogens (Cavaco et al., 2021). During the early interaction between grapevine and the biotrophic oomycete Plasmopara viticola, the polyunsaturated alpha-linolenic acids were highly accumulated in the leaves of the tolerant genotype, followed by alterations in the expression of desaturase genes, regulation of membrane fluidity, and finally JA accumulation and activation of JA biosynthetic pathway, which forms together form a complex network of disease resistance responses in grapevines. Especially, the expression patterns of FAD6 and FAD8 may be related to the timing of the JA pathway activation during the interaction of grapes with the necrotrophic pathogen B. cinerea (Cavaco et al., 2021).
In Southwest China, soybeans are often exposed to heavy rainfall and high humidity from the full pod stage (R4) to the ripening stage (R8), resulting in severe pod rot and seed decay, and largely reduced soybean yield and quality (Chang et al., 2020a). In this study, the GmFAD gene was also significantly induced by F. fujikuroi causing soybean seed decay. Specially, eleven of the representative 16 GmFAD genes were significantly up-regulated at the early infection stage of F. fujikuroi in the susceptible cultivar (CX12) seeds, whereas no significant changes were observed in resistant cultivar (ND25) seeds until 48 hpi. Our study showed that at the late stage of F. fujikuroi infection, the genes GmFAB2.1/2.2, GmFAD3.3/3-2B/7-1/8-2, and GmFAD2.3/2.5 were expressed significantly higher in resistant cultivar (ND25) than in sensitive cultivar (ST06). This suggests that some GmFAD genes are involved in pathogen defense responses. Since F. fujikuroi is one necrotrophic fungi, JA signaling can be activated as plant immunity (Song et al., 2023). In addition, F. fujikuroi is also well-known as the causal agent of rice bakanae bring big threat to rice production worldwide, and transcription profile of rice to F. fujikuroi infection found the pathways involved in bakanae resistance, such as chitin, JA-dependent signaling, and hypersensitive response (Matić et al., 2016). Furthermore, OsWRKY114 has been reported to act as a player in rice JA-mediated immunity against Fusarium fujikuroi in rice (Song et al., 2023).Therefore, we hypothesized that there may be a more persistent stimulation of unsaturated fatty acid (UFA) biosynthesis in resistant cultivar (ND25) seeds, which could lead to the induction of the JA pathway (Cavaco et al., 2021). This is also consistent with the prediction of the promoter cis-acting elements of GmFAD genes in response to JA. In conclusion, GmFAD genes play crucial roles in soybean seed resistance against the seed decay fungus F. fujikuroi, and the diversity of enzyme functions and expression patterns in the GmFAD gene family suggests the diversity of gene functions (Laureano et al., 2021). The results will enhance our understanding of the regulatory genes involved in the biosynthesis pathway of unsaturated fatty acids. In the future, gene editing technology will enable precise regulation of GmFAB2.1/2.2, GmFAD3.3/3-2B/7-1/8-2, and GmFAD2.3/2.5, allowing for the breeding of soybean varieties with enhanced resistance and good agronomic traits.
In this study, 30 full-length GmFAD genes were identified from the soybean genome. Analyses of gene structure, protein three-dimensional structure, and conserved motifs and conserved structural domains indicate that GmFAD genes are clustered into seven subfamily and evolutionarily conserved, and this is also strongly supported by the phylogenetic analysis. We found segmental duplication plays an important role in the gene amplification and subfamily generation of GmFAD in soybean genome. Most GmFAD gene promoter can be activated by light, phytohormones and other abiotic stresses, thus inducing fatty acid biosynthesis. Expression of GmFAD genes were differentially induced in the resistant and susceptible cultivars under seed decay stress caused by F. fujikuroi, some specific up-regulated genes such as GmFAB2.1/2.2, GmFAD3.3/3-2B/7-1//8-2, and GmFAD2.3/2.5 would be the potential candidate genes for seed-decay resistance breeding in soybean. Future studies will explore the roles of these genes in seed decay stress. The results obtained are crucial for researching the molecular mechanisms of fatty acid synthesis, FAD and SAD editing, and for marker-assisted and genomic selection in breeding soybean varieties with a specific fatty acid composition in their oil.
The genome data of Glycine max var. Williams 82 were retrieved from the Genome Warehouse (GWH) with the Phytozome v13 database (Annotation version: Glycine max Wm82.a4.v1, https://phytozome-next.jgi.doe.gov/) (Xu et al., 2019). The published protein sequences and gene sequences of 24 AtFADs were obtained from TAIR release 10 (http://www.arabidopsis.org) (Shaheen et al., 2023), while the published sequences of 18 OsFADs were downloaded from RGAP release 7 (http://rice.plantbiology.msu.edu) (Zhiguo et al., 2019).
The hidden Markov model (HMM) profiles for the FA_desaturase (PF00487), FA_desaturase 2 (PF03405), and TMEM189 (PF10520) domains (Cheng et al., 2022) downloaded from the Pfam protein family database (https://www.ebi.ac.uk/interpro/entry/pfam/) were searched against the soybean (Glycine max var. Williams 82) protein data using an e-value threshold of ≤1e -5. The GmFAD gene family members were then filtered to eliminate duplicates and identify potential gene family members. Subsequently, the protein physicochemical properties and subcellular localization of the GmFAD members were further analyzed using Expasy (https://www.expasy.org/) and Cell-PLoc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
We performed multiple alignments of FADs were conducted by Clustal W using full-length protein sequences from soybean (30), A. thaliana (24), and rice (18), respectively. A Neighbor-Joining (NJ) phylogenetic tree was constructed using the p-distance model by MEGA7 (http://www.megasoftware.net) (Kumar et al., 2016). The bootstrap support values were calculated from 1000 repeats. Subsequently, the evolutionary tree was refined and visualized through the iTOL online platform (https://itol.embl.de/).
The annotation file for the general feature format version 3 (GFF3) in the soybean genome database was utilized to pinpoint the chromosome locations of the GmFAD genes. Visualization of the chromosomal localization of the GmFAD genes were achieved using TBtools software, which was based on the starting position on the soybean chromosome (Chen et al., 2020). The matched sequences covered more than 80% of the length of the longer gene, exhibited over 80% similarity within their respective regions, and were products of a single duplication event (Jiang et al., 2013; Singh and Jain, 2015). To further assess the evolutionary pressure on the GmFAD gene family, the synonymous (Ks) and non-synonymous (Ka) substitution rates of the GmFAD gene pairs were computed using TBtools, along with the Ka/Ks calculator (Bailey et al., 2006; Kong et al., 2013; Kumar et al., 2016). In addition, to explore the evolutionary relationships within the soybean species, MCScanX and BLASTP were employed to detect gene pairs that were co-variantly associated with FAD members (Xue et al., 2020).
The soybean coding sequence and genome file were applied to explore the splicing phase of the GmFADs family (Zhang et al., 2021). The Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/).and TBtools software (Chen et al., 2020) were employed to map the distribution of introns, exons and non-coding regions within genes. NCBI Conserved Domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to identify the conserved structural domains of GmFAD gene family proteins, and MEME (https://meme-suite.org/meme/) to analyze the conserved motifs of FAD gene family proteins with the following parameter settings: the motif discovery was set to classical mode with a predicted motif count of 20, and each motif was allowed to occur 0 or 1 times (Bailey et al., 2006). The Pfam database was then utilized to evaluate the functions of the aforementioned motifs (Mistry et al., 2021). The conserved motifs and structural domains of the FAD gene family proteins were simultaneously visualized, and their interactions were analyzed using TBtools. The integrated structure of GmFAD was constructed using the Swiss-Model platform (https://swissmodel.expasy.org/) based on fragments of iterative templates (Wei et al., 2022), and subsequently refined and visualized by Chimera software to yield a three-dimensional structural model.
Sequences encompassing the 1500 bp region upstream of the start codon (ATG) for each GmFAD gene were retrieved from the soybean genome database using TBtools. Subsequently, the promoter sequences of these GmFAD genes were uploaded to Plant CARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify cis-regulatory elements (Lescot et al., 2002).
The functional annotation of genes was conducted using the eggNOG-mapper database (http://eggnog-mapper.embl.de/). To further understand the potential pathways that might be associated with the GmFAD genes, the TBtools eggNOG-mapper Helper tool was subsequently employed to systematically organize and process the results derived from the eggNOG-mapper. Following this, the GO-basic file and the KEGG-backend file were exported for GO enrichment analysis and KEGG Pathway enrichment, respectively (Wei et al., 2022).
The fungal isolate F. fujikuroi (No. S100) was isolated from the rot seeds of soybean in the fields and identified using sequence analysis of translation elongation factor 1 alpha (EF-1α) and DNA-directed RNA ploymerase II second largest subunit (RPB2) (Chang et al., 2020a). The cultivar CX12 exhibited susceptibility to F. fujikuroi, whereas the cultivar ND25 showed a high resistance to the fungus, and both these two cultivars were chosen to examine the expression of GmFAD genes following inoculation of soybean seeds with F. fujikuroi.
Spore suspensions of F. fujikuroi were prepared following the protocol by Chang et al. (2022). For sporulation, a mung bean liquid medium was prepared by boiling 30 g of mung bean in 1 L of sterilized water for 20 min, filtering the mixture with cheesecloth, and then autoclaving at 121℃ for 30 min (Lv and Sun, 2007). Disease-resistant and susceptible soybean seeds were inoculated with F. fujikuroi suspensions at a concentration of 1 × 106 spores per milliliter, supplemented with 0.1% Tween 20. As a control (CK), soybean seeds were treated with an equal volume of mung bean liquid medium. The treated and control seeds were then arranged on water agar medium (WA), and were incubated in the dark at a constant temperature of 25 ℃ for varying durations of 0, 6, 12, 24, and 48 h, respectively. The experiments were conducted in triplicate. At specified intervals post inoculation, soybean seeds from both control and treatment groups were collected, immediately frozen in liquid nitrogen, and subsequently stored at -80°C for RNA extraction.
The expression profiles of twenty-six GmFAD genes were analyzed using qRT-PCR with specific primer pairs listed in Supplementary Table S6. Total RNA was extracted from soybean seed samples using Fast Pure® Universal Plant Total RNA Isolation Kit (Vazyme-Bio, Chengdu, China) and subsequently assessed with a NanoDrop fluorometer (Thermo Fisher Scientific, Stuttgart, Germany) to ascertain the concentration and quality of RNA. First-strand cDNA was synthesized from 2.5 μg RNA in a 20-μL reaction volume according to the instructions of BeyoRT™II First Strand cDNA Synthesis Kit (Beyotime-Bio, Shanghai, China). The qRT-PCR assay was conducted on the Chromo4 Real-Time PCR System (Bio-Rad, CA, USA), within a 10-μL reaction mixture containing 5 μL of the SYBR qPCR Mix (2×) (Vazyme-Bio, Shanghai, China), 1 μL of each primer (10 mM), 1 μL of template DNA (10 ng), and 2 μL of ddH2O. The PCR thermal cycling conditions were set as follows: an initial heat activation at 95°C for 3 minutes, followed by 40 cycles of amplification at 10 s at 95°C for 10 s, annealing temperature for 30 s, 15 s at 95°C, 1 min at 60°C, and a final 15 s at 95°C for melting curve analysis. Relative gene expression levels were determined using the 2-ΔΔCt method (Livak and Schmittgen, 2001) with the GmActin gene serving as an internal reference for normalization. Each sample was subjected to four technical replicates and three biological replicates. Data analysis was preformed using SPSS 24 software (SPSS Software Inc., Chicago, IL, USA), and statistical analysis was conducted with Analysis of variance (ANOVA) and Duncan’s multiple range test (DMRT). The results were visualized with GraphPad Prism 10 (GraphPad Software Inc., San Diego, CA, USA).
All data generated or analyzed during this study are included in this published article and its supplementary information files. The general feature format (GFF) sequence file and the protein sequence file of soybean (Glycine max var. Williams 82) used in this study are available at Phytozome v13 database (https://phytozome-next.jgi.doe.gov/). The sequences used for the interspecific covariance homology relationships between soybean, rice and Arabidopsis, corresponding FAD sequence information are available in the rice database (http://rice.plantbiology.msu.edu) and TAIR database (https://www.arabidopsis.org/), respectively.
XL: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft. MM: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. WZ: Formal analysis, Validation, Writing – review & editing. ZS: Funding acquisition, Project administration, Writing – review & editing. XC: Funding acquisition, Project administration, Writing – review & editing. WY: Conceptualization, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Guangxi Key Research and Development Program (AB23026107), and National Key Research and Development Program of China (2023YFD1401005).
We thank Professor Xiaoling Wu’ kinds to provide the resistant and susceptible soybean cultivars.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1540003/full#supplementary-material
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Cao, J., Shi, F. (2012). Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol. Bioinform. Online. 8, 271–292. doi: 10.4137/EBO.S9652
Cavaco, A. R., Laureano, G., Cunha, J., Eiras-Dias, J., Matos, A. R., Figueiredo, A. (2021). Fatty acid modulation and desaturase gene expression are differentially triggered in grapevine incompatible interaction with biotrophs and necrotrophs. Plant Physiol. Biochem. 163, 230–238. doi: 10.1016/j.plaphy.2021.04.001
Chandra-Shekara, A. C., Venugopal, S. C., Barman, S. R., Kachroo, A., Kachroo, P. (2007). Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 7277–7282. doi: 10.1073/pnas.0609259104
Chang, X., Li, H., Naeem, M., Wu, X. L., Yong, T. W., Song, C., et al. (2020a). Diversity of the seedborne fungi and pathogenicity of Fusarium species associated with intercropped soybean. Pathogens 9, 531. doi: 10.3390/pathogens9070531
Chang, X., Li, X., Meng, H., Li, H. J., Wu, X. L., Gong, G. S., et al. (2022). Physiological and metabolic analyses provide insight into soybean seed resistance to Fusarium fujikuroi causing seed decay. Front. Plant Sci. 13, 993519. doi: 10.3389/fpls.2022.993519
Chang, X., Yan, L., Naeem, M., Khaskheli, M. I., Zhang, H., Gong, G., et al. (2020b). Maize/soybean relay strip intercropping reduces the occurrence of Fusarium root rot and changes the diversity of the pathogenic Fusarium species. Pathogens 9, 211. doi: 10.3390/pathogens9030211
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools; An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Yao, Q., Patil, G. B., Agarwal, G., Deshmukh, R. K., Lin, L., et al. (2016). Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 7, 1044. doi: 10.3389/fpls.2016.01044
Cheng, C., Liu, F., Sun, X., Wang, B., Liu, J., Ni, X., et al. (2022). Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana. Int. J. Biol. Macromol. 204, 661–676. doi: 10.1016/j.ijbiomac.2022.02.024
Chi, X., Yang, Q., Lu, Y., Wang, J., Zhang, Q., Pan, L., et al. (2011). Genome-wide analysis of fatty acid desaturases in soybean (Glycine max). Plant Mol. Biol. Rep. 29, 769–783. doi: 10.1007/s11105-010-0284-z
Dar, A. A., Choudhury, A. R., Kancharla, P. K., Arumugam, N. (2017). The FAD2 gene in plants: occurrence, regulation, and role. Front. Plant Sci. 8, 1789. doi: 10.3389/fpls.2017.01789
Dong, C. J., Cao, N., Zhang, Z. G., Shang, Q. M. (2016). Characterization of the fatty acid desaturase genes in cucumber: structure, phylogeny, and expression patterns. PloS One 11, e0149917. doi: 10.1371/journal.pone.0149917
Feng, J. Y., Dong, Y. T., Liu, W., He, Q. L., Daud, M. K., Chen, J. H., et al. (2017). Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci. Rep. 7, 45711. doi: 10.1038/srep45711
Ha, B. K., Monteros, M. J., Boerma, H. R. (2010). Development of SNP assays associated with oleic acid QTLs in N00-3350 soybean. Euphytica 176, 403–415. doi: 10.1007/s10681-010-0225-9
Hajiahmadi, Z., Abedi, A., Wei, H., Sun, W. B., Ruan, H. H., Zhuge, Q., et al. (2020). Identification, evolution, expression, and docking studies of fatty acid desaturase genes in wheat (Triticum aestivum L.). BMC Genomics 21, 778. doi: 10.1186/s12864-020-07199-1
Hashimoto, K., Yoshizawa, A. C., Okuda, S., Kuma, K., Goto, S., Kanehisa, M. (2008). The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 49, 183–191. doi: 10.1194/jlr.M700377-JLR200
He, M., He, C. Q., Ding, N. Z. (2018). Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 9, 1771. doi: 10.3389/fpls.2018.01771
Hernández, M. L., Padilla, M. N., Sicardo, M. D., Mancha, M., Martínez-Rivas, J. M. (2011). Effect of different environmental stresses on the expression of oleate desaturase genes and fatty acid composition in olive fruit. Phytochemistry 72, 178–187. doi: 10.1016/j.phytochem.2010.11.026
Horiguchi, G., Iwakawa, H., Kodama, H., Kawakami, N., Nishimura, M., Iba, K. (1996). Expression of a gene for plastid ω-3 fatty acid desaturase and changes in lipid and fatty acid compositions in light-and dark-grown wheat leaves. Physiol. Plant 96, 275–283. doi: 10.1111/j.1399-3054.1996.tb00214.x
Im, Y. J., Han, O., Chung, G. C., Cho, B. H. (2002). Antisense expression of an Arabidopsis omega-3 fatty acid desaturase gene reduces salt/drought tolerance in transgenic tobacco plants. Mol. Cells 13, 264–271.
Jiang, C. J., Shimono, M., Maeda, S., Inoue, H., Mori, M., Hasegawa, M., et al. (2009). Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Interact. 22, 820–829. doi: 10.1094/MPMI-22-7-0820
Jiang, Y., Wang, W. X., Xie, Q. J., Liu, N., Liu, L. X., Wang, D. P., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jiang, H., Wu, Q., Jin, J., Sheng, L., Yan, H., Cheng, B., et al. (2013). Genome-wide identification and expression profiling of ankyrin-repeat gene family in maize. Dev. Genes Evol. 223, 303–318. doi: 10.1007/s00427-013-0447-7
Jin, Y., Wang, Y., Lu, Q., Ren, N., Liu, L., Shen, G., et al. (2024). Genome-wide analysis of fatty acid desaturase (FAD) gene family in Camellia sinensis: identification, expression and their biochemical functions in low temperature resistance. Ind. Crops Prod. 222, 119755. doi: 10.1016/j.indcrop.2024.119755
Johnson, D. A., Thomas, M. A. (2007). The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Mol. Biol. Evol. 24, 2412–2423. doi: 10.1093/molbev/msm184
Kachroo, A., Fu, D. Q., Havens, W., Navarre, D., Kachroo, P., Ghabrial, S. A. (2008). An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant Microbe Interact. 21, 564–575. doi: 10.1094/MPMI-21-5-0564
Kachroo, A., Kachroo, P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Kachroo, P., Venugopal, S. C., Navarre, D. A., Lapchyk, L., Kachroo, A. (2005). Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol. 139, 1717–1735. doi: 10.1104/pp.105.071662
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 433. doi: 10.1186/1471-2164-14-433
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laureano, G., Cavaco, A. R., Matos, A. R., Figueiredo, A. (2021). Fatty acid desaturases: uncovering their involvement in grapevine defence against downy mildew. Int. J. Mol. Sci. 22, 5473. doi: 10.3390/ijms22115473
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Liu, A., Najeeb, U., Zhou, W. J., Liu, H., Yan, G. J., et al. (2021). Genome-wide investigation and expression analysis of membrane-bound fatty acid desaturase genes under different biotic and abiotic stresses in sunflower (Helianthus annuus L.). Int. J. Biol. Macromol. 175, 188–198. doi: 10.1016/j.ijbiomac.2021.02.013
Li, L. Y., Wang, X. L., Gai, J. Y., Yu, D. Y. (2007). Molecular cloning and characterization of a novel microsomal oleate desaturase gene from soybean. J. Plant Physiol. 164, 1516–1526. doi: 10.1016/j.jplph.2006.08.007
Li, A., Zhang, R. Z., Pan, L., Tang, L. C., Zhao, G. Y., Zhu, M. Z., et al. (2011). Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PloS One 6, e28810. doi: 10.1371/journal.pone.0028810
Liu, F., Wang, G. L., Liu, R. Y., Guan, C. Y. (2016). The promoter of fatty acid desaturase on chromosome C5 in Brassica napus drives high-level expression in seeds. Plant Biotechnol. Rep. 10, 369–381. doi: 10.1007/s11816-016-0407-6
Liu, F., Zhao, Y. P., Zhu, H. G., Zhu, Q. H., Sun, J. (2017). Simultaneous silencing of GhFAD2-1 and GhFATB enhances the quality of cottonseed oil with high oleic acid. J. Plant Physiol. 215, 132–139. doi: 10.1016/j.jplph.2017.06.001
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lou, Y., Schwender, J., Shanklin, J. (2014). FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J. Biol. Chem. 289, 17996–18007. doi: 10.1074/jbc.M114.572883
Luginbuehl, L. H., Menard, G. N., Kurup, S., Erp, H. V., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
Lv, G. Z., Sun, Y. W. (2007). Soybean pests and diseases and control of the original color atlas (Beijing: Golden Shield Publishing House).
Matić, S., Bagnaresi, P., Biselli, C., Orru’, L., Amaral Carneiro, G., Siciliano, I., et al. (2016). Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genomics 17, 608. doi: 10.1186/s12864-016-2925-6
Mikkilineni, V., Rocheford, T. R. (2003). Sequence variation and genomic organization of fatty acid desaturase-2 (fad2) and fatty acid desaturase-6 (fad6) cDNAs in maize. Theor. Appl. Genet. 106, 1326–1332. doi: 10.1007/s00122-003-1190-7
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Nayeri, F. D., Yarizade, K. (2014). Bioinformatics study of delta-12 fatty acid desaturase 2 (FAD2) gene in oilseeds. Mol. Biol. Rep. 41, 5077–5087. doi: 10.1007/s11033-014-3373-5
Saini, R., Kumar, S. (2019). Genome-wide identification, characterization and in-silico profiling of genes encoding FAD (fatty acid desaturase) proteins in chickpea (Cicer arietinum L.). Plant Gene 18), 100180. doi: 10.1016/j.plgene.2019.100180
Sayanova, O., Smith, M. A., Lapinskas, P., Stobart, A. K., Dobson, G., Christie, W. W., et al. (1997). Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of Δ 6 -desaturated fatty acids in transgenic tobacco. Proc. Natl. Acad. Sci. 94, 4211–4216. doi: 10.1073/pnas.94.8.4211
Schlueter, J. A., Vasylenko-Sanders, I. F., Deshpande, S., Yi, J., Siegfried, M., Roe, B. A., et al. (2007). The FAD2 gene family of soybean: insights into the structural and functional divergence of a paleopolyploid genome. Crop Sci. 47, S14–S26. doi: 10.2135/cropsci2006.06.0382tpg
Shaheen, N., Khan, U. M., Farooq, A., Zafar, U. B., Khan, S. H., Ahmad, S., et al. (2023). Comparative transcriptomic and evolutionary analysis of FAD-like genes of Brassica species revealed their role in fatty acid biosynthesis and stress tolerance. BMC Plant Biol. 23, 250. doi: 10.1186/s12870-023-04232-9
Singh, V. K., Jain, M. (2015). Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 6, 918. doi: 10.3389/fpls.2015.00918
Song, N., Hu, Z., Li, Y., Li, C., Peng, F. X., Yao, Y. Y., et al. (2013). Overexpression of a wheat stearoyl-ACP desaturase (SACPD) gene TaSSI2 in Arabidopsis ssi2 mutant compromise its resistance to powdery mildew. Gene 524, 220–227. doi: 10.1016/j.gene.2013.04.019
Song, G., Son, S., Nam, S., Suh, E. J., Lee, S. I., Park, S. R. (2023). OsWRKY114 is a player in rice immunity against Fusarium fujikuroi. Int. J. Mol. Sci. 24, 6604. doi: 10.3390/ijms24076604
Soria-Garcï, A. Ï. N., Rubio, M. A. C., Lagunas, B., Lï-Pez-Gomollï, N. S., de Los Ï Ngeles Lujï, N. M. A., Dï-Az-Guerra, R. L., et al. (2019). Tissue distribution and specific contribution of Arabidopsis FAD7 and FAD8 plastid desaturases to the JA- and ABA-Mediated cold stress or defense responses. Plant Cell Physiol. 60, 1025–1040. doi: 10.1093/pcp/pcz017
Suga, H., Arai, M., Fukasawa, E., Motohashi, K., Nakagawa, H., Tateishi, H., et al. (2018). Genetic differentiation associated with Fumonisin and Gibberellin production in Japanese Fusarium fujikuroi. Appl. Environ. Microbiol. 85, e02414–e02418. doi: 10.1128/AEM.02414-18
Upchurch, R. G. (2008). Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 30, 967–977. doi: 10.1007/s10529-008-9639-z
Wang, J., Ming, F., Pittman, J., Han, Y., Hu, J., Guo, B., et al. (2006). Characterization of a rice (Oryza sativa L.) gene encoding a temperature-dependent chloroplast ω-3 fatty acid desaturase. Biochem. Biophys. Res. Commun. 340, 1209–1216. doi: 10.1016/j.bbrc.2005.12.126
Wei, H., Movahedi, A., Xu, S., Zhang, Y. Y., Liu, G. Y., Aghaei-Dargiri, S., et al. (2022). Genome-wide characterization and expression analysis of fatty acid desaturase gene family in poplar. Int. J. Mol. Sci. 23, 11109. doi: 10.3390/ijms231911109
Wrather, J. A., Sleper, D. A., Stevens, W. E., Shannon, J. G., Wilson, R. F. (2003). Planting date and cultivar effects on soybean yield, seed quality, and phomopsis sp. seed infection. Plant Dis. 87, 529–532. doi: 10.1094/PDIS.2003.87.5.529
Xu, M., Li, P., Meng, H., Wu, X. L., Gong, G. S., Chen, H. B., et al. (2023). First report of Colletotrichum fructicola causing anthracnose on Glycine max in China. Plant Dis. 107, 2240. doi: 10.1094/PDIS-09-22-2222-PDN
Xu, L., Zeng, W. J., Li, J. J., Liu, H., Yan, G. J., Si, P., et al. (2019). Characteristics of membrane-bound fatty acid desaturase (FAD) genes in Brassica napus L. and their expressions under different cadmium and salinity stresses. Environ. Exp. Bot. 162, 144–156. doi: 10.1016/j.envexpbot.2019.02.016
Xue, Y. F., Chai, C. Y., Chen, B. J., Shi, X. F., Wang, B. T., Jiang, M. L., et al. (2020). Whole-genome mining and in silico analysis of FAD gene family in Brassica juncea. J. Plant Biochem. Biotechnol. 29, 149–154. doi: 10.1007/s13562-019-00516-0
Xue, Y., Chen, B. J., Wang, R., Win, A. N., Li, J. N., Chai, Y. R. (2018). Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl. Biochem. Biotechnol. 184, 582–598. doi: 10.1007/s12010-017-2563-8
Xue, H. Q., Upchurch, R. G., Kwanyuen, P. (2008). Relationships between oleic and linoleic acid content and seed colonization by Cerospora kikuchii and Diaporthe phaseolorum. Plant Dis. 92, 1038–1042. doi: 10.1094/PDIS-92-7-1038
Yaeno, T., Matsuda, O., Iba, K. (2004). Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J.: Cell Mol. Biol. 40, 931–941. doi: 10.1111/j.1365-313X.2004.02260.x
Yara, A., Yaeno, T., Hasegawa, M., Seto, H., Montillet, J. L., Kusumi, K., et al. (2007). Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of omega-3 fatty acid desaturases. Plant Cell Physiol. 48, 1263–1274. doi: 10.1093/pcp/pcm107
Zhang, M., Barg, R., Yin, M., Gueta-Dahan, Y., Leikin-Frenkel, A., Salts, Y., et al. (2005). Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 44, 361–371. doi: 10.1111/j.1365-313X.2005.02536.x
Zhang, Y., Maximova, S. N., Guiltinan, M. J. (2015). Characterization of a stearoyl-acyl carrier protein desaturase gene family from chocolate tree, Theobroma cacao L. Front. Plant Sci. 6, 239. doi: 10.3389/fpls.2015.00239
Zhang, Z., Wei, X., Liu, W., Min, X. Y., Jin, X. Y., Ndayambaza, B., et al. (2018). Genome-wide identification and expression analysis of the fatty acid desaturase genes in Medicago truncatula. Biochem. Biophys. Res. Commun. 499, 361–367. doi: 10.1016/j.bbrc.2018.03.165
Zhang, B., Xia, P., Yu, H., Li, W., Chai, W., Liang, Z. (2021). Based on the whole genome clarified the evolution and expression process of fatty acid desaturase genes in three soybeans. Int. J. Biol. Macromol. 182, 1966–1980. doi: 10.1016/j.ijbiomac.2021.05.161
Zhao, M., Wang, W., Wei, L., Chen, P., Peng, L., Qin, Z., et al. (2019). The evolution and biocatalysis of FAD2 indicate its correlation to the content of seed oil in plants. Int. J. Mol. Sci. 20, 849. doi: 10.3390/ijms20040849
Keywords: Glycine max, fatty acid desaturases, phylogenetic analysis, gene expression, Fusarium fujikuroi, seed decay
Citation: Li X, Munir M, Zeng W, Sun Z, Chang X and Yang W (2025) Characterization of fatty acid desaturase gene family in Glycine max and their expression patterns in seeds after Fusarium fujikuroi infection. Front. Plant Sci. 16:1540003. doi: 10.3389/fpls.2025.1540003
Received: 05 December 2024; Accepted: 03 February 2025;
Published: 25 February 2025.
Edited by:
Babu N. Motagi, University of Agricultural Sciences, IndiaReviewed by:
Milind Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaCopyright © 2025 Li, Munir, Zeng, Sun, Chang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Chang, eGxfY2hhbmcxNDA0MkBzaWNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.