
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 March 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1539625
 Zhaopeng Yu†
Zhaopeng Yu† Guangyi Xu†
Guangyi Xu† Keying Xie
Keying Xie Zhuang Xie
Zhuang Xie Dachuan Wang
Dachuan Wang Linlu Tan
Linlu Tan Yinghua Ling
Yinghua Ling Guanghua He
Guanghua He Fangming Zhao*
Fangming Zhao*Rice chromosome segment substitution lines (CSSLs) are ideal for creating natural variation and dissecting complex quantitative traits. In addition, it builds a bridge for molecular breeding and accurate identification of quantitative trait loci (QTLs). In this study, to construct an indica rice library of single-segment substitution lines (SSSLs) spanning the whole genome, a rice CSSL-Z691 carrying four substitution segments (4.07 Mb of average length) was identified by marker-assisted selection (MAS) from indica restorer line “Jinhui35” in the “Xihui18” genetic background. Compared with large panicle type Xihui18, seed setting ratio, grain width, and 1000-grain weight increased in Z691. In contrast, the number of primary branches, spikelet number per panicle, grain number per panicle, grain length, rate of length to width, and yield per plant decreased in Z691. Then, 11 QTLs were identified in the secondary F2 population from Xihui18/Z691. Again, four QTLs (qGW6, qGL4, qRLW4, and qGWT4) were validated by three SSSLs (S1–S3) developed in F3. In addition, 11 new QTLs were detected by the three SSSLs that were not identified in the F2 population. Moreover, the different QTLs in D1–D3 showed various genetic models. Some QTLs, e.g., qGWT6 (a = 0.96 g) and qGWT7 (a = −0.29 g), displayed independent inheritance, while others exhibited various epistatic interactions. Thus, it is vital to identify different QTLs and their genetic models. Resolving the epistasis effects among different QTLs is crucial for screening QTLs for breeding by design. Finally, qGL4 and qGW6 were fine-mapped to 160- and 240-kb intervals on chromosomes 4 and 6, and two candidate genes were determined by DNA sequencing. These results provide valuable genetic and breeding materials for cloning qGL4 and qGW6 and for future molecular breeding by design.
Rice (Oryza sativa L.) is the staple food for more than one-half of the world’s population (Yi and Li, 2022). High yield and high quality are the most essential elements in rice breeding from ancient times until now (Zhang et al., 2024a; You et al., 2022). The formation mechanism of rice yield is complex (Zuo and Li, 2014). The effective number of panicles, grain number per panicle, and grain weight mainly control rice yield (Li et al., 2021). At the same time, plant height, plant type, panicle length, grain size, and other factors are also closely related to yield (Wang et al., 2012). However, these agronomic traits are often controlled by multiple genes and influenced by the environment (Abe et al., 2012). In the post-genomic era, the concept of “breeding by design”, which aims to utilize favorable alleles in the crop genome to design and breed varieties for people’s needs, has been proposed. This concept’s core utilizes natural variation (Zhang, 2021). Notably, “breeding by design” combines theoretical research on quantitative trait locus (QTL) mapping with breeding programs, which is essential for rice molecular breeding and genetic improvement (Xu et al., 2021).
As rice chromosome segment substitution lines (CSSLs) differ from the recipient parent in only one or a few substitution segments identified by known molecular markers, they serve as a valuable resource for gene dissection and superior gene pyramiding (Zong et al., 2012). CSSLs enable the precise improvement of parental lines (maintenance and restoration lines) in hybrid rice to facilitate the strategic and efficient development of superior hybrid rice varieties tailored to specific breeding requirements. However, the first step to realizing this great strategy is to dissect and clone the genes for related traits and then study their functions and molecular mechanisms. Traits associated with rice yield are controlled by genes and environment, with genes playing a more significant role in regulation (Zhao et al., 2022). Much research has been conducted on many of the QTLs that determine rice yield by previous generations. For example, GL3.1 reduces the expression of Cyclin-T1 protein in rice through a certain regulatory pathway to shorten the length of rice grains (Fan et al., 2006). GS5 can make endocytosis difficult in rice by preventing the interaction of two related genes (Xu et al., 2015). Reduced or no expression of OsTB1 increased rice tiller number (Takeda et al., 2003). OsMPK6 is essential in regulating rice grain size by promoting cell proliferation in spikelet hulls (Bai et al., 2024; Liu et al., 2015; Xu et al., 2018). Natural variation in the promoter of qRBG1/OsBZR5 enhanced rice yield by OsGSK2–qRBG1–OsBZR1–D2–OFP1 in the Brassinolide signaling pathway (Zhang et al., 2024b). Through cloning and further study of related genes by an increasing number of researchers, our understanding of the molecular mechanism underlying rice yield formation has gradually established and deepened. It has also laid a good foundation for researching and developing molecular breeding for rice yield and quality. In addition, there is a growing awareness of the complexity and multifaceted nature of various genes and metabolic pathways in controlling the traits of rice (Lei et al., 2024). At present, although some QTLs for yield-related traits in rice have been identified, their number remains insufficient to fulfill the requirement (Wang et al., 2021). Especially because some of these genes are identified from mutants, which often pose serious challenges such as reduced plant height and reduced seed setting ratio, it is more difficult to be directly applied as breeding materials in breeding practice. However, using CSSLs, especially single-segment substitution lines (SSSLs), it is possible to find alleles of these cloned genes in them and identify new favorable genes for yield and quality traits. Moreover, CSSLs are excellent breeding materials for gene identification and for the pyramid of favorable genes (Balakrishnan et al., 2018).
In this study, we aimed to construct an indica SSSL library covering the entire rice genome and to identify QTLs for yield-related traits in rice. We used the rice CSSL-Z691 containing four substitution segments from the indica rice restoration line Jinhui35 in the genetic background of Xihui18 as experimental materials. We used the secondary F2 population constructed from the cross between Xihui18 and Z691 for the QTL mapping of important agronomic traits in rice. We further built three SSSLs and three dual-segment substitution lines (DSSLs) by marker-assisted selection (MAS). Then, we used the SSSLs and DSSLs for further QTL identification and the analysis of the epistatic effect between different QTLs. In addition, we constructed the fine-mapping population and the overlapping substitution segment mapping population of qGL4 and qGW6 and accordingly completed the fine-mapping of qGL4 and qGW6. The results of this study provide a foundation for map-based cloning of QTLs and for “breeding by design” in rice.
Z691 is a four-segment CSSL selected by MAS from the progeny (F2:4) of a cross between Xihui18 and Z403. Xihui18 is an indica rice restorer line bred by Southwest University, China. Z403 is a CSSL containing 12 segments from the indica rice restorer line Jinhui35 on the genetic background of Xihui18. The flowchart of Z691 development is shown in Supplementary Figure S1. The specific development method of Z403 was described by Xu et al. (2023). The substitution lengths of Z691 were estimated by the method of Paterson et al. (1991), and chromosome substitution segments were mapped using the MapChart 2.32 software (https://www.wur.nl/en/show/Mapchart.htm).
For the QTL mapping, a secondary F2 segregating population of 150 individuals raised from the cross between Xihui18 and Z691 was used.
Six F2 individuals with the target substitution marker and the other 0–1 heterozygous marker were screened according to the result of QTL mapping to develop SSSLs and DSSLs.
The fine-mapping population of qGL4 was an F3 population with 631 individuals. These individuals were developed from a recombinant plant of qGL4 in the F2 population. Additionally, four individuals with different genotypes were screened from the F3 population to establish an overlapping substitution mapping population of qGL4.
The fine-mapping population of qGW6 was an F3 population with 100 individuals. These individuals were developed from a recombinant plant of qGW6 in the F2 population. Two individuals with different genotypes were screened from the F3 population to develop an overlapping substitution mapping population of qGW6.
In July 2020, Xihui18 was crossed with Z691 to obtain hybrid seeds at the Chongqing experimental station of Southwest University of China (29.76°N, 106.38°E). In September of the same year, the hybrid seeds were planted to obtain F1 seeds at the Lingshui experimental base in Hainan province, China (18.42°N, 109.8°E). In March 2021, Xihui18, Z691, and the F2 population were planted at the experimental station of the Southwest University of Chongqing, China. In April, 30 plants of Xihui18 and Z691 and 150 individuals of the F2 population were transplanted in the same field. The space between plants was 16.80 cm, and that between rows was 26.50 cm. In March 2022, Xihui18, Z691, and six F2 individuals for developing SSSLs and DSSLs were transplanted, with 30 plants for each material, on April 15, 2022, together with 631 individuals for qGL4 fine-mapping and 100 individuals for qGW6 fine-mapping. In March 2023, Xihui18, Z691, three SSSLs (S1–S3), and three DSSLs (D1–D3) were planted in the same experimental field, and 30 plants were transplanted for each material in April of the same year. In addition, six secondary heterozygous F3 individuals for developing SSSLs were planted for overlapping substitution mapping of qGL4 and qGW6, each line with 100 individuals. Conventional field management practices were applied (according to the local rice management methods of the two experimental bases; the experimental fields used are representative of the local areas).
Ten plants each from Xihui18, Z691, SSSLs, and DSSLs plots, together with 150 F2 plants, were harvested at the maturity stage. Twelve yield-related traits were measured, including plant height, spikelet number per panicle, panicle length, number of primary panicles, number of secondary panicles, grain number per panicle, seed setting ratio, seed-set density, length of 10 grains, width of 10 grains, and 1000-grain weight. The measurement methods were the same as those of Ma et al. (2020). Panicle length, spikelet number per panicle, grain number per panicle, and seed setting ratio were measured using all the effective panicles of the plant. In this case, the seed setting ratio was expressed as the percentage of grain number per panicle to the spikelet number per panicle. The seed-set density was expressed as the mean of the total number of grains per 10 cm panicle length. The length and width of 10 grains for each plant were measured using a vernier caliper, and then the mean values were calculated. According to the mean value of the length and width of 10 grains, the rate of grain length to width was obtained for each plant. The 1000-grain weight of Xihui18 and Z691 were measured using an electronic balance, with three repetitions. The 1000-grain weight of each F2 plant was determined as the weight of 200 grains multiplied by 5, with three repetitions. Yield per plant was weighed using an electronic balance. Finally, various parameters such as minimum, maximum, mean, and standard deviation of the above agronomic trait measurements of 10 plants of Xihui18, Z691, and 150 F2 populations were counted and underwent a t-test using Microsoft Excel 2010.
A secondary F2 population with 150 individuals was used for QTL mapping. They were derived from a cross between Xihui18 and Z691. The total genomic DNA of Xihui18, Jinhui35, Z691, and the 150 individuals from the F2 population was extracted using the cetyltrimethylammonium bromide method (McCouch et al., 1988). PCR amplification, non-denaturing polyacrylamide gel electrophoresis, and rapid silver staining were conducted using the methods described by Zhao et al. (2016). Bands identical to Xihui18 received a score of “−1”, bands identical to Z691 received a score of “1”, heterozygous bands received a score of “0”, and the lack of marker bands received a score of “.”. For QTL mapping, the phenotypic values of each member of the F2 population and the marker assignments were utilized. QTL mapping was performed using the restricted maximum likelihood method by mixed linear models (REML) implemented in the HPMIXED procedure of SAS 9.3 (SAS Institute Inc., Cary, NC, USA), with significance determined at α = 0.05 (Zhao et al., 2016; Sun et al., 2022).
Combining the results of phenotypic analysis and preliminary QTL localization, six individual plants containing the target QTLs and 0–1 heterozygous markers were selected from the F2 population and planted as lines in the same field in 2022. Ten individual plants were taken from each line and further characterized using MAS to select single-segment substitution lines and dual-segment substitution lines containing the target QTLs.
In the fall of 2023, 10 plants of Xihui18 and SSSLs (S1–S3) were harvested to measure the above yield-related traits. As there was only one chromosome segment difference between each SSSL and Xihui18 under the same environmental conditions, it can be assumed that all the genetic traits exhibited by SSSLs that were different from Xihui18 were associated with this differential chromosome segment. Therefore, the genetic model of the recipient parent Xihui18 was P0 = μ0 + ϵ, and the genetic model of SSSLs carrying specific QTLs was Pi = μ0 + ai + ϵ, where μ0 is the average of the Xihui18 population phenotypes, ai is the additive effect of the QTLs; pi and p0 represent the phenotypic values of the SSSLs and Xihui18, respectively; and ϵ is the random error. Each SSSLi (S1, S2, and S3) was hypersized (H0) to have no QTLs in the substituted segments of SSSLi. One-way ANOVA and Duncan’s multiple comparisons in IBM SPSS Statistics 23.0 were used to statistically analyze each trait for each SSSLi with 10 plants of SSSLi and Xihui18, and QTL was considered present in SSSLi when the p-value was <0.05. Therefore, the additive effect of the QTL can be calculated as ai = (pi − p0)/2 (one-half of the phenotypic difference was considered to be caused by genetics) (Li et al., 2022a).
Similarly, 10 plants of Xihui18 and DSSL (D1–D3) were harvested to measure the above yield-related traits in the fall of 2023. In the same environment, the genetic model of DSSL was Pij = μ0 + ai + aj + Iij + ϵ, where Pij represents the phenotypic value of DSSLij, ai and aj represent the additive effect of QTL in substitution segment i and j, respectively, and Iij represents the aiaj epistasis effect between QTLs in substitution segment i and j. With this, two-way ANOVA and Duncan’s multiple comparisons in IBM SPSS Statistics 23.0 were utilized to analyze the significance between Xihui18, DSSLij, SSSLi, and SSSLj for each trait, where Xihui18, SSSLi, SSSLj, and DSSLij represent the phenotypic values of the corresponding phenotypic traits of Xihui18, SSSL, and DSSL, respectively. When the p-value was >0.05, it was considered that two QTLs were inherited independently. When the probability value was less than 0.05, it was considered that there was an epistatic effect among QTLs. Therefore, the epistatic effect between non-equivalent QTLs was estimated as [(Xihui18 + DSSLij) − (SSSLi + SSSLj)]/2 (one-half of the phenotypic difference was considered to be caused by genetics) (Li et al., 2022a; Liang et al., 2021).
Based on the qGL4 primary mapping, an F3 population of 631 plants constructed by one recombinant line of qGL4 self-fertilization was used for genetic analysis and fine-mapping of qGL4 in 2022. Among them, 151 short-grain recessive individual plants, three newly synthesized polymorphic seed setting ratio (SSR) markers, and one polymorphic SSR marker in the initial localization of qGL4 were used for linkage analysis. In addition, four single plants with different genotypes were selected to breed secondary SSSLs of qGL4 in the F3 population in 2023. All selected SSSLs and Xihui18 were then measured for grain length and used for overlapping substitution mapping of qGL4. When the grain lengths of secondary SSSLs and Xihui18 were significantly different, it indicated that a QTL for grain length was detected in the substitution segments of SSSLs. The QTL was localized in the overlapping region when multiple substitution segments with the target trait overlapped (Yang et al., 2021). Then, within the interval of qGL4, the candidate genes were predicted according to the possible gene family reported by Li et al. (2018) by the Gramene (https://www.gramene.org/), the Rice Annotation Project (https://rapdb.dna.affrc.go.jp/), and China Rice Data Center (http://www.ricedata.cn/gene/index.htm) databases; the entire DNA of these genes between S1 and Xihui18 was sequenced.
Based on the initial mapping of qGW6, in 2022, an F3 population consisting of 100 individuals was constructed from the self-fertilization of a recombinant plant of qGW6. Subsequently, polymorphic markers were synthesized within this interval, and the existing substitution markers were used to conduct MAS on the 100 individual plants. This process aimed to build secondary SSSLs with different genotypes at this locus (some markers were not homozygous). In 2023, 100 plants of each of the selected non-homozygous SSSLs were planted to continue the construction of homozygous SSSLs. Subsequently, all the bred SSSLs with the same genotype and Xihui18 were measured for grain width to be used for the overlapping substitution mapping of qGW6. Then, within the interval of qGW6, the candidate genes were predicted by the related websites involved above, and the entire DNA of these genes between S2 and Xihui18 was sequenced.
According to the previous identification of Z691 substitution segments with 236 polymorphic SSR markers between two parents screened from 429 markers covering the whole rice genome, 10 Z691 plants were further identified with 10 SSR markers on the Z691 substitution segment and more than 90 SSR markers outside the substitution segment for the further identification of substitution segments and genetic background purity testing. The results showed that the substitution segments of 10 plants of Z691 were identical, and no other residual segments from Jinhui35 were detected. This indicates that the genotype of Z691 has become homozygous. Z691 contained four substitution segments from Jinhui35, distributed on chromosomes 4, 5, 6, and 7. The total length of these substitution segments in Z691 was estimated as 16.26 Mb. Among them, the longest substitution segment on Z691 was estimated as 5.225 Mb, the shortest one was 1.755 Mb, and the average length of these substitution segments was 4.065 Mb (Figure 1).
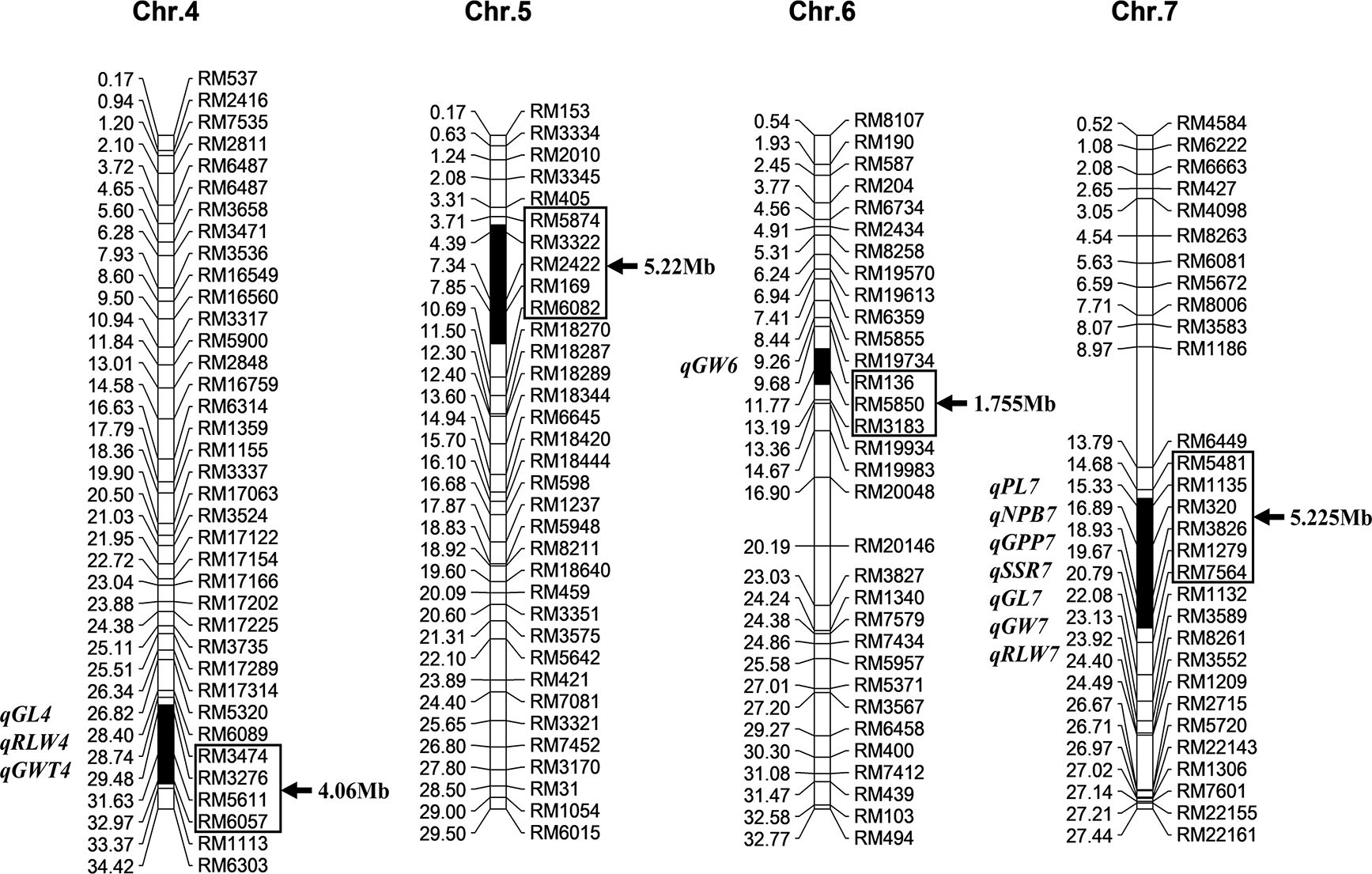
Figure 1. Substitution segments and detected quantitative trait loci (QTLs) of Z691. The left side of each chromosome is the physical distance (Mb) and the QTL of positioning; the right side is the marker name and the length of the chromosome replacement fragment. Substitution intervals (in-box markers). GL is the grain length, RLW is the rate of length to width, GWT is 1000-grain weight, GW is the grain width, PL is panicle length, NPB is number of primary branches, GPP is grain number per panicle, and SSR is seed setting ratio.
Z691 had a plant type similar to the recipient parent, Xihui18. However, there were still many traits that were different; the heading date of Z691 displayed earlier 5 d than that (115 d) of Xihui18 in Chongqing, China (Figure 2A). Compared to Xihui18, Z691 showed a significant increase in seed setting ratio (93.26%) (Figures 2B, F), grain width (3.35 mm) (Figures 2D, J), and 1000-grain weight (29.80 g) (Figure 2L), with increases of 7.35%, 11.30%, and 3.15%, respectively. However, Z691 exhibited a significant decrease in the number of primary branches (14.98) (Figure 2E), spikelet number per panicle (201.57 grains) (Figure 2G), grain number per panicle (187.48 grains) (Figure 2H), grain length (10.43 mm) (Figures 2C, I), the rate of length to width (3.12) (Figure 2K), and yield per plant (25.09 g) (Figure 2M), with reductions of 13.51%, 21.18%, 18.29%, 2.16%, 11.86%, and 21.52%, respectively.
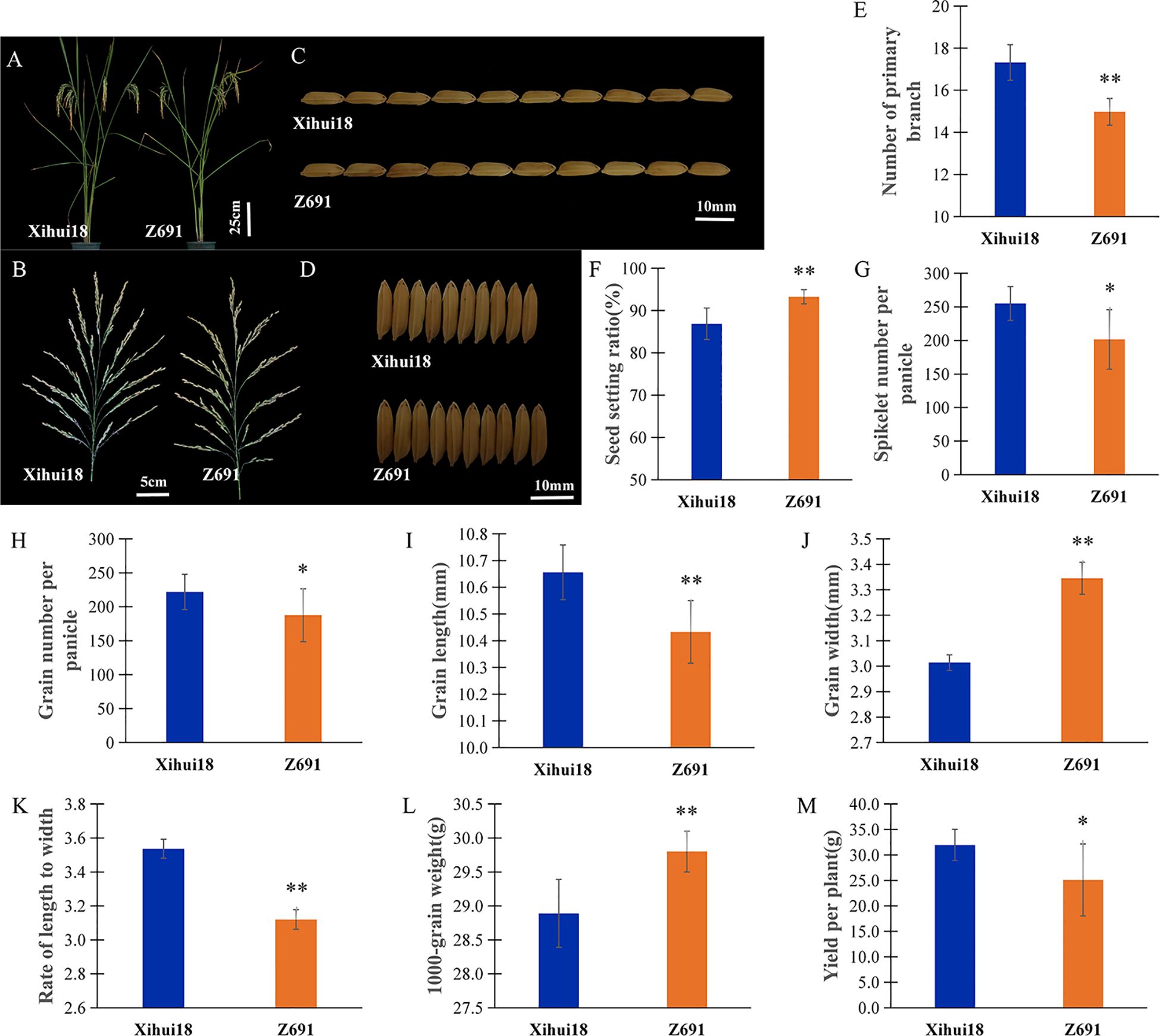
Figure 2. Phenotypic and differential traits of CSSL-Z691 and its recipient Xihui18. (A) Plant type of Xihui18 (left) and Z691 (right). (B) Main panicle of Xihui18 (left) and Z691 (right). (C) Grain length of Xihui18 (upper)and Z691 (lower). (D) Grain width of Xihui18 (upper) and Z691 (lower). (E) Number of primary branches per plant. (F) Seed setting ratio. (G) Spikelet number per panicle. (H) Grain number per panicle. (I) Grain length. (J) Grain width. (K) Rate of length to width. (L) 1000-grain weight. (M) Yield per plant of Xihui18 and Z691. Data are given as the mean and SE (n = 10). * and ** indicate significant differences of traits between Xihui18 and Z691 at p < 0.05 and p < 0.01, respectively. Bars in (A), 25 cm; (B), 5 cm; (C, D), 10 mm.
Eleven QTLs from Jinhui35 were detected in the secondary F2 population from a cross between Xihui18 and Z691. These QTLs were distributed on three substitution segments of Z691 and explained the phenotypic variation from 3.72% to 46.43% (Table 1). Among them was one QTL for panicle length, number of primary branches, grain number per panicle, grain weight, and seed setting ratio. Moreover, there were two QTLs for grain length, grain width, and the rate of length to width. Except for qGW7, the other were all major QTLs, explaining more than 10% of the variation. The additive effect of qPL7 reduced the panicle length of Z691 by 0.65 cm, explaining 11.66% of the variation in the panicle length. The additive effect of qNPB7 decreased the number of primary branches of Z691 by 0.70, explaining 35.29% of the variation in the number of primary branches. The additive effect of the qGPP7 reduced the grain number per panicle of Z691 by 14.23 grains, explaining 11.22% of the variation in grain number per panicle. The additive effect of the qSSR7 decreased the seed setting ratio of Z691 by 3.85%, explaining 19.27% of the variation in the seed setting ratio. qGL4 and qGL7-1 were major QTLs, controlling the grain length of Z691, and their additive effects reduced the grain length of Z691 by 0.16 and 0.20 mm, respectively. qGL4 and qGL7-1 explained 22.14% and 34.39% of the variation in grain length, respectively. The grain width of Z691 was mainly controlled by qGW6 and qGW7, whose additive effects increased the grain width of Z691 by 0.03 mm and decreased it by 0.02 mm, respectively. qGW6 and qGW7 explained 10.61% and 3.72% of the variation of grain length, respectively. The additive effects of qRLW4 and qRLW7 reduced the rate of length to width of Z691 by 0.05 and 0.08, respectively. qRLW4 and qRLW7 explained 18.93% and 46.43% of the variation in the rate of length to width, respectively. The additive effect of qGWT4 on the 1000-grain weight of Z691 could reduce the 1000-grain weight of Z691 by 0.75 g, explaining 16.72% of the variation of 1000-grain weight (Table 1).
Based on the QTL mapping results, the MAS was utilized in the F3:4 generation to dissect further three SSSLs (S1–S3) and three DSSLs (D1–D3). S1 harbored the substitution segment RM3474–RM3276-RM5611–RM6057 of chromosome 4. S2 harbored the substitution segment RM136–RM5850–RM3183 of chromosome 6. S3 harbored the substitution segment RM3826–RM1279–RM7564 of chromosome 7. D1 harbored one substitution segment on chromosome 4 (RM3474–RM3276-RM5611–RM6057) and one substitution segment on chromosome 7 (RM3826–RM1279–RM7564). D2 harbored one substitution segment on chromosome 4 (RM3474–RM3276-RM5611–RM6057) and one substitution segment on chromosome 6 (RM136–RM5850–RM3183). D3 harbored one substitution segment on chromosome 6 (RM136–RM5850–RM3183) and one substitution segment on chromosome 7 (RM3826–RM1279–RM7564). All these substitution segments above were from Jinhui35 (Figure 3).
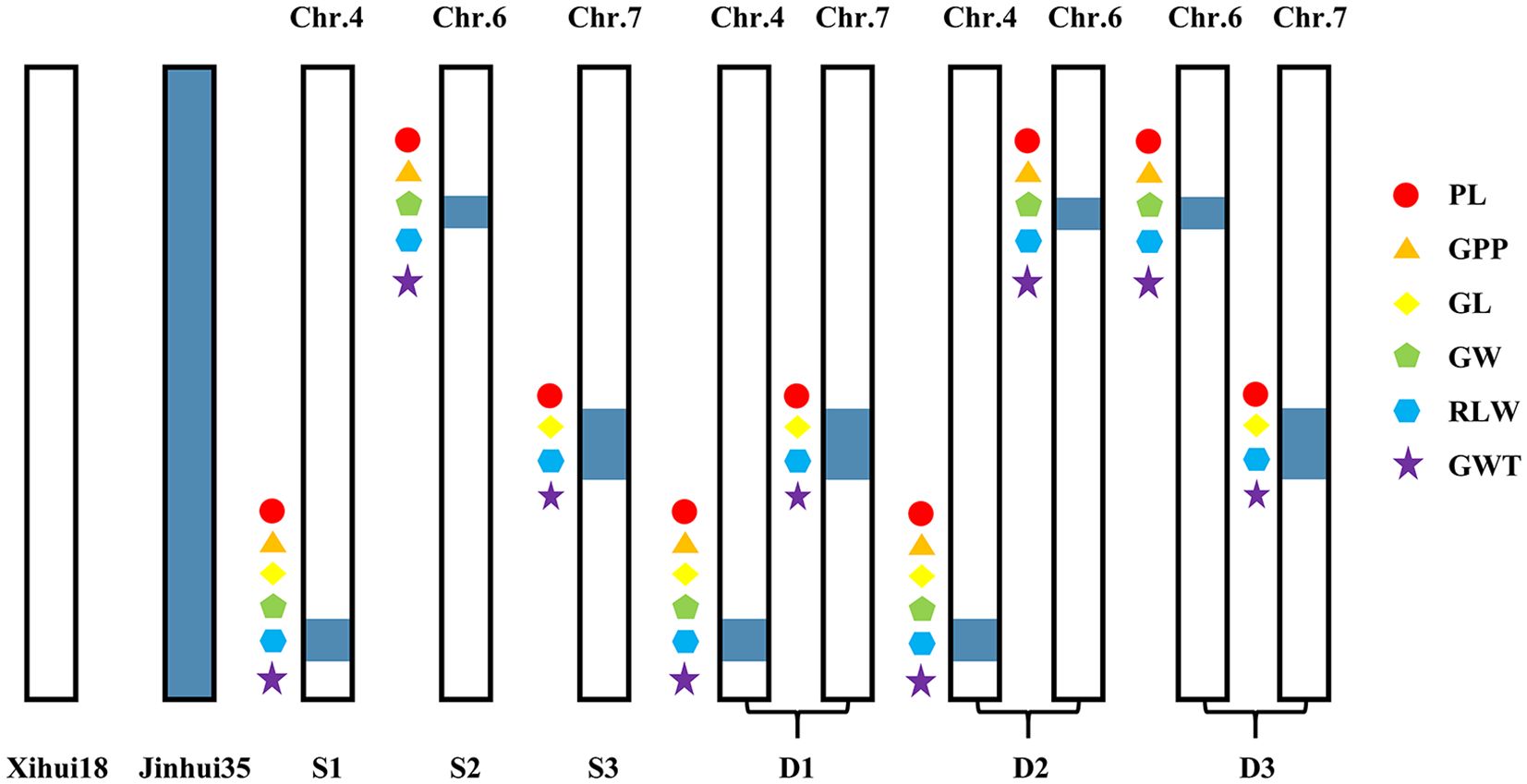
Figure 3. Map of substitution segment on secondary substitution lines. Chr, chromosome; S, single-segment substitution line; D, dual-segment substitution line. PL is panicle length, GPP is grain number per panicle, GL is the grain length, GW is the grain width, RLW is the rate of length to width, and GWT is the 1000-grain weight. S1: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb)]. S2: [Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb)]. S3: [Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)]. D1: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb), Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)]. D2: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb), Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb)]. D3: [Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb), Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)]. Internal tags connected with “-” indicate substitution fragments from donors, while tags at both ends of substitution fragments connected with “–” indicate that fragment reorganization may occur.
S1 contained the substitution segment on chromosome 4. A total of six QTLs (qGL4, qRLW4, qGWT4, qPL4, qGPP4, and qGW4) were detected in S1 (Figure 3). Among them, qGL4, qRLW4, and qGWT4 can also be detected in the F2 population, with their additive effects being −0.24 mm (Figure 4A), −0.13 (Figure 5E), and −0.35 g (Figure 4F), respectively. Additionally, three new QTLs, qPL4, qGPP4, and qGW4, were detected in S3, with their additive effects being −0.51 cm (Figure 4C), 10.32 grains (Figure 4D), and 0.05 mm (Figure 4B), respectively. However, these QTLs were not detected in the F2 population.
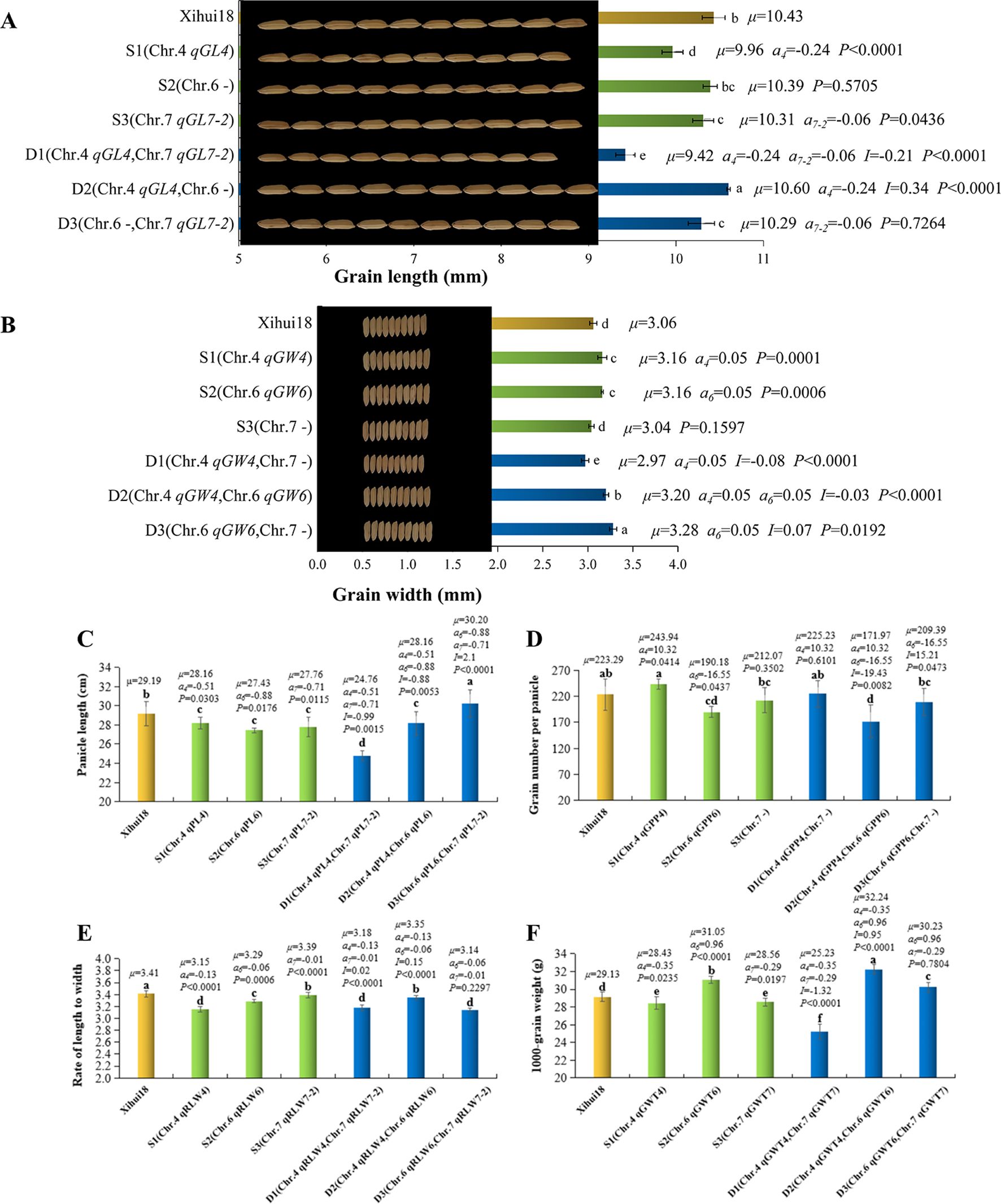
Figure 4. Analysis of additive and epistatic effects of quantitative trait loci (QTLs) for related traits of single-segment substitution lines (SSSLs) and dual-segment substitution lines (DSSLs). (A) Grain length (GL). (B) Grain width (GW). (C) Panicle length (PL). (D) Grain number per panicle (GPP). (E) Rate of length to width (RLW). (F) 1000-grain weight (GWT). The p-value of SSSL indicates the probability of a significant difference between SSSL and Xihui18 by one-way ANOVA, and p < 0.05 denotes carrying QTL in the substitution segment. The p-value of DSSL represents the probability of QTL1 × QTL2 of the DSSL, and p < 0.05 denotes existing epistatic effects between QTL1 and QTL2 of DSSL. S1: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb)]. S2: [Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb)]. S3: [Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)]. D1: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb), Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)]. D2: [Chr.4:RM3474 (28.74 Mb)-RM3276 (29.48 Mb)-RM5611 (31.63 Mb)–RM6057 (32.97 Mb), Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb)]. D3: [Chr.6:RM136 (9.68 Mb)–RM5850 (11.77 Mb)–RM3183 (13.19 Mb), Chr.7:RM3826 (18.93 Mb)–RM1279 (19.67 Mb)–RM7564 (20.79 Mb)). Internal tags connected with “-” indicate substitution fragments from donors, while tags at both ends of substitution fragments connected with “–” indicate that fragment reorganization may occur. Different small letters indicate a significant difference (P <0.05) as determined by Duncan’s multiple comparisons.
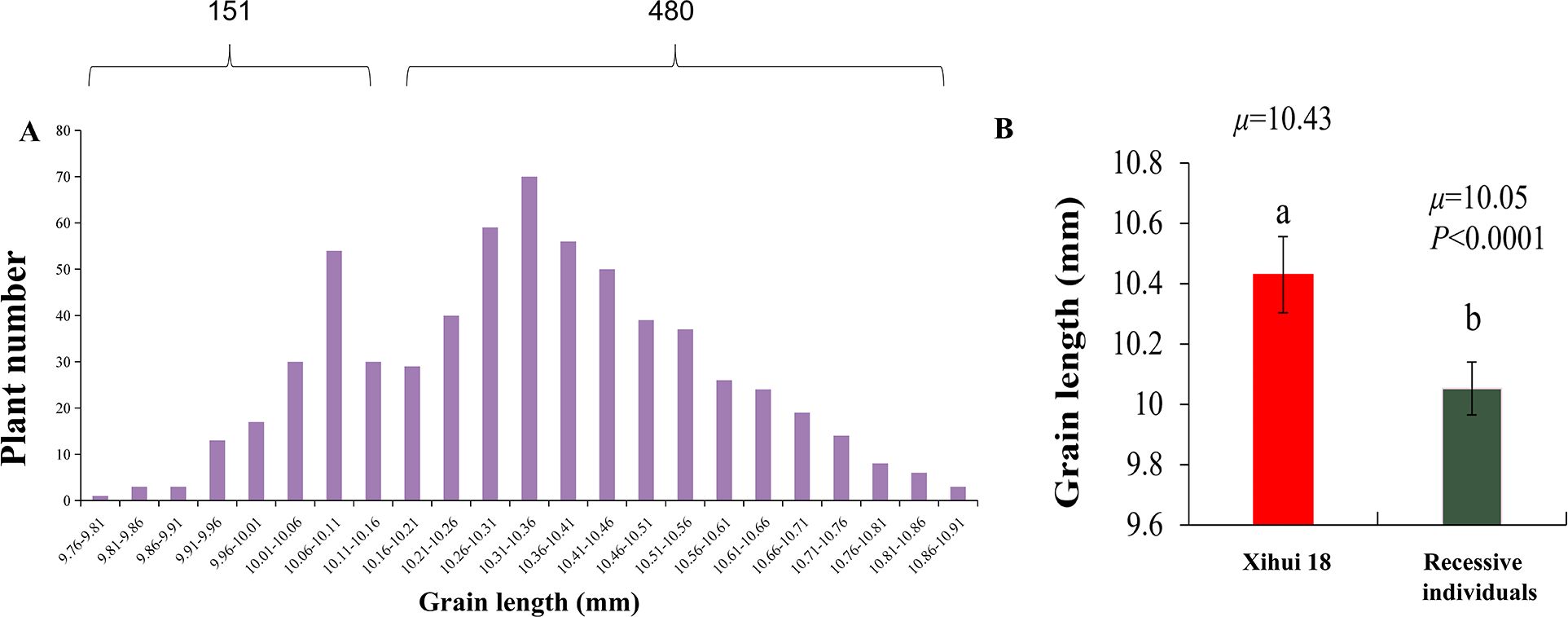
Figure 5. Genetic analysis of grain length of qGL4. (A) Distribution of grain length in F3 population. (B) Comparison of grain length between Xihui18 and recessive individuals. Different small letters indicated the sigmificant difference between Xihui18 and the recessive individuals.
S2 contained the substitution segment on chromosome 6. A total of five QTLs (qGW6, qPL6, qGPP6, qRLW6, and qGWT6) were detected in S2 (Figure 3). Among these, qGW6 can also be detected in the F2 population, with an additive effect of 0.05 mm (Figure 4E). In addition, four new QTLs, qPL6, qGPP6, qRLW6, and qGWT6, were detected in S4, with additive effects being −0.88 cm (Figure 4C), 16.55 grains (Figure 4D), −0.06 (Figure 4E), and 0.96 g (Figure 4F), respectively.
S3 contained the substitution segment on chromosome 7. A total of four QTLs (qGW7, qPL7-2, qGL7-2, qRLW7-2, and qGWT7) were detected in S3 (Figure 3). Among them, qGW7, which was detected in the F2 population, was not confirmed by S3. However, four new QTLs, qPL7-2, qGL7-2, qRLW7-2, and qGWT7, were detected in S5, with additive effects being −0.71 cm (Figure 4C), −0.06 mm (Figure 4A), −0.01 (Figure 4E), and −0.29 g (Figure 4F), respectively.
In conclusion, among the 11 QTLs (qPL7, qNPB7, qGPP7, qSSR7, qGL4, qGL7, qGW6, qGW7, qRLW4, qRLW7, and qGWT4) detected in the secondary F2 population of Xihui18/Jinhui35, four QTLs (qGL4, qRLW4, qGWT4, and qGW6) were validated by their corresponding SSSLs, with only the minor effect QTL, qGW7, not confirmed by its corresponding SSSL. The other six QTLs, qPL7, qNPB7, qGPP7, qSSR7, qGL7, and qRLW7, could not be validated due to the absence of corresponding SSSLs. Therefore, these six QTLs require further validation in the following years. In addition, within these three SSSLs (S1–S3), 11 new QTLs (qPL4, qGPP4, qGW4, qPL6, qGPP6, qRLW6, qGWT6, qPL7-2, qGL7-2, qRLW7-2, and qGWT7) were detected. These QTLs were not identified in the F2 population but in the corresponding SSSLs, suggesting that SSSLs offer a higher efficiency in QTL detection due to only one segment on Jinhui35 introgressed in each SSSL based on the Xihui18 background, eliminating the influence of other identical trait QTLs (Deng et al., 2023).
The interaction of qGL4 (a = −0.24 mm) and qGL7-2 (a = −0.06 mm) produced an epistatic effect of −0.21, which decreased grain length genetically by 0.51 mm in D1. Owing to −0.51 < −0.24 < −0.06, the grain length (9.42 mm) of D1 was significantly shorter than that (9.96, 10.31, and 10.43 mm) of S1 (qGL4), S3 (qGL7-2), and Xihui18. The interaction of qGL4 (a = −0.24 mm) and a substitution locus without GL on chromosome 6 in D2 produced an epistatic effect of 0.34, which increased grain length genetically by 0.10 mm. Thus, the grain length (10.60 mm) of D2 was significantly longer than that (9.96, 10.39, and 10.43 mm) of S1 (qGL4), S2 (without GL QTL), and Xihui18. This indicates that the substituted segment on chromosome 6, which does not contain grain length QTLs, also significantly affects the expression of qGL4, resulting in a significant increase in grain length in D2. However, qGL7-2 (a = −0.06 mm) and a substitution locus without GL on chromosome 6 belonged to independent inheritance in D3, indicating that the genetic effect of D3 is equal to the additive effect (−0.06 mm) of qGL7-2. Accordingly, the grain length (10.29 mm) of D3 displayed no significant difference compared to that (10.31 mm) of S3 containing qGL7-2 while significantly shorter than that (10.39 and 10.43 mm) of S2 and Xihui18 (Figure 4A).
The interaction of qGW4 (a = 0.05 mm) and a substitution locus without grain width (GW) on chromosome 7 in D1 produced an epistatic effect of −0.08, resulting in −0.03 mm of the genetic effect of D1 for grain width. Thus, the grain width (2.97 mm) of D1 was significantly narrower than that (3.16, 3.04, and 3.06 mm) of S1 (qGW4), S3 (without GW QTL), and Xihui18. This indicates that the substitution segment on chromosome 7, no grain width QTL, also significantly affects the expression of qGW4, resulting in a significant decrease in grain length in D1. The interaction of qGW4 (a = 0.05 mm) and qGW6 (a = 0.05 mm) produced an epistatic effect of −0.03, causing the genetic effect of 0.07 mm in D2, larger than any of the additive effects of the single QTL. Accordingly, the grain width (3.20 mm) of D2 was significantly wider than that (3.16, 3.16, and 3.06 mm) of S1 (qGW4), S2 (qGW6), and Xihui18. This indicates that the interaction of qGW4 and qGW6 creates a wider grain. The interaction of qGW6 (a = 0.05 mm) and a substitution locus without GW on chromosome 7 produced an epistatic effect of 0.07, which increased GW genetically by 0.12 mm in D3, larger than the additive effect of qGW6. Thus, the grain width (3.28 mm) of D3 was significantly broader than that (3.16, 3.04, and 3.06 mm) of S2 (qGW6), S3 (without GW QTL), and Xihui18. This indicates that the substituted segment on chromosome 7, no GW QTL, also significantly affects the expression of qGW6, resulting in a significant increase in grain width in D3 (Figure 4B).
The interaction of qPL4 (a = −0.51 cm) and qPL7-2 (a = −0.71 cm) produced an epistatic effect of −0.99 cm, resulting in −2.21 cm of the genetic effect of D1 for panicle length, less than any of the additive effect of each single QTL. Thus, the panicle length (24.76 cm) of D1 was significantly shorter than that (28.16, 27.76, and 29.19 cm) of S1 (qPL4), S3 (qPL7-2), and Xihui18. This indicates that the interaction of qPL4 and qPL7-2 resulted in a shorter panicle length. The interaction of qPL4 (a = −0.51 cm) and qPL6 (a = −0.88 cm) produced an epistatic effect of 0.88 cm, resulting in −0.51 cm of genetic effect for panicle length in D2, equal to the additive effects of qPL4 and qPL6. Thus, the panicle length (28.16 mm) of D2 displayed no significant difference compared to that (28.16 and 27.43 cm) of S1 (qPL4) and S2 (qPL6) while significantly shorter than that (3.06 mm) of Xihui18. The interaction of qPL6 (a = −0.88 cm) and qPL7-2 (a = −0.71 cm) produced an epistatic effect of 2.10 cm, causing 0.51 cm of the genetic effect for panicle length in D3, larger than all the additive effects (−0.88 and −0.71 cm) of each single QTL. Accordingly, the panicle length (30.20 cm) of D3 was significantly longer than that (27.43, 27.76, and 29.19 cm) of S2 (qPL6), S3 (qPL7-2), and Xihui18 (Figure 4C). This indicates that the pyramiding of qPL6 and qPL7-2 can produce a longer panicle length.
qGPP4 (a = 10.32 grains) and a substitution locus without grain number per panicle (GPP) on chromosome 7 in D1 belonged to independent inheritance. Thus, the genetic effect for grain number per panicle of D1 equals the additive effect of qGPP4. Accordingly, D1 displayed no significant difference in grain number per panicle (225.23 grains) from S1 (243.94 grains) containing qGPP4. The interaction of qGPP4 (a = 10.32 grains) and qGPP6 (a = −16.55 grains) produced an epistatic effect of −19.43 grains, resulting in −25.66 grains of genetic effect in D2, parallel to the additive effect of qGPP6. Thus, the grain number per panicle (171.97 grains) of D2 exhibited no significant difference from that (190.18 grains) of S2 carrying qGPP6 while significantly less than that (243.94 and 223.29 grains) of S1 (qGPP4) and Xihui18. This indicates that qGPP6 is epistatic to qGPP4. The interaction of qGPP6 (a = −16.55 grains) and a substitution locus without GPP on chromosome 7 produced an epistatic effect of 15.21 grains, resulting in −1.34 grains of the genetic effect of D3, parallel to the additive effect of qGPP6. Thus, the grain number per panicle (209.39 grains) of D3 displayed no significant difference from that (190.18, 212.07, and 223.29 grains) of S2 (qGPP6), S3 (without GPP QTL), and Xihui18. This suggests that the segment on chromosome 7 without the GPP locus has no significant effect on the expression of qGPP6 (Figure 4D).
The interaction of qRLW4 (a = −0.13) and qRLW7-2 (a = −0.01) produced an epistatic effect of 0.02, resulting in −0.12 of the genetic effect for the rate of length to width in D1, parallel to the additive effect of qRLW4. Accordingly, the rate of length to width (3.18) of D1 showed no significant difference from that (3.15) of S1 containing qRLW4 while significantly lower than that (3.41 and 3.39) of Xihui18 and S3 (qRLW7-2). The interaction of qRLW4 (a = −0.13) and qRLW6 (a = −0.06) produced an epistatic effect of 0.15, causing −0.04 of the genetic effect of D2, larger than the additive effects of each single QTL. Accordingly, the rate of length to width (3.35) of D2 was significantly higher than that (3.15 and 3.29) of S1 (qRLW4) and S2 (qRLW6) while significantly lower than that (3.41) of Xihui18. However, qRLW6 (a = −0.06) and qRLW7-2 (a = −0.01) belonged to independent inheritance in D3. Thus, the genetic effect of D3 was −0.07 for the rate of length to width. Accordingly, the rate of length to width of D3 (3.14) was significantly lower than that (3.29, 3.18, and 3.41) of S2 (qRLW6), S3 (qRLW7-2), and Xihui18 (Figure 4E).
The interaction of qGWT4 (a = −0.35 g) and qGWT7 (a = −0.29 g) in D1 produced an epistatic effect of −1.32, resulting in −1.96 g of the genetic effect of D1, smaller than any of the additive effect of each single QTL. Thus, the 1000-grain weight of D1 (25.23 g) was significantly lighter than that (28.43, 28.56, and 29.13 g) of S1 (qGWT4), S3 (qGWT7), and Xihui18. The interaction of qGWT4 (a = −0.35 g) and qGWT6 (a = 0.96 g) produced an epistatic effect of 0.95, resulting in 1.56 g of the genetic effect of D2, larger than any of the additive effects of each single QTL. Accordingly, the 1000-grain weight of D2 (32.24 g) was significantly heavier than that (28.43, 31.05, and 29.13 g) of S1 (qGWT4), S2 (qGWT6), and Xihui18. However, qGWT6 (a = 0.96 g) and qGWT7 (a = −0.29 g) were inherited independently in D3. Therefore, the genetic effect on 1000-grain weight was 0.67 g in D3, larger than the additive effect of qGWT7 whereas less than that of qGWT6. Accordingly, the 1000-grain weight of D3 (30.23 g) was significantly heavier than that (29.13 g) of Xihui18 and S3 (28.56 g) containing qGWT7 while significantly lighter than that of S2 (31.05 g) carrying qGWT6 (Figure 4F).
The recipient parent, Xihui18, has a long-grain phenotype, while the recipient parent, Jinhui35, has a short-grain phenotype. The grain length of Z691 is significantly shorter than that of Xihui18. An F3 population was further developed from a self-cross of an F2 recombinant plant containing only the heterozygous qGL4 locus. In this F3 population of 631 individual plants, the grain lengths’ frequency distribution was bimodal, with grain lengths ranging from 9.76 to 10.91 mm. The short-grain peak is mainly between 9.76 and 10.16 mm, with a total of 151 plants, and the long-grain peak is mainly between 10.16 and 10.91 mm, with a total of 480 plants. After chi-square testing, the ratio of long-grain plants (480) to short-grain plants (151) fits a 3:1 segregation ratio (χ2 = 0.36 < χ2 (0.05,1) = 3.84), indicating that the short grain controlled by qGL4 in S1 is governed by an incomplete single recessive gene (Figure 5A). Comparing the grain length of the 151 recessive plants with that of Xihui18, it was found that the grain length of the recessive plants is significantly shorter than that of Xihui18, further indicating that the long-grain trait is incompletely dominant over the short-grain trait (Figure 5B).
qGL4 is a major QTL for grain length, explaining 22.14% of the variation. Genetic analysis indicates that qGL4 from Jinhui35 is inherited as a single recessive gene in S1. Within the initial mapping interval of qGL4, RM3474–RM3276–RM6057, 12 new primer pairs were synthesized, of which five pairs were polymorphic, namely, RM17446, RM17450, RM17453, RM17468, and RM17401. However, the bands of RM17446 and RM17401 were consistent with the recipient Xihui18 in S1, leaving only three pairs of markers as substitution segment markers. Consequently, the estimated length of the substitution segment in S1 was shortened from the original 1.445 Mb to 495 kb. The maximum length of the substitution segment was shortened from the original 2.89 Mb to 600 kb (Figure 6A). Subsequently, using the dense markers on these three substitution segments and the original substitution markers, linkage analysis with qGL4 was conducted by 151 recessive plants (short grain) in the F3 population. By calculating their genetic distances, qGL4 was ultimately localized to an 80-kb interval between RM17453 and RM3276 (Figure 6B).
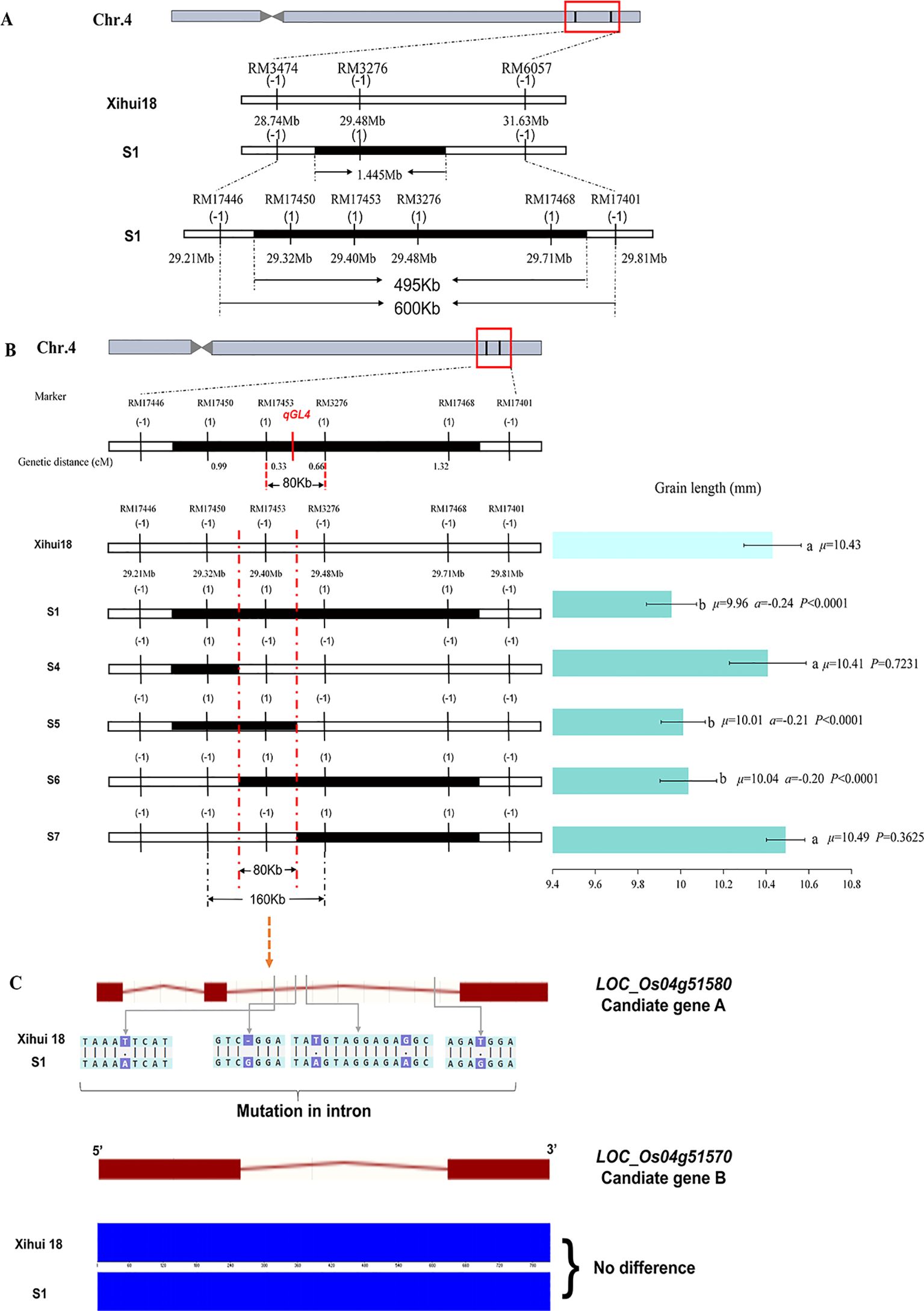
Figure 6. Fine-mapping and candidate gene analysis of qGL4. (A) Shorten the replacement fragment on Z691 by encryption marker. (B) Fine-mapping of qGL4 by linkage analysis. Diffferent small letters indicated the significant difference among the genetic materials. (C) DNA sequence of Candidate gene A and Candidate gene B for qGL4 in S1 compared with that of Xihui18. The green lines with arrowhead indicate the variation between the single-segment substitution lines (SSSLs) and Xihui18. The red rectangles indicate the exons, and the red lines indicate the introns. The blue rectangles indicate the sequencing peak map.
Furthermore, to construct an SSSL containing the shortest length of qGL4, MAS was conducted on all substitution markers for 100 plants from the F3 population, and a total of four SSSLs (S4–S7) were constructed for this locus. S1 (with a grain length of 9.96 mm), which contained the substitution segment RM17446–RM17450-RM17453-RM3276-RM17468–RM17401, S5 (with a grain length of 10.01 mm), which contained the substitution segment RM17446–RM17450-RM17453–RM3276, and S6 (with a grain length of 10.04 mm), which contained the substitution segment RM17450–RM17453-RM3276-RM17468–RM17401, all had grain lengths significantly shorter than those of Xihui18 (10.43 mm), with overlapping substitution regions at RM17453. However, S4, containing the substitution segment RM17446–RM17450–RM17453 with a grain length of 10.41 mm, and S7, containing the substitution segment RM17453–RM3276-RM17468–RM17401 with a grain length of 10.49 mm, showed no significant difference in grain length compared to Xihui18 (10.43 mm). According to the theory of overlapping substitution mapping, qGL4 was located within the overlapping intervals of S1, S5, and S6. Ultimately, qGL4 was localized to an estimated length of 80 kb within RM17450–RM17453–RM3276, with the maximum length being 160 kb (Figure 6B).
Within the interval, two prospective genes of qGL4 were determined and named Candidate gene A and Candidate gene B (Figure 6C). The Candidate gene A (LOC_Os04g51580) exhibited multiple differences between Xihui18 and S1, including four single-nucleotide polymorphism (SNP) differences and one base insertion in the intron region, which may affect the alternative splicing of this gene in S1. As for the Candidate gene B (LOC_Os04g51570), there were no differences between Xihui18 and S1. Therefore, LOC_Os04g51580 (Candidate gene A) can be considered the candidate gene for qGL4.
Furthermore, qGW6 is a major QTL for grain width located in the RM136–RM5850–RM3183 interval of S2, explaining 10.61% of variation. Furthermore, we synthesized 14 new primer pairs, of which three showed polymorphisms between Xihui18 and Jinhui35, namely, RM19834, RM3330, and RM19849. However, the bands of RM19834 and RM19849 were consistent with the recipient Xihui18 in S2, leaving only one pair of markers as substitution segment markers. Consequently, the estimated length of the substitution segment in S2 was reduced from the original 1.755 Mb to 230 kb, and the maximum length of the substitution segment was shortened from the original 3.51 Mb to 390 kb (Figure 7A).

Figure 7. Overlapping group substitution mapping and candidate gene analysis of qGW6. (A) Shorten the replacement fragment on Z691 by encryption marker. (B) Fine-mapping of qGW6 by linkage analysis. Different small letters indicated significant difference among the genetic materials. (C) DNA sequence of Candidate gene C for qGW6 in S2 compared with that of Xihui18. The red rectangles indicate the exons, and the red lines indicate the introns. The green lines with arrowhead indicate the variation between the single-segment substitution lines (SSSLs) and Xihui18.
Using an F2 recombinant plant containing only the qGW6 locus for self-crossing to develop an F3 population, we conducted MAS on 100 individual plants with substitution markers, and we constructed a total of two SSSLs (S8 and S9) for this locus. S2 (with a grain width of 3.15 mm), containing the substitution segment RM19834–RM5850-RM3330–RM19849 and S8 (with a grain width of 3.20 mm), containing the substitution segment RM19834–RM5850–RM3330, had significantly larger grain widths than Xihui18 (3.06 mm), and they included an overlapping substitution interval RM19834–RM5850–RM3330. Additionally, S9, containing the substitution segment RM5850-RM3330–RM19849, had a grain width (3.06 mm) similar to that (3.06 mm) of Xihui18. Applying the overlapping substitution mapping theory indicates that qGW6 was located within the overlapping interval of S2 and S8. Ultimately, qGW6 was localized to an estimated length of 120 kb within RM19834–RM5850–RM3330, with the maximum estimated length being 240 kb (Figure 7B).
Within the interval, LOC_Os06g19070 was predicted as the candidate gene of qGW6. It exhibited two SNP differences at the 1594th and 1598th bases of its exon; however, only one resulted in an amino acid change (Figure 7C).
Most of the agronomic traits in rice are quantitative traits controlled by multiple genes, such as plant height, grain length, grain width, and yield (Ren et al., 2023; Xing and Zhang, 2010). Accurately mapping these QTLs is fundamental to studying these important agronomic traits. However, the genetic background of primary mapping populations is very complex, which is not conducive to accurate QTL mapping and identifying individual QTL genetic effects (Xu et al., 2017). CSSLs have only a few chromosome segments from the donor parent, with the rest of the genetic background consistent with the recipient parent. Therefore, using CSSLs greatly improves the efficiency of QTL detection and lays the foundation for molecular design breeding in rice (Li et al., 2022b). In this study, the dense-grain and large-grain rice CSSL Z691 was identified as containing four substitution segments from Jinhui35 based on the genetic background of Xihui18, with a total length of substitution segments of 16.26 Mb and an average length of substitution of 4.045 Mb. However, Z691 was not still an excellent breeding material because contained some unfavorable traits, such as the grain number per panicle significantly decreased by 21.18%, the grain length decreased by 18.29%, and the yield per plant decreased by 21.52% compared with those of the large panicle type restorer line Xihui18. Thus, to create more ideal materials for direct breeding and basic study in gene functional analysis, nine SSSLs and three DSSLs were further constructed carrying various QTLs by QTL mapping and MAS through Xihui18/Z691. S1 harboring qGL4 (a = −0.24 mm), qGW4 (a = 0.05 mm), qGWT4 (a = −0.35 g), qPL4 (a = −0.51 cm), and qGPP4 (a = 10.32 grains) displayed 9.66 mm of grain length, 3.16 mm of grain width, 28.43 g of 1000-grain weight, 28.16 cm of panicle length, and 243.94 grains per panicle, which belonged to a multiple grain and large grain type breeding material. S2 containing qGW6 (a = 0.05 mm), qGWT6 (a = 0.96 g), qPL6 (a = −0.88 cm), and qGPP6 (a = −16.55 grains) exhibited 3.16 mm of grain width, 31.05 g of 1000-grain weight, 27.43 cm of panicle length, and 190.18 grains per panicle, which still belonged to a multiple grain and large grain type breeding material. S3 carrying qGL7-2 (a = −0.06 mm), qGWT7 (a = −0.29 g), and qPL7-2 (a = −0.71 cm) showed 10.31 mm of grain length, 28.56 g of 1000-grain weight, 27.76 cm of panicle length, and 212.07 grains per panicle, which also belonged to a multiple and large grain type breeding material. In comparison, S1 and S2 are better breeding materials. However, in the future, S3 can be edited using the Cas9 technique to knock out qGL7-2, qGWT7, and qPL7-2, so S3 is potentially a great breeding material. In addition, Deng et al. (2023), using Xihui18 as the recipient parent, developed four SSSLs and detected 25 QTLs, including qPN3, qGW3, qPN4, and qPL4. Liang et al. (2021) constructed four SSSLs on the genetic background of Xihui18 and detected eight QTLs for grain size, including qGL3-2, qGL3-1, and qGL7. Li et al. (2022a) developed six SSSLs on the genetic background of Xihui18 and detected 11 QTLs, including qGL11, qGL3, qGL5, and qGW5. Sun et al. (2022) developed six SSSLs on the genetic background of Xihui18 and detected 15 QTLs, including qPN5, qGL1, qGL2, and qGL5. Nevertheless, these SSSLs are all different from those constructed in this study. Thus, the new SSSLs dissected from Z691 are important to perfect the SSSL library. Notably, using SSSLs as a platform to pyramid favorable alleles for breeding new varieties has become a reality (Dai et al., 2015). Some SSSLs have been successfully used to design a series of new cultivars, such as Hua Xiaohei No. 1 and Hua Biao No. 1 (Zhang, 2021). In the future, these SSSLs can be re-sequenced to ensure a completely purified background with the price of high-throughput sequencing becoming cheaper. Therefore, these SSSLs lay a solid foundation for the genetic analysis of QTLs for grain size and other traits and establish a basis for designing breeding rice varieties that meet production needs using SSSLs as the platform.
We utilized the secondary F2 population of Xihui18/Z691 and the three dissected single-segment substitution lines (S1–S3) to identify 20 QTLs for important agronomic traits in rice. By comparing with genes previously mapped or cloned in these intervals, we found that OsHXK1 is located within the substitution intervals of qGPP7 and qSSR7. qGPP7 and qSSR7 are linked to the same substitution interval (RM5481–RM1135–RM320), and they may represent pleiotropy. OsAGO2 is also involved in the development of rice. Further research revealed that the knockdown of OsAGO2 leads to a significant upregulation of the expression level of the hexokinase gene OsHXK1 and a significant decrease in the methylation level of its promoter. That indicates that OsAGO2 directly binds to the promoter region of OsHXK1 and regulates the expression of OsHXK1 through DNA methylation, thereby affecting pollen development and significantly reducing the grain number and the seed setting ratio (Zheng et al., 2019). Therefore, OsAGO2 can be considered a candidate gene for qGPP7 and qSSR7.
qGL4, qRLW4, and qGWT4 are located within the substitution interval (RM3474–RM3276-RM5611–RM6057), where the gene FLO2 was found. The FLO2 mutation results in smaller grains and reduced starch quality. FLO2 is highly expressed in developing seeds, coinciding with the starch and storage protein synthesis period, and is also highly expressed in leaves. The overexpression of FLO2 can significantly increase grain size, and FLO2 plays a key regulatory role in grain size and starch quality by affecting the accumulation of storage substances in the endosperm (She et al., 2010). Compared to the wild type (WT), at the seedling stage, the DHT1 mutant exhibits slower growth and smaller roots, although the number of tillers at this stage does not significantly differ from that of the WT. However, at the heading stage, the DHT1 mutant has a reduced plant height due to shorter internodes and has more tillers. Histological analysis indicates that the stem cells of the DHT1 mutant are smaller than those of the wild type, with reduced cell width and length (Liu et al., 2022).
Similarly, S1 harboring qRLW4 and qGWT4 displayed shorter grain length and lighter grain weight than Xihui18. This suggests that FLO2 can be a candidate gene for qGL4, qRLW4, and qGWT4. In addition, we further fine-mapped the qGL4 into 160 kb of the largest substitution interval and found a candidate gene LOC_Os04g51580 (a leucine-rich repeat-containing protein) by DNA sequencing. However, FLO2 was not within the 160-kb interval. OsPEX1, encoding a leucine-rich repeat extension protein, regulates grain size and rice quality (Luan et al., 2021). As for whether FLO2 or LOC_Os04g51580 is the target gene of qGL4, it needs to be confirmed by a genetic complementary test in the future.
qGW6, qRLW6, and qGWT6 are located within the same substitution interval (RM136–RM5850–RM3183) and may exhibit pleiotropy. Within this interval, the gene OsGSR1 has been found to regulate cell growth rather than cell proliferation by positively regulating the gibberellin signaling pathway by modulating the response and biosynthesis of gibberellin, thereby controlling rice grain size. The knockout of OsGSR1 results in reduced grain size and weight and a decrease in gibberellin content in the young panicle, while the overexpression of OsGSR1 leads to an increase in grain size and weight (Shi et al., 2020). Additionally, gene expression analysis has shown that OsGSR1 is also regulated by brassinolide treatment and accumulates in the panicle, indicating that OsGSR1 may also be involved in the brassinosteroid-regulated pathway for controlling grain length (Tang et al., 2020). S2 containing qGW6, qRLW6, and qGWT6 from Huhan3 exhibited an increase in grain width and weight compared with Xihui18. This suggests that OsGSR1 can be a candidate gene for qGW6, qRLW6, and qGWT6.
Furthermore, qGW6 was limited to 240 kb of the largest substitution interval through overlapping substitution mapping, LOC_Os06g19070 was determined as the candidate gene by DNA sequencing, and OsGSR1 was not within the 240-kb interval. LOC_Os06g19070, also a cytochrome P450 protein, controls grain width by regulating the gibberellin (GA) content in rice spikelets (Dang et al., 2024). GNS4 also encodes cytochrome P450 protein by the brassinosteroid (BR) signaling pathway to regulate rice grain size. The promoter variation of the gene in the gns4 mutant exhibits a small grain size (Zhou et al., 2017). A genetic complementary test must also verify whether LOC_Os06g19070 or OsGSR1 is the candidate gene for qGW6.
qGW7, qGL7-2, and qGWT7 are linked to the marker RM1279 (19.67 Mb), and near this marker, OsBZR1 is a downstream signaling molecule in BR signaling. Compared to the non-transgenic control, the overexpression of OsBZR1 leads to an increase in sugar accumulation in developing anthers and seeds, as well as an increase in grain yield, with enhancements in grain length, width, thickness, 1000-grain weight, and the spikelet number per panicle. In contrast, suppressing OsBZR1 expression results in pollen maturation defects, reduced seed size and weight, and decreased starch accumulation. Furthermore, during the development of rice pollen and seeds, OsBZR1 can directly promote the expression of CSA, which triggers the expression of genes related to sugar allocation and metabolism (Zhu et al., 2015). S3 carrying qGL7-2 and qGWT7 from Jinhui35 displayed a decrease in grain length and grain weight. Therefore, OsBZR1 may be a candidate gene for qGW7.
qGPP4 is linked to the marker RM3276 (29.48 Mb), and LVPA4 and OsNPF7.4 are very close to qGPP4 in position. Compared to the recurrent parent, the near-isogenic line NIL-LVPA4LT showed an increase of 7.8% in the total area of the large vascular bundles in the neck of the panicle, an increase of 12.7% in the phloem of the large vascular bundles, and an increase of 9.4% in the number of large vascular bundles. Still, there were no significant differences in the number of small vascular bundles and the area of the xylem of the large vascular bundles. In addition, the size of the panicle; the length, width, and area of the flag leaf; and the leaf area index significantly increased, especially with a 17.2% increase in the spikelet number per panicle, a 19.7% increase in the number of secondary branches, a 22.5% increase in grain number per panicle, and an increase in the content of non-structural carbohydrates in the sheath of the stem at the heading stage, ultimately leading to a 7.6%–9.6% increase in the yield of NIL-LVPA4LT in plot trials while also improving rice quality. The study shows that the single gene LVPA4 improves rice yield and quality through the coordinated action of source, sink, and flow (Zhai et al., 2022). The overexpression of OsNPF7.1 or the knockout of OsNPF7.4 can increase seedling biomass, the number of tillers, grain number, and yield per plant; the phenotype is opposite when RNAi silencing OsNPF7.1 or overexpressing OsNPF7.4 (Huang et al., 2019). S1 harboring qGPP4 showed an increase in grain number per panicle (243.94 grains) compared to Xihui18. Therefore, based on the gene function and location, LVPA4 and OsNPF7.4 may be candidate genes for qGPP4.
The remaining seven QTLs (qPL7, qNPB7, qGL7-1, qRLW7, qPL4, qPL6, and qGPP6) have yet to be reported. Therefore, they may be newly identified QTLs. The results of this study establish a solid foundation for further functional analysis of these QTLs and “breeding by design.”
The inheritance of most crop agronomic traits is highly complex and influenced by the environment. The importance of epistasis as the genetic basis of complex traits has been confirmed on the foundation of classical quantitative genetic studies. Experiments on the changes in allele frequency of isoenzyme markers have also revealed that epistasis between multiple loci may play an important role in the evolution of complex traits in plant species (Allard, 1996; Zhuang et al., 2002). We constructed three SSSLs and three DSSLs for the target QTLs in the progeny of Xihui18/Z691. We analyzed the epistasis effects of QTLs related to six important rice agronomic traits. The results showed that even the same QTL interacting with different QTLs produces different genetic models, displaying independent inheritance or various epistatic effects. For instance, the interaction of qGW4 (a = 0.05 mm) and qGW6 in D2 (a = 0.05 mm) resulted in an epistatic effect of −0.03, whose genetic effect was 0.07 mm for grain width, greater than any additive effect of single QTL, which resulted in that the grain width (3.20 mm) of D2 being significantly higher than that of Xihui18, S3 containing qGW4, and S4 containing qGW6. That is to say, if the wide grain is a breeding goal, we can select the two QTL pyramiding. The interaction of qGWT4 (a = −0.35 g) and qGWT6 (a = 0.96 g) in D2 produced an epistatic effect of 0.95, which increased the 1000-grain weight genetically by 1.56 g, larger than the additive effect of any single QTL. If large grain is our breeding goal, qGWT4 and qGWT6 can be selected for breeding by design, which can increase 1000-grain weight by 1.56 g predictably. As per our prediction, the 1000-grain weight of D2 (32.24 g) was significantly heavier than that of S1 (qGWT4), S2 (qGWT6), and Xihui18. However, qGWT6 (a = 0.96 g) and qGWT7 (a = −0.29 g) inherited independently in D3. The genetic effect on 1000-grain weight in D3 was 0.67 g. Thus, due to the different genetic models among QTLs, it is essential to identify each QTL and its genetic models by SSSLs for breeding by design.
As each SSSL carries more than one QTL for different traits, it is necessary to evaluate the breeding value of DSSLs or SSSLs comprehensively. In D1, the genetic effect for grain length after QTL pyramiding was −0.51 mm (less than the additive effects of qGL4 and qGL7-2), that for grain width was −0.03 mm (smaller than the additive effect of qGW4), that for 1000-grain weight was −1.96 g (less than the additive effects of qGWT4 and qGWT7), that for panicle length was −2.21 cm (less than the additive effects of qPL4 and qPL7-2), and that for grain number per panicle was equal to the additive effect (10.32 grains) of qGPP4. Thus, after pyramiding these QTLs, the dual-segment substitution line (D1) displays the phenotype of multiple grains (225.23 grains) and smaller grains (25.23 g) compared to Xihui18. According to Calingacion et al. (2014), grain shapes larger than 3.1 in length to width belong to the slender type. Thus, D1 still exhibits a slender type (3.18 for length to width), although less than 3.41 for the rate of length to width of Xihui18. In D2, the genetic effect for grain number per panicle after QTL pyramiding was −19.43 grains (less than the additive effect of qGPP4, while equal to that of qGPP6), that for panicle length was −0.51 cm (equal to the additive effects of qPL4 and qPL6), that for grain length was 0.10 mm (larger than the additive of qGL4), that for grain width was 0.07 mm (larger than the additive effects of qGW4 and qGW6), and that for 1000-grain weight was 0.95 g (equal to the additive effect of qGWT6, while larger than that of qGWT4). Thus, after the pyramiding of these QTLs, the phenotype of D2 displayed less grain number (171.97 grains per panicle) and large grains (32.24 g) compared to Xihui18, a large panicle type indica restorer line. The grain type still belongs to the slender type (3.25 for the rate of length to width) compared to that (3.41) of Xihui18. In D3, the genetic effect for grain length after QTL pyramiding was equal to the additive effect (−0.06 mm) of qGL7-2, that for grain width was 0.12 mm (larger than the additive effect of qGW6), that for 1000-grain weight was 0.67 g (larger than the additive of qGWT7 while smaller than that of qGWT6), that for panicle length was 0.51 cm (larger than the additive effects of qPL6 and qPL7-2), and that for grain number per panicle was −1.34 grains (parallel to the additive effect of qGPP6). Thus, after pyramiding these QTLs, D3 displayed large grain type (30.23 g for 1000-grain weight) and large panicle type (30.20 cm) with 209.39 grains per panicle compared to Xihui18. The grain shape (3.14 for the rate of length to width) still belongs to slender, while it becomes short-wider compared with that (3.41) of Xihui18. Altogether, after pyramiding different QTLs, D1, D2, and D3 displayed various phenotypes for increasing yield, and all can be potentially useful breeding materials. Similarly, Xu et al. (2023) produced higher yield dual-segment substitution lines (D2 and D3) and triple-segment substitution lines (TSSLs; T1 and T2) by pyramiding various QTLs compared to Xihui18. All these SSSLs, DSSLs, and TSSLs will benefit the breeding by design on the platform of SSSL-Xihui18 in the future.
We dissected three rice SSSLs and DSSLs on the genetic background of Xihui18 from the progeny of Xihui18/Z691. We found 15 QTLs associated with yield-related traits. S1 harboring qGL4 (a = −0.24 mm), qGW4 (a = 0.05 mm), qGWT4 (a = −0.35 g), qPL4 (a = −0.51 cm), and qGPP4 (a = 10.32 grains) displayed multiple and large grains, which are a good high-yield breeding material. Owing to various genetic models after pyramiding different QTLs, it is essential to identify the additive effect of each single QTL and the epistatic effect between QTLs using SSSLs and DSSLs in future breeding by design. D1, D2, and D3 displayed various phenotypes for increasing yield, and all can be potentially useful breeding materials. Finally, our fine-mapping and candidate gene determination of qGL4 and qGW6 lays a good foundation for map-based cloning and functional studies of these genes, which benefits future breeding by design.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
ZY: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. GX: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. KX: Conceptualization, Investigation, Writing – review & editing. ZX: Conceptualization, Investigation, Writing – review & editing. DW: Conceptualization, Investigation, Methodology, Writing – review & editing. LT: Conceptualization, Investigation, Writing – review & editing. YL: Conceptualization, Formal Analysis, Writing – review & editing. GH: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. FZ: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was supported by NSFC-Key projects of the Joint Fund (U23A20184 to GH), Science and Technology Innovation 2030-Major Project (2023ZD0406804), the Chongqing Key Laboratory Open Fund Program (ZYZDSYSKFXM202102), and the Chongqing Rice Molecular Design Innovation Group Project (cstc2021jcyj-cxttX0004).
The authors thank Professor Shizhong Xu at the University of California, Riverside, USA, for writing the stem program for QTL mapping.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1539625/full#supplementary-material
Supplementary Figure 1 | The flow chart of Z691 development.
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Allard, R. W. (1996). Genetic basis of the evolution of adaptedness in plants. Euphytica 92, 1–11. doi: 10.1007/bf00022822
Bai, C., Wang, G. J., Feng, X. H., Gao, Q., Wang, W. Q., Xu, R., et al. (2024). OsMAPK6 phosphorylation and CLG1 ubiquitylation of GW6a non-additively enhance rice grain size through stabilization of the substrate. Nat. Commun. 15, 4300. doi: 10.1038/s41467-024-48786-0
Balakrishnan, D., Surapaneni, M., Mesapogu, S., Neelamraju, S. (2018). Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor. Appl. Genet. 132, 1–25. doi: 10.1007/s00122-018-3219-y
Calingacion, M., Laborte, A., Nelson, A., Resurreccion, A., Concepcion, J. C., Daygon, V. D., et al. (2014). Diversity of global rice markets and the science required for consumer-targeted rice breeding. PloS One 9, e85106. doi: 10.1371/journal.pone.0085106
Dai, Z., Lu, Q., Luan, X., Cai, J., Zhu, H., Liu, Z., et al. (2015). Development of a platform for breeding by design of CMS lines based on an SSSL library in rice (Oryza sativa L.). Euphytica 205, 63–72. doi: 10.1007/s10681-015-1384-5
Dang, X., Xu, Q., Li, Y., Song, S., Hu, C., Jing, C., et al. (2024). GW3, encoding a member of the P450 subfamily, controls grain width by regulating the GA4 content in spikelets of rice (Oryza sativa L.). Theor. Appl. Genet. 137, 251. doi: 10.1007/s00122-024-04751-5
Deng, K., Zhang, H., Wu, J., Zhao, Z., Wang, D., Xu, G., et al. (2023). Development of Single-Segment Substitution Lines and Fine-Mapping of qSPP4 for Spikelets Per Panicle and qGW9 for Grain Width Based on Rice Dual-Segment Substitution Line Z783. Int. J. Mol. Sci. 24, 17305. doi: 10.3390/ijms242417305
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Huang, W., Nie, H., Feng, F., Wang, J., Lu, K., Fang, Z. (2019). Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Sci. 283, 23–31. doi: 10.1016/j.plantsci.2019.01.019
Lei, B., Shao, J., Zhang, F., Wang, J., Xiao, Y., Cheng, Z., et al. (2024). Genetic analysis and fine mapping of a grain size QTL in the small-grain sterile rice line Zhuo201S. J. Integr. Agric. 23, 2155–2163. doi: 10.1016/j.jia.2023.07.026
Li, G., Tang, J., Zheng, J., Chu, C. (2021). Exploration of rice yield potential: Decoding agronomic and physiological traits. Crop J. 9, 577–589. doi: 10.1016/j.cj.2021.03.014
Li, N., Xu, R., Duan, P., Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 237–251. doi: 10.1007/s00497-018-0333-6
Li, J., Yang, H., Xu, G., Deng, K., Yu, J., Xiang, S., et al. (2022a). QTL analysis of Z414, a chromosome segment substitution line with short, wide grains, and substitution mapping of qGL11 in Rice. Rice 15, 25. doi: 10.1186/s12284-022-00571-7
Li, S., Zou, J., Fan, J., Guo, D., Tan, L. (2022b). Identification of quantitative trait loci for important agronomic traits using chromosome segment substitution lines from a japonica × indica cross in rice. Mol. Breed. 42, 73. doi: 10.1007/s11032-022-01343-3
Liang, P., Wang, H., Zhang, Q., Zhou, K., Li, M., Li, R., et al. (2021). Identification and pyramiding of QTLs for rice grain size based on Short-Wide Grain CSSL-Z563 and Fine-Mapping of qGL3–2. Rice 14, 35. doi: 10.1186/s12284-021-00477-w
Liu, S., Hua, L., Dong, S., Chen, H., Zhu, X., Jiang, J., et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84, 672–681. doi: 10.1111/tpj.13025
Liu, T., Zhang, X., Zhang, H., Cheng, Z., Liu, J., Zhuo, C., et al. (2022). Dwarf and high tillering1 represses rice tillering through mediating the splicing of D14 pre-mRNA. Plant Cell. 34, 3301–3318. doi: 10.1093/plcell/koac169
Luan, X., Ke, S., Liu, S., Tang, G., Huang, D., Wei, M., et al. (2021). OsPEX1, a leucine-rich repeat extension protein, functions in the regulation of caryopsis development and quality in rice. Crop J. 10, 704–715. doi: 10.1016/j.cj.2021.10.001
Ma, F. Y., Du, J., Wang, D. C., Wang, H., Zhao, B. B., He, G. H., et al. (2020). Identification of long-grain chromosome segment substitution line Z744 and QTL analysis for agronomic traits in rice. J. Integr. Agric. 19, 1163–1169. doi: 10.1016/s2095-3119(19)62751-6
McCouch, S. R., Kochert, G., Yu., Z. H., Wang, Z. Y., Khush, G. S., Coffman, W. R., et al. (1988). Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76, 815–829. doi: 10.1007/bf00273666
Paterson, A. H., Damon, S., Hewitt, J. D., Zamir, D., Rabinowitch, H. D., Lincoln, S. E., et al. (1991). Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127, 181–197. doi: 10.1093/genetics/127.1.181
Ren, D., Ding, C., Qian, Q. (2023). Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 68, 314–350. doi: 10.1016/j.scib.2023.01.026
She, K., Kusano, H., Koizumi, K., Yamakawa, H., Hakata, M., Imamura, T., et al. (2010). A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22, 3280–3294. doi: 10.1105/tpc.109.070821
Shi, C. L., Dong, N. Q., Guo, T., Ye, W. W., Shan, J. X., Lin, H. X. (2020). A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 103, 1174–1188. doi: 10.1111/tpj.14793
Sun, S., Wang, Z., Xiang, S., Lv, M., Zhou, K., Li, J., et al. (2022). Identification, pyramid, and candidate gene of QTL for yield-related traits based on rice CSSLs in indica Xihui18 background. Mol. Breed. 42, 19. doi: 10.1007/s11032-022-01284-x
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. doi: 10.1046/j.1365-313x.2003.01648.x
Tang, Z., Gao, X., Zhan, X., Fang, N., Wang, R., Zhan, C., et al. (2020). Natural variation in OsGASR7 regulates grain length in rice. Plant Biotechnol. J. 19, 14–16. doi: 10.1111/pbi.13436
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, D., Zhou, K., Xiang, S., Zhang, Q., Li, R., Li, M., et al. (2021). Identification, pyramid and candidate genes of QTLs for associated traits based on a dense erect panicle rice CSSL-Z749 and five SSSLs, three DSSLs and one TSSL. Rice 14, 55. doi: 10.1186/s12284-021-00496-7
Xing, Y., Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xu, G., Deng, K., Yu, J., Li, Q., Li, L., Xiang, A., et al. (2023). Genetic effects analysis of QTLs for rice grain size based on CSSL-Z403 and its dissected single and dual-segment substitution lines. Int. J. Mol. Sci. 24, 12013. doi: 10.3390/ijms241512013
Xu, R., Duan, P., Yu, H., Zhou, Z., Zhang, B., Wang, R., et al. (2018). Control of grain size and weight by the osMKKK10-osMKK4-OsMAPK6 signaling pathway in rice. Mol. Plant 11, 860–873. doi: 10.1016/j.molp.2018.04.004
Xu, Y., Li, P., Yang, Z., Xu, C. (2017). Genetic mapping of quantitative trait loci in crops. Crop J. 5, 175–184. doi: 10.1016/j.cj.2016.06.003
Xu, C., Liu, Y., Li, Y., Xu, X., Xu, C., Li, X., et al. (2015). Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 66, 2611–2623. doi: 10.1093/jxb/erv058
Xu, J., Xing, Y., Xu, Y., Wan, J. (2021). Breeding by design for future rice: Genes and genome technologies. Crop J. 9, 491–496. doi: 10.1016/j.cj.2021.05.001
Yang, W., Liang, J., Hao, Q., Luan, X., Tan, Q., Lin, S., et al. (2021). Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 14, 33. doi: 10.1186/s12284-021-00476-x
Yi, X., Li, C. (2022). Main controllers for improving the resistant starch content in cooked white rice. Food Hydrocolloids 122, 107083. doi: 10.1016/j.foodhyd.2021.107083
You, H., Zafar, S., Zhang, F., Zhu, S., Chen, K., Shen, C., et al. (2022). Genetic mechanism of heterosis for rice milling and appearance quality in an elite rice hybrid. Crop J. 10, 1705–1716. doi: 10.1016/j.cj.2022.05.001
Zhai, L., Yan, A., Shao, K., Wang, S., Wang, Y., Chen, Z., et al. (2022). Large Vascular Bundle Phloem Area 4 enhances grain yield and quality in rice via source–sink–flow. Plant Physiol. 191, 317–334. doi: 10.1093/plphys/kiac461
Zhang, G. (2021). Target chromosome-segment substitution: A way to breeding by design in rice. Crop J. 9, 658–668. doi: 10.1016/j.cj.2021.03.001
Zhang, X., Wang, Y., Liu, M., Yan, P., Niu, F., Ma, F., et al. (2024a). OsEXPA7 encoding an expansin affects grain size and quality traits in rice (Oryza sativa L.). Rice 17, 36. doi: 10.1186/s12284-024-00715-x
Zhang, Q., Wu, R., Hong, T., Wang, D., Li, Q., Wu, J., et al. (2024b). Natural variation in the promoter of qRBG1/OsBZR5 underlies enhanced rice yield. Nat. Commun. 15, 8565. doi: 10.1038/s41467-024-52928-9
Zhao, F. M., Tan, Y., Zheng, L. Y., Zhou, K., He, G. H., Ling, Y. H., et al. (2016). Identification of rice chromosome segment substitution line Z322-1-10 and mapping QTLs for agronomic traits from the F3population. Cereal Res. Commun. 44, 370–380. doi: 10.1556/0806.44.2016.022
Zhao, D., Zhang, C., Li, Q., Liu, Q. (2022). Genetic control of grain appearance quality in rice. Biotechnol. Adv. 60, 108014. doi: 10.1016/j.bioteChadv.2022.108014
Zheng, S., Li, J., Ma, L., Wang, H., Zhou, H., Ni, E., et al. (2019). OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Natl. Acad. Sci. United States America 116, 7549–7558. doi: 10.1073/pnas.1817675116
Zhou, Y., Tao, Y. J., Zhu, J. Y., Miao, J., Liu, J., Liu, Y. H., et al. (2017). GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice 10, 34. doi: 10.1186/s12284-017-0171-4
Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., et al. (2015). Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 82, 570–581. doi: 10.1111/tpj.12820
Zhuang, J. Y., Fan, Y. Y., Rao, Z. M., Wu, J. L., Xia, Y. W., Zheng, K. L. (2002). Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor. Appl. Genet. 105, 1137–1145. doi: 10.1007/s00122-002-0974-5
Zong, G., Wang, A., Wang, L., Liang, G., Gu, M., Sang, T., et al. (2012). A pyramid breeding of eight grain-yield related quantitative trait loci based on marker-assistant and phenotype selection in rice (Oryza sativa L.). J. Genet. Genomics 39, 335–350. doi: 10.1016/j.jgg.2012.06.004
Keywords: rice, chromosome segment substitution lines, single-segment substitution lines, QTL mapping, fine-mapping, grain size
Citation: Yu Z, Xu G, Xie K, Xie Z, Wang D, Tan L, Ling Y, He G and Zhao F (2025) Genetic dissection of QTL for important agronomic traits and fine-mapping of qGL4 and qGW6 based on a short-width grain rice CSSL-Z691. Front. Plant Sci. 16:1539625. doi: 10.3389/fpls.2025.1539625
Received: 04 December 2024; Accepted: 12 February 2025;
Published: 10 March 2025.
Edited by:
Satish Kumar, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Yang Xu, Yangzhou University, ChinaCopyright © 2025 Yu, Xu, Xie, Xie, Wang, Tan, Ling, He and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangming Zhao, emhhb2ZhbmdtaW5nMjAwNEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.