- 1College of Agriculture, Heilongjiang Bayi Agricultural University, Daqing, Heilongjiang, China
- 2Shandong Key Laboratory of Precision Molecular Crop Design and Breeding, Peking University Institute of Advanced Agricultural Sciences, Shandong Laboratory of Advanced Agricultural Sciences in Weifang, Weifang, Shandong, China
Background: Frigida (FRI) genes are crucial for regulating flowering time in plants. While the biological importance of the Frigida-like (FRL) gene family has been recognized in Arabidopsis, a systematic analysis of these genes in soybean is lacking. Characterizing FRL genes in soybean will help uncover their roles in flowering regulation, offering valuable insights for improving soybean adaptation.
Results: In this study, we identified 16 Frigida genes in soybean, naming them based on their relationship to the FRL genes in Arabidopsis thaliana. These genes are unevenly distributed across thirteen chromosomes. Phylogenetic analysis categorizes Frigida-like proteins from Arabidopsis, soybean, and rice into four distinct subfamilies (I–IV). Our findings indicate that eight GmFRLs arose from whole-genome duplication (WGD) events, alongside two tandem duplication events. Gene structure analysis confirmed that all GmFRL members contain Frigida domains. Additionally, promoter analysis revealed numerous cis-acting elements related to photoperiodic response, suggesting their significant role in soybean’s light response mechanisms. RNA-seq data demonstrated variable expression levels of GmFRL genes across tissues, including flower, leaf, pod, and seed, and other tissues, while subcellular localization and qPCR analyses further support their vital role in light responsiveness in soybean.
Conclusion: In summary, our comprehensive analysis offers valuable insights into the evolution and potential functions of GmFRL genes, emphasizing their significance in photoperiodic responses and establishing a foundation for further research on the GmFRL family.
1 Introduction
Seasonal flowering is fundamental to the reproductive success and survival of higher plants. Plants have evolved a complex response of endogenous clues and environmental factors, such as day length and temperature control of flowering time of genetic networks (Choi et al., 2011; Takada et al., 2019; Dean, 2002). FRI (FRIGIDA) is a key regulator of flowering time and can inhibit flowering without vernalization (Chen et al., 2020). The FRL gene encodes a novel protein that lacks structural domains indicative of immediate functions, yet it contains two potential coiled-coil domains (Goff et al., 2002), which are believed to interact with other proteins or nucleic acids. This unique domain activates the expression of FLC (FLOWERING LOCUS C), which encodes a MADS-box transcription factor that quantitatively suppresses the floral transition by inhibiting flowering pathway integrators, such as FT (Flowering Locus T) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1) in Arabidopsis (Schmitz and Amasino, 2007). FRI acts as a scaffold protein, interacting with FRL1, SUF4, FLX, FES1, UBC1, and CBP20 to form a transcriptional activation complex. This complex recruits chromatin-modifying factors, including the SWR1 complex and SET2 homologs, to epigenetically modify the histone methylation levels at the FLC locus (Choi et al., 2011; Li et al., 2018).
Recent studies have indicated that FRI mediates the activation of the floral repressor FLC, thereby negatively regulating flowering time (Zhu et al., 2021). Most rapid-cycling Arabidopsis carry loss-of-function mutations in FRL, leading to low levels of FLC and rapid flowering in the absence of vernalization (Ding et al., 2013). Studies related to FRI genes have been carried out in A. thaliana (Risk et al., 2010), Medicago sativa (Chao et al., 2013), Oryza sativa (Goff et al., 2002), Populus balsamifera (Keller et al., 2011) and Vitis vinifera (Risk et al., 2010). The AtFRL regulates flowering in Arabidopsis, but the orthologs of FRL from EjFRL (from loquat) also have the ability to influence Arabidopsis flowering. The FRL gene is not only related to the regulation of flowering time but also involved in other biological processes related to reproduction, such as embryo development and seed maturation (Hu et al., 2014; Vieira et al., 2019).They found that PmFRL may be linked to ABA signal regulators and gibberellin signal regulators, thereby exerting their biological functions during dormancy and flowering in P. mume (Li et al., 2021). In tomatoes, they found that Brassinosteroid (BR) regulate tomato flowering through the interaction between SlFRLs and SlBIN2 (Khan et al., 2022). Apple orthologs of Arabidopsis genes, FRIGIDA, exhibit similar expression patterns as reported in Arabidopsis, suggesting that functional conservation in floral signal integration and meristem determination pathways (Kumar et al., 2016a). These studies reveal the potential important functions of the FRL family genes in plant response to regulates flowering.
Soybean serves as a quintessential example of a crop that demonstrates significant sensitivity to photoperiod. The various developmental stages, including the growth, flowering, and maturity periods, as well as the resulting plant morphology, are closely regulated by photoperiodic conditions. The complete genome sequence of the Williams 82 variety (Glycine max) was finalized in 2010 and has since been widely used (Schmutz et al., 2010). The primary reference genome assembly, Wm82, which has been in use for the past decade, has undergone significant improvements. Over 3,600 gaps have been closed, adding more than 5 Mbp, and regions previously exhibiting high heterozygosity in the earlier reference assembly have been enhanced. Notably, recent updates to the high-resolution linkage map have significantly strengthened the assembly of the Wm82 reference genome (Valliyodan et al., 2019), incorporating more sequence information and fewer errors. This provides more detailed and accurate gene function annotations, establishing a stronger foundation for the identification and characterization of the FRL gene family.
The FRL gene family is a key regulatory factor in the control of flowering time in plants. It is essential to identify and analyze FRLs on a genomic scale to uncover their molecular functions, which may provide deeper insights into plant development. The identification of FRLs has been reported in Arabidopsis (Risk et al., 2010) and Oryza sativa (Goff et al., 2002), while the phylogenetic and structural characteristics of the FRL family have only been studied in Prunus mume (Li et al., 2021). Currently, there is no systematic research reporting on the GmFRL gene family in soybeans. Therefore, the functional roles of its members within the soybean genome require further investigation. In this study, we identified 16 GmFRL genes and conducted a genome-wide analysis of their evolutionary characteristics and biological functions. We examined the phylogenetic relationships, gene structures, conserved motifs, repetitive patterns, cis-element organization, and tissue-specific expression patterns of the GmFRL genes. Additionally, since soybean cultivation is not affected by vernalization, the inhibitory role of the GmFRL gene in flowering may have potential functions in soybeans. Some evidence suggests that FRI does not appear to change photoperiodic responsiveness but rather shifts the response to much later flowering times (Lee and Amasino, 1995). Therefore, we also analyzed the expression response of the GmFRL gene to photoperiod in soybeans and its subcellular localization. The systematic analysis of GmFRL gene family lays a foundation for further study of its key role in the regulation of soybean light response.
2 Materials and methods
2.1 Plant materials and growth conditions
The Glycine max Williams 82 was used in this study. The seeds were sterilized in 1% sodium hypochlorite for 1 minute, followed by three washes with sterilized water. They were then germinated in a growth chamber for 15 days under LD (16 h light/8 h dark) at 25°C and 60% relative humidity. Fifteen days later, during the seedling stage of soybeans, which is highly sensitive to changes in photoperiod, leaf samples were collected at 0, 4, 8, 12, 16, 20, and 24 hours of Zeitgeber time (ZT). The samples were immediately frozen in liquid nitrogen and stored at -80°C for gene expression analysis (Cao et al., 2015). Three biological replicates were obtained for each time point.
2.2 In silico identification of FRIGIDA family genes in Glycine max
The Hidden Markov Model (HMM) of FRIGIDA (PF07899) was obtained from PFAM (http://pfam.xfam.org/) and then used as a query to retrieve the soybean proteome sequences. Soybean proteome sequences (Wm82.a4.v1) were downloaded from the phytozome database. To avoid missing FRIGIDA family members, a new HMM based on the resulting sequence is constructed using HMMER software (http://hmmer.org/), and the model is presented as a query sequence (E values < 10-5), and the sequence data of soybean proteome were retrieved again. After removing the redundant sequences, the SMART online platform (http://smart.embl-heidelberg.de/) checks the remaining sequences to predict the full FRIGIDA domain. The gene encoding a protein with the FRIGIDA domain was identified as a member of the FRIGIDA family. TBtools-II were used to calculate the protein properties of FRIGIDA family members, such as amino acid number, isoelectric point (pI) (Chen et al., 2023). The SOPMA tool (https://www.npsa-prabi.ibcp.fr/cgi-binpsa_automat.pl?page=/NPSApsa_sopma.html) was used to predicted the secondary structure of GmFRL protein, and AlphaFold3 (http://alphafoldserver.com) was used to predicted the tertiary structures of GmFRL proteins.
2.3 Evolutionary analysis, chromosomal location, and synteny analysis
ClustalW is used for multiple sequence alignment of all GmFRL proteins, and a phylogenetic tree of GmFRL proteins was constructed using MEGA 11.0 (Hall, 2013; Thompson et al., 2003; Tamura et al., 2021), and a comprehensive phylogenetic tree that includes Arabidopsis, soybean, and rice. Both phylogenetic trees were constructed using the maximum likelihood (ML) algorithm with 1000 Bootstrap repeats (Kumar et al., 2016b), Gene duplication events and synteny analysis (Glycine max vs. Arabidopsis; Glycine max vs. Glycine max) were performed using the default parameters of Tbtools-II software (Chen et al., 2023).
2.4 Structural characterization, conserved motif analysis, and cis-acting elements
The exon/intron structure of each GmFRL gene was analyzed by TBtools-II (Chen et al., 2023). The conserved motifs of all GmFRL proteins were analyzed by MEME tool (http://meme-suite.org/tools/meme). Conserved domains within all GmFRL proteins were identified using the CD-Search tool (https://www.ncbi.nlm.nih.gov/cdd/) from the NCBI database. Cis-acting elements in the promoter sequences (upstream of 2000 bp) of the GmFRL gene family were predicted using the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Tbtools-II is used to visualize results (Chen et al., 2023).
2.5 Subcellular localization of GmFRL proteins
CELLO v.2.5 (http://cello.life.nctu.edu.tw/) was used to predict the subcellular localization of all GmFRL proteins. Subsequently, four GmFRLs were cloned and transiently overexpressed in tobacco leaves for subcellular localization experiments to validate the prediction results. The GmFRL genes were amplified and subsequently ligated into the fusion expression vector pSuper1300-MAS-EGFP following digestion, leading to the successful construction of the expression vector. The recombinant plasmids were then transformed into Agrobacterium strain GV3101. Both the Agrobacterium containing the pSuper1300-MAS-GmFRL01/04/09/13-EGFP expression vector and those harboring the empty control vector pSuper1300-MAS-EGFP were cultured, and the bacterial cells were harvested by centrifugation at 4000 rpm for 15 minutes, followed by the removal of the supernatant. Subsequently, 1 mL of tobacco transformation solution (OD600 = 0.7–1.0) was added to resuspend the Agrobacterium. After resuspension, the tobacco leaves were injected following a 2-hour incubation at room temperature or 28°C. Approximately 2–3 days post-injection, the lower epidermis of the tobacco leaves was peeled off, and the subcellular localization of the fused protein was observed using confocal microscopy, with images captured simultaneously.
2.6 Analysis of expression patterns of GmFRL genes
According to the RNA-seq data (TPM) of GmFRL extracted from SoyMD (Yang et al., 2024), the expression pattern of GmFRL genes in different tissues of soybean was studied. Transcriptomic data on soybean were obtained from the National Center for Biotechnology Information (NCBI) publicly accessible database (Accession number: GSE94228). After removing adapter sequences and low-quality reads from the RNA-seq data using fastp (v.0.23.0) (Chen et al., 2018), we aligned the cleaned RNA-seq data to the Wm82.a4.v1 genome using HISAT2 (v.2.1.2) (Kim et al., 2015) with default parameters. We then quantified and normalized the data using StringTie (v.1.3.5) (Pertea et al., 2015) with default settings. The heat map function in Tbtools-II was used for further expression analysis (Chen et al., 2023).
2.7 RNA extraction and qPCR analysis
Total RNA was extracted from 15-day-old seedlings with TRIzol Reagent (Invitrogen) and reverse transcribed by MMLV reversetranscriptase (Promega). Quantitative Real-time PCR (q-PCR) was performed with a SYBR Green PCR Master Mix kit. Analysis was performed using the Applied Biosystems StepOnePlus real-time PCR system. Whole plant seedlings from wild-type on the same place were collected separately at the same time. Three independent experiments were conducted. Relative transcript levels were normalized to GmACT11. The reaction and the calculation of relative expression levels were performed as described previously (Miura et al., 2007). The qRT-PCR was carried out as described previously (Iqbal et al., 2020; Tong et al., 2009).
3 Results
3.1 Identification of GmFRL genes in soybean
A total of 16 GmFRL genes were identified from the soybean genome (Wm82.a4.v1) within the Phytozome v13 database. Based on their homology with members of AtFRL family, the 16 GmFRL family genes were designated as GmFRL1 to GmFRL16 reflecting their homology with members of the AtFRL family (Table 1). The physicochemical properties of GmFRL genes were predicted as shown in the Table 1, including the number of amino acids, molecular weight, and theoretical isoelectric point (pI). The full lengths of the GmFRL proteins varied between 519 amino acids (GmFRL3) and 1297 amino acids (GmFRL16), with molecular weights ranging from 56.80 kDa to 152.26 kDa. Additionally, the isoelectric points varied from 5.90 (GmFRL5) to 9.13 (GmFRL10). Overall, the pI of GmFRL family proteins showed significant differences. All proteins were hydrophilic, as reflected by Grand Average of Hydropathicity less than 0.

Table 1. Analysis of the physicochemical properties and primary and secondary structure of 16 GmFRL genes.
According to secondary structure prediction by the SOPMA tool, the GmFRL proteins were found to encompass α-helix, extended strand, and random coil secondary structural elements (Table 1). The α-helix content was the highest overall, constituting 37.36%-70.01%, followed by random coil, constituting 23.52%-52.01%, while the extended strand was the lowest, constituting 6.48%-12.82%. Prediction using the Alpha Fold3 tool revealed that the tertiary structures of same subfamily of GmFRL proteins were largely similar (Figure 1), which was consistent with the gene structure analysis outcomes.
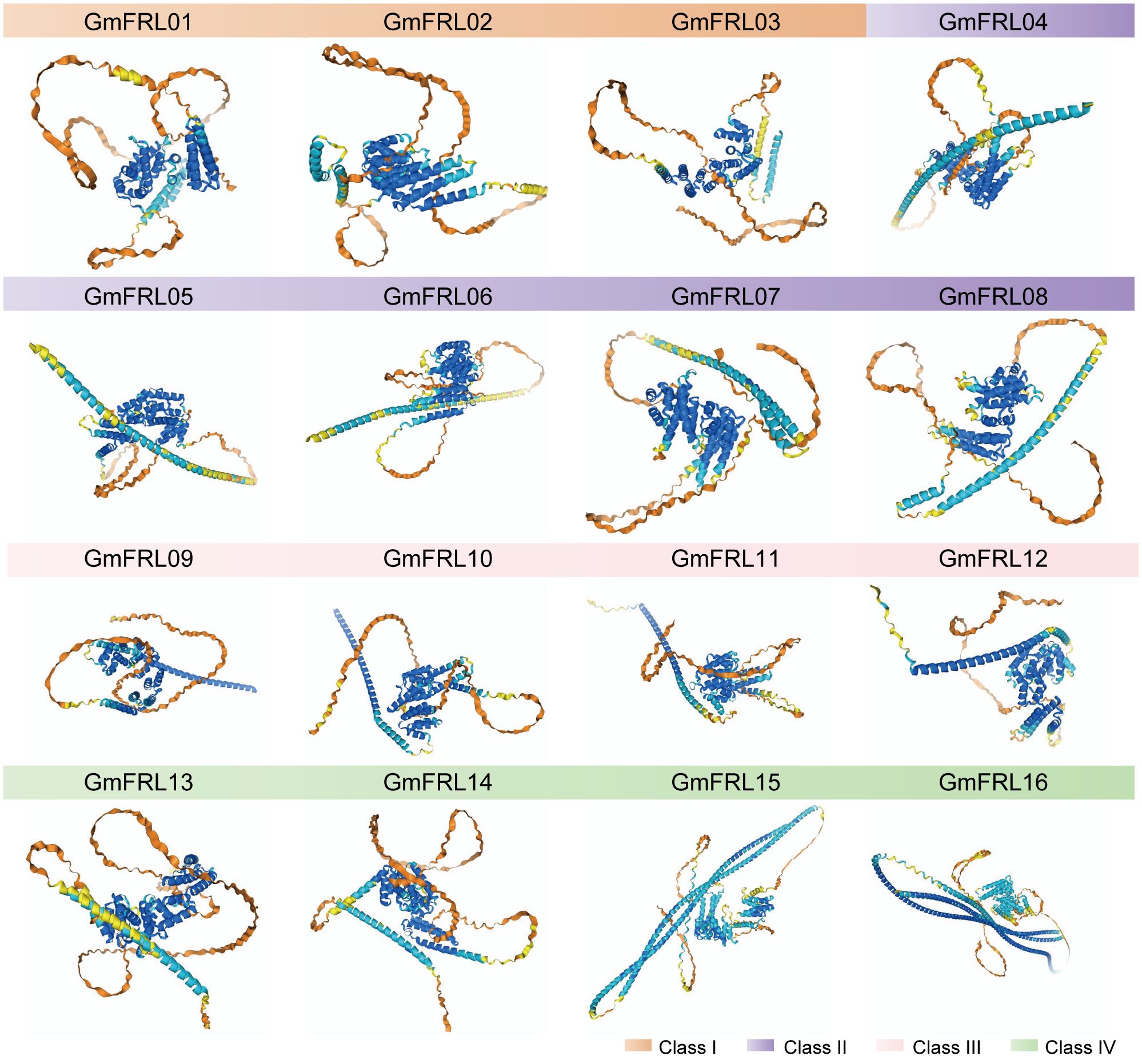
Figure 1. Tertiary structure model of GmFRL proteins. The different colored labels the different class of GmFRL proteins.
3.2 Chromosomal distribution and expansion patterns of GmFRL genes
TBtools software was used to illustrate the physical locations of GmFRL genes on soybean chromosomes. The results indicated that the 16 GmFRL genes were unevenly distributed across 13 chromosomes (Figure 2A). Specifically, chromosomes 3, 5, and 8 each contained two GmFRL genes each, while the remaining chromosomes carried only one gene. Notably, chromosomes 1, 7, 9, 13, 14, 15, and 19 did not harbor any GmFRL genes. These findings suggest that the distribution of GmFRL genes across soybean chromosomes is not uniform. Co-linearity analysis of the GmFRL genes revealed two pairs of tandemly repeated sequences among the 16 identified GmFRL genes. To investigate the evolutionary relationship between AtFRL and GmFRL, a syntenic map of the genomes of Arabidopsis and Glycine max was visualized using a circos plot (Figure 3A). The syntenic map exhibited a linear relationship between GmFRL11, GmFRL12, AtFRL4a and AtFRL4b. Furthermore, GmFRL06 and AtFRL2 displayed a linear relationship, suggesting that they share homology and may serve similar functions.

Figure 2. Genomic distribution and phylogenetic analysis of the GmFRL genes. (A) Distribution of GmFRL genes across chromosomes. The scale at the bottom indicates the length of the chromosomes, with the chromosome numbers displayed on the left side of each chromosome. (B) Phylogenetic analysis of Frigida-like proteins from soybean, Arabidopsis and rice. Phylogenetic trees were constructed using the maximum likelihood (ML) algorithm with 1000 bootstrap repetitions.
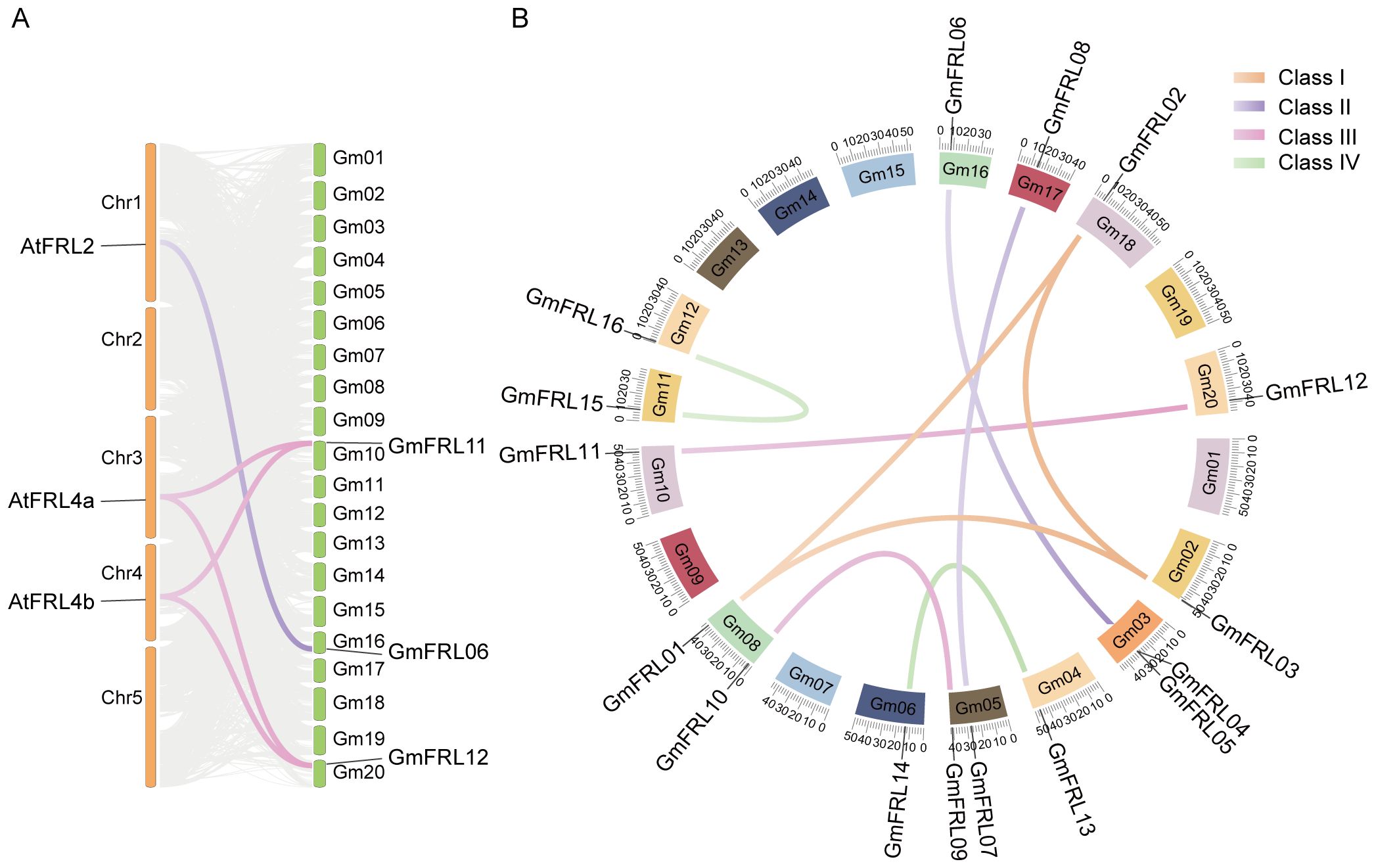
Figure 3. Collinear analysis of GmFRL proteins and FRL proteins from different plants. (A) The chromosomal distribution and Syntenic relationships prediction of Glycine Max and Arabidopsis thaliana FRL genes. (B) Distribution and synteny analysis of GmFRL genes. The soybean chromosomes are resented in different-colored partial circles. The different colored labels the different class of GmFRLs.
The duplication events among GmFRL genes were analyzed, focusing on segmental and tandem duplications, to gain insights into their expansion within the soybean genome (Figure 3B). The results indicated that the gene pair GmFRL04 and GmFRL05 expanded through tandem duplication. In contrast, the gene pairs GmFRL01/02/03, GmFRL07/08, GmFRL04/06, GmFRL09/10, GmFRL11/12, GmFRL13/14, and GmFRL15/16 underwent expansion through segmental duplication. These findings suggest that segmental duplication is the primary driving force behind the significant expansion of GmFRL genes in the soybean genome. According to the KaKs_Calculator 3.0, the Ka/Ks values of the tandemly repeated GmFRL sequences (GmFRL04/GmFRL05) were 0.1679 (Ka = 0.0584, Ks = 0.3479), which is less than 1. This suggests that purifying selection has acted on these genes during the process of evolution. In other words, GmFRL genes are highly conserved and evolve slowly.
3.3 Phylogenetic analysis of the GmFRL proteins
Phylogenetic analysis of 36 FRL proteins from 3 species of different affinities was performed that including 16 from Glycine max, 8 from Arabidopsis thaliana, and 12 from Oryza sativa. The results of the phylogenetic analysis indicated that these FRL proteins were classified into four distinct subgroups (I~IV) (Figure 2B). Each subgroup contains 3 to 5 members. The presence of both soybean FRL proteins and AtFRL proteins within the same subgroup suggests a possible conservation of function among dicot species. Generally, members within the same subgroup may have similar functions, which aids in our understanding of the potential biological functions of GmFRLs. In subgroup I, three members of the GmFRL family (GmFRL1/2/3) clustered with two members of the AtFRL family, including AtFRL1 and AtFRL2. AtFRL1 and AtFRL2 play crucial roles in regulating FLC expression in Arabidopsis, with their functions dependent on the genes that affect the chromatin structure of the FLC locus (Emami and Kempken, 2019). In subgroup II, five members of the GmFRL family clustered with an AtFRL family protein named AtFRL3, which can regulate the photoperiod pathway in Arabidopsis (Li et al., 2019). In a previous study, GmFRL07 was identified as potentially participating in the regulation of soybean growth stages, based on the strong correlation peak SNP and LD blocks of four significant SNPs (Gm5_27111367, Gm11_10629613, Gm11_10950924, Gm19_34768458) (Li et al., 2019).
3.4 Gene structures and protein motifs of GmFRL family
Through the screening of sequences and annotation files, we delineated the gene structures for the GmFRL family (Figure 4). The number of introns within the GmFRL genes spanned from 2 to 8, whereas the number of exons fluctuated between 3 and 4. Among the GmFRL genes, GmFRL03 demonstrated the maximum number of introns, totaling eight. Furthermore, eight genes possessed two introns (GmFRL01, GmFRL04, GmFRL07, GmFRL06, GmFRL08, GmFRL09, GmFRL10, GmFRL15, and GmFRL16), whereas the remaining GmFRL genes possessed either three or four introns.

Figure 4. Analysis of the intron and exon compositions of GmFRL genes, as well as the conserved domains and motifs of GmFRL proteins, is conducted in the context of their phylogenetic relationships.
Conserved domain analysis revealed that all members of four subfamily possess Frigida domains, implying a potential conservation of function. To further elucidate the structural diversity of the GmFRL proteins, we identified a total of eight conserved motifs utilizing the MEME suite (Figure 3, Supplementary Figure S1). As illustrated in Figure 3, all GmFRL proteins exhibited motif-1. Although the types and numbers of motifs among members of the same subfamily were comparable, some variations in motif patterns were observed among specific members. In summary, the members of the same subfamily displayed similar structures and conserved motifs, thereby reinforcing the reliability of the constructed phylogenetic tree.
3.5 Cis−acting elements in GmFRL promoters
Cis-acting elements in the promoter region play a crucial role in how plants respond to growth factors and environmental stresses by regulating gene expression through transcription (Walther et al., 2007). In this study, in addition to the abundant core promoter (TATA box) and enhancer elements (CAAT box), four types of cis-regulatory elements were identified: light-responsive, stress-responsive, hormone-responsive, and development-responsive elements (see Figure 5). Among these, light-responsive elements were the most abundant. The number of stress-related response elements and hormone-responsive elements was roughly equivalent, while development-related elements were the least prevalent. Nearly all GmFRL genes contained various types of light-responsive elements. For example, GmFRL01 included the GT1-motif (a light-responsive element), the AE-box (part of a module for light response), and Box 4 (a conserved DNA module associated with light responsiveness). These findings suggest that GmFRL genes may play a significant role in light responsiveness in soybeans. Further analysis of hormone-related response elements in the promoters of GmFRL family members indicates that these elements can be classified into categories associated with auxin, gibberellic acid (GA), abscisic acid (ABA), salicylic acid (SA), and methyl jasmonate (MeJA). This categorization underscores their potential significance in plant hormone signaling and stress responses.
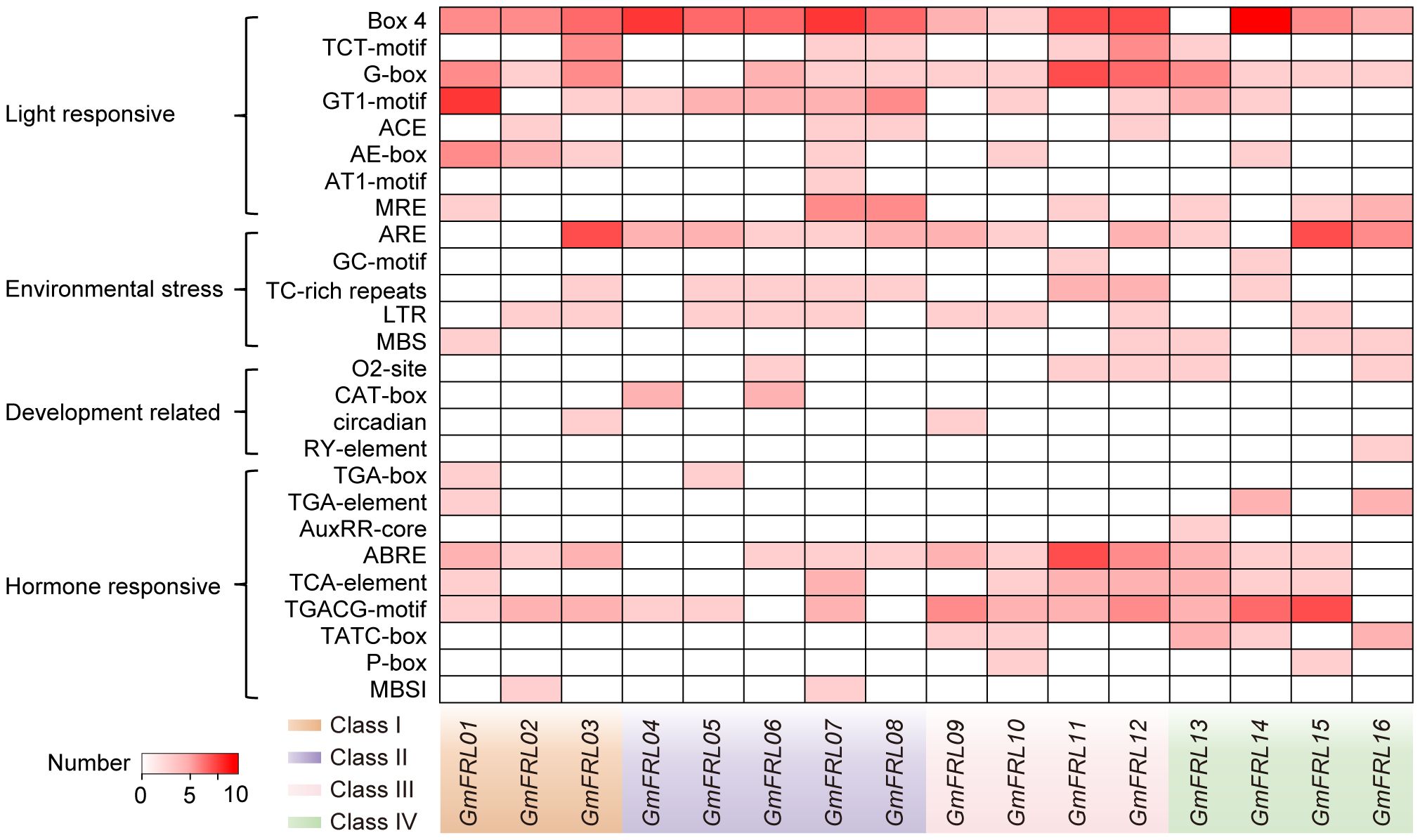
Figure 5. Predicted cis-elements in the promoters of GmFRL genes. Types and numbers of cis-elements in the promoter regions of GmFRL genes.
3.6 Expression patterns in different soybean tissues and subcellular localization of GmFRL family
To understand the potential functions of GmFRL family, The RNA-seq data (TPM) to study GmFRL gene expression patterns in different soybean tissues, including shoot apical meristem (SAM), root, root hair, stem, leaf, flower, pod, nodule, and seed (Figure 6A). The heatmap illustrates the expression of 16 GmFRL genes across 9 tissues. The results indicate that all 16 GmFRL genes are expressed in at least one tissue, with most family members showing high expression levels in flowers and seeds. There are notable differences in tissue-specific expression among the different subfamilies of GmFRLs, Class II GmFRLs exhibit specific expression in flowers and leaf buds, while Class III GmFRLs are specifically expressed in pods and seeds. Additionally, compared to other GmFRL family members, certain GmFRL genes, such as GmFRL03, and GmFRL09 are specifically expressed in leaf buds; GmFRL06, GmFRL08, and GmFRL16 are specifically expressed in leaf buds; GmFRL13 exhibits significantly higher expression levels in flowers, leaves, and leaf buds; and GmFRL15, are virtually non-expressed in all tissues except for seeds. This observation may indicate that some GmFRLs play a potential role in regulating flowering or seed development in plants.
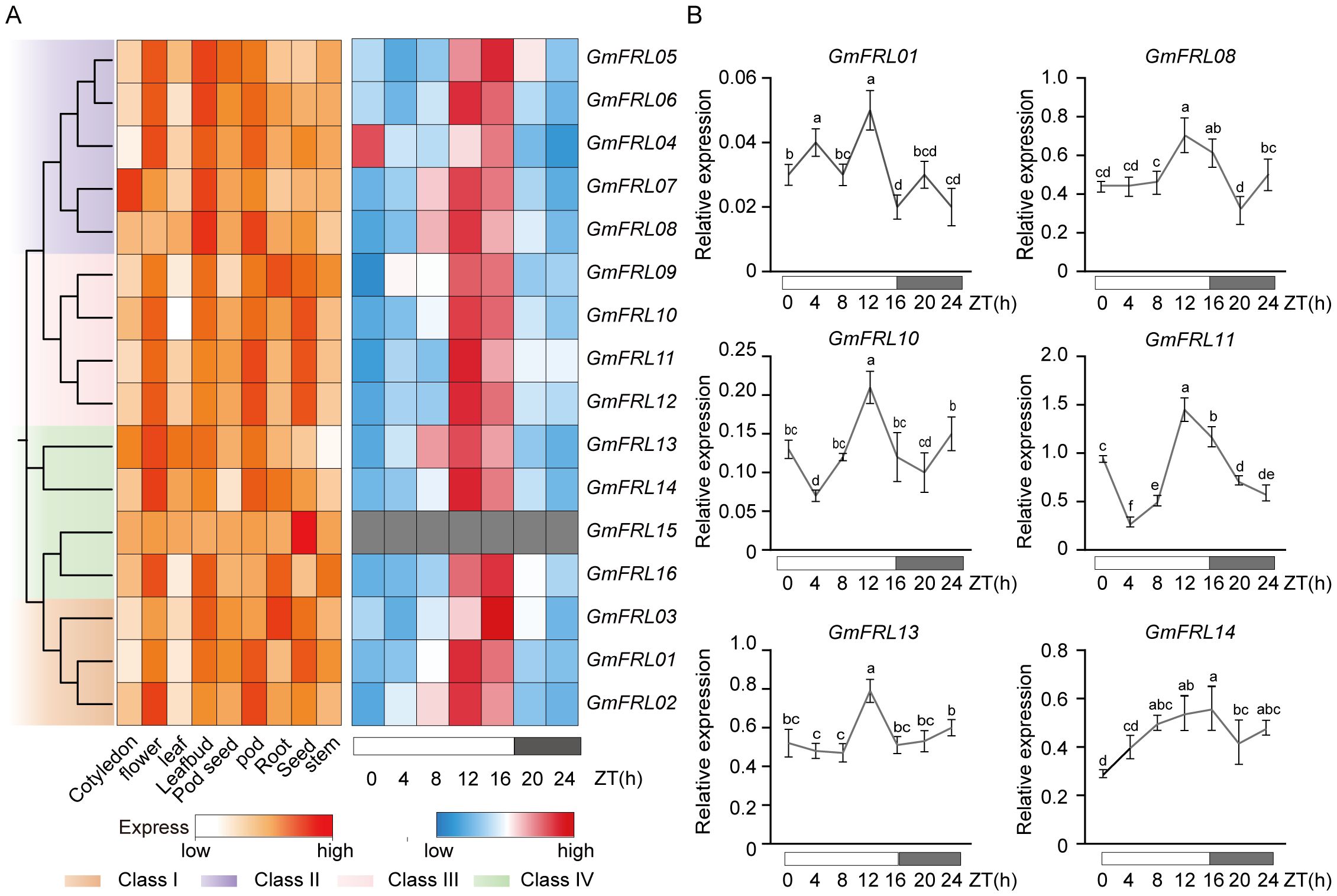
Figure 6. Expression pattern analysis of GmFRL genes. (A) Expression levels of GmFRL genes at different tissues. And the expression levels of GmFRL genes in LD. (B) Relative expression levels of GmFRL genes in response to LD light. The soybean cultivar Wm82 was cultured for 15 days, and the relative expression levels of GmFRL genes were measured at 0h, 4h, 8h, 12h, 16h, 20h, and 24h under LD. The GmACT11 gene served as the internal control. The data represented the mean ± SD of three independent biological repetitions.
In addition, we utilized CELLO to predict the subcellular localization of all members, with the results indicating that they are primarily localized in the nucleus (Table 1). To further validate the subcellular localization of GmFRL proteins, we conducted transient expression experiments with GFP-GmFRL01/04/09/13 constructs in Nicotiana benthamiana. The results indicated that the green fluorescence of GmFRL01/04/09/13 was distinctly visible in the nucleus (Figure 7). This observation leads us to hypothesize that GmFRL proteins may be primarily localized in the nucleus.
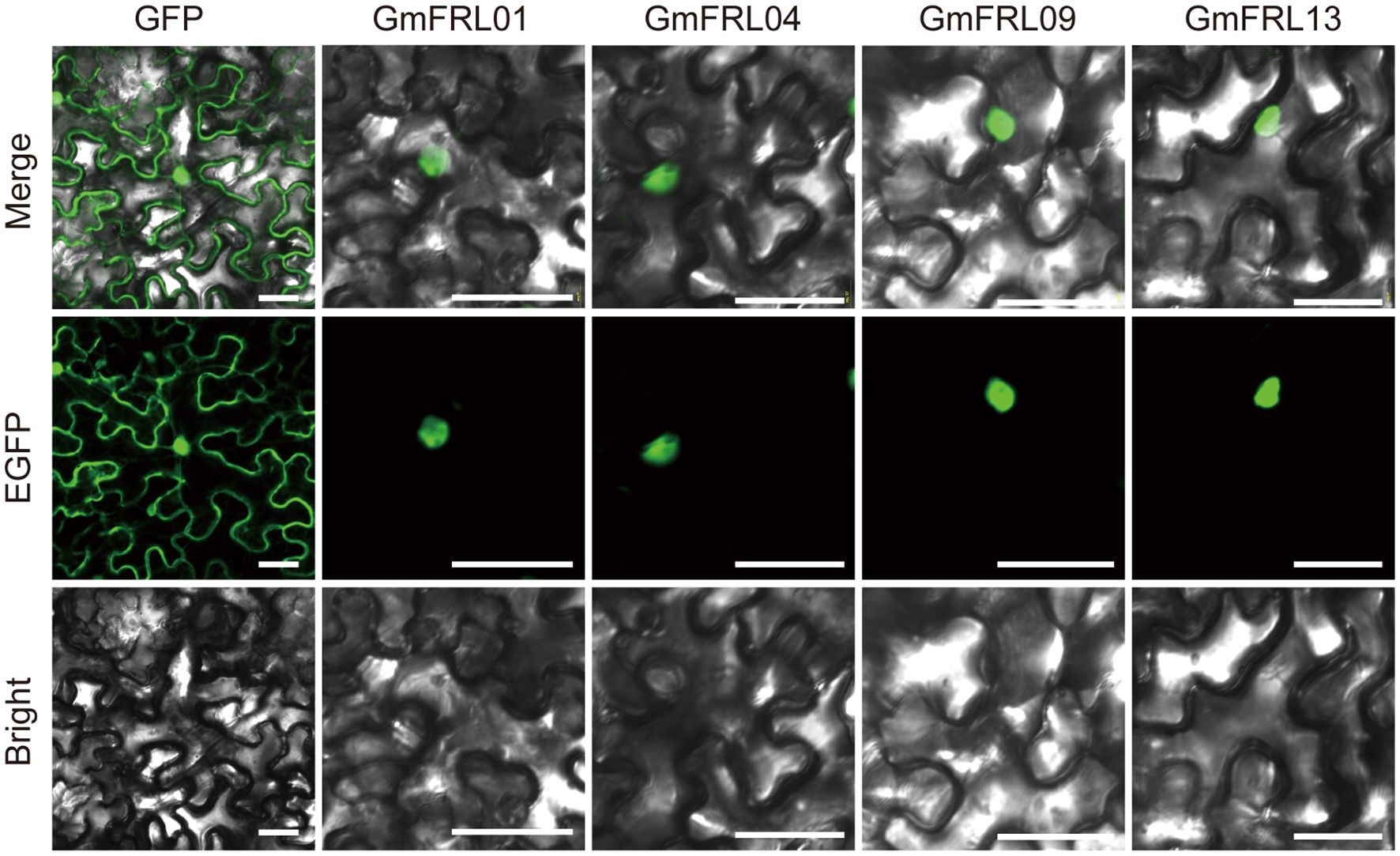
Figure 7. Subcellular localization analysis of GmFRL genes. eGFP indicates the fusion protein; Bright, bright field; Merge, merged GFP and Bright. Bars 50μm.
3.7 Expression patterns of GmFRL genes in different light condition
Previous studies have emphasized the significant role of phytochrome-mediated regulation in flowering time. RNA-seq transcriptome data from soybean leaves at multiple time points (0, 4, 8, 12, 16, 20, and 24 hours) under long-day (LD) light was analyzed. The expression of GmFRLs exhibits a rhythmic pattern across multiple cycles (Supplementary Figure S2A). The results from a single cycle demonstrate that, except for GmFRL15, which is virtually non-expressed in leaves, all other Class I-IV GmFRLs exhibit a rhythmic expression pattern, with higher expression levels observed around ZT16, approximately 12 hours after light exposure or at dusk (Figure 6A; Supplementary Figure S2B). To further investigate whether the expression of GmFRL genes is indeed influenced by circadian rhythms, RT-qPCR was performed under LD conditions to analyze the relative expression levels of GmFRL genes across different time points. The results indicated that several GmFRL genes (GmFRL01, GmFRL08, GmFRL10, GmFRL11, GmFRL13, and GmFRL14) exhibited higher expression levels at ZT12 or around dusk at ZT16 (Figure 6B). Based on these findings, we propose that certain GmFRL members may play a potential role in the response to light under long-day conditions in soybean.
4 Discussion
FRL genes play an important role in the process of plant growth and development. To date, FRL family have not been identified or analyzed in Glycine max. The continuously updated whole-genome sequencing of soybean provides a valuable resource for bioinformatics analyses of various gene families within this species (Wang et al., 2023). In this study, we identified a total of 16 GmFRLs from Glycine max, a significantly greater number than the 8 AtFRLs identified in Arabidopsis (Michaels et al., 2004). Furthermore, we conducted comparative analyses of FRL-like proteins from different plant genomes, including mosses, ferns, and various angiosperms (Supplementary Figure 3). The evolutionary analysis of the FRL family indicates that the FRL structure was established prior to the divergence of terrestrial plants and algae, as we identified proteins with FRL configurations in the unicellular alga Chlamydomonas reinhardtii. Comparative analyses of FRLs among mosses, rice, and Arabidopsis reveal that the FRL family underwent independent expansions during the early evolution of terrestrial plant lineages, following the divergence of angiosperms and bryophytes, and throughout the diversification of each angiosperm lineage. The most pronounced expansions were observed in the lineages of soybean and maize, where the number of FRL members is nearly double or even four times that found in Arabidopsis.
Gene duplication is a major driving force for the expansion of gene families and the evolution of novel functions, such as adaptation to stress and induction of disease (Cannon et al., 2004; Panchy et al., 2016; Vision et al., 2000). The presence of two or more genes on the same chromosome indicate a tandem duplication event, while two or more genes present on different chromosomes reveal a segmental duplication event. Tandem duplication and segmental duplications have been considered as the main duplication patterns for gene family expansion (Kong et al., 2007; Yu et al., 2005). Previous studies indicate that the soybean genome experienced two rounds of segmental duplication in its evolutionary history, occurring approximately 13 and 59 million years ago (Mya). This has led to a highly duplicated genome, in which nearly 75% of the genes are present in multiple copies across various genomes (Schmutz et al., 2010). The genes that arise from these genomic duplication events provide the raw material for the generation of new genes, which in turn promotes the development of new functions, thereby facilitating the expansion of gene families and functional evolution. A comprehensive review of existing literature on genomic repeat events in soybean and its ancestral species, Glycine soja, indicates that 8 (50%) of the GmFRL genes are derived from whole genome duplication (WGD) events in wild soybean (Du et al., 2012), while 4 (25%) of the GmFRL genes are classified as singletons. These findings suggest that the GmFRL gene family in soybean has undergone significant gene duplication throughout its evolutionary history and has been retained through multiple WGD events. Furthermore, this study identifies that 2 of the 16 GmFRL genes have experienced tandem duplication, which may be associated with an early legume duplication event that occurred approximately 28 million years ago. This observation underscores the contribution of tandem repeats to the expansion of the GmFRL gene family. In summary, segmental duplication emerges as the primary mechanism driving the expansion of the GmFRL family, occurring in conjunction with tandem duplication events among certain members, thereby facilitating the overall proliferation of the GmFRL gene family. The Ka/Ks ratio is a measure used to examine the mechanisms of gene duplication evolution after divergence from their ancestors (Salih et al., 2016). The Ka/Ks ratio provides insight into the selection pressure acting on amino acid substitutions: a Ka/Ks ratio < 1 indicates purifying selection, a ratio of 1 suggests neutral selection, and a ratio > 1 indicates positive selection. The Ka/Ks values for the tandem duplicate GmFRL genes are less than 1, suggesting that these genes have undergone negative selection. This result reflects a slow evolutionary rate and significant conservation within this gene family.
In addition, the molecular weights of different GmFRL proteins exhibit variability, indicating potential differences in their structure and composition, which suggests that their functions may also differ. A phylogenetic tree analysis showed that members of the GmFRL family were classified into 4 distinct subgroups. Some GmFRL proteins and AtFRL proteins belong to the same subfamily, perhaps this part of the GmFRL proteins have a genetic structures and conserved motifs, which may contribute to their crucial biological functions. In this study, we identified the number of introns for 16 GmFRL members; GmFRL03 had 8 introns, others genes had 2-4 introns (Figure 3). Introns were considered to be a necessary way to acquire new gene functions and preferred to rise at the earlier stages of gene expansion and gradually diminish over time (Roy and Gilbert, 2006; Rose, 2018; Roy and Penny, 2007; Iwamoto et al., 1998).
Expression pattern analyses can provide valuable insights into the potential functions of genes. RNA-seq data indicate that all 16 GmFRL genes were expressed in at least one tissue, with the majority of gene family members exhibiting high expression levels in flowers and seeds, suggesting their essential roles in these developmental stages. The tissue-specific expression of GmFRL genes varies among different classes, which may imply functional diversification. Some GmFRLs may have specific functions in particular tissues and could be involved in the soybean life cycle. Environmental signals are typically sensed by leaves, while flowers develop from primordia that form on the sides of the SAM (Searle et al., 2006). In Arabidopsis, the localized expression of FRI in the phloem and leaves activates FLC, thereby delaying flowering (Searle et al., 2006). At the same time, the spatial expression of FRI in roots may generate a mobile signal that is transmitted from the roots to the shoot apex, antagonizing FT signaling to further delay flowering (Kong et al., 2019). Therefore, the specific tissue expression patterns of GmFRLs may be related to the mechanism by which FLC regulates flowering in a spatially dependent manner. FRI itself encodes a large protein that cannot move over long distances, but by upregulating FLC expression, FRI can function in specific tissues, including the phloem, leaves, shoot apical meristem, and roots, to delay flowering (Kong et al., 2019).
Previous studies have identified that a major determinant of flowering time in natural variants of Arabidopsis thaliana is the AtFRI gene. AtFRI functions by upregulating the expression of the floral repressor FLC, thereby establishing a vernalization requirement and promoting a winter annual growth habit (Zhu et al., 2021). In other research, they analyzed the localization of AtFRI in vivo of the Arabidopsis. Like many other co-transcriptional regulators (Sabari et al., 2020; Bienz, 2020) they found that FRI–GFP forms nuclear condensates, which were increased in size and number after cold exposure (Zhu et al., 2021). Other research showed that FRI function is also suppressed by mutations in the FRI homologs FRL1 and FRL2, and these FRI-related proteins, therefore, may all form a complex in vivo. these FRI interactors both influence FLC capping but fall into different groups of FRI suppressors: those that are specific for FRI and those that suppress FLC up-regulation more generally. FRI suppressors with apparently different specificities might appear to influence a common mechanism through intimate connection of the co-transcriptional processes linking 5’ capping, 3’ end formation, nuclear export, and transcriptional elongation (Geraldo et al., 2009). In our study, we analyzed the subcellular localization of the GmFRL gene family. The subcellular localization results revealed that four genes (GmFRL01, GmFRL04, GmFRL09, and GmFRL13) exhibited a surprising and intriguing observation: the GmFRL01/04/09/13-GFP fusion proteins localize primarily in nuclear condensates. This finding suggests that certain GmFRL genes may share functional similarities with AtFRI in Arabidopsis, possibly acting as transcriptional activators. However, functional analysis of the GmFRL family is still in its early stages, and these results warrant further investigation through gene cloning and expression analysis in future studies.
In Arabidopsis, the winter annual growth habit is conferred by FRIGIDA (FRI) and FLC. FRI encodes a plant-specific scaffold protein and functions dominantly to upregulate FLC expression to a high level that inhibits flowering (Choi et al., 2011). RNA-seq data and qPCR results show that most GmFRL genes have an expression pattern that is upregulated during midday, and downregulated in dusk. These results reveal that GmFRL genes might play potential roles in the photoperiod pathway, in addition to responding to cold signals to regulate flowering time in soybean. However, within the molecular network regulating flowering in plants, the expression of the FLC gene is directly or indirectly regulated by various key factors through different pathways. This study focused solely on exploring and validating the photoperiodic response of GmFRLs under long-day conditions. Further research is needed on the light-responsive expression patterns of GmFRL, as well as its effects on plant flowering under combined regulatory conditions with light response.
5 Conclusions
This study presents a comprehensive and systematic analysis of the GmFRL gene family utilizing bioinformatics approaches. A total of 16 members of the soybean FRL gene family were identified, and the amplification and functional differentiation of the GmFRL gene family were explored. The expression profiles generated under varying photoperiods, along with the results from subcellular localization studies, underscore the potential roles of GmFRL genes in the photoperiodic responses of soybean. The extensive bioinformatics and expression analyses conducted on the GmFRL genes significantly enhance our understanding of their functions in light responses and the regulation of flowering time. Collectively, these findings establish a robust theoretical framework and provide a valuable reference for future investigations into the associated functions and regulatory mechanisms of these genes.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94228.
Author contributions
SY: Supervision, Writing – review & editing, Methodology, Data curation, Formal analysis, Visualization. YW: Methodology, Validation, Visualization, Writing – original draft, Conceptualization, Data curation, Formal analysis. WR: Methodology, Validation, Data curation, Software, Writing – original draft. YF: Methodology, Validation, Writing – review & editing, Funding acquisition, Software. LW: Methodology, Validation, Writing – review & editing, Software. YZ: Writing – review & editing, Methodology, Software, Validation. CS: Data curation, Methodology, Visualization, Writing – review & editing, Project administration, Software, Supervision. XL: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing, Formal analysis, Funding acquisition, Methodology.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported in part by the Taishan Scholars Program (tsqn202211301), Natural Science Foundation of Shandong Province (ZR2021YQ16, ZR2023QC085, ZR2022QC262), Weifang Science and Technology Plan (2023ZJ1063), and project SYS202206 supported by the Shandong Provincial Natural Science Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1536866/full#supplementary-material
Supplementary Figure 1 | Motif logo of GmFRL conserved domains.
Supplementary Figure 2 | The expression levels of GmFRL genes. (A). According to the RNA-seq data, the expression levels of GmFRL genes in LD within 48h. (B). The relative expression levels of the remaining GmFRL genes were measured at 0, 4, 8, 12, 16, 20, and 24 hours under long-day (LD) conditions.
Supplementary Figure 3 | Inventory of FRL genes in different plant genomes.
References
Bienz, M. (2020). Head-to-tail polymerization in the assembly of biomolecular condensates. Cell 182, 799–811. doi: 10.1016/j.cell.2020.07.037
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Cao, D., Li, Y., Lu, S., Wang, J., Nan, H., Li, X., et al. (2015). GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 56 (12), 2409–22. doi: 10.1093/pcp/pcv152
Chao, Y., Yang, Q., Kang, J., Zhang, T., Sun, Y. (2013). Expression of the alfalfa FRIGIDA-like gene, MsFRI-L delays flowering time in transgenic Arabidopsis thaliana. Mol. Biol. Rep. 40, 2083–2090. doi: 10.1007/s11033-012-2266-8
Chen, W., Wang, P., Wang, D., Shi, M., Xia, Y., He, Q., et al. (2020). EjFRI, FRIGIDA (FRI) ortholog from eriobotrya japonica, delays flowering in arabidopsis. Int. J. Mol. Sci. 21, 1087. doi: 10.3390/ijms21031087
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Choi, K., Kim, J., Hwang, H.-J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
Dean, G. G. S. A. C. (2002). Arabidopsis, the rosetta stone of flowering time. Science 296, 285–289. doi: 10.1126/science.296.5566.285
Ding, L., Kim, S. Y., Michaels, S. D. (2013). FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol. 163, 243–252. doi: 10.1104/pp.113.223958
Du, J., Tian, Z., Sui, Y., Zhao, M., Song, Q., Cannon, S. B., et al. (2012). Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24, 21–32. doi: 10.1105/tpc.111.092759
Emami, H., Kempken, F. (2019). PRECOCIOUS1 (POCO1), a mitochondrial pentatricopeptide repeat protein affects flowering time in Arabidopsis thaliana. Plant J. 100, 265–278. doi: 10.1111/tpj.14441
Geraldo, N., BäUrle, I., Kidou, S.-I., Hu, X., Dean, C. (2009). FRIGIDA delays flowering in arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150, 1611–1618. doi: 10.1104/pp.109.137448
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Hu, X., Kong, X., Wang, C., Ma, L., Zhao, J., Wei, J., et al. (2014). Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 26, 4763–4781. doi: 10.1105/tpc.114.132738
Iqbal, S., Ni, X., Bilal, M. S., Shi, T., Khalil-Ur-Rehman, M., Zhenpeng, P., et al. (2020). Identification and expression profiling of sugar transporter genes during sugar accumulation at different stages of fruit development in apricot. Gene. 742, 144584. doi: 10.1016/j.gene.2020.144584
Iwamoto, M., Maekawa, M., Saito, A., Higo, H., Higo, K. (1998). Evolutionary relationship of plant catalase genes inferred from exon-intron structures: isozyme divergence after the separation of monocots and dicots. Theor. Appl. Genet. 97, 9–19. doi: 10.1007/s001220050861
Keller, S. R., Levsen, N., Ingvarsson, P. K., Olson, M. S., Tiffin, P. (2011). Local selection across a latitudinal gradient shapes nucleotide diversity in balsam poplar, Populus balsamifera L. Genetics 188, 941–952. doi: 10.1534/genetics.111.128041
Khan, M., Luo, B., Hu, M., Fu, S., Liu, J., Jiang, M., et al. (2022). Brassinosteroid signaling downstream suppressor BIN2 interacts with SLFRIGIDA-LIKE to induce early flowering in tomato. Int. J. Mol. Sci. 23, 11264. doi: 10.3390/ijms231911264
Kim, D., Landmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–U121. doi: 10.1038/Nmeth.3317
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., Depamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Kong, X., Luo, L., Zhao, J., Chen, Q., Chang, G., Huang, J., et al. (2019). Expression of FRIGIDA in root inhibits flowering in Arabidopsis thaliana. J. Exp. Bot. 70, 5101–5114. doi: 10.1093/jxb/erz287
Kumar, G., Arya, P., Gupta, K., Randhawa, V., Acharya, V., Singh, A. K. (2016a). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 6, 20695. doi: 10.1038/srep20695
Kumar, S., Stecher, G., Tamura, K. (2016b). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, I., Amasino, R. M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162. doi: 10.1104/pp.108.1.157
Li, H., Gao, J., Shi, T., Iqbal, S., Ni, Z., Gao, Z. (2021). Genome-wide identification and expression analysis of the frigida domain gene family in Prunus mume (Prunus mume Sieb. et Zucc.). Horticult Environment Biotechnol. 62, 817–828. doi: 10.1007/s13580-021-00357-8
Li, M., Liu, Y., Tao, Y., Xu, C., Li, X., Zhang, X., et al. (2019). Identification of genetic loci and candidate genes related to soybean flowering through genome wide association study. BMC Genomics 20, 987. doi: 10.1186/s12864-019-6324-7
Li, Z., Jiang, D., He, Y. (2018). FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 4, 836–846. doi: 10.1038/s41477-018-0250-6
Michaels, S. D., Bezerra, I. C., Amasino, R. M. (2004). FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 3281–3285. doi: 10.1073/pnas.0306778101
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Panchy, N., Lehti-Shiu, M., Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Risk, J. M., Laurie, R. E., Macknight, R. C., Day, C. L. (2010). FRIGIDA and related proteins have a conserved central domain and family specific N- and C- terminal regions that are functionally important. Plant Mol. Biol. 73, 493–505. doi: 10.1007/s11103-010-9635-2
Rose, A. B. (2018). Introns as gene regulators: A brick on the accelerator. Front. Genet. 9. doi: 10.3389/fgene.2018.00672
Roy, S. W., Gilbert, W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. doi: 10.1038/nrg1807
Roy, S. W., Penny, D. (2007). A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 24, 1447–1457. doi: 10.1093/molbev/msm048
Sabari, B. R., Dall’agnese, A., Young, R. A. (2020). Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977. doi: 10.1016/j.tibs.2020.06.007
Salih, H., Gong, W., He, S., Sun, G., Sun, J., Du, X. (2016). Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 17, 129. doi: 10.1186/s12863-016-0436-8
Schmitz, R. J., Amasino, R. M. (2007). Vernalization: A model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta (BBA) - Gene Structure Expression 1769, 269–275. doi: 10.1016/j.bbaexp.2007.02.003
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Takada, S., Akter, A., Itabashi, E., Nishida, N., Shea, D. J., Miyaji, N., et al. (2019). The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 9, 13843. doi: 10.1038/s41598-019-50122-2
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson, J. D., Gibson, T. J., Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinf. 00, 2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00
Tong, Z., Gao, Z., Wang, F., Zhou, J., Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10, 71. doi: 10.1186/1471-2199-10-71
Valliyodan, B., Cannon, S. B., Bayer, P. E., Shu, S., Brown, A. V., Ren, L., et al. (2019). Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 100, 1066–1082. doi: 10.1111/tpj.14500
Vieira, N. G., Ferrari, I. F., Rezende, J. C., Mayer, J. L. S., Mondego, J. M. C. (2019). Homeologous regulation of Frigida-like genes provides insights on reproductive development and somatic embryogenesis in the allotetraploid Coffea arabica. Sci. Rep. 9, 8446. doi: 10.1038/s41598-019-44666-6
Vision, T. J., Brown, D. G., Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Walther, D., Brunnemann, R., Selbig, J. (2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PloS Genet. 3, e11. doi: 10.1371/journal.pgen.0030011
Wang, L., Zhang, M., Li, M., Jiang, X., Jiao, W., Song, Q. (2023). A telomere-to-telomere gap-free assembly of soybean genome. Mol. Plant 16, 1711–1714. doi: 10.1016/j.molp.2023.08.012
Yang, Z., Luo, C., Pei, X., Wang, S., Huang, Y., Li, J., et al. (2024). SoyMD: a platform combining multi-omics data with various tools for soybean research and breeding. Nucleic Acids Res. 52, D1639–d1650. doi: 10.1093/nar/gkad786
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The Genomes of Oryza sativa: a history of duplications. PloS Biol. 3, e38. doi: 10.1371/journal.pbio.0030038
Keywords: Glycine max, FRIGIDA-LIKE, photoperiod response, gene expression, genome-wide identification
Citation: Yu S, Wang Y, Ren W, Fang Y, Wang L, Zhang Y, Song C and Luo X (2025) Comprehensive genome-wide analysis of the GmFRIGIDA gene family in soybean: identification, characterization, and expression dynamics. Front. Plant Sci. 16:1536866. doi: 10.3389/fpls.2025.1536866
Received: 29 November 2024; Accepted: 07 February 2025;
Published: 10 March 2025.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jing Zhao, Hebei Academy of Agricultural and Forestry Sciences, ChinaJingzhe Sun, Chinese Academy of Sciences (CAS), China
Hongru Xu, Wuhan University, China
Copyright © 2025 Yu, Wang, Ren, Fang, Wang, Zhang, Song and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengyang Song, chengyang.song@pku-iaas.edu.cn; Xiao Luo, xiao.luo@pku-iaas.edu.cn
†These authors have contributed equally to this work
 Song Yu1†
Song Yu1†