
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 February 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1535880
Anagenesis accumulates favorable mutations that enable crops to adapt to continually improving artificial production environments, while cladogenesis results in the deposition of beneficial variations across diverse ecotypes. Integrating advantageous genetic variations from diverse evolutionary sources establishes the foundation for the continued genetic improvement of crops. For a long time, rice breeding practices have been guided by the established belief that the Asian cultivated rice consists of two subspecies: Oryza sativa subsp. indica and subsp. japonica. Integrating elite genetic variants from both subspecies has been a major strategy for genetic improvement. This approach has proven successful through the achievements of temperate japonica breeding programs in China, Japan, and Korea over the past decades. The genetic differentiation within the Asian cultivated rice has been successfully harnessed for heterosis breeding, thereby enhancing rice yield productivity. Genomic investigations have revealed more genetic divergences in the Asian cultivated rice, prompting the proposal of six subgroups within it. This indicates that there is greater potential for uncovering additional genetic divergences and diversity in future breeding practices. Genetic introgression and gene flow among subgroups have led to improvements in agronomic traits within the indica, temperate japonica, and tropical japonica subgroups during the modern rice breeding process. The introgression process has widened the genetic diversity within subgroups and reduced the genetic distance between them, resulting in the creation of new genetic blocks and subpopulations. Artificial introgression has accelerated the evolution process in rice breeding history. Advancements in the study of genetic divergence and diversity in rice offer valuable insights to guide breeding practices. The mini subgroups aus, basmatic, and rayada possess untapped genetic potential but have been poorly studied worldwide; more samples should be further investigated. This information will be invaluable for harnessing these advantageous variations through introgression breeding. Further studying the nature of reproductive barriers among subgroups will enhance our understanding of genetic differentiation, allow us to overcome these barriers and facilitate effective genetic exchange, and even enable us to harness heterosis among subgroups.
Genetic diversity is not only essential for crop resilience in diverse environments but also fundamental to genetic improvement. Enhancing the genetic diversity of breeding pools that carry optimum combinations of favorable alleles for targeted crop-growing regions is crucial for sustaining genetic gain (Bohra et al., 2022). The evolutionary process of crops typically manifests as dynamic changes in genetic diversity, driven by major forces such as migration, selection, genetic drift, reproductive barriers, and polyploidization (Ladizinsky, 1998; Wu et al., 2020; Cao et al., 2023). Migration is the movement of genes within and between populations. When immigrants from other populations bring unique genes, they can increase the genetic variability of the recipient population. When a plant species is domesticated and becomes a crop to sustain human populations, long-term accumulation of genetic diversity occurs through genetic mutation and migration, leading to diffusion and regional expansion of the crop (Ladizinsky, 1998; Hancock, 2012). Genetic improvement is a co-evolutionary process between human civilization and crop species, where new genetic blocks with beneficial characteristics are continuously selected to meet the evolving demands for improved cultivation conditions and higher yields, in order to support the growing human population. Artificial hybridization and selection allow favorable genetic variants dispersed across different populations to be combined into new genetic combinations, and this introgression process accelerates crop evolution. In Asian cultivated rice (Oryza sativa L.), the recognition that it consists of two subspecies or groups (Kato et al., 1928) has long been used to guide rice breeding practices aimed at exploiting beneficial genetic diversity and heterosis. The success of breeding programs aimed at genetic improvement of temperate japonica rice in Northeast China since the 1950s (Yang et al., 1962) has been demonstrated through inter-subspecific introgression, resulting in significant increases in indica-allele frequencies in the cultivars bred after 1990 (Sun et al., 2012). indica-japonica heterosis, which was expected to show great potential, has been supported by various studies (Yuan, 1987; Fu et al., 2014; Birchler, 2015; Zhang, 2020; Ouyang et al., 2022; Wang et al., 2022a). Advances in genomics have provided deeper insights into the genetic divergence and diversity within the Asian cultivated rice (Garris et al., 2005; McNally et al., 2009; Huang et al., 2012; Xie et al., 2015; Wang et al., 2018; Lin et al., 2020; Qin et al., 2021; Wang et al., 2022b; Ye et al., 2022), highlighting the role of introgression among subgroups and offering a more comprehensive understanding of the genetic makeup and evolutionary history of this crop.
However, the progress of human society and population growth are exerting increasing pressure on rice production and productivity. Therefore, accurately and effectively utilizing the dispersed and rich variations in the Asian cultivated rice to develop new varieties that can address constantly emerging problems in rice production, such as climate change, water scarcity, overuse of fertilizers, insecticides, antibiotics, and herbicides, and thus reduce the cost of rice production and environmental pollution, is a fundamental issue.
Therefore, a scenario for rice genetic improvement that involves integrating elite genetic variations into new combination blocks through inter-subgroup introgression was outlined. Additionally, how the genetic structure within and between subgroups of Asian cultivated rice has evolved and changed over time, highlighting key genetic variations and patterns that have emerged, was reviewed, too. Then, that further emphasis should be placed on mining favorable alleles in previously rarely mentioned subgroups, including aus, basmatic, and rayada (Bin Rahman and Zhang, 2013; Wang et al., 2013; Jing et al., 2023), since these subgroups contain favorable traits that have not been fully identified and utilized worldwide, such as adaptation to water-insufficient upland conditions, robust growth in low-fertilizer environments, and strong competition with weeds, which could be exploited for genetic improvement of other subgroups. Furthermore, it is crucial to conduct investigations into whether hybrid vigor exists among six subgroups of the Asian cultivated rice. Additionally, examining the nature of hybrid sterility between these subgroups will be essential for the future utilization of heterosis among them.
The Asian cultivated rice, O. sativa, was domesticated from the wild species, Oryza rufipogon and O. nivara. During the domestication process, a series of obvious changes occurred in morphological traits, physiological characteristics, and ecological adaptability. These changes include the transition from creeping to erect growth, the loss of grain shattering, the shortening or absence of awns, changes in hull and grain color, reduced grain dormancy, alterations in panicle architecture, increased grain number and weight, and improved regional adaptability. These changes have collectively contributed to the improvement of agronomic traits that were favored by our human ancestors (Xu and Sun, 2021). One typical example is the amylose content of rice grain, which is an important factor in determining the eating quality of rice. The diverse amylose content in rice is mainly attributed to mutations and selections in the Waxy gene, transitioning from the ancestral allele Wxlv in wild ancestors to the indica-type allele Wxa, the temperate japonica-type allele Wxb, the tropical japonica-type allele Wxin, and the intermediate-type allele Wxop. Furthermore, the Wxmp and wx alleles were derived from mutations of the Wxb allele (Zhang et al., 2019). For the last century, modern rice breeding activities have endowed the Asian cultivated rice with more trait improvements. For example, modern rice cultivars with a semi-dwarf plant type that adapts to higher fertilizer inputs and increased planting density without lodging have been bred. These cultivars provide more food to feed the growing population of the world. These improvements are accompanied by a series of favorable mutations and allelic variant accumulations in the crop. Consequently, some favorable alleles, such as various dwarf alleles of Sd1, have become prevalent in modern rice cultivars (Chen et al., 2020; Sha et al., 2022).
The evolution of O. sativa involves the fixation of favorable variants/alleles and the elimination of inferior ones under cultivation conditions, reflecting the dynamic genetic diversity throughout its evolution. Over time, original unique alleles are lost, while new unique alleles are added (Yonemaru et al., 2012). Over decades of breeding, the total number of alleles, the number of unique alleles, and the number of varieties with unique alleles tend to increase in O. sativa (Tang et al., 2021). When compared to O. rufipogon, the diversity of the entire O. sativa population has been reduced by only ~10% of its ancestors (Huang et al., 2012; Wang et al., 2022b).
As one of the most important crops in the world, rice (O. sativa L.) is widely distributed in a range of tropical, subtropical, and temperate climates, from 55°N in China to 36°S in Chile. This far exceeds the distribution range of its ancestors, O. rufipogon and O. nivara, which are primarily limited to swampy habitats in humid tropical Asia. It evolved various ecotypes in different ecosystems, such as irrigated, lowland, upland, and flood-prone areas, and expanded its range to high latitude and altitude temperate climate conditions. Additionally, it accumulated numerous favorable variants, for instance, glutinous rice harboring the wx haplotype, which was absent in its wild progenitors (Chatterjee, 1948; Morishima et al., 1961; Morishima, 1969; Khush, 1997). It was estimated that more than 4,120,000 rice cultivars and germplasm accessions have been recognized worldwide (Song et al., 2021). For a long time, it has been widely accepted and recognized that cultivars of O. sativa are classified into two major types: O. sativa subsp. indica and subsp. japonica, based on morphological, serological characters, as well as inter-varietal hybrid fertility (Kato et al., 1928). Based on morphological characters, some researches proposed that it could be grouped in three types, A, B, and C (Matsuo, 1952), or indica, javanica, and japonica (Morinaga, 1954), or indica, tropical japonica, and temperate japonica (Oka, 1958). One thousand six hundred and eighty-eight traditional rice varieties of the Asian cultivated rice were clustered into six groups based on isozyme polymorphic markers (Glaszmann, 1987), which roughly coincided with different ecological types and geographical distributions. Subsequent researchers proposed that the Asian cultivated rice can be classified into five subgroups: indica, aus, aromatic, temperate japonica, and tropical japonica, using SSR markers (Garris et al., 2005), SNPs, and genomic sequence data (McNally et al., 2009; Wang et al., 2018; Chen et al., 2019). Since the term ‘aromatic’ implies fragrant rice, it is not appropriate to use it to refer to a group of rice that includes both fragrant and non-fragrant varieties, which are specifically found in South Asia and its surrounding regions. Therefore, we refer to this group as “basmatic” instead of “aromatic” (Zhang et al., 2022; Zhou et al., 2022) (Figure 1). A recent investigation suggested that the Asian cultivated rice consists of two separate monophyletic groups, representing the two subspecies. The subspecies indica encompasses two subgroups: indica and aus. Meanwhile, the subspecies japonica includes four subgroups: aromatic (also known as basmatic), rayada, temperate japonica, and tropical japonica. Notably, three minor subgroups—aus, basmatic, and rayada—are genetically distinct, suggesting that they deserve further attention despite being cultivated in limited areas in South and Southeast Asia (Jing et al., 2023). Furthermore, some investigations indicated that both aus and rayada exhibit high levels of genetic diversity and genetic differentiation (Parsons et al., 1999; Cai and Morishima, 2000; Wang et al., 2013; Travis et al., 2015; Islam et al., 2018). While Civán et al. (2015) proposed that the origin of aus was parallel to that of indica and japonica rice.
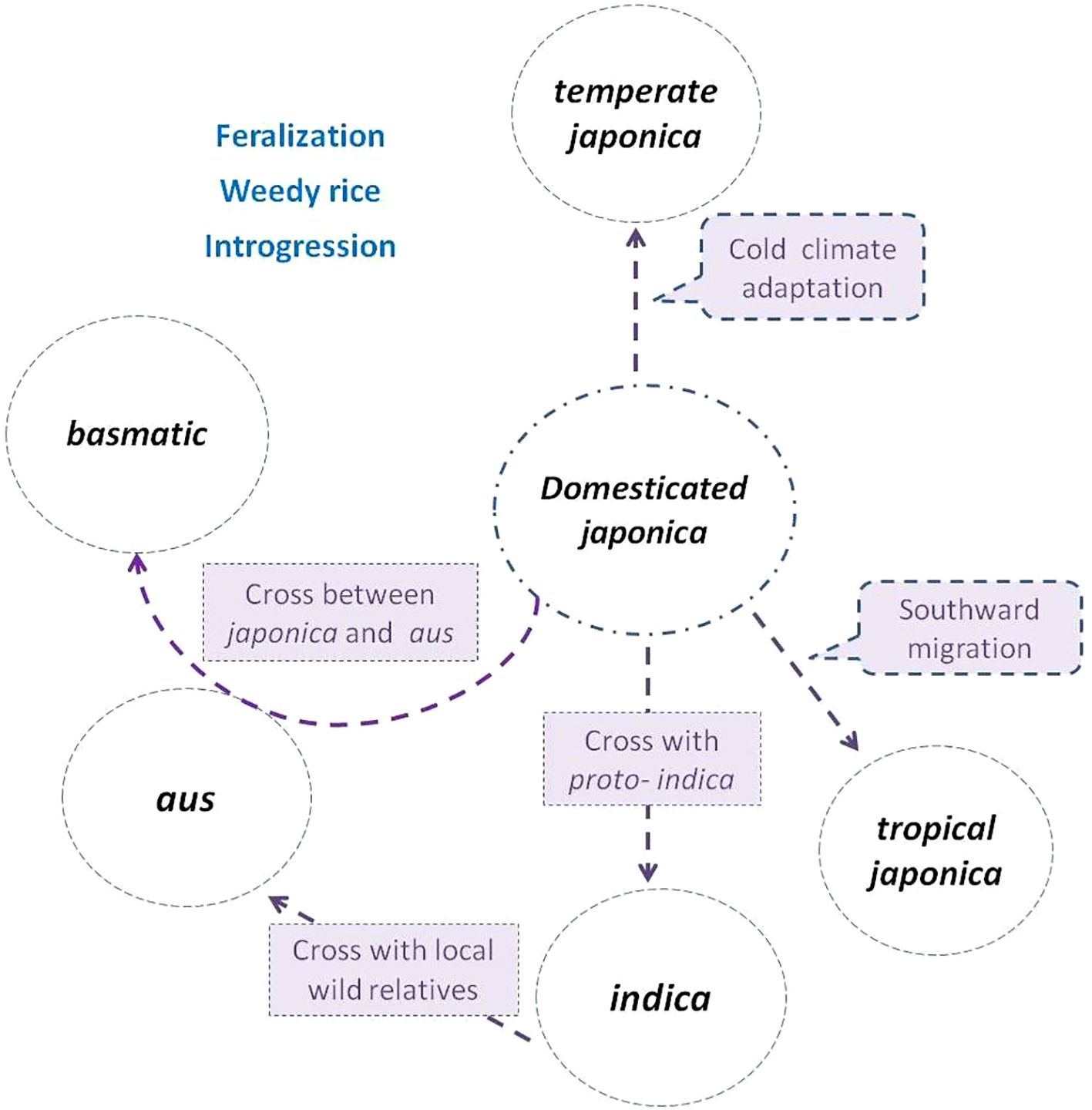
Figure 1. Subgroup differentiation of O. sativa in the evolution process (Zhou et al., 2022) japonica rice (circled in the center) was first domesticated from Oryza rufipogon, then diversified northward as temperate japonica and southward as tropical japonica (Gutaker et al., 2020). indica descended from hybridization between japonica and local wild populations or proto-indica (Fuller, 2011; Gross and Zhao, 2014). aus was derived from the hybridization between indica and local wild populations, while basmatic was derived from hybridization between japonica and aus (Civán et al., 2015; Kim et al., 2016; Wang et al., 2017; Choi et al., 2020). Weedy rice strains de-domesticated from and coexisted with cultivated subgroups, and frequently crossed with wild populations (if present) or landraces during their evolutionary process (Li and Olsen, 2020; Vigueira et al., 2020).
Generally, subgroup or subpopulation differentiation is a common phenomenon in the Asian cultivated rice, adapting to various ecological conditions at both the species level and the subspecies levels of indica and japonica. Greater genetic divergence and diversity implies further potential for unlocking available genetic variation in future breeding practices.
Artificial hybridization in crop plants, which began about 200 years ago, has allowed breeders to compile traits from various landraces and cultivars into a single plant (Ladizinsky, 1998). For example, the NRT1.1b gene, conferring nitrogen utilization efficiency (NUE), carries a beneficial allele in indica varieties. The DEP1 gene, which influences panicle architecture and yield, has a favorable allele in japonica varieties. When the nrt1.1b allele was incorporated into japonica lines carrying dep1, there was a significant increase in nitrate reductase activity, nitrogen uptake, and grain yield under low nitrogen conditions. Conversely, introducing the dep1 allele into indica lines already carrying the nrt1.1b allele resulted in enhanced glutamine synthetase activity, improved nitrogen transfer, and increased grain yield under both low and high nitrogen conditions, ultimately boosting yield potential in specific planting environments (Zhao et al., 2017).
indica-temperate japonica introgression has played an important role in temperate japonica breeding practices in Northeast China, Korea and Japan. In the early breeding stage of temperate japonica rice, trait improvement was primarily achieved through hybridization between temperate japonica varieties and subsequent artificial selection, which aimed to pyramid and fix superior alleles of key genes. However, this process resulted in a narrowing of the genetic background, compared to the prior stage (Cui et al., 2022). To broaden the genetic basis of temperate japonica cultivars, hybridization and introgression between temperate japonica and indica varieties were practiced in Northeast China in the 1950s (Yang et al., 1962). Favorable traits from indica rice genome were integrated into temperate japonica background through this method. Most of the prevalent commercial rice varieties grown in Northern China since the 1980s were bred from crosses between indica and temperate japonica (Jiang et al., 2022). The frequency of indica alleles was significantly increased in cultivars bred after 1990. These alleles were positively and significantly correlated with the number of spikelets per panicle, and negatively and significantly correlated with the number of panicles per plant. Specifically, favorable indica alleles of GN1a and GS3 were partially fixed in the genome of northern temperate japonica cultivars. In contrast, inferior indica alleles of Wx and qSH1 were eliminated during the breeding process, while favorable temperate japonica alleles of DEP1 and qSW5 were retained (Sun et al., 2012). A large introgression segment from indica, located on chromosome 12, was detected in 24% of the temperate japonica samples. This segment covers many functional genes, including two rice blast resistance genes, Pi-ta and Ptr. Another high-frequency introgression from indica, also on chromosome 12, was present in approximately 19% of the temperate japonica samples. This introgression harbors a QTL, qS12, associated with hybrid male sterility. On chromosome 11, introgressions from indica were detected in 12.1% of the temperate japonica samples, which contain a gene related to rice stripe virus resistance, STV11 (Chen et al., 2020). Favorable alleles of IPA1, SMG1, Dep3, Ghd7, GW5, OsPIN3t, xa13, Bph3, Pia, Pib, and Pi-d2 were introduced from indica rice into temperate japonica rice, This facilitated the recombination of the gene pools of temperate japonica and indica, resulting in the creation of new recombinant blocks and the improvement of agronomic traits in temperate japonica cultivars. Consequently, this contributed to the modern breeding of improved plant types, transitioning from multi-tiller plants to moderate-tiller plants with larger panicle sizes and enhanced blast resistance (Cui et al., 2022). Over the past fifty years, the indica-temperate japonica hybridization breeding process has achieved significant genetic gains in temperate japonica varieties in Northeast China. During this period, indica introgressions were detected on all 12 chromosomes. The introgression of indica segments carrying qSB2, qSB8, and qSB10 led to a 100% increase in secondary branching number per panicle and a 50% increase in grain number per panicle. In Heilongjiang Province, Northeast China, the average content of indica introgressions in a cultivar has increased markedly over time, from 8.2 Mb before 1980 to 76.6 Mb after 2010. Notably, the two adjacent rice blast resistance loci, Pi-ta and Ptr, which originate from indica and have beneficial effects, have significantly enhanced blast resistance in temperate japonica rice (Chen et al., 2023). The last century has witnessed tremendous leaps in China’s rice productivity, from 1.9 t/ha in 1949 to 7.0 t/ha in 2018 (http://faostat.fao.org/). This significant increase in rice productivity can be primarily attributed to the revolution of semi-dwarf and hybrid rice varieties. Additionally, the rapid expansion of high-yielding temperate japonica varieties that incorporate indica introgressions has also played a partial role (Wang et al., 2023a). In the national new rice varietal trials conducted in Northeast China, the average yield of new temperate japonica varieties increased from 8.3 t/ha in 2004 to 9.0 t/ha in 2018 (Fei et al., 2020). In South China, particularly in Jiangsu Province, 278 temperate japonica rice varieties with low amylose content, disease resistance, high yield, and excellent eating quality were released between 2001 and 2021. These varieties integrated favorable alleles from both japonica (Wxmp, Badh2) and indica (Gs3, Stv-bi, Pi-ta, Pi-b) sources. The breeding of temperate japonica rice with low amylose content, high yield, and multiple disease resistances has become an important direction for enhancing the taste and quality of rice in the middle and lower reaches of the Yangtze River (Wang, 2022c).
In India, indica-japonica introgression breeding program initiated in 1952 (Chakraborti et al., 2021), The International Rice Commission (IRC) was focusing to cross tall “indica” rice to dwarf “japonica” rice to evolve with shorter rice varieties, and few varieties developed by IRC made greater impact on rice growing farmers, One indica × japonica breeding line Mahsuri became popular in different countries (Patra et al., 2020).
The Tongil rice, developed in 1972, emerged as a high-yield rice variety that played a pivotal role in achieving staple food self-sufficiency in Korea during the renowned ‘Korean Green Revolution’. This groundbreaking cultivar represented the first successful example of japonica-indica hybridization in Korea. An in-depth analysis of its genome structure reveals that the Tongil genome is predominantly composed of the indica genome, with a modest yet significant proportion of japonica genome introgression (Kim et al., 2014). In Japan, high-yielding rice cultivars have been successfully developed through crosses between overseas indica varieties and domestic japonica varieties. Among these cultivars, some exhibit indica-type characteristics (Yonemaru et al., 2014). Minghui 63, a famous indica restorer parent of three-line hybrid rice from China, shows an introgression on chromosome 6 (0–4 Mb) from japonica. This introgression coincides with QTLs reported in several studies to be associated with various traits, including leaf area, vascular bundle number, root features, plant height, cooking quality, and amylose content (McNally et al., 2009). All of the indica varieties carrying the introgression of temperate japonica Wxb allele exhibit significantly lower amylose content (Zhao et al., 2010). About 63% of the indica samples carried introgression from japonica that introduced the Wxb allele, leading to a reduction in amylose content in rice grains and subsequently improving eating quality. The japonica introgression in 45% of the indica samples carried the japonica allele of the ALK gene, which was reported to reduce grain gelatinization temperature, also contributes to eating quality (Chen et al., 2020).
Breeding practices increased mutual introgression between the two subgroups, likely increasing within-population diversity and reducing subgroup differentiation. Interestingly, there was greater gene flow from japonica to indica than from indica to japonica, which was obviously due to the increased inter-subgroup crosses made in many rice breeding programs (Zhang et al., 2021).
To break the yield potential barrier, IRRI scientists proposed the high-yielding indica new plant type in the late 1980s and early 1990s. The first-generation NPT lines developed at IRRI through hybridization between indica and tropical japonica had large panicles, few unproductive tillers and resistance to lodging but low grain yield due to the limited production of biomass and poor filling of grains (Khush, 1995). The second-generation NPT lines were obtained by crossing elite indica with improved tropical japonica, in which, genes from indica parents have efficiently decreased panicle size and enhanced tiller ability. indica germplasm also helped to improve other NPT characteristics such as quality of grains and disease and insect resistance (Rahangdale et al., 2019). In Indonesia, the NPT rice architecture allows a higher yield than the green revolution rice type represented by the Ciherang variety. The yield advantage is 1.67 t ha-1 or 26% based on the best-lines average (Aswidinnoor et al., 2023).
Hybrid introgression among subgroups has renovated the genomes of improved cultivars by integrating favorable alleles from exogenous germplasms during the breeding practices of the past century. There is a highly elevated level of introgression from indica into many tropical japonica varieties near the Sd1 gene. Pi-ta is known to be of indica origin, but because tropical japonica varieties are best adapted to upland growing conditions, breeders have frequently introgressed this resistance gene to enhance the productivity of tropical japonica cultivars. An extensive segment of indica DNA located in the centromeric region on chromosome 12 is found in several tropical japonica accessions. The Pi-ta gene in the US varieties can be traced back to the cultivar, Tetep, a Vietnamese indica accession (McNally et al., 2009; Zhao et al., 2010). The tropical japonica accessions that exhibit introgression have identical haplotype with indica accessions. The tropical japonica population acquired some favorable alleles of phenotype-related loci, such as heat tolerant gene TT1, rice quality genes Wx, ALK, BAD2, and grain shape gene Gs3, from the indica through genetic introgression. Excessive genetic introgression is observed between the indica subgroup and the tropical japonica subgroup, which may be due to the overlap in their distribution areas, both located in the southern part of Southeast Asia. Thus, tropical japonica may have originated from ancient japonica through genetic introgression and developed into a unique ecotype after acquiring numerous alleles from indica to adapt to the local environment in the southern part of East Asia (Gong and Han, 2022).
Moroberekan, a tropical japonica traditional variety from Africa and a popular donor for disease resistance and drought tolerance, contains several regions on chromosome 6 introgressed from indica or aus, one of which colocalizes to a large cluster of NB-ARC-type resistance genes between 9.2 and 11.1 Mb (McNally et al., 2009).
In 3K project, within Chromosome 1, seven tropical japonica accessions from Asia share an indica/aus haplotype of more than 10 Mb (from 9.2 Mb to 19.3 Mb). The predominant japonica subgroup in Africa is tropical japonica. A 3.8 Mb introgression on chromosome 6 from aus is shared by 14 tropical japonica accessions from West Africa. Additionally, two accessions share a shorter aus haplotype between 17.9 and 20.26 Mb. On the end of chromosome 6, between 25.8 and 26.7 Mb, a block of haplotype of aus ancestry was found in half of the tropical japonica accessions from Africa. The above results suggest that hybridization and introgression between the tropical japonica accessions and the aus subgroup played a role in the generation of West African upland accessions. Out of a total of 46 genes underlying this introgression, some genes have functions directly related to responses to abiotic stresses such as salt stress and water stress. The presence of ABA receptor genes (OsPYL/RCAR7, OsPYL/RCAR8, and OsPYL7) from the introgression of aus suggests their function in water stress responses. Several studies have highlighted the high adaptability of the aus ecotype to drought or heat. African upland rice varieties, exemplified by Moroberekan, are known for their remarkable drought tolerance. An adaptive introgression on chromosome 6 derived from aus bears genes potentially involved in drought responses. This work illustrates how the evolution of genetic diversity along geographic migration can be used to enhance the corpus of genes involved in crop adaptation (Beye et al., 2023).
Genomic investigations suggestion that the basmatic rice is a result of the hybridization between japonica and aus in early evolution stage (Civán et al., 2015; Kishor et al., 2020). The japonica rice contributed the highest amount of genetic material to basmatic, while strong evidence of admixture between the basmatic and aus groups was detected (Choi et al., 2020). A unique pericarp color haplotype, rc-s, is shared only by aus and basmatic subgroups (Sweeney et al., 2007). The 3K project found that about 65% of the basmatic genome was derived from aus, while the remaining 35% from japonica, suggesting basmatic originated from crosses between aus and japonica (Wang et al., 2022b).
Historical efforts to genetically improve hybrid parental lines have led to significant increases in commercial hybrid rice productivity in China, from approximately 3.8 t/ha in the 1970s to approximately 6.8 t/ha in recent years (Yuan, 2015). The superior performance of the hybrid rice may have resulted from independent improvements in the two rice subpopulations within indica (Xie et al., 2015). Geographic adaptation and the accumulation of divergent selections in distinct breeding pools may cause differentiation within the indica subgroup. Genome clustering analysis can divide indica into several divergent subpopulations, two of which are consistently aligned with the hypothetical two heterotic groups in the germplasms of indica rice: short-statured varieties of South China origin and medium-height lines of Southeast Asia origin. ind-3B, corresponding to the maintainer lines of three-line hybrid rice, and ind-3R, the restorer lines of three-line hybrid rice, may represent two heterotic groups in Chinese indica rice (Xie et al., 2015; Li et al., 2020; Chen et al., 2020).
The genetic differences that cause hybrid vigor between male and female parents in hybrid rice breeding history in China involve exogenous genetic introgression. The male parents of hybrid rice varieties are either introduced directly from the International Rice Research Institute (IRRI) or bred using IRRI varieties as donor parents (Xie and Zhang, 2018). This means most of the restorer lines share more than 40% ancestry from IRRI varieties (Zhao et al., 2022). During hybrid breeding, regions that were divergently selected from subgroups including indica, aus, and japonica were first introgressed into hybrid parents. These introgressed regions were then selected based on their effect. The introgressed regions explained an average of 41.44% of grain yield heritability, 45.46% of biomass heritability, 45.86% of tiller number heritability, and 35.02% of panicle weight heritability in the hybrids (Lin et al., 2020). More than thirty-megabase (33.3-Mb) differentiated regions were identified between the ind-3B and ind-3R subpopulations. Many of the agronomically important genes, such as NOG1, LAX1, SD1, Wx, RFT1, Hd3a, and Hd1, fell within these differentiated regions. The heterotic loci Hd3a and TAC1, distributed differentially between the female and male parents of three-line indica hybrid rice, indicate their potential contribution to heterosis. Hd3a was present in over 98% of ind-3R samples, while this ratio was less than 30% in ind-3B subpopulation. In contrast, the introgression surrounding TAC1 on chromosome 9, from japonica, was present in 67.1% of the ind-3B samples, while only 5.1% of the ind-3R samples carried this introgression (Chen et al., 2020). Differential introgressions formatted heterotic groups (Chen et al., 2020; Lin et al., 2020). Minghui 63, the famous restorer parent of indica three-line hybrid rice, shows an introgression on chromosome 6 (0–4 Mb) from japonica, which also contains heterotic loci associated with high yield performance in heterozygous vs. homozygous individuals (McNally et al., 2009).
Landraces typically exhibit unique characteristics that make them adapt to their local environments, aligning with the interests of local farmers and consumers. However, they often contain linkage drags or detrimental traits. In breeding practice, hybridization and selection of several local varieties or accessions are used to combine favorable traits and eliminate unfavorable ones, resulting in newly combining blocks. Past breeding generally increased haplotype diversity in modern varieties compared with landraces in both indica and japonica (Zhang et al., 2021).
Most modern Japanese rice varieties were bred from crosses among a limited number of oldest varieties or landraces. However, phylogenetic analysis suggests that a strong population structure developed in Japanese cultivars during modern breeding activities. As a result, the genetic structure of modern varieties significantly differed from those bred before 1922 and landraces. Over the 75 years from 1931 to 2005, the number of new haplotype blocks gradually increased as cultivars underwent modern breeding (Yonemaru et al., 2012).
indica is the largest subgroup in O. sativa, accounting for over 80% of total rice cultivation worldwide. It is the staple food for most people in tropical and subtropical regions of South China, Southeast Asia, and South Asia (Mahesh et al., 2016). Modern breeding practices have greatly changed the indica population structure. Based on genomic sequence data, 809 worldwide indica varieties were classified into two groups: IndI and IndII. The IndI group had germplasm mainly of South China origin, while IndII mainly originated from South Asia and Southeast Asia. The differentiation between IndI and IndII might be caused by geographic adaptation and the accumulation of divergent selections in distinct breeding pools (Xie et al., 2015). In the 3K project, indica was divided into four clusters: XI-1A from East Asia, XI-1B of modern varieties of diverse origins, XI-2 from South Asia, and XI-3 from Southeast Asia (Wang et al., 2018). Based on the frequency differentiation of local haplotypes from sequencing data of 2,429 indica accessions, three groups were classified. One group mainly originated from China, another from South Asia and Southeast Asia, and the third distinct group was mainly constituted of modern varieties with diverse origins. The modern indica subgroup exhibits signatures of gathering favorable alleles for rice production (Qin et al., 2021). In a recent investigation, indica was divided into two groups: indica-I (IND-I) and indica-II (IND-II). Most of the Chinese rice varieties belonged to IND-II, which was further classified into two subpopulations. The IND-C1 subpopulation was mainly composed of land cultivars bred in the 1950s, while the IND-C2 subpopulation included modern cultivars bred after the 1980s (Ge et al., 2022). Modern varieties with diverse origins formed a separate subpopulation that strongly represented beneficial alleles related to agricultural production. This genetic differentiation did not occur on a genome-wide scale, but rather was confined to specific loci or chromosome intervals (Xu et al., 2016; Wang et al., 2018; Qin et al., 2021). Genetic divergences in the indica group demonstrate parallel evolution in ecosystems. Subsequently, breeding preferences under certain spatial and temporal conditions also impact the result of classification. This may be due to certain variety groups harboring similar allelic combination blocks by using similar backbone parents or popular alleles, such as Sd1, during a specific breeding phase. Conditions and preferences during this phase can also influence allelic combinations. Therefore, long-term regional distribution, geographical barriers, and breeding efforts are the major factors contributing to genetic divergence in the indica subgroup.
Generally, Asian cultivated rice has undergone a remarkable evolution in adapting to improved cultivation environments. Variants that were adapted to specific ecosystems were selected, leading to the formation of unique haplotypes. Favorable mutations were fixed and accumulated within these ecosystems, ultimately resulting in the formation of subgroups, subpopulations, or ecotypes (Figure 1; Zhou et al., 2022). Mutual introgression among subgroups can create new recombination and genetic blocks. Intensive artificial introgression and selection further enhance genetic migration and genetic drift, which sharply accelerates the evolution process (Figure 2).

Figure 2. Introgression impacts genetic divergence, diversity in the evolution process of O. sativa. Triangles of various colors represent special variants or mutations that have occurred during the evolutionary process. Purple solid arrows denote the direction of introgression during the domestication process and early evolution stages. These introgressions played a role in the formation of the basmatic genome by integrating genetic material from japonica and aus (Civán et al., 2015; Choi et al., 2020). Black solid arrows indicate introgression and gene flow during the modern breeding process, (1) indica introgrsssion for temperate japonica improvement in northeast China (Sun et al., 2012; Chen et al., 2020; Cui et al., 2022); (2) japonica introgression for indica improvement in China, India, Japan, and Korea (Zhao et al., 2010; Kim et al., 2014; Yonemaru et al., 2014; Patra et al., 2020; Chakraborti et al., 2021). indica/aus intrgression for tropical japonica improvement (McNally et al., 2009; Gong and Han, 2022; Beye et al., 2023); (4) aus/japonica introgression shaped heterotic group (McNally et al., 2009; Chen et al., 2020; Lin et al., 2020); (5) tropical japonica introgression for the development of NPT indica (Khush, 1995; Aswidinnoor et al., 2023). The gene flow between subgroups without literature reference is not shown in the figure. The purple dashed circle signifies further introgression among different subgroups.
Artificial hybridization in crop plants began about 200 years ago. It has enabled the assembly of traits, which individually exist in several landraces and cultivars, into one plant (Ladizinsky, 1998). In rice, the frequencies of favorable haplotypes are typically very low in modern indica and japonica varieties for most genes. However, favorable alleles at most cloned genes are present at high frequencies in landraces from specific geographic regions. Unfortunately, accessions from these populations have rarely been used as breeding parents. Specifically, the tropical japonica, subtropical japonica, and aus subgroups have high frequencies of favorable haplotypes at most genes. These haplotypes are present at low frequencies in modern varieties. As a result, most modern varieties currently grown in farmers’ fields do not have the ‘best’ alleles at most gene loci. Therefore, there is a huge potential to improve yield traits and productivity of modern varieties by pyramiding the missing favorable haplotypes at multiple loci from carefully selected breeding parents (Zhang et al., 2021).
indica-japonica introgression is progressing well in rice breeding practices and has achieved remarkable results. However, there has been limited progress in utilizing favorable variations from other subgroups, especially the mini-subgroups such as aus, basmatic, and rayada, in which distinct characters are worthy of more attention (Bin Rahman and Zhang, 2013; Jing et al., 2023). For instance, the favorable allele for higher nitrogen use efficiency (NUE) found in aus and basmatic rice can be utilized to enhance this trait in japonica and indica rice (Liu et al., 2021), thereby alleviating the increasing application of nitrogen fertilizers and corresponding environmental pollution issues. Favorable alleles that affect rice quality, identified in most basmatic rice varieties (Siddiq et al., 2012; Babar et al., 2022), could be exploited to develop new favorable genetic combinations in the backgrounds of other subgroups, thereby improving rice quality. The combination of population growth and restrictions on available arable land has intensified genetic improvement efforts and the intensive cultivation of temperate japonica rice in East Asia, particularly in Northeast China, Japan, and Korea. This has resulted in compact plant types and superior yield traits among temperate japonica varieties in this region (Fei et al., 2020; Cui et al., 2022; Wang et al., 2023a). At the same time, increased fertilizer application and management inputs are required to ensure sufficient biomass and grain production. In comparison, rice varieties from South and Southeast Asia, including indica, aus, basmatic, and tropical japonica, can achieve good biomass and exhibit resistance/tolerance to biotic and abiotic stresses even under low-fertilizer and low-input conditions (Table 1).
Apart from the heterosis observed in hybrids resulting from the genetic differentiation between two indica heterotic groups: short-statured varieties of South China origin and medium-height lines of Southeast Asia origin (Xie et al., 2015), inter-subspecific hybrids between indica and japonica rice varieties exhibited significantly superior performance and higher heterosis compared to intra-subspecific hybrids (Ouyang et al., 2022). With the gradual resolution of hybrid sterility issues (Zhang, 2020; Ouyang et al., 2022; Wang et al., 2023b), the utilization of heterosis from inter-subspecific hybrids between indica and japonica rice has emerged as a feasible strategy for developing hybrid rice with higher yield potential. In South China, a series of indica-japonica inter-subspecific hybrid rice varieties, exampled by Yongyou series of intersubspecific hybrid rice, have been bred through introgression and fixation of advantageous alleles related to hybrid sterility and key yield-determining genes into both male and female parents (Wang et al., 2022a). The effective pyramiding of rare superior alleles with positive dominance effects in hybrids has contributed to higher yields. With the discovery of favorable rare alleles in both indica and japonica rice varieties, it is anticipated that indica-japonica hybrid rice will exhibit even better performance in the future (Wei and Huang, 2019).
Genomic investigation has provided evidence of apparent introgression of heterotic loci from other subgroups into indica hybrid rice parents (Lin et al., 2020), suggesting the presence of potential heterotic loci among subgroups and indicating that hybrid vigor among the six subgroups could be expected.
Genomic advances have provided rich information on genetic diversity and subgroup divergence within the Asian cultivated rice, leading to the proposal of more subgroups beyond the two subspecies, indica and japonica. This provides valuable guidance for exploiting favorable variants for further genetic improvement. Although the six subgroups, including the ‘mini’ groups, aus, basmatic, and rayada, exhibit distinguishable genealogies, genomic information for these subgroups, except for the widely recognized indica and japonica rice, has only been obtained from a limited number of samples from the ‘mini’ subgroups. This may not be sufficient to fully reflect and represent the genetic characteristics and diversity of these subgroups.
Although genomic evidence provides suggestions on genetic divergence in the Asian cultivated rice, it could be further substantiated by additional evidences, such as reproductive barriers, which are important indicators of genetic divergence and speciation in plant evolution. Hybrid sterility is the phenomenon where the F1 plant produces partially abortive male or female gametes, while its parents possess normal gametes (Kato et al., 1928). It represents a common reproductive barrier among distant varieties and poses a major obstacle to harnessing hybrid vigor between indica and japonica rice. Severe hybrid sterility typically leads to spikelet sterility in F1 plants derived from crosses between indica and japonica rice. Therefore, studying the phenomenon of hybrid sterility is not only beneficial for understanding the degree of genetic differentiation among subgroups of Asian cultivated rice, but also crucial for overcoming hybrid sterility and achieving the utilization of their heterosis. Twenty-four hybrid sterility loci have been identified between indica and temperate japonica, representing the highest number of such loci among the subgroups compared. Eight loci were found between temperate japonica and tropical japonica, two between tropical japonica and aus, two between indica and aus, five between temperate japonica and aus, and four between tropical japonica and indica. Strikingly, three loci were also discovered within the indica subgroup, suggesting potential genetic divergence within this subgroup (Zhang et al., 2022). Although genomic evidence has suggested that mini-subgroups such as aus, basmatic, and rayada possess unique genetic lineages (Jing et al., 2023), little work has focused on hybrid sterility in these mini-subgroups. Consequently, only a few hybrid sterility loci related to aus and tropical japonica have been detected. To date, there are limited reports (Wang et al., 2023c; Lv et al., 2024) on hybrid sterility associated with basmatic, and even fewer on the rarely studied subgroup, rayada.
Besides hybrid sterility, various other reproductive barriers have been observed in rice, including hybrid weakness (Oka, 1957; Sato and Morishima, 1987), hybrid breakdown (Oka, 1957; Oka and Doida, 1962; Sato and Morishima, 1987; Fukuoka et al., 1998; Kubo and Yoshimura, 2002), transmission ratio distortion (Nakagahra, 1972), certation (Song et al., 2002), and reciprocal gene loss of duplicated genes (Mizuta et al., 2010). Genes responsible for reproductive barrier traits are termed “speciation genes,” which contribute to the cessation of gene flow between populations and can offer clues regarding the ecological settings, evolutionary forces, and molecular mechanisms that drive the divergence of populations and species (Rieseberg and Blackman, 2010).
Past efforts in utilizing rice heterosis have primarily concentrated on intra-subspecific indica-indica hybrid vigor, primarily attributed to rarely hybrid sterility among varieties within it. Advancement on inter-subspecific indica- temperate japonica hybrid vigor in China (Wang et al., 2022a) provided a way for effectively overcoming hybrid sterility, and harnessing heterosis among sub-groups in the Asian cultivated rice. However, little attention has been paid to heterosis among all six subgroups, due to a lack of comprehensive understanding of genetic differentiation and reproductive barriers among these divergent subgroups.
Therefore, future efforts should be focused on the following: 1) identifying more unique advantageous traits/alleles within each subgroup; 2) analyzing whether there exists hybrid vigor/heterotic loci among the six subgroups; 3) studying the genetic nature of reproductive barriers, particularly hybrid sterility, among subgroups in order to deeply understand genetic differentiation, overcome reproductive barriers, achieve effective introgression of target genetic variations, and ultimately utilize the hybridization vitality among subgroups or subpopulations.
Crop diversity underpins the productivity, resilience, and adaptive capacity of agricultural systems. Genetic diversity, on the other hand, represents a dynamic process in crop evolution. Plants can be considered as evolving along two dimensions: anagenesis and cladogenesis. Integrating favorable traits from both evolutionary dimensions to reconstruct new combination blocks for genetic improvement is a major and effective approach in rice breeding, and this method is proving successful in indica-japonica introgression breeding practices, especially exampled in breeding programs in Northeast China. Artificial introgression accelerated evolution. Genomic advances have enhanced our understanding of the greater genetic divergences and diversity among subgroups of the Asian cultivated rice, suggesting that there is further potential for genetic variation awaiting utilization in future breeding practices. This will benefit the targeted and effective exploitation of favorable variants within the germplasm of a specific subgroup. For the mini subgroups—aus, basmatic, and rayada—more samples should be used to further investigate and obtain more genomic and phenotypic information about their genetic differentiation, as well as to exploit favorable variants deposited within them. Research into whether there is hybrid vigor among subgroups; as well as into reproductive barrier traits, primarily manifested as hybrid sterility, will help us better understand subgroup differentiation. It will also aid in overcoming reproductive barriers to achieve effective genetic exchange among subgroups, and even harness hybrid vigor among them. Introgression breeding is also a rational approach to improving the mini subgroups for agricultural purposes.
JZ: Writing – original draft, Writing – review & editing. JL: Writing – review & editing. YZ: Writing – review & editing. YY: Writing – review & editing. YL: Writing – review & editing. QP: Writing – review & editing. XD: Writing – review & editing. DT: Conceptualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by National Natural Science Foundation of China (Grant Nos. 31991221, 32160489, and U2002202), Yunnan Provincial Science and Technology Department, China (Grant Nos. 530000210000000013809, 202201AS070072, 202205AC160057; 202205AR070001-04; 202301BD070001-051; 202401AT070090), Yunnan Provincial Government (YNWR-QNBJ-2018-359).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acuña, T., Laftte, H., Wade, L. (2008). Genotype × environment interactions for grain yield of upland rice backcross lines in diverse hydrological environments. Field Crops Res. 108, 117–125. doi: 10.1016/j.fcr.2008.04.003
Ara, I., Lewis, M., Ostendorf, B. (2017). Understanding the spatially variable effects of climate change on rice yield for three ecotypes in Bangladesh 1981-2010. Adv. Agric. 2017, 1–11. doi: 10.1155/2017/6287156
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Aswidinnoor, H., Listiyanto, R., Suwarno, W. B. (2023). Genetic architecture of new plant type rice (Oryza sativa L.) lines based on a 12-years multi-experiment. SABRAO J. Breed. Genet. 55, 1025–1037. doi: 10.54910/sabrao2023.55.4.2
Babar, A., Zaka, A., Naveed, S., Ahmad, N., Aslam, K., Asif, M., et al. (2022). Development of Basmati lines by the introgression of three bacterial blight resistant genes through marker-assisted breeding. Euphytica 218, 59. doi: 10.1007/s10681-022-03013-z
Bernier, J., Kumar, A., Venuprasad, R., Spaner, D., Verulkar, S., Mandal, N., et al. (2009). Characterization of the effect of a QTL for drought resistance in rice, qtl12.1, over a range of environments in the Philippines and eastern India. Euphytica 166 , 207–217. doi: 10.1007/s10681-008-9826-y
Beye, A., Billot, C., Ronfort, J., McNally, K., Diouf, D., Glaszmann, J. (2023). Traces of introgression from cAus into tropical japonica observed in African upland rice varieties. Rice 16, 12. doi: 10.1186/s12284-023-00625-4
Bin Rahman, A., Zhang, J. (2013). Rayada specialty: the forgotten resource of elite features of rice. Rice 6, 41. doi: 10.1186/1939-8433-6-41
Birchler, J. (2015). Heterosis: The genetic basis of hybrid vigour. Nat. Plants 1, 15020. doi: 10.1038/nplants.2015.20
Bohra, A., Kilian, B., Sivasankar, S., Caccamo, M., Mba, C., McCouch, S., et al. (2022). Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 40, 412–431. doi: 10.1016/j.tibtech.2021.08.009
Bradbury, L., Fitgerald, T., Henry, R., Jin, Q., Waters, D. (2005). The gene for fragrance in rice. Plant Biotech. J. 3, 363–370. doi: 10.1111/j.1467-7652.2005.00131.x
Cai, H., Morishima, H. (2000). Diversity of rice varieties and cropping systems in Bangladesh deepwater areas. JARQ 34, 225–231. Available online at: https://www.jircas.go.jp/sites/default/files/publication/jarq/34-4-225-231_0.pdf.
Cao, Y., Zhao, K., Xu, J., Wu, L., Hao, F., Sun, M., et al. (2023). Genome balance and dosage effect drive allopolyploid formation in Brassica. PNAS 120, e2217672120. doi: 10.1073/pnas.2217672120
Carrillo, M., Martin, F., Variar, M., Bhatt, J., Perez-Quintero, L., Leung, H., et al. (2021). Accumulating candidate genes for broadspectrum resistance to rice blast in a drought-tolerant rice cultivar. Sci. Rep. 11, 21502. doi: 10.1038/s41598-021-00759-9
Chakraborti, M., Anilkumar, C., Verma, R. L., Abdul Fiyaz, R., Reshmi Raj, K. R., Patra, B. C., et al. (2021). Rice breeding in India: eight decades of journey towards enhancing the genetic gain for yield, nutritional quality, and commodity value. Oryza 58, 69–88. doi: 10.35709/ory.2021.58.spl.2
Chatterjee, D. (1948). A modified key and enumeration of the species of Oryza Linn. Ind. Jour. Agr. Sci. 18, 185–192.
Chen, Z., Bu, Q., Liu, G., Wang, M., Wang, H., Liu, H., et al. (2023). Genomic decoding of breeding history to guide breeding-by-design in rice. Natl. Sci. Rev. 10, nwad029. doi: 10.1093/nsr/nwad029
Chen, E., Huang, X., Tian, Z., Wing, R. A., Han, B. (2019). The genomics of Oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 70, 639–665. doi: 10.1146/annurev-arplant-050718-100320
Chen, Z., Li, X., Lu, H., Gao, Q., Du, H., Peng, H., et al. (2020). Genomic atlases of introgression and differentiation reveal breeding footprints in Chinese cultivated rice. J. Genet. Genomics 47, 637–649. doi: 10.1016/j.jgg.2020.10.006
Choi, J., Lye, Z., Groen, S., Dai, X., Rughani, P., Zaaijer, S., et al. (2020). Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol. 21, 21. doi: 10.1186/s13059-020-1938-2
Civán, P., Craig, H., Cox, C. J., Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nat. Plants 1, 15164. doi: 10.1038/NPLANTS.2015.164
Cui, D., Zhou, H., Ma, X., Lin, Z., Sun, L., Han, B., et al. (2022). Genomic insights on the contribution of introgressions from xian/indica to the genetic improvement of geng/japonica rice cultivars. Plant Commun. 3, 100325. doi: 10.1016/j.xplc.2022.100325
Fei, C., Xu, Q., Xu, Z., Chen, W. (2020). Effect of rice breeding process on improvement of yield and quality in China. Rice Sci. 27, 363–367. doi: 10.1016/j.rsci.2019.12.009
Fu, D., Xiao, M., Hayward, A., Fu, Y., Liu, G., Jiang, G., et al. (2014). Utilization of crop heterosis: A review. Euphytica 197, 161–173. doi: 10.1007/s10681-014-1103-7
Fukuoka, S., Namai, H., Okuno, A. (1998). RFLP mapping of the genes controlling hybrid breakdown in rice. Theor Appl Genet 97, 446–449. doi: 10.1007/s001220050915
Fuller, D. Q. (2011). Finding plant domestication in the Indian subcontinent. Curr. Anthropol. 52, S347–S362. doi: 10.1086/658900
Gao, Z., Zeng, D., Cui, X., Zhou, Y., Yan, M., Huang, D., et al. (2003). Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China C: Life Sci. 46, 661–668. doi: 10.1360/03yc0099
Garris, A., McCouch, S., Kresovich, S. (2003). Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165, 759–769. doi: 10.1093/genetics/165.2.759
Garris, A., Tai, T., Coburn, J., Kresovich, S., McCouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi: 10.1534/genetics.104.035642
Ge, J., Wang, J., Pang, H., Li, F., Lou, D., Fan, W., et al. (2022). Genome-wide selection and introgression of Chinese rice varieties during breeding. J. Genet. Genomics 49, 492–501. doi: 10.1016/j.jgg.2022.02.025
Ghimire, K., Quiatchon, L., Vikram, P., Swamy, B., Dixit, S., Ahmed, H., et al. (2012). Identification and mapping of a QTL (qDTY1. 1) with a consistent effect on grain yield under drought. Field Crops Res. 131, 88–96. doi: 10.1016/j.fcr.2012.02.028
Glaszmann, J. (1987). Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 74, 21–30. doi: 10.1007/bf00290078
Gong, H., Han, B. (2022). Genetic introgression between different groups reveals the differential process of Asian cultivated rice. Scientifc Rep. 12, 17662. doi: 10.1038/s41598-022-22674-3
Grondin, A., Dixi, S., Torres, R., Venkateshwarlu, C., Rogers, E., Mitchell-Olds, T., et al. (2018). Physiological mechanisms contributing to the qDTY3.2 effects on improved performance of rice Moroberekan x Swarna BC2F3:4 lines under drought. Rice 11, 43. doi: 10.1186/s12284-018-0234-1
Gross, B. L., Zhao, Z. (2014). Archaeological and genetic insights into the origins of domesticated rice. Proc. Natl. Acad. Sci. U. S. A. 111, 6190–6197. doi: 10.1073/pnas.1308942110
Gutaker, R. M., Groen, S. C., Bellis, E. S., Choi, J. Y., Pires, I. S., Bocinsky, R. K., et al. (2020). Genomic history and ecology of the geographic spread of rice. Nat. Plants 6, 492–502. doi: 10.1038/s41477-020-0659-6
Hancock, J. (2012). Plant evolution and the origin of crop species. 3rd Edition (Cambridge, MA, USA: CABI)ISBN: 978-1-84593-801-7.
Hattori, Y., Nagai, K., Furukawa, S., Song, X., Kawano, R., Sakakibara, H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. doi: 10.1038/nature08258
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8, 1497. doi: 10.1038/s41467-017-01501-8
Islam, M., Khalequzzaman, M., Prince, M., Siddique, M., Rashid, E., Ahmed, M., et al. (2018). Diversity and population structure of red rice germplasm in Bangladesh. PloS One 13 , e0196096. doi: 10.1371/journal.pone.0196096
Jia, Y., Bryan, G., Farrall, L., Valent, B. (2003). Natural variation at the Pi-ta rice blast resistance locus. Phytopathology 93, 1452–1459. doi: 10.1094/PHYTO.2003.93.11.1452
Jiang, L., Wu, L., Wang, Y., Xu, Q., Xu, Z., Chen, W. (2022). Research progress on the divergence and genetic basis of agronomic traits in xian and geng rice. Crop J. 10, 924–931. doi: 10.1016/j.cj.2022.02.006
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Jing, C., Zhang, F., Wang, X., Wang, M., Zhou, L., Cai, Z., et al. (2023). Multiple domestications of Asian rice. Nat. Plants 9, 1221–1235. doi: 10.1038/s41477-023-01476-z
Kato, S., Kosaka, H., Hara, S. (1928). On the affinity of rice varieties as shown by the fertility of rice plants. Centr. Agric. Inst. Kyushu. Imp. Univ. 2, 241–276. doi: 10.15017/20773
Khush, G. S. (1995). Breaking the yield frontier of rice. Geo J. 35, 329–332. doi: 10.1007/BF00989140
Khush, G. (1997). Origin, dispersal, cultivation and variation of rice. Plant Molecul. Biol. 35, 25–34. doi: 10.1023/A:1005810616885
Kim, H., Jung, J., Singh, N., Greenberg, A., Doyle, J., Tyagi, W., et al. (2016). Population dynamics among six major groups of the Oryza rufipogon species complex, wild relative of cultivated Asian rice. Rice 9, 56. doi: 10.1186/s12284-016-0119-0
Kim, B., Kim, D., Lee, G., Seo, J., Choi, I., Choi, B., et al. (2014). Defining the genome structure of `Tongil’ rice, an important cultivar in the Korean “Green Revolution. Rice 7, 22. doi: 10.1186/s12284-014-0022-5
Kishor, D., Seo, J., Chin, J. H., Koh, H. (2020). Evaluation of whole-genome sequence, genetic diversity, and agronomic traits of basmati rice (Oryza sativa L.). Front. Genet. 11. doi: 10.3389/fgene.2020.00086
Kokaji, H., Shimizu, A. (2022). An indica rice cultivar ‘Habataki’ segment on chromosome 6 improves low-phosphorus tolerance. J. Crop Res. 67, 1–6. doi: 10.18964/jcr.67.0_1
Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., et al. (2003). LAX and SPA: major regulators of shoot branching in rice. PNAS 100, 11765–11770. doi: 10.1073/pnas.1932414100
Kubo, T., Yoshimura, A. (2002). Genetic basis of hybrid breakdown in a japonica/indica cross of rice, Oryza sativa L. Theor. Appl. Genet. 105, 906–911. doi: 10.1007/s00122-002-1059-1
Ladizinsky, G. (1998). Plant evolution under domestication (United States: Kluwer Academic Publishers). doi: 10.1007/978-94-011-4429-2
Li, X., Chen, Z., Zhang, G., Lu, H., Qin, P., Qi, M., et al. (2020). Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of Chinese rice accessions. Sci. China Life Sci. 63, 1688–1702. doi: 10.1007/s11427-019-1682-6
Li, L., Olsen, K. M. (2020). “Population genomics of weedy crop relatives: insights from weedy rice,” in Population genomics: crop plants. Ed. Rajora, O. (Springer, Cham, Switzerland: Springer Nature Switzerland AG). doi: 10.1007/13836_2020_77
Lin, Z., Qin, P., Zhang, X., Fu, C., Deng, H., Fu, X. (2020). Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. PNAS 117, 4623–4631. doi: 10.1073/pnas.1919086117
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Lou, Q., Guo, H., Li, J., Han, S., Khan, N., Gu, Y., et al. (2022). Cold-adaptive evolution at the reproductive stage in geng/japonica subspecies reveals the role of OsMAPK3 and OsLEA9. Plant J. 111 (4), 1032–1051. doi: 10.1111/tpj.15870
Lu, K., Wu, B., Wang, J., Zhu, W., Nie, H., Qian, J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 16, 1710–1722. doi: 10.1111/pbi.12907
Lv, Y., Li, J., Yang, Y., Pu, Q., Zhou, J., Deng, X., et al. (2024). Identification of a novel hybrid sterility locus S67 between temperate japonica subgroup and basmati subgroup in Oryza sativa L. Sci. Rep. 14, 28619. doi: 10.1038/s41598-024-80011-2
Mahesh, H., Shirke, M., Singh, S., Rajamani, A., Hittalmani, S., Wang, G., et al. (2016). indica rice genome assembly, annotation and mining of blast disease resistance genes. BMC Genomics 17, 242. doi: 10.1186/s12864-016-2523-7
Mao, D., Xin, Y., Tan, Y., Hu, X., Bai, J., Liu, Z., et al. (2019). Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. PNAS 116 , 3494–3501. doi: 10.1073/pnas.1819769116
Matsuo, T. (1952). Genecological studies on cultivated rice (in Japanese). Bull. Nat. Inst. Agric. Sci. Jpn. D. 3, 1–111.
McNally, K., Childs, K., Bohnert, R., Davidson, R., Zhao, K., Ulat, V., et al. (2009). Genome wide SNP variation reveals relationships among landraces and modern varieties of rice. PNAS 106, 12273–12278. doi: 10.1073/pnas.0900992106
Miura, K., Ikeda, M., Matsubara, A., Song, X., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Mizuta, Y., Harushima, Y., Kurata, N. (2010). Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. PNAS 107, 20417–20422. doi: 10.1073/pnas.1003124107
Morinaga, T. (1954). Classification of rice varieties on the basis of affinity. Stud. Rice Breed. 4, 1–14.
Morishima, H. (1969). Phenetic similarity and phylogenetic relationships among strains of Oryza perennis, estimated by methods of numerical taxonomy. Evolution 23, 429–443. doi: 10.2307/2406698
Morishima, H., Oka, H.-I., Chang, W.-T. (1961). Directions of differentiation in populations of wild rice, Oryza perennis and O. sativa f. spontanea. Evol. 15, 326–339. doi: 10.2307/2406231
Nakagahra, M. (1972). Genetic mechanism of the distorted segregation of marker genes belonging to the eleventh linkage group in cultivated rice. Jpn J. Breed 22, 232–238. doi: 10.1270/jsbbs1951.22.232
Oikawa, T., Maeda, H., Oguchi, T., Yamaguchi, T., Tanabe, N., Ebana, K., et al. (2015). The birth of a black rice gene and is local spread by introgression. Plant Cell 27, 2401–2414. doi: 10.1105/tpc.15.00310
Oka, H. I. (1957). Phylogenetic differentiation of cultivated rice. XV. Complementary lethal genes in rice. Jpn J. Genet. 32, 83–87. doi: 10.1266/jjg.32.83
Oka, H. I. (1958). Intervarietal variation and classification of cultivated rice. Indian J. Genet. Plant Breed. 18, 79–89. doi: 10.1093/aob/mcl210
Oka, H. I., Doida, Y. (1962). Phylogenetic differentiation of cultivated rice. XX. Analysis of the genetic basis of hybrid breakdown in rice. Jpn J. Genet. 37, 24–35. Available online at: https://www.jstage.jst.go.jp/article/ggs1921/37/1/37_1_24/_pdf/-char/en.
Ouyang, Y., Li, X., Zhang, Q. (2022). Understanding the genetic and molecular constitutions of heterosis for developing hybrid rice. J. Genet. Genomics 49, 385–393. doi: 10.1016/j.jgg.2022.02.022
Parsons, B., Newbury, H., Jackson, M., Ford-Lloyd, B. (1999). The genetic structure and conservation of aus, aman and boro rices from Bangladesh. Genet. Resour. Crop Evol. 46, 587–598. doi: 10.1023/A:1008749532171
Patra, B. C., Anilkumar, C., Chakraborti, M. (2020). Rice breeding in India: a journey from phenotype based pure line selection to genomics assisted breeding. Agric. Res. J. 57 , 816–825. doi: 10.5958/2395-146X.2020.00120.9
Qin, C., Guo, Y., Wu, J., Wang, L., Traw, M., Zhang, Y. (2021). Comparative population genomic analysis provides insights into breeding of modern indica rice in China. Gene 5 768, 145303. doi: 10.1016/j.gene.2020.145303
Rahangdale, S., Singh, Y., Kujur, M., Koutu, G. K. (2019). “Exploration of new plant type (NPT) rice lines for crop improvement,” in Advances in agriculture sciences, vol. 19. (New Delhi, India: AkiNik Publications New Delhi), 53–67.
Rieseberg, L., Blackman, B. (2010). Speciation genes in plants. Ann. Bot. 106, 439–455. doi: 10.1093/aob/mcq126
Sato, Y., Morishima, H. (1987). Studies on the distribution of complementary genes causing F1 weakness in common rice and its wild relatives. II. Distribution of two complementary genes, Hwc-1 and Hwc-2 gene in native cultivars and its wild relatives of tropical Asia. Euphytica 36, 425–431. doi: 10.1007/BF00041485
Schatz, M., Maron, L., Stein, J., Hernandez Wences, A., Gurtowski, J., Biggers, E., et al. (2014). New whole genome de novo assemblies of three divergent strains of rice (O. sativa) documents novel gene space of aus and indica. Genome Biol. 15, 506. Available at: http://genomebiology.com/2014/15/12/506.
Sha, H., Liu, H., Zhao, G., Han, Z., Chang, H., Wang, J., et al. (2022). Elite sd1 alleles in japonica rice and their breeding applications in Northeast China. Crop J. 10, 224–233. doi: 10.1016/j.cj.2021.05.005
Siddiq, E., Vemireddy, L., Nagaraju, J. (2012). Basmati rices: genetics, breeding and trade. Agric. Res. 1, 25–36. doi: 10.1007/s40003-011-0011-5
Song, J., Arif, M., Zi, Y., Sze, S., Zhang, M., Zhang, H. (2021). Molecular and genetic dissection of the USDA rice mini-core collection using high-density SNP markers. Plant Sci. 308, 110910. doi: 10.1016/j.plantsci.2021.110910
Song, Z., Lu, B., Zhu, Y., Chen, J. (2002). Pollen competition between cultivated and wild rice species (Oryza sativa and O. rufipogon). New Phytol. 153, 289–296. doi: 10.1046/j.0028-646X.2001.00319.x
Sun, J., Liu, D., Wang, J., Ma, D., Tang, L., Gao, H., et al. (2012). ). The contribution of intersubspecific hybridization to the breeding of super-high-yielding japonica rice in Northeast China. Theor. Appl. Genet. 125, 1149–1157. doi: 10.1007/s00122-012-1901-z
Sweeney, M., Thomson, M., Cho, Y., Park, Y., Williamson, S., Bustamante, C., et al. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PloS Genet. 3, 133. doi: 10.1371/journal.pgen.0030133
Tang, R., Cui, D., Zhou, J., Li, W., Ma, X., Han, B., et al. (2021). Comparative analysis of genetic diversity of rice (Oryza sativa L.) varieties cultivated in different periods in China. Genet. Resour Crop Evol. 68, 1439–1451. doi: 10.1007/s10722-020-01073-5
Travis, A., Norton, G., Datta, S., Sarma, R., Dasgupta, T., Savio, F., et al. (2015). Assessing the genetic diversity of rice originating from Bangladesh, Assam and West Bengal. Rice 8, 35. doi: 10.1186/s12284-015-0068-z
Vigueira, P. A., Olsen, K. M., Wagner, C. R., Chittick, Z., B. and Vigueira, C. C. (2020). Weedy rice from South Korea arose from two distinct de-domestication events. Front. Agron. 2. doi: 10.3389/fagro.2020.602612
Wang, C. (2022c). Development and enlightenment of japonica rice breeding with good eating quality in Jiangsu province (in Chinese with English abstract). China Rice 28, 92–106. doi: 10.3969/j.issn.1006-8082.2022.05.014
Wang, Y., Li, F., Zhang, F., Wu, L., Xu, N., Sun, Q., et al. (2023a). Time-ordering japonica/geng genomes analysis indicates the importance of large structural variants in rice breeding. Plant Biotechnol. J. 21, 202–218. doi: 10.1111/pbi.13938
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, P., Qi, F., Yao, X., Xu, X., Li, W., Meng, J., et al. (2022a). Fixation of hybrid sterility genes and favorable alleles of key yield-related genes with dominance contribute to the high yield of Yongyou series of intersubspecific hybrid rice. J. Genet. Genomics 49, 448–457. doi: 10.1016/j.jgg.2022.02.027
Wang, H., Vieira, F. G., Crawford, J. E., Chu, C., Nielsen, R. (2017). Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 27, 1029–1038. doi: 10.1101/gr.204800.116
Wang, C., Wang, J., Lu, J., Xiong, Y., Zhao, Z., Yu, X., et al. (2023b). A natural gene drive system confers reproductive isolation in rice. Cell 186 , 3577–3592.e18. doi: 10.1016/j.cell.2023.06.023
Wang, X., Wang, W., Tai, S., Li, M., Gao, Q., Hu, Z., et al. (2022b). Selective and comparative genome architecture of Asian cultivated rice (Oryza sativa L.) attributed to domestication and modern breeding. J. Advanced Res. 42, 1–16. doi: 10.1016/j.jare.2022.08.004
Wang, J., Zhou, J., Li, J., Yang, Y., Pu, Q., Lv, Y., et al. (2023c). Studies on F1 fertility of five different ecological types of Asian cultivated rice with indica and japonica (In Chinese with English abstract). J. Yunnan University: Natural Sci. Edition 45, 1–9. doi: 10.7540/j.ynu.20220015
Wang, M., Zhu, Z., Tan, L., Liu, F., Fu, Y., Sun, C., et al. (2013). Complexity of indica-japonica varietal differentiation in Bangladesh rice landraces revealed by microsatellite markers. Breed. Sci. 63, 227–232. doi: 10.1270/jsbbs.63.227
Wei, X., Huang, X. (2019). Origin, taxonomy, and phylogenetics of rice. In Rice. (Fourth Edition) Ed. Jinsong, Bao. (AACC International: Elsevier Inc). doi: 10.1016/B978-0-12-811508-4.00001-0
Wu, S., Han, B., Jiao, Y. (2020). Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13, 59–71. doi: 10.1016/j.molp.2019.10.012
Xie, W., Wang, G., Yuan, M., Yao, W., Lyu, K., Zhao, H., et al. (2015). Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. PNAS 112, E5411–E5419. doi: 10.1073/pnas.1515919112
Xie, F., Zhang, J. (2018). Shanyou 63: An elite mega rice hybrid in China. Rice 11, 17. doi: 10.1186/s12284-018-0210-9
Xu, R., Sun, C. (2021). What happened during domestication of wild to cultivated rice. Crop J. 9, 564–576. doi: 10.1016/j.cj.2021.02.005
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., et al. (2006). Sub1A is an ethyleneresponse-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. doi: 10.1038/nature04920
Xu, Q., Yuan, X., Wang, S., Feng, Y., Yu, H., Wang, Y., et al. (2016). The genetic diversity and structure of indica rice in China as detected by single nucleotide polymorphism analysis. BMC Genet. 17, 53. doi: 10.1186/s12863-016-0361-x
Yamashita, M., Ootsuka, C., Kubota, H., Adachi, S., Yamaguchi, T., Murata, K., et al. (2022). Alleles of high-yielding indica rice that improve root hydraulic conductance also increase flag leaf photosynthesis, biomass, and grain production of japonica rice in the paddy field. Field Crops Res. 289, 108725. doi: 10.1016/j.fcr.2022.108725
Yang, S., Shen, X., Gu, W., Cao, D. (1962). Studies on cross-breeding of japonica with indica rice (in Chinese with English abstract). Acta agronomica Sin. 1, 97–102.
Ye, C., Tenorio, F., Redoña, E., Morales–Cortezano, P., Cabrega, G., Jagadish, K., et al. (2015). Fine-mapping and validating qHTSF4.1 to increase spikelet fertility under heat stress at flowering in rice. Theor. Appl. Genet. 128, 1507–1517. doi: 10.1007/s00122-015-2526-9
Ye, J., Zhang, M., Yuan, X., Hu, D., Zhang, Y., Xu, S., et al. (2022). Genomic insight into genetic changes and shaping of major inbred rice cultivars in China. New Phytol. 236, 2311–2326. doi: 10.1111/nph.18500
Yonemaru, J., Mizobuchi, R., Kato, H., Yamamoto, T., Yamamoto, E., Matsubara, K., et al. (2014). Genomic regions involved in yield potential detected by genome-wide association analysis in Japanese high-yielding rice cultivars. BMC Genomics 15, 346. doi: 10.1186/1471-2164-15-346
Yonemaru, J., Yamamoto, T., Ebana, K., Yamamoto, E., Nagasaki, H., Shibaya, T., et al. (2012). Genome-wide haplotype changes produced by artificial selection during modern rice breeding in Japan. PloS One 7, e32982. doi: 10.1371/journal.pone.0032982
Yu, B., Lin, Z., Li, H., Li, X., Li, J., Wang, Y., et al. (2007). TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Yuan, L. (2015). Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 14, 197–205. doi: 10.1016/S2095-3119(14)60922-9
Zhang, G. (2020). Prospects of utilization of inter-subspecific heterosis between indica and japonica rice. J. Integr. Agric. 19, 1–10. doi: 10.1016/S2095-3119(19)62843-1
Zhang, F., Wang, C., Li, M., Cui, Y., Shi, Y., Wu, Z., et al. (2021). The landscape of gene-CDS-haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol. Plant 14, 787–804. doi: 10.1016/j.molp.2021.02.003
Zhang, Y., Wang, J., Pu, Q., Yang, Y., Lv, Y., Zhou, J., et al. (2022). Understanding the nature of hybrid sterility and divergence of Asian cultivated rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.908342
Zhang, C., Zhu, J., Chen, S., Fan, X., Li, Q., Lu, Y., et al. (2019). Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhao, M., Geng, X., Bi, W., Xu, Q., Sun, J., Huang, Y., et al. (2017). Recombination between DEP1 and NRT1.1B under japonica and indica genetic backgrounds to improve grain yield in rice. Euphytica 213, 265. doi: 10.1007/s10681-017-2038-6
Zhao, K., Wright, M., Kimball, J., Eizenga, G., McClung, A., Kovach, M., et al. (2010). Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PloS One 5, e10780. doi: 10.1371/journal.pone.0010780
Zhao, L., Zhou, S., Wang, C., Li, H., Huang, D., Wang, Z., et al. (2022). Breeding effects and genetic compositions of a backbone parent (Fengbazhan) of modern indica Rice in China. Rice Sci. 29, 397–401. doi: 10.1016/j.rsci.2022.07.001
Keywords: rice, genetic divergence, introgression, genetic improvement, evolution
Citation: Zhou J, Li J, Zhang Y, Yang Y, Lv Y, Pu Q, Deng X and Tao D (2025) Introgression among subgroups is an important driving force for genetic improvement and evolution of the Asian cultivated rice Oryza sativa L.. Front. Plant Sci. 16:1535880. doi: 10.3389/fpls.2025.1535880
Received: 28 November 2024; Accepted: 29 January 2025;
Published: 20 February 2025.
Edited by:
Kazuki Matsubara, Institute of Crop Science (NARO), JapanReviewed by:
Wricha Tyagi, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaCopyright © 2025 Zhou, Li, Zhang, Yang, Lv, Pu, Deng and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayun Tao, dGFvZHkxMkBhbGl5dW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.