
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 February 2025
Sec. Plant Abiotic Stress
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1535733
This article is part of the Research Topic Salinity and Drought Stress in Plants: Understanding Physiological, Biochemical and Molecular Responses Volume II View all 26 articles
 Xueyun Xuan1†
Xueyun Xuan1† Shiying Su1†
Shiying Su1† Jialu Chen1†
Jialu Chen1† Jiaqi Tan1
Jiaqi Tan1 Zhen Yu1
Zhen Yu1 Yang Jiao1
Yang Jiao1 Sijia Cai1
Sijia Cai1 Zhijun Zhang1*
Zhijun Zhang1* Muthusamy Ramakrishnan2*
Muthusamy Ramakrishnan2*Dirigent (DIR) proteins are key regulators of lignin and lignan biosynthesis and play critical roles in plant hormone responses, abiotic stress tolerance, and growth and development. This study identified and characterized 47 PeDIR genes in Moso bamboo, classifying them into three groups. Phylogenetic and comparative analyses revealed strong evolutionary conservation, with the Moso bamboo PeDIR genes being most closely related to those in rice and maize. DIR proteins within each subfamily exhibited high conservation in motif composition, domain structure, and 3D configuration. Subcellular localization and protein interaction studies further elucidated PeDIR gene functions. Specifically, PeDIR02 primarily localized to the cell membrane and was shown to be unable to form homodimers in yeast two-hybrid (Y2H) assays. Transcriptome and expression analyses revealed the involvement of PeDIR genes in rapid shoot growth, indicating roles in lignin biosynthesis and cell wall modification. Transcriptome and qRT-PCR data also demonstrated the responsiveness of these genes to hormones and abiotic stresses, such as drought and salinity. This study constructed the first comprehensive regulatory network between transcription factors (TFs) and PeDIR genes, identifying ERF, DOF, and MYB TFs as key synergistic regulators of PeDIR gene expression.
Moso bamboo (Phyllostachys edulis), the most extensively cultivated species in the Bambusoideae subfamily, holds substantial ecological and economic significance in China and Asia (Peng et al., 2013; Li et al., 2015; Yu et al., 2019; Huang et al., 2022). However, its rapid growth and development are often constrained by environmental factors such as salinity, drought, and low temperatures. These factors not only reduce timber production and shoot yield but also threaten the overall stability of Moso bamboo forests (Liu et al., 2019; Ramakrishnan et al., 2020). Understanding the genetic mechanisms underlying stress resistance is therefore crucial for developing more resilient bamboo varieties.
Dirigent (DIR) proteins, first discovered in weeping forsythia (Forsythia suspense), play key roles in lignin and lignan biosynthesis (Davin et al., 1997; Gang et al., 1999; Burlat et al., 2001). Through their highly conserved dirigent domain, DIR proteins precisely capture and orient E-coumaryl alcohol radical intermediates, enabling stereoselective coupling of lignin and lignan precursor compounds in the presence of auxiliary oxidative enzymes (Gang et al., 1999; Burlat et al., 2001). During lignin and lignan biosynthesis, they mediate site-specific radical coupling reactions, guiding the formation of specific stereoisomers, such as (+)-pinoresinol and (−)-pinoresinol, through a spatial orientation template (Kim et al., 2002; Dalisay et al., 2015). Lignans, a class of secondary metabolites widely distributed in plants, exhibit notable antifungal activity by inhibiting pathogen growth and spread, thereby enhancing plant resistance to external stresses (Harmatha and Dinan, 2003; Zhang et al., 2022a). Lignin, a major component of plant cell walls, provides structural support, participates in plant defense mechanisms, regulates growth and development, and exhibits antifungal activity, further enhancing plant resistance to external stresses. The DIR gene family has been characterized in various plants, including Arabidopsis thaliana (Paniagua et al., 2017), Oryza sativa (rice) (Liao et al., 2017), Solanum tuberosum (Jia et al., 2023), Picea spp (Ralph et al., 2006), and Gossypium spp (Zhengwen et al., 2021), and is categorized into six subfamilies: DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-g, and DIR-f (Luo et al., 2022). Among these, the DIR-a subfamily comprises proteins responsible for stereoselective lignin and lignan formation, referred to as dirigent genes. The functions of other subfamilies remain unclear and are termed DIR-like genes (Ralph et al., 2006, 2007).
DIR proteins are found in many vascular plants and play critical roles in growth, development, and resistance to environmental stresses and diseases (Jin-long et al., 2012; Thamil Arasan et al., 2013). Their defining feature is the highly conserved dirigent domain, which facilitates substrate binding and is essential for specific biochemical reactions (Pickel et al., 2012). Most DIR proteins have a dirigent domain with a three-dimensional structure composed of β-sheets (Li et al., 2024a). Some DIR proteins possess two tandem dirigent domains, while others have an additional jacalin domain (Kim et al., 2015; Gong et al., 2023; Jiang et al., 2024). At the genomic level, DIR genes exhibit structural diversity. Most DIR genes lack introns, but a subset contains one or two introns (Dong et al., 2021; Zhang et al., 2022a).
Research indicates that DIR proteins have diverse roles in plants, including tolerance to abiotic stresses and responses to environmental challenges (Chen et al., 2024). Notably, DIR genes often show significant responses to stress-related hormone induction and abiotic stress conditions. For example, in sugarcane, ScDIR11 and SHDIR16 expression are significantly upregulated by salicylic acid (SA) and jasmonic acid (JA) treatments and exhibit transcriptional increases under salinity, drought, and hydrogen peroxide (H2O2) stress (Jin-long et al., 2012). In soybeans, GmDIR22 expression is significantly upregulated in response to gibberellins (GA), SA, methyl jasmonate (MeJA), and abscisic acid (ABA) (Li et al., 2017).
Similarly, in peppers, CaDIR2, CaDIR3, CaDIR6, CaDIR7, and CaDIR11 are upregulated in response to MeJA, SA, NaCl, and mannitol treatments. Notably, the absence of CaDIR7 reduces root vitality, defense capabilities, and resistance to salinity and diseases (Khan et al., 2018). In eggplants, several DIR genes, such as SmDIR3, SmDIR6, SmDIR12, and SmDIR22, are downregulated under high temperatures, with varying expression levels at different stages. SmDIR5 and SmDIR22 are repressed under low temperatures, and SmDIR22 expression decreases under salt stress (Zhang et al., 2022a). Furthermore, DIR genes in pear (Cheng et al., 2018), alfalfa (Song and Peng, 2019), and tobacco (Li et al., 2024b) strongly respond to diverse hormones and abiotic stresses. DIR genes also contribute to plant growth and development. For example, the ESB1 protein with a dirigent domain aids in Casparian strip formation and maintains root barrier integrity (Hosmani et al., 2013).
Understanding the roles of PeDIR genes is crucial for revealing Moso bamboo’s defense mechanisms against abiotic stresses and hormone signaling (Saddique et al., 2024). This study systematically analyzed PeDIR gene structures, phylogenetic relationships, promoter elements, and collinearity across the genome using bioinformatics tools. A regulatory network linking PeDIR genes with upstream transcription factors (TFs) was established. This study also explored PeDIR gene expression under hormone treatments, abiotic stresses, and rapid growth, providing insights into their roles in hormone and stress responses in Moso bamboo. These findings lay a foundation for deeper investigation into the functional roles of PeDIR genes in stress responses.
Moso bamboo genome data were obtained from the National Center for Gene Research (http://server.ncgr.ac.cn/bamboo/index.php). The DIR protein domain HMM profile (PF03018) was downloaded from the Pfam database (http://pfam.xfam.org/) and used to search the bamboo genome with an e-value threshold of ≤1e-10 (Finn et al., 2011). Protein domains were confirmed using the NCBI-CDD database, leading to the identification of PeDIR family members (Letunic et al., 2011; Finn et al., 2015). The molecular weight, theoretical isoelectric point, and hydrophobicity of PeDIR proteins were analyzed using the Expasy ProtParam tool (https://web.expasy.org/protparam/) (Wilkins et al., 1999).
DIR protein sequences from Moso bamboo, rice, Zea mays (maize), and Brachypodium distachyon were identified using HMMER3 to search their respective local protein databases. Phylogenetic trees were constructed for both intra- and interspecies analyses using MEGA 11.0. The maximum likelihood method was employed, with 1,000 bootstrap trials to assess tree reliability (Tamura et al., 2021). TBtools was used to align Moso bamboo protein sequences with those from rice, maize, and B. distachyon. Chromosomal location data from the four species were combined, and MCScanX was used to identify both intraspecies and interspecies syntenic relationships within the PeDIR gene family. The results were visualized using Circos (Wang et al., 2012).
The conserved sequence patterns of PeDIR proteins were analyzed using the MEME suite (http://meme-suite.org/) (Bailey et al., 2015). The intron-exon structures of PeDIR genes were extracted from the GFF annotation file of the Moso bamboo genome. Using TBtools, the evolutionary relationships, motif patterns, and gene structures of PeDIR genes were integrated and visualized.
Cis-acting elements within the 1.5 kb upstream promoter regions of PeDIR genes were identified using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The types and distributions of regulatory elements were visualized with TBtools. Potential transcription factor (TF) binding sites (TFBS) in the 1.5 kb upstream regions of PeDIR genes were predicted via PlantPAN (http://plantpan.itps.ncku.edu.tw/plantpan4/index.html) (Chow et al., 2024). TFBS with correlation coefficients above 0.95 were selected for further study. A gene regulatory network based on the identified TFBS was constructed using Cytoscape, and a dynamic network heatmap depicting interactions between PeDIR genes and upstream TFs was created using the Omicshare platform (http://www.omicshare.com).
Conserved domains in PeDIR proteins were identified using the CDD-search website (https://www.ncbi.nlm.nih.gov/cdd) and visualized using IBS 2.0 (Xie et al., 2022). Homology modeling of PeDIR proteins was conducted via SWISS-MODEL (https://swissmodel.expasy.org/) (Waterhouse et al., 2018). The 3D structures were refined and optimized using Discovery Studio for improved visualization.
Transcriptome data for Moso bamboo under various treatments were obtained from NCBI to examine PeDIR gene expression. The treatments included: (i) seedling roots treated with gibberellin (GA) and naphthalene acetic acid (NAA) (PRJNA413166), (ii) roots treated with abscisic acid (ABA) and salicylic acid (SA) (PRJNA715101), and (iii) roots under drought and high-salinity stress (PRJNA413166). Transcript per million (TPM) values were log2-transformed, and heatmaps of PeDIR gene expression levels were created using TBtools (Zhijun et al., 2022; Guo et al., 2024). Additionally, temporal expression patterns of PeDIR genes across different shoot heights of Moso bamboo were analyzed using several publicly available transcriptome datasets (PRJNA414226). Short Time-series Expression Miner (STEM) was employed to identify expression trends during the rapid growth stages of bamboo shoots. A trend significance threshold of p-value < 0.05 was applied (Ernst and Bar-Joseph, 2006).
Moso bamboo seeds were sourced from Guilin, Guangxi, China. The seeds were grown in a controlled chamber maintained at 25°C and 70% relative humidity for one month. For hormone treatments, seedling leaves were sprayed with 100 μM ABA or SA solutions and sampled at 0, 3, 6, 12, 24, and 48 hours. Drought stress was induced by exposing the roots to 30% PEG for 3 or 6 hours, while salinity stress was applied by treating the roots with 200 mM NaCl for the same durations. Root samples were collected after each treatment. Untreated samples at 0 hours served as controls. Three biological replicates were collected randomly (Huang et al., 2021).
Total RNA was extracted using the FastPure Plant Total RNA Extraction Kit (Vazyme, China). cDNA synthesis was performed using the HiScript® III 1st Strand cDNA Synthesis Kit. Primers were designed using Beacon Designer 7.0. qRT-PCR was performed on a CFX-96 Real-Time System with three technical replicates per sample. Threshold cycle (CT) values were calculated using the 2^-ΔΔCT method and are presented as mean ± standard deviation (SD). Statistical significance was determined by one-way ANOVA, and data visualization was performed with GraphPad Prism 7 (Livak and Schmittgen, 2001).
Subcellular localization predictions for PeDIR proteins were performed using WoLF PSORT (https://wolfpsort.hgc.jp/) (Horton et al., 2007). To confirm this prediction, the full-length CDS of PeDIR02 was amplified using seamless PCR. The fragments were inserted into the pCAMBIA1300-35S-GFP vector containing the GFP (green fluorescent protein) reporter gene under the 35S promoter. Recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into the abaxial leaves of 5-week-old Nicotiana benthamiana plants. The GFP signals were analyzed with a confocal microscope to validate the predicted subcellular localization (Zhang et al., 2022b).
The CDS of PeDIR02 was amplified via PCR and cloned into the pGBKT7 and pGADT7 vectors. Yeast two-hybrid (Y2H) experiments were performed using the Matchmaker GAL4 system. Recombinant plasmids were introduced into the AH109 yeast strain and confirmed on SD/-Leu/-Trp medium. Protein interactions were assessed by growing yeast on SD/-Ade/-His/-Trp/-Leu/X-α-Gal medium at 28°C for 3 days. Positive interactions were indicated by colony growth and blue coloration from X-α-Gal breakdown (Ma et al., 2021). The primers used in this study are listed in Supplementary Tables S1 and S2.
A total of 47 PeDIR members were identified from the Moso bamboo genome database and were sequentially named PeDIR01 to PeDIR47 based on their scaffold arrangements (Table 1). The amino acid lengths of these proteins range from 88 for PeDIR47 to 360 for PeDIR14. Their molecular weights vary from 9,934.39 Da for PeDIR47 to 37,524.82 Da for PeDIR14. The predicted isoelectric points range from 4.58 for PeDIR05 to 10.65 for PeDIR16. Hydrophilicity analysis revealed that 17 PeDIR proteins are hydrophilic, while the remaining members are hydrophobic. Subcellular localization predictions indicate that PeDIR proteins are distributed across various subcellular structures, with the majority localized to the cell membrane (31), followed by the chloroplast (7) and the cell wall (5).
To explore the evolutionary relationships within the DIR gene family among Poaceae species, rice, maize, and B. distachyon were selected as representative species. An interspecies phylogenetic tree was constructed using clustering data from 53 OsDIR, 53 ZmDIR, 48 BdDIR (Supplementary Table S3), and 47 PeDIR proteins (Table 1). The analysis grouped the DIR genes into three distinct classes. The first class contained the most PeDIR proteins (22), followed by the second class with 10, and the third class with only 4 (Figure 1). The distribution of PeDIR proteins across subfamilies showed high similarity to DIR members from other Poaceae species, indicating strong conservation of DIR genes during Poaceae evolution.
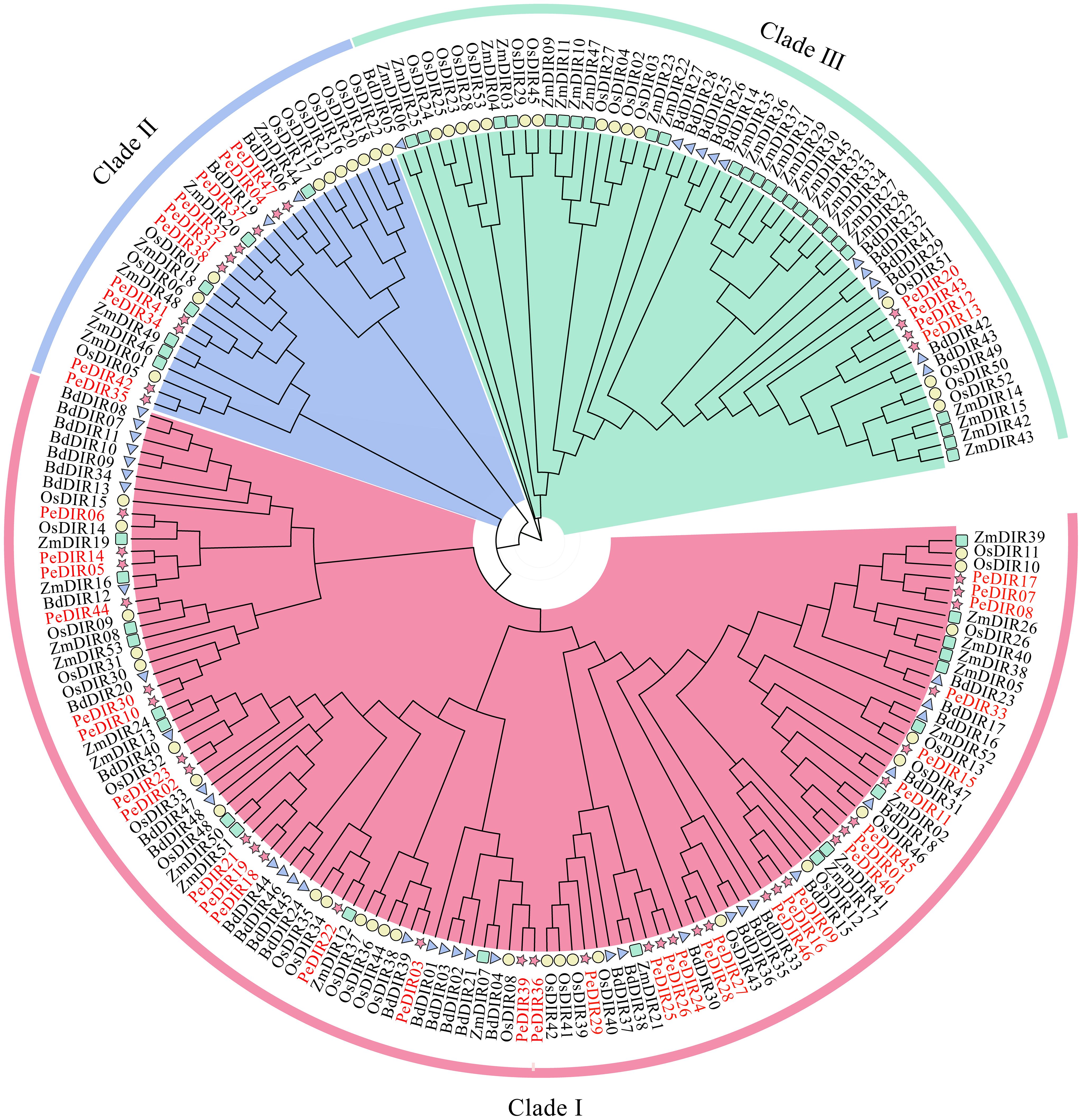
Figure 1. Phylogenetic tree of DIR proteins from Oryza sativa (Os, rice), Zea mays (Zm, maize), Brachypodium distachyon (Bd), and (Phyllostachys edulis) (Pe, Moso bamboo).
To further investigate the evolutionary dynamics of PeDIR genes, intra- and interspecies synteny analyses were performed. According to the Moso bamboo genome annotation, the 47 PeDIR genes are distributed across 16 scaffolds, with scaffold 13 containing the highest number of PeDIR genes (9). Intraspecies synteny analysis revealed 10 pairs of duplicated PeDIR genes (Figure 2A). Orthologous gene analysis with other Poaceae species identified 22 gene pairs shared between Moso bamboo and rice, 24 pairs with maize, and 12 pairs with B. distachyon (Figures 2B–D). These findings suggest that Moso bamboo shares closer evolutionary and phylogenetic relationships with maize and rice, while its relationship with B. distachyon is comparatively more distant.
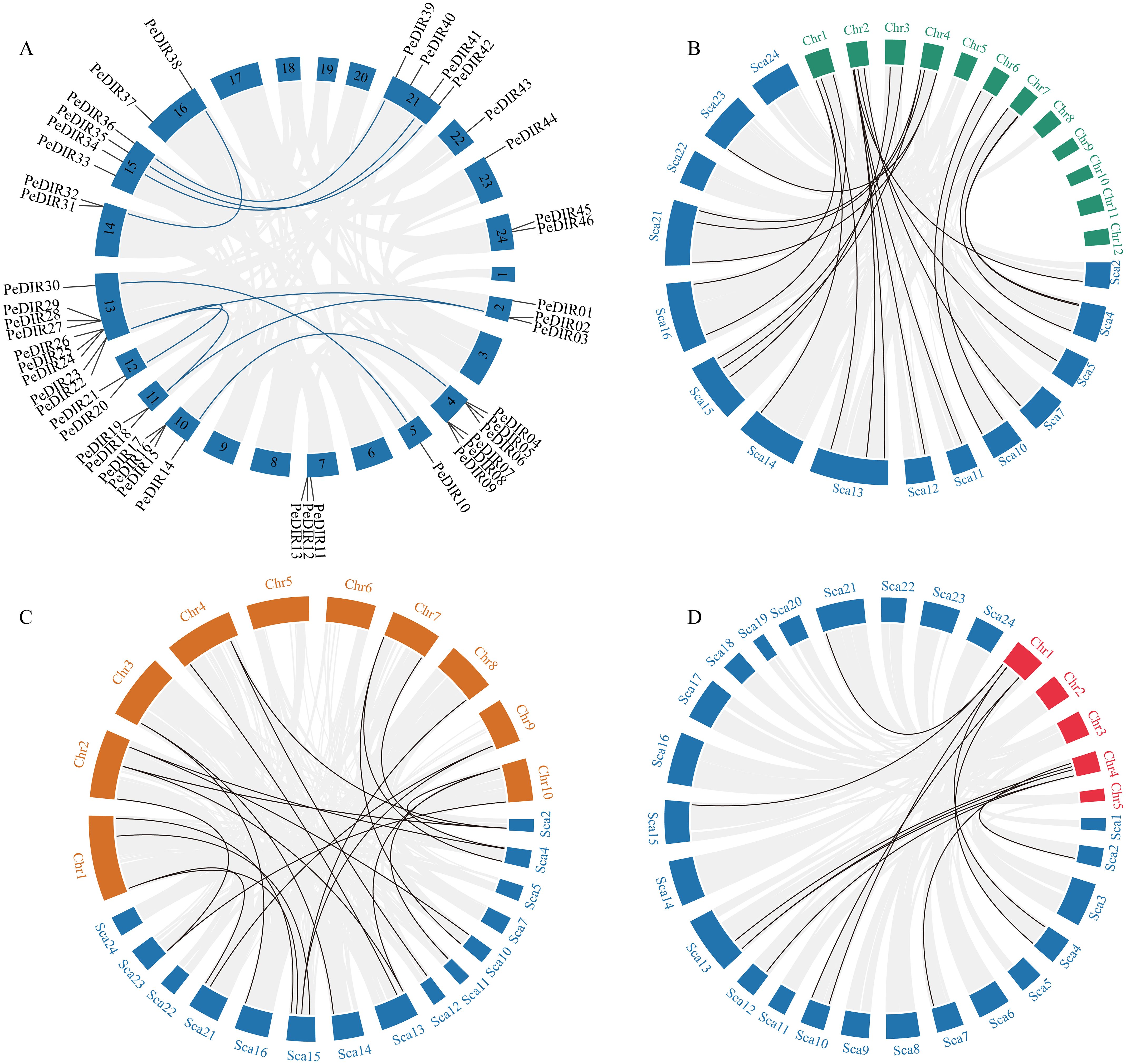
Figure 2. Synteny analysis of PeDIR genes. (A) Chromosomal distribution and interchromosomal relationships among PeDIR genes. (B–D) Interspecies collinearity analysis of DIR genes (blue for Moso bamboo, green for rice, orange for maize, and red for Brachypodium distachyon).
Intraspecies clustering analysis of PeDIR proteins revealed that Moso bamboo’s DIR genes can be grouped into three subfamilies (I, II, III), each exhibiting significant differences in gene structure. Most PeDIR genes contain 2–4 exons, except for PeDIR31, PeDIR34, PeDIR38, and PeDIR41 in subfamily III, which are intronless. The remaining PeDIR genes have 1–3 introns. Additionally, most PeDIR genes, particularly those in subfamily I, lack 5’ and 3’ untranslated regions (UTRs), whereas members of subfamilies II and III generally retain complete UTR structures (Figure 3A). Motif analysis revealed that subfamily I members commonly contain motifs 1–5, while subfamily II members are characterized by motifs 1, 4, and 5 only (Figures 3B, C). Notably, motif 6 is uniquely present in subfamily III. These findings underscore significant differences in motif distribution and number among the subfamilies. Such differences are consistent with their gene structural characteristics, suggesting that PeDIR genes have undergone structural and functional diversification during evolution.
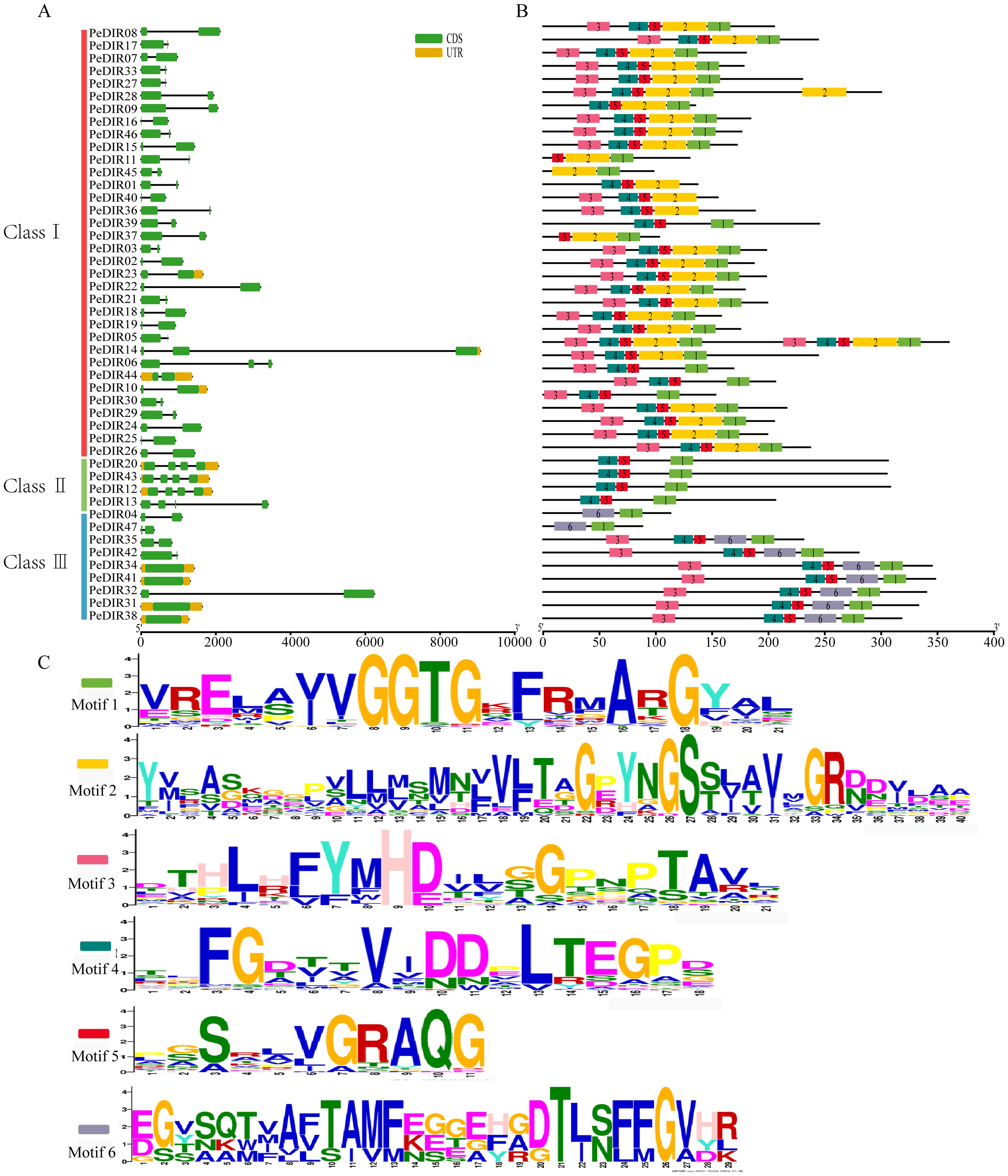
Figure 3. Gene structure and conserved motifs of PeDIR genes. (A) Gene structure of PeDIR genes, showing introns (black lines), exons (yellow rectangles), and untranslated regions (UTRs, green rectangles). (B) Conserved motifs of PeDIR genes, with motifs 1–6 represented by colored boxes. (C) Sequence logos of motifs 1–6.
The analysis of cis-acting elements in the 1500 bp promoter regions of PeDIR genes identified three primary categories: elements responsive to hormones, stress, and growth and development (Figure 4A). Hormone-responsive elements accounted for 41.2% and predominantly included ABRE (ABA response), TGACG and CGTCA (MeJA response), TCA-element (SA response), and GARE (GA response). Among these, ABRE, TGACG, and CGTCA were the most abundant, representing 37%, 25%, and 25%, respectively. Additionally, PeDIR genes contained a considerable number of stress-responsive elements, with STRE being the most prevalent, accounting for 52.2% of all stress-responsive elements. Growth and development elements, such as G-box and CAT-box, were also identified (Figures 4B, C). These findings suggest that PeDIR genes may play a significant role in the rapid growth of Moso bamboo, as well as in hormone responses and abiotic stress regulation.
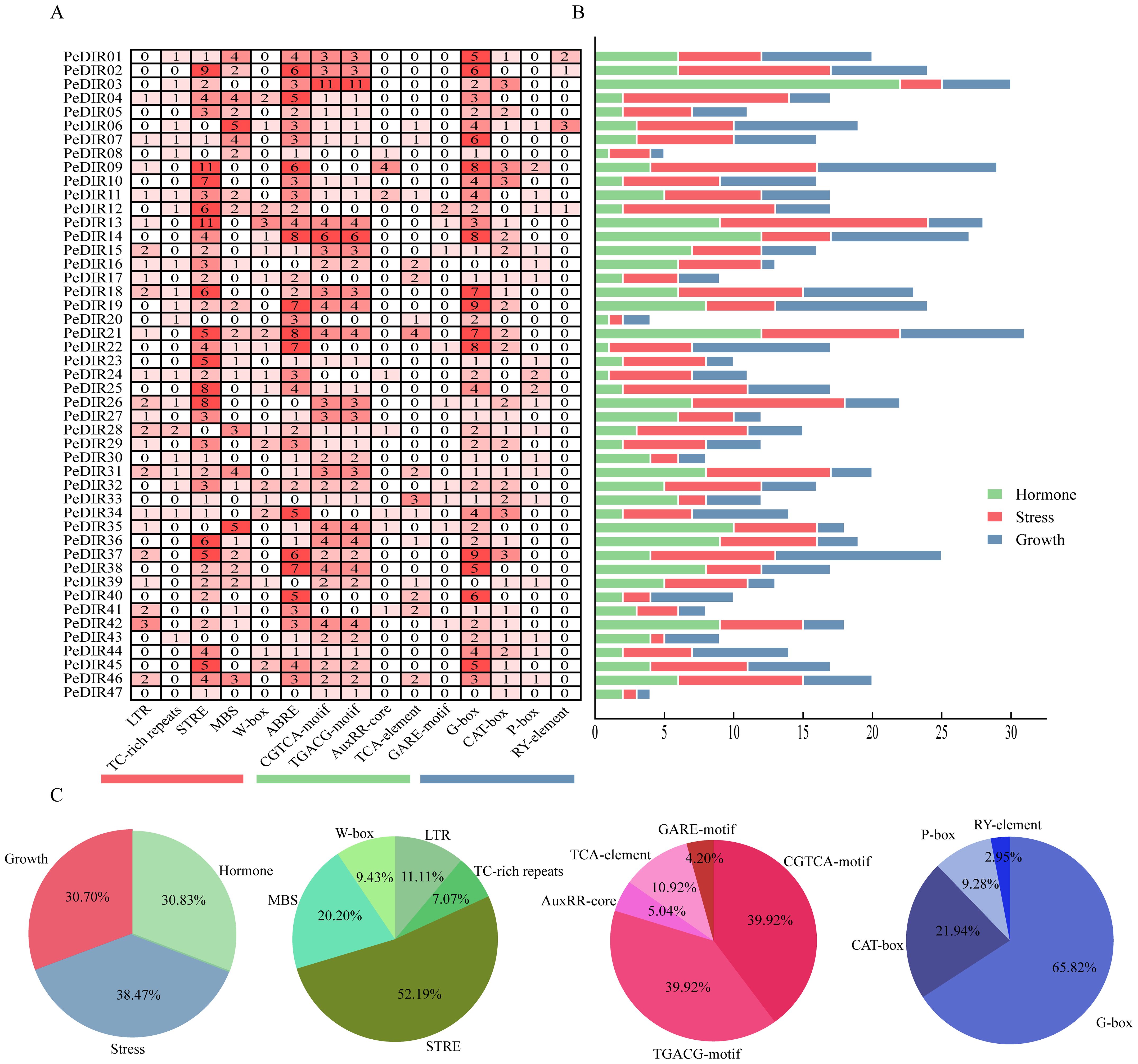
Figure 4. Cis-acting elements in the promoter regions of PeDIR genes. (A) Number of cis-acting elements identified in each PeDIR gene. (B) Distribution of the three primary types of cis-acting elements in PeDIR genes. (C) Statistical analysis of the different types of cis-acting elements.
To further explore the expression profiles of PeDIR genes under hormonal influence, transcriptome data from four different treatments (ABA, GA3, NAA, and SA) were analyzed. The results revealed distinct expression patterns among PeDIR genes in response to various hormone treatments. For instance, under ABA treatment, PeDIR07, PeDIR08, and PeDIR28 displayed upregulation, whereas PeDIR09, PeDIR12, PeDIR20, PeDIR27, and PeDIR37 were downregulated (Figure 5A). In contrast, under GA3 and SA treatments, PeDIR08, PeDIR09, PeDIR12, PeDIR20, and PeDIR37 were significantly upregulated (Figures 5B, C). Additionally, during NAA treatment, PeDIR02, PeDIR09, and PeDIR12 showed increased expression levels (Figure 5D).
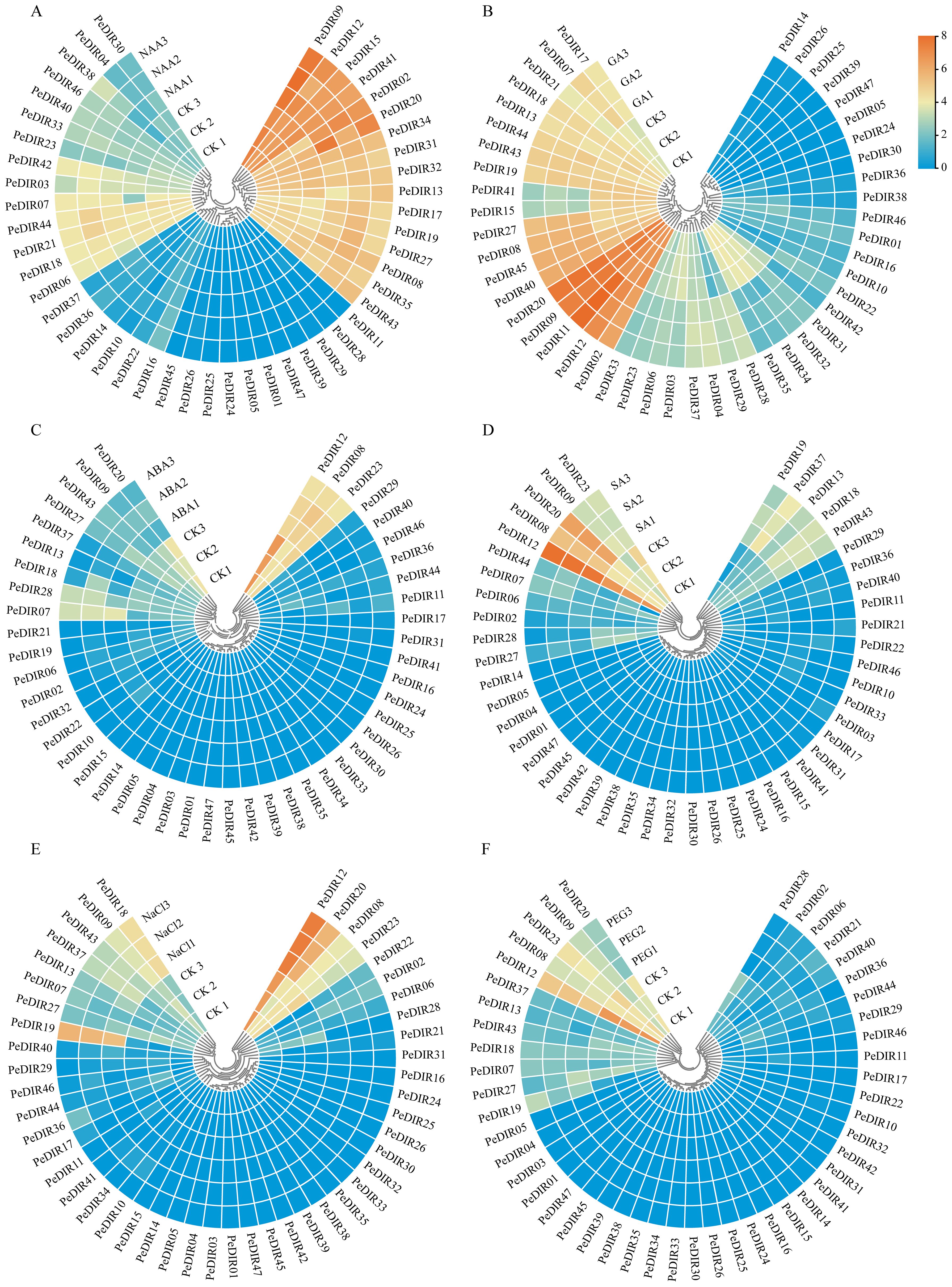
Figure 5. Heatmap of PeDIR genes expression levels under different stress treatments. (A–D) represent treatments with NAA, GA₃, ABA, and SA, respectively. (E, F) represent high-salinity and drought treatments, respectively.
Based on available transcriptome data, the expression of PeDIR genes during drought and salinity stresses was also examined. The results revealed significant expression changes in certain genes in response to these stresses. Overall, most genes exhibited relatively stable expression levels before and after treatments. However, under salinity stress, PeDIR09, PeDIR12, PeDIR20, and PeDIR43 were notably upregulated. In contrast, under drought stress, PeDIR12, PeDIR20, and PeDIR43 were significantly downregulated (Figures 5E, F). These findings suggest that these genes likely play specific regulatory roles in Moso bamboo’s responses to salinity and drought stresses.
The expression profiles of PeDIR genes in Moso bamboo shoots at varying heights (0.2–7 m) were also analyzed using STEM software. The analysis identified five PeDIR genes (PeDIR02, PeDIR03, PeDIR09, PeDIR27, and PeDIR44), highlighted in red in profile 3, as significantly expressed (Figure 6A). These genes exhibited a marked upregulation of expression at heights between 0.2 and 2 m, followed by relatively stable expression levels from 2 to 7 m (Figure 6B). These findings suggest that these genes are closely associated with the regulation of rapid growth in Moso bamboo. They likely contribute to critical processes such as lignin biosynthesis and cell wall structure modulation during early shoot development.
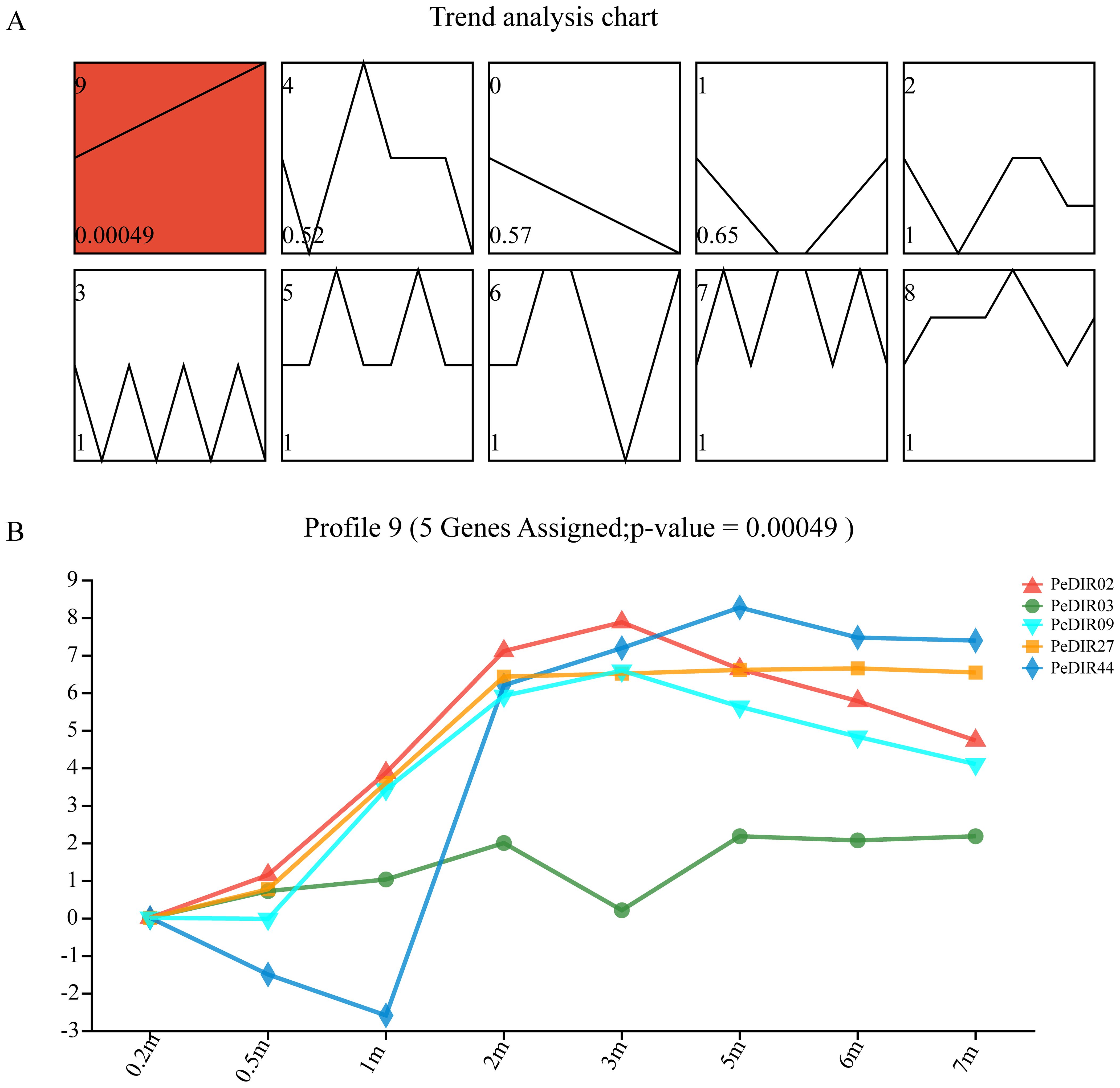
Figure 6. STEM analysis of PeDIR genes at different growth heights. (A) Red boxes indicate significantly expressed genes. All 10 profiles are shown in the top-left corner, with P-values displayed in the bottom-left corner. (B) Expression levels of significantly expressed PeDIR genes in Profile 9.
To further validate the expression profiles of PeDIR genes under different hormonal conditions, the expression responses of nine PeDIR genes were analyzed by qRT-PCR following ABA and SA treatments. During ABA treatment, PeDIR02, PeDIR11, PeDIR12, and PeDIR37 were consistently downregulated across all time points, while PeDIR08 and PeDIR20 initially exhibited downregulation followed by upregulation. PeDIR26 and PeDIR37 displayed fluctuating expression patterns (Figure 7A). Under SA treatment, most PeDIR genes were downregulated at all time points, except for PeDIR08 and PeDIR20, which were downregulated during the early stages but significantly upregulated between 24 and 48 hours (Figure 7B).
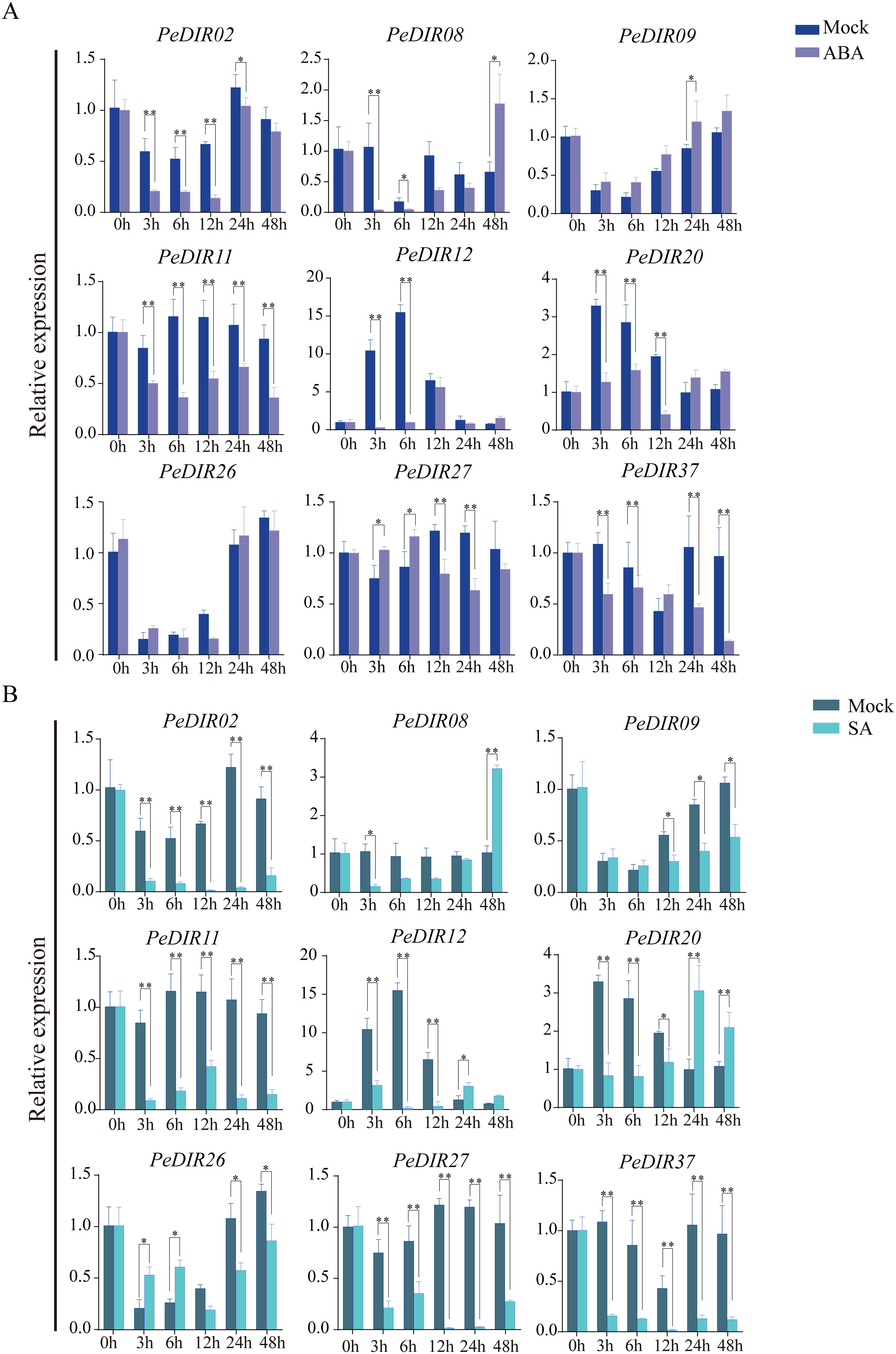
Figure 7. Expression pattern analysis of PeDIR genes under ABA (A) and SA (B) treatments. Asterisks indicate significant differences compared to the control (*P ≤ 0.05; **P ≤ 0.01).
Under drought and high-salinity stresses, qRT-PCR analysis confirmed the expression profiles of PeDIR genes, which strongly correlated with the transcriptome data. Specifically, PeDIR08 and PeDIR09 exhibited significantly reduced expression levels after 6 hours of treatment. In contrast, PeDIR20 and PeDIR43 were notably upregulated under both drought and high-salinity stresses, with PeDIR20 showing a marked increase, particularly 6 hours after salinity stress treatment (Figure 8A). Additionally, PeDIR12 and PeDIR26 were consistently downregulated under both stress conditions, whereas PeDIR34 and PeDIR41 were upregulated specifically under high-salinity stress (Figure 8B). These findings highlight the diverse expression profiles of PeDIR genes under drought and salinity stresses, underscoring the complex regulatory roles of this gene family in Moso bamboo’s abiotic stress responses.
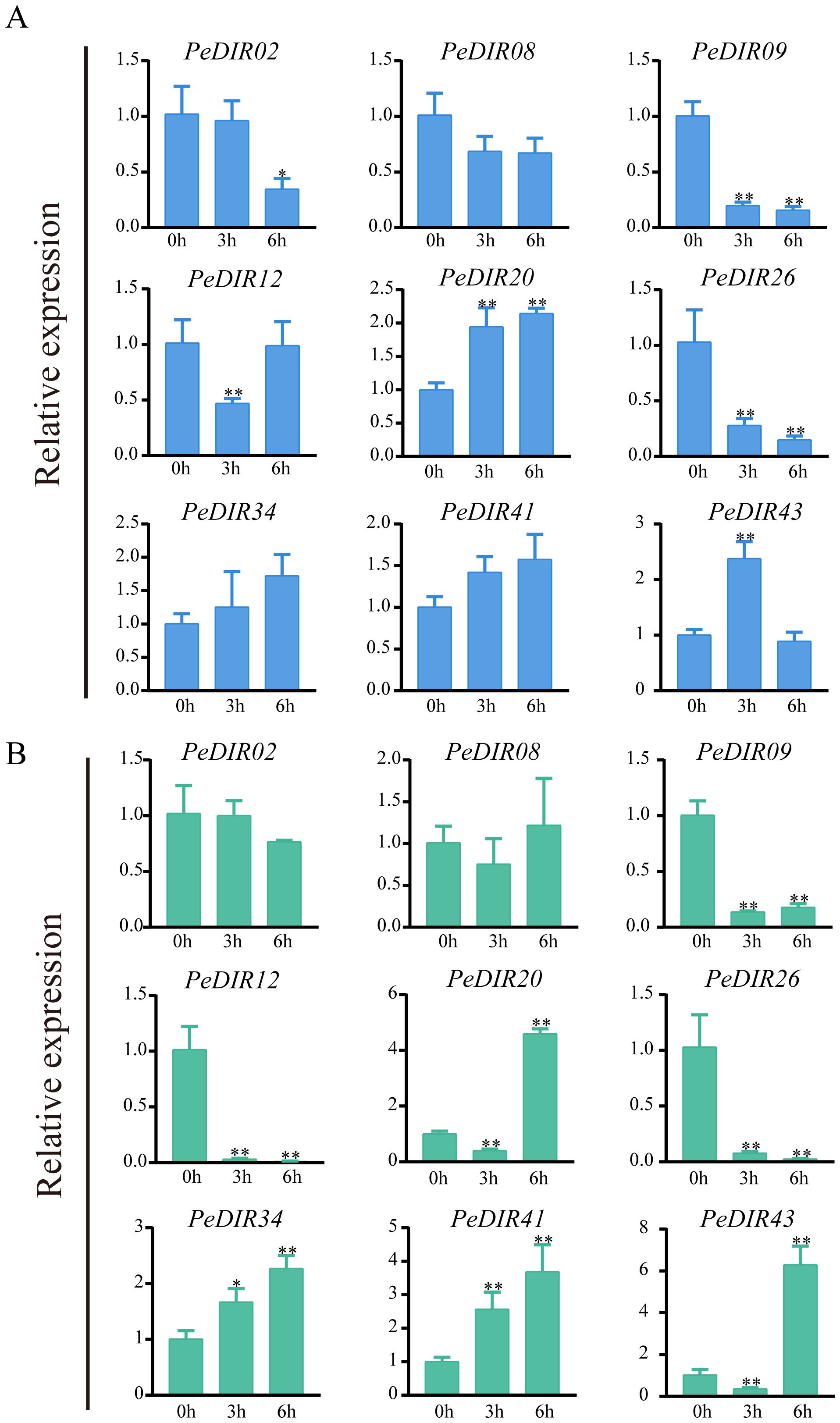
Figure 8. Expression pattern analysis of PeDIR genes under drought (A) and high-salinity (B) treatments. Asterisks indicate significant differences compared to the control (*P ≤ 0.05; **P ≤ 0.01).
To better understand the structural basis underlying the functional diversity of PeDIR proteins, domain prediction analysis was performed. This analysis revealed that all PeDIR proteins contain a Dirigent domain. Additionally, PeDIR12, PeDIR20, and PeDIR43 possess an additional Jacalin domain. Based on these domain differences, the PeDIR protein family can be categorized into two types: Type I, which contains only the Dirigent domain (e.g., PeDIR02), and Type II, which contains both Dirigent and Jacalin domains (e.g., PeDIR20) (Figure 9A). To further investigate the spatial characteristics of these domains, representative members from each type were selected for tertiary structure prediction and modeling (Figure 9B). The results revealed structural similarities between the Dirigent and Jacalin domains, both of which are composed of multiple β-strands. Specifically, the Dirigent domain consists of eight antiparallel β-strands arranged in a barrel-like structure. Notably, PeDIR20, which possesses both Dirigent and Jacalin domains, is hypothesized to exhibit functional properties characteristic of both the DIR and JRL families.
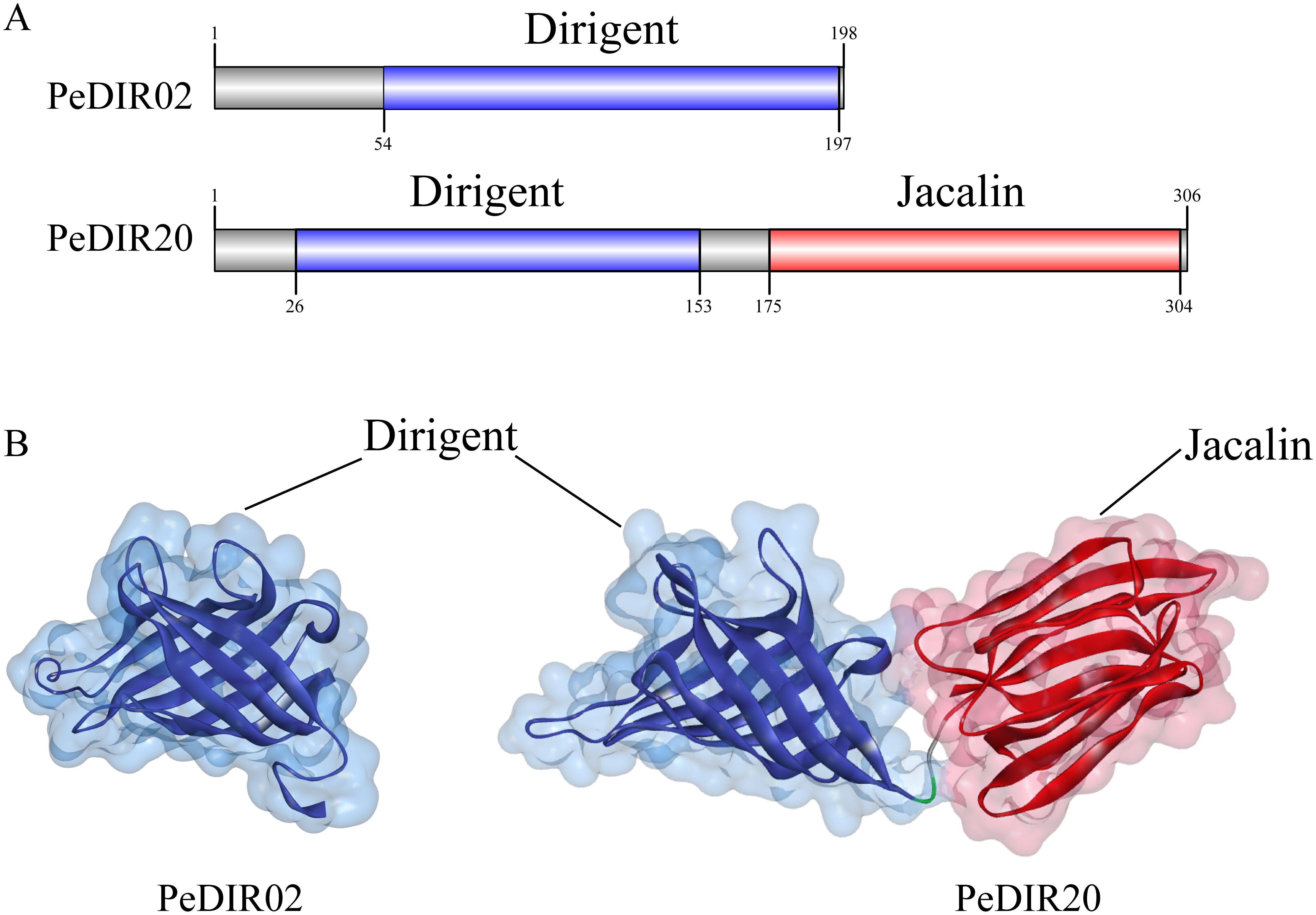
Figure 9. Secondary and tertiary structures of PeDIR02 and PeDIR20 proteins. (A) Conserved domains of PeDIR02 and PeDIR20. (B) Three-dimensional structures of PeDIR02 and PeDIR20.
To examine the subcellular localization of PeDIR02 proteins, the coding sequence (CDS) of PeDIR02 was fused with GFP, and its expression in tobacco epidermal cells was transiently driven by the CaMV35S promoter. GFP alone was used as a control. The results showed that PeDIR02 was primarily localized to the cell membrane, with minor distributions in the cytoplasm and nucleus (Figure 10). To test the self-interaction of PeDIR02 proteins, the genes were cloned into appropriate vectors and introduced into a yeast strain. Growth on SD/-Trp/-Leu medium confirmed successful transformation. However, on SD/-Trp/-Leu/-His/-Ade + X-α-Gal medium, PeDIR02 did not exhibit normal growth or a blue color reaction, indicating the absence of self-interaction (Figure 11).
To investigate the transcriptional regulation of PeDIR genes, a putative regulatory network involving transcription factors (TFs) was constructed. Twenty-eight TFs, primarily from the DOF, MYB, ERF, and WRKY families, were identified as regulators of 10 PeDIR genes. Among these, PeDIR43, PeDIR44, and PeDIR47 showed strong associations with multiple TFs, underscoring their central roles within the network (Figure 12A; Supplementary Table S4). Analysis of dynamic interactions between key TFs and PeDIR genes revealed that all TFs act as positive regulators of their target genes. Notably, aside from MYB1 and MYB3, significant positive correlations were observed among the TFs, indicating coordinated regulation of PeDIR gene expression (Figure 12B). These results suggest that the PeDIR gene family is collectively regulated by multiple TFs, contributing to Moso bamboo’s adaptation to environmental stresses and its growth processes.
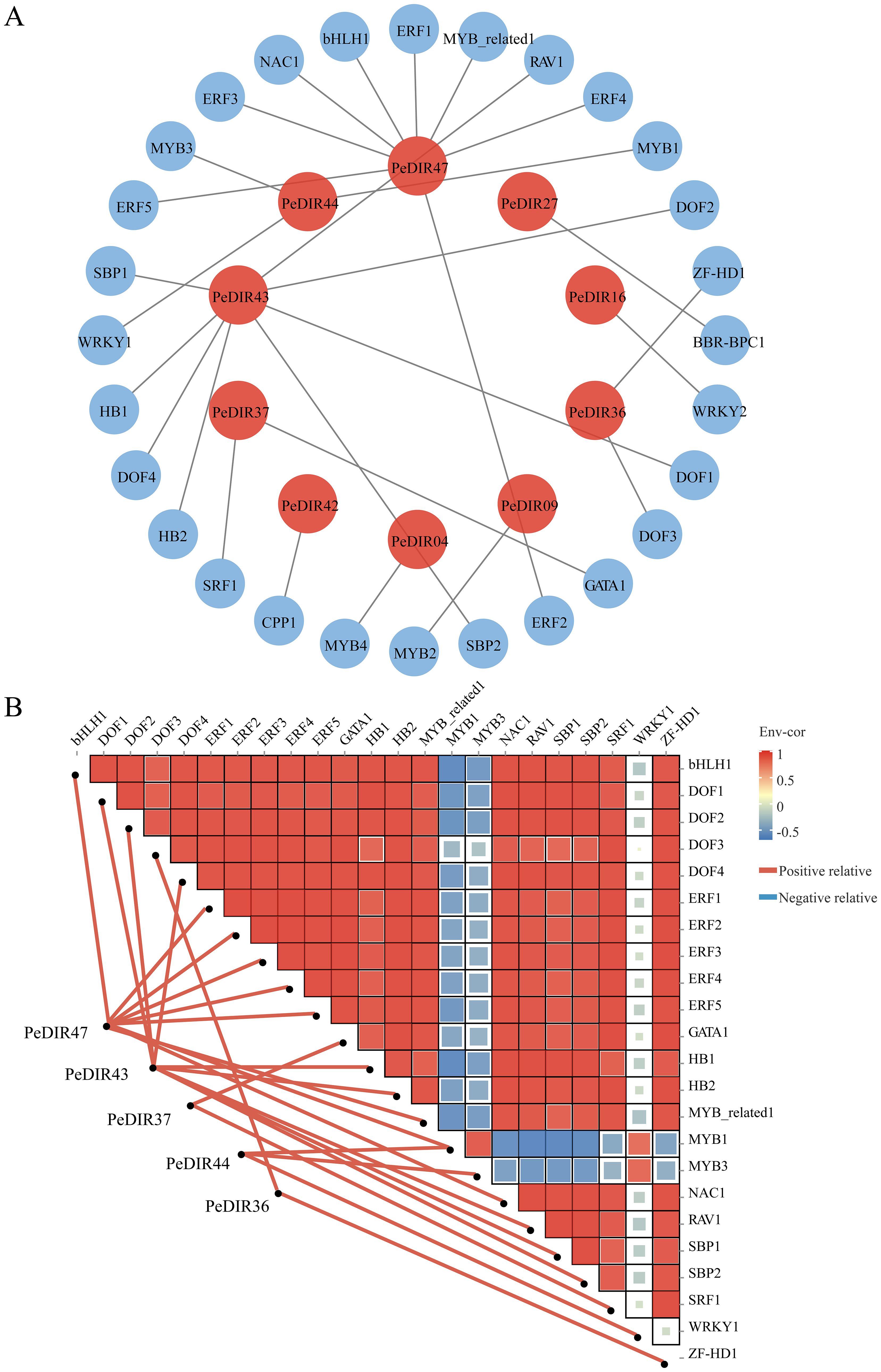
Figure 12. Regulatory network of upstream transcription factors for PeDIR genes. (A) Predicted regulatory network of upstream transcription factors for PeDIR genes. (B) Correlation heatmap between PeDIR genes and transcription factors.
The DIR gene family is widely involved in the biosynthesis of lignin and lignans and is almost ubiquitously present in vascular plants. This family has been identified in various monocots, including rice (49 members) (Liao et al., 2017), Setaria italica (38 members) (Gong et al., 2023), and Schisandra chinensis (34 members) (Dong et al., 2021). In this study, we identified a total of 47 PeDIR members in Moso bamboo. This number is comparable to other Poaceae species but significantly higher than in dicots such as Arabidopsis thaliana (25 members) (Paniagua et al., 2017), Solanum melongena (24 members) (Zhang et al., 2022a), and Isatis indigotica (19 members) (Li et al., 2014). These differences suggest that the DIR gene family may have undergone lineage-specific expansion in monocots.
Structurally, most PeDIR genes contain 2–5 introns, whereas DIR genes in dicots like Arabidopsis and poplar (Populus trichocarpa) have simpler structures, often lacking introns (Paniagua et al., 2017; Li et al., 2021). Furthermore, PeDIR and OsDIR genes are arranged in clusters within their genomes, a feature not observed in Arabidopsis. These structural differences support the hypothesis that the DIR gene family underwent distinct expansion mechanisms following the divergence of monocots and dicots. Gene duplication events, including tandem duplications, segmental duplications, and whole-genome duplications, have facilitated the formation of new genes and regulatory pathways, enabling plants to adapt to diverse environments (Freeling, 2009; Xuan et al., 2024).
In this study, 67.6% of PeDIR genes were located in tandemly duplicated regions, similar to the 72.6% observed in rice (Paniagua et al., 2017; Li et al., 2021), indicating that tandem duplication may have played a significant role in the expansion of DIR genes in monocots. Synteny analysis with other Poaceae species revealed extensive collinearity between Moso bamboo and rice or maize, but fewer syntenic relationships with B. distachyon. This suggests that PeDIR genes in Moso bamboo share closer phylogenetic relationships with those in rice and maize than with B. distachyon. These findings are consistent with previous studies on PeHAK and PeCPP genes (Guo et al., 2024; Tan et al., 2024).
Phylogenetic analysis classified the PeDIR genes into three subfamilies: DIR-b/d, DIR-a, and DIR-e. Motif and gene structure analyses further supported this classification, indicating potential functional differentiation among the subfamilies. Previous studies have shown that DIR-a genes play critical roles in the stereoselective coupling of lignin and lignan intermediates, thereby contributing to lignin biosynthesis (Gasper et al., 2016; Corbin et al., 2018).
To explore the structural and functional characteristics of PeDIR genes, three-dimensional models of two representative DIR proteins, PeDIR02 and PeDIR20, were constructed. PeDIR02, a member of the DIR-b/d subfamily, forms a single barrel-like structure composed of eight antiparallel β-strands. In contrast, PeDIR20, classified under the DIR-a subfamily, contains both a Dirigent domain and a Jacalin domain, forming two distinct barrel-like structures. Previous studies have shown that DIR proteins often function by forming homodimers or trimers (Halls et al., 2004; Kim et al., 2012). Yeast two-hybrid assays revealed that PeDIR02 cannot self-interact to form dimers, indicating that it does not function as a homodimer. Subcellular localization analysis showed that PeDIR proteins are primarily located on the cell membrane, with minor distributions in chloroplasts and the cytoplasm. Experimental validation confirmed that PeDIR02 is predominantly localized on the cell membrane. This result aligns with the subcellular localization of SiDIR proteins in foxtail millet (Gong et al., 2023), suggesting that PeDIR proteins may primarily function at the cell membrane. These insights provide valuable information on the functional mechanisms of DIR proteins in Moso bamboo, laying a foundation for further studies.
DIR genes play crucial roles in growth, development, and environmental adaptation, particularly in hormone and abiotic stress responses (Cai-Qiu et al., 2010; Peng et al., 2013). While DIR gene expression patterns vary across species, numerous studies have reported their regulation under hormonal treatments and abiotic stresses in plants such as potato (Jia et al., 2023), tomato (FengQiong et al., 2022), and barley (Luo et al., 2022). This study found that 22 PeDIR genes responded to at least two hormones, with two genes consistently upregulated under SA treatment and four genes significantly upregulated under ABA treatment. These findings align with those in rice, where hormone-responsive DIR genes are similarly regulated (Liao et al., 2017). Notably, PeDIR08 and PeDIR20 were significantly upregulated in the later stages of ABA and SA treatments, suggesting that these genes play key roles in hormonal response pathways in Moso bamboo.
Under abiotic stress conditions, PeDIR genes exhibited diverse expression patterns in response to drought and salinity stresses. Most PeDIR genes were upregulated under these stresses, with PeDIR20 and PeDIR43 showing particularly significant upregulation. Interestingly, both PeDIR20 and PeDIR43 belong to the DIR-a subfamily, which is closely associated with lignin and lignan biosynthesis. This association likely enhances protective functions under stress conditions (Cheng et al., 2018; Li et al., 2022). These findings suggest that DIR-a subfamily members, especially PeDIR20 and PeDIR43, may play critical roles in enhancing stress tolerance in Moso bamboo, contributing to its growth stability under adverse conditions.
To investigate the regulatory mechanisms of the PeDIR gene family, this study constructed and analyzed a transcriptional regulatory network for PeDIR genes. Cis-regulatory element analysis identified numerous elements associated with hormone responses (e.g., ABRE, TGACG, CGTCA, and TCA-element), abiotic stresses (e.g., STRE and MBS), and growth and development (e.g., G-box and P-box) (Kumari et al., 2025). Predictions of the transcriptional regulatory network indicated that transcription factors such as ERF, DOF, and MYB play key roles in regulating PeDIR gene expression, with evidence of positive synergistic interactions. ERF and MYB transcription factors have been shown to play critical roles in abiotic stress responses, such as drought, salinity, and cold (Xiaoxiao et al., 2022; Ziming et al., 2024). Similarly, the DOF family has been reported to positively regulate hormone signaling and abiotic stress responses in plants such as wheat (Yue et al., 2020). In Moso bamboo, these positive regulatory transcription factors likely interact with cis-elements in the promoters of PeDIR genes, coordinating their expression under hormone signaling and abiotic stress conditions to enhance environmental adaptability and regulate growth and development.
This study significantly advances our understanding of the DIR gene family in Moso bamboo, particularly highlighting the roles of specific genes such as PeDIR08 and PeDIR20 in rapid shoot growth and stress responses. The findings provide a valuable foundation for developing bamboo varieties capable of thriving in challenging environmental conditions, such as drought and high salinity. By enhancing the expression of these genes, it may be possible to improve bamboo’s growth rate and stress resistance, leading to more sustainable and efficient cultivation practices. Additionally, these insights offer a comparative framework for understanding the functions of DIR genes in other plant species and inform strategies for improving stress tolerance in crops.
Future research on the PeDIR gene family in Moso bamboo should focus on quantifying lignin to elucidate its role in cell wall development, analyzing hormone interactions to uncover regulatory mechanisms, and conducting pathway analyses to explore the complex signaling networks involving DIR genes, hormones, and stress responses. These studies will provide valuable insights into the functional roles of DIR genes and help enhance bamboo’s resilience and productivity.
This study comprehensively identified and characterized the PeDIR gene family in Moso bamboo, identifying a total of 47 members. Detailed analyses were performed on their gene structures, phylogenetic relationships, promoter cis-acting elements, and transcription factor regulatory networks. PeDIR genes demonstrated significant roles in rapid shoot growth, hormonal responses (e.g., ABA and SA), and abiotic stress responses (e.g., drought and salinity). These findings provide a crucial foundation for understanding the functions of PeDIR genes in growth and stress adaptation. Future research could leverage gene editing and biotechnological approaches to optimize the expression of key PeDIR genes, paving the way for the development of bamboo varieties with enhanced stress resistance and adaptability to climate change.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XX: Conceptualization, Writing – original draft. SS: Data curation, Writing – review & editing. JC: Data curation, Writing – review & editing. JT: Data curation, Writing – original draft. ZY: Methodology, Writing – review & editing. YJ: Methodology, Writing – review & editing. SC: Investigation, Writing – original draft. ZZ: Investigation, Writing – review & editing. MR: Investigation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Zhejiang Provincial Basic Public Welfare Research Project (Grant No. LTGN23C160002) and the National Natural Science Foundation of China (NSFC; Grant Nos. 31770721, 32171879, and 32371975).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1535733/full#supplementary-material
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Burlat, V., Kwon, M., Davin, L. B., Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. doi: 10.1016/s0031-9422(01)00117-0
Cai-Qiu, G., Gui-Feng, L., Yu-Cheng, W., Jing, J., Chuan-Ping, Y. (2010). Cloning and analysis of dirigent-like protein in gene from Tamarix androssowii. Bull. Botanical Res. 30, 81–86. doi: 10.7525/j.issn.1673-5102.2010.01.015
Chen, C., Cai, Y., He, B., Zhang, Q., Liang, D., Wang, Y., et al. (2024). Genome-wide Identification, evolution, and expression analysis of the DIR gene family in Schima superba. Int. J. Mol. Sci. 25, 7467. doi: 10.3390/ijms25137467
Cheng, X., Su, X., Muhammad, A., Li, M., Zhang, J., Sun, Y., et al. (2018). Molecular characterization, evolution, and expression profiling of the dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri). Front. Genet. 9. doi: 10.3389/fgene.2018.00136
Chow, C. N., Yang, C. W., Wu, N. Y., Wang, H. T., Tseng, K. C., Chiu, Y. H., et al. (2024). PlantPAN 4.0: updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 52, D1569–d1578. doi: 10.1093/nar/gkad945
Corbin, C., Drouet, S., Markulin, L., Auguin, D., Lainé, É., Davin, L. B., et al. (2018). A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation. Plant Mol. Biol. 97, 73–101. doi: 10.1007/s11103-018-0725-x
Dalisay, D. S., Kim, K. W., Lee, C., Yang, H., Rübel, O., Bowen, B. P., et al. (2015). Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: Integrated omics and MALDI mass spectrometry imaging. J. Nat. Prod 78, 1231–1242. doi: 10.1021/acs.jnatprod.5b00023
Davin, L. B., Wang, H. B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., et al. (1997). Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366. doi: 10.1126/science.275.5298.362
Dong, Y. Q., Qiang, T. Y., Liu, J. S., Li, B., Wei, X. P., Qi, Y. D., et al. (2021). Identification and characterization of DIR gene family in Schisandra chinensis. Zhongguo Zhong Yao Za Zhi 46, 5270–5277. doi: 10.19540/j.cnki.cjcmm.20210723.101
Ernst, J., Bar-Joseph, Z. (2006). STEM: a tool for the analysis of short time series gene expression data. BMC Bioinf. 7, 191. doi: 10.1186/1471-2105-7-191
FengQiong, C., QiuSen, C., JiaXin, L. I. N., YaTing, W., HanLin, L. I. U., BingRuoShi, L., et al. (2022). Genome-wide identification of DIR family genes in tomato and response to abiotic stress. Sci. Agric. Sin. 55, 3807–3821. doi: 10.3864/j.issn.0578-1752.2022.19.010
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2015). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Gang, D. R., Costa, M. A., Fujita, M., Dinkova-Kostova, A. T., Wang, H. B., Burlat, V., et al. (1999). Regiochemical control of monolignol radical coupling: a new paradigm for lignin and lignan biosynthesis. Chem. Biol. 6, 143–151. doi: 10.1016/s1074-5521(99)89006-1
Gasper, R., Effenberger, I., Kolesinski, P., Terlecka, B., Hofmann, E., Schaller, A. (2016). Dirigent protein mode of action revealed by the crystal structure of AtDIR6. Plant Physiol. 172, 2165–2175. doi: 10.1104/pp.16.01281
Gong, L., Li, B., Zhu, T., Xue, B. (2023). Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1243806
Guo, H., Tan, J., Jiao, Y., Huang, B., Ma, R., Ramakrishnan, M., et al. (2024). Genome-wide identification and expression analysis of the HAK/KUP/KT gene family in Moso bamboo. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1331710
Halls, S. C., Davin, L. B., Kramer, D. M., Lewis, N. G. (2004). Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 43, 2587–2595. doi: 10.1021/bi035959o
Harmatha, J., Dinan, L. (2003). Biological activities of lignans and stilbenoids associated with plant-insect chemical interactions. Phytochem. Rev. 2, 321–330. doi: 10.1023/B:PHYT.0000045494.98645.a3
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hosmani, P. S., Kamiya, T., Danku, J., Naseer, S., Geldner, N., Guerinot, M. L., et al. (2013). Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. U.S.A. 110, 14498–14503. doi: 10.1073/pnas.1308412110
Huang, B., Huang, Z., Ma, R., Ramakrishnan, M., Chen, J., Zhang, Z., et al. (2021). Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 21, 296. doi: 10.1186/s12870-021-03078-3
Huang, Z., Zhu, P., Zhong, X., Qiu, J., Xu, W., Song, L. (2022). Transcriptome analysis of Moso bamboo (Phyllostachys edulis) reveals candidate genes involved in response to dehydration and cold stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.960302
Jia, W., Xiong, Y., Li, M., Zhang, S., Han, Z., Li, K. (2023). Genome-wide identification, characterization, evolution and expression analysis of the DIR gene family in potato (Solanum tuberosum). Front. Genet. 14. doi: 10.3389/fgene.2023.1224015
Jiang, H., Peng, J., Li, Q., Geng, S., Zhang, H., Shu, Y., et al. (2024). Genome-wide identification and analysis of monocot-specific chimeric jacalins (MCJ) genes in maize (Zea mays L.). BMC Plant Biol. 24, 636. doi: 10.1186/s12870-024-05354-4
Jin-long, G., Li-ping, X., Jing-ping, F., Ya-chun, S., Hua-ying, F., You-xiong, Q., et al. (2012). A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 31, 1801–1812. doi: 10.1007/s00299-012-1293-1
Khan, A., Li, R. J., Sun, J. T., Ma, F., Zhang, H. X., Jin, J. H., et al. (2018). Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 8, 5500. doi: 10.1038/s41598-018-23761-0
Kim, M. K., Jeon, J. H., Fujita, M., Davin, L. B., Lewis, N. G. (2002). The western red cedar (Thuja plicata) 8-8’ dirigent family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 49, 199–214. doi: 10.1023/a:1014940930703
Kim, K.-W., Moinuddin, S. G. A., Atwell, K. M., Costa, M. A., Davin, L. B., Lewis, N. G. (2012). Opposite stereoselectivities of dirigent proteins in Arabidopsis and schizandra species. J. Biol. Chem. 287, 33957–33972. doi: 10.1074/jbc.M112.387423
Kim, K.-W., Smith, C. A., Daily, M. D., Cort, J. R., Davin, L. B., Lewis, N. G. (2015). Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95 Å resolution with three isolated active sites. J. Biol. Che 290, 1308–1318. doi: 10.1074/jbc.M114.611780
Kumari, A., Sopory, S. K., Joshi, R. (2025). Unraveling the intricate tapestry of bamboo transcription factors in abiotic stress signaling and resilience with special reference to moso bamboo family. Biochim. Biophys. Acta Gen. Subj. 1869, 130755. doi: 10.1016/j.bbagen.2024.130755
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P. (2011). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, Q., Chen, J., Xiao, Y., Di, P., Zhang, L., Chen, W. (2014). The dirigent multigene family in Isatis indigotica: Gene discovery and differential transcript abundance. BMC Genomics 15, 388. doi: 10.1186/1471-2164-15-388
Li, X., Liu, Z., Zhao, H., Deng, X., Su, Y., Li, R., et al. (2022). Overexpression of sugarcane ScDIR genes enhances drought tolerance in Nicotiana benthamiana. Int. J. Mol. Sci. 23, 5340. doi: 10.3390/ijms23105340
Li, T., Luo, W., Du, C., Lin, X., Lin, G., Chen, R., et al. (2024b). Functional and evolutionary comparative analysis of the DIR gene family in Nicotiana tabacum L. and Solanum tuberosum L. BMC Genomics 25, 671. doi: 10.1186/s12864-024-10577-8
Li, M., Luo, K., Zhang, W., Liu, M., Zhang, Y., Huang, H., et al. (2024a). Genome-wide identification, evolution, and expression analysis of the dirigent gene family in cassava (Manihot esculenta Crantz). Agronomy 14, 1758. doi: 10.3390/agronomy14081758
Li, L., Sun, W., Zhou, P., Wei, H., Wang, P., Li, H., et al. (2021). Genome-wide characterization of dirigent proteins in Populus: Gene expression variation and expression pattern in response to Marssonina brunnea and Phytohormones. Forests 12, 507. doi: 10.3390/f12040507
Li, N., Zhao, M., Liu, T., Dong, L., Cheng, Q., Wu, J., et al. (2017). A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01185
Li, P., Zhou, G., Du, H., Lu, D., Mo, L., Xu, X., et al. (2015). Current and potential carbon stocks in Moso bamboo forests in China. J. Environ. Manage 156, 89–96. doi: 10.1016/j.jenvman.2015.03.030
Liao, Y., Liu, S., Jiang, Y., Hu, C., Zhang, X., Cao, X., et al. (2017). Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Gene Genom. 39, 47–62. doi: 10.1007/s13258-016-0474-7
Liu, J., Cheng, Z., Xie, L., Li, X., Gao, J. (2019). Multifaceted role of PheDof12-1 in the regulation of flowering time and abiotic stress responses in Moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 20. doi: 10.3390/ijms20020424
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, R., Pan, W., Liu, W., Tian, Y., Zeng, Y., Li, Y., et al. (2022). The barley DIR gene family: An expanded gene family that is involved in stress responses. Front. Genet. 13. doi: 10.3389/fgene.2022.1042772
Ma, R., Huang, B., Chen, J., Huang, Z., Yu, P., Ruan, S., et al. (2021). Genome-wide identification and expression analysis of dirigent-jacalin genes from plant chimeric lectins in Moso bamboo (Phyllostachys edulis). PloS One 16, e0248318. doi: 10.1371/journal.pone.0248318
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301. doi: 10.1093/jxb/erx141
Peng, Z., Lu, Y., Li, L., Zhao, Q., Feng, Q., Gao, Z., et al. (2013). The draft genome of the fast-growing non-timber forest species Moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461. doi: 10.1038/ng.2569
Pickel, B., Pfannstiel, J., Steudle, A., Lehmann, A., Gerken, U., Pleiss, J., et al. (2012). A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J. 279, 1980–1993. doi: 10.1111/j.1742-4658.2012.08580.x
Ralph, S. G., Jancsik, S., Bohlmann, J. (2007). Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 68, 1975–1991. doi: 10.1016/j.phytochem.2007.04.042
Ralph, S., Park, J. Y., Bohlmann, J., Mansfield, S. D. (2006). Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol. Biol. 60, 21–40. doi: 10.1007/s11103-005-2226-y
Ramakrishnan, M., Yrjälä, K., Vinod, K. K., Sharma, A., Cho, J., Satheesh, V., et al. (2020). Genetics and genomics of Moso bamboo (Phyllostachys edulis): Current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 9, e229. doi: 10.1002/fes3.229
Saddique, M. A. B., Guan, G., Hu, B., Khan, M., Amjad, M. D., Abbas, S., et al. (2024). Genome-wide computational analysis of the dirigent gene family in Solanum lycopersicum. Proteome Sci. 22, 10. doi: 10.1186/s12953-024-00233-0
Song, M., Peng, X. (2019). Genome-wide identification and characterization of DIR genes in Medicago truncatula. Biochem. Genet. 57, 487–506. doi: 10.1007/s10528-019-09903-7
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, J., Xuan, X., Su, S., Jiao, Y., Guo, H., Zhang, Z. (2024). Comprehensive analysis of the CPP gene family in Moso bamboo: insights into their role in rapid shoot growth. BMC Genomics 25, 1173. doi: 10.1186/s12864-024-11084-6
Thamil Arasan, S. K., Park, J. I., Ahmed, N. U., Jung, H. J., Hur, Y., Kang, K. K., et al. (2013). Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 67, 144–153. doi: 10.1016/j.plaphy.2013.02.030
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–w303. doi: 10.1093/nar/gky427
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xiaoxiao, Q., Junjie, Z., Junxue, W., Kezhen, Y., Xiaoqin, W., Jie, L. (2022). A rice R2R3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC. Int. J. Mol. Sci. 23, 5873. doi: 10.3390/ijms23115873
Xie, Y., Li, H., Luo, X., Li, H., Gao, Q., Zhang, L., et al. (2022). IBS 2.0: An upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. 50, W420–W426. doi: 10.1093/nar/gkac373
Xuan, X., Su, S., Tan, J., Guo, H., Jiao, Y., Zhang, Z. (2024). Genome-wide identification, characterization, and expression pattern analysis of the JAZ gene family in Moso bamboo during rapid shoot development. Adv. Bamboo Sci. 7, 100083. doi: 10.1016/j.bamboo.2024.100083
Yu, X., Wang, Y., Kohnen, M. V., Piao, M., Tu, M., Gao, Y., et al. (2019). Large scale profiling of protein isoforms using label-free quantitative proteomics revealed the regulation of nonsense-mediated decay in Moso bamboo (Phyllostachys edulis). Cells 8, 744. doi: 10.3390/cells8070744
Yue, L., Nannan, L., Xiong, D., Dongmiao, L., Mengfei, L., Dada, C., et al. (2020). Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genomics 21, 276. doi: 10.1186/s12864-020-6691-0
Zhang, Z., Huang, B., Chen, J., Jiao, Y., Guo, H., Liu, S., et al. (2022b). Genome-wide identification of JRL genes in Moso bamboo and their expression profiles in response to multiple hormones and abiotic stresses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.809666
Zhang, K., Xing, W., Sheng, S., Yang, D., Zhen, F., Jiang, H., et al. (2022a). Genome-wide identification and expression analysis of eggplant DIR gene family in response to biotic and abiotic stresses. Horticulturae 8, 732. doi: 10.3390/horticulturae8080732
Zhengwen, L., Xingfen, W., Zhengwen, S., Yan, Z., Chengsheng, M., Bin, C., et al. (2021). Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes. BMC Plant Biol. 21, 89. doi: 10.1186/s12870-021-02859-0
Zhijun, Z., Peiyao, Y., Bing, H., Ruifang, M., Vinod, K. K., Ramakrishnan, M. (2022). Genome-wide identification and expression characterization of the DoG gene family of Moso bamboo (Phyllostachys edulis). BMC Genomics 23, 357. doi: 10.1186/s12864-022-08551-3
Keywords: DIR gene family, rapid shoot growth, hormone response, abiotic stress, TFs regulatory network, Moso bamboo
Citation: Xuan X, Su S, Chen J, Tan J, Yu Z, Jiao Y, Cai S, Zhang Z and Ramakrishnan M (2025) Evolutionary and functional analysis of the DIR gene family in Moso bamboo: insights into rapid shoot growth and stress responses. Front. Plant Sci. 16:1535733. doi: 10.3389/fpls.2025.1535733
Received: 27 November 2024; Accepted: 31 January 2025;
Published: 25 February 2025.
Edited by:
Muhammad Waseem, Hainan University, ChinaReviewed by:
Muthu Thiruvengadam, Konkuk University, Republic of KoreaCopyright © 2025 Xuan, Su, Chen, Tan, Yu, Jiao, Cai, Zhang and Ramakrishnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Zhang, emp6aGFuZ0B6YWZ1LmVkdS5jbg==; Muthusamy Ramakrishnan, cmFta3lAbmpmdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.