- 1College of Plant Science, Jilin University, Changchun, Jilin, China
- 2Institute of Agricultural Biotechnology, Jilin Academy of Agricultural Sciences (Northeast Agricultural Research Center of China), Changchun, Jilin, China
Leaf senescence is an important agronomic trait that significantly influences the quality and yield of soybeans. v-Myb avian myeloblastosis viral oncogene homolog (MYB) transcription factors are considered crucial regulators governing leaf senescence, which can be utilized to improve agronomic traits in crops. However, our knowledge regarding the functional roles of soybean MYBs in leaf senescence is extremely limited. In this study, GmMYBLJ, a CCA1-like MYB, was identified and functionally characterized with respect to leaf senescence. The GmMYBLJ protein is localized in the nucleus, and a high accumulation of its transcripts was observed in nodules and embryos. Notably, GmMYBLJ was highly expressed in soybean senescent leaves and was transcriptionally induced by dark or NaCl treatment, as confirmed by histochemical GUS staining analysis. Ectopic overexpression of GmMYBLJ in Arabidopsis not only led to earlier leaf senescence, reduced chlorophyll content, and increased MDA accumulation but also promoted the expression of several WRKY family transcription factors and senescence-associated genes, such as SAG12 and ORE1. Further investigation showed that overexpression of GmMYBLJ accelerated Arabidopsis leaf senescence under darkness and in response to Pst DC3000 infection. Moreover, transgenic soybean plants overexpressing GmMYBLJ grew faster and exhibited accelerated senescence under salt stress. DAB staining analysis showed that GmMYBLJ induced ROS accumulation in soybean hairy roots and Arabidopsis leaves. Collectively, our results provided useful information into the functional roles of GmMYBLJ in both age-dependent and stress-induced senescence.
1 Introduction
Leaf senescence is the final phase of leaf development, characterized by chloroplast degeneration and macromolecular catabolism (Lei et al., 2023). It is often accompanied by the accumulation of reactive oxygen species (ROS). Senescence is a natural process associated with aging, which can facilitates the release and recycling of nutrients for use by other developing organs or storage for the next generation or season (Zhou and Yang, 2023). However, adverse environmental factors, such as darkness, shade, nutrient deficiency, low temperature, and both abiotic and biotic stresses, often lead to premature senescence, thereby reducing the yield and quality of crop plants (Antonietta et al., 2024). Increasing evidence suggests that the process of leaf senescence is highly coordinated and exquisitely regulated by both internal and external stimuli (Sasi et al., 2022; Zhao et al., 2022; Cao et al., 2023). Thus, studying the regulators of leaf senescence and their regulatory mechanisms is agronomically important for manipulating leaf traits in crop plants.
v-Myb avian myeloblastosis viral oncogene homolog (MYB) proteins constitute a large family of transcription factors found in all eukaryotes, characterized by a varying number of MYB domains that contribute to their DNA-binding capacity. Based on the number of MYB domains, the MYB family can be classified into four categories: 1R, 2R, 3R, and 4R MYB (Biswas et al., 2023). It is well known that MYBs are involved in the regulation of many plant developmental and growth processes, as well as responses to abiotic and biotic stresses (Thakur and Vasudev, 2022; Biswas et al., 2023). Notably, several studies have shown that MYBs play positive roles in leaf senescence through hormonal pathways. For example, overexpression of MYB HYPOCOTYL ELONGATION-RELATED (AtMYBH) (a 1R-MYB gene) in Arabidopsis accelerated ethylene- and Abscisic Acid (ABA)-induced leaf senescence, while AtMYBH negatively regulated auxin homeostasis by inhibiting the expression of AtDFL1/AtGH3.6 and AtDFL2/AtGH3.10 (Huang et al., 2015). Similarly, AtMYBL (a R-R-type MYB-like gene) promoted leaf senescence in Arabidopsis, and its expression could be induced by the phytohormone ABA (Zhang et al., 2011). Likewise, overexpression of NtMYB1 in Narcissus or Arabidopsis promoted ABA production and decreased the level of gibberellin A1, leading to leaf senescence symptoms such as leaf yellowing and a decrease in photosystem II fluorescence (Yang et al., 2023). In addition, MYBs are positively involved in the regulation of dark-induced leaf senescence. A good example is that mutation of Arabidopsis GLABRA (AtGL1), which resulted in reduced susceptibility to dark-induced aging (Eckstein et al., 2019), implying its functional role in governing the initiation and progression of leaf senescence. However, a few reports have shown that some MYBs might negatively regulate leaf senescence. For example, AtMYBR1/AtMYB44 and its homolog OsMYB102 function as negative regulators of leaf senescence by affecting the ABA pathway (Jaradat et al., 2013; Piao et al., 2019). Further investigation revealed that AtMYB44 directly bound to the promoter of AtBBX14 and activated its expression, which subsequently functioned as a negative regulator of N starvation-induced senescence through AtEIN3 (Buelbuel et al., 2023). Evidently, different MYB members might exhibit functionally diverse roles in the regulation of leaf senescence. To date, several MYBs involved in the regulation of leaf senescence have been identified in various plant species, such as rice (Akhter et al., 2019; Piao et al., 2019), cottonwood (Lu et al., 2020), sunflower (Moschen et al., 2016), and Jatropha curcas (Chen et al., 2022). However, leaf senescence-associated MYBs in additional plant species need to be identified to meet the requirements of molecular breeding design.
Soybean (Glycine max) is an economically important crop due to its storage of proteins and oils. Leaf senescence is a crucial trait that significantly impacts the quality and yield of crops. Generally, delaying leaf senescence is considered to promote higher yield and quality (Antonietta et al., 2024). However, early soybean harvesting is one of the most desirable agricultural traits in high-latitude regions (Zhu et al., 2023; Gao et al., 2024), and early leaf senescence might contribute to shortening the maturation period. To date, a number of regulators have been identified that control leaf senescence in soybean, such as the GmCRY1s-GmDELLAs-GmWRKY100 regulatory cascade (Li et al., 2024), GmNACs (NAC81, NAC30, NAC65)-SAGs (Ferreira et al., 2020; Fraga et al., 2021), GmELS1 (Yamatani et al., 2021), GmBiP (Carvalho et al., 2014), GmSARK (Xu et al., 2011), GmWRKY53b, and GmCIB1 (Meng et al., 2013). Recently, it was reported that GmMYB128 might be a positive regulator of nodule senescence in soybean (Liu et al., 2024). However, little is known about the functional roles of MYBs in soybean leaf senescence.
In the current study, our primary objective is to characterize a soybean CCA1-like MYB, GmMYBLJ, and explore its functional roles in governing leaf senescence and stress response. Our results suggest that GmMYBLJ might act as a positive regulator in age-dependent and stress-induced leaf senescence by regulating WRKY and senescence-associated genes. These findings expand the understanding of the regulatory functions of MYB transcription factors in soybean senescence, which could inform breeding strategies aimed at optimizing leaf senescence and stress tolerance.
2 Materials and methods
2.1 Plant materials and growth conditions
Seedlings of Arabidopsis (Columbia-0), tobacco (Nicotiana benthamiana), and soybean (Glycine max, cv “Jilin 32”) were grown under 16 h/8 h (light/dark) photoperiod cycle at 25°C with 70%–80% relative humidity. These conditions were used for the treatments and sample collection in the study. Compound leaves were collected every 15 days (15, 30, 45, 60, and 75 days) once the fourth triple compound leaf emerged from the seedling (V4 stage). Soybean plants were grown at the experimental station of Jilin University (Changchun, China). Soybean tissues, including root, stem, leaf, flower, and nodule, were collected randomly during flowering, as well as embryos at six different developmental stages (10, 20, 30, 40, 50, and 60 days after pollination [DAP]).
2.2 Generation of transgenic Arabidopsis and soybean lines as well as transgenic hairy roots
To generate transgenic Arabidopsis and soybean (Glycine max, cv “DN50”) lines overexpressing GmMYBLJ, its full-length Coding Sequence (CDS) sequence was amplified and cloned into the YFP-infused vectors, pEarleyGate101 (pEarleyGate101-GmMYBLJ) and pTF101 (pTF101-GmMYBLJ), respectively. Transgenic Arabidopsis and soybean lines were produced using Agrobacterium-mediated floral dip (Tsuda et al., 2012) and cotyledonary-node explants (Zeng et al., 2004), respectively. Positive transgenic lines were screened on Murashige and Skoog (MS) medium with 100 mg/L of cephalosporin and 10 mg/L of glufosinate, and confirmed by RT-qPCR using gene-specific primers. Primer information is listed in Supplementary Table S1.
To generate GmMYBLJ-silenced hairy roots, specific fragments of GmMYBLJ were inserted into the RNAi vector pK7GWIGW2D(II). Meanwhile, the CDS sequence of GmMYBLJ was cloned into the overexpression vector pK7GM with a Green Fluorescent Protein (GFP) fusion, driven by an independent 35S promoter, to generate GmMYBLJ-overexpressed hairy roots. The pK7GM-GmMYBLJ- and GmMYBLJ-silenced vectors were transformed into the Agrobacterium rhizogenes strain K599. Transgenic hairy roots were generated following the method described by Li et al. (2012). The transgenic soybean hairy roots were cultured under a 16-h/8-h (light/dark) photoperiod cycle. After 6 weeks, positive hairy roots were identified by green fluorescence using LUYOR-3415CV (hand lighting) and PCR analysis.
2.3 Generation of transgenic GUS reporter lines and GUS staining analysis
To generate transgenic β-glucuronidase (GUS) reporter lines, the promoter region spanning 1,500 bp upstream of the translational start site was cloned and constructed into the Gateway vector pMDC162 to obtain pGmMYBJ-GUS. Transgenic Arabidopsis plants were generated using the Agrobacterium-mediated floral dip method (Tsuda et al., 2012). Positive transgenic lines were screened on MS with 200 mg/L of cephalosporin and 50 mg/L of hygromycin, followed by RT-PCR analysis for confirmation. Primer information is listed in Supplementary Table S1.
Histochemical GUS staining was carried out using different tissues from transgenic samples, including 7-day-old seedlings, roots, stems, cauline leaves, inflorescences, silique, rosette leaves at different positions (first to ninth), and leaves under dark condition for 5 days. After vacuum infiltration for 15 min, the samples were incubated in staining solution at 37°C overnight. The tissues were decolorized with 70% ethanol to completely remove chlorophylls, and photographs were taken with a stereomicroscope.
2.4 Treatment of soybean for the analysis of circadian rhythm
Soybean seeds were planted and grown under a 12-h light/12-h dark cycle at 22°C for 10 days, after which they were subsequently transferred to constant light conditions. Soybean samples were harvested every 4 h from 0 h to 72 h.
2.5 DAB staining analysis
Samples were collected for DAB staining analysis, including 30-day-old Arabidopsis plants overexpressing GmMYBLJ and wild-type plants, as well as GmMYBLJ-overexpressed and GmMYBLJ-silenced hairy roots. DAB staining was conducted according to the method described by Daudi and O’Brien (2012). Three biological repeats with three technical repeats were performed.
2.6 Stress treatments and measurement of MDA and chlorophyll contents
Two-week-old soybean seedlings were subjected to the following treatments as described previously (Bian et al., 2017): (1) Seedlings were grown under dark conditions, and samples were collected at 0, 1, 2, 3, 4, 5, and 6 days after dark treatment; (2) Salicylic Acid (SA) (2 mM) and ABA (100 μM) were sprayed on the leaves of seedlings, respectively, and samples were collected at 0, 3, 6, 12, 24, 48, and 72 h after SA and ABA treatments; (3) Cold treatment (4°C) and 200 mM NaCl were individually applied to the seedlings, and samples were collected at 0, 3, 6, 12, 24, 48, and 72 h after the salt and cold treatments; and (4) Water supply was withheld for drought stress, and samples were collected at 0, 2, 4, 6, 8, and 10 days of water restriction.
To investigate the effects of GmMYBLJ on dark- and stress-induced senescence, 25-day-old transgenic Arabidopsis plants overexpressing GmMYBLJ and wild-type plants were subjected to dark treatment and Pst DC3000 infection for 5 days. Samples were then harvested to measure the contents of malondialdehyde (MDA) and chlorophylls.
To investigate the effects of GmMYBLJ on salt-induced senescence, 200 mM NaCl was applied to 10-day-old transgenic soybean plants and nontransformed control for 10 days. Samples were then harvested to measure the MDA content.
The contents of MDA and chlorophylls were determined using the methods described previously (Li et al., 2020; Zhang et al., 2021). Three biological repeats with three technical repeats were performed for each sample. One-way analysis of variance (ANOVA) with Least Significant Difference (LSD) test was used to calculate the statistical significance, and significant differences are indicated by an asterisk (*p < 0.05; **p < 0.01; ***p < 0.001).
2.7 RT-qPCR analysis
Total RNA from Arabidopsis and soybean samples were extracted using the RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China). First-strand cDNA was generated using StarScript II RT Mix with gDNA Remover (GenStar, Beijing, China), following the manufacturer’s instructions. RT-qPCR was then performed using an ABI stepOnePlus PCR system and SYBR Premix Ex Taq (Takara, Beijing, China), with GmSUBI3 and AtACTIN8 used as internal controls for soybean and Arabidopsis samples, respectively. Three biological replicates with three technique repeats were performed for each sample, and data analysis was conducted using ABI StepOne Plus v2.3 software. One-way ANOVA with LSD test was used to calculate the statistical significance, with significant difference indicated by an asterisk (*p < 0.05; **p < 0.01; ***p < 0.001). The primer sequences are listed in Supplementary Table S1.
2.8 Subcellular localization of GmMYBLJ
The open reading frame (ORF) of GmMYBJ from Williams 82 was amplified and subsequently cloned into the pCAMBIA1300 vector, with YFP driven by the 35S promoter. The resulting plasmid, pCAMBIA1300-GmMYBJ-YFP, and the empty vector, pCAMBIAI1300-YFP, were transformed into Agrobacterium tumefaciens strain GV3101, and then introduced into the leaves of Nicotiana benthamiana (Sparkes et al., 2006). GFP fluorescence in the leaves was observed using a Leica confocal microscope (http://www.leica.com/).
2.9 Phylogenetic and conserved motif analysis
The sequences of GmMYBLJ and its homologous proteins were downloaded from the Phytozome website (https://phytozome-next.jgi.doe.gov). Conserved sequences were predicted on the MEME website (http://meme-suite.org/), and the conserved motifs were visualized using the TBtools software (Chen et al., 2023). Conserved domains were predicted using the tool CDD (https://www.ncbi.nlm.nih.gov/cdd). The phylogenetic tree was constructed by the Neighbor-joining method using MEGA7 (Poisson mode, complete deletion, and 1,000 bootstrap values) (Kumar et al., 2016) and visualized using the EVOLVIEW tool (https://www.evolgenius.info/evolview). The online program STRING was used to predict the interactors of GmMYBLJ (https://string-db.org/).
2.10 Yeast one-hybrid assay
To examine the binding of GmMYBLJ to its targets, the full-length CDS of GmMYBLJ was cloned and introduced into the vector pB42AD. The binding sites in the promoter region of the three WRKY genes (AtWRKY30, AtWRKY45, AtWRKY75) were predicted using the online program PlantCare (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The promoter fragments spanning the predicted binding sites were cloned as preys and constructed into the vector pLacZi (pWRKY30, 342 bp; pWRKY45-1, 372 bp; pWRKY45-2, 320 bp; pWRKY75-1, 411 bp; pWRKY75-2, 469 bp). Negative controls, i.e., pB42AD/pLacZi, pB42AD-GmMYBLJ/pLacZi, and pB42AD/pLacZi-pAtWRKY30, pB42AD/pLacZi-pAtWRKY45, pB42AD/pLacZi-pAtWRKY75, and one positive control, pB42AD-AtRVE8/placZi-AtPRR5, were also included. The primers are listed in Supplementary Table S1.
3 Results
3.1 Identification of GmMYBLJ and its subcellular localization
To investigate the CCA1-like MYBs associated with leaf senescence, a homology search was performed using the sequence of Arabidopsis MYB HYPOCOTYL ELONGATION-RELATED (MYBH) (a leaf senescence-associated regulator) against the 54 CCA1-like MYBs identified previously in soybean (Bian et al., 2017). As a result, 12 CCA1-like MYBs showed high protein identity with AtMYBH and were clustered into three clades (Figure 1A). It was further observed that the CCA1-like MYBs within the same clade exhibited similar motif compositions (Figures 1B, C). All the proteins contained a MYB domain with the conserved motif SHAQK(Y/F)F (Figures 1B–D), implying functional redundancy among these proteins. Subsequently, GmMYBLJ was selected as a representative to investigate the functional roles of soybean CCA1-like MYBs in leaf senescence. The protein identities of the abovementioned 12 MYBs with GmMYBLJ were calculated, ranging from 41% to 87%, with Glyma.04G051000 showing the maximum value (Figure 1A). GmMYBLJ is closely related to AtMYBH, with 47% protein identity, suggesting that GmMYBLJ might be involved in the regulation of leaf senescence.
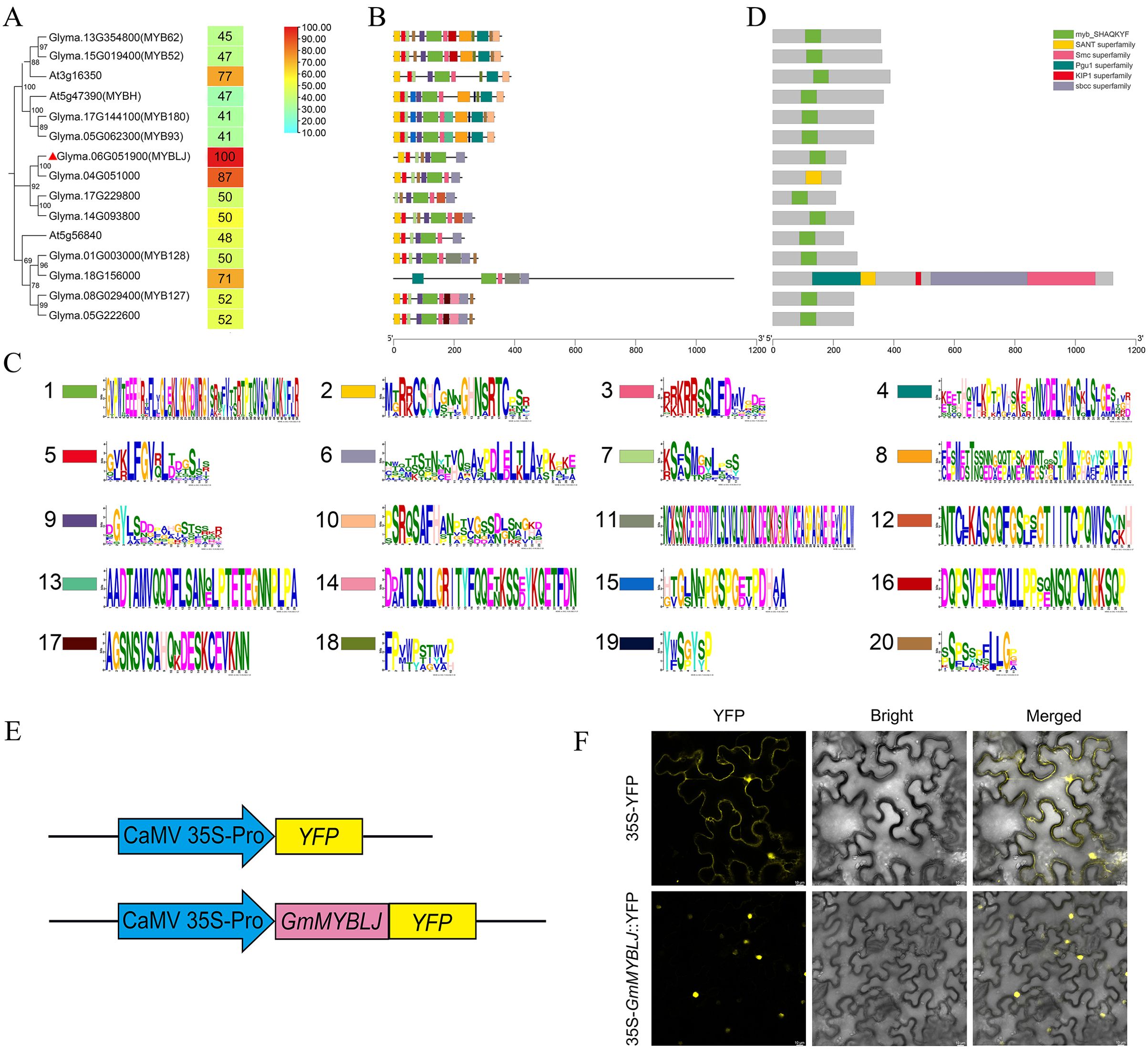
Figure 1. Characterization of GmMYBLJ and its homologous proteins in soybean and Arabidopsis. (A) Phylogenetic analysis of GmMYBLJ and its homologs in Arabidopsis (three AtMYBs) and soybean (11 GmMYBs). GmMYBLJ is marked with a red triangle. All the sequences of MYB proteins were downloaded from the phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). The protein identities of GmMYBLJ with other MYBs are listed. (B) Motif compositions of GmMYBLJ and its homologs. The online program MEME was used to predict motifs, with the maximum number set at 20. (C) Sequence logos of the 20 motifs. The height of the letters within each stack represents their relative frequency. Colored symbols correspond to the colored boxes representing motifs. (D) Diagrams of conserved domains for the 12 GmMYBs and three AtMYBs. The length of each protein sequence is represented by a gray bar, with colored boxes indicating conserved domains. The sequence length of each protein is shown by a gray line at the bottom. (E) Schematics of the two constructed vectors: p35S-GmMYBJ::YFP and p35S-YFP. (F) Subcellular localization of GmMYBLJ. A. tumefaciens carrying the plasmid p35S-GmMYBJ::YFP or p35S-YFP was infiltrated into N. benthamiana leaves and visualized using confocal microscopy. The scale bar is shown as 10 μm.
To investigate its subcellular localization, the full ORF of GmMYBLJ was fused with yellow fluorescent protein (YFP) to create p35S-GmMYBLJ::YFP, with p35S-YFP used as a negative control (Figure 1E). As shown in Figure 1F, YFP fluorescence predominantly appeared in the nucleus of tobacco cells expressing p35S-GmMYBLJ::YFP. In comparison, the p35S-YFP control exhibited YFP signals in both the nucleus and cytoplasm (Figure 1F). These results suggested that GmMYBLJ may act as a transcription factor, performing its functions in the nucleus.
3.2 Spatiotemporal expression of GmMYBLJ and its response to leaf senescence
To provide functional insights into GmMYBLJ in soybean, its expression pattern was investigated in roots, stems, leaves, flowers, nodules, and embryos at different developmental stages (10, 20, 30, 40, 50, and 60 DAP). It was found that GmMYBLJ exhibited the highest expression in nodules, followed by embryos and stems, with the lowest expression in flowers (Figure 2A). Further observation showed that the transcripts of GmMYBLJ gradually accumulated as embryos develop (Figure 2A), suggesting that GmMYBLJ might play important roles during embryo development.
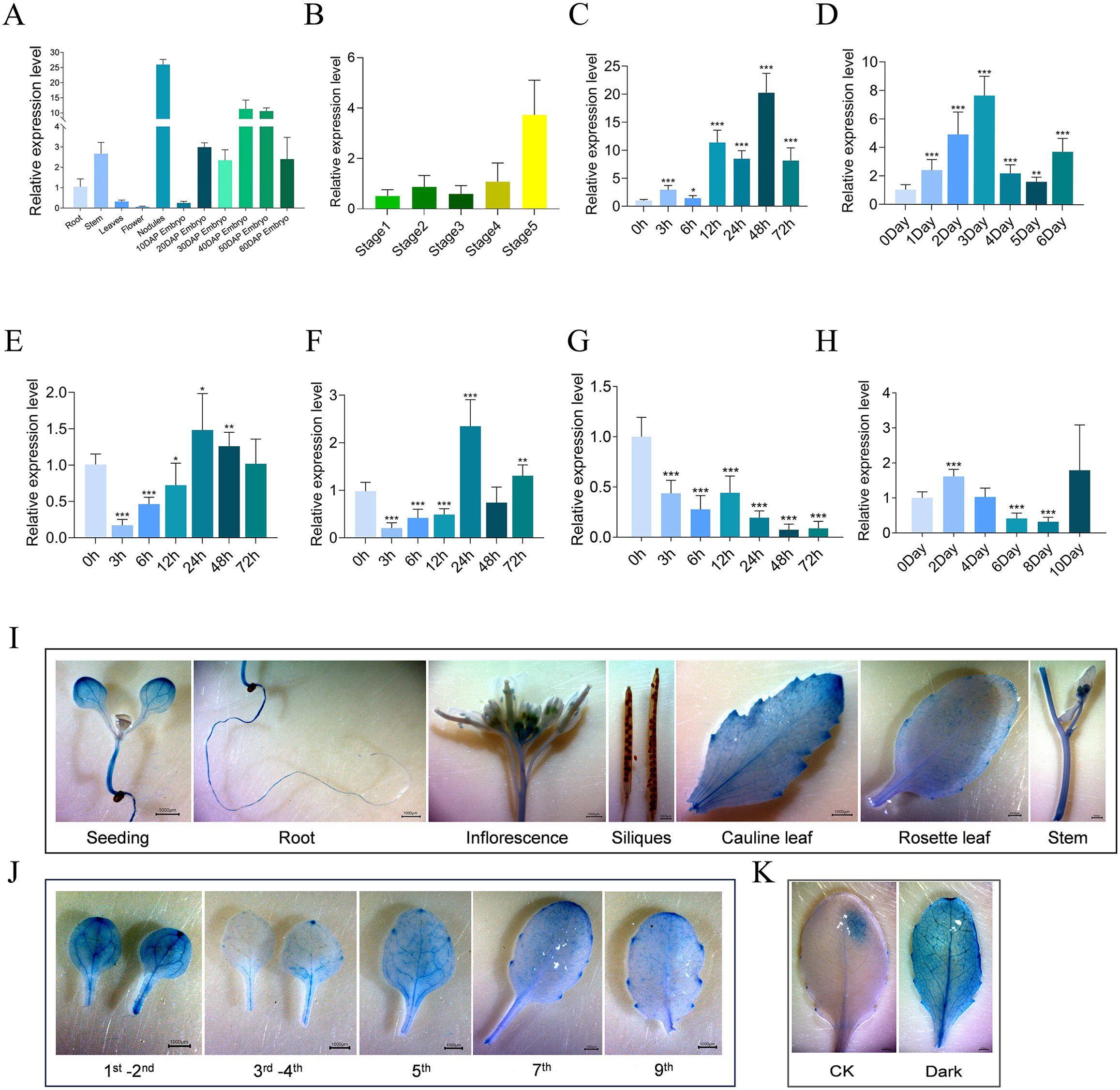
Figure 2. Spatiotemporal expression of GmMYBLJ and its response to leaf senescence. (A, B) Gene expression of GmMYBLJ in various tissues (A) and leaves at five developmental stages (B). Tissues include root, stem, leaf, flower, nodule, and embryo at different developmental stages (10, 20, 30, 40, 50, and 60 days after pollination [DAP]). The five developmental stages of leaves correspond to 15, 30, 45, 60, and 75 days after the appearance of the third compound leaves. (C–H) Expression pattern of GmMYBLJ under various treatments: salt (C), dark (D), SA (E), ABA (F), cold (G), and drought (H). Two-week-old soybean seedlings were subjected to 200 mM NaCl or cold conditions, and samples were collected at 0, 3, 6, 12, 24, 48, and 72 h after treatment. For SA and ABA treatments, 2 mM SA or 100 μM ABA was sprayed on the leaves of 2-week-old soybean seedlings, and samples were similarly collected at 0, 3, 6, 12, 24, 48, and 72 h post-treatment. Two-week-old soybean seedlings were grown under dark conditions, and samples were collected at 0, 1, 2, 3, 4, 5, and 6 days after the dark treatment. For drought stress, water supply was withheld, and samples were collected at 0, 2, 4, 6, 8, and 10 days of water restriction. RT-qPCR analysis was performed with three biological replicates, each comprising three technical replicates. The standard error of the mean is represented by error bars. Data were normalized against the GmSUBI-3 gene. Statistical significance was calculated using one-way ANOVA with the LSD test, with significant differences indicated by an asterisk (*p < 0.05; **p < 0.01; ***p < 0.001). (I–K) Histochemical analysis of pGmMYBLJ::GUS was performed using different various Arabidopsis tissues (I), rosette leaves at different positions (J), and under dark conditions (K). Materials for GUS staining include 7-day-old seedlings, roots, stems, cauline leaves, inflorescences, siliques, rosette leaves at different positions (first–ninth), and leaves exposed to dark conditions for 5 days. The scale bar is shown as 1,000 μm.
To explore whether GmMYBLJ expression changes in responding to leaf senescence, its expression level was examined in soybean leaves at five different stages (15, 30, 45, 60, and 75 days after the appearance of the third compound leaves, Supplementary Figure S1). It was shown that the expression of GmMYBLJ was relatively low at the first four stages and remarkably increased in the 75-day-old leaf (Figure 2B), suggesting that GmMYBLJ might play a functional role in the regulation of leaf senescence. Additionally, the expression patterns of GmMYBLJ were investigated under the following treatments: darkness (for 0, 1, 2, 3, 4, 5, and 6 days), NaCl (for 0, 3, 6, 12, 24, 48, and 72 h), drought (for 0, 2, 4, 8, and 10 days), cold, SA, and ABA (for 0, 3, 6, 12, 24, 48, and 72 h). The expression of GmMYBLJ was significantly increased under NaCl and dark conditions, especially at 3 days after the dark treatment (Figures 2C, D), while it was remarkably decreased shortly after the SA and ABA treatments (Figures 2E, F), suggesting that phytohormones and darkness might be involved in regulating the expression of GmMYBLJ. Additionally, it was observed that the expression of GmMYBLJ gradually decreased as the cold treatment progressed (Figure 2G), while it decreased at 6 and 8 days after the drought treatment (Figure 2H). Unexpectedly, no rhythmicity was observed in the expression of GmMYBLJ (Supplementary Figure S2).
To test the promoter activity of GmMYBLJ, we cloned the promoter region (1,500 bp upstream of the translational start site) and fused it with the GUS gene. Subsequently, transgenic Arabidopsis plants were generated as reporter lines. Histochemical staining analysis indicated that GUS signals appeared in seedlings, roots, anthers, cauline leaves, rosette leaves, and stems at varied intensities, while no signal was detected in siliques (Figure 2I). Furthermore, the promoter activity was investigated in the leaves at different positions (first–ninth) of the rosette using transgenic Arabidopsis reporter lines. It was found that cotyledons showed more GUS signals than other rosette leaves (Figure 2J), Additionally, it was observed that GUS signals in the leaves were stronger under dark conditions than normal conditions (Figure 2K). These results suggest that the promoter of GmMYBLJ might exhibit higher activity in senescent leaves or under dark conditions.
3.3 GmMYBLJ affects plant morphology and confers leaf senescence in Arabidopsis
To investigate the functional roles of GmMYBLJ, transgenic Arabidopsis lines overexpressing GmMYBLJ were generated and confirmed using RT-qPCR (Figures 3A, B). Three transgenic Arabidopsis lines (OX-9, OX-13, OX-15) exhibited multiple morphological phenotypes (such as smaller size, darker and curly leaves) as compared to wild type (WT) (Figures 3A, B). Further investigation revealed that the total chlorophyll content in the three transgenic lines (25-day-old) reached 3 mg/g, which was significantly higher than that in WT plants (Figure 3C). Additionally, transgenic plants exhibited an early flowering phenotype, with the average bolting time of 25 days, occurring 5 days earlier than in WT plants, which bolted at 30 days (Figures 3D, E).
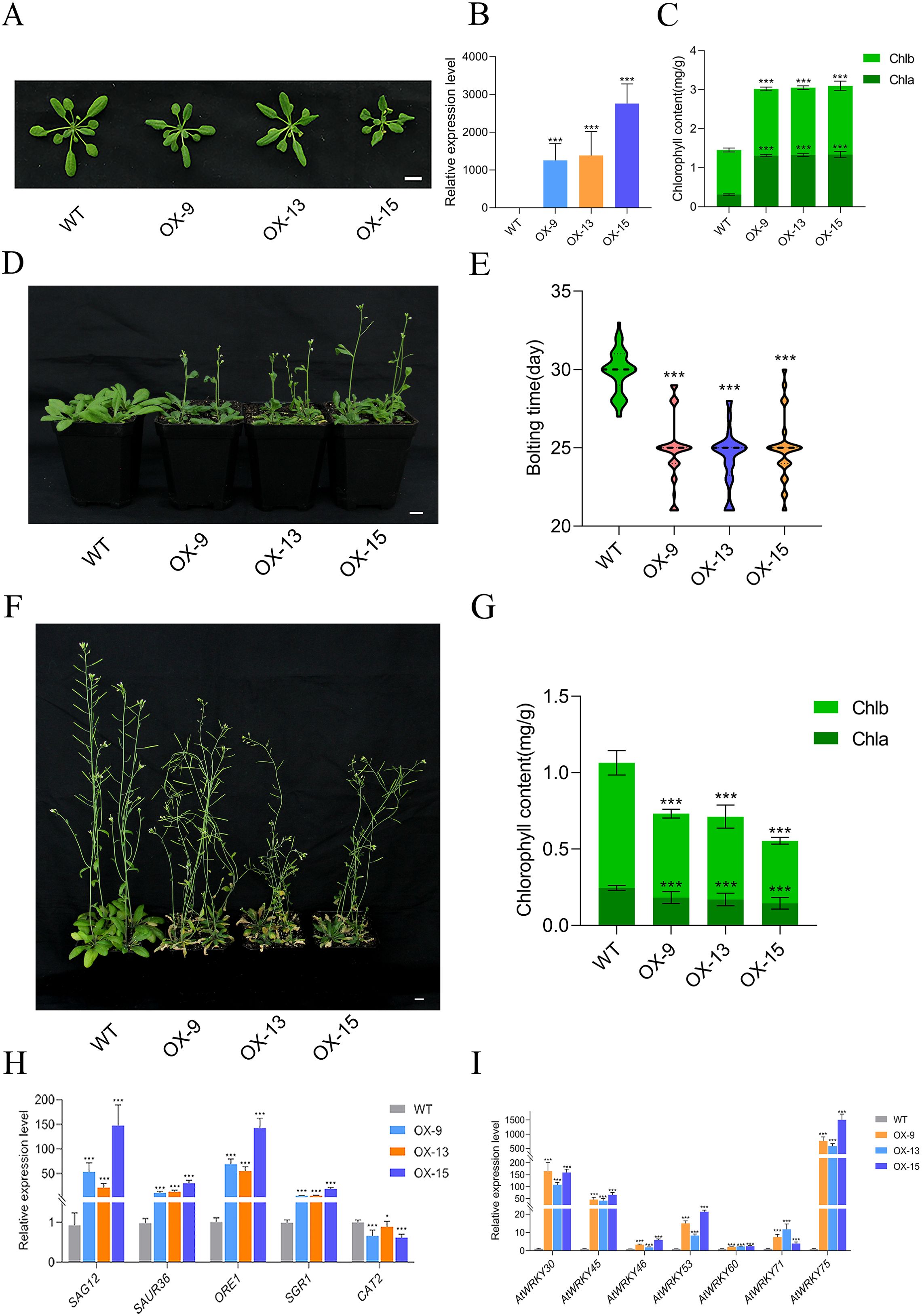
Figure 3. GmMYBLJ influences morphology and induces leaf senescence in Arabidopsis. (A) Photograph of 25-day-old transgenic Arabidopsis plants overexpressing GmMYBLJ and wild type (WT). Three transgenic lines are designated as OX-9, OX13, and OX15. (B, C) Relative expression of GmMYBLJ (B) and chlorophyll content (C) in the three transgenic Arabidopsis lines (25-day-old). (D, E) Photograph (D) and comparison (E) of bolting time among the three transgenic Arabidopsis lines and WT. (F, G) Photograph (F) and chlorophyll content (G) of 45-day-old transgenic Arabidopsis plants overexpressing GmMYBLJ and WT. (H, I) Expression patterns of senescence-associated genes in the three transgenic Arabidopsis lines and WT. The senescence-associated genes include SENESCENCE-ASSOCIATED GENE 12 (SAG12), SMALL AUXIN UP RNA 36 (SAUR36), ORESARA 1 (ORE1), STAY-GREEN 1 (SGR1), and seven WRKYs (AtWRKY53, AtWRKY75, AtWRKY60, AtWRKY71, AtWRKY45, AtWRKY6, and AtWRKY46). The data were normalized against the AtACT8 gene for all the RT-qPCR analyses. Three biological replicates and three technical replicates for each biological replicate were carried out for all the analyses. The standard error of the mean is represented by an error bar. The one-way ANOVA with LSD test was used to calculate the statistical significance, and the significant difference is indicated by an asterisk (***p < 0.001). The scale bar is shown as 1 cm.
Notably, overexpression of GmMYBLJ led to leaf senescence in 45-day-old plants, as evidenced by the dried and dead rosette leaves (Figure 3F). Chlorophyll content was then measured in the fourth and fifth rosette leaves, revealing a 31.27%–47.98% reduction in total chlorophyll content in transgenic Arabidopsis leaves as compared to WT (Figure 3G). This suggests that GmMYBLJ possibly promoted leaf senescence in Arabidopsis. Furthermore, the expression patterns of leaf senescence-associated genes, including SENESCENCE-ASSOCIATED GENE 12 (SAG12), SMALL AUXIN UP RNA 36 (SAUR36), ORESARA 1 (ORE1), STAY-GREEN 1 (SGR1), and CATALASE2 (CAT2), were investigated in the three transgenic Arabidopsis lines, as demonstrated previously (Lei et al., 2023). It was found that the expression of AtSAG12, AtSAUR36, AtORE1, and AtSGR1 was significantly upregulated by GmMYBLJ overexpression, particularly AtSAG12 and AtORE1, which showed 23.1–159.6-fold changes (Figure 3H). In contrast, AtCAT2 was transcriptionally downregulated by GmMYBLJ overexpression (Figure 3H). In addition, seven WRKY genes were selected to investigate their expression patterns, as they have been reported to play important roles in leaf senescence, including AtWRKY53, AtWRKY75, AtWRKY60, AtWRKY71, AtWRKY45, AtWRKY6, and AtWRKY46 (Cao et al., 2023). As shown in Figure 3I, all seven WRKYs were transcriptionally elevated in the three transgenic Arabidopsis lines overexpressing GmMYBLJ. In particular, AtWRKY30, AtWRKY45, and AtWRKY75 were upregulated by 40–1,500-fold (Figure 3I). Subsequently, the three WRKY genes were subjected to yeast one-hybrid (Y1H) analysis to investigate whether they could be bound by GmMYBLJ. No blue colonies were observed when GmMYBLJ was used as the bait and the promoter fragments of the three WRKY genes as prey (Supplementary Figure S3).
To investigate whether GmMYBLJ interacts with other transcription factors such as WRKYs and NACs, the online program STRING was used to predict potential interactors of GmMYBLJ. As shown in Supplementary Figure S4, no WRKY or NAC proteins were predicted to interact with GmMYBLJ. Instead, the following four GmMYBs were predicted to have the potential to interact with GmMYBLJ: Glyma.04G004100 (MYB047), Glyma.06G003800 (MYB48), Glyma.11G107100 (MYBZ2), and Glyma.12G032200 (MYB54).
3.4 GmMYBLJ accelerates darkness and Pst DC3000 infection-induced leaf senescence in Arabidopsis
A large number of studies have demonstrated that darkness promotes leaf senescence (Liebsch and Keech, 2016). To test the role of GmMYBLJ in dark-induced leaf senescence, 4-week-old transgenic Arabidopsis plants were treated in the dark for 5 days. As shown in Figure 4A, the transgenic Arabidopsis plants exhibited more severe leaf senescence compared to WT. Further investigation revealed that the chlorophyll content in transgenic Arabidopsis leaves was significantly reduced, with a 48.54%–60.86% reduction compared to WT (Figure 4B). Conversely, the MDA content increased significantly by 33.68%–71.58% (Figure 4C), suggesting that GmMYBLJ may accelerate dark-induced leaf senescence in Arabidopsis.
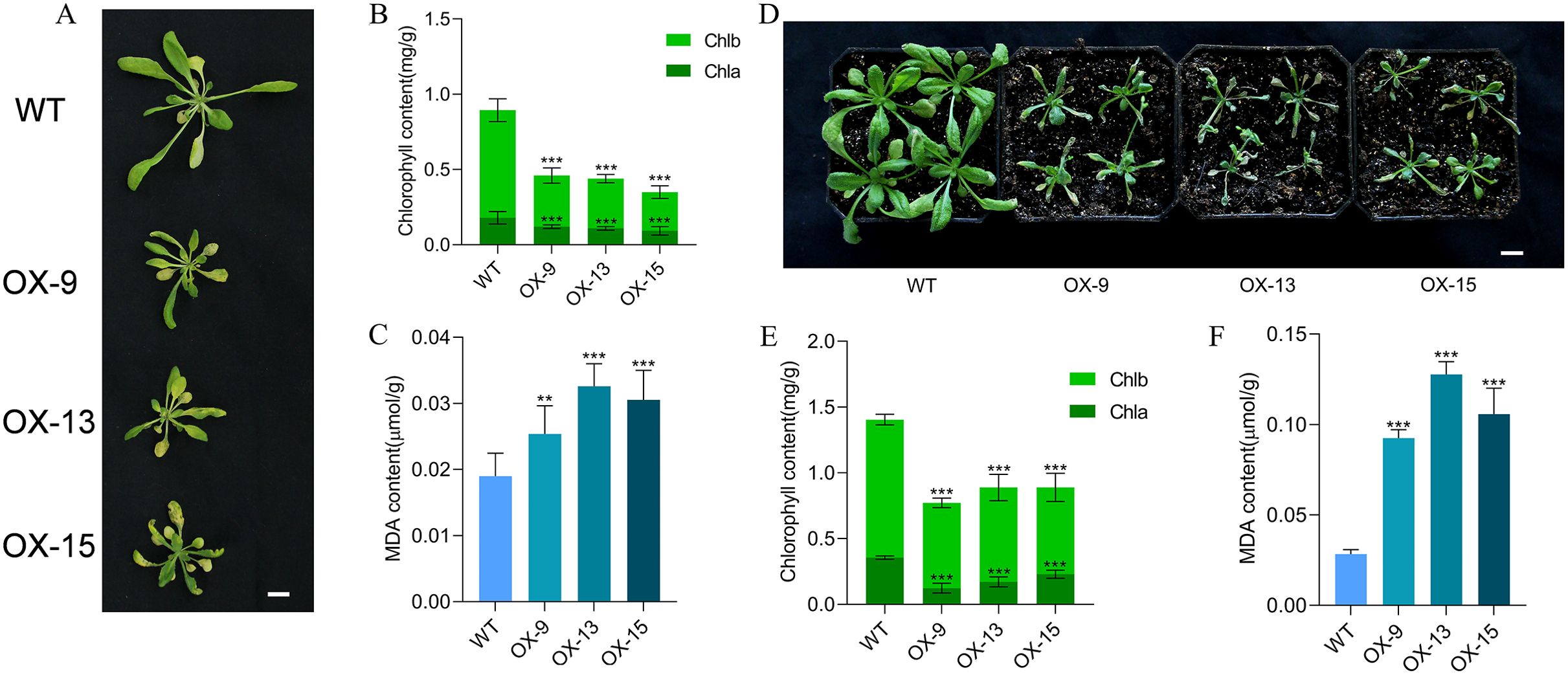
Figure 4. GmMYBLJ accelerates dark and Pst DC3000-induced leaf senescence in Arabidopsis. (A) Photograph of 4-week-old transgenic Arabidopsis plants overexpressing GmMYBLJ and wild type (WT) under dark conditions for 5 days. Three transgenic lines are designated as OX-9, OX13, and OX15. (B, C) The contents of chlorophylls (B) and MDA (C) in the three transgenic lines and WT under dark conditions. (D) Photograph of 4-week-old transgenic Arabidopsis plants overexpressing GmMYBLJ and WT under Pst DC3000-treated condition for 5 days. (E, F) The contents of chlorophylls (E) and MDA (F) in the three transgenic lines and WT under Pst DC3000 stress. Three biological replicates and three technical replicates for each biological replicate were performed for all the analyses. The standard error of the mean is represented by an error bar. A one-way ANOVA with LSD test was used to calculate statistical significance, and significant differences are indicated by an asterisk (**p < 0.01; ***p < 0.001). The scale bar is shown as 1 cm.
Biotic stress is another factor that induces plant leaf senescence (Antonietta et al., 2024). To explore the effect of GmMYBLJ on leaf senescence under biotic stress conditions, 4-week-old plants of the three transgenic Arabidopsis lines were treated with the bacterial pathogen Pst DC3000 for 5 days. As shown in Figure 4D, the transgenic Arabidopsis plants exhibited more severe leaf senescence compared to the WT. Further investigation revealed that the chlorophyll content in the transgenic Arabidopsis plants was significantly lower compared to the WT (Figure 4E), while the MDA content increased by 2.25–3.49-fold (Figure 4F), suggesting that GmMYBLJ overexpression accelerated Pst DC3000-induced leaf senescence in Arabidopsis.
3.5 GmMYBLJ causes ROS accumulation in Arabidopsis and soybean hairy roots
Reactive oxygen species (ROS) play a crucial role in the process of plant organ senescence (Asad et al., 2024). To investigate whether GmMYBLJ is involved in regulating leaf senescence through the ROS-mediated pathway, DAB staining analysis was performed using 30-day-old transgenic Arabidopsis plants overexpressing GmMYBLJ. As shown in Figure 5A, a brown color clearly appeared on the leaves of the transgenic Arabidopsis plants, indicating that overexpression of GmMYBLJ likely resulted in increased ROS production. Furthermore, GmMYBLJ-overexpressed and GmMYBLJ-silenced soybean hairy roots were individually generated using A. rhizogenes-mediated hairy root transformation. Positive transgenic hairy roots were identified by LUYOR-3415CV handheld lamp irradiation and RT-qPCR (Figures 5B, C). The transgenic roots were then collected for DAB staining analysis. As shown in Figure 5D, GmMYBLJ-overexpressed hairy roots showed more brown color after DAB staining, while GmMYBLJ-silenced hairy roots exhibited less coloration (Figure 5D). These results suggest that GmMYBLJ may positively regulate the accumulation of ROS in soybean hairy roots.
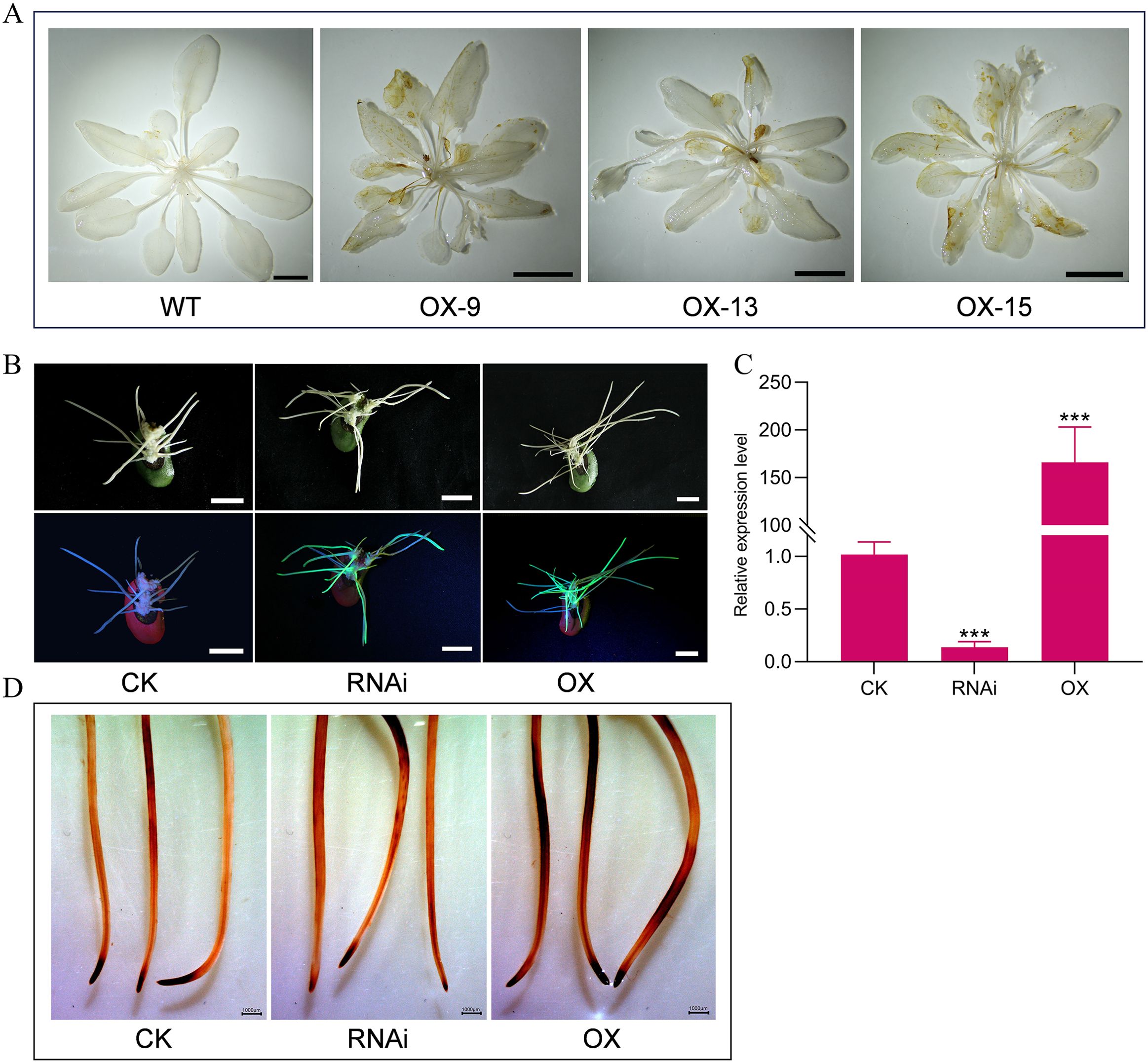
Figure 5. GmMYBLJ causes ROS accumulation in Arabidopsis and soybean hairy roots. (A) DAB staining analysis of 30-day-old Arabidopsis plants from the three transgenic lines overexpressing GmMYBLJ and wild type (WT). The scale bar is shown as 1 cm. (B) Photographs of soybean hairy roots overexpressing and silencing GmMYBLJ. The scale bar is shown as 1 cm. (C) Expression level of GmMYBLJ in soybean hairy roots overexpressing and silencing GmMYBLJ. (D) DAB staining analysis of soybean hairy roots overexpressing and silencing GmMYBLJ. The data were normalized against GmSUBI-3 for RT-qPCR analysis. The standard error of the mean is represented by an error bar. The one-way ANOVA with LSD test was used to calculate the statistical significance, and the significant difference is indicated by an asterisk (***p < 0.001). Three biological replicates and three technical replicates for each biological replicate were performed for all the analyses. The scale bar is shown as 1,000 μm.
3.6 GmMYBLJ overexpression accelerates salt-induced senescence in soybean
To investigate the effects of GmMYBLJ on leaf senescence in soybean, transgenic soybean plants overexpressing GmMYBLJ were generated and verified using RT-qPCR (Figures 6A, B). Ten-day-old transgenic soybean plants were subjected to salt stress for 10 days. Under normal conditions, the transgenic plants exhibited faster growth and greater height compared to WT (Figure 6C), with no significant difference detected in MDA levels (Figure 6D). When subjected to salt stress, transgenic soybean plants exhibited more severe leaf wilting compared to WT (Figure 6E), and the MDA content increased by 3.21–14.56-fold relative to WT (Figure 6F), suggesting that the overexpression of GmMYBLJ accelerates salt-induced senescence in soybean.
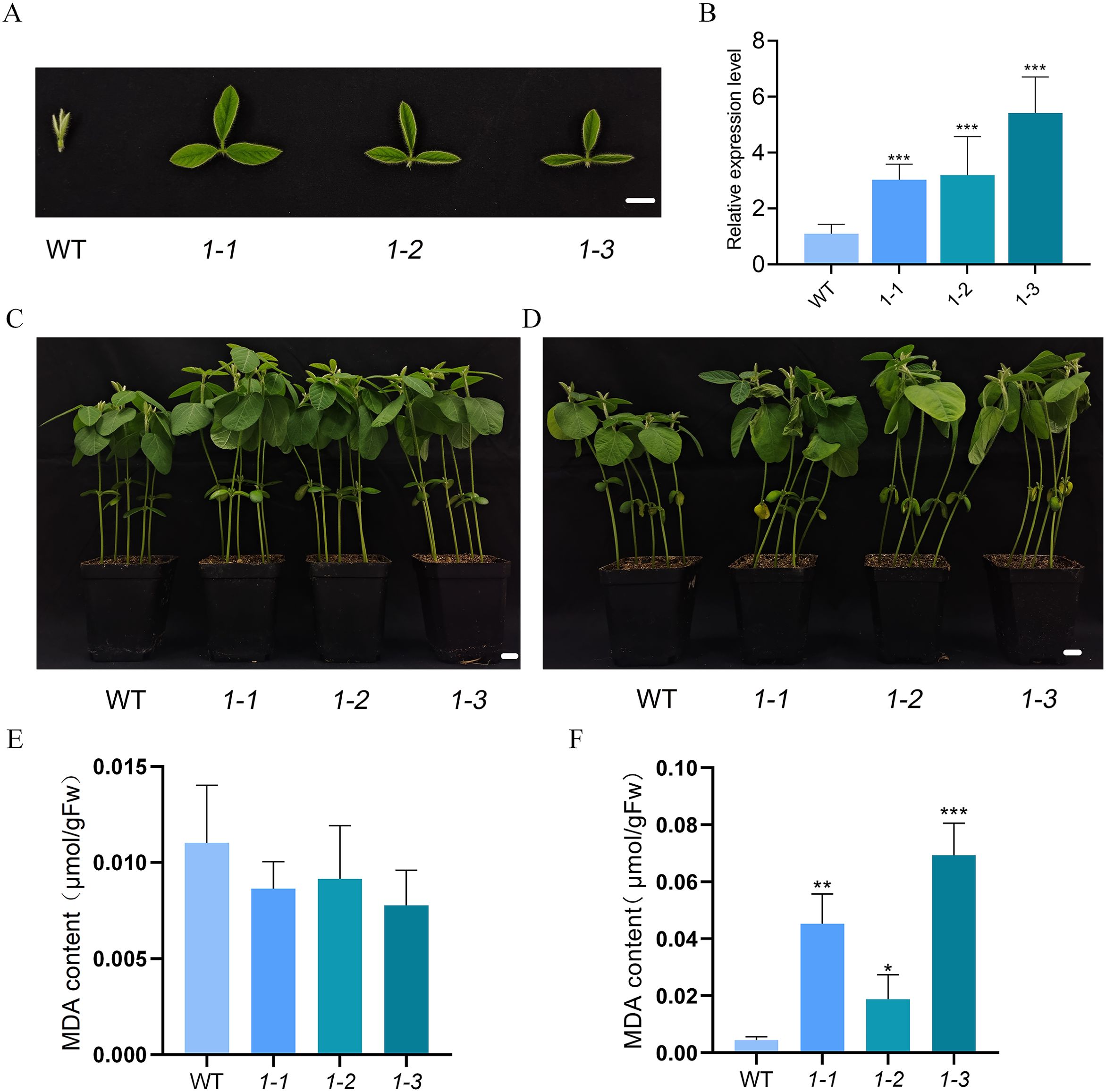
Figure 6. GmMYBLJ accelerates salt-induced senescence in soybean. (A) Photographs of the second round of the three compound leaves of 18-day-old transgenic soybean plants overexpressing GmMYBLJ. (B) The relative expression level of GmMYBLJ in the transgenic soybean plants. (C, D) Photographs of 10-day-old transgenic soybean plants and untransformed control (WT) grown under normal conditions (C) and treated with 200 mM NaCl (D) for 10 days. (E, F) MDA content of 10-day-old transgenic soybean plants and WT grown under normal conditions (E) and treated with 200 mM NaCl (F) for 10 days. The standard error of the mean is represented by an error bar. One-way ANOVA with LSD test was used to calculate statistical significance, and significant differences are indicated by an asterisk (*p < 0.05; **p < 0.01; ***p < 0.001). Three biological replicates and three technical replicates for each biological replicate were conducted for all the analyses. The scale bar is shown as 1 cm.
4 Discussion
MYB transcription factors have been considered important players in the regulation of leaf senescence. To date, genome sequence availability has allowed the identification of a few hundred MYBs in soybean (Du et al., 2012). However, little is known about their functional roles in the progression of leaf senescence. In this study, a soybean CCA1-like gene, GmMYBLJ, was identified as playing a role in the regulation of leaf senescence, laying the foundation for further investigation into its regulatory mechanisms.
MYBs constitute one of the most important transcription factor families in plants, performing functions in virtually every aspect of plant growth and development, as well as response to stresses (Thakur and Vasudev, 2022; Biswas et al., 2023). In many plant species, MYBs have been shown to positively regulate leaf senescence. For example, overexpression of AtMYBL, AtMYBH, and NtMYB1 accelerated leaf senescence (Zhang et al., 2011; Huang et al., 2015; Yang et al., 2023). However, it has also been reported that AtMYB59 and AtGLKs act as negative regulators of leaf senescence (Zhang et al., 2022; He et al., 2023). In this study, we provide evidence supporting that soybean MYBLJ might be a positive regulator in the process of leaf senescence. The most direct evidence is that overexpression of GmMYBLJ in Arabidopsis directly accelerated leaf senescence (Figures 3F, G). Meanwhile, GmMYBLJ itself exhibited an age-dependent expression pattern and promoter activity (Figures 2B, J). Moreover, AtSAG12 is considered one of the most important senescence-associated marker genes for characterizing leaf senescence (Gombert et al., 2006). Our results showed that the expression of AtSAG12 was significantly elevated by the overexpression of GmMYBLJ (Figure 3G). Notably, GmMYBLJ is closely related to AtMYBH, with 47% protein identity (Figure 1A). AtMYBH has been reported to play positive roles in both developmentally regulated and dark-induced leaf senescence (Huang et al., 2015). Therefore, we proposed that GmMYBLJ might be positively involved in the regulation of leaf senescence, similar to its homologous gene AtMYBH. Additionally, the high protein identity of GmMYBLJ with other GmMYBs, such as Glyma.04G051000 (as shown in Figure 1A), suggests that there may be other GmMYB(s) that complement the functions of GmMYBLJ in the regulation of soybean senescence.
It is well known that biotic and abiotic stresses can accelerate leaf senescence and programmed cell death in most crop plants, leading to the cessation of photosynthesis, transpiration, and other assimilatory processes (Fraga et al., 2021; Lei et al., 2023). Numerous studies have shown that delaying senescence can enhance plant resistance to stress (Zhou and Yang, 2023; Antonietta et al., 2024). However, in some cases, delayed senescence did not impact stress resistance (Antonietta et al., 2024). In this study, GUS staining and expression pattern analysis showed that GmMYBLJ might be transcriptionally induced by dark and salt stresses (Figures 2C, F, K). Its overexpression in Arabidopsis or soybean accelerated leaf senescence and MDA accumulation under dark conditions, upon Pst DC3000 infection, or salt stress (Figures 4, 6), supporting the notion that GmMYBLJ not only promotes leaf senescence under normal conditions but also accelerates stress-induced senescence. Generally, a course of action interacts with the ROS burst during aging and senescence progression (Niu et al., 2020; Shirzadian-Khorramabad et al., 2022). In this study, transgenic Arabidopsis plants overexpressing GmMYBLJ showed greater MDA accumulation compared to WT (Figures 4C, F). Moreover, brown coloration clearly appeared on transgenic Arabidopsis leaves based on DAB-mediated ROS detection (Figure 5A). These results suggest that GmMYBLJ may perform its functions through a ROS-associated pathway. Numerous studies have indicated that NACs and WRKYs play pivotal roles in the regulation of leaf senescence (Ferreira et al., 2020; Niu et al., 2020; Zhang et al., 2022). Consistently, our data showed that AtORE1/AtNAC2, AtWRKY75, AtWRKY71, AtSGR1, and AtSAG12 were all remarkably upregulated in transgenic Arabidopsis overexpressing GmMYBLJ (Figures 3H, I). Previous studies have indicated that AtORE1/AtNAC2 plays a key role in the onset of leaf senescence by affecting the ROS-mediated pathway and promoting AtSGR1 expression (Ueda et al., 2020; Doan et al., 2022). Interestingly, some evidence suggests that AtWRKY71 influences leaf senescence by directly activating AtORE1/AtNAC2 (Yu et al., 2021), while AtWRKY75 forms a regulatory loop with SA and ROS to accelerate leaf senescence (Guo et al., 2017). Thus, it is possible that GmMYBLJ may play its functional roles during leaf senescence through the WRKY-ORE1/NAC2-ROS regulatory module. Additionally, several other WRKY family members have been reported to play important roles in leaf senescence, such as AtWRKY53 (Wang et al., 2023), AtWRKY45 (Barros et al., 2022), AtWRKY75 (Zhang et al., 2022), and AtWRKY30 (Besseau et al., 2012). In this study, all these WRKYs were transcriptionally elevated by GmMYBLJ overexpression in Arabidopsis (Figure 3I). It is thus hypothesized that GmMYBLJ may promote the expression of leaf senescence-associated genes and ROS production through the WRKY genes and AtORE1/AtNAC2, thereby accelerating leaf senescence in Arabidopsis. Since ROS plays important roles in the regulation of developmental events and responses to stresses as a cellular signaling molecule (Lopez et al., 2024), it will be very interesting to investigate in the future whether a feedback interaction exists between ROS and GmMYBLJ in soybean.
Since adverse external factors often accelerate senescence, delayed senescence under stressful conditions is generally considered an indicator of crop resilience (Antonietta et al., 2024). In this study, overexpression of GmMYBLJ resulted in increased sensitivity to stresses in both Arabidopsis and soybean (Figures 4, 6). Although we did not determine whether GmMYBLJ accelerates leaf senescence in soybean, transgenic Arabidopsis plants overexpressing GmMYBLJ displayed early leaf senescence (Figure 3). Seemingly, GmMYBLJ may act as an unfavorable gene for crop resistance to stresses. It would be interesting to further investigate whether GmMYBLJ can coordinate early leaf senescence with soybean performance under stress.
In conclusion, our findings suggest that GmMYBLJ acts as a positive regulator of leaf senescence. Moreover, GmMYBLJ accelerated leaf senescence induced by dark, salt, and Pts DC3000 stresses. The effect of GmMYBLJ on leaf senescence is possibly associated with the WRKY family and senescence-associated genes, such as AtSAG12, AtORE1, and AtSGR1. Its regulatory mechanisms require further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
GB: Writing – original draft, Visualization, Investigation. XX: Writing – original draft, Data curation, Investigation. JY: Writing – original draft, Visualization, Investigation. JL: Writing – original draft, Data curation, Investigation. TS: Writing – original draft, Data curation. XZ: Writing – original draft, Data curation. SB: Writing – review & editing, Conceptualization. XL: Writing – review & editing, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the International Science & Technology Cooperation Projects from the Science and Technology Department of Jilin Province, China (20220402052 GH) to XL.
Acknowledgments
We thank the reviewers for their valuable comments and Professor Yuhai Cui (London Research and Development Centre, AAFC, Canada) for revising the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1533592/full#supplementary-material
Supplementary Figure 1 | Photographs of soybean leaves at different developmental stages. Scale bar is shown in 1 cm.
Supplementary Figure 2 | The rhythmical expression of GmMYBLJ in soybean seedlings.
Supplementary Figure 3 | Interaction of GmMYBLJ with the promoters of three GmWRKYs using Y1H assay. (A) The schematic diagrams of the promoters of three GmWRKYs. The blue and yellow downward triangles indicate the elements of MYB binding sites, and the number shows the start site of each element. The light blue bars show the promoter fragments for Y1H assay. (B) Interaction assay between GmMYBLJ and the promoters of the three GmWRKYs using Y1H assay. pB42AD-AtRVE8/placZi-AtPRR5 was set as positive control.
Supplementary Figure 4 | Predicted interactors of GmMYBLJ using the online program STRING.
References
Akhter, D., Qin, R., Nath, U. K., Eshag, J., Jin, X. L., Shi, C. H. (2019). A rice gene, OsPL, encoding a MYB family transcription factor confers anthocyanin synthesis, heat stress response and hormonal signaling. Gene 699, 62–72. doi: 10.1016/j.gene.2019.03.013
Antonietta, M., Martinez, D., Guiamet, J. J. (2024). Delayed senescence and crop performance under stress: always a functional couple? J. Exp. Bot. 75, 4244–4257. doi: 10.1093/jxb/erae174
Asad, M. A. U., Yan, Z., Zhou, L. J., Guan, X. Y., Cheng, F. M. (2024). How abiotic stresses trigger sugar signaling to modulate leaf senescence? Plant Physiol. Biochem. 210, 108650. doi: 10.1016/j.plaphy.2024.108650
Barros, J. A. S., Cavalcanti, J. H. F., Pimentel, K. G., Medeiros, D. B., Silva, J. C. F., Condori-Apfata, J. A., et al. (2022). The significance of WRKY45 transcription factor in metabolic adjustments during dark-induced leaf senescence. Plant Cell Environ. 45, 2682–2695. doi: 10.1111/pce.14393
Besseau, S., Li, J., Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. doi: 10.1093/jxb/err450
Bian, S. M., Jin, D. H., Li, R. H., Xie, X., Gao, G. L., Sun, W. K., et al. (2017). Genome-wide analysis of CCA1-Like proteins in soybean and functional characterization of GmMYB138a. Int. J. Mol. Sci. 18, 2040. doi: 10.3390/ijms18102040
Biswas, D., Gain, H., Mandal, A. (2023). MYB transcription factor: A new weapon for biotic stress tolerance in plants. Plant Stress 10, 100252. doi: 10.1016/j.stress.2023.100252
Buelbuel, S., Sakuraba, Y., Sedaghatmehr, M., Watanabe, M., Hoefgen, R., Balazadeh, S., et al. (2023). Arabidopsis BBX14 negatively regulates nitrogen starvation- and dark-induced leaf senescence. Plant J. 116, 251–268. doi: 10.1111/tpj.16374
Cao, J., Liu, H. R., Tan, S. Y., Li, Z. H. (2023). Transcription factors-regulated Leaf senescence: current knowledge, challenges and approaches. Int. J. Mol. Sci. 24, 9245. doi: 10.3390/ijms24119245
Carvalho, H. H., Silva, P. A., Mendes, G. C., Brustolini, O. J. B., Pimenta, M. R., Gouveia, B. C., et al. (2014). The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 164, 654–670. doi: 10.1104/pp.113.231928
Chen, C. J., Wu, Y., Li, J. W., Wang, X., Zeng, Z. H., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, Y. B., Wu, P. Z., Zhang, C., Guo, Y. L., Liao, B. B., Chen, Y. P., et al. (2022). Ectopic expression of JcCPL1, 2, and 4 affects epidermal cell differentiation, anthocyanin biosynthesis and leaf senescence in Aabidopsis thaliana. Int. J. Mol. Sci. 23, 1924. doi: 10.3390/ijms23041924
Daudi, A., O’Brien, J. A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis Leaves. Bio Protoc. 2, 94087. doi: 10.21769/BioProtoc.263
Doan, P. P. T., Kim, J. H., Kim, J. (2022). Rapid investigation of functional roles of genes in regulation of leaf senescence using Arabidopsis Protoplasts. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.818239
Du, H., Yang, S. S., Liang, Z., Feng, B. R., Liu, L., Huang, Y. B., et al. (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106. doi: 10.1186/1471-2229-12-106
Eckstein, A., Grzyb, J., Hermanowicz, P., Labuz, J., Banas, A. K. (2019). A role for GLABRA1 in dark-induced senescence. Acta Biochim. Polonica 66, 243–248. doi: 10.18388/abp.2018_2825
Ferreira, D. O., Fraga, O. T., Pimenta, M. R., Caetano, H. D. N., MaChado, J. P. B., Carpinetti, P. A., et al. (2020). GmNAC81 Inversely modulates leaf senescence and drought tolerance. Front. Genet. 11. doi: 10.3389/fgene.2020.601876
Fraga, O. T., de Melo, B. P., Quadros, I. P. S., Reis, P. A. B., Fontes, E. P. B. (2021). Senescence-associated Glycine max (Gm)NAC genes: integration of natural and stress-induced leaf senescence. Int. J. Mol. Sci. 22, 8287. doi: 10.3390/ijms22158287
Gao, Y., Zhang, Y. G., Ma, C. Y., Chen, Y. H., Liu, C. X., Wang, Y. L., et al. (2024). Editing the nuclear localization signals of E1 and E1Lb enables the production of tropical soybean in temperate growing regions. Plant Biotechnol. J. 22, 2145–2156. doi: 10.1111/pbi.14335
Gombert, J., Etienne, P., Ourry, A., Le Dily, F. (2006). The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J. Exp. Bot. 57, 1949–1956. doi: 10.1093/jxb/erj142
Guo, P. R., Li, Z. H., Huang, P. X., Li, B. S., Fang, S., Chu, J. F., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29, 2854–2870. doi: 10.1105/tpc.17.00438
He, S. C., Zhi, F., Min, Y. C., Ma, R., Ge, A. K., Wang, S. X., et al. (2023). The MYB59 transcription factor negatively regulates salicylic acid- and jasmonic acid-mediated leaf senescence. Plant Physiol. 192, 488–503. doi: 10.1093/plphys/kiac589
Huang, C. K., Lo, P. C., Huang, L. F., Wu, S. J., Yeh, C. H., Lu, C. A. (2015). A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 88, 269–286. doi: 10.1007/s11103-015-0321-2
Jaradat, M. R., Feurtado, J. A., Huang, D. Q., Lu, Y. Q., Cutler, A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13, 192. doi: 10.1186/1471-2229-13-192
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lei, P., Yu, F., Liu, X. Y. (2023). Recent advances in cellular degradation and nuclear control of leaf senescence. J. Exp. Bot. 74, 5472–5486. doi: 10.1093/jxb/erad273
Li, X., Chen, L., Dhaubhadel, S. (2012). 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 71, 239–250. doi: 10.1111/j.1365-313X.2012.04986.x
Li, X. Y., Hou, Y. M., Xie, X., Li, H. X., Li, X. D., Zhu, Y., et al. (2020). A blueberry MIR156a-SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. J. Exp. Bot. 71, 5976–5989. doi: 10.1093/jxb/eraa327
Li, Z., Lyu, X. G., Li, H. Y., Tu, Q. C., Zhao, T., Liu, J., et al. (2024). The mechanism of low blue light-induced leaf senescence mediated by GmCRY1s in soybean. Nat. Commun. 15, 798. doi: 10.1038/s41467-024-45086-5
Liebsch, D., Keech, O. (2016). Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol. 212, 563–570. doi: 10.1111/nph.14217
Liu, W. Y., Zhang, W. Q., Cheng, H. P., Ding, Y. X., Yao, B. H., Shangguan, Z. P., et al. (2024). Rhizobia cystathionine γ-lyase-derived H2S delays nodule senescence in soybean. Plant Physiol. 196, 2232–2250. doi: 10.1093/plphys/kiae411
Lopez, L. E., Ibeas, M. A., Diaz Dominguez, G., Estevez, J. M. (2024). Exploring the puzzle of reactive oxygen species acting on root hair cells. J. OF Exp. Bot. 75, 4589–4598. doi: 10.1093/jxb/erae260
Lu, H. W., Gordon, M., Amarasinghe, V., Strauss, S. H. (2020). Extensive transcriptome changes during seasonal leaf senescence in field-grown black cottonwood (Populus trichocarpa Nisqually-1). Sci. Rep. 10, 6581. doi: 10.1038/s41598-020-63372-2
Meng, Y. Y., Li, H. Y., Wang, Q., Liu, B., Lin, C. T. (2013). Blue light- dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25, 4405–4420. doi: 10.1105/tpc.113.116590
Moschen, S., Bengoa Luoni, S., Di Rienzo, J. A., Caro, M. D., Tohge, T., Watanabe, M., et al. (2016). Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol. J. 14, 719–734. doi: 10.1111/pbi.12422
Niu, F. F., Cui, X., Zhao, P. Y., Sun, M. T., Yang, B., Deyholos, M. K., et al. (2020). WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 104, 171–184. doi: 10.1111/tpj.14914
Piao, W., Kim, S. H., Lee, B. D., An, G., Sakuraba, Y., Paek, N. C. (2019). Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 70, 2699–2715. doi: 10.1093/jxb/erz095
Sasi, J. M., Gupta, S., Singh, A., Kujur, A., Agarwal, M., Katiyar-Agarwal, S. (2022). Know when and how to die: gaining insights into the molecular regulation of leaf senescence. Physiol. Mol. Biol. Plants 28, 1515–1534. doi: 10.1007/s12298-022-01224-1
Shirzadian-Khorramabad, R., Moazzenzadeh, T., Sajedi, R. H., Jing, H. C., Hille, J., Dijkwel, P. P. (2022). A mutation in Arabidopsis SAL1 alters its in vitro activity against IP3 and delays developmental leaf senescence in association with lower ROS levels. Plant Mol. Biol. 108, 549–563. doi: 10.1007/s11103-022-01245-0
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Thakur, S., Vasudev, P. G. (2022). MYB transcription factors and their role in Medicinal plants. Mol. Biol. Rep. 49, 10995–11008. doi: 10.1007/s11033-022-07825-z
Tsuda, K., Qi, Y. P., Nguyen, L. V., Bethke, G., Tsuda, Y., Glazebrook, J., et al. (2012). An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 69, 713–719. doi: 10.1111/j.1365-313X.2011.04819.x
Ueda, H., Ito, T., Inoue, R., Masuda, Y., Nagashima, Y., Kozuka, T., et al. (2020). Genetic interaction among phytochrome, ethylene and abscisic acid signaling during dark-Induced senescence in Arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00564
Wang, Q., Li, X. X., Guo, C., Wen, L. C., Deng, Z. C., Zhang, Z. L., et al. (2023). Senescence-related receptor kinase 1 functions downstream of WRKY53 in regulating leaf senescence in Arabidopsis. J. Exp. Bot. 74, 5140–5152. doi: 10.1093/jxb/erad240
Xu, F., Meng, T., Li, P. L., Yu, Y. Q., Cui, Y. J., Wang, Y. X., et al. (2011). A Soybean dual-specificity kinase, GmSARK, and Its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 157, 2131–2153. doi: 10.1104/pp.111.182899
Yamatani, H., Heng, T. T. R., Yamada, T., Kusaba, M., Kaga, A. (2021). Identification and characterization of an early leaf senescence gene in soybean. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.784105
Yang, J. W., Huang, J. Z., Wu, X., Xu, Y., Gu, Z., Chen, Y. J., et al. (2023). NtMYB1 and NtNCED1/2 control abscisic acid biosynthesis and tepal senescence in Chinese narcissus (Narcissus tazetta). J. Exp. Bot. 74, 6505–6521. doi: 10.1093/jxb/erad311
Yu, Y. C., Qi, Y. N., Xu, J. P., Dai, X. H., Chen, J. C., Dong, C. H., et al. (2021). Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 107, 1819–1836. doi: 10.1111/tpj.15433
Zeng, P., Vadnais, D. A., Zhang, Z., Polacco, J. C. (2004). Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill. Plant Cell Rep. 22, 478–482. doi: 10.1007/s00299-003-0712-8
Zhang, M. Y., Bao, G. Z., Yan, B. R., Chang, Y. X., Tang, W. Y., Qu, Y., et al. (2021). Physiological effects of cutting on Secale cereale L. seedlings under freeze-thaw and alkaline salt stress. Grassland Sci. 67, 299–305. doi: 10.1111/grs.12316
Zhang, X., Ju, H. W., Chung, M. S., Huang, P., Ahn, S. J., Kim, C. S. (2011). The R-R-type MYB-Like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 52, 138–148. doi: 10.1093/pcp/pcq180
Zhang, H. Y., Zhang, L. P., Ji, Y. R., Jing, Y. F., Li, L. X., Chen, Y. L., et al. (2022). Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 73, 182–196. doi: 10.1093/jxb/erab391
Zhao, W. X., Zhao, H. Y., Wang, H. S., He, Y. (2022). Research progress on the relationship between leaf senescence and quality, yield and stress resistance in horticultural plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1044500
Zhou, M., Yang, J. D. (2023). Delaying or promoting? Manipulation of leaf senescence to improve crop yield and quality. Planta 258, 48. doi: 10.1007/s00425-023-04204-1
Keywords: soybean, GmMYBLJ, leaf senescence, Arabidopsis, dark, stress
Citation: Bao G, Xu X, Yang J, Liu J, Shi T, Zhao X, Li X and Bian S (2025) Identification and functional characterization of the MYB transcription factor GmMYBLJ in soybean leaf senescence. Front. Plant Sci. 16:1533592. doi: 10.3389/fpls.2025.1533592
Received: 24 November 2024; Accepted: 06 January 2025;
Published: 24 January 2025.
Edited by:
Muhammad Waqas, Hochschule Geisenheim University, GermanyReviewed by:
Masood Jan, VIB Laboratory for Systems Biology, BelgiumKamran Shah, South China Agricultural University, China
Copyright © 2025 Bao, Xu, Yang, Liu, Shi, Zhao, Li and Bian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuyan Li, eHV5YW5saUBqbHUuZWR1LmNu; Shaomin Bian, c2htYmlhbkBqbHUuZWR1LmNu
†These authors have contributed equally to this work
 Guohua Bao1†
Guohua Bao1† Xiao Xu
Xiao Xu Juanjuan Liu
Juanjuan Liu Xuyan Li
Xuyan Li Shaomin Bian
Shaomin Bian